Abstract
Cardiovascular disease (CVD) is the leading cause of death in developed countries, and dyslipidemia is a major risk factor for CVD. We previously identified a cluster of quantitative trait loci (QTL) on baboon chromosome 11 for multiple, related quantitative traits for serum LDL-cholesterol (LDL-C). Here we report differentially regulated hepatic genes encoding an LDL-C QTL that influences LDL-C levels in baboons. We performed hepatic whole-genome expression profiling for LDL-C-discordant baboons fed a high-cholesterol, high-fat (HCHF) diet for seven weeks. We detected expression of 117 genes within the QTL 2-LOD support interval. Three genes were differentially expressed in low LDL-C responders and 8 in high LDL-C responders in response to a HCHF diet. Seven genes (ACVR1B, CALCOCO1, DGKA, ERBB3, KRT73, MYL6B, TENC1) showed discordant expression between low and high LDL-C responders. To prioritize candidate genes, we integrated miRNA and mRNA expression profiles using network tools and found that four candidates (ACVR1B, DGKA, ERBB3, TENC1) were miRNA targets and that the miRNAs were inversely expressed to the target genes. Candidate gene expression was validated using QRT-PCR and Western blotting. This study reveals candidate genes that influence variation in LDL-C in baboons and potential genetic mechanisms for further investigation.
Keywords: dyslipidemia, cardiovascular disease, diet-responsive liver gene expression, low density lipoprotein-cholesterol
Cardiovascular disease (CVD), the leading cause of death in developed countries (1), is commonly due to the development of atherosclerosis. Atherogenesis is a complex, multifactorial process attributable to the interaction of genotype and environmental factors, including diet. CVD is characterized by accumulation of lipoproteins, inflammatory cells, and fibrous tissues in the walls of the arteries, resulting in development of lesions (2, 3). Upon rupture of advanced arterial plaques, occlusion of narrow arteries by thrombosis may lead to heart attack or stroke (4). Thus, CVD has profound economic and social impact.
Dyslipidemia is a major variable in the etiology of atherosclerosis; high serum levels of LDL-cholesterol (LDL-C) and low serum levels of HDL-cholesterol (HDL-C) are associated with high risk of developing atherosclerosis in humans and experimental animals (3, 5, 6). Because LDL-C is positively correlated with the extent and severity of atherosclerotic lesions (2), genetic variation underlying differing individual LDL-C serum concentrations is an important determinant of atherosclerosis. Significant information is available on a few genes that influence dyslipidemia (7–9), and microRNAs (miRNA) regulate their expression (10); however, these genes account for only a small percentage of LDL-C variation (11). In addition, despite the enormous economic and social constraints caused by dyslipidemia, there is still lack of a comprehensive understanding of underlying molecular genetic mechanisms regulating variation in LDL-C serum concentrations that, consequently, impairs efforts to develop genetically informed, personalized treatment. A systems biology approach has the potential to uncover gene networks underlying dyslipidemia-related genetic variation and provide insights to candidate therapeutic targets.
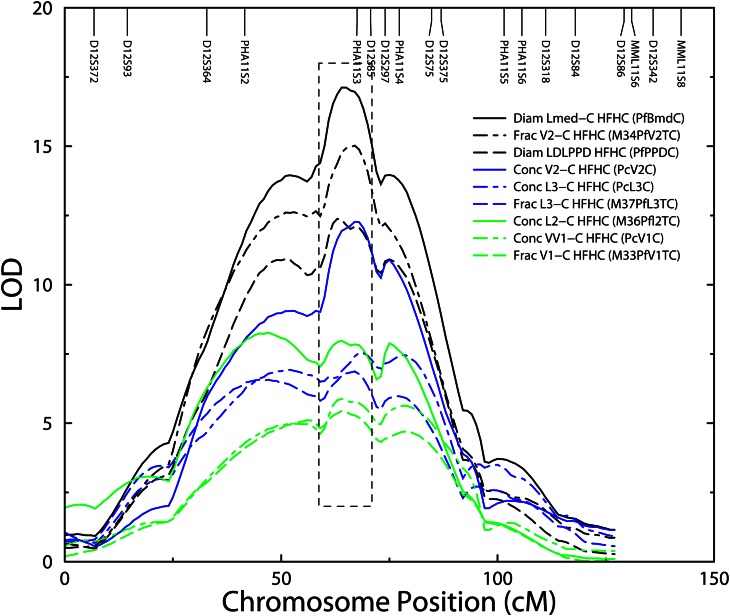
Previously we used the baboon, a well-characterized model for atherosclerosis (2, 12), to identify a cluster of QTLs on chromosome (chr) 11 (homolog of human chr 12, localized at genomic region, 12q13.13-q14.1) that encodes variation in multiple quantitative traits related to serum LDL-C (13). In a pedigreed baboon population (n = 2,044), we phenotyped individuals for LDL-C serum concentrations and genotyped the baboons for microsatellite markers (n = 287) localized in a baboon genome linkage map (14). We performed genome scans and identified a QTL on chr 11 for LDL-C serum concentration. Using additional markers and LDL-C traits, we identified additional QTLs for multiple LDL-C related traits overlapping the LDL-C serum concentration QTL (Fig. 1). The identification of a chr 11 cluster of QTLs lead us to posit that a pleiotropic gene(s), discordantly expressed between low and high LDL-C responders, is responsible for variation in LDL-C serum concentration and that this gene(s) plays a role in lipid metabolism.
Fig. 1.
Cluster of QTLs for multiple LDL-C traits on baboon chr 11, the ortholog of human chr 12q13.13-q14.1. The lower x axis denotes the chromosome in centmorgans (cM), and the upper x axis shows the order and location of microsatellite markers in the baboon genome (14). The LOD are shown on the y axis. Lines in the legend represent peaks of detected QTLs for each LDL-C trait. The box indicates the 2-LOD drop interval for the predominant QTL (Lmed-C). This figure has been modified from Ref. 13.
The goal of this study was to identify candidate genes encoding variation in the chr 11 LDL-C serum concentration QTL. To augment detection of genetic variation influencing variation in LDL-C levels, we selected three pairs of half-sib baboons discordant for LDL-C serum concentrations and discordant for genotypes of markers within the QTL. That is, we selected related animals at extremes of a population of LDL-C measures in order to minimize overall genetic variation and maximize variation in the region of the genome encoding the QTL. The discordant baboons (low LDL-C, n = 3; high LDL-C, n = 3) were challenged with a high-cholesterol, high-fat (HCHF) diet for seven weeks. Biopsies were collected from liver, the primary organ for lipid metabolism, before and after the diet challenge. We performed whole-genome expression profiling to identify pathways and genes encoded within the QTL interval responsive to HCHF diet. Gene expression profiles that differed between baseline and HCHF diets and were discordant between low and high LDL-C baboons were considered candidates encoding LDL-C phenotypic variation. To further prioritize the candidate genes, we integrated expression profiles of genes discordant between low and high LDL-C baboons with miRNA expression profiles. Genes and miRNAs were integrated based on miRNA target sites located in the gene and inverse expression between the miRNA and targeted gene. Differential expression of prioritized candidate genes was validated by QRT-PCR and Western blot. We identified four candidate genes that influence variation in LDL-C levels. All four genes are involved in regulation of protein kinase B α/glycogen synthetase 3-β (AKT1/GSK3β) and activin A receptor/mothers against decapentaplegic (ACVR/SMAD) signaling pathways. In addition, the identified miRNA targets provide bases for identification of miRNA-related functional mechanism underlying baboon LDL-C variation in future studies.
METHODS
Baboons and blood samples
Samples from 951 pedigreed baboons were previously analyzed to identify LDL-C QTLs. These animals had been genotyped (14, 15) and are organized into 11 three- to four-generation pedigrees that have a rich diversity of relative pairs. On the basis of phenotypic and genotypic analysis of the pedigreed baboon population, three pairs of half-sib baboons with contrasting phenotypes for LDL-C were chosen for this study. Each pair differed by at least two standard deviations for LDL-C serum concentrations. In addition, members of each selected sibling pair were discordant for at least one marker loci within the chromosomal region of interest (16). By applying this study design strategy, we augmented the detection of genes influencing LDL-C variation by minimizing genetic background.
For collection of serum samples, baboons were immobilized with ketamine, blood was taken from the femoral artery and serum was prepared by low-speed centrifugation. Samples were frozen at −80°C until use. Animals were maintained at Southwest National Primate Research Center (SNPRC) of the Texas Biomedical Research Institute (Texas Biomed), a facility accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International. The Institutional Animal Care and Use Committee approved all experimental procedures.
Measurement of lipid and lipoprotein traits
Lipid and lipoprotein traits were measured as described (13, 17, 18).
Diet challenge and tissue collection
Baboons were maintained on a commercial monkey chow diet (basal diet; Teklad). Baboons were fed a HCHF (i.e., 40% of calories from lard, 1.7 mg/kcal from cholesterol) diet for seven weeks. Biopsies were collected from the left lobe of the liver using Temno Evolution Needle, gauge 14 (CareFusion, San Diego, CA) before and at the end of the seven-week HCHF diet. For liver biopsy collection, baboons were sedated with ketamine (10 mg/kg), given atropine (0.025 mg/kg), and intubated. Anesthesia was induced and maintained with isoflurane (1–2%). Blood pressure was measured by automated arm cuff (Coulin), and oxygen saturation, heart, and respiration rates were monitored by pulse oximetry. During postbiopsy recovery, analgesia was provided in the form of Stadol (0.15 mg/kg twice daily for three days) and ampicillin (25 mg/day for ten days). Liver biopsies were quick frozen in liquid nitrogen at the time of collection and stored at −80°C for later RNA extractions.
All animal surgical procedures were conducted by staff veterinarians at SNPRC in conformity with the Public Health Service Policy on Humane Care and Use of Laboratory Animals, and conducted in facilities approved by the Association for Assessment and Accreditation of Laboratory Animal Care International. The Texas Biomed Institutional Animal Care and Use Committee approved all procedures.
RNA isolation from liver
RNA was isolated from liver tissue using Trizol Reagent (Ambion Inc., Austin, TX) according to the manufacturer's instructions. Approximately 20–30 mg of frozen liver was cut on an aluminum plate on dry ice and homogenized in 1ml Trizol Reagent using a PowerGen 125 Homogenizer (Thermal Scientific, Pittsburgh, PA). Genomic DNA in the sample was sheared by passing the homogenate three times through a 22 gauge needle attached to a 1 ml syringe. The homogenized samples were incubated for 5 min at 25°C. Chloroform (200 μl) was added to resulting lysate, shaken vigorously by hand for 15 s, and incubated at room temperature for 3 min. Samples were then centrifuged at 14,000 g for 10 min at 4°C. The upper aqueous phase containing RNA was carefully aspirated and transferred to a cleaning column from an RNeasy MinElute Kit (Qiagen Valencia, CA). RNA was precipitated and stored at −80°C.
Whole-genome expression profiling
cRNA was synthesized and biotin labeled using Illumina TotalPrep RNA Amplification Kit (Ambion Inc., Austin, TX) according to manufacturer's instructions. Briefly, total RNA was used for first- and second-strand cDNA synthesis followed by in vitro transcription to synthesize biotin-labeled cRNA. cRNA was quality checked and then hybridized to Human Genome-6 BeadChips (Illumina Inc., San Diego, CA). Individual cRNA samples were used to interrogate each BeadChip (LDL-C low responders, n = 3; LDL-C high responders, n = 3; for chow and HCHF diets). Gene expression was detected and cleaned using GenomeStudio software (Illumina Inc.) and filtered using quality score (>0.95). Array data from each sample were all-median normalized and log2 transformed. Box plots were inspected to ensure that the median for each group was zero and variance among groups was similar. Statistical analyses of array data were performed by t-test using GeneSifter software (GeneSifter.Net, VizX Labs, Seattle, WA) for pairwise comparisons.
Pathway analysis
Array data for differently expressed genes were overlaid onto Gene Ontological (GO) pathways (http://www.geneontology.org/8) and KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways (www.genome.jp/kegg.org9) using GeneSifter. Pathways were considered significantly different between groups if the z-score for that pathway was greater than 2. Z-scores were calculated in GeneSifter using the formula: z-score = [r−n(R/N)]/[√((n(R/N))(1−R/N)(1−((n−1)/(N−1)))], where R = total number of genes meeting selection criteria, N = total number of genes measured, r = number of genes meeting selection criteria with specified GO term, and n = total number of genes measured with specific GO term.
Network analysis
Network analysis was performed using Ingenuity Pathway Analysis (IPA; Ingenuity Systems, Redwood City, CA) by incorporating differentially expressed genes (P < 0.05) from each pairwise comparison. Networks were built using the IPA knowledgebase, using expression profiles from this dataset and LDL-C miRNA expression profiles (19) and requiring direct connections between molecules based on experimental evidence. Network significance was calculated in IPA using Fisher exact t-test.
Validation of gene expression for targeted genes
mRNA expression levels of candidate genes were quantified using Taqman probes (Applied Biosystems). We optimized cDNA concentrations for QRT-PCR by titrating a range of RNA concentrations from a pool of representative RNAs for the low and high LDL-C responder baboons. After optimization, individual cDNAs were synthesized and expression was determined by QRT-PCR using AB7900 Real-Time PCR System. We quantified mRNA according to manufacturer's instructions. In brief, total RNA (200 ng) was reverse transcribed in a 10 μl reaction using a SuperScript III Kit (Invitrogen). For real-time PCR, target cDNA, gene-specific primers, and 1X TaqMan Universal PCR master mix (Applied Biosystems) were employed. 18S rRNA (Hs99999901_s1) was quantified as an endogenous control. Expression abundance was quantified for v-erb-b2 erythroblastic leukemia viral oncogene homolog 3, also known as epidermal growth factor receptor 3 (ERBB3; Applied Biosystems catalog number Hs00951456_g1), calcium binding and coiled-coil domain 1 (CALCOCO1; Hs01548317_m1), and tensin-like C1 domain containing phosphatase (TENC1; Hs00539259_g1). All samples were assayed in triplicate. The relative expression of each gene was determined using the formula ΔΔCt by subtracting the ΔCt of the calibrator (optimized pool of RNA from low and high LDL-C) from ΔCt of target gene. Fold difference was calculated using the expression 2−ΔΔCt.
Protein quantification
Cell lysate was prepared by homogenizing 1–3 mg of frozen liver tissue in a Biomasher microhomogenizer (ISC Bio Express, UT) in ice-cold RIPA lysis buffer (12.5 mM Tris-HCl at pH 7.6, 0.5% Np-40, 0.5% sodium deoxycholic acid, 0.1% sodium dodecylsulfate, 75 mM sodium chloride). RIPA buffer contained 1X Halt protease inhibitor cocktail (Thermal Scientific). Cell debris was removed by centrifugation at 15, 000 g for 20 s at 4°C. Supernatants were assayed for total protein concentration by Bio-Rad protein assay and diluted to a working concentration (3 μg/μl) using the lysing buffer.
Protein analysis was performed using SDS-PAGE and Western blot. After boiling for 5 min at 95°C in 1× SDS sample buffer (50 mM Tris-HCl at pH 6.8, 12.5% glycerol, 1% SDS, 0.01% bromophenol blue) containing 5% β-mercaptoethanol, the lysates (18 μg) were run in 10% polyacrylamide gel under reducing conditions and then transferred onto PVDF membrane (Bio-Rad Systems, Hercules, CA) overnight at 4°C. The membrane was washed with double distilled water three times for 5 min each, and then incubated with a StartingBlock blocking buffer (Thermal Scientific) for 20 min at room temperature. Subsequently, the membrane was incubated with TENC1, ERBB3, DGKA, or ACVR1B primary antibodies or β-actin (control) in blocking solution overnight at 4°C. After washing three times with 1× TBST buffer (20 mM Tris, 500 mM NaCl at pH 7.5, 0.05% Tween 20) for 10 min each, the membrane was incubated with goat anti-rabbit secondary antibody for 90 min at room temperature. The membrane was washed three times with TBST and three times with 1× TBS buffer (20 mM Tris, 500 mM NaCl at pH 7.5) for 5 min each and the bound antibody visualized using ECL Plus Western Blotting Detection reagents (GE Healthcare, Buckinghamshire, UK). Polyclonal antibodies were obtained from the following sources: anti-TENC1 from Dr. Hafizi, University of Portsmouth, United Kingdom, and Abcam (ab96381), Cambridge, MA; anti-ERBB3 (c-17:sc-285), β-actin (sc-130659), and secondary rabbit polyclonal antibody containing HRP conjugate (sc-2004) from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA); anti-ACVR1B (AF1477) from Bio-Rad Systems; anti-DGKA (H00001606-B01P) from Abnova (Taiwan); anti-CALCOCOL1 (ab-70564) from Abcam. The membrane was scanned using Storm 480 (GE Healthcare, Waukesha, WI) and membrane-bound proteins were quantified by ImageQuant TL v2003.02 (Amersham Biosciences, UK) following the manufacturer's instructions. Intensity scores for the protein bands of interest were normalized using β-actin intensity scores, and mean ± SD was calculated for each treatment group. Significance was tested by Student t-test (P ≤ 0.05).
miRNA-mRNA integration to prioritize candidate genes
Previously we reported miRNAs expressed in low and high LDL-C livers that were identified using a high-throughput sequencing approach (19). To prioritize LDL-C QTL candidate genes, we used the MiRNA Targets Filter tool embedded in IPA to integrate the 28 differentially expressed hepatic miRNAs in response to the HCHF diet with the expression profiles of the seven LDL-C QTL candidate genes to identify miRNA targets with expression profiles that are negatively correlated with miRNA expression.
RESULTS
LDL-C serum concentrations for baboon panel
The mean ± SD LDL-C serum concentrations for the low and high LDL-C responders, before and at the end of the seven-week diet challenge, are shown in Table 1.
TABLE 1.
Low and high LDL-C responders’ serum LDL-C values for chow and HCHF diets
| Serum LDL-C Value on Chow Diet | Serum LDL-C Value on HCHF Diet | |
| Low LDL-C responders | 19.3 (8.1) | 31.3 (2.5) |
| High LDL-C responders | 43.7 (15.0) | 114.3 (19.5) |
Standard deviations are indicated in parenthesis.
Whole-genome expression profiling
For low LDL responders, we detected 9,904 genes. Of these genes, 83 were upregulated and 87 were downregulated in response to the HCHF diet challenge. Pathway analysis revealed 61 biological pathways that were upregulated and 59 downregulated in response to the HCHF diet challenge. Upregulated pathways included lysine biosynthesis, ether lipid metabolism, and bile acid biosynthesis. Downregulated pathways included valine, leucine, and isoleucine biosynthesis; starch and sucrose metabolism; circadian rhythm; basal transcription factors; and citrate cycle (TCA cycle) (supplementary Table I). For high LDL-C responders, we detected 9,992 genes. Of these genes, 96 were upregulated and 117 were downregulated in response to the HCHF diet challenge. Pathway analysis revealed that an equal number of biological pathways (n = 16) were upregulated or downregulated in response to the HCHF diet challenge. Upregulated pathways included basal transcription factors, bile acid biosynthesis, notch signaling, and glycerolipid metabolism. Downregulated pathways included ribosome, glycolysis/gluconeogenesis, and urea cycle and metabolism of amino groups (supplementary Table II). Overall, a greater number of genes (N = 213) were differentially expressed in the livers of high LDL-C responders in response to the HCHF diet challenge compared with livers of low LDL-C responders (N = 170) (χ2 test, P = 0.04).
Expression of genes in the QTL 2-LOD support interval
We identified candidate genes encoding LDL-C concentration QTL in baboons. The 2-LOD support interval of the QTL includes 229 genes. Among these genes, 117 passed quality filter for expression in our LDL-C discordant baboons. Three genes were differentially expressed (P ≤ 0.05) in response to HCHF diet in low LDL-C baboons, while 8 genes were differentially expressed in response to HCHF diet in high LDL-C responders (Table 2). Two genes, keratin 80 (KRT80) and glutaminase 2 (GLS2), were upregulated in both low and high LDL-C responders; while 7 genes, including activin A receptor, type IB (ACVR1B), diacylglycerol kinase α (DGKA), keratin 73 (KRT73), myosin light chain 6B (MYL6B), TENC1, CALCOCO1, and ERBB3, showed a differential response to the HCHF diet between low and high LDL-C responders (Table 2).
TABLE 2.
Expression change in response to diet for nine chr 11 QTL candidate genes in low and high LDL-C responders
| Direction of Change of Gene Expression |
|||
| Gene | Physical location (Mb) [Feb. 2009 (GRCh37/hg 19)] | Low LDL-C liver | High LDL-C liver |
| ACVR1B | 52.3 | — | ▾ |
| KRT80 | 52.5 | ▴ | ▴ |
| KRT73 | 53.0 | — | ▾ |
| TENC1 | 53.4 | ▴ | — |
| CALCOCO1 | 54.1 | — | ▴ |
| DGKA | 56.3 | — | ▴ |
| ERBB3 | 56.4 | — | ▾ |
| MYL6B | 56.5 | — | ▾ |
| GLS2 | 56.8 | ▴ | ▴ |
▴ (triangle up filled) indicates upregulation, ▾ (triangle down filled) indicates downregulation, and — indicates no detectable change in gene expression.
miRNA prioritization of candidate genes
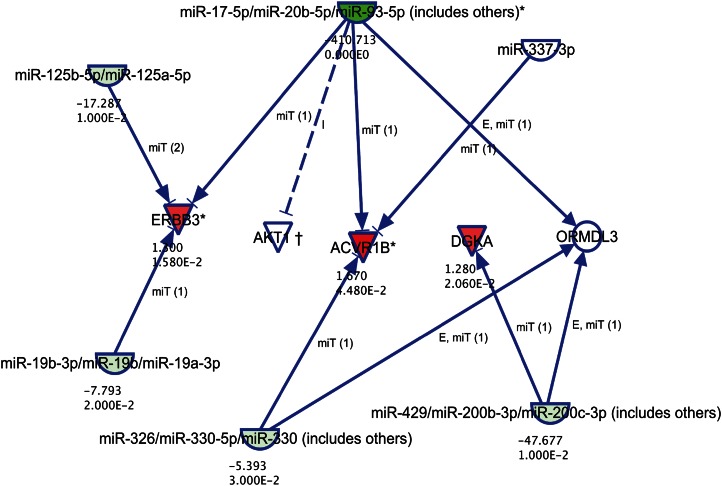
To further prioritize the seven discordant candidate genes, we identified those targeted by at least one miRNA that exhibited differential response to HCHF in low and high LDL-C baboons (19) and that showed inverse expression between the miRNA and targeted gene. Differential expression of these genes and the corresponding targeting miRNAs are shown in Table 3. For example, TENC1, which is upregulated in response to HCHF diet in low LDL-C baboon livers, is targeted by miR-34 family that is downregulated in response to the diet challenge. A miRNA-mRNA interaction module for high LDL-C baboon responders is shown in Fig. 2. ERBB3 and ACVR1B, which are upregulated in response to HCHF diet in high LDL-C baboon livers, are targeted by two or more miRNAs that are downregulated in response to the diet challenge. The miR-17 family, which is downregulated by 410-fold in response to the diet challenge, is predicted to target both ACVR1B and ERBB3.
TABLE 3.
Candidate genes and targeting miRNAs: differential expression in response to HCHF diet
| Candidate Gene | Fold Change | P | Differentially Expressed Liver miRNA Targeting Candidate Gene | Fold Changea | Pa |
| TENC1 | 1.40 | 0.04 | miR-34b | −1.10 | 0.00 |
| miR-377 | −1.58 | 0.04 | |||
| ACVR1B | 1.67 | 0.04 | miR-34b | −1.10 | 0.00 |
| miR-93 | −410.71 | 0.00 | |||
| miR-326 | −5.39 | 0.03 | |||
| DGKA | 1.28 | 0.004 | miR-200b | −47.67 | 0.01 |
| ERBB3 | 1.30 | 0.01 | miR-125b | −17.28 | 0.01 |
| miR-19b | −7.79 | 0.02 | |||
| miR-93 | −410.71 | 0.00 |
Taken from Ref. 48.
Fig. 2.
miRNA-mRNA interaction module in high-LDL-C responder baboons. Shown are candidate genes and miRNAs differentially expressed in response to HCHF diet challenge. Genes and miRNAs are represented as nodes. Red nodes indicate upregulated genes, and green nodes denote downregulated miRNAs. The intensity of the color indicates the degree of differential expression. Numerals below each colored node represent expression fold-change and P-values. The molecular relationship between nodes is represented as a line (edge); arrows indicate the direction of interaction.
Validation of gene expression by QRT-PCR
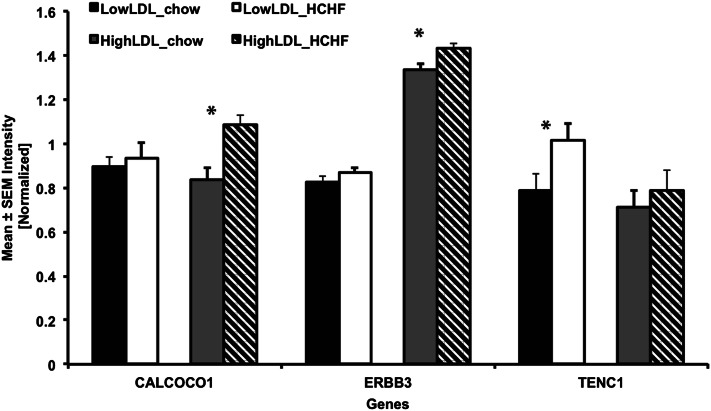
Gene array expression was confirmed by QRT-PCR (Fig. 3). TENC1 was upregulated in low LDL-C responders in response to the HCHF diet (P < 0.05) with no change in high LDL-C responders. CALCOCO1 and ERBB3 were upregulated in high LDL-C responders (P < 0.05) with no change in low LDL-C responders.
Fig. 3.
Validation of gene expression in response to HCHF diet. The x axis shows gene IDs, and the y axis denotes gene expression normalized to 18S rRNA. Bars represent standard errors. *P ≤ 0.05.
Quantification of protein expression
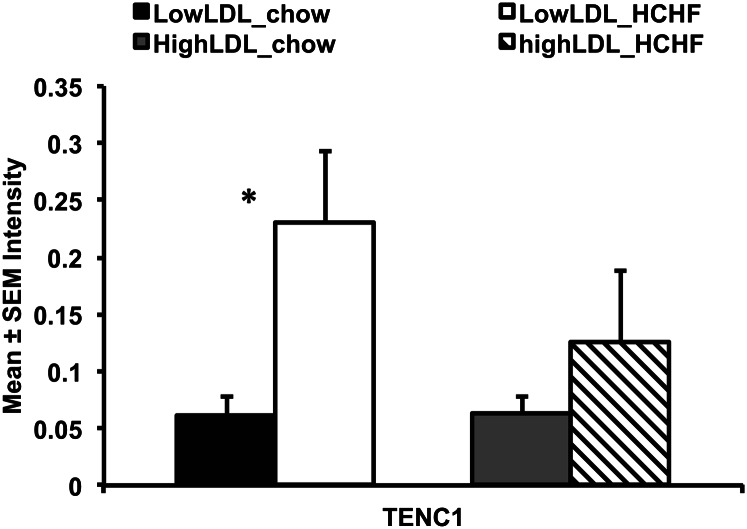
TENC1 protein expression was upregulated in low LDL-C responders in response to the HCHF diet but not in high LDL-C responders (Fig. 4). Baboon ACVR1B, DGKA, CALCOCO1, and ERBB3 proteins were not detected using commercially available antibodies.
Fig. 4.
Quantification of TENC1 protein expressed in response to HCHF diet in low and high LDL-C baboon livers. The y axis denotes protein expression normalized to β-actin; bars represent standard errors. *P ≤ 0.05.
Candidate genes and pathways
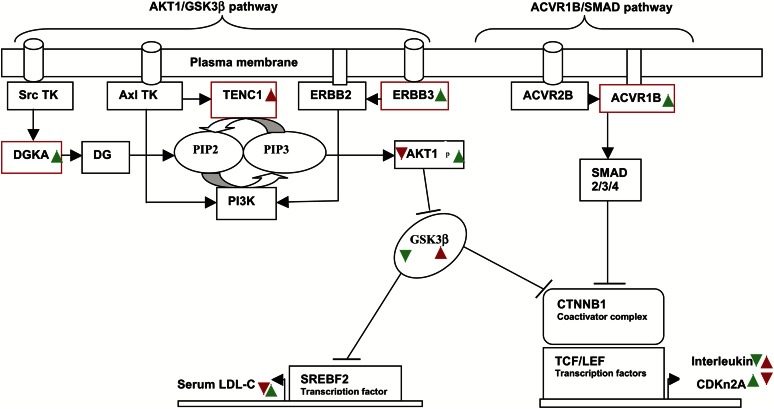
ERBB3, DGKA, TENC1, and ACVR1B are involved in AKT1/GSK3β and ACVR/SMAD pathways, respectively (Fig. 5).
Fig. 5.
Schematic representation of AKT1/GSK-3β and ACVR1B/SMAD signaling pathways regulation in response to HCHF diet. Major genes in the pathways are shown. Red boxes represent candidate genes in the LDL-C QTL interval. RTK, receptor tyrosine kinase; TENC1, tensin-like C1 domain containing phosphatase; DGKA, diacylglycerol kinase α; DG, diacylglycerol; ERBB2/3, epidermal growth factor receptor 2/3; PIP2/3, phosphatidylinositol bi/triphosphate; PI3K, phosphoinositide 3-kinase; AKT1, protein kinase B α; GSK3β, glycogen synthase kinase 3-β; ACVR1B/2B, activin A receptor, type IB/IIB; SMAD, mothers against decapentaplegic; CTNNB1, β-catennin; TCF/LEF, T-cell-specific/lymphoid enhancer factor; CDKN2A, cyclin-dependent kinase inhibitor 2A; SREBP2, sterol regulatory element-binding transcription factor 2. Red arrows indicate gene expression response to the HCHF diet in low LDL-C responders, and green arrows indicate gene expression response to the HCHF diet in high LDL-C responders.
DISCUSSION
Previously we reported a cluster of multiple LDL-C-related QTLs on baboon chr 11, including one QTL influencing variation in LDL-C serum concentration (LOD = 12) (13). In the human genome, 229 genes are encoded within the homologous baboon 2-LOD support interval for the LDL-C concentration QTL (UCSC Genome Browser, Feb. 2009, GRCH37/hg19). In this study, we used hepatic whole-genome expression profiling of half-sib baboons discordant for serum LDL-C and genotypes of markers within baboon chr 11 LDL-C concentration QTL to discover candidate genes encoding the LDL-C serum concentration QTL. This approach augmented detection of genes influencing LDL-C variation by maximizing genetic differences in the QTL interval and minimizing genetic background.
We identified 9,945 genes expressed in the baboon liver transcriptome. Among these genes, 146 were differentially expressed in low LDL-C baboons in response to the HCHF diet and 213 were differentially expressed in high LDL-C baboons in response to the HCHF diet. The significant difference in number of diet-responsive genes in low LDL-C responders compared with high LDL-C responders suggests that different molecular mechanisms underlie the HCHF response between the two phenotypes.
We hypothesized that the candidate gene(s) encoding the LDL-C concentration QTL would be discordantly expressed between low and high LDL-C responders. Within the 2-LOD support interval of the LDL-C QTL, 117 of the 229 genes were expressed, of which 9 were differentially expressed in response to HCHF diet in at least one of the two LDL-C responder groups. Seven of the 9 genes were discordant between low and high LDL-C responders and therefore were candidates encoding the LDL-C concentration QTL. Notably, Sp1 transcription factor (SP1), apolipoprotein F (APOF), sterol O-acyltransferase 2 (SOAT2), and LDL receptor-related protein 1 (LRP1), implicated in lipoprotein metabolism (20–24), were not differentially expressed in response to the HCHF diet challenge in our study. Our findings suggest we have identified novel candidate genes involved in lipid metabolism-related pathways. Moreover, the identification of multiple related LDL-C trait QTLs overlapping the LDL-C concentration QTL suggests that candidate genes underlying LDL-C concentration may be pleiotropic and play a role in networks related to lipid metabolism.
We further hypothesized that the candidate genes were regulated by miRNAs. miRNAs are small, endogenous, noncoding RNAs that posttranscriptionally regulate gene expression. Dysregulation of miRNAs has been shown to result in diseases, including cancer and atherosclerosis (25, 26). Thus, we reasoned that candidate genes targeted by differentially expressed miRNAs in response to the diet challenge might be vital in providing insights on miRNA-related mechanisms influencing dyslipidemia in baboons. Therefore, we prioritized the LDL-C QTL candidate genes by identifying those with an miRNA target site in which the miRNAs also exhibited differential response to HCHF diet in low and high LDL-C responders. Furthermore, we required inverse expression profiles between gene expression and miRNA expression. Using this strategy, we narrowed the number of candidate genes encoding the LDL-C concentration QTL from 229 to 4 genes: TENC1, ERBB3, DGKA, and ACVR1B. We validated candidate gene expression using QRT-PCR. Further, we validated expression of candidate proteins using Western blot and observed that TENC1 expression is in concordance with mRNA levels. The baboon TENC1 protein localized to approximately 148 kDa on PVDF membrane, similar to the human isoform_1 or isoform_2 protein (27). TENC1 isoforms 1 and 2 have been shown to inhibit cell migration (28, 29). We were not successful in detecting baboon ERBB3, CALCOCOL1, DGKA, or ACVR1B proteins, probably due to sequence differences between baboon and commercially targeted human epitopes.
The resulting miRNA-mRNA interaction module from high LDL-C responders indicates that a single miRNA regulates more than one candidate gene and, conversely, that a single gene is targeted by more than one miRNA. For example, the miR-125, miR-19, and miR-17 families are predicated to target ERBB3, and the miR-17 and miR-326 families regulate ACVR1B. This observation is consistent with previous findings (30, 31) and suggests that these miRNAs are master coordinators of the cellular processes influenced by the candidate genes. Most of the miRNAs in the module have been implicated in cell proliferation-related diseases, such as cancer and chemoresistance. For example, miR-17 is associated with retinoblastoma (32) and miR-125 and miR-19 with cancer chemoresistance (33, 34). To our knowledge, there are no previous publications that implicate these miRNAs in dyslipidemia, suggesting novel targets for therapeutic intervention.
Although a few genes have previously been associated with LDL-C variation, it is plausible that well-coordinated signaling pathways regulate dyslipidemia. The four prioritized candidate genes in our study play central roles in AKT1/GSK3β and ACVR/SMAD pathways. These pathways influence key cell biological processes, including cell growth and proliferation, cholesterol homeostasis, and inflammation. Further, deregulation of these pathways is associated with complex diseases, including various types of cancers (35, 36).
For the AKT1/GSK3β pathway, key molecules include three candidate genes (TENC1, ERBB3, DGKA). The switching of the phosphatidylinositol biphosphate (PIP2) lipid molecule to phosphatidylinositol triphosphate (PIP3) and vice versa through phosphorylation and dephosphorylation, respectively, regulates the pathway. For the high LDL-C responder baboons, DGKA and ERRB3 were upregulated in response to HCHF diet, with no significant change in expression of TENC1. Previous studies indicate that stimulation of Src tyrosine kinase receptor by growth factors influences receptor binding to DGKA, which phosphorylates DG lipid molecule to yield PIP2 (37). To generate AKT1 substrate, PIP3, an ERBB3 receptor, upon stimulation by an epidermal growth factor recruits and forms a heteromeric complex with ERBB2, which possesses intrinsic kinase activity (38). The complex binds and activates phosphoinositide 3-kinase (PI3K), which phosphorylates and converts PIP2 to PIP3. PIP3 activates AKT1, which phosphorylates and inactivates GSK3β (39). Inactivated GSK3β promotes stability and enhances transcription activity of primarily SREBF2 and lipid production (36, 40). SREBF2 is a transcription factor that activates expression of genes involved in lipid and cholesterol synthesis (41). GSK3β inhibits SREBF activity by phosphorylation, triggering targeted ubiquitination and proteosomal degradation (42). In contrast, TENC1 phosphotase, upregulated in low LDL-C baboon responders, dephosphorylates PIP3 to PIP2, inhibiting AKT1 activation (43). Consequently, the ability of GSK3β to inhibit SREBF transcription activity is enhanced, and the result is reduced lipid production and low serum LDL-C levels. The involvement of TENC1 and ERBB3 in the AKT1/GSK3β pathway appears to be agonistic, which is consistent with their discordant expression in low and high LDL-C baboons.
For the ACVR/SMAD pathway, activin is a key stimulator of signal transduction through type I and II activin receptors. ACVR1B (type I receptor) was upregulated in response to diet challenge in high LDL-C responders, whereas there was no significant expression change in low LDL-C animals. Evidence indicates that dietary cholesterol induces hepatic inflammation (44), and our study suggests that the ACVR/SMAD pathway could be involved and modulated by ACVR1B to trigger an anti-inflammatory response. ACVR1B lacks an active tyrosine kinase domain; thus, when bound by activin, the receptor recruits type II activin receptor to activate SMAD2/3 complex, which interacts with SMAD4 to regulate transcription activity of CTNNB1 (β-catennin), a coactivator for TCF/LEP transcription factors that control expression of interleukin cytokines and cell-cycle control genes, such as CDK activators and inhibitors (45). Thus, the ACVR/SMAD pathway may influence a plethora of biological processes depending on cell type and context. ACVR1B is a member of the transforming growth factor-β receptor (TGFR) family of receptors that are master regulators of inflammation, cell proliferation, and differentiation (46). Moreover, both ACVR1B and TGFR are transducers of ACVR/SMAD pathway and are anti-inflammatory, which explains the alleviation of TGFR in atherosclerosis patients (45, 47).
Conclusions
Using whole-genome expression profiling, QRT-PCR and Western blots for expression profile validation, and integration of miRNA and mRNA expression profiles, we identified four candidate genes encoding an LDL-C concentration QTL on baboon chr 11 that may regulate lipid metabolism via the AKT1/GSK3β and ACVR/SMAD pathways. Discordant expression between low and high LDL animals with inverse expression profiles of miRNAs targeting these candidate genes supports their role in LDL-C metabolism. Of the four candidate genes identified in this study, none are reportedly associated with lipid metabolism, suggesting these are novel genes involved in lipid metabolism-related pathways. Taken together, our results suggest that the chr 11 QTL for LDL-C concentration is encoded by multiple genes and that miRNAs regulate these genes. miRNA-mRNA interaction studies are required to validate the network modules. By deciphering the miRNA-mRNA interactions and the mechanisms by which the HCHF diet regulate these miRNAs and mRNAs, we will gain new insights into dyslipidemia. The interconnectedness of the candidate genes underlines potential molecular genetic complexities leading to dyslipidemia and atherosclerosis and suggests that a systems biology approach will be required for disease intervention. For example, alleviation of LDL-C levels may demand control of various checkpoints in the AKT1/GSK3β and ACVR/SMAD pathways, including increasing TENC1 and GSK3β proteins and inactivation of ERBB3 and DGKA. Emerging evidence indicates that biological processes leading to distinct complex diseases are interlinked; thus, candidate gene and/or single miRNA approaches for the development of therapeutic agents are vulnerable to drawbacks due to limited incorporation of information on molecule interactions in the biological system of interest.
Supplementary Material
Acknowledgments
The authors thank Dr. Sassan Hafizi, University of Portsmouth, United Kingdom, for providing an in-house-generated TENC1 polyclonal antibody and lysates from HEK 293 cells expressing TENC1 for experimental control.
Footnotes
Abbreviations:
- chr
- chromosome
- CVD
- cardiovascular disease
- HCHF
- high-cholesterol, high-fat
- HDL-C
- HDL-cholesterol
- LDL-C
- LDL-cholesterol
- LOD
- logarithm of odds
- miRNA
- microRNA
- QTL
- quantitative trait locus
This work was supported by National Institutes of Health Grants P01 HL-028972-27, P01 HL-028972-Supplement, and P51 OD011133. This work was conducted in part in facilities constructed with support from Research Facilities Improvement Program Grant Numbers C06 RR013556 and C06 RR015456 from the National Center for Research Resources (now Office of Research Infrastructure Programs), National Institutes of Health.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of two tables.
REFERENCES
- 1.Ross R. 1993. The pathogenesis of atherosclerosis: a perspective for the 1990s. Nature. 362: 801–809 [DOI] [PubMed] [Google Scholar]
- 2.McGill H. C., Jr, McMahan C. A., Herderick E. E., Tracy R. E., Malcom G. T., Zieske A. W., Strong J. P. 2000. Effects of coronary heart disease risk factors on atherosclerosis of selected regions of the aorta and right coronary artery. PDAY Research Group. Pathobiological Determinants of Atherosclerosis in Youth. Arterioscler. Thromb. Vasc. Biol. 20: 836–845 [DOI] [PubMed] [Google Scholar]
- 3.Kushwaha R. S., McGill H. C., Jr 1998. Diet, plasma lipoproteins and experimental atherosclerosis in baboons (Papio sp.). Hum. Reprod. Update. 4: 420–429 [DOI] [PubMed] [Google Scholar]
- 4.Libby P. 2006. Atherosclerosis: disease biology affecting the coronary vasculature. Am. J. Cardiol. 98: 3Q–9Q [DOI] [PubMed] [Google Scholar]
- 5.Barter P. J., Rye K. A. 1996. Molecular mechanisms of reverse cholesterol transport. Curr. Opin. Lipidol. 7: 82–87 [DOI] [PubMed] [Google Scholar]
- 6.McGill H. C., Jr, McMahan C. A., Kruski A. W., Kelley J. L., Mott G. E. 1981. Responses of serum lipoproteins to dietary cholesterol and type of fat in the baboon. Arteriosclerosis. 1: 337–344 [DOI] [PubMed] [Google Scholar]
- 7.Brown M. S., Goldstein J. L. 1983. Lipoprotein metabolism in the macrophage: implications for cholesterol deposition in atherosclerosis. Annu. Rev. Biochem. 52: 223–261 [DOI] [PubMed] [Google Scholar]
- 8.Rust L. 1999. Tobacco prevention advertising: lessons from the commercial world. Nicotine Tob. Res. 1(Suppl. 1): S81–S89 [DOI] [PubMed] [Google Scholar]
- 9.Willer C. J., Sanna S., Jackson A. U., Scuteri A., Bonnycastle L. L., Clarke R., Heath S. C., Timpson N. J., Najjar S. S., Stringham H. M., et al. 2008. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat. Genet. 40: 161–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rayner K. J., Esau C. C., Hussain F. N., McDaniel A. L., Marshall S. M., van Gils J. M., Ray T. D., Sheedy F. J., Goedeke L., Liu X., et al. 2011. Inhibition of miR-33a/b in non-human primates raises plasma HDL and lowers VLDL triglycerides. Nature. 478: 404–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Konigsberg L. W., Blangero J., Kammerer C. M., Mott G. E. 1991. Mixed model segregation analysis of LDL-C concentration with genotype-covariate interaction. Genet. Epidemiol. 8: 69–80 [DOI] [PubMed] [Google Scholar]
- 12.McGill H. C., Jr, McMahan C. A., Kruski A. W., Mott G. E. 1981. Relationship of lipoprotein cholesterol concentrations to experimental atherosclerosis in baboons. Arteriosclerosis. 1: 3–12 [DOI] [PubMed] [Google Scholar]
- 13.Rainwater D. L., Cox L. A., Rogers J., VandeBerg J. L., Mahaney M. C. 2009. Localization of multiple pleiotropic genes for lipoprotein metabolism in baboons. J. Lipid Res. 50: 1420–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cox L. A., Mahaney M. C., Vandeberg J. L., Rogers J. 2006. A second-generation genetic linkage map of the baboon (Papio hamadryas) genome. Genomics. 88: 274–281 [DOI] [PubMed] [Google Scholar]
- 15.Rogers J., Mahaney M. C., Witte S. M., Nair S., Newman D., Wedel S., Rodriguez L. A., Rice K. S., Slifer S. H., Perelygin A., et al. 2000. A genetic linkage map of the baboon (Papio hamadryas) genome based on human microsatellite polymorphisms. Genomics. 67: 237–247 [DOI] [PubMed] [Google Scholar]
- 16.Kammerer C. M., Rainwater D. L., Cox L. A., Schneider J. L., Mahaney M. C., Rogers J., VandeBerg J. L. 2002. Locus controlling LDL cholesterol response to dietary cholesterol is on baboon homologue of human chromosome 6. Arterioscler. Thromb. Vasc. Biol. 22: 1720–1725 [DOI] [PubMed] [Google Scholar]
- 17.Rainwater D. L., Mitchell B. D., Mahaney M. C., Haffner S. M. 1997. Genetic relationship between measures of HDL phenotypes and insulin concentrations. Arterioscler. Thromb. Vasc. Biol. 17: 3414–3419 [DOI] [PubMed] [Google Scholar]
- 18.Rainwater D. L., Kammerer C. M., Mahaney M. C., Rogers J., Cox L. A., Schneider J. L., VandeBerg J. L. 2003. Localization of genes that control LDL size fractions in baboons. Atherosclerosis. 168: 15–22 [DOI] [PubMed] [Google Scholar]
- 19.Karere G. M., Glenn J. P., VandeBerg J. L., Cox L. A. 2010. Identification of baboon microRNAs expressed in liver and lymphocytes. J. Biomed. Sci. 17: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Llorente-Cortes V., Badimon L. 2005. LDL receptor-related protein and the vascular wall: implications for atherothrombosis. Arterioscler. Thromb. Vasc. Biol. 25: 497–504 [DOI] [PubMed] [Google Scholar]
- 21.Paromov V. M., Morton R. E. 2003. Lipid transfer inhibitor protein defines the participation of high density lipoprotein subfractions in lipid transfer reactions mediated by cholesterol ester transfer protein (CETP). J. Biol. Chem. 278: 40859–40866 [DOI] [PubMed] [Google Scholar]
- 22.Rudel L. L., Davis M., Sawyer J., Shah R., Wallace J. 2002. Primates highly responsive to dietary cholesterol up-regulate hepatic ACAT2, and less responsive primates do not. J. Biol. Chem. 277: 31401–31406 [DOI] [PubMed] [Google Scholar]
- 23.Rudel L. L., Lee R. G., Parini P. 2005. ACAT2 is a target for treatment of coronary heart disease associated with hypercholesterolemia. Arterioscler. Thromb. Vasc. Biol. 25: 1112–1118 [DOI] [PubMed] [Google Scholar]
- 24.Lu S., Archer M. C. 2010. Sp1 coordinately regulates de novo lipogenesis and proliferation in cancer cells. Int. J. Cancer. 126: 416–425 [DOI] [PubMed] [Google Scholar]
- 25.Bonauer A., Carmona G., Iwasaki M., Mione M., Koyanagi M., Fischer A., Burchfield J., Fox H., Doebele C., Ohtani K., et al. 2009. MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science. 324: 1710–1713 [DOI] [PubMed] [Google Scholar]
- 26.Chen L. J., Lim S. H., Yeh Y. T., Lien S. C., Chiu J. J. 2012. Roles of microRNAs in atherosclerosis and restenosis. J. Biomed. Sci. 19: 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hafizi S., Alindri F., Karlsson R., Dahlback B. 2002. Interaction of Axl receptor tyrosine kinase with C1-TEN, a novel C1 domain-containing protein with homology to tensin. Biochem. Biophys. Res. Commun. 299: 793–800 [DOI] [PubMed] [Google Scholar]
- 28.Yam J. W., Ko F. C., Chan C. Y., Yau T. O., Tung E. K., Leung T. H., Jin D. Y., Ng I. O. 2006. Tensin2 variant 3 is associated with aggressive tumor behavior in human hepatocellular carcinoma. Hepatology. 44: 881–890 [DOI] [PubMed] [Google Scholar]
- 29.Hafizi S., Ibraimi F., Dahlback B. 2005. C1-TEN is a negative regulator of the Akt/PKB signal transduction pathway and inhibits cell survival, proliferation, and migration. FASEB J. 19: 971–973 [DOI] [PubMed] [Google Scholar]
- 30.Joung J. G., Hwang K. B., Nam J. W., Kim S. J., Zhang B. T. 2007. Discovery of microRNA-mRNA modules via population-based probabilistic learning. Bioinformatics. 23: 1141–1147 [DOI] [PubMed] [Google Scholar]
- 31.Peng X., Li Y., Walters K. A., Rosenzweig E. R., Lederer S. L., Aicher L. D., Proll S., Katze M. G. 2009. Computational identification of hepatitis C virus associated microRNA-mRNA regulatory modules in human livers. BMC Genomics. 10: 373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Conkrite K., Sundby M., Mukai S., Thomson J. M., Mu D., Hammond S. M., MacPherson D. 2011. miR-17∼92 cooperates with RB pathway mutations to promote retinoblastoma. Genes Dev. 25: 1734–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liang Z., Li Y., Huang K., Wagar N., Shim H. 2011. Regulation of miR-19 to breast cancer chemoresistance through targeting PTEN. Pharm. Res. 28: 3091–3100 [DOI] [PubMed] [Google Scholar]
- 34.Shaham L., Binder V., Gefen N., Borkhardt A., Izraeli S. 2012. MiR-125 in normal and malignant hematopoiesis. Leukemia. 26: 2011–2018 [DOI] [PubMed] [Google Scholar]
- 35.Bellacosa A., Kumar C. C., Di Cristofano A., Testa J. R. 2005. Activation of AKT kinases in cancer: implications for therapeutic targeting. Adv. Cancer Res. 94: 29–86 [DOI] [PubMed] [Google Scholar]
- 36.Sundqvist A., Bengoechea-Alonso M. T., Ye X., Lukiyanchuk V., Jin J., Harper J. W., Ericsson J. 2005. Control of lipid metabolism by phosphorylation-dependent degradation of the SREBP family of transcription factors by SCF(Fbw7). Cell Metab. 1: 379–391 [DOI] [PubMed] [Google Scholar]
- 37.Flores G., Wood G. K., Liang J. J., Quirion R., Srivastava L. K. 1996. Enhanced amphetamine sensitivity and increased expression of dopamine D2 receptors in postpubertal rats after neonatal excitotoxic lesions of the medial prefrontal cortex. J. Neurosci. 16: 7366–7375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sithanandam G., Smith G. T., Masuda A., Takahashi T., Anderson L. M., Fornwald L. W. 2003. Cell cycle activation in lung adenocarcinoma cells by the ErbB3/phosphatidylinositol 3-kinase/Akt pathway. Carcinogenesis. 24: 1581–1592 [DOI] [PubMed] [Google Scholar]
- 39.Cross D. A., Alessi D. R., Cohen P., Andjelkovich M., Hemmings B. A. 1995. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature. 378: 785–789 [DOI] [PubMed] [Google Scholar]
- 40.Manning B. D., Cantley L. C. 2007. AKT/PKB signaling: navigating downstream. Cell. 129: 1261–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fernandez-Hernando C., Moore K. J. 2011. MicroRNA modulation of cholesterol homeostasis. Arterioscler. Thromb. Vasc. Biol. 31: 2378–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sundqvist A., Ericsson J. 2003. Transcription-dependent degradation controls the stability of the SREBP family of transcription factors. Proc. Natl. Acad. Sci. USA. 100: 13833–13838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hafizi S., Gustafsson A., Oslakovic C., Idevall-Hagren O., Tengholm A., Sperandio O., Villoutreix B. O., Dahlback B. 2010. Tensin2 reduces intracellular phosphatidylinositol 3,4,5-trisphosphate levels at the plasma membrane. Biochem. Biophys. Res. Commun. 399: 396–401 [DOI] [PubMed] [Google Scholar]
- 44.Kleemann R., Verschuren L., van Erk M. J., Nikolsky Y., Cnubben N. H., Verheij E. R., Smilde A. K., Hendriks H. F., Zadelaar S., Smith G. J., et al. 2007. Atherosclerosis and liver inflammation induced by increased dietary cholesterol intake: a combined transcriptomics and metabolomics analysis. Genome Biol. 8: R200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Toma I., McCaffrey T. A. 2012. Transforming growth factor-beta and atherosclerosis: interwoven atherogenic and atheroprotective aspects. Cell Tissue Res. 347: 155–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feng X. H., Derynck R. 2005. Specificity and versatility in tgf-beta signaling through Smads. Annu. Rev. Cell Dev. Biol. 21: 659–693 [DOI] [PubMed] [Google Scholar]
- 47.Wang J., Luo Z. G. 2008. The role of Wnt/beta-catenin signaling in postsynaptic differentiation. Commun. Integr. Biol. 1: 158–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karere G. M., Glenn J. P., Vandeberg J. L., Cox L. A. 2012. Differential microRNA response to a high-cholesterol, high-fat diet in livers of low and high LDL-C baboons. BMC Genomics. 13: 320 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.