Abstract
Malignant gliomas rely on the production of certain critical growth factors including VEGF, interleukin (IL)-6 and IL-8, to fuel rapid tumor growth, angiogenesis, and treatment resistance. Post-transcriptional regulation through adenine and uridine-rich elements (ARE) of the 3′ untranslated region (UTR) is one mechanism for upregulating these and other growth factors. In glioma cells, we have shown that the post-transcriptional machinery is optimized for growth factor upregulation secondary to overexpression of the mRNA stabilizer, HuR. The negative regulator, tristetraprolin (TTP), on the other hand, may be suppressed because of extensive phosphorylation. Here we test that possibility by analyzing the phenotypic effects of a mutated form of TTP (mt-TTP) in which 8 phosphoserine residues were converted to alanines. We observed a significantly enhanced negative effect on growth factor expression in glioma cells at the post-transcriptional and transcriptional levels. The protein became stabilized and displayed significantly increased antiproliferative effects compared to wild-type TTP. Macroautophagy was induced with both forms of TTP, but inhibition of autophagy did not affect cell viability. We conclude that glioma cells suppress TTP function through phosphorylation of critical serine residues which in turn contributes to growth factor upregulation and tumor progression.
1. Introduction
Tristetraprolin (TTP), a Cys-Cys-Cys-His (CCCH) tandem zinc finger protein, negatively regulates many cytokine and growth factor mRNAs linked to glioma progression, including vascular endothelial growth factor (VEGF), tumor necrosis factor (TNF)-α, interleukin (IL) -8, and IL-6 [1-10]. These cytokines promote angiogenesis, proliferation, glioma stem cell phenotype, and chemoresistance. TTP binds to AU-rich elements (ARE) in the 3′ untranslated regions (UTR) of these cytokine mRNAs with a high affinity for the nonamer, UUAUUUAUU, and destabilizes the transcript by recruiting factors that initiate deadenylation and exonuclease activity [1, 2]. The importance of ARE-mediated gene regulation in glioma biology was recently highlighted when knockdown of the ARE-binding protein, HuR, a positive regulator of RNA stability and translation, produced significant attenuation of tumor growth in vivo [11]. Ectopic TTP expression in glioma cells leads to a similar phenotype, with downregulation of VEGF and IL-8 [12]. TTP is extensively regulated post-translationally, mainly through phosphorylation of numerous serine residues [13, 14]. Phosphorylation has been linked to suppressed function possibly by reducing TTP binding to target mRNAs or sequestration by 14-3-3 proteins [1, 15-18]. We previously showed that TTP is heavily phosphorylated in glioblastoma multiforme tumors and postulated that this modification keeps TTP in check. In the current report, we further investigated this hypothesis by mutating select phosphoserines from TTP and testing the molecular and cellular impact in glioma cells.
2. Materials and Methods
2.1. DNA constructs, cell culture, transfection, and transgene expression
A FLAG-tagged mutant form of TTP (mt-TTP) (S88A, S90A, S93A, S197A, S214A, S218A, S228A, and S296A) [14] was stably transfected into U251MG Tet On cells as previously described for wild type TTP [12]. Other mutants were produced by a kit (Stratagene): M2 (S88A, S90A, S93A) or M1 (S88A, S90A, S93A, S214A, 218A). For transfection, DNA doses were: 100 ng (mt-TTP and M1), 200 ng (M2) and 350 ng (wt-TTP). Myc-hnRNP C was used as a control [19]. The pGL2-IL-8 3′ UTR reporter was described previously [20] and the 3′ UTR control consisted of a non-ARE region in the TNF-α 3′ UTR (nucleotides 1450-1565; GenBank M10988). The full length IL-8 promoter (-1480 to +104) was a gift from Dr. Hector Wong (Cincinnati Children’s Medical Center). The pSV-β-galactosidase was purchased (Promega). U251 malignant glioma (MG) and U251 Tet-On MG cells were grown as previously described [21]. Unless otherwise specified, cells were induced with doxycycline (0.25-0.5 μg/mL) for 6h followed by stimulation with TNF-α (10 ng/mL) or diluent as indicated.
2.2. RNA quantification, decay kinetics and RNA immunoprecipitation
RNA levels were assessed by qRT-PCR (Applied Biosystems). For RNA kinetics, cells were treated with actinomycin D (0-4h) and RNA degradation plots were generated as previously described [12]. RNA immunoprecipitation were performed on cytosolic extracts (250 μg) as described elsewhere [22]. One aliquot was used to quantitate ectopic TTP expression by Western blot.
2.3. ELISA, Western analysis, and immunocytochemistry
ELISAs were performed as described previously [12]. For Western analysis, 25-50 μg of extracts were immunoblotted. For LC3, whole cell lysates were sonicated prior to extract separation. Chloroquine (50 μM) was added to some wells as a control. Cells were fixed in 4% paraformaldehyde. LC3 immunostaining was detected using Tyramide Signal Amplification system (Perkin Elmer). The following antibodies were used: Atg7 (Cell Signaling/Millipore, 1:1,000, cat. # 2631S); FLAG (M2) (Sigma-Aldrich, 1:2000, cat. # F3165); LC3-I/II (Western blot, Abgent, 1:400, cat. # AP1802a; immunocytochemistry, Cell Signaling/Millipore, 1:500, cat. # 2775); α-Tubulin (Sigma-Aldrich, 1:2000, cat. # T9026); HuR 3A2 (Santa Cruz, 1.2 μg/mL, cat. # SC-5261); mouse IgG (Santa Cruz, 1.2 ug/mL, cat. # sc-2025); PARP (Cell Signaling/Millipore, 1:500, cat. # 9542); cleaved PARP (Cell Signaling/Millipore, 1:500, cat. # 9541S); lamin (Santa Cruz, 1:400, cat. # sc-6215); caspase-3 (Cell Signaling/Millipore, 1:500, cat. # 9662) and caspase-8 (Cell Signaling/Millipore, 1:1000; cat. # 9496S).
2.4. Cell viability, soft agar, chemosensitivity assays
Cell viability was assessed by Trypan Blue Exclusion [12]. Colony formation in soft agar was assessed as previously described [11]. For chemosensitivity, cells were induced with doxycycline (0.25 μg/mL) and treated with temozolomide (Sigma) for 24 h. Cells were stimulated with TNF-α (5 ng/mL) or vehicle (PBS). Cell viability was assessed by the Calcein AM assay (Invitrogen).
2.5. Protein stability and luciferase assays
For protein stability, TTP clones were treated with cycloheximide (10 μg/mL). Some cells were treated with p38 MAP kinase inhibitor, SB202190 (5 μM), and ERK1/2 inhibitor, UO126 (10 μM) or vehicle (DMSO). For the 3′ UTR luciferase reporter assay, pGL2-IL-8 3′ UTR was cotransfected with TTP plasmids and pSV-β-galactosidase. Luciferase and β-galactosidase measurements were performed as previously described [22]. For translational efficiency, cells were co-transfected with plasmids as described above, and mRNA levels were measured by qRT-PCR. A translation efficiency value was calculated based on published methods and normalized to Tet On control cells [23]. For IL-8 promoter assays, U251MG cells were cotransfected with pGL2-IL-8 promoter, pSV-ß-galactosidase, and TTP plasmids. Some cells were stimulated with TNF-α (10 ng/mL). Luciferase activity was normalized to β-galactosidase activity.
2.6. Analysis of autophagy and apoptosis
TTP clones were induced with doxycycline (0.25 μg/mL) and some cells were stimulated with TNF-α (2 ng/mL) for 12 h. LC3-II, caspase-3, caspase-8 and cleaved PARP expression were assessed by Western blot. Some cells were pre-incubated for 6 h with wortmannin (WM), 3-methyladenine (3-MA), or DMSO prior to induction. For silencing experiments, cells were transfected with Atg7 siRNA (Dharmacon) or a scrambled control by electroporation. After 24 h, doxycycline and TNF-α were added as above. Cell viability/proliferation was assessed by the Vialight assay (Lonza).
2.7. Statistics
All statistics were determined using a Student’s t test.
3. Results
3.1 Mutant TTP potently suppresses IL-8, VEGF and IL-6 expression in malignant glioma cells
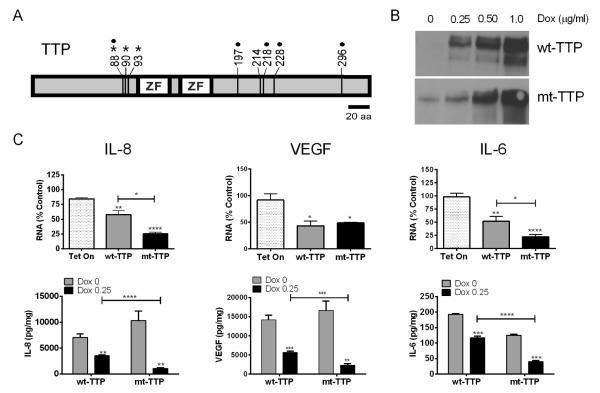
TTP is extensively phosphorylated in malignant glioma [12], and we hypothesized that this modification suppresses TTP function in glioma cells. We examined a mutant form of TTP (mt-TTP) in which eight serine residues were changed to alanines (Fig. 1A). Three of the serines, (S88, S90, and S93) are putative p38 phosphorylation sites [24], and others have been shown to be phosphorylated in vivo [14]. We used a doxycycline-inducible system (Tet-On) in U251 glioma cells to assess the impact of mt-TTP at low levels of expression. We observed comparable expression of wild type (wt) and mt-TTP (Fig. 1B). RNA and protein levels of three critical glioma growth factors, VEGF, IL-8 and IL-6, were assessed following TTP induction [4, 5, 25]. Both TTP forms produced a significant decline in mRNA levels (Fig. 1C). For IL-6 and IL-8, mt-TTP attenuated mRNA expression by an additional two-fold (P <0.05). Even minimal induction with very low doses of doxycycline suppressed IL-8 mRNA levels in mt-TTP cells (Supplementary Fig. 1A), indicating that the phenotype was not an artifact of overexpression [26]. GAPDH mRNA expression was not altered (Supplementary Fig. 1B) indicating that the attenuation was target specific. The effect on protein expression was assessed by ELISA (Fig. 1C). Wt-TTP suppressed IL-8 and VEGF by approximately 2-fold (P <0.005). IL-6 expression was attenuated by 1.7 fold (P <0.005). Mutant TTP produced a much larger decrease for all targets (3 to 10-fold, P < 0.0005) with IL-8 showing the greatest change. RNA kinetic analysis showed that each target mRNA was destabilized by both forms of TTP (Supplementary Fig. 2A). IL-8 mRNA half-life showed the greatest change (from > 4.0 h in control cells to ~0.8 h), but there was no difference between wt- and mt-TTP. VEGF and IL-6 followed a similar pattern with reduction in half-life by 2 to 2.5-fold. To assess target mRNA association with TTP, we performed RNA immunoprecipitation on cytoplasmic extracts (Supplementary Fig. 2B). RNA levels for the FLAG antibody precipitates were adjusted to ectopic TTP expression as determined by Western blot (not shown). We observed robust binding of wt- and mt-TTP to VEGF and IL-8 mRNA with a statistically non-significant trend of increased recovery with wt-TTP. With HuR, there was significantly less binding to VEGF and IL-8 in mt-TTP cells (p < 0.05). In summary, mt-TTP exerted significantly more potent suppression of IL-8, VEGF and IL-6, without significant differences in mRNA half-lives or target RNA binding.
Fig. 1. Mutant TTP produces significantly greater suppression of growth factors in malignant glioma cells.
A) Diagram of wild-type TTP (wt-TTP) highlighting the serine residues which were converted to alanines to make mutant TTP (mt-TTP). •, confirmed phosphorylation sites in vivo [14]; *, putative p38 phosphorylation sites not sensitive to MK2 expression [24]. B) Western blot of extracts from Tet-On U251 malignant glioma clones expressing either FLAG-wt-TTP or FLAG-mt-TTP. Cells were induced with doxycycline (Dox) and the blot was probed with an anti-FLAG antibody. C) mt-TTP and wt-TTP glioma clones were assessed for IL-8, VEGF and IL-6 expression after induction with doxycycline (0.25μg/ml) and TNF-α stimulation. Upper row: qRT-PCR analysis of IL-8, VEGF and IL-6 mRNA in TTP clones or the parent glioma cell line (Tet-On). Data were normalized to house-keeping gene, S9 and results are expressed as a percent of control (no doxycycline). Lower row: ELISA analysis of protein concentrations for these targets in the growth media. ELISA values were adjusted to total protein content in the culture dish. Data for RNA and protein analysis are derived from at least 4 independent experiments. *, P <0.05; **, P <0.005; ***, P < 0.0005 ; ****, P <0.0001 comparing to control unless otherwise indicated.
3.2 mt-TTP shows greater inhibition of gene expression at the post-transcriptional and transcriptional levels
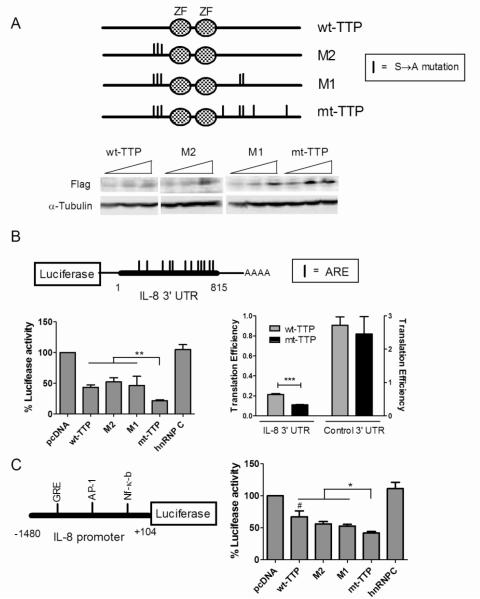
FLAG-tagged TTP expression plasmids (Fig. 2A) were cotransfected with a luciferase reporter fused to the ARE elements in the IL-8 3′ UTR (Fig. 2B). We also assessed two intermediate mutants, M1 and M2, in which certain phosphoserine residues were restored. Expression of the proteins was confirmed by Western blot (Fig. 2A). We consistently observed higher expression of mt-TTP and M1, and to a lesser degree, M2, compared to wt-TTP. We adjusted the DNA transfection doses to correct for these differences. We observed inhibition of luciferase activity with all forms of TTP, but mt-TTP had the greatest negative effect, reducing luciferase activity by ~80% compared to control. This reduction exceeded that of wt-TTP (P <0.002). The two intermediate phosphomutants were similar to wt-TTP indicating that mt-TTP required all 8 serine mutations for maximal effect. The control, hnRNP C, did not have an effect on reporter expression. For translational efficiency, we co-transfected the IL-8 and β-galactosidase reporters into the TTP glioma clones (Fig. 2B). A translational efficiency was calculated as described elsewhere [23]. We observed a ~50% reduction in translational efficiency with the IL-8 3′UTR for mt-TTP compared to wt-TTP (P <0.0005). With the control 3′ UTR, there was an overall increase in translational efficiency, but no significant difference between wt- and mt-TTP. Since the RNA half-lives were not differentially affected between wt-TTP and mt-TTP, we looked at transcriptional effects. Glioma cells were transiently transfected with a luciferase reporter containing the human IL-8 promoter [27] along with TTP expression plasmids (Fig. 2C). This promoter contains AP-1 and NF-κB sites which are targets of TTP repressor activity [28-30]. Both forms of TTP inhibited IL-8 promoter activity, but mt-TTP exceeded wt-TTP (41% versus 68%, P <0.05). Maximal suppression required all 8 serine mutations. In summary, mt-TTP had enhanced negative effects on transcriptional and post-transcriptional gene regulation.
Fig. 2. mt-TTP shows significantly greater suppression of gene expression through the IL-8 3′ UTR and promoter.
A) Schematic of TTP expression plasmids with serine-to-alanine mutations highlighted (see Fig. 1 for more detail). Below is a Western blot of U251 cell extract following transient transfection of the plasmids at different doses (100, 200 and 500 ng). Antibodies are shown to the left. B) Top: diagram of the IL-8 3′ UTR used in the reporter assay. AU-rich elements (ARE) are shown. Left panel: U251 cells were transiently transfected with TTP expression plasmids and the reporter. DNA doses of the TTP plasmids were adjusted to correct for differences in protein expression (see Materials and Methods). A β-galactosidase plasmid served as a transfection control. Luciferase activity was normalized to β-galactosidase and then expressed as a percentage of vector (pcDNA) control. ** P < 0.002. Right panel: translational efficiency was assessed by cotransfecting the luciferase IL-8 3′ UTR or control 3′ UTR reporter and a β-galactosidase plasmid into wt-TTP, mt-TTP or Tet-On U251 cells followed by induction with doxycycline. Luciferase and β-galactosidase activities and mRNA levels were measured and translation efficiency was calculated by the following equation: [Luciferase activity/Luciferase mRNA]/[β-galactosidase activity/ β-galactosidase mRNA] (see Materials and Methods). Values for the control 3′ UTR reporter are based off the right Y axis. ***, P < 0.0005. Data points represent the mean (± SEM) of at least three independent tests. C) U251 cells were cotransfected with TTP expression plasmids and IL-8 full length promoter luciferase reporter containing NF-kB, AP-1 and GRE binding sites. A β-galactosidase plasmid served as a transfection control. Luciferase activity was adjusted to β-galactosidase activity and expressed as a percentage of vector (pcDNA). Data points represent the mean (± SEM) of at least three independent tests. #, P <0.05; comparing vector and hnRNP C controls to wt-TTP; *P <0.05.
3.3 mt-TTP protein is stabilized in glioma cells
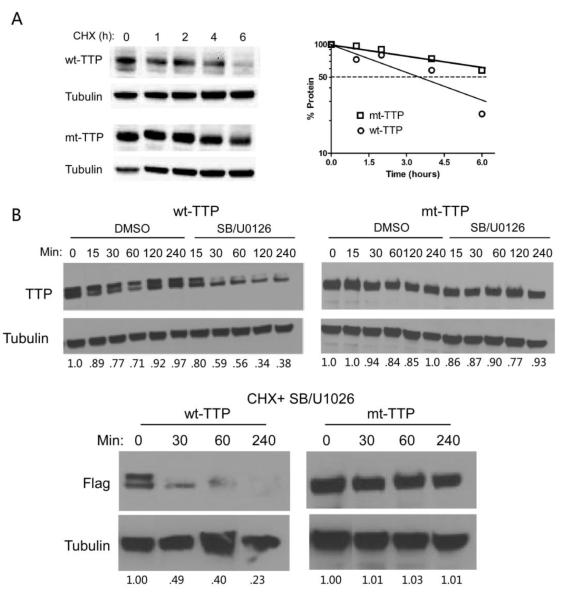
Phosphorylation has been linked to TTP stability [16, 26] which prompted us to evaluate protein kinetics (Fig. 3A). Following treatment with cycloheximide (CHX), we observed a differential decline in protein levels, with mt-TTP showing greater stability. The estimated half-life was > 6 h for mt-TTP versus ~3.5 h for wt-TTP. In the presence of p38 and ERK MAPK inhibitors, wt-TTP rapidly declined with the upper band nearly disappearing after 60 min (Fig. 3B). Mt-TTP remained stable at 4 h. Destabilization of wt-TTP in the presence of the inhibitors was further demonstrated following treatment with CHX. There was near loss of signal at 4 h while mt-TTP did not change (Fig.3B). These findings are consistent with a prior report with wild-type TTP where p38 MAPK and ERK inhibition led to marked protein destabilization [26]. The stability of mt-TTP likely explains the higher expression observed in transiently transfected U251 cells (Fig. 2). We also observed higher expression of mt-TTP in PC-3 (prostate) and MCF7 (breast) cells following transient transfection (Supplementary Fig. 3), suggesting that the stabilizing effect is not limited to glioma cells.
Fig. 3. mt-TTP protein is stabilized in glioma cells.
A) Western blot of wt-TTP and mt-TTP glioma clones induced with doxycycline (0.25 μg/mL), stimulated with TNF-α, and pulsed at various time intervals with cycloheximide (CHX). The blot was probed with anti-FLAG (wt-TTP and mt-TTP) and α-tubulin antibodies. Protein degradation curves shown to the right were derived from densitometry of the bands normalized to tubulin. The value at time 0 represents 100%. B) Western blot of clones, as above, treated with either DMSO control or SB202190 (5 μM) and U0126 (10 μM), with or without CHX at the indicated time intervals. Densitometry measurements normalized to α-tubulin are shown below the blots. Experiments were repeated 2 times with similar results.
Since phosphorylation of TTP modulates cellular localization [26], we assessed cellular location of TTP. Nuclear and cytosolic extracts were assessed by Western blot (Supplementary Fig. 4). Mutant TTP predominantly localized to the nuclear compartment in the unstimulated state, whereas wt-TTP was more abundant in the cytosol. After TNF-α stimulation, mt-TTP translocated to the cytosol, whereas wt-TTP did not change substantially. In summary, the 8 serine mutations led to TTP stabilization and a change in cellular location.
3.4. mt-TTP has an enhanced antiproliferative effect on glioma cells
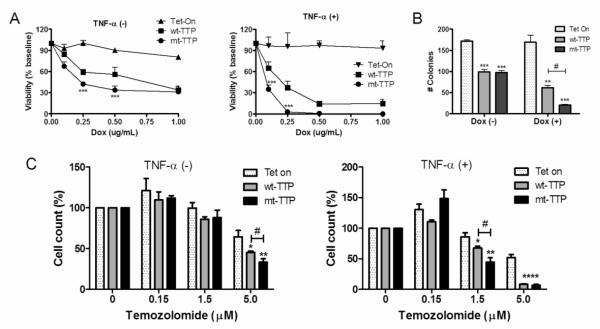
Because mt-TTP enhanced the downregulation of growth factors, we hypothesized that it would produce greater attenuation of the cancer phenotype. We assessed cell viability (Fig. 4A) and found that mt-TTP showed greater cell loss at lower doxycycline doses (0.25-0.50 μg/mL; P <0.008). Stimulation with TNF-α shifted the toxicity curves to the left, again with mt-TTP showing greater cytotoxicity (P < 0.005). We examined anchorage independent growth using a soft agar assay (Fig. 4B). Wt-TTP or mt-TTP cells were grown in soft agar in the presence or absence of doxycycline. In the absence of doxycycline, we observed a significant but equivalent decrease in the number of colonies for wt-TTP and mt-TTP (~40% versus control, P < 0.0005). This finding may be related to basal expression in the Tet-On system [12, 31]. With doxycycline induction, the number of colonies for mt-TTP dropped to 10% of control cells versus 36% for wt-TTP (P <0.002). Lastly, we examined the effect of temozolomide, a chemotherapeutic used routinely in the treatment of malignant gliomas (Fig. 4C) [32]. Cells were induced with doxycycline (0.25 μg/mL) and treated with low doses of temozolomide for 24 h. At 5 μM, both forms of TTP showed a significant reduction in cell count (P < 0.01) compared to control cells, but more so in mt-TTP (P < 0.01). With TNF-α stimulation, a decline in cell count was observed at a lower dose of temozolomide (1.5 μM), again with mt-TTP being greater than wt-TTP (P < 0.01). Taken together, these results indicate that mt-TTP exerts a greater antiproliferative effect than wt-TTP.
Fig. 4. mt-TTP has enhanced antiproliferative effects in glioma cells.
A) Cell viability was determined after doxycycline induction of wt-TTP or mt-TTP by Trypan Blue exclusion counting (at baseline or after TNF-α stimulation). Data points are expressed as a percentage of uninduced cells and are the mean ± SD of three independent experiments. ***P <0.0005 comparing mt-TTP to wt-TTP cells. B) mt-TTP significantly reduces colony formation in soft agar compared to wt-TTP. Colonies were allowed to grow for 3 weeks and then counted. Data points represent the mean ± SD of three independent experiments. **P < 0.005, and *** P < 0.001 compared to Tet-On control cells; #P < 0.002. C) mt-TTP enhances chemosensitivity of glioma cells. Cells were induced with 0.25 μg/mL of doxycycline and treated with varying doses of temozolomide for 24 h and then assessed by an ATP-based luminescence assay. Data points are expressed as a percentage of untreated, doxycycline-induced cells (dose 0) and represent the mean ± SEM of at least 4 independent experiments. *P < 0.02; ** P < 0.008; **** P < 0.0001 comparing wt-TTP or mt-TTP to Tet-On control cells; #P < 0.01.
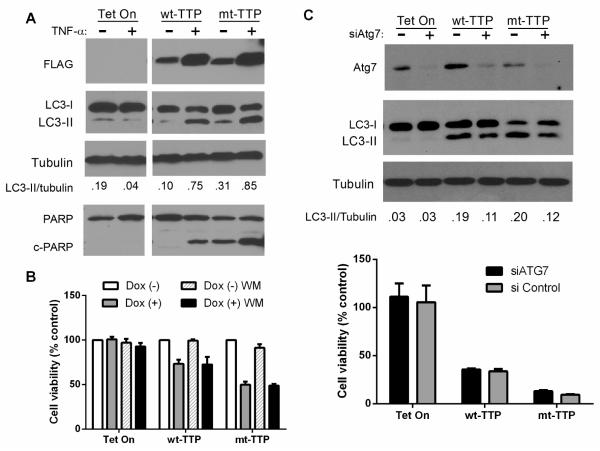
3.5. TTP causes the accumulation of autophagic vacuoles and induction of apoptosis in glioma cells
We postulated that depletion of trophic factors by TTP may provide a stimulus for macroautophagy induction. To assess for autophagy, we initially performed immunocytochemistry of doxycycline-induced TTP cells using an antibody to LC3-I/II, and found abundant immunoreactivity (Supplementary Fig. 5). Under conditions of cell stress that lead to macroautophagy induction, soluble LC3-I becomes LC3-II upon its cleavage and lipidation, which allows for its insertion into autophagosome membranes [33]. Thus, LC3-II is used widely as a selective biochemical marker of autophagosomes [34]. To detect this conversion, we performed Western blot analysis and found an increase in LC3-II for both TTP forms which was accentuated by TNF-α stimulation (Fig. 5A). To assess apoptosis in TTP-induced cell death we analyzed cleavage of PARP. In unstimulated cells, cleaved PARP (c-PARP) was barely detectable in wt-TTP cells but was robust in mt-TTP cells. With TNF-α stimulation, levels of c-PARP increased with both TTP forms, but more so with mt-TTP. We further assessed activation of apoptosis by investigating cleavage of caspase 3 and 8, two important components of the extrinsic pathway (Supplementary Fig. 6). We found cleavage of both caspases, visibly greater with the mutant form. Even at the lowest dose of TTP expression, (0.01 μg/ml of doxycycline), there was caspase cleavage indicating that this biological effect is not an artifact of TTP overexpression. We assessed whether TTP-induced cell death is altered after suppressing autophagy. Inhibition of autophagy with wortmannin had no effect on cell viability (100 nM, Fig. 5B and 1 μM, Supplementary Fig. 7). Similarly, cell viability did not change with another autophagy inhibitor, 3-methyladenine (Supplementary Fig. 7). We next silenced Atg7 a protein that critically regulates autophagy induction (Fig. 5C). Compared to control siRNA, there was a marked decrease in levels of Atg7 and LC3-II, together indicating the effective knockdown of functional autophagy. Cell viability, however, was not altered under those conditions, confirming that macroautophagy induction per se is not responsible for TTP-induced cell death.
Fig. 5. mt-TTP induces autophagic vacuole accumulation and apoptosis.
A) Western blot analysis of glioma cells induced with doxycycline (0.25μg/ml) with or without TNF-α stimulation. Antibodies are shown to the left of the blot. LC3-II/LC3-I ratios were determined by densitometry. B) Cell viability assessment in glioma clones after pharmacological inhibition of autophagy with wortmannin (100 nM) for 4 h prior to doxycycline induction and TNF-α stimulation. Cell viability was determined at 24 h with an ATP-based luminescence assay, and values were expressed as a percent of DSMO-treated control cells. C) Cell viability assessment in glioma clones after silencing Atg7. Upper panel: Western blot analysis of glioma cells following transfection with Atg7 or control siRNA. Lower panel: assessment of cell viability as described above.
4. Discussion
TTP is a negative regulator of growth factor signaling and its induction downregulates key transcriptional and trophic factors that promote cell proliferation [30]. Many tumors have depressed levels of TTP [30, 35], but in malignant glioma, TTP is readily detected in high-grade tumors, but is extensively phosphorylated [12]. Since TTP phosphorylation, in general, has been associated with suppressed function [36], we tested the antiproliferative activity of a mutant form of TTP which lacked 8 serine residues known to be phosphorylated in vivo [13, 14, 24]. Mutant TTP showed enhanced trophic factor suppression, apoptosis, chemosensitivity and decreased anchorage-independent growth. Similar phenotypic effects were produced by knockdown of HuR in glioma cells [11]. HuR binds to IL-6, IL-8 and VEGF 3′ UTRs but produces the opposite effect of mRNA stabilization in glioma and other cells [11, 20, 21, 37-40]. This overlap in mRNA binding and target regulation has prompted the observation that the two proteins compete and that trophic factor overexpression in cancer cells requires both suppression of TTP function and gain of HuR function [12, 37, 41]. Three findings in our study suggest that the deleted phosphoserines allowed mt-TTP to escape the regulatory environment in glioma cells. First, mt-TTP protein was stable compared to wt-TTP, indicating that the 8 mutated serines play an integral role in regulating protein degradation in glioma cells. P38/MAPK has been reported to induce TTP stabilization through MK2 phosphorylation of S52 and S178 in the mouse orthologue (equivalent to S60 and S186 in human TTP) [16, 26]. The rapid degradation of wt-TTP following p38/MAPK and ERK inhibition observed here is consistent with these prior reports. S60 and S186, however, were preserved in mt-TTP indicating additional determinants of protein stability in the 8 mutated serine residues. Interestingly, mt-TTP showed higher expression in other non-glioma cancer cell lines (breast and prostate) suggesting that protein stabilization may not be limited to glioma cells.
The second finding was the cellular distribution of mt-TTP. Phosphorylation of TTP via MK2 at S52 and S178 has been linked to cytoplasmic translocation [26, 42]. In U251 and other malignant glioma cells, endogenous and ectopic wild-type TTP predominantly localizes to the cytosol ([26] and unpublished data). Our finding that mt-TTP was more prominent in the nuclear compartment suggests that cellular location is also regulated by one or more of the 8 mutated phosphoserines. TTP can associate with the p65 subunit of NF-κB and HDACs to inhibit transcription, and the nuclear redistribution of mt-TTP may explain the enhanced transcriptional repressor activity [28, 29]. This function is independent of its RNA binding/destabilizing capacity [29]. Dephosphorylated TTP also represses the TNF-α promoter in LPS-stimulated RAW macrophage cells [43].
The third finding of enhanced function centers on translational suppression of ARE mRNAs by mt-TTP. The fold decline in protein levels of growth factors was significantly higher in mt-TTP, which suggests translational silencing (Fig. 1). Reporter studies with the IL-8 3′ UTR supported this possibility (Fig. 2). TTP typically binds to AU-rich elements in the 3′ UTR and promotes degradation through recruitment of degrading enzymes (e.g. deadenylases, exonucleases and decapping enzymes) that reside in P bodies or the exosome [44-46]. TTP can silence translation without increasing degradation by sequestering the mRNA away from polysomes [47]. The decrease in binding of the targets to cytoplasmic HuR (Supplementary Fig. 2) would be further supportive as this RNA binding protein is generally linked to enhanced translation of ARE-containing mRNAs [48, 49]. Our experimental conditions included TNF-α stimulation which maximizes cytoplasmic accumulation of mt-TTP thus promoting its negative effect on translational efficiency.
While this study of a single glioma cell line precludes overarching conclusions to gliomas, the mRNA targets examined here play an integral role at many levels of glioma tumor progression [3-10, 30, 50]. All three targets, especially VEGF, are produced in high amounts and are linked to angiogenesis [3, 4]. More recently, IL-6 has been characterized as a paracrine factor involved in cellular cross-talk within glioma tumors to promote cell proliferation and tumor cell heterogeneity through activation of epidermal growth factor receptor [5]. Both IL-6 and IL-8 promote a glioma stem-like cell phenotype, invasion and possibly chemoresistance [6-10, 50]. Because many pro-tumor mRNAs have AREs in the 3′ UTR, there are likely additional targets affected by increased TTP activity [30, 35]. Suppression of transcriptional factors, (e.g. NF-kB and AP-1), substantially broadens the pool of affected targets beyond ARE-containing mRNAs. Therefore, the anti-proliferative phenotype observed here may be related to a number of altered gene targets. Although autophagy did not appear to affect cell viability, the lack of complete attenuation of LC3-II induction with Atg7 silencing suggests that TTP may be inhibiting lysosomal degradation of LC3-II. Thus, alteration of autophagy by lysosomal inhibition may be contributing to the deleterious effects of TTP. This possibility warrants future investigation.
Supplementary Material
Acknowledgments
Funding Grant support: Department of Veterans Affairs Research, Merit Review grants (PHK and JJS), National Institutes of Health grants (RO1 NS064133 to PHK, CA131468 to EAS, and NS057664 to LL).
The sponsors played no role in the design of experiments, interpretation of results, or writing this manuscript.
Footnotes
Conflict of Interest statement The authors report no conflict of interest
References
- 1.Sanduja S, Blanco FF, Dixon DA. The roles of TTP and BRF proteins in regulated mRNA decay. Wiley Interdiscip Rev RNA. 2011;2:42–57. doi: 10.1002/wrna.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blackshear PJ. Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover. Biochem Soc Trans. 2002;30:945–952. doi: 10.1042/bst0300945. [DOI] [PubMed] [Google Scholar]
- 3.Fischer I, Gagner J-P, Law M, Newcomb EW, Zagzag D. Angiogenesis in Gliomas: Biology and Molecular Pathophysiology. Brain Pathology. 2005;15:297–310. doi: 10.1111/j.1750-3639.2005.tb00115.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brat DJ, Bellail AC, Van Meir EG. The role of interleukin-8 and its receptors in gliomagenesis and tumoral angiogenesis. Neuro-oncol. 2005;7:122–133. doi: 10.1215/S1152851704001061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Inda MM, Bonavia R, Mukasa A, Narita Y, Sah DW, Vandenberg S, Brennan C, Johns TG, Bachoo R, Hadwiger P, Tan P, Depinho RA, Cavenee W, Furnari F. Tumor heterogeneity is an active process maintained by a mutant EGFR-induced cytokine circuit in glioblastoma. Genes Dev. 2010;24:1731–1745. doi: 10.1101/gad.1890510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jin X, Yin J, Kim S-H, Sohn Y-W, Beck S, Lim YC, Nam D-H, Choi Y-J, Kim H. EGFR-AKT-Smad Signaling Promotes Formation of Glioma Stem-like Cells and Tumor Angiogenesis by ID3-Driven Cytokine Induction. Cancer Res. 2011;71:7125–7134. doi: 10.1158/0008-5472.CAN-11-1330. [DOI] [PubMed] [Google Scholar]
- 7.Beier D, Schulz J, Beier C. Chemoresistance of glioblastoma cancer stem cells - much more complex than expected. Molecular Cancer. 2011;10:128. doi: 10.1186/1476-4598-10-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de la Iglesia N, Konopka G, Lim KL, Nutt CL, Bromberg JF, Frank DA, Mischel PS, Louis DN, Bonni A. Deregulation of a STAT3-interleukin 8 signaling pathway promotes human glioblastoma cell proliferation and invasiveness. J Neurosci. 2008;28:5870–5878. doi: 10.1523/JNEUROSCI.5385-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raychaudhuri B, Vogelbaum M. IL-8 is a mediator of NF-κB induced invasion by gliomas. J Neuro-Onc. 2011;101:227–235. doi: 10.1007/s11060-010-0261-2. [DOI] [PubMed] [Google Scholar]
- 10.Liu Q, Li G, Li R, Shen J, He Q, Deng L, Zhang C, Zhang J. IL-6 promotion of glioblastoma cell invasion and angiogenesis in U251 and T98G cell lines. J Neurooncol. 2010;100:165–176. doi: 10.1007/s11060-010-0158-0. [DOI] [PubMed] [Google Scholar]
- 11.Filippova N, Yang X, Wang Y, Gillespie GY, Langford C, King PH, Wheeler C, Nabors LB. The RNA-Binding Protein HuR Promotes Glioma Growth and Treatment Resistance. Molecular Cancer Research. 2011;9:648–659. doi: 10.1158/1541-7786.MCR-10-0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suswam E, Li Y, Zhang X, Gillespie GY, Li X, Shacka JJ, Lu L, Zheng L, King PH. Tristetraprolin Down-regulates Interleukin-8 and Vascular Endothelial Growth Factor in Malignant Glioma Cells. Cancer Res. 2008;68:674–682. doi: 10.1158/0008-5472.CAN-07-2751. [DOI] [PubMed] [Google Scholar]
- 13.Cao H, Deterding LJ, Blackshear PJ. Phosphorylation site analysis of the anti-inflammatory and mRNA-destabilizing protein tristetraprolin. Expert Rev Proteomics. 2007;4:711–726. doi: 10.1586/14789450.4.6.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao H, Deterding LJ, Venable JD, Kennington EA, Yates JR, Tomer KB, Blackshear PJ. Identification of the anti-inflammatory protein tristetraprolin as a hyperphosphorylated protein by mass spectrometry and site-directed mutagenesis. Biochem J. 2006;394:285–297. doi: 10.1042/BJ20051316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carballo E, Cao H, Lai WS, Kennington EA, Campbell D, Blackshear PJ. Decreased Sensitivity of Tristetraprolin-deficient Cells to p38 Inhibitors Suggests the Involvement of Tristetraprolin in the p38 Signaling Pathway. J Biol Chem. 2001;276:42580–42587. doi: 10.1074/jbc.M104953200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hitti E, Iakovleva T, Brook M, Deppenmeier S, Gruber AD, Radzioch D, Clark AR, Blackshear PJ, Kotlyarov A, Gaestel M. Mitogen-Activated Protein Kinase-Activated Protein Kinase 2 Regulates Tumor Necrosis Factor mRNA Stability and Translation Mainly by Altering Tristetraprolin Expression, Stability, and Binding to Adenine/Uridine-Rich Element. Mol Cell Biol. 2006;26:2399–2407. doi: 10.1128/MCB.26.6.2399-2407.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Y-L, Huang Y-L, Lin N-Y, Chen H-C, Chiu W-C, Chang C-J. Differential regulation of ARE-mediated TNFα and IL-1β mRNA stability by lipopolysaccharide in RAW264.7 cells. Biochem Biophys Res Commun. 2006;346:160–168. doi: 10.1016/j.bbrc.2006.05.093. [DOI] [PubMed] [Google Scholar]
- 18.Qian X, Ning H, Zhang J, Hoft DF, Stumpo DJ, Blackshear PJ, Liu J. Posttranscriptional Regulation of IL-23 Expression by IFN-{gamma} through Tristetraprolin. Journal of Immunology. 2011 doi: 10.4049/jimmunol.1002672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meng Z, Jackson NL, Choi H, King PH, Emanuel PD, Blume SW. Alterations in RNA-binding activities of IRES-regulatory proteins as a mechanism for physiological variability and pathological dysregulation of IGF-IR translational control in human breast tumor cells. Journal of Cellular Physiology. 2008;217:172–183. doi: 10.1002/jcp.21486. [DOI] [PubMed] [Google Scholar]
- 20.Nabors LB, Gillespie GY, Harkins L, King PH. HuR, an RNA stability factor, is expressed in malignant brain tumors and binds to adenine and uridine-rich elements within the 3′ untranslated regions of cytokine and angiogenic factor mRNAs. Cancer Res. 2001;61:2154–2161. [PubMed] [Google Scholar]
- 21.Nabors LB, Suswam E, Huang Y, Yang X, Johnson MJ, King PH. Tumor Necrosis Factor-α Induces Angiogenic Factor Up-Regulation in Malignant Glioma Cells: A Role for RNA Stabilization and HuR. Cancer Res. 2003;63:4181–4187. [PubMed] [Google Scholar]
- 22.Suswam EA, Nabors LB, Huang Y, Yang X, King PH. IL-1β induces stabilization of IL-8 mRNA in malignant breast cancer cells via the 3′ untranslated region: Involvement of divergent RNA-binding factors HuR, KSRP and TIAR. Int J Cancer. 2005;113:911–919. doi: 10.1002/ijc.20675. [DOI] [PubMed] [Google Scholar]
- 23.Vasudevan S, Steitz JA. AU-Rich-Element-Mediated Upregulation of Translation by FXR1 and Argonaute 2. Cell. 2007;128:1105–1118. doi: 10.1016/j.cell.2007.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chrestensen CA, Schroeder MJ, Shabanowitz J, Hunt DF, Pelo JW, Worthington MT, Sturgill TW. MAPKAP Kinase 2 Phosphorylates Tristetraprolin on in Vivo Sites Including Ser178, a Site Required for 14-3-3 Binding. J Biol Chem. 2004;279:10176–10184. doi: 10.1074/jbc.M310486200. [DOI] [PubMed] [Google Scholar]
- 25.Maher EA, Furnari FB, Bachoo RM, Rowitch DH, Louis DN, Cavenee WK, DePinho RA. Malignant glioma: genetics and biology of a grave matter. Genes Dev. 2001;15:1311–1333. doi: 10.1101/gad.891601. [DOI] [PubMed] [Google Scholar]
- 26.Brook M, Tchen CR, Santalucia T, McIlrath J, Arthur JSC, Saklatvala J, Clark AR. Posttranslational Regulation of Tristetraprolin Subcellular Localization and Protein Stability by p38 Mitogen-Activated Protein Kinase and Extracellular Signal-Regulated Kinase Pathways. Mol Cell Biol. 2006;26:2408–2418. doi: 10.1128/MCB.26.6.2408-2418.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fiedler MA, Wernke-Dollries K, Stark JM. Mechanism of RSV-induced IL-8 gene expression in A549 cells before viral replication. Am J Physiol Lung Cell Mol Physiol. 1996;271:L963–971. doi: 10.1152/ajplung.1996.271.6.L963. [DOI] [PubMed] [Google Scholar]
- 28.Liang J, Lei T, Song Y, Yanes N, Qi Y, Fu M. RNA-destabilizing factor tristetraprolin negatively regulates NF-kappaB signaling. J Biol Chem. 2009;284:29383–29390. doi: 10.1074/jbc.M109.024745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schichl YM, Resch U, Hofer-Warbinek R, de Martin R. Tristetraprolin impairs NF-kappaB/p65 nuclear translocation. J Biol Chem. 2009;284:29571–29581. doi: 10.1074/jbc.M109.031237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amit I, Citri A, Shay T, Lu Y, Katz M, Zhang F, Tarcic G, Siwak D, Lahad J, Jacob-Hirsch J, Amariglio N, Vaisman N, Segal E, Rechavi G, Alon U, Mills GB, Domany E, Yarden Y. A module of negative feedback regulators defines growth factor signaling. Nat Genet. 2007;39:503–512. doi: 10.1038/ng1987. [DOI] [PubMed] [Google Scholar]
- 31.Meyer-Ficca ML, Meyer RG, Kaiser H, Brack AR, Kandolf R, Kupper J-H. Comparative analysis of inducible expression systems in transient transfection studies. Anal Biochem. 2004;334:9–19. doi: 10.1016/j.ab.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 32.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJB, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, Gorlia T, Allgeier A, Lacombe D, Cairncross JG, Eisenhauer E, Mirimanoff RO. Radiotherapy plus Concomitant and Adjuvant Temozolomide for Glioblastoma. NEJM. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 33.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brennan SE, Kuwano Y, Alkharouf N, Blackshear PJ, Gorospe M, Wilson GM. The mRNA-destabilizing protein tristetraprolin is suppressed in many cancers, altering tumorigenic phenotypes and patient prognosis. Cancer Res. 2009;69:5168–5176. doi: 10.1158/0008-5472.CAN-08-4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ronkina N, Menon MB, Schwermann J, Tiedje C, Hitti E, Kotlyarov A, Gaestel M. MAPKAP kinases MK2 and MK3 in inflammation: complex regulation of TNF biosynthesis via expression and phosphorylation of tristetraprolin. Biochem Pharmacol. 2010;80:1915–1920. doi: 10.1016/j.bcp.2010.06.021. [DOI] [PubMed] [Google Scholar]
- 37.Young LE, Sanduja S, Bemis-standoli K, Pena EA, Price RL, Dixon DA. The mRNA Binding Proteins HuR and Tristetraprolin Regulate Cyclooxygenase 2 Expression During Colon Carcinogenesis. Gastroenterology. 2009;136:1669–1679. doi: 10.1053/j.gastro.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dixon DA, Tolley ND, King PH, Nabors LB, McIntyre TM, Zimmerman GA, Prescott SM. Altered expression of the mRNA stability factor HuR promotes cyclooxygenase-2 expression in colon cancer cells. J Clin Invest. 2001;108:1657–1665. doi: 10.1172/JCI12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levy NS, Chung S, Furneaux H, Levy AP. Hypoxic Stabilization of Vascular Endothelial Growth Factor mRNA by the RNA-binding Protein HuR. J Biol Chem. 1998;273:6417–6423. doi: 10.1074/jbc.273.11.6417. [DOI] [PubMed] [Google Scholar]
- 40.Spooren A, Mestdagh P, Rondou P, Kolmus K, Haegeman G, Gerlo S. IL-1[beta] potently stabilizes IL-6 mRNA in human astrocytes. Biochem Pharmacol. 2011;81:1004–1015. doi: 10.1016/j.bcp.2011.01.019. [DOI] [PubMed] [Google Scholar]
- 41.Ming X-F, Stoecklin G, Lu M, Looser R, Moroni C. Parallel and Independent Regulation of Interleukin-3 mRNA Turnover by Phosphatidylinositol 3-Kinase and p38 Mitogen-Activated Protein Kinase. Mol Cell Biol. 2001;21:5778–5789. doi: 10.1128/MCB.21.17.5778-5789.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnson BA, Stehn JR, Yaffe MB, Blackwell TK. Cytoplasmic Localization of Tristetraprolin Involves 14-3-3-dependent and -independent Mechanisms. J Biol Chem. 2002;277:18029–18036. doi: 10.1074/jbc.M110465200. [DOI] [PubMed] [Google Scholar]
- 43.Zhu W, Brauchle MA, Di Padova F, Gram H, New L, Ono K, Downey JS, Han J. Gene suppression by tristetraprolin and release by the p38 pathway. Am J Physiol Lung Cell Mol Physiol. 2001;281:L499–508. doi: 10.1152/ajplung.2001.281.2.L499. [DOI] [PubMed] [Google Scholar]
- 44.Sandler H, Stoecklin G. Control of mRNA decay by phosphorylation of tristetraprolin. Biochem Soc Trans. 2008;36:491–496. doi: 10.1042/BST0360491. [DOI] [PubMed] [Google Scholar]
- 45.Anderson P, Kedersha N. RNA granules. J Cell Biol. 2006;172:803–808. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clement SL, Scheckel C, Stoecklin G, Lykke-Andersen J. Phosphorylation of Tristetraprolin by MK2 Impairs AU-Rich Element mRNA Decay by Preventing Deadenylase Recruitment. Mol Cell Biol. 2011;31:256–266. doi: 10.1128/MCB.00717-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Franks TM, Lykke-Andersen J. TTP and BRF proteins nucleate processing body formation to silence mRNAs with AU-rich elements. Genes Dev. 2007;21:719–735. doi: 10.1101/gad.1494707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brennan CM, Steitz JA. HuR and mRNA stability. Cell Mol Life Sci. 2001;58:266–277. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abdelmohsen K, Yuki K, Kim HH, Gorospe M. Posttranscriptional gene regulation by RNA-binding proteins during oxidative stress: implications for cellular senescence. Biol Chem. 2008;389:243–255. doi: 10.1515/BC.2008.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meads MB, Hazlehurst LA, Dalton WS. The Bone Marrow Microenvironment as a Tumor Sanctuary and Contributor to Drug Resistance. Clin Cancer Res. 2008;14:2519–2526. doi: 10.1158/1078-0432.CCR-07-2223. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.