Abstract
Genetic changes required for the formation and progression of human Schwann cell tumors remain elusive. Using a Sleeping Beauty forward genetic screen, we identified several genes involved in canonical Wnt signaling as potential drivers of benign neurofibromas and malignant peripheral nerve sheath tumors (MPNSTs). In human neurofibromas and MPNSTs, activation of Wnt signaling increased with tumor grade and was associated with down-regulation of β-catenin destruction complex members or overexpression of a ligand that potentiates Wnt signaling, R-spondin 2 (RSPO2). Induction of Wnt signaling was sufficient to induce transformed properties to immortalized human Schwann cells, and down-regulation of this pathway was sufficient to reduce the tumorigenic phenotype of human MPNST cell lines. Small molecule inhibition of Wnt signaling effectively reduced viability of MPNST cell lines, and synergistically induced apoptosis when combined with an mTOR inhibitor, RAD-001, suggesting that Wnt inhibition represents a novel target for therapeutic intervention in Schwann cell tumors.
Keywords: Malignant peripheral nerve sheath tumors, Schwann cells, neurofibromatosis type 1 syndrome, neurofibromin 1, neurofibromatosis 1, Wnt signaling, β-catenin, targeted therapies, Sleeping Beauty transposon system, forward genetic screen, murine models
Introduction
Malignant peripheral nerve sheath tumors (MPNSTs) are soft tissue sarcomas that are believed to originate in the Schwann cell or Schwann cell precursors (1). These tumors can occur in the context of Neurofibromatosis Type 1 Syndrome (NF1), which occurs in 1 in 3,000 live births, but can also occur spontaneously in the general population (2,3). Due to the incomplete understanding of the genes and pathways driving MPNST development and progression, the current treatment for patients is surgical resection of the tumor, if possible, followed by non-specific, high-dose chemotherapy (4,5). These therapies often prove ineffective, and subsequently, patients with MPNSTs suffer very poor 5-year survival rates of less than 25% (3,4). This demonstrates the urgent need for a more complete understanding of the genetic events that drive these tumors, in order to develop novel targeted therapies to treat these patients.
It is known that biallelic loss of the Neurofibromin 1 gene (NF1) in Schwann cells is the pathological cause of the benign neurofibromas seen in NF1 patients, but secondary genetic changes, many of which remain unknown, must occur for these benign tumors to transform into MPNSTs (1,5–8). Ten percent of NF1-associated neurofibromas will undergo malignant transformation, the leading cause of death in adult NF1 patients (4, 5). MPNSTs can also form spontaneously, in the absence of NF1 loss, and the genes responsible for spontaneous MPNST formation are also largely unknown (3,4). Loss of Phosphatase and Tensin Homolog (PTEN) expression and overexpression of Epidermal Growth Factor Receptor (EGFR) are two changes often seen in both spontaneous MPNSTs and NF1-associated MPNSTs, but there are likely many other important genetic changes and signaling pathways yet to be identified (5,8–14).
Canonical Wnt/β-catenin signaling has been shown to play a role in many types of cancer, including colorectal, lung, breast, ovarian, prostate, liver and brain tumors (15). However, this pathway has not been directly implicated in neurofibromas or MPNSTs. In other cell types, Wnt signaling can be activated in cancer through a variety of mechanisms, including activating mutations in β-catenin (CTNNB1), overexpression of Wnt ligand genes, inactivating mutations in AXIN1, GSK3B, and APC (all members of the β-catenin destruction complex), and promoter hypermethylation of negative regulators of Wnt signaling (16). Another mechanism of Wnt pathway activation recently shown in colorectal cancer is overexpression of R-spondins due to gene fusions (17). R-spondins are secreted ligands that can potentiate Wnt signaling in the presence of Wnt ligands (18). Wnt signaling can also be activated via crosstalk with other signaling pathways including the PI3K/AKT/mTOR pathway, where loss of PTEN can activate PKB/AKT, causing the phosphorylation and inactivation of GSK3B, resulting in stabilization of β-catenin protein (19). Growth factor signaling pathways can also activate Wnt signaling; such as the case of Epidermal Growth Factor (EGF) stimulation of the receptor (EGFR), which results in the activation of β-catenin/TCF/LEF-dependent transcription of genes such as CyclinD1 (CCND1), C-Myc (MYC), and Survivin (BIRC5) (20,21). Notably, some human MPNSTs have been shown to have loss of PTEN expression and/or express high levels of active EGFR (9,11,12,14). β-catenin-dependent transcription can promote progression through the cell cycle, stem cell self-renewal, and epithelial to mesenchymal transition, all of which play a role in tumor initiation and progression (15,16,22). Due to the importance of this pathway in driving tumorigenesis in many types of cancer, the development of small molecule inhibitors that target Wnt signaling is rapidly underway and has the potential for profound clinical benefits for patients with Wnt-driven tumors (16,23,24).
We have implicated canonical Wnt signaling in the development and progression of peripheral nerve tumors by using a murine Sleeping Beauty (SB) forward genetic screen (Rahrmann, et al., submitted). Additionally, we demonstrate that several well-established murine models of neurofibromas and MPNST development also exhibit activation of the Wnt signaling pathway. Activation of this pathway has been confirmed in human patient samples by gene expression microarray analysis and tissue microarray (TMA) studies. Activation of Wnt signaling occurs in human tumors through multiple mechanisms including down-regulation of β-catenin destruction complex members and overexpression of R-spondin 2 (RSPO2). We demonstrate that activating canonical Wnt signaling is sufficient to induce transformed properties in immortalized human Schwann cells in vitro. We also show that down-regulating canonical Wnt signaling reduces the oncogenic and tumorigenic phenotypes observed in human NF1-associated and sporadic MPNST cell lines as measured by cell viability, colony formation, and xenograft tumor growth. Further, we find that small molecule inhibitors of Wnt signaling inhibit MPNST cell viability with little effect on normal human Schwann cells. These inhibitors show marked synergistic effects when combined with the mammalian target of rapamycin (mTOR) inhibitor, RAD-001, which has previously shown to be moderately effective in preclinical MPNST models (25–27). These results suggest that Wnt/β-catenin signaling is a novel drug target for patients with MPNSTs.
Results
Diverse mouse models implicate canonical Wnt signaling in Schwann cell tumor development and progression
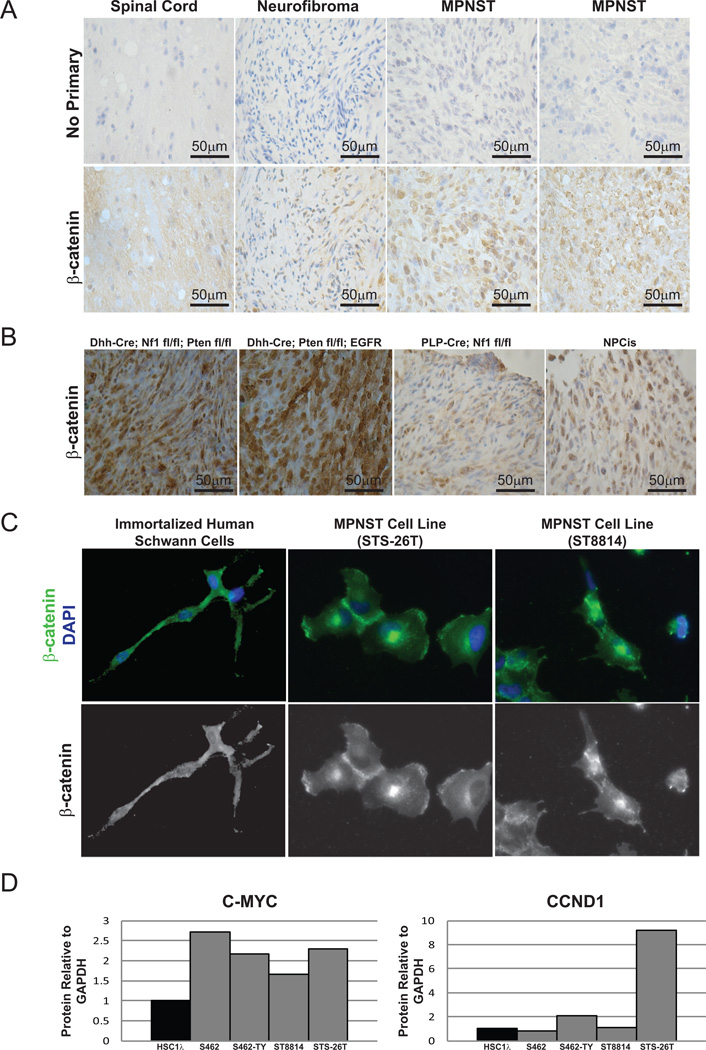
To identify genes and pathways that drive Schwann cell tumor development and progression, a forward genetic screen using the SB transposon system was conducted in mice, similar to many that have been previously published (28). The full details of this screen will be published separately (Rahrmann, et al., submitted). This screen identified several members of the canonical Wnt/β-catenin signaling pathway in the development of benign neurofibromas and MPNSTs (Supplementary Table 1). This screen was conducted using a CNPase promoter driving Cre expression which, in combination with a Cre/Lox-regulated Rosa26-SB11LSL transgene, allows for T2/Onc mutagenic transposon mobilization in Schwann cells and their precursors, the cell of origin for peripheral nerve sheath tumors (29). A common transposon insertion site (CIS) associated gene list was generated independently for benign neurofibromas and MPNSTs, as diagnosed by histopathological examination (Rahrmann, et al., submitted). We required that integrations map uniquely to the murine genome with a sequence length that would preclude random mapping. Further, we required that each integration be present at greater than 1/10,000 of the total sequences present to exclude potential non-driver mutations that are found in minor sub-clones or due to artifacts (30)(30). When the entire CIS gene list for MPNSTs was analyzed using Ingenuity Pathway Analysis (IPA, Ingenuity ® Systems, www.ingenuity.com), we found enrichment for genes that are present in the Wnt/β-catenin pathway (p= 3.93E-4). Further, when immunohistochemistry (IHC) analysis was performed on murine tumors from this SB screen at various stages of progression, the level and nuclear localization of β-catenin increased with tumor progression (Figure 1A). In addition to tumors from mice induced with SB, four other well-established mouse models of MPNST showed nuclear β-catenin expression (Figure 1B) (13,14,31–33). These mouse models represent both NF1-associated and sporadic MPNST development by a variety of genetic mechanisms. Interestingly, tumors from the rapidly forming, highly aggressive models, Dhh-Cre; Nf1fl/fl; Ptenfl/fl and Dhh-Cre; Ptenfl/fl;CNP-EGFR stained very strongly for nuclear β-catenin, while the slower forming NPcis and PLP-Cre; Nf1fl/fl tumors had less total β-catenin, but still highly nuclear staining. Thus, the intensity of nuclear β-catenin staining may correlate with and indeed drive tumor aggressiveness.
Figure 1. Murine peripheral nerve tumors and human MPNST cell lines show an increase in nuclear β-catenin and Wnt pathway outputs.
A. Mouse tumors from the SB screen (Rahrmann, et al., submitted) were stained by IHC for β-catenin. High grade tumors showed an increased level and nuclear localization of β-catenin compared to normal nerve or benign neurofibromas. B. Tumor sections from 4 established mouse models demonstrate nuclear β-catenin staining by IHC. C. Immortalized human Schwann cells (iHSC2λ) stain positive for β-catenin by immunofluorescence, but it is mainly localized in the cytoplasm. In contrast, MPNST cell lines (STS-26T and ST8814) show more nuclear-localized β-catenin. D. Compared to immortalized human Schwann cells (iHSC1λ), some MPNST cell lines show higher levels of C-MYC and CCND1 protein by western blot.
To determine the role of Wnt/β-catenin signaling in human Schwann cell tumors, we assayed expression and localization of β-catenin and expression of known Wnt target genes in human Schwann cells and MPNST cell lines. Activated β-catenin is found in the nucleus where it can be transcriptionally active, while inactive β-catenin is cytoplasmically localized or membrane-bound (22). In a TERT/CDK4R24C immortalized human Schwann cell line (iHSC2λ, Margaret Wallace, manuscript in preparation), β-catenin was expressed, but localized primarily in the cytoplasm (Figure 1C). In contrast, both sporadic and NF1-associated MPNST cell lines (STS-26T and ST8814, respectively) expressed a similar level of β-catenin, but with predominantly nuclear localization. Western blot analysis showed that in all 4 MPNST cell lines analyzed, the level of MYC protein was increased, and in 2 out of 4 MPNST cell lines, CCND1 protein was increased compared to the immortalized human Schwann cell line (Figure 1D). These data suggest that known β-catenin outputs are increased in MPNST cell lines compared to immortalized human Schwann cells.
Canonical Wnt signaling is activated in a subset of human neurofibromas and MPNSTs
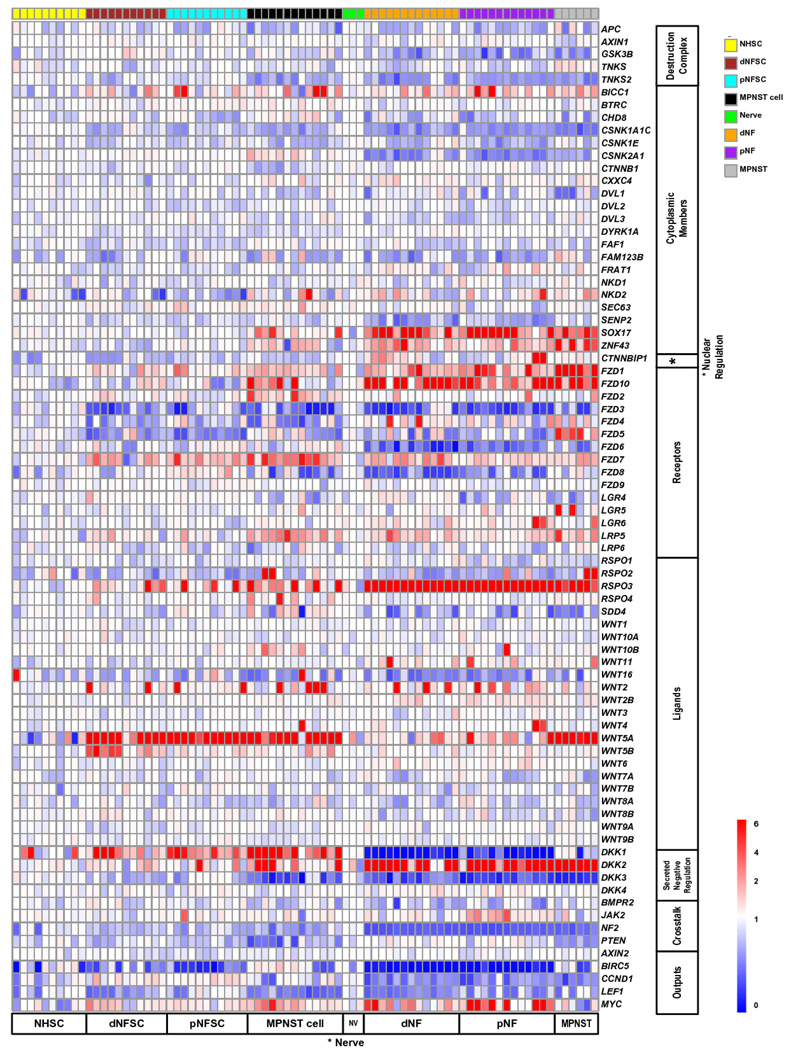
To characterize whether alterations in regulators of Wnt signaling underlie pathway activation in MPNST cell lines, we assayed Wnt regulators by gene expression microarray in purified Schwann cells taken from human peripheral nerve, neurofibromas, and MPNSTs, as well as solid tumors at various stages of disease (34) (Figure 2). Several members of the β-catenin destruction complex, including APC and GSK3B, showed down-regulation with tumor progression, in a subset of cases. The Wnt ligands WNT2, WNT5A, and WNT5B showed an increase in many or most samples. These data suggest that multiple mechanisms activate canonical Wnt signaling in human Schwann cell tumors.
Figure 2. Microarray expression of Wnt signaling regulators and pathway members.
Heat-map depicting microarray expression analysis for Wnt signaling regulators and pathway members. NHSC = purified normal human Schwann cells, dNFSC= purified dermal neurofibroma Schwann cells, pNFSC = purified plexiform-neurofibroma Schwann cells, MPNST cell = human MPNST cell lines, NV = normal human nerve, dNF = dermal neurofibroma, pNF = plexiform-neurofibroma, MPNST = bulk MPNST tumor.
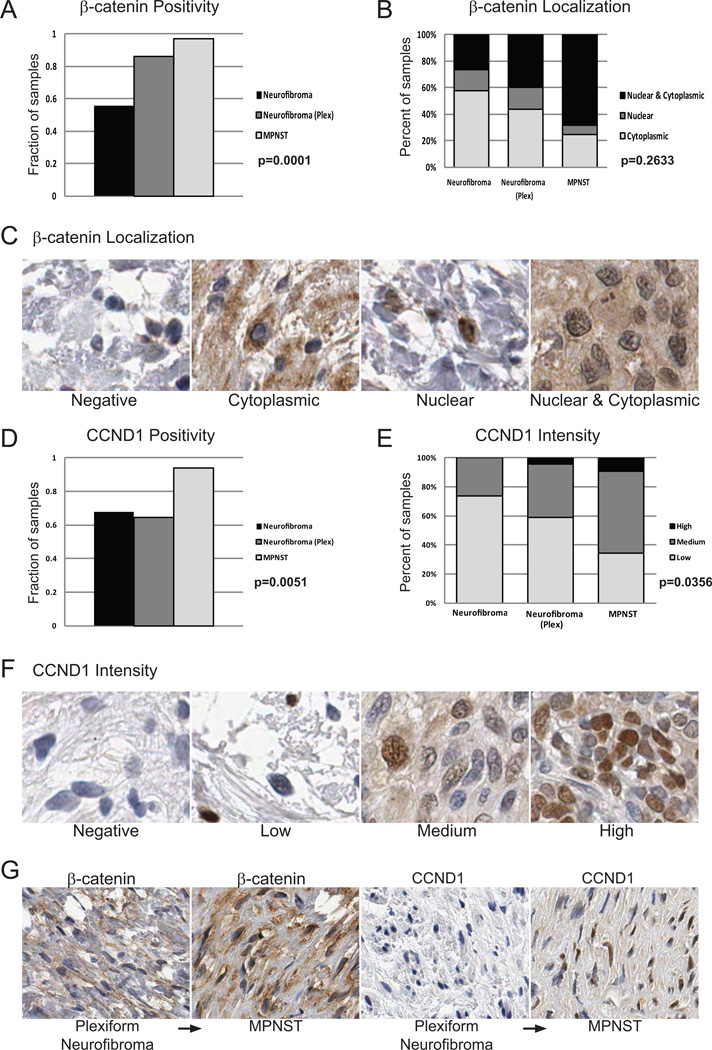
To further investigate activation of Wnt signaling in human tumors, and to account for regulation of Wnt signaling by post-translational mechanisms such as protein degradation or localization, we constructed a human tissue microarray (TMA) composed of 30 benign dermal neurofibromas (dNF), 32 plexiform neurofibromas (pNF), and 31 MPNSTs. The fraction of tumors positive for β-catenin increased with tumor progression (55.9% dNF, 86.2% pNF, 96.9% MPNSTs, p=0.0001 Fisher’s Exact Test) (Figure 3A). Additionally, the percentage of tumors with nuclear β-catenin was higher in the MPNSTs than in the neurofibromas (Figure 3B & C). As a transcriptional output of Wnt signaling, we also quantified the percentage of CCND1 positive tumors and found that it also increased in the higher grade tumors (67.6% dNF, 64.7% pNF, 94.1% MPNST, p=0.0051 Fisher’s Exact Test) (Figure 3D). Further, the intensity of CCND1 staining increased with tumor progression (p=0.0356 Fisher’s Exact Test) (Figure 3E & F). As an additional transcriptional output of Wnt signaling, we assessed C-MYC staining intensity and found that it too showed a trend towards increased expression with tumor progression (p=0.1603 Fisher’s Exact Test) (Supplementary Figure 1A & B). This suggests that the acquisition of nuclear β-catenin in these tumors is activating known β-catenin-targets, and correlates with tumor progression. To confirm this, we assessed β-catenin and CCND1 staining on a plexiform neurofibroma which transformed into an MPNST. While there was only a single patient represented on our TMA with a paired plexiform neurofibroma and MPNST, we found that indeed, β-catenin level and nuclear localization increased in the MPNST. Further, while the plexiform neurofibroma was negative for CCND1, the MPNST was positive for CCND1 (Figure 3G).
Figure 3. Human tissue microarray shows a subset of human neurofibromas and MPNSTs have activated Wnt/β-catenin signaling.
A. A large subset of Schwann cell tumors stain positive for β-catenin by IHC. The fraction of β-catenin positive tumors increases from benign neurofibromas (neurofibroma) to plexiform neurofibromas (plex), to MPNSTs. B. The fraction of tumors staining positive for nuclear β-catenin increases from benign neurofibroma to MPNSTs as well. C. Example staining of β-catenin localization. D. Large subsets of Schwann cell tumors are positive for CCND1, an output of canonical Wnt/β-catenin signaling. A larger fraction of MPNSTs are CCND1 positive compared to the benign neurofibromas and plexiform neurofibromas. E. The level of CCND1 increases from benign neurofibromas to plexiform neurofibromas and is the greatest in MPNSTs. F. Example of CCND1 staining intensity. G. A patient-matched plexiform neurofibroma which transformed into an MPNST shows higher total and nuclear β-catenin staining with transformation. The plexiform neurofibroma stained negative for CCND1, while the MPNST was CCND1 positive.
Activation of Wnt/β-catenin signaling is sufficient to induce transformed phenotypes in immortalized Schwann cells
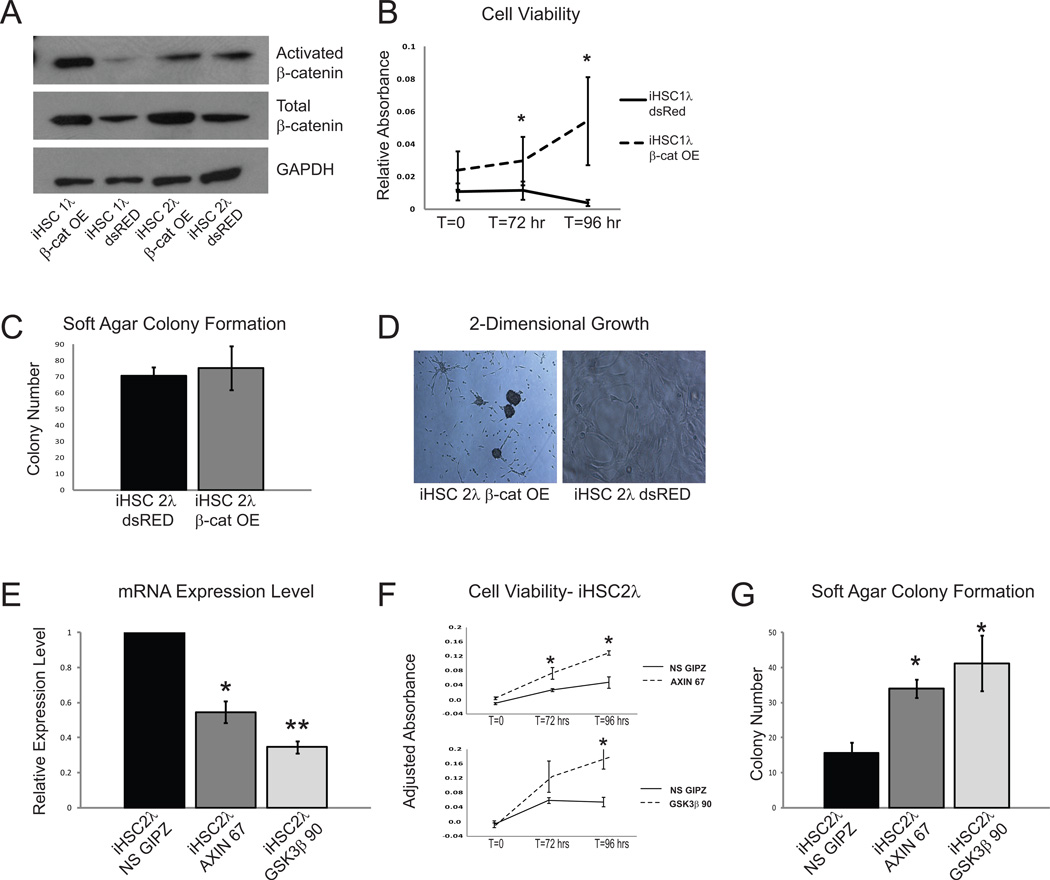
We next functionally validated a subset of the β-catenin regulatory genes identified in our Sleeping Beauty screen that were also implicated in human tumors (Supplementary Table 1 & Figure 2). We hypothesized that if Wnt/β-catenin signaling played a role in the development of peripheral nervous system tumors, then activation of this pathway in immortalized human Schwann cells may be sufficient to drive a more transformed phenotype in these cells. To activate Wnt signaling, we expressed an activated form of CTNNB1 (CTNNB1S33Y) in immortalized human Schwann cells (iHSC1λ and iHSC2λ). Cells overexpressing CTNNB1S33Y showed increased levels of total β-catenin protein, but interestingly, only the iHSC1λ cells showed increased activated β-catenin (Figure 4A). Expression of CTNNB1S33Y resulted in increased expression of MYC, CCND1, AXIN2, LEF1, and BIRC5 in both iHSC1λ and iHSC2λ cell lines, demonstrating activation of genes that have been shown to be β-catenin targets (Supplementary Figure 2A & B). CTNNB1S33Y expression also resulted in increased cell viability (Figure 4B), but did not change soft agar colony formation (Figure 4C). However, we did observe that cells overexpressing CTNNB1S33Y stopped growing as a monolayer, as seen in non-transformed cells, and began growing as three-dimensional colonies (Figure 4D).
Figure 4. Immortalized human Schwann cells show an increase in transformed properties when Wnt/β-catenin signaling is activated.
A. Overexpression of a CTNNB1S33Y construct in the immortalized human Schwann cell line, iHSC1λ, results in an increase in total and activated β-catenin, while expression in iHSC2λ cells results in only an increase in total β-catenin. B. Expression of activated β-catenin increases cell viability in immortalized human Schwann cells. C. Expression of CTNNB1S33Y has no effect on soft agar colony formation in iHSC2λ cells. D. Expression of CTNNB1S33Y results in altered morphology when iHSC2λ cells are grown in 2-dimensional culture. E. The immortalized human Schwann cell line, iHSC2λ, has reduced AXIN1 and GSK3B transcript levels, respectively, when treated with shRNA constructs against these genes (AXIN 67 and GSK3β 90). F. Knockdown of AXIN1 and GSK3B in iHSC2λ cells increases cell viability. G. Reducing the expression of AXIN1 and GSK3B in iHSC2λ cells is sufficient to increase soft agar colony formation.* p<0.05, **p<0.0001 unpaired T-test. Error bars represent SEM.
In addition to activating Wnt signaling by overexpressing CTNNB1S33Y, we used shRNA to knock down AXIN1 and GSK3B, both members of the β-catenin destruction complex identified in our SB screen (Supplementary Table 1) and known tumor suppressor genes in other types of cancers (15). These genes were knocked down in both immortalized human Schwann cell lines (iHSC1λ and iHSC2λ) using short hairpin RNAs (shRNAs), and validated by QPCR analysis (Figure 4E & Supplementary Figure 2I) and western blot analysis (Supplementary Figure 2C & F). We show that knockdown of either AXIN1 or GSK3B activated genes that have been shown to be to be targets of β-catenin including CCND1, MYC, AXIN2, LEF1, and BIRC5 (Supplementary Figure 2D, E, G & H). Knockdown of AXIN1 or GSK3B was sufficient to induce oncogenic properties in immortalized human Schwann cells as demonstrated in vitro by a significant increase in cell viability (Figure 4F & Supplementary Figure 2J) and anchorage-independent growth (Figure 4G & Supplementary Figure 2K). When the immortalized human Schwann cells that expressed either AXIN1 or GSK3B shRNAs were injected into immunodeficient mice, they were unable to induce tumor formation (data not shown), suggesting that while activation of Wnt signaling is sufficient to induce some oncogenic properties in Schwann cells in vitro, it is not sufficient to induce tumorigenic properties in vivo.
Inhibition of Wnt signaling can reduce the tumorigenic phenotype of MPNST cells
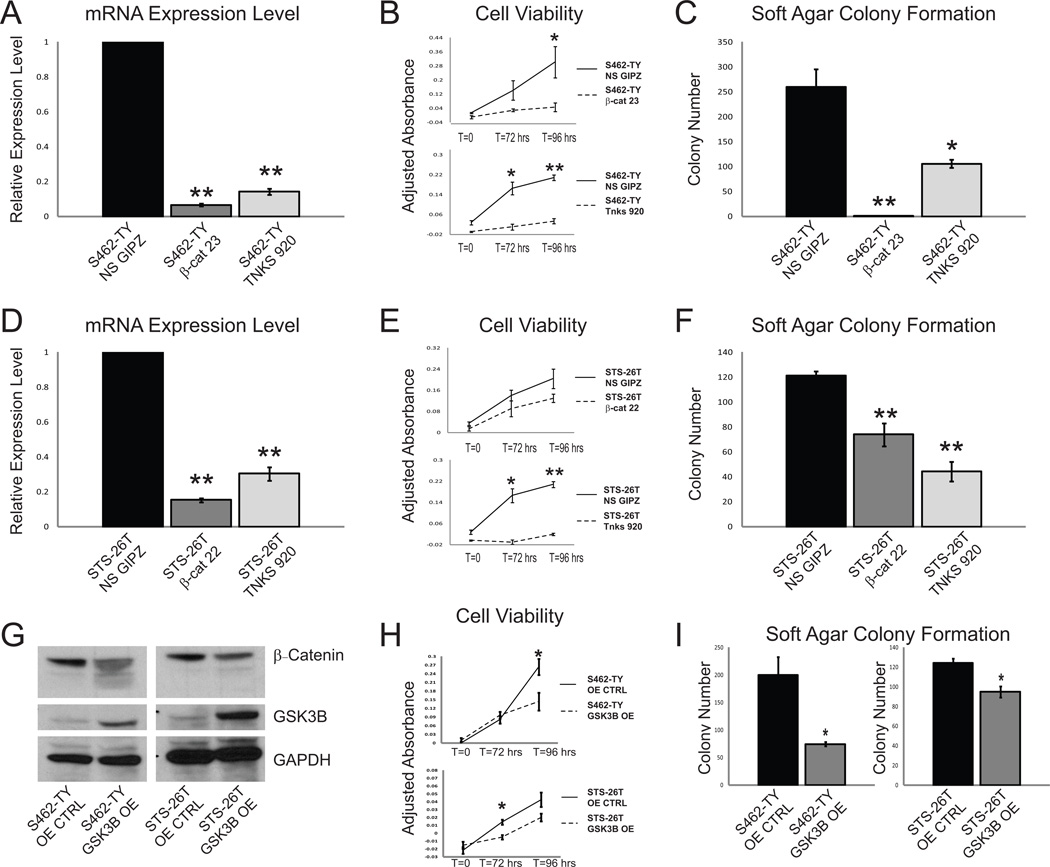
We next sought to determine whether inhibition of Wnt/β-catenin signaling in MPNST cell lines was sufficient to reduce cellular viability and anchorage-independent growth. To reduce Wnt signaling, we knocked down CTNNB1 and TNKS in two MPNST cell lines, S462-TY (NF1-associated MPNST cell line (35)) and STS-26T (sporadic MPNST cell line (36)) using shRNA vectors. We chose to knockdown CTNNB1 since it plays a direct role in the transcription of Wnt-dependent genes by binding TCF/LEF in the nucleus and acting as a transcriptional activator (22,23). TNKS was also knocked down because it was identified in our forward genetic screen and inhibition of TNKS is known to stabilize AXIN1, leading to the degradation of β-catenin protein (37). TNKS is also the target of many small molecule inhibitors of the Wnt pathway (37). Knockdown of CTNNB1 and TNKS by shRNA was confirmed by QPCR (Figure 5A & D). Reduction in either β-catenin or TNKS was sufficient to decrease expression of β-catenin transcriptional targets, as demonstrated by a reduction in the expression of MYC, CCND1, AXIN2, LEF1, and BIRC5 (Supplementary Figure 3A, B, C & D). Knockdown of either CTNNB1 or TNKS reduced cell viability (Figure 5B & E) and anchorage-independent growth (Figure 5C & F) in both S462-TY and STS-26T cells.
Figure 5. NF1-associated and sporadic MPNST cell lines show a decrease in cell viability, soft agar colony formation, and xenograft tumor growth when Wnt/β-catenin signaling is down-regulated.
A. shRNA reduces β-catenin (β-cat 23) and TNKS (TNKS 920) expression levels in an NF1-associated MPNST cell line (S462-TY) compared to non-silencing shRNA (NS GIPZ) treated cells. B. Knockdown of β-catenin or TNKS results in a reduction in cell viability in the S462-TY cell line. C. Soft agar colony formation is reduced in an NF1-associated cell line (S462-TY) when β-catenin or TNKS levels are reduced. D. shRNA treatment results in a reduction of β-catenin (β-cat 22) and TNKS (TNKS 920) levels in a cell line derived from a sporadic MPNST (STS-26T) compared to non-silencing shRNA (NS GIPZ) treated controls. E. Knockdown of β-catenin or TNKS is sufficient to reduce cell viability in STS-26T cells. F. A reduction in β-catenin or TNKS expression reduces soft agar colony formation in this spontaneous MPNST cell line. G. Western blot analysis shows that overexpression of GSK3B results in the degradation of β-catenin protein in both an NF1-associated and sporadic MPNST cell lines (S462-TY and STS-26T). H. Overexpression of GSK3B decreases cell viability in S462-TY and STS-26T MPNST cell lines. I. Overexpression of GSK3B reduces soft agar colony formation in S462-TY and STS-26T MPNST cell lines. * p<0.05, **p<0.0001 unpaired T-test. Error bars represent SEM.
Overexpression of GSK3B, a member of the β-catenin destruction complex, also was sufficient to reduce the oncogenic properties of both the NF1-associated and sporadic MPNST cell lines. GSK3B overexpression was confirmed by western blot and QPCR and resulted in reduced β-catenin total protein levels (Figure 5G, Supplementary Figure 3E & F). The reduction in known Wnt signaling outputs was also shown by a decrease in MYC, CCND1, AXIN2, LEF1, and BIRC5 transcript levels (Supplementary Figure 3E & F). GSK3B overexpression resulted in a decrease in cell viability (Figure 5H) and a significant decrease in soft agar colony formation in both cell lines (Figure 5I). These results suggest that these MPNST cell lines depend on Wnt signaling for rapid proliferation and robust colony forming abilities, and reduction in Wnt signaling can reduce these oncogenic properties.
To investigate the role of Wnt/β-catenin signaling in vivo, the S462-TY and STS-26T cell lines expressing either the CTNNB1 shRNA or GSK3B cDNA were injected into immunodeficient mice, which were monitored for the rate of tumor formation and growth. Reduction in CTNNB1 by shRNA delayed the rate of tumor growth in the NF1-asociated cell line (Supplementary Figure 4A & B). Tumor sections derived from cells expressing the CTNNB1 shRNA showed fewer Ki67 positive cells compared to tumors derived from NS shRNA expressing cells (Supplementary Figure 4C). Overexpression of GSK3B resulted in reduction in tumor growth in the spontaneous MPNST cell line (Supplementary Figure 4D & E). Tumor sections from cells overexpressing GSK3B also showed fewer Ki67 positive cells compared to tumors derived from non-silencing (NS) shRNA expressing cells (Supplementary Figure 4F). Despite a significant delay in tumor onset, tumor volume at 40 days post-injection was similar in control and experimental groups. Evaluation of gene expression in these tumors indicated that knockdown of CTNNB1 and overexpression of GSK3B was not maintained over the course of the experiment (data not shown).
R-spondin 2 is overexpressed in a subset of Schwann cell tumors and is a driver of Wnt signaling and MPNST cell growth
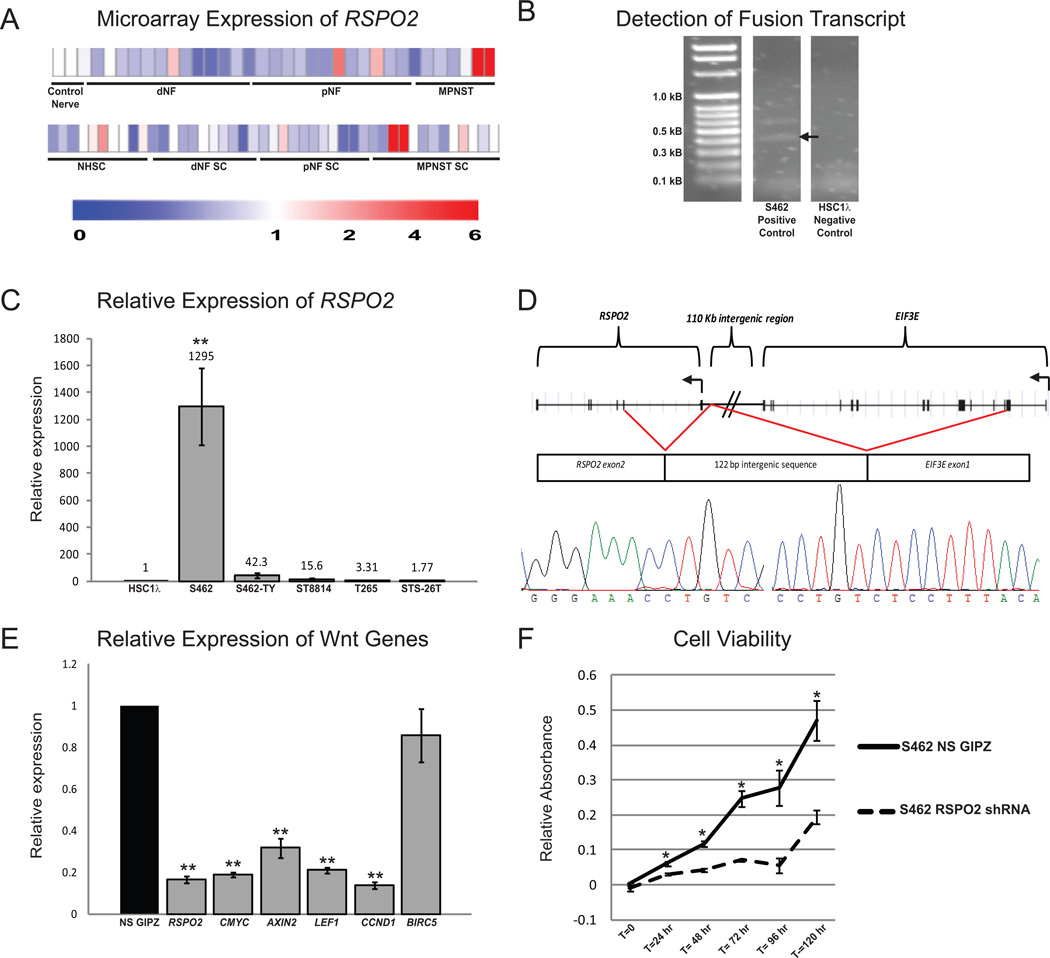
R-spondins are secreted ligands that potentiate Wnt signaling when Wnt ligand is available (18). Microarray analysis showed that R-spondin 2 (RSPO2) was highly expressed in 2 of 6 MPNST cell lines and 2 of 13 primary MPNSTs evaluated, compared with purified normal human Schwann cells or normal nerve (Figure 6A). Further, it has been shown that one mechanism for RSPO2 overexpression is a deletion-mediated gene fusion between exon 1 of EIF3E and exon 2 of RSPO2, resulting in the overexpression of native RSPO2 protein (17). To determine whether similar gene fusions occur in Schwann cell tumors, primers were designed to amplify any fusion transcripts between EIF3E and RSPO2 in cDNA libraries made from 5 human MPNST cell lines and 2 immortalized human Schwann cell lines. We identified a fusion transcript expressed by the S462 MPNST cell line that was not present in any of the other MPNST or immortalized human Schwann cell lines (Figure 6B). Expression of this fusion correlated with a 1295-fold increase in RSPO2 expression in the S462 cell line (Figure 6C). The fusion transcript that was identified included exon 1 of EIF3E fused to a 122 base pair region of chromosome 8 located between EIF3E and RSPO2, fused to exon 2 of RSPO2 (Figure 6D). To determine if S462 cells require RSPO2, we used an shRNA construct to knock down its expression. Depletion of RSPO2 significantly reduced expression of Wnt target genes and cell viability (Figure 6E & F). These data suggest that overexpression of RSPO2 may drive a subset of human MPNSTs, and further support the model that Wnt signaling is activated in Schwann cells by multiple mechanisms.
Figure 6. The secreted Wnt/β-catenin activator RSPO2 is highly expressed in a subset of human Schwann cell tumors and can be detected as a fusion transcript with the upstream EIF3E gene.
A. Gene expression microarray shows that a subset of plexiform neurofibromas and MPNSTs from both purified Schwann cells (top) and primary tumors (bottom) have high RSPO2 expression. B. cDNA libraries derived from human MPNST cell lines were screened for EIF3E-RSPO2 fusion transcripts (fusion transcripts verified by sequencing are denoted by an arrow). C. 5 MPNST cell lines were screened for RSPO2 expression levels and normalized to an immortalized human Schwann cell line (iHSC1λ). The S462 cell line showed very high RSPO2 expression and was found to contain a fusion between EIF3E and RSPO2. D. The S462 cell line has a fusion between EIF3E exon 1, followed by a 122 base pair chromosome 8 intergenic sequence and RSPO2 exon 2. This fusion results in the production of a native RSPO2 protein. E. The S462 cell line was transduced with a lentivirus expressing an RSPO2 shRNA. This resulted in a decrease in RSPO2 expression, as well as a decrease in expression of known Wnt signaling outputs MYC, AXIN2, LEF1, CCND1, and BIRC5, as compared to S462 cells treated with a non-silencing shRNA (NS GIPZ). F. Knockdown of RSPO2 in S462 cells significantly reduces cell viability. Error bars represent standard error of the mean. * p<0.05, **p<0.0001 unpaired T-test. Error bars represent SEM.
Wnt inhibitors effectively reduce cellular viability in vitro
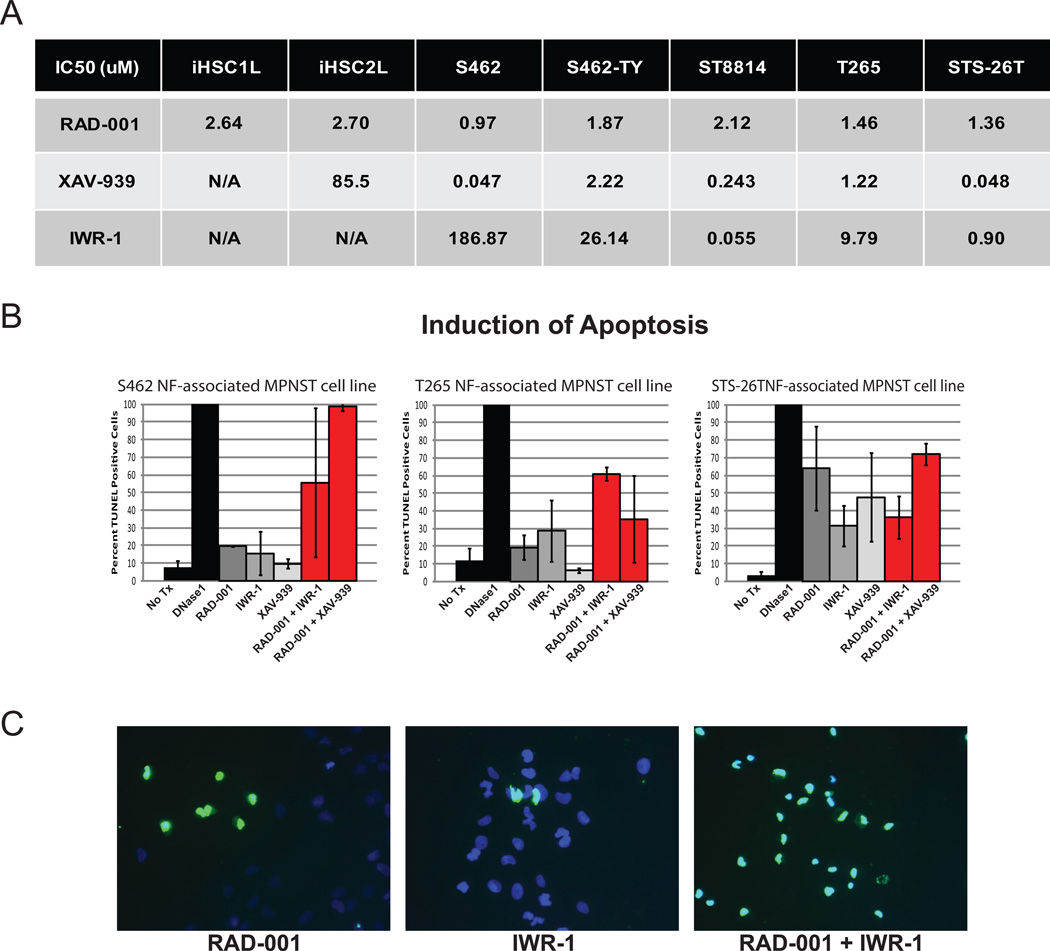
Our functional data suggested that targeting Wnt/β-catenin signaling with small molecule inhibitors, might be effective at reducing MPNST cell viability and tumor forming properties. To test this hypothesis, we exposed a panel of 5 MPNST cell lines (S462, S462-TY, ST8814, T265, and STS-26T) and 2 immortalized human Schwann cell lines (iHSC1λ and iHSC2λ) to two compounds that inhibit Wnt signaling, XAV-939 and IWR-1. These compounds both function to stabilize AXIN1 by inhibiting TNKS, resulting in an increase in β-catenin phosphorylation and subsequent degradation (37). We find normal Schwann cells are highly resistant to proliferation inhibition and cell death by these two compounds as demonstrated by high IC50 values, while the MPNST cell lines were quite sensitive with relatively low IC50s of 0.047uM to 2.22uM for XAV-939 and 0.055uM to 186.87uM for IWR-1 (Figure 7A) (CalcuSyn Version 2.1, BioSoft).
Figure 7. β-catenin inhibitors are effective in vitro and synergize with RAD-001.
A. IC50 concentrations (uM) of two immortalized human cell lines (iHSC1λ and iHSC2λ) and 5 human MPNST cell lines (S462, S462-TY, ST8814, T265, STS-26T) for the mTOR inhibitor RAD-001 (Everolimus) and two Wnt signaling inhibitors, IWR-1 and XAV-939. B. Induction of apoptosis in NF1-associated MPNST cell lines (S462, T265) and the sporadic MPNST cell line (STS-26T) as determined by the percentage of TUNEL positive cells that were treated with IC50 concentrations of RAD-001, IWR-1 and XAV-939 shows that these single drug treatments are largely cytostatic. When treated with IC25 concentrations of RAD-001 in combination with IWR-1 or XAV-939, a synergistic increase in apoptosis is often observed. No treatment (No Tx) and 50uM DNase1 were used as positive and negative controls, respectively. C. Representative images of S462 cells treated with RAD-001, IWR-1 or combination of RAD-001 and IWR-1. Cells were stained with TUNEL (green) and DAPI (blue), images taken at 20X magnification. Error bars represent SEM.
Wnt inhibitors synergize with an mTOR inhibitor to induce apoptosis of MPNST cells
We next conducted a screen to identify targeted therapies that synergized with Wnt signaling inhibitors to induce apoptosis in MPNST cell lines. Several drugs were screened, including inhibitors of PI3K (PI-103), mTOR (RAD-001 and Rapamycin), and MEK (PD-901 and AZD-6244). Of these drugs, the most significant synergism was seen with RAD-001. It has been shown that RAD-001 is modestly effective in several preclinical models of peripheral nerve tumors, but the effects of this drug are largely cytostatic (25,26). In most cases, treatment of the S462, T265, and STS-26T MPNST cell lines with RAD-001, IWR-1, or XAV-939 as single agents at their IC50 concentrations resulted in less than 50% TUNEL-positive cells, demonstrating that while 50% of proliferation was inhibited, only a minority of cells underwent apoptosis (Figure 7B & C). In contrast, when the MPNST cell lines were treated with their IC25 dose of RAD-001 in combination with either IWR-1 or XAV-939 also at their IC25 dose, the percentage of TUNEL positive cells often went above 50%, and in the S462 cell line, even approached 100%, demonstrating that co-targeting the Wnt and mTOR pathways is synergistic in inducing apoptosis.
Discussion
Analysis of MPNST tumor genomes has demonstrated a wide variety of genetic changes ranging from point mutations to entire chromosome gains and losses (38). Determining which of these changes contribute to tumorigenesis and which changes are simply “passenger” mutations is crucial in identifying targets for therapeutic intervention (1,38,39). Prior studies have focused on NF1 and p53/Rb regulated pathways in MPNSTs, yet there remains a need to broaden our understanding of the genetic alterations that contribute to Schwann cell tumorigenesis. The SB transposon system is an unbiased, powerful tool to identify oncogenes and tumor suppressor genes (28). In an SB screen conducted to find genetic drivers of MPNSTs and neurofibromas, we uncovered many alterations that affect canonical Wnt/β-catenin signaling, suggesting this pathway is a likely driver of Schwann cell tumor development, progression and maintenance (Supplementary Table 1) (Rahrmann, et al., submitted). We show that murine tumors from both the SB screen and from four other established MPNST models show nuclear-localized β-catenin, suggesting that they have activated canonical Wnt signaling (Figure 1). Additionally, we identified activation of Wnt signaling in a majority of human MPNST cell lines and primary human tumors investigated (Figure 1–3).
In this study, we show that multiple genetic alterations correlate with activation of the canonical Wnt/β-catenin pathway, and that these alterations can induce oncogenic properties in human Schwann cells, and are required for tumor maintenance in MPNST cells. A subset of human Schwann cell tumors show down-regulation of β-catenin destruction complex components, such as APC and GSK3B (Figure 2). Microarray analysis also demonstrated overexpression of several Wnt ligands and RSPO2, a secreted ligand that can potentiate Wnt signaling, in a subset of human tumors (Figure 6). Interestingly, we find that while CCND1 protein levels correlate with tumor progression, CCND1 mRNA expression is not as convincing (Figure 2 & 3). It should be noted that the microarray data suggests that in whole tumors CCND1 may be decreased, but in purified cells it seems to be unchanged or slightly up-regulated. This may be due to the effects of contaminating cells within the whole tumor such as fibroblasts, macrophages, and endothelial cells. In addition, we believe that the discrepancy seen between the TMA and the microarray gene expression of CCND1 may be due to the fact that the CCND1 transcript is highly regulated at the translational level. Due to the long 5’ untranslated region and secondary structure of CCND1 mRNA, it is often poorly translated, although many factors, including PI3K/mTOR signaling can increase the translation of this transcript (40). Based on the mRNA expression data, MYC may be a more consistent marker for activated Wnt signaling, while CCND1 may be a better marker of Wnt signaling activation by immunohistochemistry.
We tested the functional importance of Wnt pathway activation in Schwann cell tumorigenesis through gain- and loss-of-function studies in vitro. Using gene overexpression and knockdown in immortalized human Schwann cells and MPNST cell lines, we showed that aberrant expression of Wnt regulators resulted in phenotypic and functional changes in these cells. Down-regulation of GSK3B and AXIN1 and overexpression of CTNNB1 were sufficient to induce transformed properties in immortalized human Schwann cells and down-regulation of CTNNB1 and TNKS, as well as overexpression of GSK3B reduced the oncogenic properties of MPNST cell lines and delayed tumor growth in vivo (Figure 4, Figure 5, Supplementary Figure 2 & Supplementary Figure 4).
Recently, a novel mechanism for activating Wnt signaling was demonstrated in colorectal cancer in which gene fusions occurred between EIF3E and RSPO2 due to a deletion on chromosome 8, resulting in the overexpression of native RSPO2 protein (17). We identified a similar fusion transcript in a human MPNST cell line with elevated RSPO2 expression, and showed that knockdown of RSPO2 was sufficient to reduce Wnt signaling outputs and cellular viability in this cell line (Figure 6). Further work is needed to determine the frequency of R-spondin fusion transcript expression in primary human Schwann cell tumors. It will also be necessary to identify whether expression of R-spondin fusion transcripts in MPNSTs is mediated by chromosome 8 deletions similar to those seen in colorectal cancer, or if there is another mechanism by which this occurs, such as transcription-mediated gene fusion. Based on our results, a drug that could block the function of RSPO2 may be therapeutically beneficial for patients with MPNSTs overexpressing RSPO2.
In addition to the mechanisms described above, there are likely other mechanisms by which this pathway is activated in human tumors. For example, activation of PKB/AKT, a common phenomenon in MPNSTs, leads to the phosphorylation and inactivation of GSK3B, resulting in stabilization of β-catenin protein (19,41). Loss of PTEN expression has been demonstrated in human MPNSTs, and this could activate Wnt signaling through activation of AKT (12,13). It has also been shown that in cells expressing EGFR, EGF stimulation can result in the activation of β-catenin/TCF/LEF-dependent transcription, and EGFR expression has been implicated in MPNST development as well (9,11,20). Recently, Mo et al. demonstrated a novel mechanism of NF1-associated MPNST progression in which autocrine activation of CXCR4 by CXCL12 mediates tumor progression through PI3K and β-catenin (42). Although no insertions were identified in Cxcr4 or Cxcl12 in our SB screen, gene expression data from human tumors showed statistically significant differential regulation of CXCR4 in MPNST tumor-to-nerve comparison (fc=5.3×, p-value = 0.016), but not in MPNST cell-to-NHSC comparison (fc=0.85×, p-value=0.16). In contrast, CXCL12 showed statistically differential expression pattern in MPNST cell-to-NHSC comparison (fc=2.7×, p-value=0.02), but not in MPNST tumor-to-nerve comparison (fc=4.7×, p-value = 0.15). This study by Mo et al. compliments our work in showing yet another mechanism by which human Schwann cell-derived tumors activate Wnt signaling, leading to tumor progression. Additional work will be required to determine the contribution of each of these pathways to Wnt activation in human MPNSTs.
It is unclear from this study whether Wnt pathway activation is important for tumor initiation or for tumor progression, but it seems likely that it may play a role in both processes. It is clear that a subset of benign human neurofibromas exhibit activated Wnt signaling, which may play a role in the initiation of these benign tumors (Figure 3 & Supplementary Figure 1). A larger percentage of high grade tumors (MPNSTs) have activation of this signaling pathway, making it plausible that this pathway also plays a role in progression from a benign tumor to malignancy. It is also likely that this pathway may cooperate with other pathways that play a role in the development or progression of MPNSTs. For example, Wnt signaling activation is an initiating event for colorectal cancer, leading to hyperplasia, but other genetic changes must occur for a tumor to form (43)(43). Future work will need to focus on identifying cooperating mutations or signaling pathways that function in concert with Wnt signaling in MPNST formation. Indeed, loss of NF1 expression is a good candidate cooperating alteration, and future experiments in which NF1 is lost and the Wnt pathway is activated in normal Schwann cells will be critical for our understanding of cooperating genes and pathways.
While the Wnt pathway is known to play a critical role in oncogenesis, the development of clinically beneficial targeted therapies has lagged. The discovery of small molecule inhibitors of the Wnt/β-catenin pathway is being aggressively pursued, as these therapies have the potential to benefit patients suffering from a wide variety of cancers (16,23,24). Wnt/β-catenin signaling is critical for many normal developmental and cellular processes, thus, inhibition of this pathway may have many unwanted and negative side effects (16,44). To this end, inhibitors targeting the specific Wnt pathway molecules (such as RSPO2) and functions in tumorigenesis may be the key to identifying a successful targeted therapy (16,44). We show that inhibiting Wnt signaling in vitro is an effective cytostatic therapy, but when combined with RAD-001, they create a potent cytotoxic therapy (Figure 7). Further work will need to be done to investigate the use of Wnt pathway inhibitors using in vivo models of MPNST, as these may be a valuable new class of targeted therapies for patients with MPNSTs.
Methods
Mouse Tumor Immunohistochemistry (IHC)
Formalin fixed, paraffin embedded tissues were sectioned at 5 microns, mounted, and heat-fixed onto glass slides to be used for IHC analyses. Briefly, the glass section slides were de-waxed and rehydrated through a gradual decrease in ethanol concentration. The antigen epitopes on the tissue sections were then unmasked using a commercially available unmasking solution (Vector Laboratories) according to the manufacturer’s instructions. The tissue section slides were then treated with 3% hydrogen peroxide to remove endogenous peroxidases. Blocking was performed at room temperature in normal goat serum (5% serum in PBS) in a humidified chamber for one hour. Sections were then incubated overnight at 4°C in a humidified chamber with primary antibody (β-catenin, 1:100, Cell Signaling). After primary incubation, sections were washed thoroughly in PBS before incubating with goat anti-rabbit horseradish peroxidase conjugated-secondary antibody (Santa Cruz Biotechnology). After 3 washes with PBS, the sections were treated with freshly prepared DAB substrate (Vector Laboratories) and allowed to develop before stopping the reaction in water once adequate signal was obtained. Finally, sections were then lightly counter-stained with hematoxylin, dehydrated through gradual increase in ethanol concentration, cleared in Citrosol (Fisher Scientific) and mounted in Permount (Fisher Scientific).
Tissue culture reagents and cell lines
Cultured immortalized Schwann cells (iHSC1λ and iHSC2λ) were both derived from a patient’s normal sciatic nerve, are NF1 wild-type, and were immortalized by hTERT and CDK4R24C to allow in vitro studies (Dr. Margaret Wallace, manuscript in preparation). Immortalized human Schwann cell and MPNST cell lines (S462 (45), S462-TY (35), ST8814 (46), T265 (47), and STS-26T (36)) were maintained in Dulbecco’s Modification of Eagle Medium (DMEM) supplemented with 10% fetal bovine serum and penicillin/streptomycin (Cellgro) and cultured on tissue culture-treated plates under standard conditions of 37 degrees Celsius and 5% CO2. No authentication of cell lines were done by the authors.
Immunofluorescence and TUNEL staining
For immunofluorescence assays, cells were grown to 80% confluency on 8 chambered slides (Lab-TekII). Cells were fixed in 10% formalin and washed with phosphate-buffered saline (PBS) with 0.1% Tween-20 (PBST). Cells were incubated in β-catenin primary antibody (1:100, Cell Signaling) at 4 degrees Celsius overnight, followed by 1 hour room temperature incubation in anti-rabbit AlexaFluor 488 secondary antibody (Invitrogen). TUNEL staining was performed using the In Situ Cell Death Detection Kit, POD (Roche). Slides were mounted using Prolong Gold Antifade Reagent with DAPI (Invitrogen) and images using a Zeiss Axiovert 25 inverted microscope. For analysis of cell death, total cells and TUNEL positive cells were counted and averaged over three independent frames with 50–100 cells per frame.
Western blot analysis
1 million cells were lysed using an NP-40 buffer (50mM Tris-HCl pH 7.6, 150mM NaCl, 1% NP-40, 5mM NaF, 1mM EDTA) containing a protease inhibitor (Roche) and phosphatase inhibitors (Sigma). Whole cell lysates were cleared by centrifugation. Protein samples were prepared in an SDS solution with reducing agent (Invitrogen) and run on 10% Bis-Tris pre-made gels (NuPage, Invitrogen). Gels were transferred onto PVDF membranes using the iBlot system (Invitrogen) and activated in 100% methanol. Membranes were blocked in filtered 5% Bovine Serum Albumin (BSA) for 2 hours at room temp followed by a 4 degrees Celsius overnight incubation in primary antibody. The primary antibodies used in this study were activated β-catenin (1:1000), total β-catenin (1:100), GSK3B (1:1000), AXIN1 (1:1000), and GAPDH (1:2000) (Cell Signaling). Following primary antibody incubation, membranes were thoroughly washed in TBS with 0.1% Tween-20 (TBST) and incubated in goat anti-rabbit IgG-HRP conjugated secondary antibody (Santa Cruz, 1:4000 in 0.5% BSA, 1 hour at room temperature). Blots were thoroughly washed in TBST and developed using the SuperSignal WestPico Chemiluminescence Detection Kit (Thermo Scientific). Densitometry quantification was done using ImageJ software and normalized to GAPDH (48)(48).
Microarray gene expression analysis
We used the published data (GEO accession #:GSE14038, Affymetrix GeneChip HU133 Plus 2.0) [38] for gene microarray analysis. This dataset used custom CDF (custom GeneChip library file) based on RefSeq target definitions (Hs133P REFSEQ Version 8) for accurate interpretation of GeneChip data (49). The dataset in Figure 2 includes 86 microarrays on purified human Schwann cells and primary tissues taken from a normal sciatic nerve, dermal neurofibromas, plexiform neurofibromas, and MPNST cell lines. The heatmap was generated using CRAN's pheatmap package (http://cran.r-project.org/web/packages/pheatmap/index.html) and Refseq IDs were replaced with corresponding HGNC official gene symbols. Statistical comparisons were done using R/Bioconductor's Limma package (http://www.bioconductor.org) and GeneSpring GXv7.3.1 (Agilent Technologies). Differentially expressed (DE)-genes were defined as genes with expression levels at least three-fold higher or lower in target groups (MPNST) compared to NHSC after applying Benjamini and Hochberg false discovery rate correction (FDR/BH p ≤ 0.05).
Tumor microarray (TMA) construction and immunohistochemistry
Representative areas of disease were identified on hematoxylin and eosin-stained sections for 30 neurofibromas, 32 plexiform neurofibromas, and 31 MPNSTs. TMA blocks consisting of duplicate 1.0 mm core samples were constructed with a manual tissue arrayer (MTA-1, Beecher Inc, WI) and limited to 64 cores per recipient block. Immunohistochemistry for CCND1 (SP4) monoclonal antibody (Neomarkers, Thermo Scientific, 1:50) and β-catenin (6B3) monoclonal antibody (Cell Signaling; 1:200) was performed utilizing an automated immunohistochemical staining platform (Nemesis 7200, Biocare) following standard IHC protocols (50). Digital images of IHC stained TMA slides were obtained as previously described by Rizzardi et al. 2012 (51)(51). P-values were determined using a 3×2 (staining positivity) or 3×3 (staining intensity or localization) Fisher’s Exact Test to determine differences in staining between the three tumor types.
Gene knockdown and overexpression
GSK3B and Luciferase cDNA (Invitrogen) were each cloned into a vector containing a CAGGS promoter to drive cDNA expression followed by an IRES-GFP. These plasmid vectors were transfected into cells using the NEON transfection system, according to manufacturer’s protocol (Invitrogen). Three independent shRNAs targeting AXIN1, GSK3B, CTNNB1, TNKS, RSPO2, and a non-silencing (NS) control shRNA were purchased from OpenBiosystems. Lentiviral particles containing the shRNAs were produced in 293T cells using the Trans-Lentiviral Packaging Kit (Thermo Scientific). A dsRed control and CTNNB1S33Y were cloned into a vector containing a CAGGS promoter to drive cDNA expression followed by an IRES-GFP and also transduced into cells using lentiviral particles. Viral supernatant was collected after 24 hours of viral production, cleared, and applied directly to the cells for 6 hours. Following viral transduction, cells underwent selection in 4ug/mL puromycin (Invitrogen). shRNA expression was validated by GFP expression, western blot and QPCR. Data is shown for the one shRNA construct that gave the greatest knockdown as determined by western blot analysis and/or QPCR.
Quantitative Real-Time PCR (QPCR)
RNA was extracted from 1 million cells using the High Pure RNA Isolation Kit (Roche). RNA was analyzed by nanodrop (Thermo Scientific) and by agarose gel electrophoresis for quantification and quality control. 1ug of RNA was used to synthesize cDNA using the Transcriptor First Strand Synthesis (Roche) with both random hexamer and oligo dT primers. QPCR reactions were conducted using LightCycler 480 SYBR I Green (Roche) and run on an Eppendorf Mastercycler ep gradient S. Primer sequences can be found in Supplementary Table 2 (21,52,53). Data were analyzed using RealPlex software, calibrated to ACTB levels and normalized to either over-expression control cells (expressing CAGGS>Luciferase-IRES-GFP or dsRed) or NS shRNA expressing cells and averaged over three experimental replicates.
Cellular Viability assays
Cellular viability assays were set up in a 96-well format with 100 cells plated per well in DMEM full media containing 4ug/mL puromycin (Invitrogen). Readings were taken every 24 hours over 6 days by the MTS assay (Promega). Absorbance was read at 490nm to determine viability and 650nm to account for cellular debris on a BioTek Synergy Mx automated plate reader.
Anchorage-independent growth assay
6-well plates were prepared with bottom agar composed of 3.2% SeaPlaque Agar (Lonza) in DMEM full media) and allowed to solidify before 10,000 cells in top agar (0.8% SeaPlaque Agar in DMEM full media) were plated and allowed to solidify. DMEM full media with 4ug/mL puromycin (Invitrogen) was plated over the cells and cells were incubated under standard conditions (5% CO2, 37 degrees Celsius) for 14 days. Top media was removed and cells were fixed in 10% formalin (Fisher Scientific) containing 0.005% crystal violet (Sigma) for 1 hour at room temperature. Formalin was removed and colonies were imaged on a Leica S8 AP0 microscope. 12 images per cell line were taken and automated colony counts were done using ImageJ software (48)(48). Results shown are a representative example of at least three independent experiments.
Xenografts
1.5 million cells in serum-free DMEM with 33% Matrigel Basement Membrane (BD Biosciences) were injected into the back flanks of athymic Foxn1nu/nu mice (Charles Rivers Laboratory). Each mouse was injected on the right flank with cells expressing a specific shRNA or a cDNA and on the left flank with control cell lines expressing a non-silencing (NS) shRNA or a Luciferase cDNA with at least 4 mice per experiment. Tumor volume was measured bi-weekly using calipers and mice were sacrificed when the tumor reached approximately 10% of the total body weight or 40 days post injection. Tumors were harvested for immunohistochemistry and western blot analysis. All animal work was conducted according to the University of Minnesota’s approved animal welfare protocol.
In Vitro Drug Studies to Determine IC50
RAD-001, IWR-1, and XAV-939 were solubilized in DMSO and then subsequently diluted in sterile PBS. 1,200 cells per well of a 96-well plate were treated with varying concentration of drug in quadruplicate and assayed for cell viability using the MTS assay (Promega) to determine the IC50 values. All data analysis was done using Calcusyn software (CalcuSyn Version 2.1, BioSoft).
Supplementary Material
Statement of Significance.
We demonstrate canonical Wnt/β-catenin signaling as a novel genetic driver of Schwann cell tumor development and progression, due to down-regulation of β-catenin destruction complex members and overexpression of R-spondin 2. Inhibitors of Wnt signaling alone, or in combination with RAD-001, may have therapeutic value for patients with MPNSTs or neurofibromas.
Acknowledgments
This work was supported by the National Institutes of Health (P50 NS057531), The Children’s Tumor Foundation, The Zachary NF Fund, and The Jacqueline Dunlap NF Fund. A.L.W is funded by the 2011 Children’s Tumor Foundation Young Investigators Award (2011-01-018). K.C. is funded by the National Cancer Institute Training Grant 5T32CA059268-15. C.B.C is funded by NIH F30 CA171547 and NIH MSTP grant T32 GM008244. These studies utilized BioNet histology and digital imaging core facilities which are supported by NIH grants P30-CA77598 (D Yee), P50-CA101955 (D Buchsbaum), and KL2-RR033182 (B Blazar), and by the University of Minnesota Academic Health Center.
Footnotes
Conflict of interest statement: Dr. Largaespada is the co-founder and part owner of two biotechnology companies, NeoClone Biotechnology, Inc. and Discovery Genomics, Inc. No company resources or personnel were involved in this research, which is unrelated to the goals of these companies.
References
- 1.Carroll SL, Ratner N. How does the Schwann cell lineage form tumors in NF1? Glia. 2008;56:1590–1605. doi: 10.1002/glia.20776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Uhlmann EJ, Plotkin SR. Neurofibromatoses. Adv Exp Med Biol. 2012;724:266–277. doi: 10.1007/978-1-4614-0653-2_20. [DOI] [PubMed] [Google Scholar]
- 3.Widemann BC. Current status of sporadic and neurofibromatosis type 1-associated malignant peripheral nerve sheath tumors. Curr Oncol Rep. 2009;11:322–328. doi: 10.1007/s11912-009-0045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Katz D, Lazar A, Lev D. Malignant peripheral nerve sheath tumour (MPNST): the clinical implications of cellular signalling pathways. Expert Rev Mol Med. 2009;11:e30. doi: 10.1017/S1462399409001227. [DOI] [PubMed] [Google Scholar]
- 5.Gregorian C, Nakashima J, Dry SM, Nghiemphu PL, Smith KB, Ao Y, et al. PTEN dosage is essential for neurofibroma development and malignant transformation. Proc Natl Acad Sci U S A. 2009;106:19479–19484. doi: 10.1073/pnas.0910398106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kourea HP, Cordon-Cardo C, Dudas M, Leung D, Woodruff JM. Expression of p27(kip) and other cell cycle regulators in malignant peripheral nerve sheath tumors and neurofibromas: the emerging role of p27(kip) in malignant transformation of neurofibromas. Am J Pathol. 1999;155:1885–1891. doi: 10.1016/S0002-9440(10)65508-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kourea HP, Orlow I, Scheithauer BW, Cordon-Cardo C, Woodruff JM. Deletions of the INK4A gene occur in malignant peripheral nerve sheath tumors but not in neurofibromas. Am J Pathol. 1999;155:1855–1860. doi: 10.1016/S0002-9440(10)65504-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carroll SL. Molecular mechanisms promoting the pathogenesis of Schwann cell neoplasms. Acta Neuropathol. 2012;123:321–348. doi: 10.1007/s00401-011-0928-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holtkamp N, Malzer E, Zietsch J, Okuducu AF, Mucha J, Mawrin C, et al. EGFR and erbB2 in malignant peripheral nerve sheath tumors and implications for targeted therapy. Neuro Oncol. 2008;10:946–957. doi: 10.1215/15228517-2008-053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tawbi H, Thomas D, Lucas DR, Biermann JS, Schuetze SM, Hart AL, et al. Epidermal growth factor receptor expression and mutational analysis in synovial sarcomas and malignant peripheral nerve sheath tumors. Oncologist. 2008;13:459–466. doi: 10.1634/theoncologist.2007-0166. [DOI] [PubMed] [Google Scholar]
- 11.Ling BC, Wu J, Miller SJ, Monk KR, Shamekh R, Rizvi TA, et al. Role for the epidermal growth factor receptor in neurofibromatosis-related peripheral nerve tumorigenesis. Cancer Cell. 2005;7:65–75. doi: 10.1016/j.ccr.2004.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mawrin C. Critical role of PTEN for development and progression of nerve sheath tumors in neurofibromatosis type 1. Future Oncol. 2010;6:499–501. doi: 10.2217/fon.10.29. [DOI] [PubMed] [Google Scholar]
- 13.Keng VW, Rahrmann EP, Watson AL, Tschida BR, Moertel CL, Jessen WJ, et al. PTEN and NF1 Inactivation in Schwann Cells Produces a Severe Phenotype in the Peripheral Nervous System That Promotes the Development and Malignant Progression of Peripheral Nerve Sheath Tumors. Cancer Res. 2012 doi: 10.1158/0008-5472.CAN-11-4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keng VW, Watson AL, Rahrmann EP, Li H, Tschida BR, Moriarity BS, et al. Conditional inactivation of Pten together with EGFR overexpression in Schwann cells models sporadic MPNST. Sarcoma. 2012 doi: 10.1155/2012/620834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Curtin JC, Lorenzi MV. Drug discovery approaches to target Wnt signaling in cancer stem cells. Oncotarget. 2010;1:563–577. doi: 10.18632/oncotarget.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seshagiri S, Stawiski EW, Durinck S, Modrusan Z, Storm EE, Conboy CB, et al. Recurrent R-spondin fusions in colon cancer. Nature. 2012;488:660–664. doi: 10.1038/nature11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin YR, Yoon JK. The R-spondin family of proteins: emerging regulators of WNT signaling. Int J Biochem Cell Biol. 2012;44:2278–2287. doi: 10.1016/j.biocel.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gehrke I, Gandhirajan RK, Kreuzer KA. Targeting the WNT/beta-catenin/TCF/LEF1 axis in solid and haematological cancers: Multiplicity of therapeutic options. Eur J Cancer. 2009;45:2759–2767. doi: 10.1016/j.ejca.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 20.Hu T, Li C. Convergence between Wnt-beta-catenin and EGFR signaling in cancer. Mol Cancer. 2010;9:236. doi: 10.1186/1476-4598-9-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Emami KH, Nguyen C, Ma H, Kim DH, Jeong KW, Eguchi M, et al. A small molecule inhibitor of beta-catenin/CREB-binding protein transcription [corrected] Proc Natl Acad Sci U S A. 2004;101:12682–12687. doi: 10.1073/pnas.0404875101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barker N. The canonical Wnt/beta-catenin signalling pathway. Methods Mol Biol. 2008;468:5–15. doi: 10.1007/978-1-59745-249-6_1. [DOI] [PubMed] [Google Scholar]
- 23.Yao H, Ashihara E, Maekawa T. Targeting the Wnt/beta-catenin signaling pathway in human cancers. Expert Opin Ther Targets. 2011;15:873–887. doi: 10.1517/14728222.2011.577418. [DOI] [PubMed] [Google Scholar]
- 24.Ischenko I, Seeliger H, Schaffer M, Jauch KW, Bruns CJ. Cancer stem cells: how can we target them? Curr Med Chem. 2008;15:3171–3184. doi: 10.2174/092986708786848541. [DOI] [PubMed] [Google Scholar]
- 25.Johansson G, Mahller YY, Collins MH, Kim MO, Nobukuni T, Perentesis J, et al. Effective in vivo targeting of the mammalian target of rapamycin pathway in malignant peripheral nerve sheath tumors. Mol Cancer Ther. 2008;7:1237–1245. doi: 10.1158/1535-7163.MCT-07-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu J, Dombi E, Jousma E, Scott Dunn R, Lindquist D, Schnell BM, et al. Preclincial testing of sorafenib and RAD001 in the Nf(flox/flox) ;DhhCre mouse model of plexiform neurofibroma using magnetic resonance imaging. Pediatr Blood Cancer. 2012;58:173–180. doi: 10.1002/pbc.23015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johannessen CM, Johnson BW, Williams SM, Chan AW, Reczek EE, Lynch RC, et al. TORC1 is essential for NF1-associated malignancies. Curr Biol. 2008;18:56–62. doi: 10.1016/j.cub.2007.11.066. [DOI] [PubMed] [Google Scholar]
- 28.Largaespada DA. Transposon-mediated mutagenesis of somatic cells in the mouse for cancer gene identification. Methods. 2009;49:282–286. doi: 10.1016/j.ymeth.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lappe-Siefke C, Goebbels S, Gravel M, Nicksch E, Lee J, Braun PE, et al. Disruption of Cnp1 uncouples oligodendroglial functions in axonal support and myelination. Nat Genet. 2003;33:366–374. doi: 10.1038/ng1095. [DOI] [PubMed] [Google Scholar]
- 30.Sarver AL, Erdman J, Starr T, Largaespada DA, Silverstein KA. TAPDANCE: an automated tool to identify and annotate transposon insertion CISs and associations between CISs from next generation sequence data. BMC Bioinformatics. 2012;13:154. doi: 10.1186/1471-2105-13-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vogel KS, Klesse LJ, Velasco-Miguel S, Meyers K, Rushing EJ, Parada LF. Mouse tumor model for neurofibromatosis type 1. Science. 1999;286:2176–2179. doi: 10.1126/science.286.5447.2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cichowski K, Shih TS, Schmitt E, Santiago S, Reilly K, McLaughlin ME, et al. Mouse models of tumor development in neurofibromatosis type 1. Science. 1999;286:2172–2176. doi: 10.1126/science.286.5447.2172. [DOI] [PubMed] [Google Scholar]
- 33.Mayes DA, Rizvi TA, Cancelas JA, Kolasinski NT, Ciraolo GM, Stemmer-Rachamimov AO, et al. Perinatal or adult Nf1 inactivation using tamoxifen-inducible PlpCre each cause neurofibroma formation. Cancer Res. 2011;71:4675–4685. doi: 10.1158/0008-5472.CAN-10-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller SJ, Jessen WJ, Mehta T, Hardiman A, Sites E, Kaiser S, et al. Integrative genomic analyses of neurofibromatosis tumours identify SOX9 as a biomarker and survival gene. EMBO Mol Med. 2009;1:236–248. doi: 10.1002/emmm.200900027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mahller YY, Vaikunth SS, Ripberger MC, Baird WH, Saeki Y, Cancelas JA, et al. Tissue inhibitor of metalloproteinase-3 via oncolytic herpesvirus inhibits tumor growth and vascular progenitors. Cancer Res. 2008;68:1170–1179. doi: 10.1158/0008-5472.CAN-07-2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dahlberg WK, Little JB, Fletcher JA, Suit HD, Okunieff P. Radiosensitivity in vitro of human soft tissue sarcoma cell lines and skin fibroblasts derived from the same patients. Int J Radiat Biol. 1993;63:191–198. doi: 10.1080/09553009314550251. [DOI] [PubMed] [Google Scholar]
- 37.Huang SM, Mishina YM, Liu S, Cheung A, Stegmeier F, Michaud GA, et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009;461:614–620. doi: 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- 38.Kobayashi C, Oda Y, Takahira T, Izumi T, Kawaguchi K, Yamamoto H, et al. Chromosomal aberrations and microsatellite instability of malignant peripheral nerve sheath tumors: a study of 10 tumors from nine patients. Cancer Genet Cytogenet. 2006;165:98–105. doi: 10.1016/j.cancergencyto.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 39.Brekke HR, Ribeiro FR, Kolberg M, Agesen TH, Lind GE, Eknaes Y, et al. Genomic changes in chromosomes 10, 16, and X in malignant peripheral nerve sheath tumors identify a high-risk patient group. J Clin Oncol. 2010;28:1573–1582. doi: 10.1200/JCO.2009.24.8989. [DOI] [PubMed] [Google Scholar]
- 40.Gera JF, Mellinghoff IK, Shi Y, Rettig MB, Tran C, Hsu JH, et al. AKT activity determines sensitivity to mammalian target of rapamycin (mTOR) inhibitors by regulating cyclin D1 and c-myc expression. J Biol Chem. 2004;279:2737–2746. doi: 10.1074/jbc.M309999200. [DOI] [PubMed] [Google Scholar]
- 41.Persad S, Troussard AA, McPhee TR, Mulholland DJ, Dedhar S. Tumor suppressor PTEN inhibits nuclear accumulation of beta-catenin and T cell/lymphoid enhancer factor 1-mediated transcriptional activation. J Cell Biol. 2001;153:1161–1174. doi: 10.1083/jcb.153.6.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mo W, Chen J, Patel A, Zhang L, Chau V, Li Y, et al. CXCR4/CXCL12 Mediate Autocrine Cell- Cycle Progression in NF1-Associated Malignant Peripheral Nerve Sheath Tumors. Cell. 2013;152:1077–1090. doi: 10.1016/j.cell.2013.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fearon ER. Molecular genetics of colorectal cancer. Annu Rev Pathol. 2011;6:479–507. doi: 10.1146/annurev-pathol-011110-130235. [DOI] [PubMed] [Google Scholar]
- 44.Gonsalves FC, Klein K, Carson BB, Katz S, Ekas LA, Evans S, et al. An RNAi-based chemical genetic screen identifies three small-molecule inhibitors of the Wnt/wingless signaling pathway. Proc Natl Acad Sci U S A. 2011;108:5954–5963. doi: 10.1073/pnas.1017496108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Frahm S, Mautner VF, Brems H, Legius E, Debiec-Rychter M, Friedrich RE, et al. Genetic and phenotypic characterization of tumor cells derived from malignant peripheral nerve sheath tumors of neurofibromatosis type 1 patients. Neurobiol Dis. 2004;16:85–91. doi: 10.1016/j.nbd.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 46.Fletcher JA, Kozakewich HP, Hoffer FA, Lage JM, Weidner N, Tepper R, et al. Diagnostic relevance of clonal cytogenetic aberrations in malignant soft-tissue tumors. N Engl J Med. 1991;324:436–442. doi: 10.1056/NEJM199102143240702. [DOI] [PubMed] [Google Scholar]
- 47.Badache A, De Vries GH. Neurofibrosarcoma-derived Schwann cells overexpress platelet-derived growth factor (PDGF) receptors and are induced to proliferate by PDGF BB. J Cell Physiol. 1998;177:334–342. doi: 10.1002/(SICI)1097-4652(199811)177:2<334::AID-JCP15>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 48.Rasband WS. ImageJ. 1997–2012 [Google Scholar]
- 49.Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33:e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.DeRycke MS, Andersen JD, Harrington KM, Pambuccian SE, Kalloger SE, Boylan KL, et al. S100A1 expression in ovarian and endometrial endometrioid carcinomas is a prognostic indicator of relapse-free survival. Am J Clin Pathol. 2009;132:846–856. doi: 10.1309/AJCPTK87EMMIKPFS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rizzardi AE, Johnson AT, Vogel RI, Pambuccian SE, Henriksen J, Skubitz AP, et al. Quantitative comparison of immunohistochemical staining measured by digital image analysis versus pathologist visual scoring. Diagn Pathol. 2012;7:42. doi: 10.1186/1746-1596-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cui W, Taub DD, Gardner K. qPrimerDepot: a primer database for quantitative real time PCR. Nucleic Acids Res. 2007;35:D805–D809. doi: 10.1093/nar/gkl767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Biechele TL, Adams AM, Moon RT. Transcription-based reporters of Wnt/beta-catenin signaling. Cold Spring Harb Protoc. 2009;2009 doi: 10.1101/pdb.prot5223. pdb prot5223. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.