Abstract
Vitamin A or retinol is arguably the most multifunctional vitamin in the human body, as it is essential from embryogenesis to adulthood. The pleiotropic effects of vitamin A are exerted mainly by one active metabolite, all-trans retinoic acid (atRA), which regulates the expression of a battery of target genes through several families of nuclear receptors (RARs, RXRs, and PPARβ/δ), polymorphic retinoic acid (RA) response elements, and multiple coregulators. It also involves extranuclear and nontranscriptional effects, such as the activation of kinase cascades, which are integrated in the nucleus via the phosphorylation of several actors of RA signaling. However, vitamin A itself proved recently to be active and RARs to be present in the cytosol to regulate translation and cell plasticity. These new concepts expand the scope of the biologic functions of vitamin A and RA.
Keywords: nuclear receptor, retinoic acid receptor, retinoid X receptor, transcription, kinase cascade
Vitamins are essential organic molecules that an organism depends on for survival. The main characteristics of a vitamin are that it cannot be synthesized by the organism and thus must be obtained from the diet. Vitamin A is arguably the most multifunctional vitamin in the human body and is provided as preformed vitamin A in foods of animal origin (liver) and as provitamin A carotenoids found in plant-derived foods (1).
Vitamin A or retinol is essential for human survival at every point, from embryogenesis to adulthood (2, 3). Such an important role has been revealed by both experimental approaches and clinical observations. Indeed deficiency of vitamin A leads to neonatal growth retardation and a large array of congenital malformations collectively referred to as the fetal “vitamin A deficiency” (VAD) syndrome. Moreover, in adults, vitamin A is well known to be essential for several functions, such as vision, immunity, and reproduction. However, new biologic functions for vitamin A are continuously being discovered in new fields such as lipid metabolism, insulin response, energy balance and the nervous system.
It is well documented that most of these pleiotropic functions are not exerted by retinol itself but by active metabolites. Among these metabolites is 11-cis-retinaldehyde (11cRAL), which is involved in phototransduction through binding opsin to form rhodopsin and cone pigments. In fact, the predominant endogenous active metabolite is all-trans retinoic acid (atRA), which regulates the expression of a battery of target genes involved in cell growth and differentiation, development, and homeostasis. Moreover, in keeping with its antiproliferative activity, retinoic acid (RA) is used in the treatment of certain cancers (4). Basically, RA mobilizes to the nucleus through binding to small intracellular lipid-binding proteins. The resulting complex channels RA to specific nuclear receptors, the so-called retinoic acid receptors (RAR), which work as ligand-dependent regulators of transcription and transduce the RA signal in vivo as heterodimers with retinoic X receptors (RXR) (5, 6). Therefore, vitamin A/RA is considered as the paradigm of the link between vitamin, nutrition, homeostasis, and development via the regulation of gene expression.
During the last decade, this scenario became more complicated with the discovery that RA also has extranuclear, nontranscriptional effects, such as the activation of the mitogen-activated protein kinase (MAPK) signaling pathway, which influences the expression of RA target genes via phosphorylation processes. Moreover, other studies revealed that RA activates not only RARs but also other nuclear receptors, such as the peroxisome proliferator-activated receptors (PPAR). Finally, vitamin A/retinol proved recently to be active and to activate the Janus kinase/STAT5 signaling pathway. Remarkably, these two last novel effects result in the regulation of genes that are not direct RAR targets, thus increasing the widespread nature of the biologic functions of vitamin A/RA, especially in the field of energy balance. Finally, there is unexpected role of RARs in translation, out of the nucleus.
In this review, we highlight that the spectrum of activities of vitamin A and RA is much broader and more complex than previously suspected due to the diversity of their receptors and to the large spectrum of their extranuclear and nontranscriptional effects. We also attempt to recapitulate how all these effects crosstalk and/or cooperate with each other to expand the scope of vitamin A and RA functions.
DIVERSITY OF THE FUNCTIONAL EFFECTORS OF VITAMIN A SIGNALING
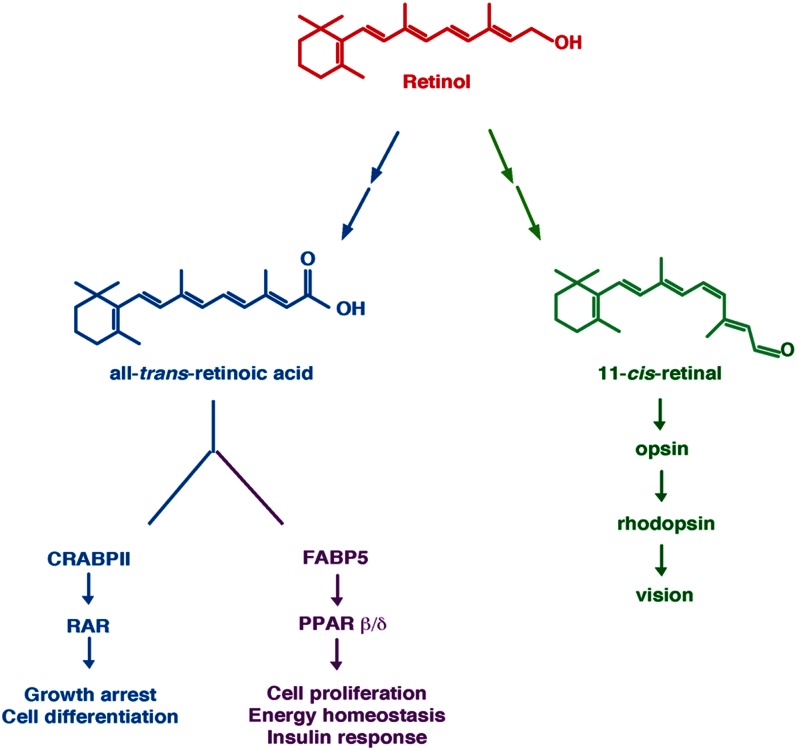
Vitamin A or retinol is composed of a β-ionone ring, a polyunsaturated side chain, and a polar end group (7) (Fig. 1). This chemical structure makes it poorly soluble in water but easily transferable through membrane lipid bilayers. In fact, vitamin A exerts its effects via oxidized metabolites, and the pleiotropicity of its effects is generated through the multiplicity of its active metabolites and of its targets.
Fig. 1.
Retinol and its two main metabolites, atRA and 11cRAL. AtRA can bind the intracellular lipid binding proteins CRABPII or FBP5. Binding to CRABPII channels RA to RARs, which then regulate genes involved in cell growth and differentiation. In contrast, binding to FABP5 channels RA to PPARβ/δ, which controls the expression of other subsets of genes involved in energy homeostasis and insulin response. 11cRAL serves as a cofactor for rhodopsin and is critical for vision. Adapted from Ref. 127.
Diversity of the active metabolites
11-cis retinaldehyde and vision.
Vitamin A is essential for vision. In the eye, its active metabolite is 11-cis-retinaldehyde (11cRAL) (for review, see Ref. 8). It is present in the photoreceptor cells of the retina and belongs to the visual pigment responsible for sensing light (Fig. 1). 11cRAL works as a chromophore and binds opsin, a G protein. Visual perception starts with the absorption of a photon, which induces isomerization of 11cRAL to 11-trans retinaldehyde (11tRARL). Then 11tRAL is released from opsin and the 11-cis chromophore is regenerated for sustained vision.
Retinoic acid and other retinoids.
The most important biologically active metabolite of vitamin A is atRA (Fig. 1). Today, several compounds that do not fit with the chemical structure of RA but are much more active in several assays have been synthesized. Now, retinol, RA, other active metabolites and active synthetic compounds are grouped as “retinoids” (9).
Today, it is well admitted that RA binds and activates RARs. However, the existence of a physiological RXR ligand is still being investigated. Indeed RXRs cannot bind atRA, and although its 9-cis isomer (9cRA) was initially considered as a bona fide RXR ligand (10, 11), it is now controversial due to the inability to detect this compound in vivo (12–14). Note, however, that 9cRA has been detected recently in pancreatic cells and found to have a function in regulating glucose-stimulated insulin secretion (15). Thus endogenous 9cRA exists and is physiological relevant. Nevertheless, RXR can bind a series of flexible fatty acids, including the unsaturated fatty acid docosahexanoic acid (DHA), oleic acid, phytanic acid, and honokiol, strongly suggesting the involvement of this receptor as a sensor of the cellular metabolic status (16). Moreover synthetic “rexinoids” have been designed and have confirmed that binding of a ligand to the RXR partner modulates the functions of the RXR/RAR heterodimers (16).
Diversity of receptors
RA binding to fatty acid-binding proteins in the cytosol for RA delivery to nuclear receptors.
In the cytosol, RA is well known to bind the cellular retinoic acid-binding protein CRABPII, which belongs to the intracellular lipid binding proteins iLBP family. It is a small cytosolic protein, which specifically associates with RA with subnanomolar affinities. Upon binding of RA, CRABPII mobilizes to the nucleus, where it associates with RARs through direct protein-protein interactions (17–19). The resulting complex channels RA from the binding protein to the RAR and markedly facilitates the formation of the liganded receptor. It was demonstrated that by directly delivering RA to RARs, CRABPII significantly enhances the transcriptional activity of RARs and sensitizes the cells to RA biological activities.
Recent studies have shown that in certain cell types, RA also binds FABP5, another fatty acid-binding protein of the iLBP family. Classically, FABPs are known to bind a variety of fatty acids and to cooperate with PPARs in a manner similar to that found for CRABPII and RAR (20). Most interestingly, RA binding activates the nuclear translocation of FABP5, which then delivers the ligand to the PPARβ/δ subtype (but not to the other PPARs) (21–23). These observations raised a novel paradigm, according to which RA can alternatively activate two different types of nuclear receptors, RARs and PPARβ/δ, depending on the relative expression levels of CRABPII and FABP5 (Fig. 1).
RARs: the canonical nuclear RA receptors.
RARs consist of three subtypes, α (NR1B1), β (NR1B2), and γ (NR1B3), which are encoded by separate genes. For each subtype, there are at least two isoforms generated by differential promoter usage and alternative splicing and differing only in their N-terminal regions. In vivo, RARs transduce the RA signal as heterodimers with RXRs, which also consist of three subtypes, α (NR2B1), β (NR2B2), and γ (NR2B3) (14, 24, 25).
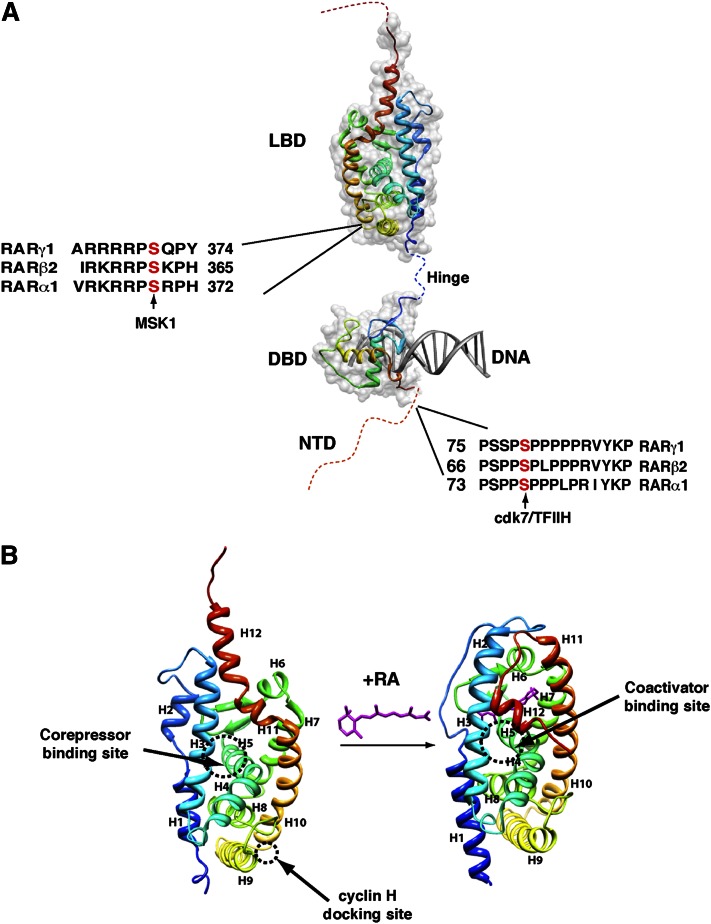
RARs and RXRs have a well-defined domain organization and structure, consisting of a variable N-terminal domain (NTD) and two well-structured and highly conserved domains, a central DNA-binding domain (DBD) and a C-terminal ligand-binding domain (LBD) linked by a flexible hinge peptide (26–29) (Fig. 2A). RARs, but not RXRs, also depict a poorly conserved and not structured region extending C-terminal to H12. However, the function of this region is poorly understood.
Fig. 2.
RAR structure. (A) RARs depict a domain organization with an unstructured NTD and two well-structured domains: a central DBD and a C-terminal LBD. The phosphorylation sites located in the NTD and the LBD are shown. (B) Structural changes induced upon RA binding. The crystal structures of the unliganded RXRα and liganded RARγ LBDs are shown. Helices are represented as ribbons and labeled from H1 to H12. The binding domains for corepressors/coactivators and for cyclin H are shown. Adapted from Protein Data Bank 1lbd and 2lbd.
The LBD is formed by 12 conserved α-helices and a β-turn, which are separated by loops and folded into a three-layered, parallel helical sandwich (27, 30, 31) (Fig. 2B). One of the most important characteristics of the LBD is the conformational flexibility of the C-terminal helix, H12, which adopts different conformations from one subtype to the other and also after ligand binding (Fig. 2B). The other characteristic is its functional complexity as it is involved in ligand binding, dimerization, and interaction with multiple coregulators via numerous interaction surfaces. The ligand-binding pocket comprises hydrophobic residues mainly from helices H3, H5, and H11 and the β-hairpin s1–s2 (32). Over 75% of the heterodimerization surface is composed by helices H9 and H10 (33). The coregulator binding surfaces include mainly the well-known hydrophobic surface involved in corepressor/coactivator binding (34, 35) as well as a recently evidenced binding domain for cyclin H, a subunit of the CAK subcomplex of the general transcription factor TFIIH (36) (Fig. 2B).
The DBD (Fig. 2A), which confers sequence-specific DNA recognition, is composed of two zinc-nucleated modules and two α-helices that cross at right angles and fold into a single globular domain, which has been determined by nuclear magnetic resonance and crystallographic studies (37). The DBD includes several highly conserved sequence elements, referred to as P, D, T, and A boxes (38, 39), which contribute to a dimerization interface and which are involved in the contacts with DNA.
The NTD is also functionally important, but in contrast to the DBD and the LBD, it is not conserved between RARs and RXRs or even between the different subtypes and isoforms. Moreover, there are still no high-resolution structures available (29). Several biochemical and structural studies coupled to structure prediction algorithms suggest that the NTDs of RARs, as well as of any member of the nuclear receptor family, are of naturally disordered structure (40, 41). Most importantly, it has recently emerged that disordered domains provide the flexibility that is needed for modification by enzymes such as kinases and ubiquitin-ligases (42, 43). Such modifications may induce changes in the structural properties of the domain, with profound impacts on its interactions with coregulators and/or on the dynamics of adjacent structural domains. In line with this, it is important to note that the NTDs of RARs and RXRs contain phosphorylation sites, which are conserved between RARs (Fig. 2A) (44).
PPARβ/δ: a novel nuclear receptor for RA.
Like RARs, PPARs belong to the superfamily of nuclear receptors and regulate the expression of target genes as heterodimers with RXRs. There are three main subtypes, PPARα (NR1C1), PPARγ (NR1C3), and PPARβ/δ (NR1C2), which are encoded by separate genes and display structural characteristics very similar to those of RARs (45, 46).
PPARs are lipid sensors and as such are activated by various fatty acids and fatty acid derivatives. Most importantly, PPARβ/δ (but not the other PPARs) can also bind RA, but with an affinity that is around 15 nM and thus over an order of magnitude weaker than that of RARs, which is in the sub-nM range. Accordingly, the PPARβ/δ subtype depicts a ligand-binding pocket, which is considerably larger than that of other nuclear receptors, consistent with the promiscuous ligand binding displayed by this receptor (47).
Though ubiquitously expressed, PPARβ/δ is preferentially expressed in brain, adipose tissue, skeletal muscle, and skin. Most interestingly, keratinocytes and adipocytes also express high levels of FABP5 compared with CRABPII, and in these cells RA activates PPARβ/δ. This finding would explain why RA has a proproliferative effect in keratinocytes instead of the expected antiproliferative one (21). It also revealed new functions of RA in the regulation of energy homeostasis and insulin responses (48).
Some recent in vitro studies suggest that RA signaling could be mediated by other receptors, such as RORβ (NR1F2) (49), COUP-TFII (NR2F2) (50), or TR2/4 (51). However, the in vivo relevance of such observations remains to be determined.
Multiple coregulators
Over many years, a plethora of RAR coregulator molecules have been identified. These coregulators include the well-known classical coactivators and corepressors (27, 34, 52–55), which interact with a hydrophobic surface of the LBD generated by H3 and H4. At the molecular point of view, the discrimination between corepressors and coactivators is governed by the ligand, which reorientates H12 and thus contributes in a critical manner to change the interaction surface. Interestingly, most of these corepressors and coactivators are common for RARs and PPARs. However, during the last years, other coregulators interacting specifically with RARs but with other surfaces of the LBD and with the NTD have been identified.
“Classical” corepressors.
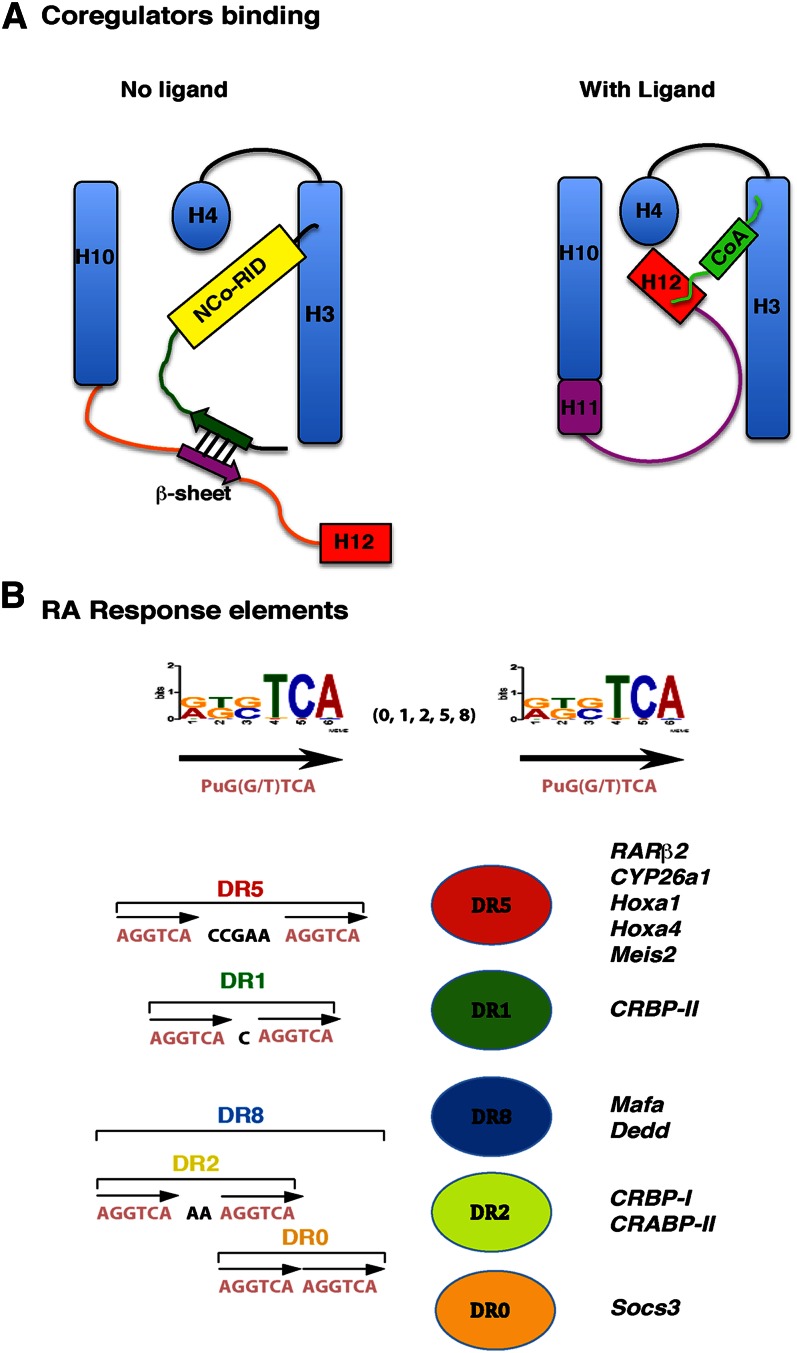
In the unliganded state, the hydrophobic surface of the RARα subtype can bind corepressors, such as nuclear receptor corepressor (NCoR) or silencing mediator of retinoic acid and thyroid hormone receptor (SMRT), which are genetic paralogs. In fact, SMRT would be the RAR-favored corepressor, and multiple protein variants are produced by alternative splicing. The majority of the SMRT variants possess at least two receptor interaction domains (RID), which contain an extended α helical box with an LxxI/HIxxxI/L motif (55). Through one of these domains, SMRT docks with the hydrophobic groove found at the surface of the RARα LBD and generated by H3 and H4. However, according to recent structural studies, efficient interaction also requires an antiparallel β-sheet interface involving N-terminal flanking residues of the corepressor RID and an extended β-strand conformation of the receptor H11 (56) (Fig. 3A). This interface removes H12 and unmasks the hydrophobic groove for binding of the RID α helix. Note that the region that maps C-terminal to H12 would also help stabilize H12 in an open conformation competent for corepressor binding (57).
Fig. 3.
Coregulator binding surfaces and RA response elements. (A) Schematic models for the recruitment of corepressors and coactivators. Adapted from Ref. 56. (B) The classical retinoid response elements are composed of a direct repeat of the motif 5′-Pu G (G/T) TCA spaced by 0 (DR0), 1 (DR1), 2 (DR2), 5 (DR5), or 8 (DR8) base pairs. DR8 comprises three half-sites with DR2 and DR0 spacing. Some RARE-associated genes are shown.
It has been generally accepted that each partner of the RAR/RXR heterodimer contacts a separate RID on a single corepressor. However, according to recent studies, it seems that SMRT would be recruited to the heterodimer only through the RAR partner. Indeed the hydrophobic groove of RXR has been shown to be masked by H12 (58). Moreover, SMRTe, a corepressor variant interacting specifically with RARs, depicts only one RID, excluding the possibility of an interaction with the RXR partner (59).
Note, however, that the RARγ and RARβ subtypes poorly interact with corepressors (54, 60, 61). It has been proposed that in these receptors, H12 interacts with H3 even in the absence of ligand, thus occluding the corepressor-docking site.
“Classical” coactivators.
In contrast, in the liganded state, RARα binds coactivators, which include essentially the p160 subfamily of steroid receptor coactivators (SRC), namely, SRC-1 (also referred to as NCoA-1), SRC-2 (TIF-2, GRIP-1), and SRC-3 (pCIP, ACTR, AlB1, TRAM1, RAC3) (52). Their main characteristic is the presence of three copies of a highly conserved LxxLL motif, which forms a short α-helix. Through one of these motifs, the coactivators bind to a surface that is topologically related to that involved in corepressor interaction but with H12, subsequent to RA-induced structural changes.
When RA is locked in the cavity of the ligand-binding pocket through hydrophobic interactions, there is a β-strand-to-α-helix secondary structure switch (56), which induces the repositioning of H11 in the continuity of H10. Concomitantly, H12 swings in a mousetrap model, sealing the “lid” of the LBP and tightly packing against H3 and H4. The new hydrophobic cleft formed among H3, H4, and H12 generates a defined interaction surface for the p160 coactivators (27) (Fig. 3A). Indeed, a charge clamp formed between a conserved glutamate residue in H12 and a lysine in H3 can specifically grip the ends of a helix of the specific length specified by the LxxLL motif of the p160 coactivators, thereby allowing the leucine side chains to pack into the hydrophobic cavity.
In the liganded state, RARs can also recruit at the same surface other unconventional coregulators with LxxLL motifs, such as a specific subunit of the Mediator, which was identified as DRIP205/TRAP220 (52), the receptor interacting protein of 140 kDa (RIP140/NRIP1) (62), the preferentially expressed antigen in melanoma (PRAME) (63), the transcription intermediary factor-1 α (TIF1α/Trim24) (64), and the thyroid receptor interacting protein-1 (TRIP1/SUG-1) (65), which is a subunit of the 19S regulatory subcomplex of the proteasome with an ATPase activity.
Other coregulators.
Several other molecules have been shown to interact with the LBD of RARs but through other surfaces. Among them is cyclin H, which interacts with the LBD at a surface involving L8–9 and the beginning of H9 (36) (Fig. 2B), allowing the recruitment of TFIIH (see below). There is also CRABPII, which in association with cyclin D3 (66) serves as an RA-channeling molecule to the receptor (17, 18) (see above).
Finally, a few proteins interacting with the NTD of RARs have been more recently identified, among which are Acinus-S′, a nuclear protein implicated in apoptotic chromatin condensation and mRNA processing, and HACE1, an HECT domain and ankyrin repeat-containing E3 ubiquitin-protein ligase (67, 68).
In fact, the interesting feature of the NTD of RARs (but not RXRs nor the other nuclear receptors) is the presence of a proline-rich motif (PRM), which contains phosphorylation sites (Fig. 2A). Such PRMs bind proteins with SH3 or WW domains, the phosphorylation preventing or favoring the interaction (69–71). In line with this, the phosphorylated PRM of RARα has been shown to bind peptidyl-prolyl cis-trans isomerase never in mitosis A (NIMA)-interacting 1 (Pin1) (72, 73), a WW domain-containing protein. In contrast, according to a recent study performed in our laboratory, the nonphosphorylated PRM of RARγ interacts with vinexinß (74), an adaptor characterized by the presence of three SH3 domains. Remarkably, vinexinß dissociates upon phosphorylation of the PRM of RARγ (75) (see below).
Polymorphism of the RA response elements and new RAR binding loci
RARs heterodimerized with RXRs bind to specific retinoic acid response elements (RARE) located in the promoters of target genes. Classically, RAREs are composed of two direct repeats (DR) of a core hexameric motif (A/G) G (G/T) TCA separated by 1, 2, or 5 nucleotides and referred as DR1, DR2, and DR5 (26, 76, 77) (Fig. 3B). Such RAREs have been identified in the promoters of a large number of RA target genes involved in a wide variety of functions, such as transcription, cell signaling, development, neuronal functions, and tumor suppression. For example, the classical DR5 elements are found in the promoters of the RARβ2 gene itself, the cytochrome 450, family 26, subfamily a, polypeptide 1 (CYP26A1) gene, and several homeobox (Hox) and hepatocyte nuclear factor (Hnf) genes. However, new RARE-associated genes have been recently revealed by high-throughput and in silico studies (78). Moreover, in some cases, several RAREs were found to be associated to the same gene (Cyp26A1, RARβ2, and Meis2) (78). Comparison of the different studies highlighted similarities in the sets of target genes but also major differences due to species and/or cell type specificities (79).
Most interestingly, the recent genome wide ChIP-seq (chromatin immunoprecipitation coupled with deep sequencing) technology allowed the identification of new RAR-binding loci and expanded the repertoire of potential high-affinity response elements (80–82). These studies revealed an unexpected diversity in the spacing and topology of the DNA elements. They also revealed that RARs can occupy a larger repertoire of sites with noncanonical IR0 (inverted repeats), DR0, and DR8 spacing, most DR8 elements comprising three half-sites with DR2 and DR0 spacing (81) (Fig. 3B). Intriguingly, dissection of these new components revealed that DR8 elements would be a new signature, whereas DR0 would be less favored, indicating that spacing would regulate RAR function.
Note that PPAR/RXR heterodimers also bind response elements that are composed of two direct repeats of a core hexameric motif with DR1 spacing and that are located in other subsets of genes involved in energy balance (45, 46).
Different architecture of the RAR/RXR heterodimers depending on the RARE spacing
On DR2 and DR5 elements, RXR occupies the 5′ hexameric motif, whereas the RAR partner occupies the 3′ motif (5′-RXR-RAR-3′). In contrast, on DR1 elements, the polarity is reversed, with the RAR DBD binding upstream and the RXR DBD binding downstream (5′-RAR-RXR-3′), switching the activity of the heterodimer from an activator to a repressor of target genes in the presence of RA (27, 83, 84).
Each DBD interacts with the DNA major groove at the level of a half-site through the P box of the first helix responsible for discrimination between different half-site sequences. Then they arrange head to tail with cooperative contacts between the DBDs, leading to a mutual reinforcement of protein-protein and protein-DNA interactions. Depending on the DR spacing, different regions of the DBD of each receptor are used to create the dimerization interface to achieve the required binding to the response elements (38, 39).
The first crystal structures to be solved were those of RAR-RXR and RXR-RXR DBDs in complex with DR1 elements (85, 86). Recently, integrative, complementary structural biology approaches provided the structure of full-length RAR-RXR heterodimers on DR1 and DR5 elements, but without the NTD, which is disordered in solution (87, 88). In both cases, the structures showed an extended asymmetric shape, the LBD dimers being positioned at the 5′ end side of the response element with an orientation orthogonal to the DNA axis. They also demonstrated that the spacing of the repeats modulates the relative positions of the domains, the RAR-RXR-DR1 complex showing a larger connecting volume between the DBDs and LBDs than the RXR-RAR-DR5 complex. Finally, these studies pointed out the flexibility and the essential role of the hinge domain in the positioning of the LBDs and in maintaining the integrity of the functional structures. Similar conclusions were also reached for the full-length PPAR/RXR heterodimers bound on DR1 elements (46).
CANONICAL MODEL OF REGULATION OF TRANSCRIPTION BY RAR-RXR HETERODIMERS
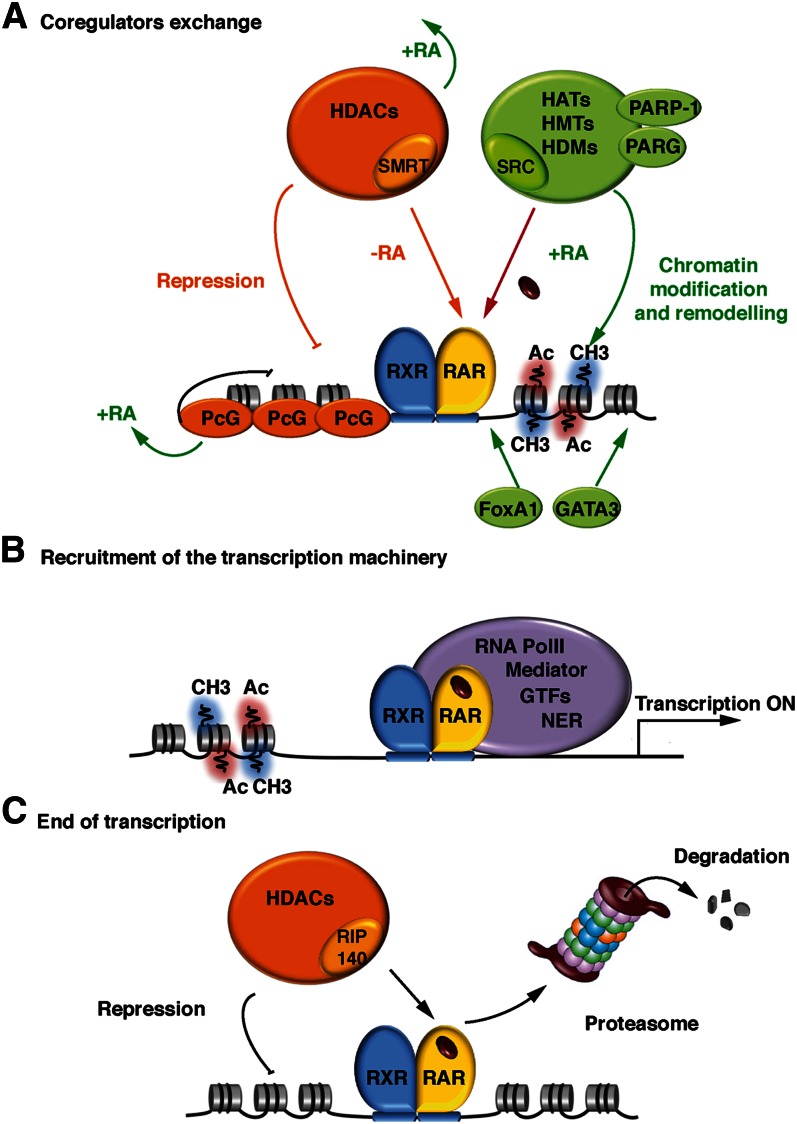
The transcriptional regulation of RA target genes by DNA-bound RAR-RXR heterodimers is a complex process that involves several dynamic, sequential, and coordinated steps. At the molecular point of view, the “on” switch to transcription activation relies on ligand-induced conformational changes that direct coregulator exchanges (Fig. 4).
Fig. 4.
Coregulator exchange at RXR/RAR heterodimers. (A) In the absence of ligand, RARα/RXR heterodimers bound to DNA are associated with corepressor complexes. Upon ligand binding, the corepressors dissociate, allowing the recruitment of coactivators and large complexes with enzymatic activities that decompact repressive chromatin. (B) When chromatin is decompacted, the transcriptional machinery, consisting of the Mediator, RNA PolII, the general transcription factors (GTF), and the nuclear excision repair (NER) factors, is recruited to the promoter, resulting in the initiation of transcription. (C) Transcription ends with the recruitment of nonconventional coactivators, such as RIP140, associated to large complexes with chromatin-repressing activity and/or through the degradation of RARs by the ubiquitin-proteasome system.
In the absence of ligand, transcription is turned “off” by RXR/RAR heterodimers bound to DNA and associated to corepressor complexes
According to the current model of gene regulation by RARs (89), in the absence of ligand, the DNA-bound RARα subtype is associated with corepressors, which serve as adaptors recruiting high molecular weight complexes endowed with histone deacetylase (HDAC) activity (90). These complexes deacetylate lysine residues in the N-terminal tails of histones and maintain chromatin in a condensed, repressed state over the target promoter (35, 89) (Fig. 4A). The corepressor complexes also contain other components, such as transducin β-like 1 (TBL1) and TBL1-related protein 1 (TBLR1), which serve as adaptors regulating corepressor assembly and function (91).
Note that transcription repression in the absence of RA also involves a large number of other proteins that form complexes with RAR/RXR heterodimers and bind specific regions of DNA. As an example, repression has been shown to involve interactions with polycomb group (PcG) proteins (92, 93) and the multiple LIM domain-containing protein ajuba (94). These proteins dissociate after RA addition via a mechanism that is still ill-defined (Fig. 4A).
Transcription is turned “on” subsequent to ligand binding
According to recent ChIP-seq experiments, the occupancy of many RAREs was increased in response to RA (95, 96). In general, there was a significant correlation between transcription activation and the binding of the heterodimers to DNA (82), but how RA promotes RAR/RXR recruitment to DNA is still an open question.
Nevertheless, upon ligand binding, RARα undergoes conformational changes that provoke corepressor release and coactivator binding (Fig. 3B). According to recent studies (87, 97), only one coactivator molecule is recruited by the heterodimer through the RAR partner, and the architecture of the DNA-bound receptor complexes orients the coactivator on one side of the DNA opposite to the RXR LBD and DBD so that it can reach its targets, the histone tails. This raises the question how the RXR partner participates in the transcriptional activity of the heterodimers in addition to increasing DNA binding efficiency (98). The question is still the focus of investigations, although a number of studies proposed that RXR is subordinated to the RAR partner, which is in fact the “active” partner (99).
Then like corepressors, the coactivators initiate an ordered and coordinated recruitment of large complexes with different enzymatic activities, such as histone acetyl transferases (HAT), histone methyl transferases (HMT), histone demethylases (HDM), and DNA-dependent ATPases (34, 35, 90, 91) (Fig. 4A). All these complexes alter the chromatin structure surrounding the promoter of target genes and create tags or binding sites that form a “histone code” read by particular effectors, which in turn mediate distinct outcomes (100). In some examples, these chromatin marks function in a combined manner. This code coordinates the recruitment of additional HATs, HMTs, or HDMs for further chromatin decompaction. It also orchestrates the recruitment of chromatin remodelers, which use the energy of ATP-hydrolysis to reposition nucleosomes through sliding them in cis or displacing them in trans, allowing the formation of nucleoside-free or nucleosome-spaced regions at the promoter. According to recent studies, optimal chromatin remodeling at some RA target genes also requires the recruitment by the RXR/RXR heterodimer complexes of the poly (ADP-ribose) polymerase (PAPR-1) (101) and of a poly (ADP-ribose) glycohydrolase (PARG) (102).
Then, activated RARs recruit the transcription machinery, including the multisubunit Mediator complex, RNA polymerase II, and the general transcription factors (35, 89) (Fig. 4B). Recent studies demonstrated that the transcription machinery assembles sequentially with the nucleotide excision repair (NER) factors at the promoter to achieve optimal histone modifications and thus efficient RNA synthesis (103). Optimal transcription also involves other proteins that form complexes with RAR/RXR heterodimers and bind specific regions of DNA, such as the zinc-finger protein Zfp423 (104), FoxA1, and Gata3 (105, 106). Finally, according to recent chromatin conformation capture experiments, when bound at several RAREs of the same gene, RARs can form loops (96). One emerging view is that RARs, as other transcription factors, would create long-range chromatin loops, bridging genomic loci located even on different chromosomes, thus creating hot spots of transcription (107).
The end of transcription
At the end of the transcriptional response, liganded RARs have been shown to recruit unconventional coregulators with LxxLL motifs, such as RIP140 and PRAME (Fig. 4C). In contrast to the classical p160 coregulators, RIP140 and PRAME inhibit the transcriptional activity of RARs through the recruitment of HDACs (108) and PcG proteins (63) respectively. It has been suggested that they would constitute a functional negative feedback mechanism limiting and/or ending RAR activity. TIF1α also represses RARα-mediated transcription (109), but through a mechanism that is still unknown.
RARs are also degraded by the ubiquitin-proteasome system, through the recruitment of TRIP1/SUG-1, which is a subunit of the 19S regulatory subcomplex of the proteasome with an ATPase activity (110–112). It has been proposed that, as for most “activators” of transcription, this degradation provides an efficient way to limit RAR function and/or to signal the end of the transcriptional process (113).
ANOTHER CONCEPT: RA AND RETINOL HAVE EXTRANUCLEAR AND NONTRANSCRIPTIONAL EFFECTS AND ACTIVATE KINASE CASCADES
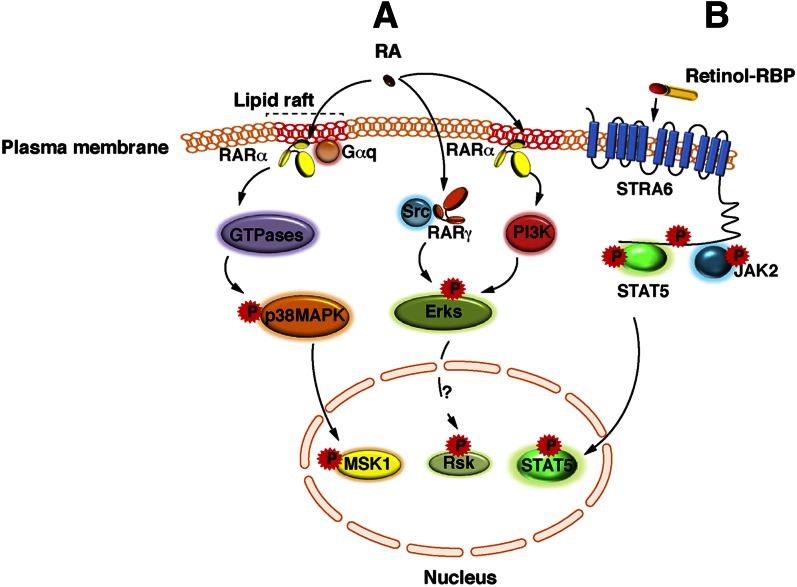
Today, it is admitted that, in addition to the above classical genomic effects, RA also has a number of nongenomic effects. Indeed, studies from several laboratories demonstrated that RA activates rapidly (within minutes after RA addition) and transiently several kinase cascades (Fig. 5A). RA activates the p38 mitogen-activated protein kinase (p38MAPK) in fibroblasts, mouse embryo carcinoma cells, mammary breast tumor cells, and leukemia cells (96, 111, 114, 115). Subsequently, p38MAPK activates the downstream mitogen- and stress-activated kinase 1 (MSK1) (96). However, in neuronal cells, Sertoli cells, and embryonic stem cells, RA instead activates the p42/p44 extracellular signal-regulated kinases (Erk) or classical MAPKs (116–122). Then, whether Erks activate a downstream kinase, such as MSK1 or ribosomal protein S6 kinase 3 (RSK2) requires further investigation. Nevertheless, the RA-induced activation of the two MAPK pathways is subsequent to the activation of upstream cascades involving Rho-GTPases (114, 115, 117), phosphoinositide 3-kinase (PI3K) and/or protein kinase B (PKB)/Akt (120, 121, 123), suggesting an atypical, nongenomic activation event similar to that described for steroid nuclear receptors (124, 125).
Fig. 5.
Extranuclear and nontranscriptional effects of RA and vitamin A. (A) A subpopulation of RARα is present in membrane lipid rafts and initiates cascades of kinase activations upon RA binding. In various epithelial and fibroblast cells, in response to RA, RARα localized in lipid rafts interacts with Gαq proteins and activates Rho-GTPases, p38MAPK, and MSK1. However, in neuronal cells, in response to RA, RARα activates Erks through the activation of the PI3K/Akt pathway. Whether Erks activate downstream effectors is unknown, but RSK2 would be an interesting candidate. Strikingly, in neuronal cells, RA has been also shown to activate Erks via RARγ in association with the Src kinase. (B) Vitamin A bound to RBP binds to the extracellular moiety of Stra6 and triggers the phosphorylation of its cytosolic domain. Phosphorylated STRA6 recruits and activates JAK2, which in turns phosphorylates STAT5. Then STAT5 translocates to the nucleus to regulate gene expression.
In line with this concept, though classically known to reside in the nucleus, RARs have been reported to be present in membranes. Indeed, in cells that respond to RA via the activation of p38MAPK, RARα is present in lipid rafts where it forms complexes with Gαq proteins (115) (Fig. 5A). However, in neuronal cells, the activation of Erks has been shown to involve either RARα localized in membranes (121) or RARγ in association with the sarcome (Src) kinase (117). Altogether, these results indicate that depending on the cell type, different RAR subtypes and kinase cascades may be involved in the activation of the MAPK pathways.
Unexpectedly, recent studies ruled out the current concept, according to which retinol functions only through active metabolites such as RA. Indeed retinol that was associated to the “retinol-binding protein (RBP)” and that was bound to the cell surface signaling receptor stimulated by retinoic acid 6 (Stra6) was found to activate the Janus kinases (JAK)/signal transducer and activator of transcription 5 (STAT5) signaling cascade (126–128) (Fig. 5B).
NUCLEAR INTEGRATION OF THE SIGNALING PATHWAYS INDUCED BY RA
Today it is admitted that signaling pathways are integrators dispensing decisions to the downstream cellular and transcriptional machineries that coordinately manage major cellular fates. In line with this concept, the RA- and retinol-induced kinases have been reported to cross talk with the transcriptional activity of RARs and other transcription factors. This nuclear integration involves mainly phosphorylation processes that influence their DNA recruitment and the timing of the sequential recruitment of the different classes of coregulators, as well as the stability of the phosphorylated proteins.
RAR phosphorylation
For several years, RARs and RXRs have been known to be phosphoproteins (129). Studies from our laboratory demonstrated that RARs are phosphorylated at two serine residues, one in the LBD (serine 369 in RARα) and the other one in the NTD (serine 77 in RARα) (130, 131) (Fig. 2A). Interestingly, serine 369 is an exposed residue located in a loop between helices 9 and 10 within the LBD. It belongs to an arginine-lysine-rich motif that corresponds to a consensus phosphorylation motif for several kinases, including the cyclic AMP-dependent protein kinase and MSK1. In contrast, serine 77 located in the NTD belongs to a proline-rich motif. The kinase responsible for phosphorylating this site has been identified as cdk7 (130), the activity of which depends on its association with cyclin H and MAT1 to form the ternary cyclin-dependent kinase (CDK)-activating kinase (CAK) complex of the general transcription factor TFIIH. The correct positioning of the cdk7 kinase and thereby the efficiency of the NTD phosphorylation by cdk7 rely on the docking of cyclin H at a specific site of the LBD located in loop L8–9 and the N-terminal part of H9 (36) (Fig. 2B).
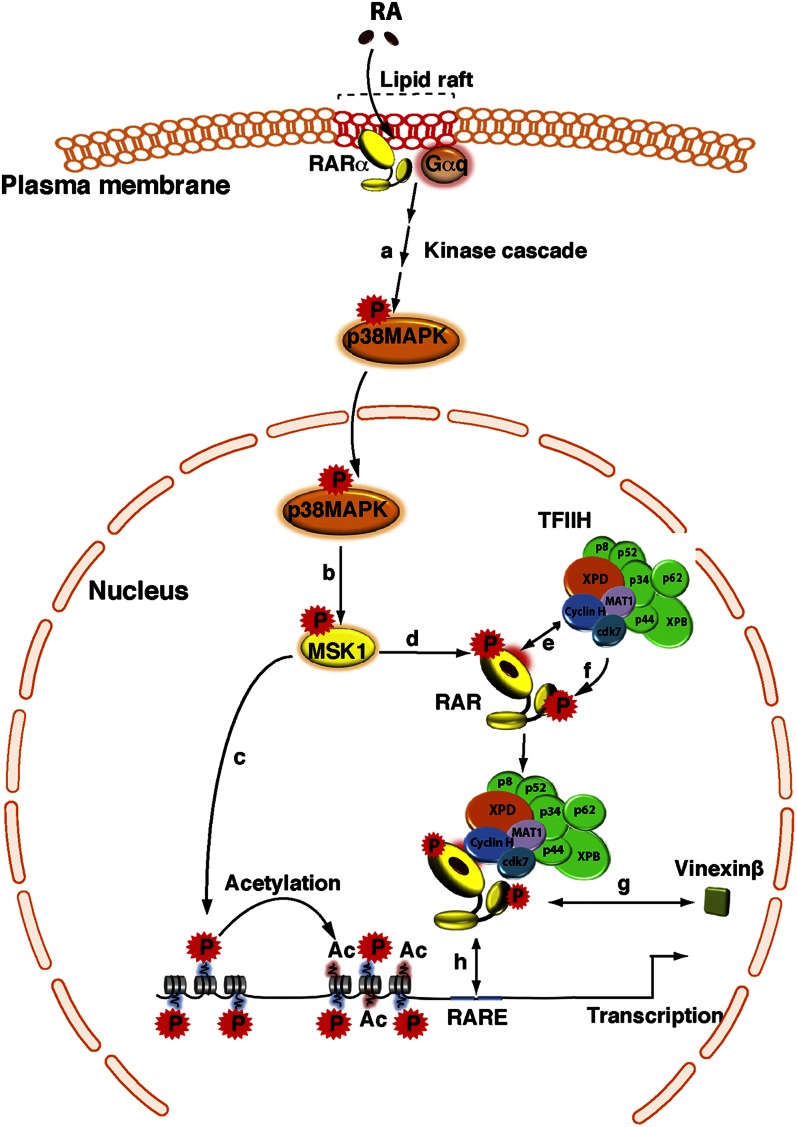
How these residues become phosphorylated was ill defined until our laboratory demonstrated that, in the case of RARα, phosphorylation results from a coordinated cascade and must be understood in terms of structural features (Fig. 6). Indeed RA-activated MSK1 phosphorylates RARα at S369 (96), and phosphorylation of this residue induces an allosteric network affecting the structural conformation/flexibility of the adjacent loop L8–9 (44) and the subsequent binding of the cyclin H/cdk7subcomplex of TFIIH (132). Consequently, cdk7 is well positioned and can phosphorylate the NTD at serine 77 (Fig. 6). The phosphorylation sites are conserved between RARs (44). The RARγ subtype is also phosphorylated at the LBD and at the NTD (131, 133), but whether it results from the same cascade as above is under investigation.
Fig. 6.
Crosstalk between the RA-activated p38MAPK pathway and the expression of RAR target genes. In response to RA, p38MAPK is activated (a), and then translocates into the nucleus and phosphorylates MSK1 (b). Activated MSK1 phosphorylates histones (c) and RARα at a serine located in the LBD (d). Subsequent to conformational changes, the cyclin H subunit of the CAK subcomplex of TFIIH is recruited to an adjacent domain (e), allowing the formation of a RARα/TFIIH complex and the phosphorylation of the NTD by the cdk7 kinase (f). In the case of the RARγ subtype, phosphorylation of the NTD promotes the dissociation of coregulators, such as vinexinß (g). Finally, phosphorylated RARα is recruited to response elements located in the promoter of target genes (h).
Phosphorylation and RAR recruitment to DNA
The interesting feature of the phosphorylation site located in the NTD is that it belongs to a PRM and is in the vicinity of the DBD. Recently our laboratory showed that when nonphosphorylated, the PRM of the RARγ subtype interacts with vinexinß, an adaptor protein with SH3 domains. As such, vinexinß represses RARγ-mediated transcription via inhibiting the binding of the receptor to DNA and its sequestration out of chromatin (75). Remarkably, phosphorylation of the serine residue located in the PRM induces changes in its conformation and decreases its propensity to bind SH3 domains (B. Kieffer et al., unpublished results). Consequently, vinexinß dissociates from RARγ (75), allowing the binding of the receptor to DR5 RAREs (Fig. 6). Thus, phosphorylation of the NTD appears to promote DNA binding via the dissociation of proteins that occlude the DBD. Such a mechanism might be at the basis of the observation made by several laboratories that the occupancy of several promoters by RAR/RXR heterodimers is increased in response to RA (80, 82, 96).
The next question is the in vivo relevance of RARs phosphorylation. Unfortunately, the genetic approaches in the animal (6, 134) are not appropriate yet to study the role of RAR and RXR phosphorylation due to their complexity and that of the signaling pathways. In fact, as cell differentiation is one of the most crucial steps during development, mouse embryo carcinoma cells (F9 cell line) (135) and embryonic stem (ES) cells (136) provided interesting tools to study the influence of RAR phosphorylation. F9 cells markedly resemble embryonic cells from the blastocyst and differentiate into endoderm-like cells in response to RA. Concerning ES cells, they are pluripotent cells that self-renew indefinitely and can differentiate in vitro into a larger variety of cell types (137), such as neuronal cells, in response to RA (138).
Invalidation experiments from our laboratory revealed that the differentiation of F9 cells in primitive endoderm (139) and that of ES cells into neurons (Al Tanoury et al., unpublished observations) involves the RARγ2 subtype. Most interestingly, the generation of stable rescue cell lines expressing RARγ2 phosphomutants in a RARγ null background argued that phosphorylation of the RARγ2 NTD is critically required for the RA-induced differentiation of these cells (Ref. 139 and Al Tanoury et al., unpublished observations). Moreover, recent genome-wide RNA-seq (high-throughput sequencing) analysis experiments performed with ES cells highlighted some direct target genes, the expression of which is controlled by the phosphorylated form of RARγ2 (Al Tanoury et al., unpublished observations). These data suggest an important role for RAR phosphorylation in RA signaling during cell differentiation, and they pave the way for further investigation during embryonic and tissue development.
Phosphorylation signals RAR degradation
At the end of the RA signal, the proteasomal degradation of RARγ has been shown to depend on the phosphorylation by p38MAPK of an additional serine residue located in the NTD (110, 111). How phosphorylation of this residue controls the recruitment of the ubiquitin-proteasome system is still ill defined (111), but it is modulated by the RXRα partner (140).
In contrast, it is still unclear whether the proteasomal degradation of RARα is phosphorylation and ubiquitination dependent. Note, however, that the degradation of RARα has been correlated to the recruitment of Pin1 to the phosphorylated N-terminal PRM (72, 73). Pin1 is well known to induce cis-trans isomerization of the proline residues that follow the phosphorylated serines in order to create new specific recognition sites for interacting factors (141), but the mechanism of the Pin1-mediated degradation of RARα remains to be defined.
Other phosphorylation targets: histones, coregulators, and other transcription factors
The kinases activated in response to RA phosphorylate not only RARs but also RXRα and other actors of RA signaling.
As an example, upon recruitment to RARα target promoters, MSK1 phosphorylates histones H3 (96). According to several reports (96, 142, 143), histone phosphorylation would contribute to transcription as a chromatin mark accounting for, in cooperation with other histone modifications, chromatin remodeling and promoter recruitment of RXR/RAR heterodimers and the transcriptional machinery (Fig. 6).
Remarkably, not only MSK1 but also the upstream kinase p38MAPK phosphorylates several targets. Indeed, p38MAPK phosphorylates RXRα at three residues located in the NTD (144, 145), but the consequences on transcription remain ill defined. P38MAPK also phosphorylates corepressors and coactivators to modulate their interaction with RARs and the dynamics of their exchanges. This is exemplified by the phosphorylation of SMRT, which induces its release from RARs and/or disturbs the organization of the overall corepressor complex (146, 147). In fine, it helps corepressors exchange for coactivators. The coactivator SRC-3 is also phosphorylated (148), resulting in its dissociation from RARs. Then phosphorylation marks SRC-3 for ubiquitination and degradation by the proteasome (149). It has been proposed that this ubiquitination-degradation process would facilitate the dynamics of RAR-mediated transcription via allowing other coregulators to bind.
It is important to highlight that the signaling pathways activated by retinoids can also phosphorylate other factors involved in the regulation of genes that are not RAR targets, thus expanding the spectrum of their biological activities. As an example, in RA-treated P19 cells, the activated Erks phosphorylate testicular nuclear receptor 2 (TR2) that then becomes a repressor of the octamer-binding transcription factor 4 (Oct4) gene, thus facilitating the differentiation program that is expected to take place in response to RA (116). There is also the recent discovery that the retinol-activated JAKs phosphorylate the transcription factor STAT5, which then translocates in the nucleus (Fig. 5B) to regulate the expression of target genes, such as the suppressor of cytokine signaling 3 (SOCS3) and PPARγ, resulting in the inhibition of insulin signaling and lipid accumulation (126, 128).
Finally, as RA can also activate PPARβ/δ, one can speculate that this receptor would be also a target for phosphorylation. PPARs are well known to be phosphoproteins (150), but the β/δ isotope is the least studied in terms of phosphorylation. Whether it becomes phosphorylated in response to RA requires further investigation.
UNCONVENTIONAL PICTURE IN RA SIGNALING: CYTOSOLIC LOCALIZATION OF RARS IN SERTOLI AND NEURONAL CELLS
It is classically believed that RARs are nuclear proteins. However, several recent articles highlighted an unconventional presence of RARs in the cytosol of Sertoli cells, hepatic stellate cells, and neuronal cells. However the mechanism and the role of this cytosolic capture or sequestration are still ill defined.
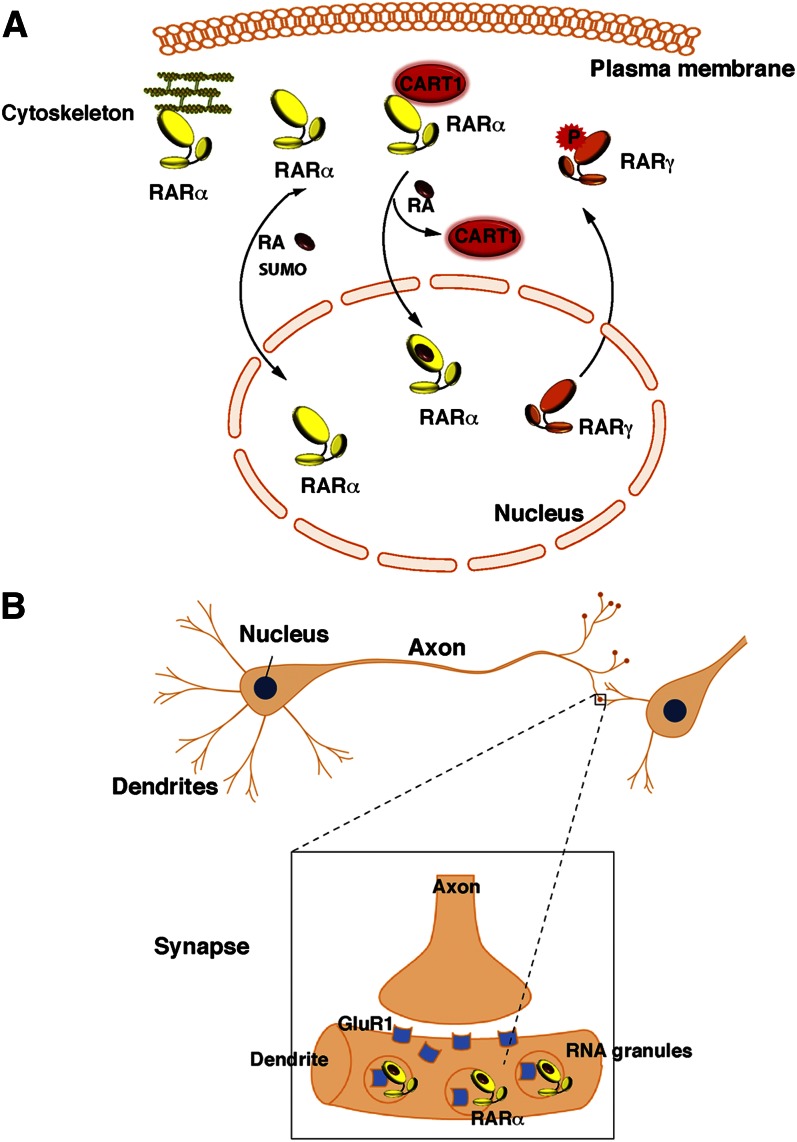
Several hypotheses have been proposed for this unconventional localization, among which is posttranslational modification (Fig. 7A). It has been proposed that phosphorylation would shift RARs out of nuclei (151, 152). Other studies demonstrated that, in Sertoli cells, RARα is a substrate for small ubiquitin-like modifier-2 (SUMO-2) and that a dynamic process of sumoylation and desumoylation influences the cytosolic/nuclear localization of the receptor (153). In Sertoli cells and hepatic stellate cells, a cytosolic localization of RARα has been also observed upon interaction with the cytoplasmic adaptor for RAR and TR (CART1) protein or with cytoskeleton proteins that sequester the receptor out of nuclei (154, 155). Given that sumoylation and protein interactions can be regulated by phosphorylation processes, one cannot exclude that a dynamic process of phosphorylation, sumoylation, and protein interaction influences the cytosolic/nuclear localization of RARs. Nevertheless, such a cytosolic capture has been correlated to a decrease in the nuclear pool of RARs for gene transcription, and changes in the modifications or interactions that sequester RARs in the cytosol restore their nuclear localization and transcriptional activity.
Fig. 7.
Unconventional presence of RARs out of the nucleus. (A) In certain cell types (Sertoli and hepatic stellate cells), RARs are sequestered in the cytosol upon phosphorylation, sumoylation, or interaction with proteins such as CART1 or cytoskeleton proteins. (B) RARα is present in the dentrites of neuronal cells, where it is associated to mRNA granules and controls translation.
Finally, two laboratories (156–158) independently demonstrated that in hippocampus neurons, RARα is exported to dendritic RNA granules and associated with a subset of mRNAs and RNA-binding proteins (Fig. 7B). Interestingly, in response to RA, this extranuclear pool of RARα triggers rapid local translation of the postsynaptic glutamate receptor GluR1 and subsequently an increase in synaptic strength. It remains to be determined whether this nongenomic effect occurs through the MAPK signaling pathway that is concomitantly activated or through a novel mechanism (157, 159). Nevertheless, it highlights a novel pool of extranuclear RARα as a sensor to the compensatory increased levels of RA generated upon neuronal activity for safeguarding information and homeostatic plasticity (160, 161). It also opens new avenues in the field of special learning and memory with a novel extranuclear role of RA.
CONCLUSION
Through its main active RA metabolite, vitamin A has pleiotropic effects, basically through genomic effects, namely, the control of a battery of target genes. This review highlights that the diversity of these effects relies on the multiplicity of the target genes and of the actors, which include not only the canonical RARs and RXRs but also other nuclear receptors, such as PPARβ/δ. Upon their activation, these receptors regulate an ever-growing spectrum of functions from cell growth and differentiation to lipid and sugar metabolism. The fact that RA can activate both RARs and PPARβ/δ provides a rationale for the long-noted but poorly understood function of vitamin A in regulating energy balance.
However the function of RA is not restricted to genomic effects. Today it is accepted that RA in conjunction with RARs also has extranuclear and nontranscriptional effects such as the activation of kinase cascades, which influence gene expression through phosphorylation processes. Moreover, new concepts are now arising, and vitamin A/retinol have proved to be active and also to activate kinase pathways, resulting in the activation of other subsets of genes involved in lipid homeostasis and insulin responses, increasing again the spectrum of action of retinoids.
Consequently, one can speculate that the integrity of all these pathways would be required for “correct” RA and vitamin A signaling. As an example, diverting RA to PPARβ/δ might be a critical component to facilitate tumor proliferation (21, 22). Moreover, deregulation of the “kinome” would have deleterious downstream effects. In line with this hypothesis, in xeroderma pigmentosum patients, who are characterized by mutations affecting subunits of the core of the general transcription factor TFIIH, RARα is not efficiently phosphorylated by cdk7, with characteristic downstream consequences on the expression of RAR target genes (162). This deficient phosphorylation has been correlated at least in part to the clinical abnormalities of the patients. Another example is that of several cancers characterized by amplified or deregulated cytosolic kinase cascades (163), ending at Akt or at different MAPKs (Erk, JNK, p38MAPK). These cancers are generally resistant to the antiproliferative action of RA (164, 165). Most interestingly, in these cancers and others, the RA-induced activation of the MAPK pathway is abrogated (115), and RARα and RXRα are aberrantly phosphorylated (166–168). Thus one can postulate that aberrant RAR phosphorylation and activity would correlate with tumoral growth and/or RA resistance (169).
Finally, the observation that RARs are present in neuronal dendrites to control translation and synaptic plasticity expands the scope of their biologic functions. Further insights into the effects of vitamin A/RA will likely continue to reveal new targets and mechanisms that will help explain their pleiotropicity and that might be manipulated in the treatment of metabolic disorders.
Footnotes
Abbreviations:
- atRA
- all-trans-retinoic acid
- RA
- retinoic acid
- RAR
- retinoic acid receptor
- RXR
- retinoid X receptor
- DBD
- DNA-binding domain
- DR
- direct repeat
- HDAC
- histone deacetylase
- LBD
- ligand-binding domain
- MAPK
- mitogen-activated protein kinase
- MSK1
- mitogen- and stress-activated kinase 1
- NTD
- N-terminal domain
- PPAR
- peroxisome proliferator-activated receptor
- RARE
- retinoic acid response elements
The work was supported by CNRS, INSERM, Agence Nationale pour la Recherche (ANR-05-BLAN-0390-02 and ANR-09-BLAN-0297-01), Foundation pour la Recherche Medicale (FRM, DEQ20090515423), Institut National du Cancer (INCA-PLO7-96099 and PL09-194), and Association pour la Recherche sur le Cancer (ARC 3169 and ARC SL220110603474). Z.A. was supported by INCA, and A.P. was supported by FRM and the TATA Memorial Trust.
REFERENCES
- 1.Harrison E. H. 2012. Mechanisms involved in the intestinal absorption of dietary vitamin A and provitamin A carotenoids. Biochim. Biophys. Acta. 1821: 70–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clagett-Dame M., DeLuca H. F. 2002. The role of vitamin A in mammalian reproduction and embryonic development. Annu. Rev. Nutr. 22: 347–381 [DOI] [PubMed] [Google Scholar]
- 3.Clagett-Dame M., Knutson D. 2011. Vitamin A in reproduction and development. Nutrients. 3: 385–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altucci L., Gronemeyer H. 2001. The promise of retinoids to fight against cancer. Nat. Rev. Cancer. 1: 181–193 [DOI] [PubMed] [Google Scholar]
- 5.McKenna N. J. 2012. EMBO retinoids 2011: mechanisms, biology and pathology of signaling by retinoic acid and retinoic acid receptors. Nucl. Recept. Signal. 10: e003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mark M., Ghyselinck N. B., Chambon P. 2009. Function of retinoic acid receptors during embryonic development. Nucl. Recept. Signal. 7: e002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Curley R. W., Jr 2012. Retinoid chemistry: synthesis and application for metabolic disease. Biochim. Biophys. Acta. 1821: 3–9 [DOI] [PubMed] [Google Scholar]
- 8.Kusakabe T. G., Takimoto N., Jin M., Tsuda M. 2009. Evolution and the origin of the visual retinoid cycle in vertebrates. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364: 2897–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sporn M. B., Roberts A. B., Goodman D. S. 1994. The Retinoids: Biology, Chemistry, and Medicine. 2nd edition. Raven Press, New York. [Google Scholar]
- 10.Heyman R. A., Mangelsdorf D. J., Dyck J. A., Stein R. B., Eichele G., Evans R. M., Thaller C. 1992. 9-cis retinoic acid is a high affinity ligand for the retinoid X receptor. Cell. 68: 397–406 [DOI] [PubMed] [Google Scholar]
- 11.Levin A. A., Sturzenbecker L. J., Kazmer S., Bosakowski T., Huselton C., Allenby G., Speck J., Kratzeisen C., Rosenberger M., Lovey A., et al. 1992. 9-cis retinoic acid stereoisomer binds and activates the nuclear receptor RXR alpha. Nature. 355: 359–361 [DOI] [PubMed] [Google Scholar]
- 12.Wolf G. 2006. Is 9-cis-retinoic acid the endogenous ligand for the retinoic acid-X receptor? Nutr. Rev. 64: 532–538 [DOI] [PubMed] [Google Scholar]
- 13.Calléja C., Messaddeq N., Chapellier B., Yang H., Krezel W., Li M., Metzger D., Mascrez B., Ohta K., Kagechika H., et al. 2006. Genetic and pharmacological evidence that a retinoic acid cannot be the RXR-activating ligand in mouse epidermis keratinocytes. Genes Dev. 20: 1525–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Germain P., Chambon P., Eichele G., Evans R. M., Lazar M. A., Leid M., De Lera A. R., Lotan R., Mangelsdorf D. J., Gronemeyer H. 2006. International Union of Pharmacology. LXIII. Retinoid X receptors. Pharmacol. Rev. 58: 760–772 [DOI] [PubMed] [Google Scholar]
- 15.Kane M. A., Folias A. E., Pingitore A., Perri M., Obrochta K. M., Krois C. R., Cione E., Ryu J. Y., Napoli J. L. 2010. Identification of 9-cis-retinoic acid as a pancreas-specific autacoid that attenuates glucose-stimulated insulin secretion. Proc. Natl. Acad. Sci. USA. 107: 21884–21889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pérez E., Bourguet W., Gronemeyer H., de Lera A. R. 2012. Modulation of RXR function through ligand design. Biochim. Biophys. Acta. 1821: 57–69 [DOI] [PubMed] [Google Scholar]
- 17.Budhu A. S., Noy N. 2002. Direct channeling of retinoic acid between cellular retinoic acid-binding protein II and retinoic acid receptor sensitizes mammary carcinoma cells to retinoic acid-induced growth arrest. Mol. Cell. Biol. 22: 2632–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delva L., Bastie J. N., Rochette-Egly C., Kraiba R., Balitrand N., Despouy G., Chambon P., Chomienne C. 1999. Physical and functional interactions between cellular retinoic acid binding protein II and the retinoic acid-dependent nuclear complex. Mol. Cell. Biol. 19: 7158–7167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dong D., Ruuska S. E., Levinthal D. J., Noy N. 1999. Distinct roles for cellular retinoic acid-binding proteins I and II in regulating signaling by retinoic acid. J. Biol. Chem. 274: 23695–23698 [DOI] [PubMed] [Google Scholar]
- 20.Tan N. S., Shaw N. S., Vinckenbosch N., Liu P., Yasmin R., Desvergne B., Wahli W., Noy N. 2002. Selective cooperation between fatty acid binding proteins and peroxisome proliferator-activated receptors in regulating transcription. Mol. Cell. Biol. 22: 5114–5127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schug T. T., Berry D. C., Shaw N. S., Travis S. N., Noy N. 2007. Opposing effects of retinoic acid on cell growth result from alternate activation of two different nuclear receptors. Cell. 129: 723–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schug T. T., Berry D. C., Toshkov I. A., Cheng L., Nikitin A. Y., Noy N. 2008. Overcoming retinoic acid-resistance of mammary carcinomas by diverting retinoic acid from PPARbeta/delta to RAR. Proc. Natl. Acad. Sci. USA. 105: 7546–7551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berry D. C., Noy N. 2007. Is PPARbeta/delta a retinoid receptor? PPAR Res. 2007: 73256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Germain P., Chambon P., Eichele G., Evans R. M., Lazar M. A., Leid M., De Lera A. R., Lotan R., Mangelsdorf D. J., Gronemeyer H. 2006. International Union of Pharmacology. LX. Retinoic acid receptors. Pharmacol. Rev. 58: 712–725 [DOI] [PubMed] [Google Scholar]
- 25.Germain P., Staels B., Dacquet C., Spedding M., Laudet V. 2006. Overview of nomenclature of nuclear receptors. Pharmacol. Rev. 58: 685–704 [DOI] [PubMed] [Google Scholar]
- 26.Bastien J., Rochette-Egly C. 2004. Nuclear retinoid receptors and the transcription of retinoid-target genes. Gene. 328: 1–16 [DOI] [PubMed] [Google Scholar]
- 27.Chambon P. 1996. A decade of molecular biology of retinoic acid receptors. FASEB J. 10: 940–954 [PubMed] [Google Scholar]
- 28.Laudet V., Gronemeyer H., 2002. The Nuclear Receptor FactsBook. Academic Press, London [Google Scholar]
- 29.Rochette-Egly C., Germain P. 2009. Dynamic and combinatorial control of gene expression by nuclear retinoic acid receptors. Nucl. Recept. Signal. 7: e005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bourguet W., Ruff M., Chambon P., Gronemeyer H., Moras D. 1995. Crystal structure of the ligand-binding domain of the human nuclear receptor RXR-alpha. Nature. 375: 377–382 [DOI] [PubMed] [Google Scholar]
- 31.Renaud J. P., Rochel N., Ruff M., Vivat V., Chambon P., Gronemeyer H., Moras D. 1995. Crystal structure of the RAR-gamma ligand-binding domain bound to all-trans retinoic acid. Nature. 378: 681–689 [DOI] [PubMed] [Google Scholar]
- 32.Bourguet W., Germain P., Gronemeyer H. 2000. Nuclear receptor ligand-binding domains: three-dimensional structures, molecular interactions and pharmacological implications. Trends Pharmacol. Sci. 21: 381–388 [DOI] [PubMed] [Google Scholar]
- 33.Bourguet W., Vivat V., Wurtz J. M., Chambon P., Gronemeyer H., Moras D. 2000. Crystal structure of a heterodimeric complex of RAR and RXR ligand-binding domains. Mol. Cell. 5: 289–298 [DOI] [PubMed] [Google Scholar]
- 34.Glass C. K., Rosenfeld M. G. 2000. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 14: 121–141 [PubMed] [Google Scholar]
- 35.Rosenfeld M. G., Lunyak V. V., Glass C. K. 2006. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 20: 1405–1428 [DOI] [PubMed] [Google Scholar]
- 36.Bour G., Gaillard E., Bruck N., Lalevée S., Plassat J. L., Busso D., Samama J. P., Rochette-Egly C. 2005. Cyclin H binding to the RARalpha activation function (AF)-2 domain directs phosphorylation of the AF-1 domain by cyclin-dependent kinase 7. Proc. Natl. Acad. Sci. USA. 102: 16608–16613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee M. S., Kliewer S. A., Provencal J., Wright P. E., Evans R. M. 1993. Structure of the retinoid X receptor alpha DNA binding domain: a helix required for homodimeric DNA binding. Science. 260: 1117–1121 [DOI] [PubMed] [Google Scholar]
- 38.Zechel C., Shen X. Q., Chambon P., Gronemeyer H. 1994. Dimerization interfaces formed between the DNA binding domains determine the cooperative binding of RXR/RAR and RXR/TR heterodimers to DR5 and DR4 elements. EMBO J. 13: 1414–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zechel C., Shen X. Q., Chen J. Y., Chen Z. P., Chambon P., Gronemeyer H. 1994. The dimerization interfaces formed between the DNA binding domains of RXR, RAR and TR determine the binding specificity and polarity of the full-length receptors to direct repeats. EMBO J. 13: 1425–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lavery D. N., McEwan I. J. 2005. Structure and function of steroid receptor AF1 transactivation domains: induction of active conformations. Biochem. J. 391: 449–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wärnmark A., Treuter E., Wright A. P., Gustafsson J. A. 2003. Activation functions 1 and 2 of nuclear receptors: molecular strategies for transcriptional activation. Mol. Endocrinol. 17: 1901–1909 [DOI] [PubMed] [Google Scholar]
- 42.Liu J., Perumal N. B., Oldfield C. J., Su E. W., Uversky V. N., Dunker A. K. 2006. Intrinsic disorder in transcription factors. Biochemistry. 45: 6873–6888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dyson H. J., Wright P. E. 2005. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 6: 197–208 [DOI] [PubMed] [Google Scholar]
- 44.Samarut E., Amal I., Markov G., Stote R., Dejaegere A., Laudet V., Rochette-Egly C. 2011. Evolution of nuclear retinoic acid receptors alpha (RARa) phosphorylation sites. Serine gain provides fine-tuned regulation. Mol. Biol. Evol. 28: 2125–2137 [DOI] [PubMed] [Google Scholar]
- 45.Hihi A. K., Michalik L., Wahli W. 2002. PPARs: transcriptional effectors of fatty acids and their derivatives. Cell. Mol. Life Sci. 59: 790–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chandra V., Huang P., Hamuro Y., Raghuram S., Wang Y., Burris T. P., Rastinejad F. 2008. Structure of the intact PPAR-gamma-RXR- nuclear receptor complex on DNA. Nature. 456: 350–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu H. E., Lambert M. H., Montana V. G., Parks D. J., Blanchard S. G., Brown P. J., Sternbach D. D., Lehmann J. M., Wisely G. B., Willson T. M., et al. 1999. Molecular recognition of fatty acids by peroxisome proliferator-activated receptors. Mol. Cell. 3: 397–403 [DOI] [PubMed] [Google Scholar]
- 48.Berry D. C., Noy N. 2009. All-trans-retinoic acid represses obesity and insulin resistance by activating both peroxisome proliferation-activated receptor beta/delta and retinoic acid receptor. Mol. Cell. Biol. 29: 3286–3296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stehlin-Gaon C., Willmann D., Zeyer D., Sanglier S., Van Dorsselaer A., Renaud J. P., Moras D., Schule R. 2003. All-trans retinoic acid is a ligand for the orphan nuclear receptor ROR beta. Nat. Struct. Biol. 10: 820–825 [DOI] [PubMed] [Google Scholar]
- 50.Kruse S. W., Suino-Powell K., Zhou X. E., Kretschman J. E., Reynolds R., Vonrhein C., Xu Y., Wang L., Tsai S. Y., Tsai M. J., et al. 2008. Identification of COUP-TFII orphan nuclear receptor as a retinoic acid-activated receptor. PLoS Biol. 6: e227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou X. E., Suino-Powell K. M., Xu Y., Chan C. W., Tanabe O., Kruse S. W., Reynolds R., Engel J. D., Xu H. E. 2011. The orphan nuclear receptor TR4 is a vitamin A-activated nuclear receptor. J. Biol. Chem. 286: 2877–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lefebvre P., Martin P. J., Flajollet S., Dedieu S., Billaut X., Lefebvre B. 2005. Transcriptional activities of retinoic acid receptors. Vitam. Horm. 70: 199–264 [DOI] [PubMed] [Google Scholar]
- 53.Westin S., Rosenfeld M. G., Glass C. K. 2000. Nuclear receptor coactivators. Adv. Pharmacol. 47: 89–112 [DOI] [PubMed] [Google Scholar]
- 54.Privalsky M. L. 2004. The role of corepressors in transcriptional regulation by nuclear hormone receptors. Annu. Rev. Physiol. 66: 315–360 [DOI] [PubMed] [Google Scholar]
- 55.Perissi V., Staszewski L. M., McInerney E. M., Kurokawa R., Krones A., Rose D. W., Lambert M. H., Milburn M. V., Glass C. K., Rosenfeld M. G. 1999. Molecular determinants of nuclear receptor-corepressor interaction. Genes Dev. 13: 3198–3208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.le Maire A., Teyssier C., Erb C., Grimaldi M., Alvarez S., de Lera A. R., Balaguer P., Gronemeyer H., Royer C. A., Germain P., et al. 2010. A unique secondary-structure switch controls constitutive gene repression by retinoic acid receptor. Nat. Struct. Mol. Biol. 17: 801–807 [DOI] [PubMed] [Google Scholar]
- 57.Farboud B., Privalsky M. L. 2004. Retinoic acid receptor-alpha is stabilized in a repressive state by its C-terminal, isotype-specific F domain. Mol. Endocrinol. 18: 2839–2853 [DOI] [PubMed] [Google Scholar]
- 58.Ghosh J. C., Yang X., Zhang A., Lambert M. H., Li H., Xu H. E., Chen J. D. 2002. Interactions that determine the assembly of a retinoid X receptor/corepressor complex. Proc. Natl. Acad. Sci. USA. 99: 5842–5847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mengeling B. J., Goodson M. L., Bourguet W., Privalsky M. L. 2012. SMRTepsilon, a corepressor variant, interacts with a restricted subset of nuclear receptors, including the retinoic acid receptors alpha and beta. Mol. Cell. Endocrinol. 351: 306–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Farboud B., Hauksdottir H., Wu Y., Privalsky M. L. 2003. Isotype-restricted corepressor recruitment: a constitutively closed helix 12 conformation in retinoic acid receptors beta and gamma interferes with corepressor recruitment and prevents transcriptional repression. Mol. Cell. Biol. 23: 2844–2858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hauksdottir H., Farboud B., Privalsky M. L. 2003. Retinoic acid receptors beta and gamma do not repress, but instead activate target gene transcription in both the absence and presence of hormone ligand. Mol. Endocrinol. 17: 373–385 [DOI] [PubMed] [Google Scholar]
- 62.Hu X., Chen Y., Farooqui M., Thomas M. C., Chiang C. M., Wei L. N. 2004. Suppressive effect of receptor-interacting protein 140 on coregulator binding to retinoic acid receptor complexes, histone-modifying enzyme activity, and gene activation. J. Biol. Chem. 279: 319–325 [DOI] [PubMed] [Google Scholar]
- 63.Epping M. T., Wang L., Edel M. J., Carlee L., Hernandez M., Bernards R. 2005. The human tumor antigen PRAME is a dominant repressor of retinoic acid receptor signaling. Cell. 122: 835–847 [DOI] [PubMed] [Google Scholar]
- 64.Le Douarin B., Zechel C., Garnier J. M., Lutz Y., Tora L., Pierrat P., Heery D., Gronemeyer H., Chambon P., Losson R. 1995. The N-terminal part of TIF1, a putative mediator of the ligand-dependent activation function (AF-2) of nuclear receptors, is fused to B-raf in the oncogenic protein T18. EMBO J. 14: 2020–2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.vom Baur E., Zechel C., Heery D., Heine M. J., Garnier J. M., Vivat V., Le Douarin B., Gronemeyer H., Chambon P., Losson R. 1996. Differential ligand-dependent interactions between the AF-2 activating domain of nuclear receptors and the putative transcriptional intermediary factors mSUG1 and TIF1. EMBO J. 15: 110–124 [PMC free article] [PubMed] [Google Scholar]
- 66.Despouy G., Bastie J. N., Deshaies S., Balitrand N., Mazharian A., Rochette-Egly C., Chomienne C., Delva L. 2003. Cyclin D3 is a cofactor of retinoic acid receptors, modulating their activity in the presence of cellular retinoic acid-binding protein II. J. Biol. Chem. 278: 6355–6362 [DOI] [PubMed] [Google Scholar]
- 67.Vucetic Z., Zhang Z., Zhao J., Wang F., Soprano K. J., Soprano D. R. 2008. Acinus-S’ represses RAR-regulated gene expression through interaction with the B-domain of RARs. Mol. Cell. Biol. 28: 2549–2558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhao J., Zhang Z., Vucetic Z., Soprano K. J., Soprano D. R. 2009. HACE1: a novel repressor of RAR transcriptional activity. J. Cell. Biochem. 107: 482–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Macias M. J., Wiesner S., Sudol M. 2002. WW and SH3 domains, two different scaffolds to recognize proline-rich ligands. FEBS Lett. 513: 30–37 [DOI] [PubMed] [Google Scholar]
- 70.Kay B. K., Williamson M. P., Sudol M. 2000. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 14: 231–241 [PubMed] [Google Scholar]
- 71.Zarrinpar A., Lim W. A. 2000. Converging on proline: the mechanism of WW domain peptide recognition. Nat. Struct. Biol. 7: 611–613 [DOI] [PubMed] [Google Scholar]
- 72.Brondani V., Schefer Q., Hamy F., Klimkait T. 2005. The peptidyl-prolyl isomerase Pin1 regulates phospho-Ser77 retinoic acid receptor alpha stability. Biochem. Biophys. Res. Commun. 328: 6–13 [DOI] [PubMed] [Google Scholar]
- 73.Giannì M., Boldetti A., Guarnaccia V., Rambaldi A., Parrella E., Raska I., Jr, Rochette-Egly C., Del Sal G., Rustighi A., Terao M., et al. 2009. Inhibition of the peptidyl-prolyl-isomerase Pin1 enhances the responses of acute myeloid leukemia cells to retinoic acid via stabilization of RARalpha and PML-RARalpha. Cancer Res. 69: 1016–1026 [DOI] [PubMed] [Google Scholar]
- 74.Bour G., Plassat J. L., Bauer A., Lalevée S., Rochette-Egly C. 2005. Vinexin beta interacts with the non-phosphorylated AF-1 domain of retinoid receptor gamma (RARγ) and represses RARγ-mediated transcription. J. Biol. Chem. 280: 17027–17037 [DOI] [PubMed] [Google Scholar]
- 75.Lalevée S., Bour G., Quinternet M., Samarut E., Kessler P., Vitorino M., Bruck N., Delsuc M. A., Vonesch J. L., Kieffer B., et al. 2010. Vinexinss, an atypical “sensor” of retinoic acid receptor gamma signaling: union and sequestration, separation, and phosphorylation. FASEB J. 24: 4523–4534 [DOI] [PubMed] [Google Scholar]
- 76.Balmer J. E., Blomhoff R. 2005. A robust characterization of retinoic acid response elements based on a comparison of sites in three species. J. Steroid Biochem. Mol. Biol. 96: 347–354 [DOI] [PubMed] [Google Scholar]
- 77.Germain P., Altucci L., Bourguet W., Rochette-Egly C., Gronemeyer H. 2003. Nuclear receptor superfamily: principles of signaling. Pure Appl. Chem. 75: 1619–1664 [Google Scholar]
- 78.Lalevée S., Anno Y. N., Chatagnon A., Samarut E., Poch O., Laudet V., Benoit G., Lecompte O., Rochette-Egly C. 2011. Genome-wide in silico identification of new conserved and functional retinoic acid receptor response elements (direct repeats separated by 5 bp). J. Biol. Chem. 286: 33322–33334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Delacroix L., Moutier E., Altobelli G., Legras S., Poch O., Choukrallah M. A., Bertin I., Jost B., Davidson I. 2010. Cell-specific interaction of retinoic acid receptors with target genes in mouse embryonic fibroblasts and embryonic stem cells. Mol. Cell. Biol. 30: 231–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mahony S., Mazzoni E. O., McCuine S., Young R. A., Wichterle H., Gifford D. K. 2011. Ligand-dependent dynamics of retinoic acid receptor binding during early neurogenesis. Genome Biol. 12: R2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Moutier E., Ye T., Choukrallah M. A., Urban S., Osz J., Chatagnon A., Delacroix L., Langer D., Rochel N., Moras D., et al. 2012. Retinoic acid receptors recognise the mouse genome through binding elements with diverse spacing and topology. J. Biol. Chem. 287: 26328–26341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mendoza-Parra M. A., Walia M., Sankar M., Gronemeyer H. 2011. Dissecting the retinoid-induced differentiation of F9 embryonal stem cells by integrative genomics. Mol. Syst. Biol. 7: 538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kurokawa R., DiRenzo J., Boehm M., Sugarman J., Gloss B., Rosenfeld M. G., Heyman R. A., Glass C. K. 1994. Regulation of retinoid signalling by receptor polarity and allosteric control of ligand binding. Nature. 371: 528–531 [DOI] [PubMed] [Google Scholar]
- 84.Leid M., Kastner P., Chambon P. 1992. Multiplicity generates diversity in the retinoic acid signalling pathways. Trends Biochem. Sci. 17: 427–433 [DOI] [PubMed] [Google Scholar]
- 85.Khorasanizadeh S., Rastinejad F. 2001. Nuclear-receptor interactions on DNA-response elements. Trends Biochem. Sci. 26: 384–390 [DOI] [PubMed] [Google Scholar]
- 86.Rastinejad F., Wagner T., Zhao Q., Khorasanizadeh S. 2000. Structure of the RXR-RAR DNA-binding complex on the retinoic acid response element DR1. EMBO J. 19: 1045–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Brélivet Y., Rochel N., Moras D. 2012. Structural analysis of nuclear receptors: from isolated domains to integral proteins. Mol. Cell. Endocrinol. 348: 466–473 [DOI] [PubMed] [Google Scholar]
- 88.Rochel N., Ciesielski F., Godet J., Moman E., Roessle M., Peluso-Iltis C., Moulin M., Haertlein M., Callow P., Mely Y., et al. 2011. Common architecture of nuclear receptor heterodimers on DNA direct repeat elements with different spacings. Nat. Struct. Mol. Biol. 18: 564–570 [DOI] [PubMed] [Google Scholar]
- 89.Dilworth F. J., Chambon P. 2001. Nuclear receptors coordinate the activities of chromatin remodeling complexes and coactivators to facilitate initiation of transcription. Oncogene. 20: 3047–3054 [DOI] [PubMed] [Google Scholar]
- 90.Perissi V., Jepsen K., Glass C. K., Rosenfeld M. G. 2010. Deconstructing repression: evolving models of co-repressor action. Nat. Rev. Genet. 11: 109–123 [DOI] [PubMed] [Google Scholar]
- 91.Perissi V., Aggarwal A., Glass C. K., Rose D. W., Rosenfeld M. G. 2004. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell. 116: 511–526 [DOI] [PubMed] [Google Scholar]
- 92.Gillespie R. F., Gudas L. J. 2007. Retinoic acid receptor isotype specificity in F9 teratocarcinoma stem cells results from the differential recruitment of coregulators to retinoic response elements. J. Biol. Chem. 282: 33421–33434 [DOI] [PubMed] [Google Scholar]
- 93.Gillespie R. F., Gudas L. J. 2007. Retinoid regulated association of transcriptional co-regulators and the polycomb group protein SUZ12 with the retinoic acid response elements of Hoxa1, RARbeta(2), and Cyp26A1 in F9 embryonal carcinoma cells. J. Mol. Biol. 372: 298–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hou Z., Peng H., White D. E., Negorev D. G., Maul G. G., Feng Y., Longmore G. D., Waxman S., Zelent A., Rauscher F. J., 3rd 2010. LIM protein Ajuba functions as a nuclear receptor corepressor and negatively regulates retinoic acid signaling. Proc. Natl. Acad. Sci. USA. 107: 2938–2943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Biddie S. C., John S., Hager G. L. 2010. Genome-wide mechanisms of nuclear receptor action. Trends Endocrinol. Metab. 21: 3–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bruck N., Vitoux D., Ferry C., Duong V., Bauer A., de The H., Rochette-Egly C. 2009. A coordinated phosphorylation cascade initiated by p38MAPK/MSK1 directs RARalpha to target promoters. EMBO J. 28: 34–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Osz J., Brelivet Y., Peluso-Iltis C., Cura V., Eiler S., Ruff M., Bourguet W., Rochel N., Moras D. 2012. Structural basis for a molecular allosteric control mechanism of cofactor binding to nuclear receptors. Proc. Natl. Acad. Sci. USA. 109: E588–E594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Leid M., Kastner P., Lyons R., Nakshatri H., Saunders M., Zacharewski T., Chen J. Y., Staub A., Garnier J. M., Mader S., et al. 1992. Purification, cloning, and RXR identity of the HeLa cell factor with which RAR or TR heterodimerizes to bind target sequences efficiently. Cell. 68: 377–395 [DOI] [PubMed] [Google Scholar]
- 99.Germain P., Iyer J., Zechel C., Gronemeyer H. 2002. Co-regulator recruitment and the mechanism of retinoic acid receptor synergy. Nature. 415: 187–192 [DOI] [PubMed] [Google Scholar]
- 100.Sims R. J., 3rd, Reinberg D. 2008. Is there a code embedded in proteins that is based on post-translational modifications? Nat. Rev. Mol. Cell Biol. 9: 815–820 [DOI] [PubMed] [Google Scholar]
- 101.Pavri R., Lewis B., Kim T. K., Dilworth F. J., Erdjument-Bromage H., Tempst P., de Murcia G., Evans R., Chambon P., Reinberg D. 2005. PARP-1 determines specificity in a retinoid signaling pathway via direct modulation of mediator. Mol. Cell. 18: 83–96 [DOI] [PubMed] [Google Scholar]
- 102.Le May N., Iltis I., Amé J., Zhovner A., Biard D., Egly J. M., Schreiber V., Coin F. 2012. Poly (ADP-ribose) glycohydrolase regulates retinoic acid receptor-mediated gene expression. Mol. Cell. 48: 785–798 [DOI] [PubMed] [Google Scholar]
- 103.Le May N., Mota-Fernandes D., Velez-Cruz R., Iltis I., Biard D., Egly J. M. 2010. NER factors are recruited to active promoters and facilitate chromatin modification for transcription in the absence of exogenous genotoxic attack. Mol. Cell. 38: 54–66 [DOI] [PubMed] [Google Scholar]
- 104.Huang S., Laoukili J., Epping M. T., Koster J., Holzel M., Westerman B. A., Nijkamp W., Hata A., Asgharzadeh S., Seeger R. C., et al. 2009. ZNF423 is critically required for retinoic acid-induced differentiation and is a marker of neuroblastoma outcome. Cancer Cell. 15: 328–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hua S., Kittler R., White K. P. 2009. Genomic antagonism between retinoic acid and estrogen signaling in breast cancer. Cell. 137: 1259–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ross-Innes C. S., Stark R., Holmes K. A., Schmidt D., Spyrou C., Russell R., Massie C. E., Vowler S. L., Eldridge M., Carroll J. S. 2010. Cooperative interaction between retinoic acid receptor-alpha and estrogen receptor in breast cancer. Genes Dev. 24: 171–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Schoenfelder S., Sexton T., Chakalova L., Cope N. F., Horton A., Andrews S., Kurukuti S., Mitchell J. A., Umlauf D., Dimitrova D. S., et al. 2010. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat. Genet. 42: 53–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wei L. N., Hu X., Chandra D., Seto E., Farooqui M. 2000. Receptor-interacting protein 140 directly recruits histone deacetylases for gene silencing. J. Biol. Chem. 275: 40782–40787 [DOI] [PubMed] [Google Scholar]
- 109.Khetchoumian K., Teletin M., Tisserand J., Mark M., Herquel B., Ignat M., Zucman-Rossi J., Cammas F., Lerouge T., Thibault C., et al. 2007. Loss of Trim24 (Tif1alpha) gene function confers oncogenic activity to retinoic acid receptor alpha. Nat. Genet. 39: 1500–1506 [DOI] [PubMed] [Google Scholar]
- 110.Kopf E., Plassat J. L., Vivat V., de The H., Chambon P., Rochette-Egly C. 2000. Dimerization with retinoid X receptors and phosphorylation modulate the retinoic acid-induced degradation of retinoic acid receptors alpha and gamma through the ubiquitin-proteasome pathway. J. Biol. Chem. 275: 33280–33288 [DOI] [PubMed] [Google Scholar]
- 111.Giannì M., Bauer A., Garattini E., Chambon P., Rochette-Egly C. 2002. Phosphorylation by p38MAPK and recruitment of SUG-1 are required for RA-indced RARγ degradation and transactivation. EMBO J. 21: 3760–3769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ferry C., Giannì M., Lalevée S., Bruck N., Plassat J. L., Raska I., Jr, Garattini E., Rochette-Egly C. 2009. SUG-1 plays proteolytic and non-proteolytic roles in the control of retinoic acid target genes via its interaction with SRC-3. J. Biol. Chem. 284: 8127–8135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tansey W. P. 2001. Transcriptional activation: risky business. Genes Dev. 15: 1045–1050 [DOI] [PubMed] [Google Scholar]
- 114.Alsayed Y., Uddin S., Mahmud N., Lekmine F., Kalvakolanu D. V., Minucci S., Bokoch G., Platanias L. C. 2001. Activation of Rac1 and the p38 mitogen-activated protein kinase pathway in response to all-trans-retinoic acid. J. Biol. Chem. 276: 4012–4019 [DOI] [PubMed] [Google Scholar]
- 115.Piskunov A., Rochette-Egly C. 2012. A retinoic acid receptor RARalpha pool present in membrane lipid rafts forms complexes with G protein alphaQ to activate p38MAPK. Oncogene. 31: 3333–3345 [DOI] [PubMed] [Google Scholar]
- 116.Gupta P., Ho P. C., Huq M. M., Ha S. G., Park S. W., Khan A. A., Tsai N. P., Wei L. N. 2008. Retinoic acid-stimulated sequential phosphorylation, PML recruitment, and SUMOylation of nuclear receptor TR2 to suppress Oct4 expression. Proc. Natl. Acad. Sci. USA. 105: 11424–11429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dey N., De P. K., Wang M., Zhang H., Dobrota E. A., Robertson K. A., Durden D. L. 2007. CSK controls retinoic acid receptor (RAR) signaling: a RAR-c-SRC signaling axis is required for neuritogenic differentiation. Mol. Cell. Biol. 27: 4179–4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bost F., Caron L., Marchetti I., Dani C., Le Marchand-Brustel Y., Binetruy B. 2002. Retinoic acid activation of the ERK pathway is required for embryonic stem cell commitment into the adipocyte lineage. Biochem. J. 361: 621–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Stavridis M. P., Collins B. J., Storey K. G. 2010. Retinoic acid orchestrates fibroblast growth factor signalling to drive embryonic stem cell differentiation. Development. 137: 881–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cheung Y. T., Lau W. K., Yu M. S., Lai C. S., Yeung S. C., So K. F., Chang R. C. 2009. Effects of all-trans-retinoic acid on human SH-SY5Y neuroblastoma as in vitro model in neurotoxicity research. Neurotoxicology. 30: 127–135 [DOI] [PubMed] [Google Scholar]
- 121.Masiá S., Alvarez S., de Lera A. R., Barettino D. 2007. Rapid, nongenomic actions of retinoic Acid on phosphatidylinositol-3-kinase signaling pathway mediated by the retinoic Acid receptor. Mol. Endocrinol. 21: 2391–2402 [DOI] [PubMed] [Google Scholar]
- 122.Pellegrini M., Filipponi D., Gori M., Barrios F., Lolicato F., Grimaldi P., Rossi P., Jannini E. A., Geremia R., Dolci S. 2008. ATRA and KL promote differentiation toward the meiotic program of male germ cells. Cell Cycle. 7: 3878–3888 [DOI] [PubMed] [Google Scholar]
- 123.Pan J., Kao Y. L., Joshi S., Jeetendran S., Dipette D., Singh U. S. 2005. Activation of Rac1 by phosphatidylinositol 3-kinase in vivo: role in activation of mitogen-activated protein kinase (MAPK) pathways and retinoic acid-induced neuronal differentiation of SH-SY5Y cells. J. Neurochem. 93: 571–583 [DOI] [PubMed] [Google Scholar]
- 124.Lösel R., Wehling M. 2003. Nongenomic actions of steroid hormones. Nat. Rev. Mol. Cell Biol. 4: 46–56 [DOI] [PubMed] [Google Scholar]
- 125.Piskunov A., Rochette-Egly C. 2012. MSK1 and nuclear receptors signaling. In MSKs. J. S. C. Arthur, editor. Landes Bioscience, Austin. [Google Scholar]
- 126.Berry D. C., Jin H., Majumdar A., Noy N. 2011. Signaling by vitamin A and retinol-binding protein regulates gene expression to inhibit insulin responses. Proc. Natl. Acad. Sci. USA. 108: 4340–4345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Berry D. C., Noy N. 2012. Signaling by vitamin A and retinol-binding protein in regulation of insulin responses and lipid homeostasis. Biochim. Biophys. Acta. 1821: 168–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Berry D. C., O'Byrne S. M., Vreeland A. C., Blaner W. S., Noy N. 2012. Cross-talk between signaling and vitamin A transport by the retinol-binding protein receptor STRA6. Mol. Cell. Biol. 32: 3164–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rochette-Egly C. 2003. Nuclear receptors: integration of multiple signalling pathways through phosphorylation. Cell. Signal. 15: 355–366 [DOI] [PubMed] [Google Scholar]
- 130.Rochette-Egly C., Adam S., Rossignol M., Egly J. M., Chambon P. 1997. Stimulation of RAR alpha activation function AF-1 through binding to the general transcription factor TFIIH and phosphorylation by CDK7. Cell. 90: 97–107 [DOI] [PubMed] [Google Scholar]
- 131.Rochette-Egly C., Oulad-Abdelghani M., Staub A., Pfister V., Scheuer I., Chambon P., Gaub M. P. 1995. Phosphorylation of the retinoic acid receptor-alpha by protein kinase A. Mol. Endocrinol. 9: 860–871 [DOI] [PubMed] [Google Scholar]
- 132.Gaillard E., Bruck N., Brelivet Y., Bour G., Lalevée S., Bauer A., Poch O., Moras D., Rochette-Egly C. 2006. Phosphorylation by PKA potentiates retinoic acid receptor alpha activity by means of increasing interaction with and phosphorylation by cyclin H/cdk7. Proc. Natl. Acad. Sci. USA. 103: 9548–9553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bastien J., Adam-Stitah S., Riedl T., Egly J. M., Chambon P., Rochette-Egly C. 2000. TFIIH interacts with the retinoic acid receptor gamma and phosphorylates its AF-1-activating domain through cdk7. J. Biol. Chem. 275: 21896–21904 [DOI] [PubMed] [Google Scholar]
- 134.Samarut E., Rochette-Egly C. 2012. Nuclear retinoic acid receptors: conductors of the retinoic acid symphony during development. Mol. Cell. Endocrinol. 348: 348–360 [DOI] [PubMed] [Google Scholar]
- 135.Bour G., Taneja R., Rochette-Egly C. 2006. Mouse embryocarcinoma F9 cells and retinoic acid: a model to study the molecular mechanisms of endodermal differentiation. In Nuclear Receptors in Development. R. Taneja, editor. Elsevier Press, San Diego, CA. 211–253. [Google Scholar]
- 136.Gudas L. J., Wagner J. A. 2011. Retinoids regulate stem cell differentiation. J. Cell. Physiol. 226: 322–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wilson V., Olivera-Martinez I., Storey K. G. 2009. Stem cells, signals and vertebrate body axis extension. Development. 136: 1591–1604 [DOI] [PubMed] [Google Scholar]
- 138.Bibel M., Richter J., Lacroix E., Barde Y. A. 2007. Generation of a defined and uniform population of CNS progenitors and neurons from mouse embryonic stem cells. Nat. Protoc. 2: 1034–1043 [DOI] [PubMed] [Google Scholar]
- 139.Taneja R., Rochette-Egly C., Plassat J. L., Penna L., Gaub M. P., Chambon P. 1997. Phosphorylation of activation functions AF-1 and AF-2 of RAR alpha and RAR gamma is indispensable for differentiation of F9 cells upon retinoic acid and cAMP treatment. EMBO J. 16: 6452–6465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Giannì M., Tarrade A., Nigro E. A., Garattini E., Rochette-Egly C. 2003. The AF-1 and AF-2 domains of RAR gamma 2 and RXR alpha cooperate for triggering the transactivation and the degradation of RAR gamma 2/RXR alpha heterodimers. J. Biol. Chem. 278: 34458–34466 [DOI] [PubMed] [Google Scholar]
- 141.Wulf G., Finn G., Suizu F., Lu K. P. 2005. Phosphorylation-specific prolyl isomerization: is there an underlying theme? Nat. Cell Biol. 7: 435–441 [DOI] [PubMed] [Google Scholar]
- 142.Vicent G. P., Ballare C., Nacht A. S., Clausell J., Subtil-Rodriguez A., Quiles I., Jordan A., Beato M. 2006. Induction of progesterone target genes requires activation of Erk and Msk kinases and phosphorylation of histone H3. Mol. Cell. 24: 367–381 [DOI] [PubMed] [Google Scholar]
- 143.Vicent G. P., Zaurin R., Nacht A. S., Li A., Font-Mateu J., Le Dily F., Vermeulen M., Mann M., Beato M. 2009. Two chromatin remodeling activities cooperate during activation of hormone responsive promoters. PLoS Genet. 5: e1000567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bruck N., Bastien J., Bour G., Tarrade A., Plassat J. L., Bauer A., Adam-Stitah S., Rochette-Egly C. 2005. Phosphorylation of the retinoid x receptor at the omega loop, modulates the expression of retinoic-acid-target genes with a promoter context specificity. Cell. Signal. 17: 1229–1239 [DOI] [PubMed] [Google Scholar]
- 145.Tarrade A., Bastien J., Bruck N., Bauer A., Gianni M., Rochette-Egly C. 2005. Retinoic acid and arsenic trioxide cooperate for apoptosis through phosphorylated RXR alpha. Oncogene. 24: 2277–2288 [DOI] [PubMed] [Google Scholar]
- 146.Varlakhanova N., Hahm J. B., Privalsky M. L. 2011. Regulation of SMRT corepressor dimerization and composition by MAP kinase phosphorylation. Mol. Cell. Endocrinol. 332: 180–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Jonas B. A., Varlakhanova N., Hayakawa F., Goodson M., Privalsky M. L. 2007. Response of SMRT (silencing mediator of retinoic acid and thyroid hormone receptor) and N-CoR (nuclear receptor corepressor) corepressors to mitogen-activated protein kinase kinase kinase cascades is determined by alternative mRNA splicing. Mol. Endocrinol. 21: 1924–1939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Giannì M., Parrella E., Raska I., Gaillard E., Nigro E. A., Gaudon C., Garattini E., Rochette-Egly C. 2006. P38MAPK-dependent phosphorylation and degradation of SRC-3/AIB1 and RARalpha-mediated transcription. EMBO J. 25: 739–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ferry C., Gaouar S., Fischer B., Boeglin M., Paul N., Samarut E., Piskunov A., Pankotai-Bodo G., Brino L., Rochette-Egly C. 2011. Cullin 3 mediates SRC-3 ubiquitination and degradation to control the retinoic acid response. Proc. Natl. Acad. Sci. USA. 108: 20603–20608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Burns K. A., Vanden Heuvel J. P. 2007. Modulation of PPAR activity via phosphorylation. Biochim. Biophys. Acta. 1771: 952–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Santos N. C., Kim K. H. 2010. Activity of retinoic acid receptor-alpha is directly regulated at its protein kinase A sites in response to follicle-stimulating hormone signaling. Endocrinology. 151: 2361–2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Han Y. H., Zhou H., Kim J. H., Yan T. D., Lee K. H., Wu H., Lin F., Lu N., Liu J., Zeng J. Z., et al. 2009. A unique cytoplasmic localization of retinoic acid receptor-gamma and its regulations. J. Biol. Chem. 284: 18503–18514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zhu L., Santos N. C., Kim K. H. 2009. Small ubiquitin-like modifier-2 modification of retinoic acid receptor-alpha regulates its subcellular localization and transcriptional activity. Endocrinology. 150: 5586–5595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Park U. H., Kim E. J., Um S. J. 2010. A novel cytoplasmic adaptor for retinoic acid receptor (RAR) and thyroid receptor functions as a derepressor of RAR in the absence of retinoic acid. J. Biol. Chem. 285: 34269–34278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Mezaki Y., Yamaguchi N., Yoshikawa K., Miura M., Imai K., Itoh H., Senoo H. 2009. Insoluble, speckled cytosolic distribution of retinoic acid receptor alpha protein as a marker of hepatic stellate cell activation in vitro. J. Histochem. Cytochem. 57: 687–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Chen N., Onisko B., Napoli J. L. 2008. The nuclear transcription factor RARalpha associates with neuronal RNA granules and suppresses translation. J. Biol. Chem. 283: 20841–20847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Chen N., Napoli J. L. 2008. All-trans-retinoic acid stimulates translation and induces spine formation in hippocampal neurons through a membrane-associated RARalpha. FASEB J. 22: 236–245 [DOI] [PubMed] [Google Scholar]
- 158.Maghsoodi B., Poon M. M., Nam C. I., Aoto J., Ting P., Chen L. 2008. Retinoic acid regulates RARalpha-mediated control of translation in dendritic RNA granules during homeostatic synaptic plasticity. Proc. Natl. Acad. Sci. USA. 105: 16015–16020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Poon M. M., Chen L. 2008. Retinoic acid-gated sequence-specific translational control by RARalpha. Proc. Natl. Acad. Sci. USA. 105: 20303–20308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Aoto J., Nam C. I., Poon M. M., Ting P., Chen L. 2008. Synaptic signaling by all-trans retinoic acid in homeostatic synaptic plasticity. Neuron. 60: 308–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Groth R. D., Tsien R. W. 2008. A role for retinoic acid in homeostatic plasticity. Neuron. 60: 192–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Keriel A., Stary A., Sarasin A., Rochette-Egly C., Egly J. M. 2002. XPD mutations prevent TFIIH-dependent transactivation by nuclear receptors and phosphorylation of RARalpha. Cell. 109: 125–135 [DOI] [PubMed] [Google Scholar]
- 163.Blume-Jensen P., Hunter T. 2001. Oncogenic kinase signalling. Nature. 411: 355–365 [DOI] [PubMed] [Google Scholar]
- 164.Slamon D. J., Clark G. M., Wong S. G., Levin W. J., Ullrich A., McGuire W. L. 1987. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 235: 177–182 [DOI] [PubMed] [Google Scholar]
- 165.Tari A. M., Lim S. J., Hung M. C., Esteva F. J., Lopez-Berestein G. 2002. Her2/neu induces all-trans retinoic acid (ATRA) resistance in breast cancer cells. Oncogene. 21: 5224–5232 [DOI] [PubMed] [Google Scholar]
- 166.Srinivas H., Juroske D. M., Kalyankrishna S., Cody D. D., Price R. E., Xu X. C., Narayanan R., Weigel N. L., Kurie J. M. 2005. c-Jun N-terminal kinase contributes to aberrant retinoid signaling in lung cancer cells by phosphorylating and inducing proteasomal degradation of retinoic acid receptor alpha. Mol. Cell. Biol. 25: 1054–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Srinivas H., Xia D., Moore N. L., Uray I. P., Kim H., Ma L., Weigel N. L., Brown P. H., Kurie J. M. 2006. Akt phosphorylates and suppresses the transactivation of retinoic acid receptor alpha. Biochem. J. 395: 653–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Matsushima-Nishiwaki R., Okuno M., Adachi S., Sano T., Akita K., Moriwaki H., Friedman S. L., Kojima S. 2001. Phosphorylation of retinoid X receptor alpha at serine 260 impairs its metabolism and function in human hepatocellular carcinoma. Cancer Res. 61: 7675–7682 [PubMed] [Google Scholar]
- 169.Duong V., Rochette-Egly C. 2011. The molecular physiology of nuclear retinoic acid receptors. From health to disease. Biochim. Biophys. Acta. 1812: 1023–1031 [DOI] [PubMed] [Google Scholar]