Abstract
Introduction
Nuclear EGFR (nEGFR) has been identified in various human tumor tissues, including cancers of the breast, ovary, oropharynx, and esophagus, and has predicted poor patient outcomes. We sought to determine if protein expression of nEGFR is prognostic in early stage non-small cell lung cancer (NSCLC).
Methods
Resected stage I and II NSCLC specimens were evaluated for nEGFR protein expression using immunohistochemistry (IHC). Cases with at least one replicate core containing ≥5% of tumor cells demonstrating strong dot-like nucleolar EGFR expression were scored as nEGFR positive.
Results
Twenty-three (26.1% of the population) of 88 resected specimens stained positively for nEGFR. Nuclear EGFR protein expression was associated with higher disease stage (45.5% of stage II vs. 14.5% of stage I; p=0.023), histology (41.7% in squamous cell carcinoma vs. 17.1% in adenocarcinoma; p=0.028), shorter progression-free survival (PFS) (median PFS 8.7 months [95% CI 5.1–10.7 mo] for nEGFR positive vs. 14.5 months [95% CI 9.5–17.4 mo] for nEGFR negative; hazard ratio (HR) of 1.89 [95% CI 1.15–3.10]; p=0.011), and shorter overall survival (OS) (median OS 14.1 months [95% CI 10.3–22.7 mo] for nEGFR positive vs. 23.4 months [95% CI 20.1–29.4 mo] for nEGFR negative; HR of 1.83 [95% CI 1.12–2.99]; p=0.014).
Conclusions
Expression of nEGFR protein was associated with higher stage and squamous cell histology, and predicted shorter PFS and OS, in this patient cohort. Nuclear EGFR serves as a useful independent prognostic variable and as a potential therapeutic target in NSCLC.
Keywords: non-small cell lung cancer, nuclear, epidermal growth factor receptor, prognosis, biomarker, survival analysis
1. Introduction
Non-small cell lung cancer is a heterogeneous malignancy, comprised of multiple histologic subtypes. Predicting the course of disease based upon staging is suboptimal. The identification of biological markers of aggressive clinical behavior is needed in an effort to individualize treatment and develop novel therapeutic targets.
Protein expression of membrane bound EGFR was neither prognostic nor predictive of efficacy with the use of erlotinib, gefitinib, or cetuximab in NSCLC (1,2). However, emerging preclinical and clinical evidence supports the role of nEGFR in enhancing tumor cell growth, survival, and resistance to systemic and radiation therapies (3–10). Herein, we report identification of nEGFR protein expression as an independent prognostic variable in early stage NSCLC.
2.0 Materials and Methods
2.1 Patients and specimen collection
For this retrospective analysis of patients who underwent curative intent resections, de-identified tumor specimens from 88 deceased patients with stages I and II NSCLC were collected from the University of Wisconsin Hospitals and Clinics (UWHC; Madison, WI) and from the Gundersen Lutheran Medical Center (GLMC; LaCrosse, WI). Patients did not receive either pre- or post-operative anti-cancer therapy. We also collected: age, sex, histology, smoking history, pathologic stage (AJCC Staging 6th edition), type of resection, date of relapse, and date of death. Approval for this research was obtained from the IRBs of UW-Madison and the GLMC.
2.2 Tissue microarray construction and protein expression analyses
Tumor tissue quality and pathology were confirmed by the study pathologist (DTY). Tissues were harvested within 30 minutes of resection, fixed with 10% neutral buffered formalin and embedded in paraffin. Areas of tumor and adjacent benign tissue were marked on a representative H & E stained section. Duplicate 0.6 mm cores from the corresponding paraffin block were punched out and assembled with a Manual Tissue Arrayer (Beecher Instruments, Sun Prairie, WI).
For nEGFR protein expression analyses, tissue sections were de-paraffinized and antigen retrieval was performed in citrate buffer (pH.6.0) with 0.05% Tween-20. Samples were incubated with EGFR polyclonal antibody (sc-03, Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) overnight at 4C. Samples were washed and incubated in secondary antibody for 1 hour followed by incubation with Vectastain ABC Elite kit (Vector Laboratories, Burlingame, CA, USA). 3,3-diaminobenzidine staining was used as the color-developing reagent. Slides were counterstained with Mayer hematoxylin, dehydrated through a graded series of ethanol washes to xylene, and coverslipped with Permount (Fisher, Spring- field, NJ).
We initially hypothesized that assessment of nEGFR protein would require the quantitative and subcellular localization capacity of automated quantitative analysis (AQUA). When we observed that the nuclear staining of EGFR protein revealed a distinct, robust nucleolar pattern (Figure 1A) that clearly contrasted with negative cases (Figure 1B) using routine IHC staining, we switched to the IHC methodology due to its easier translation to clinical practice. The nEGFR staining pattern was scored by the study pathologist at 5% increments by visual estimation at 20X magnification. Accordingly, cases with at least one replicate core containing at least 5% of tumor cells demonstrating strong dot-like nucleolar EGFR IHC protein expression were scored as nEGFR positive.
Figure 1. Nuclear EGFR (nEGFR) is detected in early stage NSCLC specimens.
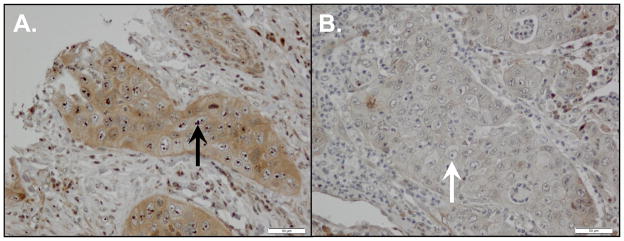
We analyzed 88 primary NSCLC tumors for nEGFR protein expression using immunohistochemistry. (A) Representative case demonstrating nEGFR expression. All positive cases had a similar distinctive pattern of strong nucleolar staining (black arrow). (B) Representative case demonstrating a lack of nEGFR protein expression. Despite the presence of prominent nucleoli, no nEGFR protein is detected (white arrow).
2.3 Statistical analyses
Our endpoints were protein expression of nEGFR and PFS and OS. Originally this study had an approximate power of 0.902, 0.747 and 0.477 to detect a hazard ratio of 2, 1.75 and 1.5, respectively, using a two-sided log-rank test at a significance level 0.05, given the sample size of 88 when the AQUA score was dichotomized using its median. The prognostic impact of nEGFR was assessed using the log-rank test and Cox proportional hazards regression models for PFS and OS. Kaplan-Meier method was used to summarize PFS and OS for patients per nEGFR IHC. Association between nEGFR protein expression and sex, histology, smoking history and pathologic stage was assessed using Fisher’s exact test.
3.0 Results
3.1 Patient characteristics
Table 1 summarizes the characteristics of the 88 patient samples studied. None of the patients received either pre- or post-operative anti-cancer therapy. The median PFS and OS for our population were 11.3 months (95% CI 9.1–16.2 mo) and 22.0 months (95% CI 15.9–24.7 mo), respectively, shorter than expected. Fifty-nine patients experienced disease relapse. Since only four patients were non-smokers, and seven underwent a pneumonectomy, these two clinical characteristics were dropped from further analyses.
Table 1.
Patient Characteristics
| N | |
|---|---|
|
| |
| Number of patients | 88 |
| Median age (range) | 73 (43–96 yrs) |
| Sex | |
| Male | 55 (62.5%) |
| Female | 33 (37.5%) |
| Histology | |
| Adenocarcinoma | 41 (46.6%) |
| Squamous cell | 36 (40.9%) |
| Bronchioloalveolar | 4 (4.5%) |
| Large cell | 3 (3.4%) |
| Non-small cell, NOS | 2 (2.3%) |
| Adenosquamous carcinoma | 2 (2.3%) |
| Smoking history | |
| Current or former | 84 (95.5%) |
| Type of surgery | |
| Lobectomy | 80 (90.9%) |
| Pneumonectomy | 7 (8%) |
| Bilobectomy | 1 (1.1%) |
| Disease Stage | |
| IA | 23 (26.1%) |
| IB | 32 (36.4) |
| IIA | 9 (10.2%) |
| IIB | 24 (27.3%) |
| T Stage | |
| T1 | 31 (35.2%) |
| T2 | 52 (59.1%) |
| T3 | 5 (5.7%) |
| N Stage | |
| N0 | 60 (68.2%) |
| N1 | 28 (31.8%) |
| Nuclear EGFR protein expression | |
| Positive | 23 (26.1%) |
Twenty-three (26.1% of the population) of 88 patients had specimens that stained positively for nEGFR (Figure 1A). When nEGFR expression was seen, greater than 40% of tumor cells were positive in most cases. Nuclear EGFR was seen in between 1% and 4% of tumor cells very rarely (4/165 tumor cores). Control cores comprised of EGFR positive ductal carcinoma of the breast and matched adjacent normal lung from each tumor were represented on the TMA as external and internal controls, respectively. Cytoplasmic and membrane EGFR staining were confirmed in the breast control, and no nEGFR expression was observed in any of the adjacent normal lung tissue. Table 2 depicts the distribution of nEGFR positivity per IHC staining across our tumor samples.
Table 2.
Distribution of nuclear EGFR protein staining per IHC across all tumor specimens
| Patient number | Percent of cells with positive nuclear EGFR protein staining per IHC | |||
|---|---|---|---|---|
| Cores (all specimens run in duplicate when tissue available) | ||||
| Tumor 1 | Tumor 2 | Adjacent normal lung 1 | Adjacent normal lung 2 | |
| 1 | 50 | NC | 0 | 0 |
| 2 | 80 | 60 | 0 | 0 |
| 3 | 80 | 20 | 0 | 0 |
| 4 | 50 | 75 | 0 | 0 |
| 5 | 95 | 50 | 0 | 0 |
| 6 | 25 | 25 | 0 | 0 |
| 7 | 0 | 5 | 0 | 0 |
| 8 | 60 | 20 | NC | NC |
| 9 | 10 | 5 | 0 | 0 |
| 10 | 60 | 50 | NC | NC |
| 11 | 20 | 30 | 0 | 0 |
| 12 | 30 | 80 | 0 | 0 |
| 13 | 5 | 10 | 0 | 0 |
| 14 | 80 | 90 | 0 | 0 |
| 15 | 15 | 5 | 0 | 0 |
| 16 | 30 | 100 | 0 | 0 |
| 17 | 20 | NC | 0 | 0 |
| 18 | 60 | 70 | 0 | 0 |
| 19 | 40 | 70 | 0 | 0 |
| 20 | 90 | 90 | 0 | 0 |
| 21 | 30 | NC | NC | NC |
| 22 | 40 | 60 | 0 | 0 |
| 23 | 30 | 5 | 0 | 0 |
| Specimens from remaining 65 patients | 0 | 0 | 0 | 0 |
NC = no core available
3.2 Nuclear EGFR protein expression and survival
According to the logrank test, nEGFR protein positivity was associated with shorter PFS (median PFS 8.7 months [95% CI 5.1–10.7 mo] for nEGFR positive vs. 14.5 months [95% CI 9.5–17.4 mo] for nEGFR negative; HR=1.89 [95% CI 1.15–3.10]; p=0.011), and shorter OS (median OS 14.1 months [95% CI 10.3–22.7 mo] for nEGFR positive vs. 23.4 months [95% CI 20.1–29.4 mo] for nEGFR negative; HR=1.83 [95% CI 1.12–2.99]; p=0.014).
3.3 Nuclear EGFR protein expression and prognosis
According to Fisher’s exact test, nEGFR protein positivity was associated with squamous cell histology, compared to adenocarcinoma (nEGFR positive in 41.7% of patients’ samples with squamous cell vs. 17.1% in adenocarcinoma specimens, p=0.028), and with higher disease stage (nEGFR positive in 45.5% of stage II vs. 14.5% of stage I, p=0.023). Nuclear EGFR protein expression was not associated with patient’s sex, or T or N status.
According to Cox proportional hazard models, of the baseline clinical characteristics (sex, disease stage, histology, T, N, and age), only age was at least marginally associated with PFS (p=0.073), but was not associated with OS. Also nEGFR protein positivity in patients’ specimens was associated with shorter PFS, after controlling for age, with an HR of 1.68 (95% CI 1.01–2.81, p=0.046), and with shorter OS with an HR of 1.83 (95% CI 1.12–2.99, p=0.016).
4.0 Discussion
Nuclear EGFR was first observed in hepatocytes during liver regeneration. Translocation from the cell membrane to the nucleus has been reported with numerous receptor tyrosine kinases (RTKs), including all HER family receptors, MET, and VEGFR2 (3,4). Protein expression of nEGFR has correlated with shortened survival in cancers of the breast, ovary, and oropharyngeal and esophageal squamous cells. Approximately 25–50% of the tumor cells expressed nEGFR (5–8).
Nuclear translocation of full length EGFR can be initiated by ligand binding, irradiation, cetuximab, and cisplatin (4,9,10). Early events for movement of EGFR from the plasma membrane to the nucleus include phosphorylation of the dimerized receptor by SRC family kinases and AKT (10,11). These stimuli induce internalization to endocytic vesicles. EGFR then undergoes retrograde translocation through the Golgi apparatus to the endoplasmic reticulum, whereupon it moves from the outer nuclear membrane to the inner nuclear membrane via interaction between importin β and the nuclear pore complex. In the inner nuclear membrane, EGFR can interact with Sec61 for removal from the membrane and release into the nucleus (4,12).
Within the nucleus three functions have been identified for the EGFR. First, EGFR associates with STAT3, STAT5 and E2F1 to act as a transcriptional co-activator, independent of its kinase activity, to increase the expression of target genes that worsen the malignant phenotype (cyclin D1, iNOS, B-myb, c-Myc, Aurora kinase A, Breast Cancer Resistance Protein, and COX-2) (3,4,13). Second, nEGFR phosphorylates proliferating cell nuclear antigen, promoting DNA replication (14). Third, it activates DNA-dependent protein kinase within the nucleus, stimulating DNA repair following exposure to irradiation and cisplatin (15).
This study demonstrates that a distinct nucleolar pattern of EGFR protein was associated with significantly shorter PFS and OS, higher stage and squamous histology in patients with early stage NSCLC. These correlations were not confounded by exposure to additional anti-cancer therapies. A limitation of our study is our shorter than expected overall survival; this is most certainly related to the fact that all samples were selected from patients who had expired by the time of our analyses. Within our patient cohort, however, nEGFR protein expression was detected in just over a quarter of our samples and was statistically associated with higher stage and squamous histology. These results are consistent with findings from other disease sites (5–8).
Our group, and others, have shown in experimental models that nEGFR contributes to treatment resistance with cetuximab, gefitinib, erlotinib, and irradiation (10,11,15). For example, we demonstrated that NSCLC cells that developed acquired resistance to cetuximab expressed increased levels of nEGFR, and that forced expression of nEGFR rendered cetuximab-sensitive cells resistant to cetuximab, both in vitro and in vivo (3,10). Similarly, Liccardi and colleagues showed that cells expressing EGFR with mutations that impair nuclear transport demonstrated reduced repair of DNA strand breaks following ionizing radiation and reduced repair of interstrand cross-links following exposure to cisplatin, as compared to cells capable of directing EGFR to the nucleus (15). Conversely, sensitivity in cetuximab-resistant NSCLC cells was re-established after blocking nuclear translocation of EGFR by co-exposing cells to either dasatinib, a SRC family kinase inhibitor, or MK2206, an AKT inhibitor (10, 11).
Investigating the functions of nuclear RTKs in untreated cancer cells also serves as a focus of research (16). Using sequential immunoprecipitation and immunoelectron microscopy assays, Li and colleagues demonstrated that ErbB2 co-localizes with β-actin and RNA polymerase-I (RNA Pol I) to the nucleoli in multiple breast cancer cell lines. Activation of this complex enhanced binding of RNA Pol I to rDNA, expediting rRNA synthesis and protein translation. These authors proposed that localization of ErbB2 to the nucleus and nucleoli contributed to tumorigenesis by increasing rRNA synthesis and protein translation. Nuclear EGFR has been identified in multiple tumor types in patients who did not undergo prior EGFR inhibiting therapy (5–8), as was the case with our population. Biological mechanisms that signal localization of EGFR to the nucleolus in untreated patients, as well as the potential role of such localization in tumor development, are under study in our laboratory.
5.0 Conclusion
We have identified nEGFR as a predictor of shortened survival in patients with early stage NSCLC. Preclinical data highlights the kinase dependent and independent processes by which nEGFR stimulates tumor cell growth, progression, and survival (3,4,10,11). This raises the question of whether or not nEGFR represents not only a useful prognostic factor in NSCLC, but also a potential therapeutic target. The biological functions of nEGFR, and strategies to improve the efficacy of cetuximab, cisplatin and radiation by disrupting nuclear translocation of EGFR, remain the subjects of our translational research efforts.
Acknowledgments
This work was supported in part by the University of Wisconsin Carbone Cancer Center 2P30 CA014520-34, the University of Wisconsin Foundation Creating Hope Campaign for Lung Cancer Research, the Gundersen Lutheran Medical Foundation, the Clinical and Translational Science Award program, previously through the National Center for Research Resources grant 1UL1RR025011, and now through the National Center for Advancing Translational Sciences grant 9U54TR000021, grant RSG-10-193-01-TBG from the American Cancer Society (DLW), and by NIH grant T32 GM08.1061-01A2 from the Graduate Training in Cellular and Molecular Pathogenesis of Human Diseases (TMB). This manuscript was written solely by the authors; the funding sources for this project did not assist in the writing or reviewing of this submission and did not pay the authors for the conduct or writing of this work. The funding sources exerted no role in the design of this project, nor in the data collection, analyses, or interpretation.
Footnotes
Conflict of Interest
No author of this article had any financial or personal relationships with other people or organizations that could inappropriately influence or bias this article.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hirsch FR, Varella-Garcia M, Bunn PA, Jr, et al. Epidermal growth factor receptor in non-small-cell lung carcinomas: Correlation between gene copy number and protein expression and impact on prognosis. J Clin Oncol. 2003;21:3798–3807. doi: 10.1200/JCO.2003.11.069. [DOI] [PubMed] [Google Scholar]
- 2.Khambata-Ford S, Harbison CT, Hart LL, et al. Analysis of potential predictive markers of cetuixmab benefit in BMS099, a phase III study of cetuximab and first-line taxane/carboplatin in advanced non-small-cell lung cancer. J Clin Oncol. 2010;28:918–927. doi: 10.1200/JCO.2009.25.2890. [DOI] [PubMed] [Google Scholar]
- 3.Han W, Lo H-W. Landscape of EGFR signaling network in human cancers: Biology and therapeutic response in relation to receptor subcellular locations. Cancer Lett. 2012;318:124–134. doi: 10.1016/j.canlet.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brand TM, Iida M, Li C, et al. The nuclear epidermal growth factor receptor signaling network and its role in cancer. Discov Med. 2011;12:419–432. [PMC free article] [PubMed] [Google Scholar]
- 5.Lo H-W, Xia W, Wei Y, et al. Novel prognostic value of nuclear epidermal growth factor receptor in breast cancer. Cancer Res. 2005;65:338–348. [PubMed] [Google Scholar]
- 6.Psyrri A, Yu Z, Weinberger PM, et al. Quantitative determination of nuclear and cytoplasmic epidermal growth factor receptor expression in oropharyngeal squamous cell cancer by using automated quantitative analysis. Clin Cancer Res. 2005;11:5856–5862. doi: 10.1158/1078-0432.CCR-05-0420. [DOI] [PubMed] [Google Scholar]
- 7.Hishino M, Fukui H, Ono Y, et al. Nuclear expression of phosphorylated EGFR is associated with poor prognosis in patients with esophageal squamous cell carcinoma. Pathobiology. 2007;74:15–21. doi: 10.1159/000101047. [DOI] [PubMed] [Google Scholar]
- 8.Xia W, Wei Y, Du Y, et al. Nuclear expression of epidermal growth factor receptor is a novel prognostic value in patients with ovarian cancer. Mol Carcinog. 2009;48:610–617. doi: 10.1002/mc.20504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dittmann K, Mayer C, Fehrenbacher B, et al. Radiation-induced epidermal growth factor receptor nuclear import is linked to activation of DNA-dependent protein kinase. J Biol Chem. 2005;280:31182–31189. doi: 10.1074/jbc.M506591200. [DOI] [PubMed] [Google Scholar]
- 10.Li C, Iida M, Dunn EF, et al. Nuclear EGFR contributes to acquired resistance to cetuximab. Oncogene. 2009;28:3801–3813. doi: 10.1038/onc.2009.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang W-C, Chen Y-J, Li L-Y, et al. Nuclear translocation of epidermal growth factor receptor by Akt-dependent phosphorylation enhances breast cancer-resistant protein expression in gefintib-resistant cells. J Biol Chem. 2011;286:20558–20568. doi: 10.1074/jbc.M111.240796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang YN, Yamaguchi H, Hui L, et al. The translocon Sec61β localized in the inner nuclear membrane transports membrane-embedded EGF receptor to the nucleus. J Biol Chem. 2010;285:38720–38729. doi: 10.1074/jbc.M110.158659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanada N, Lo H-W, Day C-P, et al. Co-regulation of B-myb expression by E2F1 and EGF receptor. Mol Carcinog. 2006;45:10–17. doi: 10.1002/mc.20147. [DOI] [PubMed] [Google Scholar]
- 14.Wang SC, Nakajima Y, Yu YL, et al. Tyrosine phosphorylation controls PCNA function through protein stability. Nat Cell Biol. 2006;8:1359–1368. doi: 10.1038/ncb1501. [DOI] [PubMed] [Google Scholar]
- 15.Liccardi G, Hartley JA, Hochhauser D. EGFR nuclear translocation modulates DNA repair following cisplatin and ionizing radiation. Cancer Res. 2011;71:1103–1114. doi: 10.1158/0008-5472.CAN-10-2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li LY, Chen H, Hsieh YH, et al. Nuclear erbB2 enhances translation and cell growth by activating transcription of ribosomal RNA genes. Cancer Res. 2011;71:4269–79. doi: 10.1158/0008-5472.CAN-10-3504. [DOI] [PMC free article] [PubMed] [Google Scholar]


