Abstract
Hexanucleotide repeat expansions in C9ORF72 are a common cause of familial and apparently sporadic amyotrophic lateral sclerosis (ALS) and frontal temporal dementia (FTD). The mechanism by which expansions cause neurodegeneration is unknown, but current evidence supports both loss-of-function and gain-of-function mechanisms. We used pooled next-generation sequencing of the C9ORF72 gene in 389 ALS patients to look for traditional loss-of-function mutations. Although rare variants were identified, none were likely to be pathogenic, suggesting that mutations other than the repeat expansion are not a common cause of ALS, and providing supportive evidence for a gain-of-function mechanism. We also show by repeat-primed PCR genotyping that the C9ORF72 expansion frequency varies by geographical region within the United States, with an unexpectedly high frequency in the Mid-West. Finally we also show evidence of somatic instability of the expansion size by Southern blot, with the largest expansions occurring in brain tissue.
Keywords: Amyotrophic lateral sclerosis, genetics, C9ORF72 hexanucleotide repeat, C9ORF72
1. INTRODUCTION
Expansions of an intronic hexanucleotide repeat (GGGGCC) in the C9ORF72 gene are the most common genetic cause of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) in Caucasian populations, explaining 37% of familial ALS, 21% of familial FTD, and 6% of cases with sporadic ALS or FTD (DeJesus-Hernandez et al., 2011; Majounie et al., 2012b; Mok et al., 2012; Rademaker 2012; Renton et al., 2011; Smith et al., 2012). Although the clinical phenotype of ALS and FTD with C9ORF72 repeat expansion patients is generally typical for these diseases, some statistical differences have emerged in some studies: a lower age of onset, shorter survival, increased incidence of neurodegenerative disease in relatives, and a propensity toward psychosis/hallucinations as a feature of dementia(Byrne et al., 2012; Snowden et al., 2012). Rare expansion carriers meeting diagnostic criteria for a growing list of neurodegenerative syndromes have also been reported, including Alzheimer’s type dementia, corticobasal syndrome, spinocerebellar ataxia, and parkinsonism(Harms et al., 2012; Lindquist et al., 2012; Majounie et al., 2012a; Rollinson et al., 2012).
Although the prevalence of C9ORF72 expansions and the resulting phenotypes have been quickly elucidated on clinical, cognitive, imaging, and neuropathologic levels, the mechanism by which C9ORF72 repeat expansions cause neurodegeneration is unknown at this point. Limited functional data and analogy to other neurological disorders caused by non-coding repeat expansions (e.g. myotonic dystrophy, fragile X, spinocerebellar ataxia type 36) have led to proposed loss-of-function and gain-of-function mechanisms (DeJesus-Hernandez et al., 2011; Renton et al., 2011). The discovery of pathogenic mutations in C9ORF72 other than the repeat expansion (single nucleotide substitutions or short insertions/deletions producing splicing or missense/nonsense changes) would shed significant light on this issue. Loss-of-function mechanisms for two other non-coding repeat disorders, fragile X mental retardation syndrome and Friedreich’s ataxia, were in fact confirmed and solidified by the identification of such mutations in a minority of patients (Campuzano et al., 1996; Cossee et al., 1999; De Boulle et al., 1993; Lugenbeel et al., 1995). In this study, we characterize the frequency and phenotype of ALS patients in the United States with C9ORF72 repeat expansions, and sequence the C9ORF72 gene in a cohort of ALS patients using next generation sequencing.
2. METHODS
2.1. Study Subjects
The familial ALS (FALS) cohort included 51 probands evaluated and followed at Washington University in St. Louis, Missouri (n=40) or Virginia Mason Medical Center in Seattle, Washington (n=11). All probands had been diagnosed with probable or definite ALS according to El Escorial Criteria (Brooks, 1994) and reported at least one first or second degree relative also diagnosed with motor neuron disease. The sporadic ALS (SALS) cohort included 797 North American cases diagnosed with possible, probable, or definite ALS but without a family history of motor neuron disease. 174 patients were evaluated at Washington University, 164 at Virginia Mason, and 459 came from DNA panels NDPT026, NDPT100, NDPT103, NDPT106, and NDPT116 that were obtained from the NINDS Human Genetics Resource Center DNA and Cell Line Repository (http://ccr.coriell.org/ninds). An additional 26 subjects (19 from panel NDPT025 and 7 from Washington University) had been diagnosed with primary lateral sclerosis (PLS) and were analyzed separately. 526 North American Caucasian neurologically normal controls without a personal or family history of motor neuron disease were drawn from Washington University (n=66) and NINDS DNA panels (n=460 from NDPT020, NDPT079, NDPT82, NDPT095, and NDPT096). Demographic and clinical information was derived from chart review for all Washington University participants and retrieved from database information for both Virginia Mason and NINDS participants. Longitudinal data was available for Washington University participants only, with survival defined as the disease duration from the time from symptom onset to death or the need for fulltime ventilatory support. All participants had provided signed informed consent for studies approved by local Institutional Review Boards.
2.2. Molecular Genetics
C9ORF72 repeat expansion genotyping
ALS cases and controls were screened for the C9ORF72 hexanucleotide expansions using repeat-primed PCR primers and methods as previously published (DeJesus-Hernandez et al., 2011). Identification of a decrementing saw-tooth pattern with 6bp periodicity and >30 peaks was considered positive for an expanded repeat (Figure 1a) as in previous applications of this assay. In practice however, all samples considered positive in this study showed >50 peaks. In a subset of 30 samples, we also performed PCR across the hexanucleotide repeat region (DeJesus-Hernandez et al., 2011) and the number of repeats was calculated by amplicon size determination using capillary gel electrophoresis. Sanger-based sequencing of these amplicons was also performed to directly confirm the number of hexanucleotide units. By correlating sequencing, PCR, and repeat-primed PCR assays, we were able to determine the number of hexanucleotide units present on individual’s longest allele (excluding those with full hexanucleotide repeat expansions) directly from the repeat-primed PCR results. We were able to validate our assay by comparing genotyping results for a subset of 210 Coriell SALS samples independently analyzed in a separate study (Harms et al., 2012, Rutherford et al., 2012a).
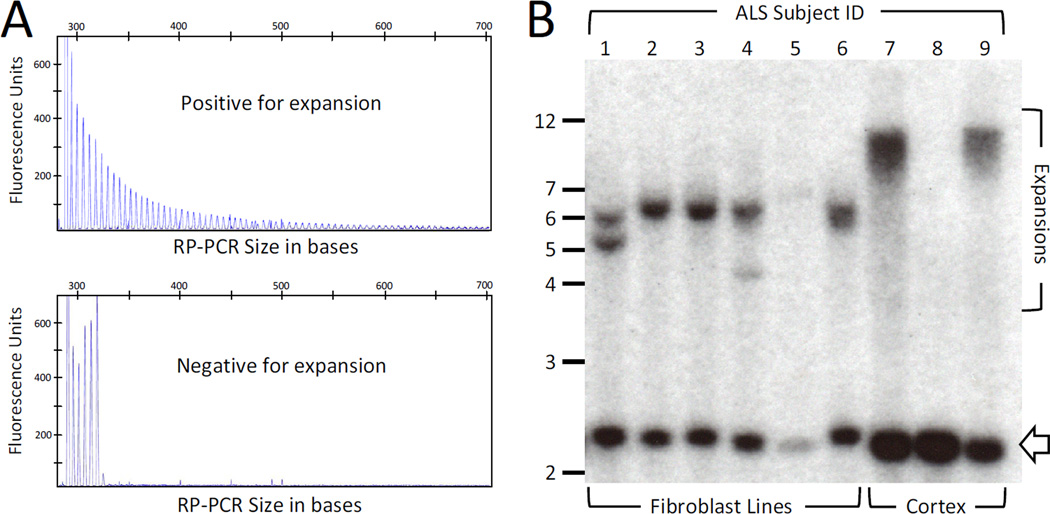
Figure 1. Identification and sizing of C9ORF72 repeat expansions.
A) A sample was considered positive for a C9ORF72 repeat expansion when repeat-primed PCR (RP-PCR) showed >30 decrementing peaks with 6bp periodicity and deemed negative in all other cases. However, all expansion positive cases in this study showed >60 repeats as shown in this example tracing.
B) Southern blot analysis of C9ORF72 repeat sizes in nine unrelated patients with ALS. Fibroblast-derived DNA was available for subjects 1-6 (lanes 1-6) and brain-derived DNA for subjects 7-9 (lanes 7–9). All samples except the negative control in lane 8 were positive for the C9ORF72 expansion by repeat-primed PCR (RP-PCR). Fibroblasts show dominant bands at 6-7kb (600-800 GGGGCC units), two with multiple bands, while brain-derived samples show a smear of sizes near 12 kb (1600 GGGGCC units). Open arrow-head marks the location of non-expanded alleles. Clinical characteristics of subjects 1-9 are found in the Supplementary Table.
C9ORF72 coding region sequencing
Pooled-sample sequencing (Supplementary Figure) was used to screen C9ORF72 coding exons for mutations in the familial and sporadic cohorts from Washington University and the Virginia Mason Medical Center. The pooled-sample sequencing followed previously published work-flow and technique (Vallania et al., 2010). Genomic DNA (gDNA) from individual patients was carefully quantified using Sybr-Gold (LifeTechnologies/Invitrogen) and pools were generated by combining equivalent amounts of gDNA from individual subjects. Six separate pools were created, each containing gDNA from 43 to 80 individuals. C9ORF72 exons were amplified directly on each gDNA pool using Pfu DNA polymerase (Agilent Technologies), a high-fidelity enzyme with a very low misincorporation rate. Primers for PCR were designed using Primer3Plus software (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi) and targeted the 10 coding exons of C9ORF72 annotated by RefSeq (NM_018325.3) along with 25–100 bp of flanking sequence (primer sequences available on request). All C9ORF72 amplicons from a given pool were combined in equimolar amounts and supplemented with positive and negative control amplicons. Combined amplicons were ligated into concatemers that subsequently fractionated and prepared for single-end, 42-bp read Illumina sequencing. Libraries from each of the 6 pools were indexed, combined, and sequenced on a single lane of an Illumina HiSeq. The SPLINTER software package was used to align the sequencing reads from each pool to the Hg19 reference sequence for C9ORF72, as well as to positive and negative control sequences (Vallania et al., 2010). The SPLINTER algorithm utilizes sequencing reads of the negative control to generate an error model and sequencing reads covering the positive control to determine sensitivity cut-offs for variant calling. As a result, the algorithm is capable of calling variants on a single allele in larger allele pools than used in this experiment (Cruchaga et al., 2012; Haller et al., 2012; Vallania et al., 2010). Targeted bases were sequenced to an average depth of 350× per individual and all bases across all pools were covered at ≥16× per individual, producing an average sensitivity for detecting a single allele in each pool of 94.5% (range 92–100%). Sequencing reads covering all variants called by SPLINTER were visualized using IGV (http://www.broadinstitute.org/igv/) to identify false positives from low-quality base calls. The remaining variants were subjected to a bioinformatic analysis, including annotation using SeattleSeq135 (http://snp.gs.washington.edu/SeattleSeqAnnotation135/), SIFT (http://sift.jcvi.org/), Exome Variant Server (http://evs.gs.washington.edu/EVS/), and the 1000 Genomes Project (http://www.1000genomes.org). Potentially deleterious mutations (rare and novel) were then genotyped in all individuals from the pool(s) in which the variant had been found. Genotyping methods included custom Sequenom assays or direct Sanger sequencing of amplicons followed by analysis using DNAStar Lasergene.
Other gene sequencing
Familial cases included in this study were also sequenced for SOD1 TARDBP, and FUS using several techniques: direct Sanger sequencing of all SOD1 coding exons as well as pooled-sample sequencing of all SOD1, TARDBP, and FUS coding exons (as above).
Southern blot quantification of C9ORF72 expansion size
10 µg of genomic DNA was isolated from either patient-derived fibroblast cell lines or frozen occipital cortex and digested overnight with XbaI in the presence of 2 mM spermidine. Fragment electrophoresis at 20V for 24 hours was performed on a 0.8% SeaKem GTG agarose gel (Takara) with 1× TBE, with subsequent transfer to GeneScreen Plus nylon membranes (PerkinElmer). A 590-bp probe containing the smaller published probe gave improved sensitivity and was generated by PCR using the following primers: forward 5’-AAATTGCGATGACTTTGCAGGGGACCGTGG and reverse 5’GCTCTCACAGTACTCGCTGAGGGTGAACAA). After gel purification, the probe was labeled with 32P-dCTP (PerkinElmer) using the Random Primed DNA labeling kit (Roche) and purified using Ambion NucAway spin columns (Life Technologies). Hybridization was carried out overnight at 68°C in Perfect Hyb Plus buffer (Sigma) containing 100µg/ml salmon sperm DNA (Life Technologies/Invitrogen). Filters were washed twice for 5 minutes at room temperature with 2× SSC+0.1% SDS, and twice for 20 minutes at 68°C with 0.2× SSC+0.1% SDS. BioMax MS film (Kodak) was exposed with an intensifying screen at −80°C for 5 days.
2.3. Statistical Analysis
Categorical clinical and demographic characteristics were reported as percentages, while continuous variables were represented as a mean (if normally distributed) or median (if not normally distributed). Fisher’s exact test was used to compare categorical variables. Comparisons of the mean ages of symptom onset utilized independent samples T-tests. Between group comparisons of median survival and hexanucleotide unit numbers were performed using Mann-Whitney U tests. Kaplan-Meier analysis was used to investigate survival differences between patients with and without C9ORF72 expansions. All tests were two-tailed and performed with SPSS software (Armonk, MY USA). Significance was set at P=0.05.
3. RESULTS
3.1. C9ORF72 repeat expansions in North American sporadic ALS
Repeat expansion frequencies in ALS vary widely across various populations and generally in proportion to the degree of Northern European admixture (Majounie et al., 2012b). However, some regions show unexpectedly high prevalence, including the island of Sardinia and in Greece (Mok et al., 2012; Sabatelli et al., 2012). To determine if different geographic areas within the United States show differences in expansion frequencies, we genotyped SALS cases from two regional ALS clinics- Washington University in St. Louis, Missouri (Mid-West) and Virginia Mason Medical Center in Seattle, Washington (Pacific Northwest). Despite similar cohort characteristics at the two centers, the frequency of expansions was significantly higher in the Mid-West compared to the Pacific Northwest (9.2% vs 3.0%, p=0.023). For a nationwide comparison, we genotyped an additional 459 SALS cases drawn from centers distributed across the United States as part of the NINDS Coriell collection (Gwinn et al., 2007), 210 of which were recently published in an independent study (Rutherford et al., 2012a). We found a repeat expansion frequency of 7.4%, which is intermediate between the two regional cohorts. Overall, hexanucleotide repeat expansions were found in 55 (6.9%) of 797 SALS cases from the United States. Demographic and clinical characteristics of the entire SALS cohort of 797 patients are shown in Table 1. We also screened 26 patients diagnosed with primary lateral sclerosis, but did not find any carrying the C9ORF72 repeat expansion. Interestingly, two of 526 neurologically normal Caucasian controls were also found to carry repeat expansions. Both individuals were elderly at the time of DNA collection and had no family history of dementia or ALS. This rate (0.4%) is within the range reported for other control populations of European or North American origin (0–0.6%) (DeJesus-Hernandez et al., 2011; Renton et al., 2011; Rollinson et al., 2012; Sabatelli et al., 2012; Simon-Sanchez et al., 2012; Smith et al., 2012).
Table 1.
Clinical and genetic characteristics of sporadic ALS patients
| Overall (n=797) |
Expansion carriers (n=55) |
p-valueb | Non-carriers (n=742) |
|
|---|---|---|---|---|
| % of SALS cohort | 6.9 | 93.1 | ||
| Sex, % male (n)a | 54.3 (431) | 41.8 (23) | 0.068 | 55.2 (408) |
| Mean age of onset, years ± SDa | 59.2 ± 11.5 | 55.9 ± 7.3 | 0.027 | 59.4 ± 11.7 |
| Age of onset, range | 14–88 | 41–72 | nc | 14–88 |
| Bulbar onset, % (n)a | 23.4 (186) | 16.4 (9) | 0.249 | 23.9 (177) |
| Dementia, % (n)a | 3.9 (31) | 1.8 (1) | 0.717 | 4.0 (30) |
| Family history dementia, % (n)a | 16 (97) | 34.7 (32) | 0.001 | 14.3 (80) |
| Median survival, months (IQR, n) | 29 (21–43, 108) | 29 (24–39, 10) | 0.832 | 29 (21–43, 98) |
Calculations reflect the following missing data: gender for 3 non-carriers; age of onset for 1 carrier and 6 non-carriers; site of onset in 1 non-carrier; presence of dementia in 1 non-carrier; family history for 6 carriers and 184 non-carriers.
The p-value column refers to the comparison of the columns on either side of it. Differences meeting statistical significance are italicized.
Abbreviations: n=number; IQR=inter-quartile range; nc=not compared
SALS patients carrying a C9ORF72 repeat expansion showed an earlier age of symptom onset and were more likely to have a family history of dementia (OR=3.2, CI 1.6–6.2; p=0.001). However, they were not more likely to experience bulbar symptom onset or be diagnosed with dementia. Longitudinal data was available for patients evaluated at Washington University only. Analysis of the 108 deceased patients showed no difference in the median disease duration for expansion carriers compared to noncarriers. Kaplan-Meier analysis of disease duration in all 174 patients also failed to demonstrate a difference between expansion carriers and non-carriers (p=0.118).
3.2. C9ORF72 repeat expansions in North American familial ALS
GGGGCC hexanucleotide repeat expansions were present in 22 of 51 unrelated FALS probands- all in self-reported Caucasian families. All C9ORF72 expansion carriers had normal sequencing for SOD1, TARDBP, and FUS. C9ORF72 repeat expansions were more than twice as common as SOD1 mutations (22 vs 8 of 51), and explained more families than mutations in SOD1 (n=8), TARDBP (n=3), and FUS (n=2) combined. Overall, a genetic diagnosis was made in 69% of pedigrees by screening for mutations in these three genes and for C9ORF72 repeat expansions.
Clinical features of C9ORF72 repeat expansion carriers were compared to probands with mutations in other genes (SOD1, TARDBP, or FUS) and to probands without mutations or repeat expansions (Table 2). Bulbar symptom onset was substantially more common in probands with repeat expansions (54.5% vs 13.8% in non-carriers; p=0.003; OR=7.5, 95% CI 1.66–36.74), as was a personal diagnosis of dementia (40.9% vs 3.4%; p= 0.001; OR=19.4, 95% CI 2.07–453), and a family history of dementia (52.6% vs 7.4%, p=0.001; OR=13.9, 95% CI 2.15–114.8]. Sub-typing of patients’ cognitive impairment was incomplete because formal neuropsychological testing was unavailable for most probands. However, in all cases, the clinical diagnosis recorded in the medical record was frontotemporal dementia. In contrast, the dementia diagnosis reported for relatives was often Alzheimer’s type. The average age of symptom onset was similar between probands with repeat expansions and those with unknown genetic cause, but later than probands with mutations in other genes.
Table 2.
Clinical Characteristics of FALS with differing etiologies
| Overall | Point mutation carriersb |
p- valuec |
C9ORF72 expansion carriers |
p- valuec |
Unknown genetic cause |
|
|---|---|---|---|---|---|---|
| Number of probands | 51 | 13 | 22 | 16 | ||
| Sex, % male (n) | 49 (25) | 46 (6) | 1.0 | 50 (11) | 1.0 | 50 (8) |
| Mean age of onset, years ± SD | 56.3 ± 13.0 | 47.4 ± 14.2 | 0.0083 | 59.7 ±11.6 | 0.83 | 58.9 ± 11.1 |
| Bulbar onset, % (n) | 31.4 (16) | 0.0 (0) | 0.0009 | 54.5 (12) | 0.1 | 25.0 (4) |
| Dementia, % (n) | 19.6 (10) | 0.0 (0) | 0.0131 | 40.9 (9) | 0.025 | 6.3 (1) |
| Family history dementia, % (n)a | 26.1 (12) | 7.7 (1) | 0.011 | 52.6 (10) | 0.009 | 7.1 (1) |
| Mean duration, months±SD (n) | 26.5 ± 16.7 (17) | 14.0 ± 4.1 (4) | 0.144 | 28 ± 16.2 (4) | 0.763 | 31.3 ± 18.6 (9) |
Calculations reflect missing family data for 3 expansion carriers and 2 without known genetic cause.
Point mutation carriers included 8 with SOD1, 3 with TARDBP, and 2 with FUS mutations.
The p-value column refers to comparison of the two columns on either side of it. Differences meeting statistical significance are italicized.
Abbreviations: SD=standard deviation
3.2. C9ORF72 repeat expansion sizes
To determine the range of expansion sizes in our cohort, we performed Southern blot analysis on those expansion carriers with available fibroblast cell lines (Figure 1b). Predominant expanded alleles were seen in all samples at 6–7 kb (normal size is ~2.3kb), translating into 600–800 repeat units. Two subjects showed additional smaller expanded alleles, supporting somatic instability of the repeat and suggesting clonal populations with different expansion sizes in cell culture. DNA derived from occipital cortex was available for an additional 2 subjects and showed much larger expansions (~12kb, or 1600 repeat units), supporting the possibility that expansions are larger within neuronal tissues and may underlie the neurodegenerative phenotypes observed with C9ORF72 expansions. Clinical characteristics of subjects studied by Southern blot are in the Supplementary Table.
3.4. Associations of non-expanded C9ORF72 repeats
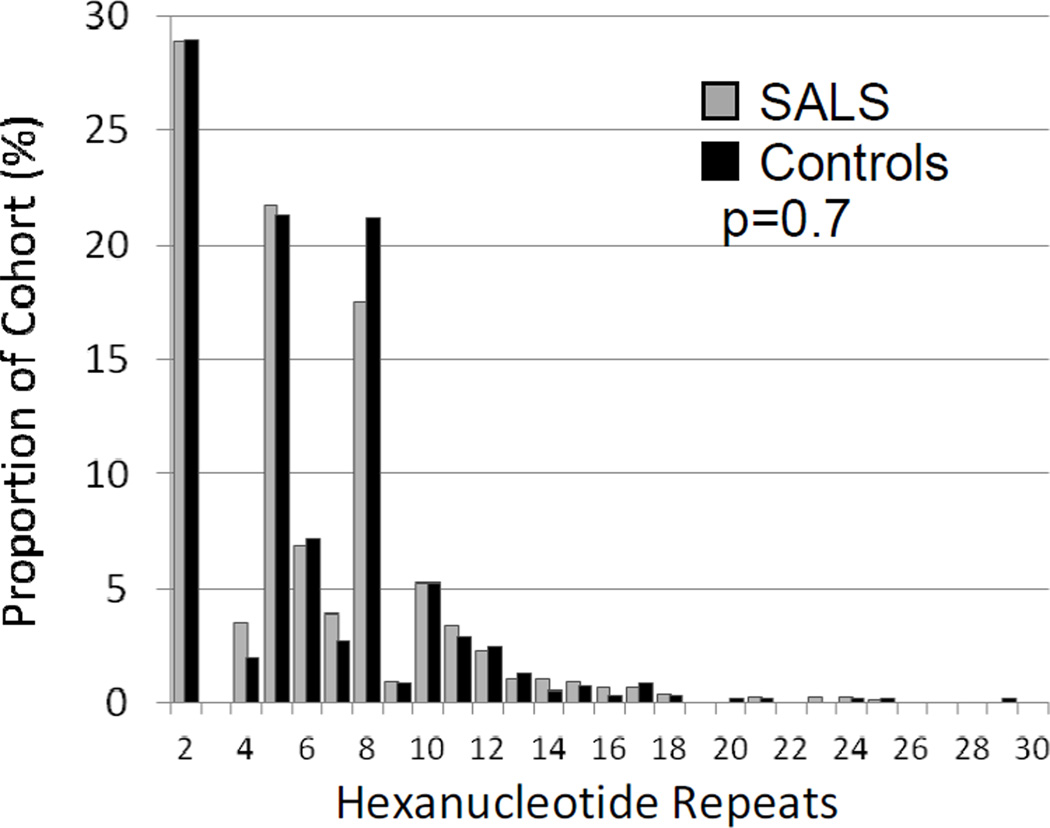
Previous studies have demonstrated that the number of GGGGCC repeats in the normal population is highly polymorphic, ranging from 2 to 23 repeat units. However, alleles are heavily biased toward fewer than 10 repeats (Byrne et al., 2012; DeJesus-Hernandez et al., 2011). A similarly skewed distribution has been observed for CAG repeats in the Ataxin-2 gene, where intermediate repeat lengths are enriched in patients with SALS and therefore considered a risk factor for disease (Elden et al., 2010; Lee et al., 2011). To see if intermediate length alleles of the C9ORF72 GGGGCC repeats behave in a similar fashion, we assessed whether “normal” alleles with higher numbers of repeat units were a risk factor for ALS by comparing the distribution of an individuals’ longest allele in SALS and controls. After excluding the 2 controls and 55 SALS patients with full repeat expansions by repeat-primed PCR, the median number of GGGGCC repeat units was 5 (IQR 2-8) in both groups and no significant difference was seen in the distribution of alleles (Figure 2, p=0.72). Additionally, in SALS patients without full repeat expansions, the highest number of repeat units showed no correlation with age of onset, site of symptom onset, or disease duration (data not shown). Because our genotyping method reliably identified only the longest allele, we were unable to explore the effect of combinations of alleles.
Figure 2. Distribution of longest normal C9ORF72 allele in SALS and controls.
All non-C9ORF72 SALS patients and controls had their longest repeat allele measured by repeat-primed PCR (RP-PCR). The distribution of alleles was identical between the two groups, p=0.7.
3.5. C9ORF72 gene sequencing
Concurrent with our repeat expansion testing, we sequenced all C9ORF72 coding exons in 389 ALS patients from the Washington University and Virginia Mason cohorts (338 with SALS and 51 with FALS). Identified variants are described in Table 3. We identified a missense variant (T49R) in a single FALS patient. This variant was also identified in 2 of 4300 Caucasian exomes reported in the Exome Variant Server. The patient carrying this T49R mutation was subsequently found to also harbor a frequent disease-associated mutation in FUS (R521G) (Kwiatkowski et al., 2009). DNA was not available to test segregation of the C9ORF72 T49R variant, but an affected family member was later confirmed by commercial sequencing to carry the FUS R521G mutation. Two novel C9ORF72 variants were identified that were not found in the Exome Variant Server (despite excellent coverage at the relevant location) or 1000 Genomes Project: a silent substitution (T352T) was seen in a patient with FALS, while two SALS patients carried an intronic variant 4bp from a canonical splice donor site. Each of these variants occurred in patients without other genetic explanations for disease, but bioinformatic analysis predicted that neither would alter splicing.
Table 3.
C9ORF72 sequence variants identified in ALS patients
| Locationa | Variantb | Variant Effect | ALS Frequency | SNP IDc | Control MAFd |
|---|---|---|---|---|---|
| 9:27566973 | c.146G>C | T49R | 1 FALS | NA | 0.023% |
| 9:27561628 | c.620T>C | N207S | 79 SALS | rs17769294 | 14.4% |
| 9:27556780 | c.870C>T | S290S | 13 FALS, 111 SALS | rs10122902 | 19.7% |
| 9:27556594 | c.1056G>T | T352T | 1 FALS | NA | 0% |
| 9:27548549 | c.1259+6T>G | near splice donor | 2 SALS | NA | 0% |
| 9:27548276 | c.1404C>T | F468F | 1 FALS, 2 SALS | rs141063383 | 0.56% |
Referenced to hg19 (GRCh37).
Variants noted in Human Genome Variation Society nomenclature utilizing NM_018325.
SNP ID as assigned in dbSNP135.
Control MAF(minor allele frequency) derived from Caucasian exomes included in the ESP6500 dataset release v.0.0.14.
4. DISCUSSION
In the short time since C9ORF72 repeat expansions were discovered in patients with ALS, FTD, or an ALS-FTD overlap, studies have begun to address the epidemiology and clinical spectrum of neurodegeneration caused by this genetic mutation. However, how repeat expansions cause disease remains unclear and both loss-of-function and gain-of-function mechanisms have been proposed (DeJesus-Hernandez et al., 2011; van Blitterswijk et al., 2012). To inform this question, we looked for other pathogenic coding mutations in C9ORF72 in our FALS and SALS cohort, but did not identify any. This absence indicates that C9ORF72 coding mutations are not a common cause of familial or sporadic ALS. While indirect, the fact that we did not find loss of function variants (premature stop codons, etc.) on comprehensive sequence analysis in ALS patients tentatively argues for a toxic gain of function mechanism for C9ORF72 repeat expansions, rather than allele suppression and haploinsufficiency. This conclusion is limited by the fact that we did not assess copy number variation in our cohort, the presence of which in ALS patients would favor a loss-of-function mechanism. Furthermore, larger cohorts should be analyzed in future to assess the possible contribution of rare variants or copy number variation.
To expand our understanding of expansion sizes in disease, we determined the repeat size from 8 unrelated patients by Southern blot. Although the sample size is small and samples were not paired from the same patient, it is interesting that cell-line expansions appear smaller than those derived from post-mortem brain. If borne out in larger groups of patients and with multiple tissues from the same individuals, this finding suggests that non-neuronal tissues preferentially contract GGGGCC repeats or alternatively, that neuronal tissues are predisposed to repeat expansion growth. Understanding the stability of GGGGCC repeats (including how, when, and in which tissues expansions or contractions occur) will help clarify whether approaches aimed at repeat stabilization may have therapeutic implications.
Our study also demonstrates geographic variability in the frequency of C9ORF72 ALS in the United States. Our screening shows that while C9ORF72 expansions cause ~7% of apparently sporadic ALS in the U.S., the frequency is higher in the Mid-West (9.2%) and much lower in the Pacific Northwest (3.0%). Although a selection bias between the two collection centers cannot be entirely excluded, it is notable that the SALS carrier frequency in British Columbia (just north of Seattle, Washington) was recently shown to be 3.6% (Stewart et al., 2012). The difference in expansion frequency we have observed may reflect the different immigration or settlement histories of these two geographical regions but has important implications for expansion screening strategies in the clinic. Specifically, in our Mid-West population, where with nearly 1 in 10 apparently sporadic patients is an expansion carrier, universal genetic screening for C9ORF72 expansions should be considered. In our cohort, SALS patients carrying C9ORF72 expansions showed an earlier onset of symptoms and were more likely have relatives diagnosed with dementia, echoing findings from several other studies (Byrne et al., 2012; Cooper-Knock et al., 2012; Stewart et al., 2012).
We found that 43% of FALS pedigrees in our North American cohort carry repeat expansions, which is similar to the frequency reported elsewhere (reviewed in van Blitterswijk et al., 2012). This frequency makes C9ORF72 repeat expansions considerably more common than other gene mutations combined (SOD1, TARDBP, and FUS) and suggests that genetic testing algorithms should begin with analysis of C9ORF72 before moving on to the sequencing of SOD1 or other ALS-associated genes. Our data also confirms that concurrent FTD or a family history of any dementia increases the chance of a finding a C9ORF72 repeat expansion (Stewart et al., 2012). Interestingly, among expansion carriers in our study, bulbar onset was considerably more common in those with a family history of ALS than in those without (54% vs 16%, p=0.0014). We cannot exclude the possibility that this discrepancy results from our modest sample size of expansion carriers. However, review of the literature for other populations where FALS and SALS cases were ascertained and described in a fashion comparable to ours discloses a similar, but not statistically significant, difference in Italy (42% vs 21%) (Chio et al., 2012; Sabatelli et al., 2012). If this discrepancy is seen in other cohorts, it will be important to determine if the site of onset correlates with repeat expansion size, which in turn might differ between those patients with and without a family history of ALS.
We also assessed whether larger hexanucleotide repeats in the normal range influence ALS phenotype, but found no association with age at symptom onset, site of symptom onset, or survival time. Furthermore, in this population, larger normal repeat numbers were not a risk factor for developing ALS. Together with similar findings by others (Rutherford et al., 2012b, Garcia-Redondo et al., 2013), these data suggest that a threshold number of repeats may exist before C9ORF72 expansions cause disease. Determining the pathogenic threshold will require Southern blot analysis of expansion sizes across a large cohort of patients and controls and should be a top priority in ALS/FTD research.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by National Institutes of Health (NIH) Grants NS069669 (R.H.B) and K08NS075094 (M.B.H.), as well as the Hope Center for Neurological Disorders. R.H.B. holds a Career Award for Medical Scientists from the Burroughs Wellcome Fund. This publication was made possible by Grant Number UL1 RR024992 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH), and NIH Roadmap for Medical Research. Its contents are solely the responsibility of the authors and do not necessarily represent the official view of NCRR or NIH. The authors would like to thank the Transgenic Vectors Core at the Hope Center for Neurological Disorders for assistance in developing the Southern blot method, the Genome Technology Access Center for assistance with library preparation and sequencing, and Dr. Todd Druley laboratory for their technical expertise and assistance in implementing SPLINTER. We would also like to thank the NHLBI GO Exome Sequencing Project and its ongoing studies which produced and provided exome variant calls for comparison: the Lung GO Sequencing Project (HL-102923), the WHI Sequencing Project (HL-102924), the Broad GO Sequencing Project (HL-102925), the Seattle GO Sequencing Project (HL-102926) and the Heart GO Sequencing Project (HL-103010). DNA panels from the NINDS Human Genetics Resource Center DNA and Cell Line Repository (http://ccr.coriell.org/ninds) were used in this study, as well as clinical data. The submitters that contributed samples are acknowledged in detailed descriptions of each panel: NDPT020, NDPT025, NDPT026, NDPT079, NDPT82, NDPT095, NDPT096, NDPT100, NDPT103, NDPT106, and NDPT116.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DISCLOSURE STATEMENTS:
Matthew Harms receives research support from the NIH and the Millstone Family Foundation, participates in studies funded by the Jain Foundation, Ultragenyx, and Biogen-Idec, and has provided expert testimony in medical-legal proceedings unrelated to this topic.
Janet Cady has nothing to disclose.
Craig Zaidman has nothing to disclose.
Paul Cooper has nothing to disclose.
Taha Bali has nothing to disclose.
Peggy Allred has nothing to disclose.
Carlos Cruchaga has nothing to disclose.
Michael Baughn has nothing to disclose.
Alan Pestronk receives revenue related to antibody patent licenses & speaker honoraria from Athena; Owns stock in Johnson & Johnson; Directs the Washington University Neuromuscular Clinical Laboratory which performs antibody testing; Receives research support from the NIH, Muscular Dystrophy Association, Neuromuscular Research Fund; Insmed, Knopp, Cytokinetics, Biogen Idec, ISIS, Genzyme, GSK, Sanofi & Ultragenyx.
Alison Goate receives revenue from expert testimony, consulting, or speaking from Pfizer, Genentech, Finnegan, and Amgen. She receives royalties from a patent license to Taconic and research support from Genentech, Pfizer, and the Barnes Jewish Foundation.
John Ravits receives research support from the ALS Association, Microsoft Research, P2ALS, serves as an unpaid consultant to the Muscular Dystrophy Association, and consults for Isis Pharmaceuticals, Inc.
Robert H. Baloh has nothing to disclose.
All participants had provided signed informed consent for studies approved by local Institutional Review Boards.
Contributor Information
Matthew B. Harms, Email: harmsm@neuro.wustl.edu.
Janet Cady, Email: cadyj@neuro.wustl.edu.
Craig Zaidman, Email: zaidmanc@neuro.wustl.edu.
Paul Cooper, Email: cooperp@neuro.wustl.edu.
Taha Bali, Email: balit@neuro.wustl.edu.
Peggy Allred, Email: allredp@neuro.wustl.edu.
Carlos Cruchaga, Email: cruchagc@psychiatry.wustl.edu.
Michael Baughn, Email: mwbaughn@ucsd.edu.
Alan Pestronk, Email: pestronka@neuro.wustl.edu.
Alison Goate, Email: goatea@psychiatry.wustl.edu.
John Ravits, Email: jravits@ucsd.edu.
Robert H. Baloh, Email: Robert.Baloh@csmc.edu.
REFERENCES
- Brooks BR. El Escorial World Federation of Neurology criteria for the diagnosis of amyotrophic lateral sclerosis. Subcommittee on Motor Neuron Diseases/Amyotrophic Lateral Sclerosis of the World Federation of Neurology Research Group on Neuromuscular Diseases and the El Escorial "Clinical limits of amyotrophic lateral sclerosis" workshop contributors. J Neurol Sci. 1994;124(Suppl):96–107. doi: 10.1016/0022-510x(94)90191-0. [DOI] [PubMed] [Google Scholar]
- Byrne S, Elamin M, Bede P, Shatunov A, Walsh C, Corr B, Heverin M, Jordan N, Kenna K, Lynch C, McLaughlin RL, Iyer PM, O'Brien C, Phukan J, Wynne B, Bokde AL, Bradley DG, Pender N, Al-Chalabi A, Hardiman O. Cognitive and clinical characteristics of patients with amyotrophic lateral sclerosis carrying a C9orf72 repeat expansion: a population-based cohort study. Lancet Neurol. 2012;11(3):232–240. doi: 10.1016/S1474-4422(12)70014-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campuzano V, Montermini L, Molto MD, Pianese L, Cossee M, Cavalcanti F, Monros E, Rodius F, Duclos F, Monticelli A, Zara F, Canizares J, Koutnikova H, Bidichandani SI, Gellera C, Brice A, Trouillas P, De Michele G, Filla A, De Frutos R, Palau F, Patel PI, Di Donato S, Mandel JL, Cocozza S, Koenig M, Pandolfo M. Friedreich's ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science. 1996;271(5254):1423–1427. doi: 10.1126/science.271.5254.1423. [DOI] [PubMed] [Google Scholar]
- Chiò A, Borghero G, Restagno G, Mora G, Drepper C, Traynor BJ, Sendtner M, Brunetti M, Ossola I, Calvo A, Pugliatti M, Sotgiu MA, Murru MR, Marrosu MG, Marrosu F, Marinou K, Mandrioli J, Sola P, Caponnetto C, Mancardi G, Mandich P, La Bella V, Spataro R, Conte A, Monsurrò MR, Tedeschi G, Pisano F, Bartolomei I, Salvi F, Lauria Pinter G, Simone I, Logroscino G, Gambardella A, Quattrone A, Lunetta C, Volanti P, Zollino M, Penco S, Battistini S, ITALSGEN consortium, Renton AE, Majounie E, Abramzon Y, Conforti FL, Giannini F, Corbo M, Sabatelli M. Clinical characteristics of patients with familial amyotrophic lateral sclerosis carrying the pathogenic GGGGCC hexanucleotide repeat expansion of C9ORF72. Brain. 2012;135(Pt 3):784–793. doi: 10.1093/brain/awr366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper-Knock J, Hewitt C, Highley JR, Brockington A, Milano A, Man S, Martindale J, Hartley J, Walsh T, Gelsthorpe C, Baxter L, Forster G, Fox M, Bury J, Mok K, McDermott CJ, Traynor BJ, Kirby J, Wharton SB, Ince PG, Hardy J, Shaw PJ. Clinico-pathological features in amyotrophic lateral sclerosis with expansions in C9ORF72. Brain. 2012;135(Pt 3):751–764. doi: 10.1093/brain/awr365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossee M, Durr A, Schmitt M, Dahl N, Trouillas P, Allinson P, Kostrzewa M, Nivelon-Chevallier A, Gustavson KH, Kohlschutter A, Muller U, Mandel JL, Brice A, Koenig M, Cavalcanti F, Tammaro A, De Michele G, Filla A, Cocozza S, Labuda M, Montermini L, Poirier J, Pandolfo M. Friedreich's ataxia: point mutations and clinical presentation of compound heterozygotes. Ann Neurol. 1999;45(2):200–206. doi: 10.1002/1531-8249(199902)45:2<200::aid-ana10>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- Cruchaga C, Haller G, Chakraverty S, Mayo K, Vallania FL, Mitra RD, Faber K, Williamson J, Bird T, Diaz-Arrastia R, Foroud TM, Boeve BF, Graff-Radford NR, St Jean P, Lawson M, Ehm MG, Mayeux R, Goate AM. Rare variants in APP, PSEN1 and PSEN2 increase risk for AD in late-onset Alzheimer's disease families. PLoS ONE. 2012;7(2):e31039. doi: 10.1371/journal.pone.0031039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boulle K, Verkerk AJ, Reyniers E, Vits L, Hendrickx J, Van Roy B, Van den Bos F, de Graaff E, Oostra BA, Willems PJ. A point mutation in the FMR-1 gene associated with fragile X mental retardation. Nat Genet. 1993;3(1):31–35. doi: 10.1038/ng0193-31. [DOI] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, Kouri N, Wojtas A, Sengdy P, Hsiung GY, Karydas A, Seeley WW, Josephs KA, Coppola G, Geschwind DH, Wszolek ZK, Feldman H, Knopman DS, Petersen RC, Miller BL, Dickson DW, Boylan KB, Graff-Radford NR, Rademakers R. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72(2):245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elden AC, Kim HJ, Hart MP, Chen-Plotkin AS, Johnson BS, Fang X, Armakola M, Geser F, Greene R, Lu MM, Padmanabhan A, Clay-Falcone D, McCluskey L, Elman L, Juhr D, Gruber PJ, Rub U, Auburger G, Trojanowski JQ, Lee VM, Van Deerlin VM, Bonini NM, Gitler AD. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature. 2010;466(7310):1069–1075. doi: 10.1038/nature09320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exome Variant Server. Seattle, WA: NHLBI GO Exome Sequencing Project (ESP); [accessed October 2012]. (URL: http://evs.gs.washington.edu/EVS/) [Google Scholar]
- García-Redondo A, Dols-Icardo O, Rojas-García R, Esteban-Pérez J, Cordero-Vázquez P, Muñoz-Blanco JL, Catalina I, González- Muñoz M, Varona L, Sarasola E, Povedano M, Sevilla T, Guerrero A, Pardo J, de Munain AL, Márquez-Infante C, de Rivera FJ, Pastor P, Jericó I, de Arcaya AÁ, Mora JS, Clarimón J, C9ORF72 Spanish Study Group. Clarimon J, Gonzalo-Martínez JF, Juárez-Rufián A, Atencia G, Jiménez-Bautista R, Morán Y, Mascías J, Hernández-Barral M, Kapetanovic S, García-Barcina M, Alcalá C, Vela A, Ramírez-Ramos C, Galán L, Pérez-Tur J, Quintáns B, Sobrido MJ, Fernández-Torrón R, Poza JJ, Gorostidi A, Paradas C, Villoslada P, Larrodé P, Capablo JL, Pascual-Calvet J, Goñi M, Morgado Y, Guitart M, Moreno-Laguna S, Rueda A, Martín-Estefanía C, Cemillán C, Blesa R, Lleó A. Analysis of the C9orf72 gene in patients with amyotrophic lateral sclerosis in Spain and different populations worldwide. Hum Mutat. 2013 Jan;34(1):79–82. doi: 10.1002/humu.22211. [DOI] [PubMed] [Google Scholar]
- Gwinn K, Corriveau RA, Mitsumoto H, Bednarz K, Brown RH, Jr, Cudkowicz M, Gordon PH, Hardy J, Kasarskis EJ, Kaufmann P, Miller R, Sorenson E, Tandan R, Traynor BJ, Nash J, Sherman A, Mailman MD, Ostell J, Bruijn L, Cwik V, Rich SS, Singleton A, Refolo L, Andrews J, Zhang R, Conwit R, Keller MA. Amyotrophic lateral sclerosis: an emerging era of collaborative gene discovery. PLoS ONE. 2007;2(12):e1254. doi: 10.1371/journal.pone.0001254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller G, Druley T, Vallania FL, Mitra RD, Li P, Akk G, Steinbach JH, Breslau N, Johnson E, Hatsukami D, Stitzel J, Bierut LJ, Goate AM. Rare missense variants in CHRNB4 are associated with reduced risk of nicotine dependence. Hum Mol Genet. 2012;21(3):647–655. doi: 10.1093/hmg/ddr498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms MB, Neumann D, Benitez BA, Cooper B, Carrell D, Racette BA, Perlmutter JS, Goate A, Cruchaga C. Parkinson disease is not associated with C9ORF72 repeat expansions. Neurobiol Aging. 2012 doi: 10.1016/j.neurobiolaging.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms MB, Benitez B, Cairns N, Cooper B, Cooper P, Mayo K, Carrell D, Faber K, Williamson J, Bird T, Diaz-Arrastia R, Roroud TM, Boeve BF, Graff-Radford NR, Mayeux R, Chakraverty S, Goate A, Cruchaga C. C9ORF72 hexanucleotide repeat expansions in clinical Alzheimer's disease. Archives of Neurology. 2013 doi: 10.1001/2013.jamaneurol.537. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski TJ, Jr, Bosco DA, Leclerc AL, Tamrazian E, Vanderburg CR, Russ C, Davis A, Gilchrist J, Kasarskis EJ, Munsat T, Valdmanis P, Rouleau GA, Hosler BA, Cortelli P, de Jong PJ, Yoshinaga Y, Haines JL, Pericak-Vance MA, Yan J, Ticozzi N, Siddique T, McKenna-Yasek D, Sapp PC, Horvitz HR, Landers JE, Brown RH., Jr Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323(5918):1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- Lee T, Li YR, Ingre C, Weber M, Grehl T, Gredal O, de Carvalho M, Meyer T, Tysnes OB, Auburger G, Gispert S, Bonini NM, Andersen PM, Gitler AD. Ataxin-2 intermediate-length polyglutamine expansions in European ALS patients. Hum Mol Genet. 2011;20(9):1697–1700. doi: 10.1093/hmg/ddr045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist S, Duno M, Batbayli M, Puschmann A, Braendgaard H, Mardosiene S, Svenstrup K, Pinborg L, Vestergaard K, Hjermind L, Stokholm J, Andersen B, Johannsen P, Nielsen J. Corticobasal and ataxia syndromes widen the spectrum of C9ORF72 hexanucleotide expansion disease. Clin Genet. 2012;9999(9999) doi: 10.1111/j.1399-0004.2012.01903.x. [DOI] [PubMed] [Google Scholar]
- Lugenbeel KA, Peier AM, Carson NL, Chudley AE, Nelson DL. Intragenic loss of function mutations demonstrate the primary role of FMR1 in fragile X syndrome. Nat Genet. 1995;10(4):483–485. doi: 10.1038/ng0895-483. [DOI] [PubMed] [Google Scholar]
- Majounie E, Abramzon Y, Renton AE, Perry R, Bassett SS, Pletnikova O, Troncoso JC, Hardy J, Singleton AB, Traynor BJ. Repeat expansion in C9ORF72 in Alzheimer's disease. N Engl J Med. 2012a;366(3):283–284. doi: 10.1056/NEJMc1113592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majounie E, Renton AE, Mok K, Dopper EG, Waite A, Rollinson S, Chio A, Restagno G, Nicolaou N, Simon-Sanchez J, van Swieten JC, Abramzon Y, Johnson JO, Sendtner M, Pamphlett R, Orrell RW, Mead S, Sidle KC, Houlden H, Rohrer JD, Morrison KE, Pall H, Talbot K, Ansorge O, Hernandez DG, Arepalli S, Sabatelli M, Mora G, Corbo M, Giannini F, Calvo A, Englund E, Borghero G, Floris GL, Remes AM, Laaksovirta H, McCluskey L, Trojanowski JQ, Van Deerlin VM, Schellenberg GD, Nalls MA, Drory VE, Lu CS, Yeh TH, Ishiura H, Takahashi Y, Tsuji S, Le Ber I, Brice A, Drepper C, Williams N, Kirby J, Shaw P, Hardy J, Tienari PJ, Heutink P, Morris HR, Pickering-Brown S, Traynor BJ. Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study. Lancet Neurol. 2012b;11(4):323–330. doi: 10.1016/S1474-4422(12)70043-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok K, Traynor BJ, Schymick J, Tienari PJ, Laaksovirta H, Peuralinna T, Myllykangas L, Chio A, Shatunov A, Boeve BF, Boxer AL, DeJesus-Hernandez M, Mackenzie IR, Waite A, Williams N, Morris HR, Simon-Sanchez J, van Swieten JC, Heutink P, Restagno G, Mora G, Morrison KE, Shaw PJ, Rollinson PS, Al-Chalabi A, Rademakers R, Pickering- Brown S, Orrell RW, Nalls MA, Hardy J. Chromosome 9 ALS and FTD locus is probably derived from a single founder. Neurobiol Aging. 2012;33(1):209, e3–e8. doi: 10.1016/j.neurobiolaging.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok KY, Koutsis G, Schottlaender LV, Polke J, Panas M, Houlden H. High frequency of the expanded C9ORF72 hexanucleotide repeat in familial and sporadic Greek ALS patients. Neurobiol Aging. 2012;33(8):1851, e1–e5. doi: 10.1016/j.neurobiolaging.2012.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademaker R. Advances in understanding the molecular basis of frontotemporal dementia. Nat Rev Neurol. 2012;8(8):423–434. doi: 10.1038/nrneurol.2012.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, Kalimo H, Paetau A, Abramzon Y, Remes AM, Kaganovich A, Scholz SW, Duckworth J, Ding J, Harmer DW, Hernandez DG, Johnson JO, Mok K, Ryten M, Trabzuni D, Guerreiro RJ, Orrell RW, Neal J, Murray A, Pearson J, Jansen IE, Sondervan D, Seelaar H, Blake D, Young K, Halliwell N, Callister JB, Toulson G, Richardson A, Gerhard A, Snowden J, Mann D, Neary D, Nalls MA, Peuralinna T, Jansson L, Isoviita VM, Kaivorinne AL, Holtta- Vuori M, Ikonen E, Sulkava R, Benatar M, Wuu J, Chio A, Restagno G, Borghero G, Sabatelli M, Heckerman D, Rogaeva E, Zinman L, Rothstein JD, Sendtner M, Drepper C, Eichler EE, Alkan C, Abdullaev Z, Pack SD, Dutra A, Pak E, Hardy J, Singleton A, Williams NM, Heutink P, Pickering-Brown S, Morris HR, Tienari PJ, Traynor BJ. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72(2):257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollinson S, Halliwell N, Young K, Callister JB, Toulson G, Gibbons L, Davidson YS, Robinson AC, Gerhard A, Richardson A, Neary D, Snowden J, Mann DM, Pickering-Brown SM. Analysis of the hexanucleotide repeat in C9ORF72 in Alzheimer's disease. Neurobiol Aging. 2012;33(8):1846, e5–e6. doi: 10.1016/j.neurobiolaging.2012.01.109. [DOI] [PubMed] [Google Scholar]
- Rutherford NJ, Dejesus-Hernandez M, Baker MC, Kryston TB, Brown PE, Lomen-Hoerth C, Boylan K, Wszolek ZK, Rademakers R. C9ORF72 hexanucleotide repeat expansions in patients with ALS from the Coriell Cell Repository. Neurology. 2012a;79(5):482–483. doi: 10.1212/WNL.0b013e31826170f1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford NJ, Heckman MG, Dejesus-Hernandez M, Baker MC, Soto-Ortolaza AI, Rayaprolu S, Stewart H, Finger E, Volkening K, Seeley WW, Hatanpaa KJ, Lomen-Hoerth C, Kertesz A, Bigio EH, Lippa C, Knopman DS, Kretzschmar HA, Neumann M, Caselli RJ, White CL, 3rd, Mackenzie IR, Petersen RC, Strong MJ, Miller BL, Boeve BF, Uitti RJ, Boylan KB, Wszolek ZK, Graff-Radford NR, Dickson DW, Ross OA, Rademakers R. Length of normal alleles of C9ORF72 GGGGCC repeat do not influence disease phenotype. Neurobiol Aging. 2012b;33(12):2950, e5–e7. doi: 10.1016/j.neurobiolaging.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatelli M, Conforti FL, Zollino M, Mora G, Monsurro MR, Volanti P, Marinou K, Salvi F, Corbo M, Giannini F, Battistini S, Penco S, Lunetta C, Quattrone A, Gambardella A, Logroscino G, Simone I, Bartolomei I, Pisano F, Tedeschi G, Conte A, Spataro R, La Bella V, Caponnetto C, Mancardi G, Mandich P, Sola P, Mandrioli J, Renton AE, Majounie E, Abramzon Y, Marrosu F, Marrosu MG, Murru MR, Sotgiu MA, Pugliatti M, Rodolico C, Moglia C, Calvo A, Ossola I, Brunetti M, Traynor BJ, Borghero G, Restagno G, Chio A. C9ORF72 hexanucleotide repeat expansions in the Italian sporadic ALS population. Neurobiol Aging. 2012;33(8):1848, e15–e20. doi: 10.1016/j.neurobiolaging.2012.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon-Sanchez J, Dopper EG, Cohn-Hokke PE, Hukema RK, Nicolaou N, Seelaar H, de Graaf JR, de Koning I, van Schoor NM, Deeg DJ, Smits M, Raaphorst J, van den Berg LH, Schelhaas HJ, De Die-Smulders CE, Majoor-Krakauer D, Rozemuller AJ, Willemsen R, Pijnenburg YA, Heutink P, van Swieten JC. The clinical and pathological phenotype of C9ORF72 hexanucleotide repeat expansions. Brain. 2012;135(Pt 3):723–735. doi: 10.1093/brain/awr353. [DOI] [PubMed] [Google Scholar]
- Smith BN, Newhouse S, Shatunov A, Vance C, Topp S, Johnson L, Miller J, Lee Y, Troakes C, Scott KM, Jones A, Gray I, Wright J, Hortobagyi T, Al-Sarraj S, Rogelj B, Powell J, Lupton M, Lovestone S, Sapp PC, Weber M, Nestor PJ, Schelhaas HJ, Asbroek AA, Silani V, Gellera C, Taroni F, Ticozzi N, Van den Berg L, Veldink J, Van Damme P, Robberecht W, Shaw PJ, Kirby J, Pall H, Morrison KE, Morris A, de Belleroche J, Vianney de Jong JM, Baas F, Andersen PM, Landers J, Brown RH, Jr, Weale ME, Al-Chalabi A, Shaw CE. The C9ORF72 expansion mutation is a common cause of ALS+/-FTD in Europe and has a single founder. Eur J Hum Genet. 2013 Jan;21(1):102–108. doi: 10.1038/ejhg.2012.98. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowden JS, Rollinson S, Thompson JC, Harris JM, Stopford CL, Richardson AM, Jones M, Gerhard A, Davidson YS, Robinson A, Gibbons L, Hu Q, DuPlessis D, Neary D, Mann DM, Pickering-Brown SM. Distinct clinical and pathological characteristics of frontotemporal dementia associated with C9ORF72 mutations. Brain. 2012;135(Pt 3):693–708. doi: 10.1093/brain/awr355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart H, Rutherford NJ, Briemberg H, Krieger C, Cashman N, Fabros M, Baker M, Fok A, DeJesus-Hernandez M, Eisen A, Rademakers R, Mackenzie IR. Clinical and pathological features of amyotrophic lateral sclerosis caused by mutation in the C9ORF72 gene on chromosome 9p. Acta Neuropathol. 2012;123(3):409–417. doi: 10.1007/s00401-011-0937-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallania FL, Druley TE, Ramos E, Wang J, Borecki I, Province M, Mitra RD. High-throughput discovery of rare insertions and deletions in large cohorts. Genome Res. 2010;20(12):1711–1718. doi: 10.1101/gr.109157.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Blitterswijk M, Dejesus-Hernandez M, Rademakers R. How do C9ORF72 repeat expansions cause amyotrophic lateral sclerosis and frontotemporal dementia: can we learn from other noncoding repeat expansion disorders? Curr Opin Neurol. 2012;25(6):689–700. doi: 10.1097/WCO.0b013e32835a3efb. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.