Abstract
Siglec-F is highly expressed on mouse eosinophils and plays an important role in regulating levels of eosinophilic lung inflammation. In this study we investigated the mechanism of constitutive and inducible Siglec-F ligand expression by lung airway epithelial cells and inflammatory cells in WT and genetically altered mice (ST3Gal-III heterozygotes; Fuc-TIV/VII double null; STAT6 null). Flow cytometry demonstrated that Siglec-F ligands are constitutively expressed in vitro and in vivo in selected lung cell types (epithelial cells, eosinophils, macrophages, mast cells, but not CD4, CD8, or B cells) and induced in response to divergent stimuli including innate stimuli (TLR ligands, Alternaria), Th2 cytokines (IL-4, IL-13), and adaptive immune stimuli (OVA allergen). Furthermore studies of deficient mice demonstrated the greater importance of the sialyltransferase ST3Gal-III compared to fucosyltransferases Fuc-TIV/VII in the synthesis of the constitutive and inducible Siglec-F ligands by lung epithelial and non-epithelial cells. In keeping with this, ST3Gal-III heterozygote mice (deficient in expression of Siglec-F ligands) also had significantly enhanced OVA-induced eosinophilic airway inflammation associated with reduced eosinophil apoptosis. Reduced eosinophil apoptosis in the lung of ST3Gal-III deficient mice is likely mediated by reduced epithelial expression of Siglec-F ligands as WT eosinophils (which highly express Siglec-F) cultured with ST3Gal-III deficient epithelial cells (which do not express Siglec-F ligand) showed reduced eosinophil apoptosis compared to WT eosinophils cultured with WT epithelial cells. Overall, these studies demonstrate that ST3Gal-III plays an important role in Siglec-F ligand formation and eosinophil apoptosis with resultant effects on eosinophilic inflammation in the lung.
INTRODUCTION
Siglecs (sialic acid-binding, immunoglobulin-like lectins) are a family of single-pass type I transmembrane receptors that are found predominantly on innate immune cells (1). Among CD33-related Siglecs, Siglec-F is highly expressed on eosinophils (2). Siglec-F possesses an immunoreceptor tyrosine-based inhibitory motif (ITIM) that can mediate inhibitory functions including induction of eosinophil apoptosis (3–8). The importance of Siglec-F in regulating levels of eosinophilic inflammation in vivo has been demonstrated in studies of Siglec-F deficient mice (4), as well as in studies of WT mice administered anti-Siglec-F Ab in models of eosinophilic inflammation (5). Studies have also investigated expression of Siglec-F ligands. Siglec-F binds preferentially to α2–3-linked Sias including 6′-sulfated Sialyl Lewis X (6′-sulfo-SLex) suggesting that it is the best glycan ligand for Siglec-F (9). Using immunohistochemistry, we previously reported that Siglec-F ligands are expressed on bronchial epithelium, and that expression is upregulated on both bronchial epithelium and peribronchial inflammatory cells upon OVA allergen challenge (4). In addition, Th2 cytokines, IL-4 and IL-13, induce upregulation of Siglec-F ligand expression on airway epithelial cells following in vivo administration (10).
In this study, we have utilized ST3Gal-III heterozygous deficient and Fuc-TIV/VII deficient mice to examine Siglec-F ligand synthesis as these enzymes have been implicated in the synthesis of the tetrasaccharide 6′-sulfo-SLex, a Siglec-F ligand (11, 12). Biosynthesis of a sulfated, sialylated Lex structure would require a disaccharide substrate (a terminal precursor N-acetyllactosamine, Galβ1–4GlcNAc), and the action of three enzymes including a sulfotransferase, a sialyltransferase, and a fucosyltransferase (11). In terms of the fucosyl transferase only two fucosyltransferases in mice (Fuc-TIV or Fuc-TVII) are known to be able to synthesize sLeX, and Fuc-TIV is highly expressed in airway epithelial cells the site where we have noted constitutive and inducible expression of Siglec-F ligands (13). Alpha2–3-Sialyltransferases must also involved in the synthesis of sLeX to catalyze the transfer of sialic acid to an acceptor glycan (14–16). Of the six known α2–3-sialyltransferases in mice, ST3Gal-II, III, IV and V are noted to be expressed in lungs (17). In addition, ST3Gal-III, IV, and VI are known to sialylate type II (Galβ1–4GlcNAc) oligosaccharides in vitro, consistent with involvement in the formation of 6′-sulfo-SLex (18, 19). Guo et al recently reported a role of ST3Gal-III in constitutive expression of Siglec-F ligands in mouse lungs using immunohistochemistry (20), but did not investigate their role in Siglec-F ligand expression induced by allergen, nor the effect ST3Gal-III deficiency on allergen induced levels of lung eosinophilic inflammation in vivo. In this study, we demonstrate the greater importance of ST3Gal-III compared to Fuc-TIV/VII in the synthesis of constitutive Siglec-F ligand synthesis in lung epithelia and non-epithelial cells (eosinophils, macrophages, mast cells). Moreover, ST3Gal-III deficient mice (deficient in expression of Siglec-F ligand) had significantly enhanced OVA allergen-induced eosinophilic airway inflammation associated with reduced eosinophil apoptosis.
MATERIALS AND METHODS
Mice
C57BL/6 mice, ST3Gal-III+/− (hereafter referred to as ST3Gal-III deficient), and STAT6−/− mice on a C57BL/6 background were purchased from The Jackson Laboratory (Bar Harbor, ME), while Fuc-TIV−/−/VII−/− mice were kindly provided by Dr JB Lowe (21). All animals were kept under specific-pathogen-free conditions in an environmentally controlled clean room at UCSD. All animal experimental protocols were approved by the University of California, San Diego Animal Subjects Committee.
Induction of eosinophilic airway inflammation
We investigated the effect of an adaptive immune stimulus (OVA), an innate immune stimulus (Alternaria), and Th2 cytokines (IL-4, IL-13) on induction of Siglec-F ligand expression by lung epithelial and non-epithelial cells.
(a) OVA
Mice were sensitized with OVA (Sigma-Aldrich Corporate, St. Louis, MO) as previously described (22). In brief, mice were sensitized intraperitoneallly with 100 μg OVA and 2 mg alum (Imject Alum, Thermo Fisher Scientific Inc., Waltham, MA) in a total volume of 200 μl PBS on days 0 and 10 followed by intranasal administration of 200 μg OVA in 20 μl PBS on days 21, 23 and 25. Control groups of mice were sensitized and challenged with PBS only. Twenty four hours after the last challenge, bronchoalveolar lavage (BAL) fluid, lungs, blood, and bone marrow were collected as previously described in this laboratory (23). The total number of BAL cells was counted and BAL differential cell counts were quantitated on cytospun slides stained with May-Grunwald-Giemsa.
(b) Alternaria
Naïve WT or STAT6−/− mice were intranasally challenged once with 100 μg/mouse of Alternaria alternata extract (Greer Laboratories, Inc., Lenoir, NC) in 80 μl PBS, or control PBS, and sacrificed 24 hours after the challenge. This protocol induces a robust innate eosinophil response within 24 hours that is not dependent on adaptive immune responses as in the OVA protocol (24).
(c) Th2 cytokines (IL-4, IL-13)
Mice were challenged intranasally once with 0.6 μg of either rmIL-4, or rmIL-13 (R&D Systems, Minneapolis, MN) in 40 μl PBS or control PBS, and sacrificed 24 hours after the challenge.
Lung Histology, Immunohistochemistry, and Tunel Assay
Twenty four hours after the last challenge, lungs were harvested and fixed in 4% PFA (Electron Microscopy Sciences, Hatfield, PA). The fixed tissues were embedded in paraffin and sliced into 5-μm sections, followed by either hematoxylin-eosin staining to assess peribronchial inflammation, or immunostaining to detect eosinophils and apoptotic eosinophils.
For MBP (major basic protein) immunostaining, an anti-mouse MBP antibody (kindly provided by James Lee, PhD, Mayo Clinic, Scottsdale, AZ) and the immunoperoxidase method were used followed by DAPI staining as previously described (6).
For some experiments, TUNEL staining was combined with MBP immunostaining to detect apoptotic eosinophils. For TUNEL staining, terminal deoxynucleotidyl transferase (TdT) was used to catalyze polymerization of labeled nucleotides to free 3′-OH DNA ends of apoptotic cells as described by the manufacturer (Roche Applied Science, Mannheim, Germany). As a negative control in the TUNEL assay, the enzymatic reaction with TdT was omitted, and as a positive control, lung sections were incubated with DNase I (Roche Applied Science) to induce DNA strand breaks prior to the labeling process. Lung sections were scanned using a Leica DM2500 (Leica Microsystems, Wetzlar, Germany). Images were captured and analyzed using Image-Pro Plus (Media Cybernetics, Inc., Bethesda, MD).
Lung single cells for FACS
To obtain lung single cell suspensions, lungs were minced and agitated vigorously in RPMI 1640 with 2 mg/ml collagenase D (Roche Applied Science) and 1 mg/ml DNAse I (Roche Applied Science) for 30 minutes at 37°C. Lung single cells were isolated using a 70-μm cell strainer and used for flow cytometry.
BAL IL-4, IL-5, and CCL24
The levels of IL-4, IL-5, and CCL24 in BAL fluid were determined using respective mouse IL-4, IL-5, and CCL24 DuoSets (R&D Systems) which each have a sensitivity of 7 pg/ml.
Primary Airway Epithelial Cell Isolation and Co- Culture with Leukocytes
Mouse primary airway epithelial cell isolation was performed as previously described (25). In brief, mouse tracheas were aseptically removed and digested with 1.4 mg/ml pronase (Roche Applied Sciences) and 0.1 mg/ml deoxyribonuclease I (Sigma, St. Louis, MO) in calcium and magnesium-free MEM overnight. The single tracheal cells recovered were incubated at 37°C for 3 hours in complete medium (DMEM/F12, insulin 120 mU/ml, and 1x non-essential amino acids) to remove fibroblasts that attach to the culture dish. The non-adherent cells were collected and transferred to collagen-coated plates (BD BioCoat Collagen I Cellware, BD Biosciences, San Jose, CA) and incubated in serum free PCT Airway Epithelial Medium Complete (Millipore #CnT-17, Temecula, CA) until they reached the desired confluency. Cultured epithelial cells were detached from the culture plates with pre-warmed trypsin-EDTA for 10 minutes prior to analysis by flow cytometry. To assess the purity of cytokeratin positive epithelial cells in the cultures, cells grown on an 8-well slide (VWR International, Radnor, PA) were fixed with cold acetone and incubated with 2 μg/ml rabbit anti-cytokeratin or control Ab (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) and detected with the alkaline phosphatase method (Vector Laboratories, Burlingame, CA).
In some experiments, mouse peripheral blood leukocytes (1–4 % eosinophils) were co-cultured with mouse primary epithelial cells isolated as described above. Briefly, after epithelial cells grown in 24-well culture plate reached full confluency, the wells were washed with PBS and WT blood leukocytes in RPMI medium supplemented with 10% FCS were co-incubated. Twenty four hours later, the number of apoptotic and viable eosinophils were counted by FACS. For eosinophil apoptosis detection by flow cytometry, cells were stained with FITC-conjugated anti-CCR3 to distinguish eosinophils and then stained with allophycocyanin-conjugated Annexin V and PI according to the manufacturer’s instructions (Annexin V Apoptosis Detection Kit APC, eBioscience).
Th2 cytokine and TLR ligand regulation of Siglec-F ligand expression by lung epithelial cells in vitro
To investigate regulation of Siglec-F ligand expression on epithelial cells, we used a murine lung epithelial cell line (MLE12) (American Type Culture Collection, Mamassas, VA). 1 × 105 MLE-12 cells/ml were incubated with or without 10 mU/ml of sialidase (Roche Applied Sciences) in 24-well culture plates for 24 hours. Sialidases hydrolyze alpha-glycosidic linkages of terminal sialic residues in structures such as 6′-sulfo-SLex, the ligand for Siglec-F, and would thus remove any constitutive Siglec-F ligand expressed. Epithelial cells were then stimulated with either Th2 cytokines i.e. 10 ng/ml rmIL-4 (R&D Systems), 10 ng/ml rmIL-13 (R&D Systems), TLR ligands i.e. 10 μg/ml CpG (TriLink, San Diego, CA), 100 ng/ml LPS (Sigma-Aldrich Corporate), 100 ng/ml Pam3 (Sigma-Aldrich Corporate), or 10 ng/ml rmGM-CSF (R&D Systems), 10 ng/ml rmIL-10 (R&D Systems), 10 ng/ml rmIL-17 (R&D Systems) for 0, 1, 3, 6, or 12 hours. Cells were trypsinized and collected for FACS detection of Siglec-F ligand expression as described below.
Flow cytometry to detect Siglec-F ligand expression on lung epithelial and non-epithelial cells
Siglec-F ligand expression on lung epithelial cells, inflammatory cells (eosinophils, macrophages, mast cells, neutrophils) and immune cells (CD4+ cells, CD8+ cells, B cells) was analyzed by flow cytometry. Individual cell types in these lung single cell digests and peripheral blood were not purified prior to FACS analysis. Briefly, lung cells (single cell suspensions, primary epithelial cells, or MLE12 cell line) were first incubated with a mAb to CD16/CD32 (clone 24G.2) for 10 minutes to block Fc receptors. Then cells were stained with either 1 μg/ml of recombinant Siglec-F human IgG Fc fusion protein (R&D Systems) or control human IgG at 4°C for 30 minutes followed by staining with 0.5 μg/ml of PE-conjugated goat anti-human IgG (eBiosciences, San Diego, CA) with cell-specific markers indicated below at 4°C for 30 minutes. For lung cells, cells were stained with PerCP-conjugated anti-CD45.2 mAb to distinguish hematopoietic cells from other cell types. For mouse primary epithelial cell identification, cells were fixed and permeabilized by BD Cytofix/Cytoperm Fixation/Permeabilization kit (BD Biosciences) as instructed by the manufacturer and stained with 2 μg/ml of rabbit anti-cytokeratin Ab (Santa Cruz Biotechnology, Inc.) followed by staining with FITC-conjugated goat anti-rabbit IgG (eBioscience). CD45.2-negative cytokeratin-positive cells were gated as epithelial cells and further analyzed for Siglec-F ligand expression. Similarly, CD4 positive T cells stained positive for both CD45.2 and FITC-conjugated CD4 (clone RM4–5, eBioscience). CD8 positive T cells stained positive for both CD45.2 and FITC-conjugated CD8 (clone 53–6.7, BD Biosciences). Mast cells stained positive for CD45.2, FITC-conjugated anti-FcεRI (clone MAR-1, eBioscience) and Alexa fluor 647-conjugated CD117 (clone 2B8, Life Technologies, Grand Island, NY). Neutrophils stained positively with CD45.2 and allophycocyanin-conjugated anti-Gr-1 (clone RB6–8C5, eBioscience). Macrophages were auto-fluorescent and stained positive for CD45.2 and allophycocyanin-conjugated CD11c (clone N418, eBioscience). B cells stained positive with CD45.2 and biotinylated anti-B220 (clone RA3–6B2, eBioscience) followed by streptavidin-allophycocyanin. Eosinophils stained positive for FITC-conjugated anti-CCR3 (clone 83101, R&D Systems) or Alexa fluor 647-conjugated anti-CCR3 (clone 83103, BD Biosciences).
In selected experiments Alexa Fluor 488-conjugated anti-mouse IL-5 receptor antibody (clone T21, BD Biosciences) was used to analyze IL-5 receptor expression on PE-conjugated anti-Siglec F (clone E50–2440, BD Biosciences) -positive and allophycocyanin-conjugated anti-CD11c-negative cells.
Stained cells were analyzed using Accuri C6 flow cytometer (Accuri Cytometers, Inc., Ann Arbor, MI) and analyzed with FlowJo software (Tree Star, Ashland, OR)
qPCR detection of levels of ST3Gal-III, Fuc-TIV, and Fuc-VII in lung epithelial and non-epithelial cells
Total RNA was extracted from purified populations (> 95% pure) of epithelial cells (MLE-12), and non-epithelial cells (macrophages, eosinophils). For this experiment macrophages were derived from culture of mouse bone marrow as previously described (26), and eosinophils from culture of mouse bone marrow as previously described (27). cDNA was prepared as described previously (28). The expression levels of ST3Gal-III, Fuc-TIV, and Fuc-TVII were determined by qPCR using an Mx3000P QPCR System (Agilent Technologies, Santa Clara, CA). The primers and probes were designed by Origene Technologies (Rockville, MD). Data were quantitated by the comparative Ct method after normalization with the value of endogenous GAPDH gene expression as described elsewhere (28). Data are expressed as the RQ values compared to the expression without stimulation.
Airway responsiveness
Airway responsiveness to methacholine (MCh) was assessed 24 h after the final OVA challenge in intubated and ventilated mice (flexiVent ventilator; Scireq, Montreal, PQ, Canada) anesthetized with ketamine (100 mg/kg) and xylazine (10 mg/kg) intraperitoneally as previously described (10). The dynamic airway resistance (Raw) was determined using Scireq software in mice exposed to nebulized PBS and MCh (3, 24, 48 mg/ml). The following ventilator settings were used: tidal volume (10 ml/kg), frequency (150/min), positive end-expiratory pressure (3 cmH2O).
Statistics
Unless otherwise specified, ANOVA and unpaired Student’s t-test, two-tailed, were used for statistical evaluation of the results. A p value <0.05 was considered statistically significant. All results are shown as means ± SEM.
RESULTS
FACS analysis of constitutive Siglec-F ligand expression in WT lung cells
We have previously used immunohistochemistry to demonstrate that Siglec-F ligands are expressed in the WT lung in epithelial cells and peribronchial inflammatory cells (10). In this study we have used FACS as an improved quantitative method (compared to immunohistochemistry) to assess whether Siglec-F ligands are constitutively or inducibly expressed on cell types in the lung other than epithelial cells and macrophages important to asthma (i.e. mast cells, eosinophils, T cells, neutrophils), and whether stimuli (allergen, Th2 cytokines, TLR ligands) and enzymes (ST3Gal-III, Fuc-TIV/VII) regulating Siglec-F ligand formation in these cells are similar or different.
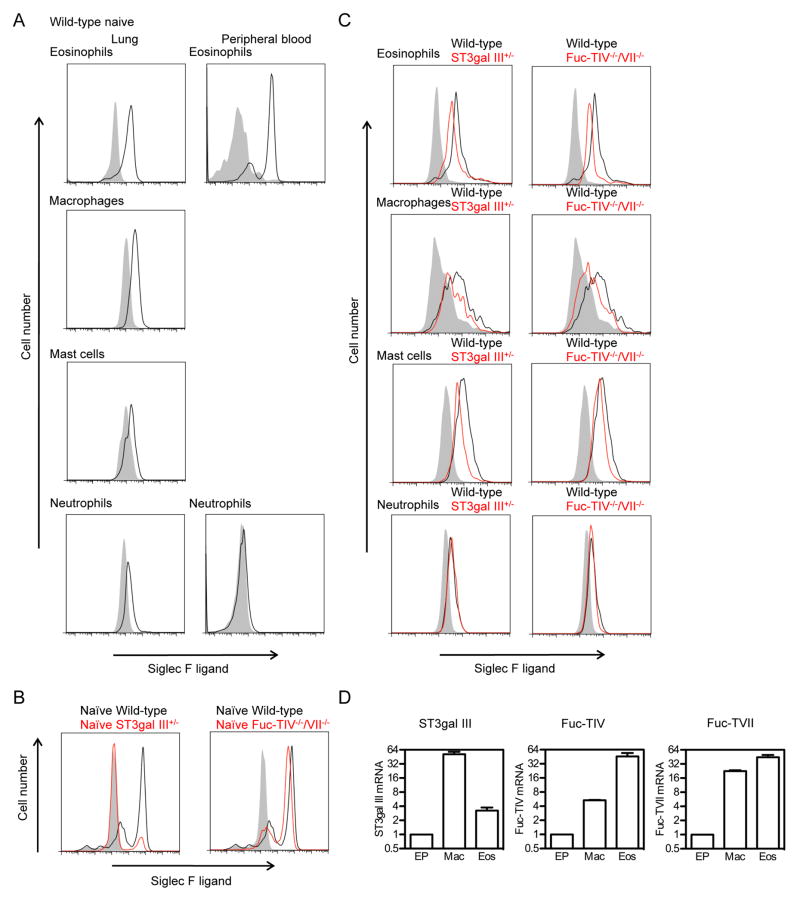
Using single cell suspensions derived from lungs of naïve WT mice, we demonstrated by FACS that non-epithelial cells such as eosinophils, macrophages, and mast cells all expressed significant levels of constitutive Siglec-F ligand (Fig 1A left panel) while neutrophils expressed much lower levels of constitutive Siglec-F ligand (Fig 1A left panel). Among the naïve WT mouse lung cells tested, eosinophils showed the highest level of constitutive expression. Naïve WT mouse lung CD4+ cells, CD8+ cells, and B cells did not express Siglec-F ligands (data not shown). Peripheral blood eosinophils from naïve WT mice also expressed significant levels of constitutive Siglec-F ligand (Fig 1A right panel). In contrast, neither peripheral blood neutrophils (Fig 1A right panel), CD4+ cells, CD8+ cells, or B cells (data not shown) from naïve WT mice expressed Siglec-F ligand.
FIGURE 1. FACS detection of Siglec-F ligands in naïve WT, ST3Gal-III+/−, and Fuc-TIV−/−/VII−/− lung cells.
A. Expression of Siglec-F ligands on naive WT lung and blood cells. Inflammatory cells from both mouse lungs (eosinophils, macrophages, mast cells, and neutrophils) and peripheral blood (eosinophils and neutrophils) were analyzed by flow cytometry for Siglec-F ligand expression. Individual cell types in these lung single cell digests and peripheral blood were not purified prior to FACS analysis. Shaded area shows the staining with control human IgG and black lines show the staining on cells from WT mice. Data are representative from three separate mice showing similar results.
B. Expression of Siglec-F ligands on naive WT, ST3Gal-III+/−, and Fuc-TIV−/−/VII−/− mouse primary airway epithelial cells. Shaded area shows the staining with control human IgG and black lines show the staining on cells from WT mice. Red lines show the expression on ST3Gal-III+/− or Fuc-TIV−/−/VII−/− mice as indicated in the figure. Data are representative from three separate experiments using three different lots of epithelial cells showing similar results.
C. The expression of Siglec-F ligands on cell surface of mouse lung cells. Shaded area show the staining with control human IgG and black lines show the staining on cells from WT mice. Red lines show the expression on ST3Gal-III+/− or Fuc-TIV−/−/VII−/− cells. Data are representative from three separate mice showing similar results.
D. Relative quantitation values for ST3Gal-III, Fuc-TIV, Fuc-TVII mRNAs assessed by qPCR in WT primary airway epithelial cells (EP), bone marrow derived macrophages (Mac), and bone marrow derived eosinophils (Eos). Data are the mean ± SEM (n = 3).
Influence of fucosyltransferases and sialyltransferases on constitutive levels of Siglec-F ligand expression in lung cells as assessed by FACS
To examine whether either sialyltransferases and/or fucosyl transferases were important to the expression of constitutive Siglec-F ligand in lung epithelial cells, we isolated primary lung tracheal epithelial cells (> 98% pure as assessed by cytokeratin immunostaining) from naïve mice (WT, ST3Gal-III+/− and Fuc-TIV−/−/VII−/−) (Fig 1B). Naïve WT mouse epithelial cells had significant surface expression of Siglec-F ligand as assessed by flow cytometry (Fig 1B). In contrast, Siglec-F ligand expression on epithelial cells from naïve ST3Gal-III+/− heterozygote mice was almost completely absent (Fig 1B left panel). Homozygous ST3Gal-III null mice were not used because they are difficult to breed and develop neurodegenerative disease. Naïve Fuc-TIV−/−/VII−/− epithelial cells showed a slight decrease in levels of surface Siglec-F ligand expression (Fig 1B right panel). These studies suggest that in naïve mouse epithelial cells, ST3Gal-III plays a major role in constitutive Siglec-F ligand synthesis, whereas Fuc-TIV/VII partially contributes to constitutive Siglec-F ligand synthesis. It is particularly notable that even a heterozygous haplo-insufficiency of ST3Gal-III is sufficient to markedly reduce constitutive Siglec-F ligand production.
In contrast to epithelial cells derived from naïve ST3Gal-III+/− mice in which levels of constitutive Siglec-F ligand were near completely absent (Fig 1B), levels of constitutive Siglec-F ligand were only partially reduced in lung eosinophils, macrophages, and mast cells derived from naïve ST3Gal-III+/− mice (Fig 1C left panel). Levels of constitutive Siglec-F ligand expression were also partially reduced in naïve Fuc-TIV−/−/VII−/− mouse lung macrophages, mast cells, and eosinophils (Fig 1C right panel). These studies suggest that there are differences between lung epithelial cells compared to eosinophils, macrophages, and mast cells in the requirement for ST3Gal-III in constitutive Siglec-F ligand synthesis in heterozygous ST3Gal-III+/− mice, with lung epithelial cells not expressing any constitutive Siglec-F ligand in heterozygous ST3Gal-III+/− mice, while lung eosinophils, macrophages, and mast cells had reduced but not absent constitutive Siglec-F ligand synthesis in heterozygous ST3Gal-III+/− mice. Lung epithelial cells as well as lung eosinophils, macrophages, and mast cells utilize Fuc-TIV/VII to a similar degree to partially contribute to constitutive Siglec-F ligand synthesis. Similar results were noted with peripheral blood eosinophils compared to lung eosinophils in their partial requirements for ST3Gal-III and Fuc-TIV/VII to synthesize constitutive Siglec-F ligands (data not shown).
Levels of expression of ST3Gal-III, Fuc-TIV, and Fuc-TVII mRNA in WT mouse epithelial cells, macrophages, and eosinophils
We examined whether differences in levels of expression of the enzyme ST3Gal-III in naïve WT mouse lung epithelial cells compared to macrophages and eosinophils could explain the difference in requirement for ST3Gal-III in constitutive Siglec-F ligand synthesis in the different cell types. Levels of ST3Gal-III as assessed by qPCR were highest in macrophages, with lower levels in eosinophils and the lowest levels in epithelial cells (Fig 1D). Similarly, levels of Fuc-TIV and Fuc-TVII were highest in eosinophils and macrophages compared to epithelial cells (Fig 1D). Although WT naïve epithelial cells expressed the lowest level of ST3Gal-III and had the greatest reduction in expression of Siglec-F ligand detected in ST3Gal-III+/− mice (compared to macrophages and eosinophils), these correlative studies are unable to determine whether this accounts for the continued Siglec-F ligand expression in ST3Gal-III+/− mice by macrophages and eosinophils who have higher levels of ST3Gal-III expression.
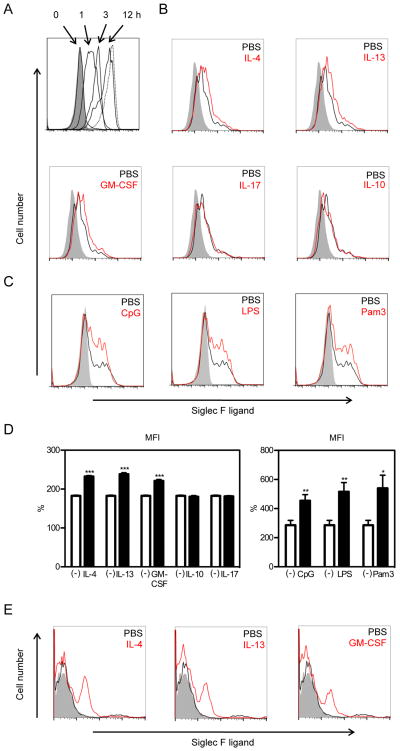
Cytokines and TLR ligands accelerate the recovery of Siglec-F ligand expression by the lung epithelial MLE-12 cell line in vitro
Since the constitutive expression of Siglec-F ligands by the lung epithelial MLE-12 cell line was very high and stable (Fig 2A), we used this lung epithelial cell line to examine whether incubating MLE-12 cells with sialidase would reduce Siglec-F ligand expression, and whether recovery of Siglec-F ligand expression would be influenced by cytokines and TLR ligands. Incubation of MLE-12 cells with sialidase significantly reduced Siglec-F ligand expression as assessed by FACS (Fig 2A). Siglec-F ligand expression recovered gradually several hours after removing the sialidase, and by 12 hours, Siglec-F ligand expression levels approximated that without sialidase treatment (Fig 2A). Incubation of MLE-12 cells with either Th2 cytokines (IL-4, IL-13 (Fig 2B, 2D), GM-CSF (Fig 2B, 2D), or TLR ligands (CpG, LPS, Pam3) (Fig 2C) accelerated the recovery of Siglec-F ligand expression on MLE-12 cells at six hours after incubation with sialidase was ended. In contrast, neither IL-10 nor IL-17 influenced levels of Siglec-F ligand expression (Fig 2B, 2D). Similar to results with the MLE-12 cell line, Th2 cytokines (IL-4, IL-13) and GM-CSF enhanced expression of Siglec-F ligand in studies of primary mouse lung epithelial cells (Fig 2E).
FIGURE 2. Siglec-F ligand expression on MLE-12 cells is upregulated by IL-4, IL-13, GM-CSF, and TLR ligands.
A. The expression of Siglec-F ligands on cell surface of MLE-12 cells at indicated times after sialidase treatment was analyzed by flow cytometry. Shaded area shows the staining with control human IgG and dotted lines show the staining on MLE-12 without sialidase pretreatment. Data are representatives from three separate experiments showing similar results. B, C. 24 hours after sialidase treatment, MLE-12 were stimulated with indicated cytokines or TLR ligands for 6 hours and analyzed for Siglec-F ligand expression. Shaded area show the staining with control human IgG, black lines show the staining on cells without stimulation, and red lines show the staining on cells after stimulation. Data are representatives from three separate experiments showing similar results. D. Quantitation of MFI of Siglec-F ligand expression in MLE-12 cells incubated either without stimulus [depicted as (-) symbol], or incubated with individual cytokines (IL-4, IL-13, GM-CSF, IL-10, IL-17) or TLR ligands (CpG, LPS, Pam3). Specific MFI calculated as % of FL2 MFI compared to control stain. Results expressed as mean ± SEM. *p<0.05, **p<0.01, and ***P<0.001 vs unstimulated (n=6). E. 24 hours after sialidase treatment, primary lung epithelial cells were stimulated with indicated cytokines for 6 hours and analyzed for Siglec-F ligand expression. Shaded area shows the staining with control human IgG, black lines show the staining on cells without stimulation, and red lines show the staining on cells after stimulation. Data are representatives from three separate experiments showing similar results.
Effect of in vivo administration of IL-4 and IL-13 on upregulation of Siglec-F ligand expression in lung epithelial and non-epithelial cells in WT mice
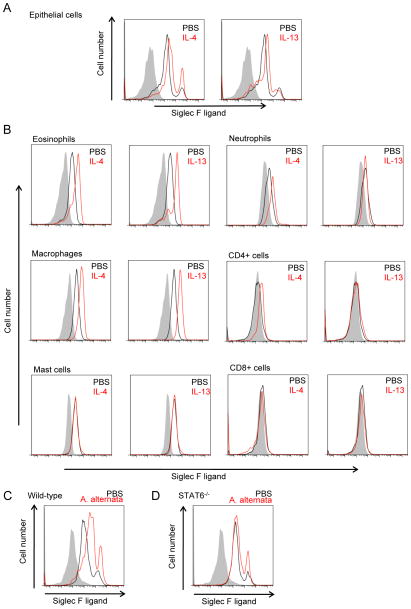
We have previously demonstrated that in vivo administration of IL-4 or IL-13 to WT mice induces expression of Siglec-F ligands on airway epithelium and peribronchial cells as assessed by immunohistochemistry (10). These immunohistochemistry studies were not designed to phenotype the peribronchial cells expressing Siglec-F ligands which we performed in this study utilizing FACS. Similar to the results of our in vitro experiments with IL-4 and IL-13 (Fig 2B), these cytokines when administered in vivo upregulated Siglec-F ligand expression on lung epithelial cells (Fig 3A). The level of Siglec-F ligand upregulation was equivalent in response to either IL-4 or IL-13.
FIGURE 3. Siglec-F ligand expression on mouse lung cells is upregulated by intranasal administration of IL-4, IL-13, and Alternaria alternata in STAT6-dependent manner.
A, B. WT mice were intranasally challenged with control PBS, IL-4, or IL-13, and 24 hours later, their lungs were digested and analyzed for Siglec-F ligand expression. Expression on (A) epithelial cells and (B) indicated cell types from mouse lungs is shown. Shaded area show the staining with control human IgG, black lines show the staining on cells from mice with control PBS administration, and red lines show the staining on cells from mice with indicated cytokine challenges. Data are representatives from three separate experiments showing similar results. C, D. Both WT (C) and STAT6−/− (D) mice were intranasally administered Alternaria alternata extract 100 μg/mouse and 24 hours later, their airway epithelial cells were analyzed for Siglec-F ligand expression. Shaded area show the staining with control human IgG, black lines show the staining on cells from mice with control PBS administration, and red lines show the staining on airway epithelial cells from mice with A. alternata challenge. Data are representatives from two separate experiments showing similar results.
We also examined whether IL-4 and IL-13 regulated expression of Siglec-F ligand expression in peribronchial cells in vivo. In naïve WT mice, lung eosinophils and macrophages express constitutive high levels of Siglec-F ligands (Fig 1A). Administration of either IL-4 or IL-13 significantly increased levels of Siglec-F ligands in both WT mouse lung eosinophils and macrophages (Fig 3B). In naïve WT mice, lung neutrophils express low levels of Siglec-F ligands (Fig 1A), and lung T cells and B cells do not express significant levels of Siglec-F ligand (data not shown). However, administration of IL-4 in vivo slightly increased levels of Siglec-F ligand expression in WT mouse lung neutrophils as well as CD4+ T cells (Fig 3B), but not lung CD8+ T cells (Fig 3B) or B cells (data not shown). In contrast to the ability of IL-4 or IL-13 to upregulate expression of Siglec-F ligands in epithelial cells, macrophages, eosinophils and CD4+ T cells, these cytokines did not upregulate expression of Siglec-F ligands in mast cells (which express significant constitutive levels of Siglec-F ligands in the lungs of naïve WT mice) (Fig 3B).
Lung airway epithelial expression of Siglec-F ligands is STAT-6 dependent
As Th2 cytokines such as IL-4 and IL-13 signal through STAT6 (29, 30), we next examined whether STAT-6 deficient mice had reduced expression of Siglec-F ligands. In these studies we challenged naïve WT mice on one occasion with a fungal allergen, Alternaria alternata, which induces a significant innate airway eosinophil and Th2 response within 24 hrs (31). In comparison to control PBS-treated mice, Alternaria alternata-challenged WT mouse airway epithelial cells showed increased Siglec-F ligand expression (Fig 3C). In contrast, STAT6−/− mice challenged with Alternaria alternata did not significantly upregulate Siglec-F ligand expression (Fig 3D).
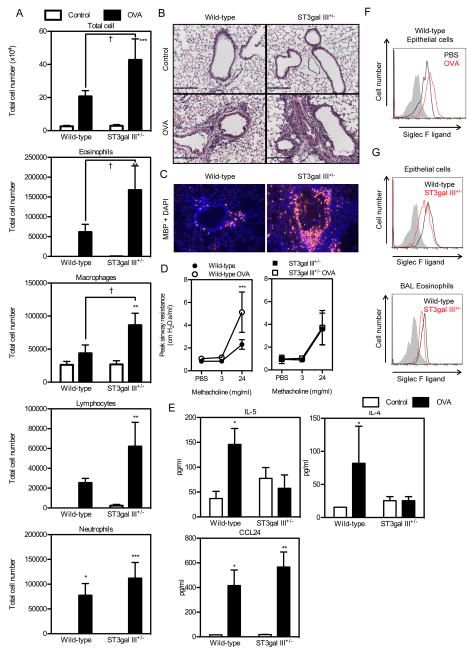
ST3Gal-III+/− mice challenged with OVA have increased lung eosinophilic inflammation
As naïve ST3Gal-III+/− mice exhibit significant reductions in levels of Siglec-F ligand expression (Fig 1B and C), we examined whether these mice would have alterations in levels of lung eosinophilic inflammation as noted in Siglec-F deficient mice challenged with allergen (4). OVA allergen challenged ST3Gal-III+/− mice exhibited a significant greater increase in total BAL cells compared to WT mice (p < 0.05), as well as in BAL eosinophils (2.7 fold) (p < 0.05 vs WT), and BAL macrophages (2 fold) (p < 0.05 vs WT) (Fig 4A). There was a modest increase in BAL lymphocytes (2.5 fold) and BAL neutrophils (1.4 fold) in ST3Gal-III+/− mice (Fig 4A). In addition to increased BAL inflammation in ST3Gal-III+/− mice, OVA allergen challenged ST3Gal-III+/− mice exhibited a significant increase in peribronchial inflammation assessed in hematoxylin-eosin stained lung sections (Fig 4B). MBP immunostaining of the lung sections showed significantly increased peribronchial eosinophil accumulation in ST3Gal-III+/− mice compared to WT mice (Fig 4C).
FIGURE 4. OVA- induced eosinophilic lung inflammation is exaggerated in ST3gal III+/− mice.
A. The numbers of BAL cells in WT mice (PBS, n = 4; OVA, n =6) and ST3Gal-III+/− mice (PBS, n = 8; OVA, n = 7) immunized i.p. with OVA with alum and sacrificed at 24 hours after the last challenge are shown. Data show the mean ± SEM. *p < 0.05, **p < 0.01, and ***p < 0.001 versus the corresponding values for control mice, and †p < 0.05 versus the indicated group. B. Hematoxylin-eosin staining of the lung sections from WT mice and ST3Gal-III+/− mice. C. MBP staining with DAPI staining of the lung sections from WT mice and ST3Gal-III+/− mice. Representative data for the mice used in (A) are shown. Scale bars = 100 μm. D. Airway responsiveness to methacholine was measured in non-OVA and OVA challenged WT mice (no OVA, n=4; OVA n=7) and ST3Gal-III+/− mice (no OVA, n=3; OVA, n=6). **p<0.01 WT OVA vs WT no OVA, p< 0.05 WT OVA vs ST3Gal-III+/− OVA. E. Levels of BAL IL-5, IL-4, and CCL24 from WT mice (PBS, n = 8; OVA, n =16) and ST3Gal-III+/− mice (PBS, n = 11; OVA, n = 14) are shown. *p < 0.05, ** p<0.01 vs control PBS group. F. Siglec F ligand expression on lung epithelial cells from WT mice are shown. Shaded area show the staining with control human IgG, black lines show the staining on cells from control mice and red lines show the staining on cells from OVA-treated mice. Data are representatives from four separate mice showing similar results. G. Siglec F ligand expression on lung epithelial cells and BAL eosinophils are shown. Shaded area show the staining with control human IgG, black lines show the staining on cells from OVA-treated WT mice and red lines show the staining on OVA-treated ST3Gal-III+/− mice. Data are representatives from four separate mice showing similar results.
The increased levels of BAL and peribronchial eosinophils in ST3Gal-III+/− mice were not due to increases in IL-5 as levels of IL-5 were less in ST3Gal-III+/− mice compared to WT mice (Fig 4E). Levels of the eosinophil chemoattractant CCL24 were also not significantly different in ST3Gal-III+/− mice compared to WT mice (Fig 4E). Although OVA challenged ST3Gal-III+/− mice had increased lung eosinophilic inflammation compared to WT mice (Fig 4A, 4B), they had less of an increase in AHR compared to WT mice challenged with OVA (Fig 4D). As OVA challenged ST3Gal-III+/− mice have reduced levels of Th2 cytokines (IL-4, IL-5) (Fig 4E), ST3Gal-III+/− mice may have immunological defects in T cells or non-T cells which through reduced expression of cytokines/mediators effects smooth muscle contractility. Alternatively, ST3Gal-III+/− mice may have intrinsic smooth muscle defects in contractility.
Expression levels of Siglec-F ligands on WT airway epithelial cells from lungs of OVA-treated mice were higher than after PBS challenge (Fig 4F). However, Siglec-F ligand levels were reduced in ST3Gal-III+/− lung epithelial cells after OVA-challenge (Fig 4G left panel). Similarly, OVA-challenged ST3Gal-III+/− mouse eosinophils in BAL had reduced levels of Siglec-F ligand expression compared to WT eosinophils (Fig 4G right panel).
Effect of ST3Gal-III on eosinophil apoptosis
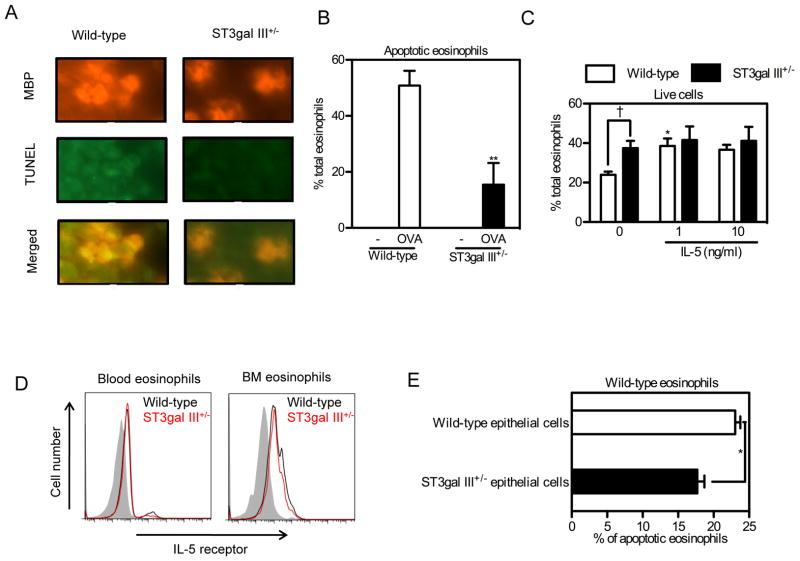
Since we have previously observed that the Siglec-F interaction with Siglec-F ligands mediates eosinophil apoptosis (4), we examined whether the decreased expression of Siglec-F ligands in ST3Gal-III+/− mice reduced the levels of apoptosis of lung eosinophils as assessed by TUNEL assay combined with MBP staining. OVA challenged ST3Gal-III+/− mice had fewer peribronchial TUNEL positive lung cells compared to WT mice (Fig 5A). The number of peribronchial cells that were TUNEL+ and MBP+ (expressed per total number of MBP+ peribronchial cells) were significantly lower in ST3Gal-III+/− mice challenged with OVA compared to OVA challenged WT mice (Fig 5B). These studies suggest there are fewer apoptotic eosinophils in ST3Gal-III compared to WT mice challenged with allergen, although TUNEL staining may detect both apoptotic and necrotic cells. In addition to studies of lung eosinophil apoptosis, we also examined the in vitro viability of peripheral blood eosinophils from ST3Gal-III+/− mice compared to WT mouse eosinophils. The percentage of live eosinophils (stained negative for both Annexin V and PI), was significantly higher in peripheral blood leukocyte cultures incubated with media alone in ST3Gal-III+/− compared to WT mice (Fig 5C). Interestingly, addition of IL-5 to peripheral blood leukocyte cultures increased the % of live WT eosinophils, but did not further increase the viability of ST3Gal-III+/− eosinophils (Fig 5C). IL-5 receptor expression as assessed by FACS was equivalent in WT and ST3Gal-III+/− eosinophils from both peripheral blood and bone marrow (Fig 5D). Finally, we co-cultured naïve WT peripheral blood leukocytes with primary airway epithelial cells from naïve WT or ST3Gal-III+/− mice (Fig 5E). Naïve WT leukocytes cultured with naïve ST3Gal-III+/− epithelial cells had significantly fewer apoptotic eosinophils compared to naïve WT leukocytes cultured with naïve WT epithelial cells (Fig 5E). These data suggest that naïve ST3Gal-III+/− epithelial cells (which have reduced Siglec-F ligand expression) induce less eosinophil-apoptosis (Fig 5E) and may thus account for the increased peribronchial eosinophilic inflammation we have noticed in vivo in ST3Gal-III+/− mice (Fig 4C). However, in vivo lung epithelial cells expressing Siglec-F ligand may not be the only cell interacting with eosinophils to mediate Siglec-F induced eosinophil apoptosis, as non-epithelial cells (i.e. eosinophils, macrophages, mast cells, neutrophils) which express Siglec-F ligands in vivo may also interact with eosinophils to induce their apoptosis.
FIGURE 5. ST3Gal-III+/− eosinophils showed less apoptosis compared to WT eosinophils.
A. Lungs from OVA sensitized and challenged mice were stained using anti-mouse MBP antibody followed by TUNEL staining. Representative data for the mice used in Fig. 4A are shown. B. Apoptotic eosinophils in (A) are quantitated and shown as percentages of total eosinophils. **p < 0.01 versus WT OVA group. C. Peripheral blood cells from WT and ST3Gal-III+/− mice were incubated for 24 hours with indicated concentrations of rmIL-5 followed by FACS analysis for live eosinophils (n = 4). Eosinophils stained for Annexin V-negative and PI-negative cells were counted as live eosinophils and the percentages of live eosinophils are shown. Data show the mean + SEM. *p < 0.05 versus the control eosinophils without rmIL-5, and †p < 0.05 versus the indicated group. D. IL-5 receptor expression on blood and bone marrow eosinophils are shown. Shaded area show the staining with control stain, black lines show the staining on cells from WT mice and red lines show the staining on cells from ST3Gal-III+/− mice. Data are representatives from three separate experiments showing similar results. E, WT blood cells were co-cultured with primary mouse epithelial cells isolated and cultured from WT or ST3Gal-III+/− mice. 24 hours later, cells were collected and stained to detect live eosinophils and apoptotic eosinophils as described above. Data show the mean ± SEM (n = 3). *p < 0.05.
DISCUSSION
In this study we have demonstrated by FACS that Siglec-F ligands are constitutively and inducibly expressed in vitro and in vivo in selected cell types in the lung (epithelial cells, eosinophils, macrophages, mast cells, but not CD4, CD8, or B cells) in response to divergent stimuli including innate stimuli (TLR ligands, Alternaria), Th2 cytokines (IL-4, IL-13), GM-CSF, and adaptive immune stimuli (OVA allergen). In addition, we demonstrated the greater importance of the sialyltransferases, ST3Gal-III compared to the fucosyltransferase (Fuc-TIV/VII) in the synthesis of the constitutive and inducible Siglec-F ligands by lung epithelial and non-epithelial cells (eosinophils, macrophages, mast cells). Finally, we demonstrated that ST3Gal-III deficient mice (deficient in expression of Siglec-F ligands) have significantly enhanced OVA-induced allergic airway inflammation associated with reduced eosinophil apoptosis. The reduced eosinophil apoptosis in the lung is likely mediated by reduced epithelial expression of Siglec-F ligands as WT eosinophils (which highly express Siglec-F) when cultured with ST3Gal-III deficient epithelial cells (which do not express Siglec-F ligand) have reduced eosinophil apoptosis compared to WT eosinophils cultured with WT epithelial cells.
We previously reported using immunohistochemistry that airway epithelia and peribronchial mononuclear leukocytes expressed Siglec-F ligands (4, 10). In this study, we have extended these results using an alternate method, flow cytometry, to phenotype and quantitate whether additional immune and inflammatory cells in the lung important to asthma (i.e. mast cells, T cells, B cells, neutrophils) express Siglec-F ligands. Our FACS studies of WT naïve lung cells demonstrated that not only epithelial cells, and peribronchial mononuclear leukocytes as previously demonstrated (4, 10), but also lung macrophages, eosinophils, and mast cells expressed significant levels of constitutive Siglec-F ligands. In contrast, T cells (CD4, CD8) and B cells did not express significant constitutive Siglec-F ligands, while neutrophils in the lung but not peripheral blood expressed low levels of Siglec-F ligands. These studies demonstrate that like constitutive Siglec-F expression, constitutive Siglec-F ligand expression is restricted to particular cell types. Interestingly, there are thus mouse lung cells important to allergic inflammation and asthma that constitutively co-express both Siglec-F and Siglec-F ligands (macrophages, eosinophils). In contrast, there are also lung cells that either constitutively only express the Siglec-F ligand (epithelial cells, mast cells, and low levels by neutrophils), or alternatively constitutively express neither Siglec-F or its ligand (CD4, CD8, B cells). In addition to constitutive Siglec-F ligand expression, in this study we demonstrate that Siglec-F ligand expression can be regulated not only by adaptive stimuli (OVA allergen) and Th2 cytokines (IL-4, IL-13) as previously demonstrated (4, 10), but also within 24 hours by innate immune stimuli (Alternaria), TLR ligands (TLR 2, 4, and 9), and GM-CSF. Interestingly, CD4 cells which do not express constitutive Siglec-F could be induced to express low levels of Siglec-F ligands following stimulation with IL-4.
Enzyme(s) responsible for Siglec-F ligand formation have not yet been clearly delineated and it seems likely that a series of enzymatic reactions are necessary for its formation. Sequential steps in the biosynthesis of 6-sulfo-SLeX, a structurally resembling glycan to the Siglec-F ligand 6′-sulfo-SLex, have been investigated using human bronchial mucosa (12). These studies demonstrated the following sequential steps: 1) sulfation by 6-sulfotransferases, 2) β-galactosylation, 3) α2–3 sialylation by sialyltransferases, and 4) α1–3 fucosylation by fucosyltransferases (12). In this study we have utilized ST3Gal-III+/− and FucTIV/VII deficient mice to examine the role of these enzymes in the 3rd and 4th steps of Siglec-F ligand formation. ST3Gal-III has previously been identified based on immunohistochemistry of lungs derived from non-allergen challenged naïve ST3Gal-III deficient mice as important in constitutive Siglec-F ligand formation (20). Our study extends these observations regarding the role of ST3Gal-III in Siglec-F ligand formation to demonstrate that different cell types (epithelial cells vs eosinophils, macrophages, mast cells) differ in their requirement for levels of ST3Gal-III needed for constitutive Siglec-F ligand formation in ST3Gal-III+/− mice. For example, lung epithelial cells from mice heterozygously deficient in ST3Gal-III, exhibit complete loss of expression of Siglec-F ligands as assessed by FACS. In contrast, eosinophils, macrophages, and mast cells from mice heterozygously deficient in ST3Gal-III have only a partial reduction in Siglec-F ligand formation. Several factors may account for the difference in requirements for ST3Gal-III in Siglec-F ligand formation in the cell types we have studied, including differences in levels of expression of ST3Gal-III, differences in levels of sialyltransferases other than ST3Gal-III, as well as the fact that we have studied ST3Gal-III heterozygotes rather than homozygous deficient mice. In terms of expression levels of ST3Gal-III as assessed by qPCR, they were lowest in epithelial cells the cell type in which constitutive Siglec-F ligand formation was most reduced in heterozygous deficient ST3Gal-III mice. While both macrophages (> 32 fold increase in ST3Gal-III mRNA compared to epithelium) and eosinophils (> 3 fold increase in ST3Gal-III mRNA compared to epithelium) had increased levels of ST3Gal-III mRNA compared to epithelium, it is possible that these differences in endogenous ST3Gal-III mRNA may or may not explain the difference in requirement for constitutive Siglec-F ligand formation in ST3Gal-III heterozygotes. Alternatively, a sialyltransferase other than ST3Gal-III could be important in Siglec-F ligand formation in macrophages, eosinophils and mast cells, but not epithelial cells. In this regard, there are more than 15 sialyltransferases in mice, with ST3Gal-II, III, IV and V among the α2–3 sialyltransferases found to be expressed in mouse lungs (17). We utilized ST3Gal-III+/− heterozygous mice rather than ST3Gal-III−/− mice as the homozygous ST3Gal-III−/− are difficult to breed and develop neurodegenerative disease. Nevertheless, our studies with ST3Gal-III+/− mice demonstrated an interesting difference in requirement for this enzyme in constitutive Siglec-F ligand synthesis in epithelium compared to non-epithelial cells which will require further investigation. In contrast to constitutive Siglec-F ligand synthesis, OVA induced Siglec-F ligand synthesis in ST3Gal-III+/− mice was significantly but not completely inhibited in epithelial cells. We also examined the role of fucosyltransferases in the expression of Siglec-F ligands. These studies demonstrated that mice deficient in both Fuc-TIV and Fuc-TVII had a very modest reduction in Siglec-F ligand formation in both epithelial cells and non-epithelial cells. Further studies are thus needed in identifying the enzymes necessary for Siglec-F ligand formation.
In addition to demonstrating an important role for ST3Gal-III in constitutive and induced Siglec-F ligand formation, we also demonstrated that ST3Gal-III+/− mice showed exaggerated eosinophilic lung inflammation in a model of OVA induced eosinophilic lung inflammation. As crosslinking Siglec-F induces eosinophil apoptosis (4), we explored whether ST3Gal-III+/− mice (which express reduced levels of Siglec-F ligands) had reduced levels of eosinophil apoptosis. These studies demonstrated that OVA challenged ST3Gal-III+/− mice had a reduced number of apoptotic eosinophils and an increased number of live eosinophils. The reduced eosinophil apoptosis in the lungs of ST3Gal-III+/− mice may be mediated by reduced epithelial expression of Siglec-F ligands as WT eosinophils (which highly express Siglec-F) when cultured with ST3Gal-III deficient epithelial cells (which do not express Siglec-F ligand) have reduced eosinophil apoptosis compared to WT eosinophils cultured with WT epithelial cells.
In summary, our studies demonstrated that Siglec-F ligands are constitutively expressed in vitro and in vivo in selected cell types in the lung (epithelial cells, eosinophils, macrophages, mast cells, neutrophils, but not CD4, CD8, or B cells) and induced in response to divergent stimuli including innate stimuli (TLR ligands, Alternaria), Th2 cytokines (IL-4, IL-13), GM-CSF, and adaptive immune stimuli (OVA allergen). In addition, studies of mutant mice (ST3Gal-III; Fuc-TIV/VII) demonstrated the greater importance of the sialyltransferases, ST3Gal-III compared to fucosyltransferases (Fuc-TIV/VII) in the synthesis of the constitutive and inducible Siglec-F ligands by lung epithelial and non-epithelial cells. ST3Gal-III deficient mice (deficient in expression of Siglec-F ligands) had significantly enhanced OVA-induced eosinophilic airway inflammation associated with reduced eosinophil apoptosis. The reduced eosinophil apoptosis in the lung of ST3Gal-III deficient mice may be mediated by reduced epithelial expression of Siglec-F ligands as WT eosinophils (which highly express Siglec-F) when cultured with ST3Gal-III deficient epithelial cells (which do not express Siglec-F ligand) had reduced eosinophil apoptosis compared to WT eosinophils cultured with WT epithelial cells. Overall, these studies demonstrate that ST3Gal-III plays an important role in Siglec-F ligand formation and eosinophil apoptosis with resultant effects on eosinophilic inflammation in the lung. However, additional as yet undetermined enzymes are also likely to play an important role in Siglec-F ligand formation.
Acknowledgments
This work was supported by NIH grants AI 38425, AI 70535, AI 72115 (D.H.B), P01HL107150 (A.V), and Grants-in-Aid for Young Scientists (B) (M.S.), from the Ministry of Education, Culture, Sports, Science and Technology, Japan. M.S. is a Postdoctoral Fellow supported by a Postdoctoral Fellowships for Research Abroad from Japan Society for the Promotion of Science. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations used in this manuscript
- AHR
airway hyperreactivity
- Alum
aluminum hydroxide
- BAL
broncoalveolar lavage
- CD33rSiglecs
CD33-related siglecs
- Cts
threshold cycles
- FITC-OVA
FITC-conjugated OVA
- PAS
periodic acid-Schiff
- RQ
relative quantitation
References
- 1.Crocker PR, Paulson JC, Varki A. Siglecs and their roles in the immune system. Nature Reviews. 2007;7:255–266. doi: 10.1038/nri2056. [DOI] [PubMed] [Google Scholar]
- 2.Angata T, Hingorani R, Varki NM, Varki A. Cloning and characterization of a novel mouse Siglec, mSiglec-F: differential evolution of the mouse and human (CD33) Siglec-3-related gene clusters. J Biol Chem. 2001;276:45128–45136. doi: 10.1074/jbc.M108573200. [DOI] [PubMed] [Google Scholar]
- 3.Ravetch JV, Lanier LL. Immune inhibitory receptors. Science. 2000;290:84–89. doi: 10.1126/science.290.5489.84. [DOI] [PubMed] [Google Scholar]
- 4.Zhang M, Angata T, Cho JY, Miller M, Broide DH, Varki A. Defining the in vivo function of Siglec-F, a CD33-related Siglec expressed on mouse eosinophils. Blood. 2007;109:4280–4287. doi: 10.1182/blood-2006-08-039255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Song DJ, Cho JY, Lee SY, Miller M, Rosenthal P, Soroosh P, Croft M, Zhang M, Varki A, Broide DH. Anti-Siglec-F antibody reduces allergen-induced eosinophilic inflammation and airway remodeling. J Immunol. 2009;183:5333–5341. doi: 10.4049/jimmunol.0801421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song DJ, Cho JY, Miller M, Strangman W, Zhang M, Varki A, Broide DH. Anti-Siglec-F antibody inhibits oral egg allergen induced intestinal eosinophilic inflammation in a mouse model. Clin Immunol. 2009;131:157–169. doi: 10.1016/j.clim.2008.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nutku E, Aizawa H, Hudson SA, Bochner BS. Ligation of Siglec-8: a selective mechanism for induction of human eosinophil apoptosis. Blood. 2003;101:5014–5020. doi: 10.1182/blood-2002-10-3058. [DOI] [PubMed] [Google Scholar]
- 8.Nutku E, Hudson SA, Bochner BS. Mechanism of Siglec-8-induced human eosinophil apoptosis: role of caspases and mitochondrial injury. Biochem Bio Research Commun. 2005;336:918–924. doi: 10.1016/j.bbrc.2005.08.202. [DOI] [PubMed] [Google Scholar]
- 9.Tateno H, Crocker PR, Paulson JC. Mouse Siglec-F and human Siglec-8 are functionally convergent paralogs that are selectively expressed on eosinophils and recognize 6′-sulfo-sialyl Lewis X as a preferred glycan ligand. Glycobiology. 2005;15:1125–1135. doi: 10.1093/glycob/cwi097. [DOI] [PubMed] [Google Scholar]
- 10.Cho JY, Song DJ, Pham A, Rosenthal P, Miller M, Dayan S, Doherty TA, Varki A, Broide DH. Chronic OVA allergen challenged Siglec-F deficient mice have increased mucus, remodeling, and epithelial Siglec-F ligands which are up-regulated by IL-4 and IL-13. Resp Research. 2010;11:154. doi: 10.1186/1465-9921-11-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsuboi S, Isogai Y, Hada N, King JK, Hindsgaul O, Fukuda M. 6′-Sulfo sialyl Lex but not 6-sulfo sialyl Lex expressed on the cell surface supports L-selectin-mediated adhesion. J Biol Chem. 1996;271:27213–27216. doi: 10.1074/jbc.271.44.27213. [DOI] [PubMed] [Google Scholar]
- 12.Degroote S, Ducourouble MP, Roussel P, Lamblin G. Sequential biosynthesis of sulfated and/or sialylated Lewis x determinants by transferases of the human bronchial mucosa. Glycobiology. 1999;9:1199–1211. doi: 10.1093/glycob/9.11.1199. [DOI] [PubMed] [Google Scholar]
- 13.Allahverdian S, Wang A, Singhera GK, Wong BW, Dorscheid DR. Sialyl Lewis X modification of the epidermal growth factor receptor regulates receptor function during airway epithelial wound repair. Clin Exp Allergy. 2010;40:607–618. doi: 10.1111/j.1365-2222.2010.03455.x. [DOI] [PubMed] [Google Scholar]
- 14.Paulson JC, Colley KJ. Glycosyltransferases. Structure, localization, and control of cell type-specific glycosylation. J Biol Chem. 1989;264:17615–17618. [PubMed] [Google Scholar]
- 15.Traving C, Schauer R. Structure, function and metabolism of sialic acids. Cell Mol Life Sci. 1998;54:1330–1349. doi: 10.1007/s000180050258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takashima S. Characterization of mouse sialyltransferase genes: their evolution and diversity. Bioscience, Biotech, and Biochem. 2008;72:1155–1167. doi: 10.1271/bbb.80025. [DOI] [PubMed] [Google Scholar]
- 17.Takashima S, Tachida Y, Nakagawa T, Hamamoto T, Tsuji S. Quantitative analysis of expression of mouse sialyltransferase genes by competitive PCR. Biochem Bio Research Commun. 1999;260:23–27. doi: 10.1006/bbrc.1999.0794. [DOI] [PubMed] [Google Scholar]
- 18.Sasaki K, Watanabe E, Kawashima K, Sekine S, Dohi T, Oshima M, Hanai N, Nishi T, Hasegawa M. Expression cloning of a novel Gal beta (1–3/1–4) GlcNAc alpha 2,3-sialyltransferase using lectin resistance selection. J Biol Chem. 1993;268:22782–22787. [PubMed] [Google Scholar]
- 19.Okajima T, Fukumoto S, Miyazaki H, Ishida H, Kiso M, Furukawa K, Urano T, Furukawa K. Molecular cloning of a novel alpha2,3-sialyltransferase (ST3Gal VI) that sialylates type II lactosamine structures on glycoproteins and glycolipids. J Biol Chem. 1999;274:11479–11486. doi: 10.1074/jbc.274.17.11479. [DOI] [PubMed] [Google Scholar]
- 20.Guo JP, Brummet ME, Myers AC, Na HJ, Rowland E, Schnaar RL, Zheng T, Zhu Z, Bochner BS. Characterization of expression of glycan ligands for Siglec-F in normal mouse lungs. Am J Resp Cell Mol Biol. 2011;44:238–243. doi: 10.1165/rcmb.2010-0007OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Homeister JW, Thall AD, Petryniak B, Maly P, Rogers CE, Smith PL, Kelly RJ, Gersten KM, Askari SW, Cheng G, Smithson G, Marks RM, Misra AK, Hindsgaul O, von Andrian UH, Lowe JB. The alpha(1,3)fucosyltransferases FucT-IV and FucT-VII exert collaborative control over selectin-dependent leukocyte recruitment and lymphocyte homing. Immunity. 2001;15:115–126. doi: 10.1016/s1074-7613(01)00166-2. [DOI] [PubMed] [Google Scholar]
- 22.Nakae S, Ho LH, Yu M, Monteforte R, Iikura M, Suto H, Galli SJ. Mast cell-derived TNF contributes to airway hyperreactivity, inflammation, and TH2 cytokine production in an asthma model in mice. J Allergy Clin Immunol. 2007;120:48–55. doi: 10.1016/j.jaci.2007.02.046. [DOI] [PubMed] [Google Scholar]
- 23.Miller M, Cho JY, Pham A, Ramsdell J, Broide DH. Adiponectin and functional adiponectin receptor 1 are expressed by airway epithelial cells in chronic obstructive pulmonary disease. J Immunol. 2009;182:684–691. doi: 10.4049/jimmunol.182.1.684. [DOI] [PubMed] [Google Scholar]
- 24.Doherty TA, Khorram N, Sugimoto K, Sheppard D, Rosenthal P, Cho JY, Pham A, Miller M, Croft M, Broide DH. Alternaria induces STAT6-dependent acute airway eosinophilia and epithelial FIZZ1 expression that promotes airway fibrosis and epithelial thickness. J Immunol. 2012;188:2622–2629. doi: 10.4049/jimmunol.1101632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brockman-Schneider RA, Amineva SP, Bulat MV, Gern JE. Serial culture of murine primary airway epithelial cells and ex vivo replication of human rhinoviruses. J Immunol Methods. 2008;339:264–269. doi: 10.1016/j.jim.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cho JY, Miller M, Baek KJ, Han JW, Nayar J, Lee SY, McElwain K, McElwain S, Raz E, Broide DH. Immunostimulatory DNA reverses established allergen-induced airway remodeling. J Immunol. 2004;173:7556–7564. doi: 10.4049/jimmunol.173.12.7556. [DOI] [PubMed] [Google Scholar]
- 27.Dyer KD, Percopo CM, Rosenberg HF. Generation of eosinophils from unselected bone marrow progenitors: wild-type, TLR- and eosinophil-deficient mice. Open Immunol J. 2009;2:163–167. doi: 10.2174/1874226200902010163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suzukawa M, Nagase H, Ogahara I, Han K, Tashimo H, Shibui A, Koketsu R, Nakae S, Yamaguchi M, Ohta K. Leptin enhances survival and induces migration, degranulation, and cytokine synthesis of human basophils. J Immunol. 2011;186:5254–5260. doi: 10.4049/jimmunol.1004054. [DOI] [PubMed] [Google Scholar]
- 29.Kaplan MH, Schindler U, Smiley ST, Grusby MJ. Stat6 is required for mediating responses to IL-4 and for development of Th2 cells. Immunity. 1996;4:313–319. doi: 10.1016/s1074-7613(00)80439-2. [DOI] [PubMed] [Google Scholar]
- 30.Palmer-Crocker RL, Hughes CC, Pober JS. IL-4 and IL-13 activate the JAK2 tyrosine kinase and Stat6 in cultured human vascular endothelial cells through a common pathway that does not involve the gamma c chain. J Clin Invest. 1996;98:604–609. doi: 10.1172/JCI118829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Havaux X, Zeine A, Dits A, Denis O. A new mouse model of lung allergy induced by the spores of Alternaria alternata and Cladosporium herbarum molds. Clin Exp Immunol. 2005;139:179–188. doi: 10.1111/j.1365-2249.2004.02679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]