Abstract
Lipidome profile of fluids and tissues is a growing field as the role of lipids as signaling molecules is increasingly understood, relying on an effective and representative extraction of the lipids present. A number of solvent systems suitable for lipid extraction are commonly in use, though no comprehensive investigation of their effectiveness across multiple lipid classes has been carried out. To address this, human LDL from normolipidemic volunteers was used to evaluate five different solvent extraction protocols [Folch, Bligh and Dyer, acidified Bligh and Dyer, methanol (MeOH)-tert-butyl methyl ether (TBME), and hexane-isopropanol] and the extracted lipids were analyzed by LC-MS in a high-resolution instrument equipped with polarity switching. Overall, more than 350 different lipid species from 19 lipid subclasses were identified. Solvent composition had a small effect on the extraction of predominant lipid classes (triacylglycerides, cholesterol esters, and phosphatidylcholines). In contrast, extraction of less abundant lipids (phosphatidylinositols, lyso-lipids, ceramides, and cholesterol sulfates) was greatly influenced by the solvent system used. Overall, the Folch method was most effective for the extraction of a broad range of lipid classes in LDL, although the hexane-isopropanol method was best for apolar lipids and the MeOH-TBME method was suitable for lactosyl ceramides.
Keywords: lipidomics, orbitrap, dual polarity, polarity switching, ANOVA simultaneous component analysis, liquid-liquid extraction
In recent years it has become clear that lipids and phospholipids have a plethora of bioactivities and signaling functions, in addition to structural roles (1); consequently lipidomic studies have become fundamental for understanding their contribution to health and disease. Mass spectrometry (MS), with its capability of providing structural information, has been the main method of choice in lipidomic studies. Recent technological advances in mass spectrometers, including increased sensitivity, higher mass accuracy, higher scan speeds, and the ability to acquire in both positive and negative mode in one run have resulted in the increased popularity of MS as a detection technique for biomolecules in recent years. This has enabled the mapping of lipids present in fluids and cells, leading to better understanding of the role of the different lipid classes in the pathophysiology of diseases.
A critical step in lipidomic analysis is lipid extraction with an appropriate organic solvent mixture (solvent system) prior to MS detection. The solvent system should be capable of effectively extracting lipids representative of the sample under study without bias, inducing or promoting the degradation of lipids, or introducing contamination by nonlipid components such as sugars, peptides, and amino acids. Therefore, the success in the identification and profiling of lipids is critically dependent on the efficiency of the extraction step. The performance of the lipid extraction for a given sample (tissue, cell, or fluid) with a particular solvent system depends on the partitioning of the different lipids into the organic phase used and consequently on the sample lipid composition. In the particular case of biological fluids, the differences in reported lipid compositions may result from the variety of solvent systems that have been used, though the most commonly used solvent systems for lipid profiling are undoubtedly the ones described over 50 years ago by Folch, Lees, and Sloane Stanley (2) and Bligh and Dyer (3). These involve the partition of lipids into a binary mixture of chloroform and methanol (MeOH) (in different ratios), where MeOH is added to the aqueous phase to disrupt the hydrogen bonding networks or electrostatic forces between lipids and proteins followed by the addition of nonpolar solvents such as chloroform to allow the formation of a two-phase system. Even though the methods were initially developed for the extraction of fat from animal tissues (3), the simplicity and efficiency of the solvent systems in extracting a broad range of lipid classes soon made them popular. Currently, these solvent systems are routinely applied for a wide range of biological fluid samples such as blood, tears (4, 5), urine (6, 7), saliva (8), cerebrospinal fluid (9, 10), human milk (11), bronchoalveolar lavage fluid (12, 13), and sperm (14). Over the years some modifications to the original solvent system composition have been introduced, tested, and evaluated; although these alterations are not always fully described by the authors, which makes comparison of the lipid content and composition of fluids in similar pathological conditions somewhat difficult. A comparison of the Folch method and the Bligh and Dyer method has shown similar recoveries in the extraction of the predominant phospholipid classes (15), and similar conclusions were reported in other studies comparing the Folch method with alternative extraction solvent systems (16–18). Other solvent mixtures that include, for example, hexane (19, 20), butanol (18, 21), ethyl acetate (21), and more recently tert-butyl methyl ether (TBME) (17, 22–24) are also described in the literature. TBME, which was reported to give comparable results to the Bligh and Dyer method in the profile of human plasma (16), has become particularly popular for sphingolipidomic studies and the extraction of lipids in fluids (21, 23). Generally, the alternative solvent systems were not reported to exhibit significant differences in the extraction of predominant lipid classes when compared with the “traditional” solvent systems (15–18). Rather, the advantage of these extraction systems lies in the avoidance of chlorinated solvents, which are environmentally harmful. Despite these limited pairwise comparisons, no broader analysis of the effects of solvents in the extraction of lipids in fluids (blood plasma) has been conducted. Moreover, the previous studies tended to focus on major phospholipid classes such as phosphatidylcholines (PCs) and phosphatidylethanolamines (PEs), and have overlooked the influence of solvent composition on extractability of less abundant lipid classes.
LDL is a much studied and important component of plasma, and various solvent systems can be employed for the profiling of its lipids; but as yet no evaluation of the extractability of the different lipid classes in LDL has been conducted. We have addressed this by studying the performance of five different solvent systems on the extraction of lipids from LDL particles isolated from pooled plasma of healthy volunteers. LC-MS was used to identify the lipids extracted, with statistical analysis to evaluate differences in the large number of species found.
MATERIALS AND METHODS
Chemicals
All chemicals used were analytical grade or the highest purity commercially available. All organic solvents were of HPLC grade or the highest purity commercially available (Fisher Scientific, Loughborough, UK). TBME was purchased from Sigma (Poole, UK). Nitrogen was purchased from BOC (Worsley, Manchester, UK). Lipid standards [PC 13:0/13:0 and dehydroepiandrosterone sulfate (DHEA)] were purchased from Avanti Polar Lipids (Alabaster, AL). Synthetic lipid compounds were considered as lipid standards when exhibiting an m/z value that did not match any of the ions present in the unspiked sample with the various solvent systems (endogenously present in the sample or exogenously introduced as contaminants by the solvent system).
Blood collection and LDL separation
Blood samples from three healthy normolipidemic female volunteers (age range 26–46 years, BMI <25, plasma cholesterol <250 mg/dl) were collected after overnight (12 h) fasting into EDTA-containing tubes (Vacuette, Greiner Bio-One, Gloucester, UK) after voluntary written consent was obtained. The study was approved by the University of Strathclyde ethics committee. Plasma was separated immediately by low speed centrifugation (2,000 g) at 4°C. Plasma samples were pooled and sterile 60% sucrose (w/v) solution was added as a cryopreservative to a final concentration of 0.6%. Pooled plasma aliquots were stored at −80°C in the dark (up to 4 months) until further use. The LDL population was isolated as described previously (25). LDL was desalted by gel filtration with an Econo-Pac 10 DG column (Bio-Rad, Hemel Hempstead, Hertfordshire, UK) using autoclaved phosphate-buffered saline (150 mM NaCl, 10 mM NaH2PO4, pH 6.0) as eluent.
Protein and cholesterol assay
The LDL protein concentration was determined by a microplate Bradford assay in triplicate as described elsewhere (26). The absorbance was read in a microplate reader (Sunrise Tecan) set at 595 nm and the protein amount in each sample calculated using the standard curve.
LDL cholesterol content (<180 mg/dl) was determined by the CHOD-PAD method using the cholesterol kit supplied by Boehringer-Mannheim (Mannheim, Germany) according to manufacturer's instructions.
SDS-PAGE electrophoresis
The purity of LDL population isolated was confirmed by sodium dodecyl sulfate/polyacrylamide gel electrophoresis (SDS-PAGE) resolved in 5% SDS-PAGE according to Laemmli's method (27). Samples (10 μl) were mixed with Laemmli buffer in 1:1 ratio (v/v) and after heat denaturing loaded into the wells and run at constant voltage. Prestained marker (15 μl) of known molecular weights in the range of 30–460 kDa (HiMArk prestained, Invitrogen, Paisley, UK) was loaded together with the samples. Gels were stained with colloidal Coomassie blue solution followed by destaining with solution containing 40% MeOH and 10% acetic acid. The gel (supplementary Fig. I) shows one main band above the top marker corresponding to ApoB-100 which is the predominant protein (>95%) in LDL population (28).
Lipid extraction protocols
Extraction of LDL lipids was performed in triplicate using 25 μg of protein (46 μl) by the five different solvent systems as described below.
Solvent system 1 [Folch method (2)]: LDL lipids were extracted by addition of 160 μl ice-cold MeOH (containing 50 μg/ml of BHT) followed by the addition of 320 μl of ice-cold CHCl3 and incubation for 20 min on ice with occasional vortex mixing. High purity water (150 μl) was added and the sample kept on ice for an additional 10 min with occasional mixing. The sample was centrifuged for 5 min at 2,000 g and the upper (aqueous) phase was removed and reextracted by addition of 250 μl of ice-cold CHCl3:MeOH (2:1, v/v) as above. The upper phase was discarded and both organic phases were combined, dried under nitrogen stream, and stored at −70°C.
Solvent system 2 [Bligh and Dyer method (3)]: LDL lipids were extracted by addition of 160 μl ice-cold chloroform (CHCl3) together with 320 μl of ice-cold of MeOH containing BHT (50 μg/ml) with occasional vortex mixing for 20 min, followed by 150 μl of water, at which point a biphasic mixture was formed. The sample was centrifuged for 5 min at 2,000 g and the upper (aqueous) phase removed and retained. A second extraction step was performed on the upper aqueous phase and both organic phases were combined and washed with 150 μl water as above. The organic phase was dried under stream of nitrogen and stored at −70°C until MS analysis.
Solvent system 3 [acidified Bligh and Dyer method (29)]: LDL lipids were extracted as described for solvent system 2, except that 2 μl of 3M hydrochloric acid was added after the 150 μl of ice-cold chloroform at the second extraction step for the aqueous phase, and the combined organic phases were washed by addition of 200 μl of water.
Solvent system 4 [MeOH-TBME method described by Matyash et al. (16)]: LDL lipids were extracted by addition of 400 μl ice-cold MeOH containing BHT (50 μg/ml) and incubated for 10 min, followed by addition of 500 μl of TBME and further incubation on ice for 1 h with occasional vortex mixing. After this, 500 μl of water were slowly added to induce phase separation and incubated for 15 min with occasional mixing, followed by centrifugation and collection of the upper organic phase. To the aqueous bottom layer, an additional extraction step was performed with 200 μl of ice-cold TBME and 15 min incubation on ice as above, followed by centrifugation at 2,000 g for 5 min. The organic phases were combined and washed by the addition of 150 μl of water.
Solvent system 5 [hexane:isopropanol method (19, 20)]: LDL lipids were extracted by the addition of 800 μl of ice-cold hexane:2-propanol (3:2, v/v) and left to incubate for 20 min on ice with occasional vortex mixing. The hexane contained BHT (50 μg/ml). The mixture was centrifuged at 2,000 g for 5 min and the upper organic phase removed and retained. The lower aqueous phase was reextracted with 200 μl hexane:MeOH (3:2, v/v) for 15 min as above. After final centrifugation both organic phases were combined in and dried under nitrogen stream.
Analysis of lipid extracts by HPLC-MS
Lipid extracts obtained from each solvent system were solubilized in 100 μl CHCl3:MeOH (1:1, v/v), further diluted in MeOH and the vials placed randomly in the autosampler tray kept at 4°C. Lipids were analyzed in duplicate, totaling six injections per solvent system. Separation of LDL lipid classes was performed using a Dionex Ultimate 3000 HPLC system (Thermo Scientific, Hemel Hempstead) by injection of 10 μl sample onto a silica gel column (150 mm × 3 mm × 3 μm; HiChrom, Reading, UK) used in hydrophilic interaction chromatography (HILIC) mode (30). Two solvents were used: solvent A [20% isopropyl alcohol (IPA) in acetonitrile] and solvent B [20% IPA in ammonium formate (20 mM)]. Elution was achieved using the following gradient at 0.3 ml/min: 0–1 min 8% B, 5 min 9% B, 10 min 20% B, 16 min 25% B, 23 min 35% B, and 26–40 min 8% B. Detection of lipids was performed in a Exactive mass spectrometer (Thermo Fisher Scientific Inc., Hemel Hempstead, UK) equipped with polarity switching. The instrument was calibrated according to the manufacturer's specifications to give an rms mass error <1 ppm. The following electrospray ionization settings were used: source voltage, ±4.50 kV; capillary voltage, 25 V; capillary temp, 275°C; sheath gas flow, 60 arbitrary units (AU); auxiliary gas flow, 25 AU; sweep gas flow, 0 AU. All LC-MS spectra were recorded in the range 100–1,200 at 50,000 resolution (FWHM at m/z 500). Three microscans were collected per data point with the injection time limited by either an automatic gain control target ion density of 106 or a maximum inject time of 250 ms. Tandem mass spectra (LC-MS2) were acquired on an LTQ Orbitrap instrument (ThermoElectron, Hemel Hempstead, UK) controlled by Xcalibur (version 2.0, Thermo Fisher Corporation) in either positive or negative ion modes as appropriate for the best detection of the parent ion. The source voltage was set at 4.5 kV, capillary temperature at 275°C, with sheath gas and auxiliary gas flow rates set at 30 and 10 AU, respectively. Collision energy was set according to the ion of interest, typically between 25 and 35 AU.
Some of the lipid classes were detected in both positive and negative ion mode, namely ceramides (Cer) ([M+H]+/[M+HCOO]−), PEs ([M+H]+/[M-H]−), choline class ([MH]+/[M+HCOO]−), and lipoamino acids (LAAs) ([M+H]+/[M-H]−), and other lipid classes were detected in the negative ion mode as different adducts ([M-H]−)/[M+HCOO]−), namely Cer, lactosyl ceramides (LacCer), and sphingomyelins (SMs). All lipid classes were manually cross-referenced to prevent overestimation of the number of lipid species identified.
Determination of percent recovery
The percent recoveries of two different standards were determined by spiking 15 μl of LDL before extraction with the following amounts: 307.5 pmol PC (13:0/13:0) and 135.8 pmol DHEA sulfate. The lipids were then extracted using the five different solvent systems described above. Lipid extracts of spiked LDL were reconstituted in 100 μl CHCl3/MeOH (1:1, v/v) and further diluted in MeOH prior to injection and separation by LC-MS (using the same column and elution program as described above) using an Ultimate 3000 system (Dionex, Camberley) controlled by Chromeleon Xpress in conjunction with Analyst software (1.5.1; ABSciex, Warrington, UK) coupled to a 5600 TripleTOF (ABSciex). The detection of lipid standards was achieved with source temperature set at 550°C, spray voltage set at ±4.5 kV, and declustering potential set at 50 V. TOF MS survey scans were collected in the mass range of 100–1,200 Da for 250 ms. The extracted ion chromatograms for each of the ions (m/z 650.4953 for 13:0/13:0 PC and m/z 367.1595 for DHEA sulfate) were generated using options available in Analyst software (1.5.1; ABSciex) and areas under the curve were used to estimate the mean recoveries (calculated in triplicate) and the matrix effects on the extraction of lipids using the different solvent systems. Limit of detection (LOD) and limit of quantitation (LOQ) were estimated by the blank method.
Data processing and statistical analysis
Data processing was carried out using a combination of the open-source tools: Proteowizard, XCMS (31, 32), and MZMatchR (33) within the R environment (34). The MSConvert tool from Proteowizard was used to convert the .raw files acquired by the mass spectrometer into .mzXML format. Files were then further processed using centWave (31, 35) from XCMS (https://xcmsonline.scripps.edu) to detect and extract features in centroid mode from individual spectra followed by analysis using the tools available through MZMatchR (alignment; peak matching; RSD filter, 0.5; noise filtering, 0.8; gap-filling; intensity filter, 104; combination of replicates, minimum of three) and conversion to text. The raw mass spectrometric data was also manually interpreted using accurate mass and retention time to identify individual lipids, with reference to an in-house database and the Human Metabolome project (36). These identified lipids were cross matched to peaks that appeared in the MZMatch filtered data to generate data set 1. While more than 350 lipids could be identified from the raw data coming from 19 lipid or lipid-related subclasses, only 169 of these were represented in the MZMatch filtered dataset, representing 12 lipid subclasses. Alternatively, for data set 2 the same .mzXML files were analyzed using the full XCMS analysis pipeline (XCMSOnline version 0.0.83, Scripps Center for Metabolomics, https://xcmsonline.scripps.edu/). Data set 2 corresponds to features extracted in centroid mode by the XCMS processing analysis (peak width between 10 and 60 s; mass tolerance in consecutive runs, 3 ppm; fold change, 1.5 for P < 0.05) appearing within the retention time windows that represented the different lipid classes, as described in Table 1, totaling 1,567 features. Some of the retention time windows (shown in Fig. 1 and given in Table 1) contain overlapped lipid classes that are not distinguished by direct analysis of chromatogram, for example FA coeluted with hexosyl ceramides (HexCer). Hence there are only 10 lipid classes formally represented in data set 2, annotated by the major species identified in the window.
TABLE 1.
List of lipid classes and sub-classes identified in LDL population with dual polarity detection in high-resolution mass spectrometer
| Lipid Classes | Number of Molecular Ions | Search Criteria (Adducts Searched in the Detection Mode) | Retention Times (min) |
| Glycerolipids | 73 | — | — |
| TAGs | 73 | CnHmNO6 ([M+NH4]+) | 2.7–3.4 |
| Sterols and steroids | 18 | — | — |
| Cholesterol | 1 | CnHmO ([M+H-H2O]+) | 2.9–3.0 |
| CSs | 4 | CnHmO4S ([M-H]−) | 1.8–1.9 |
| CEs | 10 | CnHmNO2 ([M+NH4]+) | 2.7–2.9 |
| Steroid conjugates | 3 | CnHmOz ([M+H]+)/([M-H]−) | — |
| FAs and conjugates | 32 | — | — |
| FFAs | 25 | CnHmO2 ([M-H]−) | 3.4–3.6 |
| FA conjugates | 6 | CnHmO3-5 ([M-H]−) | — |
| Eicosanoids | 1 | CnHmO3 ([M-H]−) | — |
| Sphingolipids | 102 | — | — |
| SMs | 41 | CnHmN2O6P ([MH]+) | 22.2–23.1 |
| Cer | 15 | CnHmNO5 ([M+HCOO]−) | 2.7–3.3 |
| HexCer | 11 | CnHmNO8 ([M-H]−) | 2.9–3.7 |
| LacCer | 6 | CnHmNO13 ([M-H]−) | 3.6–4.0 |
| Acidic glycosphingolipids (STs) | 29 | CnHmNO11-13S ([M-H]−) | 1.8–1.9 |
| Glycerophospholipids | 138 | — | — |
| PIs | 21 | CnHmO13P ([M-H]−) | 5.1–6.2 |
| PGs | 6 | CnHmO10P ([M-H]−) | 14.0–14.10 |
| PSs | — | CnHmNO10P ([M-H]−) | — |
| PEs | 48 | — | 12.5–13.4 |
| Diacyl-PEs | 16 | CnHmNO8P ([M-H]−) | — |
| Plasmenyl-PEs | 32 | CnHmNO7P ([M-H]−) | — |
| LysoPEs | 5 | CnHmNO7P ([M-H]−) | 14.9–15.2 |
| PCs | 54 | — | 20.3–21.9 |
| Diacyl-PCs | 34 | CnHmNO8P ([MH]+) | — |
| Plasmenyl/plasmanyl-PCs | 20 | CnHmNO7P ([MH]+) | — |
| LysoPCs | 4 | CnHmNO7P ([MH]+) | 23.4–23.9 |
| Lipid-related compounds | 10 | — | — |
| Prenols | 3 | CnHmOz ([M+H]+)/([M-H]−) | — |
| LAAs | 5 | CnHmNO4S ([M+H]+)/([M-H]−) | 2.2–2.3 |
| Total | 361 | — | — |
The total number of individual lipid species identified and the time windows in which each of the classes were detected in the chromatogram are also indicated.
Fig. 1.
Typical normal-phase LC-MS chromatograms of lipid extract from normolipidemic LDL in positive (+) (top) and negative (−) ion mode (bottom). LDL was extracted using solvent system 3 (acidified Bligh and Dyer method). Dashed boxes represent the elution time windows for lipid classes eluting (in order of appearance): 1, TAGs+CEs; 2, PCs; 3, SMs; 4, lysoPCs; 5, CS; 6, LAAs; 7, Cer+HexCer; 8, FAs+LacCer; 9, PIs; 10, PEs. Inset in the LC-MS chromatogram in negative ion mode depicts zoomed region for the elution of CS, LAAs, Cer and FAs.
Processed LC-MS data from MzMatch R was transferred to an excel data sheet and the ion intensities of the identified lipids (data set 1, 169 features) were used to evaluate the comparative efficiency of extraction (referred to as extractability) of individual lipids and different lipid classes for each of the extraction solvent systems assessed. Data from the full XCMS pipeline (data set 2) was also used to evaluate the extractability of the solvent systems by summing total ion intensities in retention time windows representing different lipid classes, as indicated in Table 1. Statistical analysis on the extractability between the different solvent systems was performed using ANOVA simultaneous component analysis (ASCA); this method merges ANOVA and principal component analysis, and was developed specifically for analyzing large instrumental data sets with underlying experimental design measured on a relatively small number of samples (37, 38). A detailed description of the ASCA model used can be found in the supplementary data online The significance of the effect of the different solvent systems was assessed using a permutation test (39–41), and ASCA with permutation test was run on data sets 1 and 2. The ASCA algorithm and permutation test (38) were implemented in MATLAB, v5.3. After significance testing of the effect of the solvent systems, post hoc comparison of the pairs of methods was performed using a P value of 0.05. The sum of the peak intensities for the ions in individual lipid classes was used as the criterion for selection of the optimal extraction protocol. The 10 lipid classes included in these analyses are representative of lipids as a whole, as they include lipids with a broad range of structures and physicochemical properties.
RESULTS
Lipid profiling of LDL particles
Figure 1 shows normal phase chromatograms for the elution of lipids extracted from LDL by the Folch method (2), with detection by both positive and negative ionization electrospray mass spectrometry. As polarity switching was used, the chromatograms are directly comparable in terms of the species eluted at each time. It can be seen that the majority of lipids eluted in the first 25 min, and some classes of lipid could be detected in both positive and negative ion modes.
The averaged LC-MS mass spectra from the various time windows of the chromatogram corresponding to the elution of different lipid classes are shown in Fig. 2, showing LC-MS spectra in positive ion mode for triacylglycerides (TAGs), cholesteryl esters (CEs), PCs, SMs, and lyso-phosphatidylcholines (lysoPCs), and the LC-MS spectra in negative ion mode for Cer, sulfatides (STs), free fatty acids (FFAs) and conjugates, LacCer, phosphatidylinositols (PIs), PEs, and LAAs. Cholesterol, cholesterol sulfate (CS), HexCer, lyso-phosphatidylethanolamines (lysoPEs), glycero-phosphatidylglycerols (PGs), and other lipid-like molecules, such as steroid conjugates and prenols, were also identified in negative ion mode (data not shown). Structures of the predominant and other lipid classes are depicted in supplementary Fig. II. The mass spectra (Fig. 2) show a large number of individual lipid species present within each lipid class. The nature of lipids and adducts identified in the mass spectra was confirmed by using specific formula search criteria, as summarized in Table 1, based on matching the data from accurate mass analysis to mass prediction for the formulas and retention time data. Table 1 also shows the number of individual lipid species identified in each of the classes, and it can be seen that glycerophospholipids are the class with the largest number of species, closely followed by sphingolipids, and in total more than 350 distinct molecular species were identified. The full list of lipid identifications obtained from accurate mass data and prediction software is shown in supplementary Table I.
Fig. 2.
Average LC-MS mass spectra of 12 classes of the 19 individual classes identified in LDL population. LC-MS spectra of lipid extracts were acquired in a high-resolution instrument (orbitrap analyzer) equipped with simultaneous detection in both positive (+) and negative (−) ion modes. Lipids classes were extracted using solvent system 3 (acidified Bligh and Dyer) with TAGs, CEs, PCs, SMs, and lysoPC detected in positive ion mode and with Cer, STs, FAs, LacCer, PIs, PEs, and LAAs, detected in the negative ion mode.
The TAG and CE classes were detected in the positive ion mode as [M+NH4]+ adducts, while PC, SM, and lysoPC classes occurred as [MH]+ molecular ions. Lipids from the choline class were also detected in the negative ion mode as formate adducts, [M+HCOO]−, as was the case of SMs, which could additionally be observed as deprotonated molecular ions [M-H]−. Attention was taken to ensure that species that ionized in several forms were only counted once in terms of the number of lipid species observed. In positive ion mode it was noted that there was a predominance of linoleoyl-containing lipid species in triacylglycerols, CEs, and choline class (TAG m/z 876.61, tri-linoleoyl; CE m/z 666.62, linoleoyl; PC m/z 758.56, palmitoyl-linoleoyl; and 786.56, stearoyl-linoleoyl) in LDL.
Lipid classes profiled in the negative ion mode (Cer, CS, STs, FAs, HexCer, LacCer, PIs, PEs, PGs, and LAAs) ionized either as deprotonated molecules [M-H]− or as formate adducts [M+HCOO]−. HexCer and LacCer were composed of the d16:1 chain and the d18:1 chain (supplementary Table I) while the PI class was mostly composed of the arachidonoyl-containing PI (18:0/20:4, m/z 885.55). Other anionic phospholipids such as the glycero-phosphatidylserine (PS) class were not found. Sulfur-containing compounds, such as CSs and STs, were detected in negative mode with the predominant ST species corresponding to the oleoyl hydroxylated chain (d18:1/16:0, m/z 794.51) and nonhydroxylated (d18:1/16:0, m/z 778.51), though the hydroxylated d24:1/16:0 (m/z 904.62) was also observed (supplementary Table I). Other lipid-like molecules were also identified, namely prenols (lipophilic antioxidants), secosteroids (vitamins), and steroid derivatives (hormones), and are summarized in supplementary Table I.
While the HILIC separation step coupled to high-resolution instrument allowed differentiation of molecular lipid species with very similar molecular mass but different molecular formulas (isobaric species), it should be noted that it is often not possible to discriminate isomeric species (the same molecular formula) within each of the lipid classes, for example due to variability of carbon chain length between the esterified FAs. Hence the structural variability in lipids isolated from LDL is likely to be even greater than indicated by Table 1 and supplementary Table I. The occurrence of isomeric species is shown in supplementary Fig. III for a 36:3 PC and a 36:4 PE species. Fragmentation of the PC ion at m/z 784.6 in positive ion mode clearly shows the presence of two species (16:0/20:3 and 18:1/18:2) from the four different pairs of fragments corresponding to loss of the fatty acyl chains, whereas the fragmentation of the PE ion at m/z 738.6 shows the presence of different carboxylate ions (m/z 255.3, 279.3, and 303.3), indicating the contribution of isomeric structures.
Influence of solvent systems on the extraction of LDL lipids
Based on the lipids identified in LDL, we conducted an evaluation of the extractability of each of the classes present in LDL using 5 different solvent systems. Supplementary Fig. IV depicts the chromatograms corresponding to the replicate injections for one solvent system (acidified Bligh and Dyer) in positive (supplementary Fig. IVA) and negative ion mode (supplementary Fig. IVB) showing reproducibility within the injections. Supplementary Fig. IV also depicts the chromatograms obtained in one injection for each of the five solvent systems tested in positive (supplementary Fig. IVC) and negative ion mode (supplementary Fig. IVD). The different colors and line forms in the chromatograms give a pictorial representation of the areas of the chromatograms showing that significant differences could be seen in some peak intensities between the different extraction methods (supplementary Fig. IV). In order to characterize the effectiveness of different extraction protocols for LDL lipids, LC-MS data were processed using two different approaches, as described in the Materials and Methods section. Data set 1 contains 169 ions from 10 lipid classes that passed the filters during processing using MzMatch (peak matching, noise filtering, and gap filling) and were evaluated for their extractability in the five solvent systems. While this is not an exhaustive assessment of all the different lipid species identified, each class of lipids is represented by the predominant ions, as shown in Fig. 2, and therefore the data is likely to be representative of the behavior for the whole class. Lipid classes and ions not included corresponded to single ions in the lipid class (LAAs) and identified lipid classes (STs, HexCer, lysoPEs, and glycero-PGs) excluded by the processing filters due to low relative abundance.
Data set 2 was generated by a second approach using the features extracted using the full XCMS platform, and included all ions identified as features in the delimited time windows (as shown in Fig. 1) corresponding to the retention times of lipid classes. This approach ensured that compounds with low relative abundance or those not identified as lipids based on their accurate mass, and thus excluded from data set 1, would be taken into account in the analysis. The disadvantage of this approach is the potential inclusion of organic-extractable ions that are not lipid related.
For both data sets, the intensities of all of the individual ions (169 for data set 1 and 1,567 for data set 2) were summed either within the retention time windows relating to individual lipid classes to obtain data for the individual classes, or for all of the classes summed across the whole chromatogram, to provide an indication of the extractability of lipids in the different solvent systems. The total lipids in each class and total lipids were used as optimization criteria. Ion intensities or relative abundances cannot be used for the estimation of the absolute molar abundances of the different lipid classes due to the differences in ionization efficiency between the different lipid classes and between different lipids within the same lipid class (42, 43). However, comparisons of ion intensities for the same lipid between samples can be employed in this study for comparing different solvent systems, as the same volume of sample was used for all extraction protocols, the same dilution factor was applied before injection, and the same MS detection parameters were used, and therefore peak intensities should be comparable and related to amount of lipids extracted using the different solvent systems. The following two sections discuss the findings using each of the data sets.
Evaluation of lipid extractability using data set 1 (data from MzMatch in R platform)
Figure 3 shows the effect of the extraction protocols on the total lipid (across the whole chromatogram) for all identified lipids in data set 1. It can be seen that solvent systems 1 (Folch method) and 3 (acidified Bligh and Dyer method) show higher extractability for total lipid than the alternative extraction protocols. Solvent systems 2 (Bligh and Dyer method) and 4 (MeOH-TBME method) exhibited similar extractability while solvent system 5 (hexane-isopropanol method) extracted the lowest overall total amount of lipid.
Fig. 3.
Evaluation of extractability of total lipids in five different solvent systems using data set 1. Data set 1 consists of 169 lipid species identified in five different solvent systems following processing of LC-MS data through MzMatch R, as described in the Materials and Methods. Graph depicts intensity in counts per second (cps); bars represent standard deviation, ±SD.
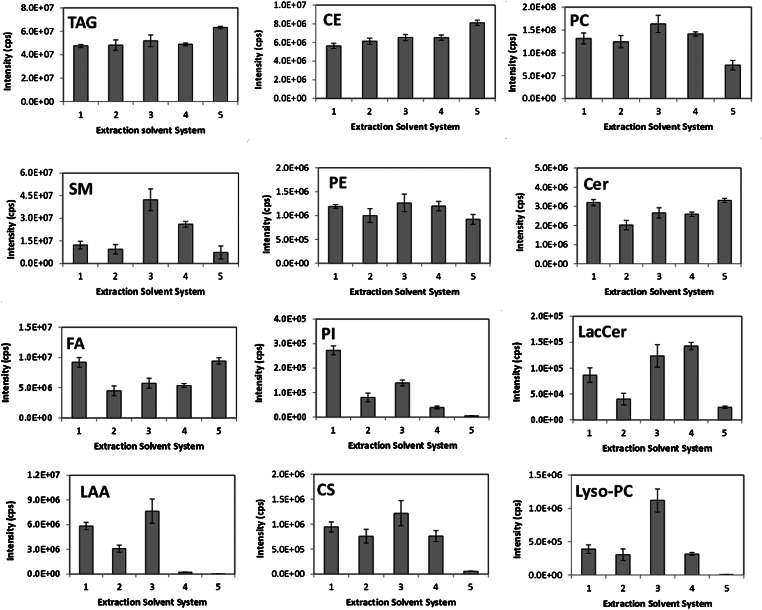
To ascertain whether the different solvent extraction protocols had any effect on the individual classes, the intensities for the identified lipids in individual lipid classes were summed and used to evaluate the various solvent systems (Fig. 4). Small differences in the extractability of the predominant lipid classes in LDL were observed, namely for PCs, CEs, and TAGs, with substantial changes observed in the extractability of minor lipid classes. Methods 1 (Folch method) and 2 (Bligh and Dyer method) show similar extractability for PCs, CEs, and TAGs, whereas method 2 shows lower extractability for all other lipid classes compared with the method 1, particularly for FFAs, PIs, LacCer, and Cer. Interestingly, the acidified Bligh and Dyer method (method 3) resulted in the increased extractability of SMs, lysoPCs, CSs, LAAs, and LacCer compared with extraction method 2 (Fig. 4). The higher apparent extractability of lysoPCs for method 3 is unlikely to be due to the acidic solution causing degradation of acid-labile bonds especially in ester phospholipids, as the intensity of FA is similar to that observed for method 2. Method 4 (MeOH-TBME method) appeared to be particularly suitable for the extraction of sphingolipids, as it resulted in nearly a two-fold increase in SMs and LacCer when compared with the more popular methods 1 and 2 (Fig. 4). Method 5 (hexane-isopropanol) was particularly suitable for the extraction of FFAs, TAGs, and CEs, probably because of their greater hydrophobicity and hence solubility in this apolar solvent system. However, method 5 was found to be inappropriate for the extraction of more polar lipid classes namely lysoPC, PI, CS, and LAA classes (Fig. 4). The differences observed in the lipid classes caused by the effect of the different solvent extraction systems were found to be significant for all lipid classes according to the permutation test (Table 2).
Fig. 4.
Evaluation of extractability of individual lipid classes using data set 1. Data set 1 consists of 169 lipid species identified in five different solvent systems, as described in Materials and Methods, following processing of LC-MS data through MzMatch R. Graphs depict intensity in counts per second (cps) for individual classes in five solvent system; bars represent standard deviation, ±SD.
TABLE 2.
Significance (P values) of the effect of solvent systems on the extraction of lipids calculated using data set 1 (identified compounds)
| Solvent Systems | Lipid Classes | ||||||||||
| All | CSs | TAGs | CEs | FAs | LacCer | PIs | PEs | PCs | SMs | LysoPCs | |
| All five | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | 0.045 | <0.001 | <0.001 | <0.001 |
| 1 versus 2 | 0.003 | 0.087 | 0.002 | 0.006 | 0.001 | <0.001 | <0.001 | 0.062 | 0.353 | 0.138 | 0.141 |
| 1 versus 3 | 0.002 | 0.088 | 0.395 | 0.026 | 0.001 | 0.008 | 0.001 | 0.313 | 0.025 | <0.001 | 0.002 |
| 1 versus 4 | <0.001 | 0.061 | 0.082 | <0.001 | <0.001 | <0.001 | 0.001 | 0.436 | 0.074 | 0.002 | 0.018 |
| 1 versus 5 | 0.002 | 0.001 | 0.034 | <0.001 | <0.001 | <0.001 | 0.001 | 0.001 | 0.001 | <0.001 | <0.001 |
| 2 versus 3 | <0.001 | 0.014 | 0.005 | 0.014 | 0.143 | <0.001 | 0.001 | 0.072 | 0.03 | <0.001 | 0.002 |
| 2 versus 4 | 0.003 | 0.484 | 0.003 | 0.006 | 0.005 | <0.001 | 0.002 | 0.701 | 0.092 | 0.001 | 0.154 |
| 2 versus 5 | 0.008 | <0.001 | 0.001 | <0.001 | <0.001 | 0.006 | <0.001 | 0.235 | <0.001 | 0.01 | 0.005 |
| 3 versus 4 | <0.001 | 0.016 | 0.235 | 0.427 | 0.01 | 0.127 | 0.001 | 0.369 | 0.086 | 0.016 | 0.002 |
| 3 versus 5 | 0.0001 | 0.001 | 0.133 | 0.003 | 0.001 | <0.001 | 0.001 | 0.02 | 0.001 | <0.001 | <0.001 |
| 4 versus 5 | 0.003 | 0.002 | 0.205 | <0.001 | 0.001 | <0.001 | 0.001 | 0.006 | <0.001 | <0.001 | 0.002 |
P values were estimated using permutation test with 1,000 permutations. Bold text indicates a statistically significant effect (P < 0.05).
The efficiency of extraction of two synthetic lipid standards [PC (13:0/13:0) and DHEA sulfate] with the five extraction protocols was also tested (supplementary Fig. V). It can be seen that the extraction system had a substantial effect on the recovery of the individual standards, which essentially paralleled the profiles observed for extraction of the corresponding lipid classes from LDL. The recovery of PCs varied from close to 100% for extraction methods 1 and 3 to approximately 50% with method 5, while the highest recoveries were around 80% for DHEA sulfate, and the lowest (for method 5) was less than 20%.
The effect of the different solvent extraction systems on the extractability of lipid is also demonstrated in the ASCA score and loading plots (Fig. 5A and B, respectively). The more polar lipid classes are situated on the right side of the loading plot (Fig. 5B), i.e., PC, PE, CS, SM, LacCer, and lysoPC, and are more effectively extracted using method 3 (filled triangles on the right side of the score plot Fig. 5A), which also gave the most efficient total lipid extraction. Likewise, the more hydrophobic lipid classes situated on the left side of the loading plot (Fig. 5B), such as FA2 and CEs, are more effectively extracted with method 5 (filled diamonds on the left side in Fig. 5A), which also gave the lowest total lipid extraction. Lipid classes positioned in the lower section of the loading plot (Fig. 5B), such as PI and some compounds from FA (FA1), TAG, and CE classes, are more effectively extracted with method 1 (filled circles in Fig. 5A). Interestingly, some lipid classes appear to display heterogeneous behavior with respect to their extraction efficiency with the different methods, shown for example by three distinct clusters for FAs (filled triangles, delimited by dashed line) and two distinct clusters for CEs (squares, delimited by solid line) on the loading plot (Fig. 5B). Examination of the mass spectrometric data indicates that saturated FAs, clustered in the region labelled FA1, are better extracted using solvent system 3, monounsaturated FAs clustered in FA2 are better extracted using solvent system 5, and polyunsaturated FAs, clustered in FA3 are best extracted using solvent system 3.
Fig. 5.
Principal component analysis plots for lipids in five solvent systems for data set 1 using ASCA model. A: Principal component analysis score plot of total lipids using ASCA model. B: Principal component analysis loading plot for the individual lipid classes in the five solvent systems.
Evaluation of lipid extractability using data set 2 (data from XCMS platform)
As can be seen from supplementary Fig. III, the extraction protocol influenced the extractability of the different lipids noticed for each individual chromatographic region. The ion intensities of lipid species in the individual chromatographic region were summed (representing data set 2). This data set contains all features identified by XCMS platform in each chromatographic window regardless of whether they could be identified as lipids. The inclusion of data that were not initially identified as lipids gives a more robust analysis; at the same time the inclusion of ions not specifically identified as lipids or the inclusion of nonlipid species could be misleading. Methods 1 and 3 gave better total extraction (supplementary Fig. VI), and in general the effectiveness of the extraction protocol demonstrated by data set 2 was similar to that of data set 1. However, summed signal intensities for the TAG and CE regions were significantly higher using method 1 suggesting higher extraction of these classes, whereas for the FA region only method 2 showed lower extractability, while in the CS region methods 2 and 5 performed better than indicated by data set 1. Overall, the extractability in between individual lipid regions for the different solvent systems was significantly different except for PE and PC (Table 3). In the ASCA score plot, solvent systems were aligned along the first principal component in order of increasing total sum of the peak areas, from method 5 on the right to method 1 on the left (Fig. 6). It can be seen that method 3 (acidified Bligh and Dyer) showed a high dispersion of points, and therefore was the least reproducible protocol. The corresponding ASCA loading plot is not shown, as it contained 1,567 points and visualization of individual points was difficult.
TABLE 3.
Significance (P values) of the effect of solvent systems on the extraction of lipids calculated using data set 2 (all compounds in the elution zones)
| Solvent Systems | Lipid Classes | ||||||||||
| All | CSs | TAGs | CEs | FAs | LacCer | PIs | PEs | PCs | SMs | LysoPCs | |
| All five | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | 0.422 | 0.508 | <0.001 | <0.001 |
| 1 versus 2 | <0.001 | <0.001 | <0.001 | 0.002 | <0.001 | 0.01 | <0.001 | — | — | 0.294 | 0.672 |
| 1 versus 3 | 0.008 | 0.004 | 0.002 | <0.001 | 0.002 | 0.001 | <0.001 | — | — | <0.001 | 0.004 |
| 1 versus 4 | 0.008 | 0.598 | 0.002 | 0.002 | 0.012 | 0.001 | 0.004 | — | — | <0.001 | 0.442 |
| 1 versus 5 | 0.002 | 0.004 | 0.006 | <0.001 | 0.006 | 0.003 | 0.004 | — | — | 0.002 | 0.004 |
| 2 versus 3 | 0.062 | 0.632 | 0.034 | 0.082 | 0.528 | 0.001 | 0.02 | — | — | 0.002 | <0.001 |
| 2 versus 4 | 0.002 | 0.004 | 0.006 | <0.001 | 0.002 | 0.001 | <0.001 | — | — | <0.001 | 0.688 |
| 2 versus 5 | 0.002 | 0.004 | 0.004 | 0.004 | 0.002 | 0.002 | <0.001 | — | — | 0.014 | <0.001 |
| 3 versus 4 | 0.002 | 0.004 | <0.001 | <0.001 | 0.014 | 0.015 | 0.004 | — | — | 0.032 | 0.002 |
| 3 versus 5 | 0.006 | 0.002 | 0.002 | 0.002 | 0.01 | <0.001 | 0.002 | — | — | 0.002 | 0.002 |
| 4 versus 5 | 0.002 | 0.006 | <0.001 | 0.004 | 0.012 | <0.001 | 0.044 | — | — | 0.002 | 0.004 |
P values were estimated using permutation test with 1,000 permutations. Bold text indicates a statistically significant effect (P < 0.05).
Fig. 6.
Principal component analysis score plot for total lipids in five solvent systems for data set 2 using ASCA model.
DISCUSSION
The aim of this study was to investigate the effectiveness of five popular lipid extraction protocols, namely the Folch (2), Bligh and Dyer (3), acidified Bligh & Dyer (29), MeOH-TBME (16), and hexane-isopropanol (19, 20) methods, designated methods 1 to 5 respectively, for the extraction of lipid classes from LDL. The lipids extracted by each method were analyzed by MS in a high-resolution instrument, which allowed identification based on accurate mass formula prediction. Evaluation of solvent systems and statistical analysis of lipid extractability was assessed by two approaches; data set 1 included only lipid species identified by processing using MzMatch and manual identification, whereas data set 2 included all ions within delimited time windows in the chromatograms. Together, these approaches provided a robust method for determining differences in the profile of extraction by the various protocols, and gave very comparable results. Although several previous studies have reported similar extractability for predominant lipid classes between three of the solvent systems here studied, we found that while the solvent systems were indeed similar for extraction of the predominant lipid classes, there were striking differences in the extraction of minor lipid classes. Based on the difference in extractability of individual lipid classes by the solvent systems tested, the optimum solvent systems for each class can be summarized (Table 4). For instance, method 4 (MeOH-TBME method) is the most suitable for the sphingolipidomic studies, whereas method 5 (hexane-isopropanol) is optimal for the extraction of apolar lipids such as CEs and FAs, though it has limitations as some lipid classes (PIs, LAAs, CS, and lysoPCs) were almost completely absent using this method. Methods 1 (Folch method) and 2 (Bligh and Dyer method) showed similar extractability for PCs, CEs, and TAGs based on total lipid intensities, which is in accordance with previously published reports (15), but for other lipid classes methods 2 and 4 gave lower lipid extractability than methods 1 and 3, apart from for LacCer where method 4 was best. Overall the data suggest that methods 1 and 3, corresponding to the Folch and acidified Bligh and Dyer methods, respectively, are most suitable for broad-based lipidomic studies of LDL, whereas other methods would be best for studies directed at some specific lipid species. While the analysis here has been derived from data on LDL lipid extraction, it seems likely that it would also be useful for selecting the most suitable solvent system for targeted detection of particular lipid classes in other sample types.
TABLE 4.
Evaluation of the most and least effective solvent system for the extraction of lipid classes in LDL
| Lipid Classes | Data Set 1 | Data Set 2 | ||
| Best | Worst | Best | Worst | |
| Total | 3 | 5 | 1 | 2 |
| CSs | 3, 1a | 5 | 3, 2 | 5 |
| TAGs | 1, 3, 4 | 2 | 1 | 2 |
| CEs | 5 (1) | 2 | 1 | 2, 3 |
| FAs | 5 (1) | 2, 3 | 5 (4) | 2, 3 |
| LacCer | 4, 3 | 5 | 4 | 5 |
| PIs | 1 | 5 | 1 | 5 |
| PEs | 3, 1, 2, 4 | 5 | — | — |
| PCs | 3,4 | 5 | — | — |
| SMs | 3 | 5 | 3 | 5 |
| LysoPCs | 3 | 5 | 3 | 5 |
Statistical analysis of data set 1 and data set 2 was performed using the ASCA model with test permutations (details in supplementary data online). Numbers in parentheses indicated the second best solvent system for extraction.
When no statistical difference is observed, all of them are indicated.
While the main aim of this work was to determine the best solvent for lipid extraction from LDL, an inevitable consequence of the study was an extensive catalog of the lipid species present in LDL. The variety of lipid structures identified in the LDL population in one run (Table 1 and supplementary Table I) was substantially increased as a consequence of acquiring data in a high-resolution analyzer operating in both positive and negative ion modes (polarity switching); this also facilitated matching of specific lipid species that occurred in different ionized forms in the two modes and reduced the possibility of double counting. LDL of normolipidemic individuals shows considerable structural variability in its lipid species, with more than 350 individual molecular species from a broad range of lipid subclasses being identified in this study. In fact, the number of individual lipid species present is likely to be higher than suggested by the numbers identified in this study, as the identification is based on the accurate m/z value of the ion, and only the total number of carbons and double bonds can be determined, so isomers with different carbon chain lengths in acyl and in ether-linked phospholipids are not distinguished. Interestingly, glycero-PSs were not identified in the LDL extracts, despite the fact that they have previously been identified in plasma (44–46), suggesting that they could be confined to other lipoproteins. The absence of PSs in LDL may not be surprising given that they are generally only localized in the external lipid leaflet during cellular apoptosis, and their inclusion in the lipid monolayer surrounding the LDL particle would expose them to the plasma.
The roles and localization of predominant lipid classes in LDL such as CEs, PCs, SMs, free cholesterol, and TAGs are known. Choline phospholipids located at the surface of LDL particles provide compartmentalization for CEs and TAGs and support for the ApoB-100 protein. SMs are thought to act as adhesive molecules, interacting favorably with cholesterol and promoting stabilization of the liquid-ordered phase (47–49). The high structural variability of SMs that we observed in LDL, with carbon chains of 33:1; 35:1, 35:0, 37:0; 39:1, 41:0, and 43:0 (supplementary Table I) agree with previously published data (23, 50). We also characterized a number of minor phospholipid classes in the LDL. The PEs observed were predominantly ether-linked PE species, with less than one-third corresponding to acyl-PE, in agreement with previous findings in LDL from normolipidemic individuals (51). In tissues, ether-PEs have been suggested to act as antioxidants by chelating cupric ions and protecting other PLs from oxidation (52, 53), but their function in LDL is as yet unknown. The major PI species detected in this study was 18:0/20:4 PI (m/z 885.5489), which agrees with the PI composition of plasma (44, 45). Glycosylated lipids, such as LacCer and HexCer with d16:1, d18:1, and d18:2 chains, have been reported previously in normolipidemic LDL particles at micromolar levels (50). LDL particles are known carriers of FFAs, which in our study were found to be predominantly stearoyl chains (18:0); these may be involved in the formation and stabilization of the lipid droplet, as recently reported for PLA2-modified LDL particles (54). Lyso-phospholipids are also known components of LDL (23) and plasma (44, 45), and may increase as result of oxidative damage (55).
CS species even though they have been previously reported in LDL (56) and at a submicromolar concentration range in the plasma (57), their localization within the particle remains uncertain. Likewise, other sulfur-containing lipids such as STs, previously observed in mammalian plasma (58, 59) and interestingly found at elevated levels in ovarian tissue cancer (60) but lowered in patients with end-stage renal disease (59), are likely localized at the surface of lipoproteins. The STs identified in this study, together with LacCer and HexCer contribute to the galactose and glucose residues quantified previously in extracts of LDL samples (61). Additionally, we identified LAAs in the lipid extracts (Table 1) that have not previously been reported in LDL particles. Acyl-glycines, acyl-carnitines, acyl-taurines, and acyl-dopamines are typical components of mammalian brain and spinal cord extracts (62) and may be substrates of fatty acid amide hydrolase (63, 64). Overall, the complexity of lipid species detected in the LDL extracts is quite remarkable and even though most of the lipid classes (TAG, CE, PC, SM, PE, PI, and Cer) have previously been identified as LDL components, the study reported here represents an in-depth exploration of the lipidome in one analysis, including some minor lipids being profiled for the first time as components in LDL.
CONCLUDING REMARKS
In summary, more than 350 individual molecular lipid species in 19 lipid subclasses were identified in the LC-MS spectra of LDL lipid extracts. While all five solvent systems evaluated showed only small differences in the extractability of the predominant lipid classes, major differences were observed in the extraction of minor and less abundant lipid classes. Based on our findings, the most suitable single solvent systems for lipidomic studies of LDL particles are the Folch method and the acidified Bligh and Dyer method for optimum lipidome coverage. This comprehensive study provides guidance for choosing the best solvent system for profiling a particular lipid class, although choice of optimal extraction conditions is always a compromise, and it is clear that a preliminary study on the effectiveness of different solvent systems and data processing procedures is useful prior to implementing routine lipidome analysis.
Supplementary Material
Acknowledgments
G.J.B. acknowledges the Scottish Metabolomics Facility (SCOTMET). C.M.S. would like to acknowledge the 08-EuroMEMBRANES-FP-021 consortium.
Footnotes
Abbreviations:
- ASCA
- ANOVA simultaneous component analysis
- AU
- arbitrary units
- BHT
- butylated hydroxytoluene
- BMI
- body mass index
- Cer
- ceramides
- CE
- cholesteryl ester
- CS
- cholesterol sulfate
- DHEA
- dehydroepiandrosterone sulfate
- FWHM
- full width half maximum
- HexCer
- hexosyl ceramides
- HILIC
- hydrophilic interaction chromatography
- IPA
- isopropyl alcohol
- LAA
- lipoamino acid
- LacCer
- lactosyl ceramides
- lysoPC
- lyso-phosphatidylcholine
- lysoPE
- lyso-phosphatidylethanolamine
- MeOH
- methanol
- TBME
- tert-butyl methyl ether
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- PG
- phosphatidylglycerol
- PI
- phosphatidylinositol
- PS
- phosphatidylserine
- ST
- sulfatide
- TAG
- triacylglyceride
This research was supported by a Marie Curie Intra-European Fellowship within the 7th European Community Framework Program (IEF 255076). Work of A.Rudnitskaya was supported by Portuguese Science and Technology Foundation, through the European Social Fund (ESF) and Programa Operacional Potencial Humano (POPH).
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of six figures and one table.
REFERENCES
- 1.Greig F. H., Kennedy S., Spickett C. M. 2012. Physiological effects of oxidized phospholipids and their cellular signaling mechanisms in inflammation. Free Radic. Biol. Med. 52: 266–280 [DOI] [PubMed] [Google Scholar]
- 2.Folch J., Lees M., Sloane Stanley G. H. 1957. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226: 497–509 [PubMed] [Google Scholar]
- 3.Bligh E. G., Dyer W. J. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917 [DOI] [PubMed] [Google Scholar]
- 4.Saville J. T., Zhao Z., Wilcox M. D., Blanksby S. J., Mitchell T. W. 2010. Detection and quantification of tear phospholipids and cholesterol in contact lens deposits: the effect of contact lens material and lens care solution. Invest. Ophthalmol. Vis. Sci. 51: 2843–2851 [DOI] [PubMed] [Google Scholar]
- 5.Rantamäki A. H., Seppänen-Laakso T., Oresic M., Jauhiainen M., Holopainen J. M.2011. Human tear fluid lipidome: from composition to function. PLoS ONE. 6: e19553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Touboul D., Roy S., Germain D. P., Baillet A., Brion F., Prognon P., Chaminade P., Laprévote O.2005. Fast fingerprinting by MALDI-TOF mass spectrometry of urinary sediment glycosphingolipids in Fabry disease. Anal. Bioanal. Chem. 382: 1209–1216 [DOI] [PubMed] [Google Scholar]
- 7.Kim H., Min H. K., Kong G., Moon M. H. 2009. Quantitative analysis of phosphatidylcholines and phosphatidylethanolamines in urine of patients with breast cancer by nanoflow liquid chromatography/tandem mass spectrometry. Anal. Bioanal. Chem. 393: 1649–1656 [DOI] [PubMed] [Google Scholar]
- 8.Palmerini C. A., Saccardi C., Ferracci F., Arienti S. 2011. Lipid patterns in the saliva of smoking young adults. Hum. Exp. Toxicol. 30: 1482–1488 [DOI] [PubMed] [Google Scholar]
- 9.Pasvogel A. E., Miketova P., Moore I. M. 2010. Differences in CSF phospholipid concentration by traumatic brain injury outcome. Biol. Res. Nurs. 11: 325–331 [DOI] [PubMed] [Google Scholar]
- 10.Miketova P., Kaemingk K., Hockenberry M., Pasvogel A., Hutter J., Drull K., Moore I. M. 2005. Oxidative changes in cerebral spinal fluid phosphatidylcholine during treatment for acute lymphoblastic leukemia. Biol. Res. Nurs. 6: 187–195 [DOI] [PubMed] [Google Scholar]
- 11.Morera S., Castellote A. I., Jauregui O., Casals I., López-Sabater M. C.2003. Triacylglycerol markers of mature human milk. Eur. J. Clin. Nutr. 57: 1621–1626 [DOI] [PubMed] [Google Scholar]
- 12.Schiller J., Hammerschmidt S., Wirtz H., Arnhold J., Arnold K. 2001. Lipid analysis of bronchoalveolar lavage fluid (BAL) by MALDI-TOF mass spectrometry and 31P NMR spectroscopy. Chem. Phys. Lipids. 112: 67–79 [DOI] [PubMed] [Google Scholar]
- 13.Georas S. N., Berdyshev E., Hubbard W., Gorshkova I. A., Usatyuk P. V., Usatyuk P. V., Saatian B., Myers A. C., Williams M. A., Xiao H. Q., Liu M., et al. 2007. Lysophosphatidic acid is detectable in human bronchoalveolar lavage fluids at baseline and increased after segmental allergen challenge. Clin. Exp. Allergy. 37: 311–322 [DOI] [PubMed] [Google Scholar]
- 14.Feki N. C., Thérond P., Couturier M., Liméa G., Legrand A., Jouannet P., Auger J.2004. Human sperm lipid content is modified after migration into human cervical mucus. Mol. Hum. Reprod. 10: 137–142 [DOI] [PubMed] [Google Scholar]
- 15.Iverson S. J., Lang S. L., Cooper M. H. 2001. Comparison of the Bligh and Dyer and Folch methods for total lipid determination in a broad range of marine tissue. Lipids. 36: 1283–1287 [DOI] [PubMed] [Google Scholar]
- 16.Matyash V., Liebisch G., Kurzchalia T. V., Shevchenko A., Schwudke D. 2008. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J. Lipid Res. 49: 1137–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Byeon S. K., Lee J. Y., Moon M. H. 2012. Optimized extraction of phospholipids and lysophospholipids for nanoflow liquid chromatography-electrospray ionization-tandem mass spectrometry. Analyst. 137: 451–458 [DOI] [PubMed] [Google Scholar]
- 18.Löfgren L., Ståhlman M., Forsberg G. B., Saarinen S., Nilsson R., Hansson G. I.2012. The BUME method: a novel automated chloroform-free 96-well total lipid extraction method for blood plasma. J. Lipid Res. 53: 1690–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hara A., Radin N. S. 1978. Lipid extraction of tissues with a low-toxicity solvent. Anal. Biochem. 90: 420–426 [DOI] [PubMed] [Google Scholar]
- 20.Zhang, R., M. L. Brennan, Z. Shen, J. C. MacPherson, D. Schmitt, C. E. Molenda, and S. L. Hazen 2002. Myeloperoxidase functions as a major enzymatic catalyst for initiation of lipid peroxidation at sites of inflammation. J. Biol. Chem. 277: 46116–46122 [DOI] [PubMed] [Google Scholar]
- 21.Hammad, S. M., J. S. Pierce, F. Soodavar, K. J. Smith, M. M. Al Gadban, B. Rembiesa, R. L. Klein, Y. A. Hannun, J. Bielawski, and A. Bielawska 2010. Blood sphingolipidomics in healthy humans: impact of sample collection methodology. J. Lipid Res. 51: 3074–3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kosicek, M., S. Kirsch, R. Bene, Z. Trkanjec, M. Titlic, L. Bindila, J. Peter-Katalinic, and S. Hecimovic 2010. Nano-HPLC-MS analysis of phospholipids in cerebrospinal fluid of Alzheimer's disease patients–a pilot study. Anal. Bioanal. Chem. 398: 2929–2937 [DOI] [PubMed] [Google Scholar]
- 23.Wiesner, P., K. Leidl, A. Boettcher, G. Schmitz, and G. Liebisch 2009. Lipid profiling of FPLC-separated lipoprotein fractions by electrospray ionization tandem mass spectrometry. J. Lipid Res. 50: 574–585 [DOI] [PubMed] [Google Scholar]
- 24.Graessler, J., D. Schwudke, P. E. Schwarz, R. Herzog, A. Shevchenko, and S. R. Bornstein 2009. Top-down lipidomics reveals ether lipid deficiency in blood plasma of hypertensive patients. PLoS ONE. 4: e6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jerlich, A., A. R. Pitt, R. J. Schaur, and C. M. Spickett 2000. Pathways of phospholipid oxidation by HOCl in human LDL detected by LC-MS. Free Radic. Biol. Med. 28: 673–682 [DOI] [PubMed] [Google Scholar]
- 26.Yue, H., S. A. Jansen, K. I. Strauss, M. R. Borenstein, M. F. Barbe, L. J. Rossi, and E. Murphy 2007. A liquid chromatography/mass spectrometric method for simultaneous analysis of arachidonic acid and its endogenous eicosanoid metabolites prostaglandins, dihydroxyeicosatrienoic acids, hydroxyeicosatetraenoic acids, and epoxyeicosatrienoic acids in rat brain tissue. J. Pharm. Biomed. Anal. 43: 1122–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laemmli U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 227: 680–685 [DOI] [PubMed] [Google Scholar]
- 28.Mahley, R. W., T. L. Innerarity, S. C. Rall, Jr., and K. H. Weisgraber 1984. Plasma lipoproteins: apolipoprotein structure and function. J. Lipid Res. 25: 1277–1294 [PubMed] [Google Scholar]
- 29.Retra, K., O. B. Bleijerveld, R. A. van Gestel, A. G. Tielens, J. J. van Hellemond, and J. F. Brouwers 2008. A simple and universal method for the separation and identification of phospholipid molecular species. Rapid Commun. Mass Spectrom. 22: 1853–1862 [DOI] [PubMed] [Google Scholar]
- 30.Zheng, L., R. T'Kind, S. Decuypere, S. J. von Freyend, G. H. Coombs, and D. G. Watson 2010. Profiling of lipids in Leishmania donovani using hydrophilic interaction chromatography in combination with Fourier transform mass spectrometry. Rapid Commun. Mass Spectrom. 24: 2074–2082 [DOI] [PubMed] [Google Scholar]
- 31.Tautenhahn, R., G. J. Patti, D. Rinehart, and G. Siuzdak 2012. XCMS online: a web-based platform to process untargeted metabolomic data. Anal. Chem. 84: 5035–5039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith, C. A., E. J. Want, G. O'Maille, R. Abagyan, and G. Siuzdak 2006. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 78: 779–787 [DOI] [PubMed] [Google Scholar]
- 33.Scheltema, R. A., A. Jankevics, R. C. Jansen, M. A. Swertz, and R. Breitling 2011. PeakML/mzMatch: a file format, Java library, R library, and tool-chain for mass spectrometry data analysis. Anal. Chem. 83: 2786–2793 [DOI] [PubMed] [Google Scholar]
- 34.R Development Core Team. 2008. R: a language and environment for statistical computing. Accessed January 1, 2011, at http://www.R-project.org
- 35.Tautenhahn R., Bottcher C., Neumann S. 2008. Highly sensitive feature detection for high resolution LC/MS. BMC Bioinformatics. 9: 504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wishart, D. S., C. Knox, A. C. Guo, R. Eisner, N. Young, B. Gautam, D. D. Hau, N. Psychogios, E. Dong, S. Bouatra, et al 2009. HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res. 37(Database issue): D603–D610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smilde, A. K., J. J. Jansen, H. C. Hoefsloot, R. J. Lamers, J. van der Greef, and M. E. Timmerman 2005. ANOVA-simultaneous component analysis (ASCA): a new tool for analyzing designed metabolomics data. Bioinformatics. 21: 3043–3048 [DOI] [PubMed] [Google Scholar]
- 38.Jansen, J. J., H. C. J. Hoefsloot, J. van der Greef, M. E. Timmerman, J. A. Westerhuis, and A. K. Smilde 2005. ASCA: analysis of multivariate data obtained from an experimental design. J. Chemom. 19: 469–481 [Google Scholar]
- 39.Westerhuis, J. A. H. C. J. Hoefsloot, S. Smit, D. J. Vis, A. K. Smilde, E. J. J. van Velzen, J. P. M. van Duijnhoven, and F. A. van Dorsten 2008. Assessment of PLSDA cross validation. Metabolomics. 4: 81–89 [Google Scholar]
- 40.Vis, D. J. J. A. Westerhuis, A. K. Smilde, and J. van der Greef 2007. Statistical validation of megavariate effects in ASCA. BMC Bioinformatics. 8: 322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anderson M. J., Legendre P. 1999. An empirical comparison of permutation methods for tests of partial regression coefficients in a linear model. J. Stat. Comput. Sim. 62: 271–303 [Google Scholar]
- 42.DeLong, C. J. P. R. Baker, M. Samuel, Z. Cui, and M. J. Thomas 2001. Molecular species composition of rat liver phospholipids by ESI-MS/MS: the effect of chromatography. J. Lipid Res. 42: 1959–1968 [PubMed] [Google Scholar]
- 43.Koivusalo, M., P. Haimi, L. Heikinheimo, R. Kostiainen, and P. Somerharju 2001. Quantitative determination of phospholipid compositions by ESI-MS: effects of acyl chain length, unsaturation, and lipid concentration on instrument response. J. Lipid Res. 42: 663–672 [PubMed] [Google Scholar]
- 44.Quehenberger, O., A. M. Armando, A. H. Brown, S. B. Milne, D. S. Myers, A. H. Merrill, S. Bandyopadhyay, K. N. Jones, S. Kelly, R. L, Shaner, et al 2010. Lipidomics reveals a remarkable diversity of lipids in human plasma. J. Lipid Res. 51: 3299–3305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sandra, K., S. Pereira Ados, G. Vanhoenacker, F. David, and P. Sandra 2010. Comprehensive blood plasma lipidomics by liquid chromatography/quadrupole time-of-flight mass spectrometry. J. Chromatogr. A. 1217: 4087–4099 [DOI] [PubMed] [Google Scholar]
- 46.Schuhmann, K., R. Almeida, M. Baumert, R. Herzog, S. R. Bornstein, and A. Shevchenko 2012. Shotgun lipidomics on a LTQ Orbitrap mass spectrometer by successive switching between acquisition polarity modes. J. Mass Spectrom. 47: 96–104 [DOI] [PubMed] [Google Scholar]
- 47.Aittoniemi, J., P. S. Niemelä, M. T. Hyvönen, M. Karttunen, and I. Vattulainen 2007. Insight into the putative specific interactions between cholesterol, sphingomyelin, and palmitoyl-oleoyl phosphatidylcholine. Biophys. J. 92: 1125–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Róg T., Pasenkiewicz-Gierula M. 2006. Cholesterol-sphingomyelin interactions: a molecular dynamics simulation study. Biophys. J. 91: 3756–3767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oborina E. M., Yappert M. C. 2003. Effect of sphingomyelin versus dipalmitoylphosphatidylcholine on the extent of lipid oxidation. Chem. Phys. Lipids. 123: 223–232 [DOI] [PubMed] [Google Scholar]
- 50.Scherer, M., A. Böttcher, G. Schmitz, and G. Liebisch 2011. Sphingolipid profiling of human plasma and FPLC-separated lipoprotein fractions by hydrophilic interaction chromatography tandem mass spectrometry. Biochim. Biophys. Acta. 1811: 68–75 [DOI] [PubMed] [Google Scholar]
- 51.Bräutigam, C., B. Engelmann, D. Reiss, U. Reinhardt, J. Thiery, W. O. Richter, and T. Brosche 1996. Plasmalogen phospholipids in plasma lipoproteins of normolipidemic donors and patients with hypercholesterolemia treated by LDL apheresis. Atherosclerosis. 119: 77–88 [DOI] [PubMed] [Google Scholar]
- 52.Kawai, Y., H. Kiyokawa, Y. Kimura, Y. Kato, K. Tsuchiya, and J. Terao 2006. Hypochlorous acid-derived modification of phospholipids: characterization of aminophospholipids as regulatory molecules for lipid peroxidation. Biochemistry. 45: 14201–14211 [DOI] [PubMed] [Google Scholar]
- 53.Wang G., Wang T. 2010. The role of plasmalogen in the oxidative stability of neutral lipids and phospholipids. J. Agric. Food Chem. 58: 2554–2561 [DOI] [PubMed] [Google Scholar]
- 54.Jayaraman S., Gantz D. L., Gursky O. 2011. Effects of phospholipase A(2) and its products on structural stability of human LDL: relevance to formation of LDL-derived lipid droplets. J. Lipid Res. 52: 549–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ravandi, A., S. Babaei, R. Leung, J. C. Monge, G. Hoppe, H. Hoff, H. Kamido, and A. Kuksis 2004. Phospholipids and oxophospholipids in atherosclerotic plaques at different stages of plaque development. Lipids. 39: 97–109 [DOI] [PubMed] [Google Scholar]
- 56.Nakamura, T., Y. Matsuzawa, M. Okano, Y. Kitano, T. Funahashi, S. Yamashita, and S. Tarui 1988. Characterization of low-density lipoproteins from patients with recessive X-linked ichthyosis. Atherosclerosis. 70: 43–52 [DOI] [PubMed] [Google Scholar]
- 57.Muskiet, F. A., G. Jansen, B. G. Wolthers, A. Marinkovic-Ilsen, and P. C. van Voorst Vader 1983. Gas-chromatographic determination of cholesterol sulfate in plasma and erythrocytes, for the diagnosis of recessive X-linked ichthyosis. Clin. Chem. 29: 1404–1407 [PubMed] [Google Scholar]
- 58.Kyogashima M. 2004. The role of sulfatide in thrombogenesis and haemostasis. Arch. Biochem. Biophys. 426: 157–162 [DOI] [PubMed] [Google Scholar]
- 59.Hu, R., G. Li, Y. Kamijo, T. Aoyama, T. Nakajima, T. Inoue, K. Node, R. Kannagi, M. Kyogashima, and A. Hara 2007. Serum sulfatides as a novel biomarker for cardiovascular disease in patients with end-stage renal failure. Glycoconj. J. 24: 565–571 [DOI] [PubMed] [Google Scholar]
- 60.Liu, Y., Y. Chen, A. Momin, R. Shaner, E. Wang, N. J. Bowen, L. V. Matyunina, L. D. Walker, J. F. McDonald, M. C. Sullards, et al 2010. Elevation of sulfatides in ovarian cancer: an integrated transcriptomic and lipidomic analysis including tissue-imaging mass spectrometry. Mol. Cancer. 9: 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tertov, V. V., A. N. Orekhov, I. A. Sobenin, J. D. Morrisett, A. M. Gotto, Jr., and J. G. Guevara, Jr 1993. Carbohydrate-composition of protein and lipid components in sialic acid-rich and acid-poor low-density lipoproteins from subjects with and without coronary-artery disease. J. Lipid Res. 34: 365–375 [PubMed] [Google Scholar]
- 62.Tan B., O'Dell D. K., Yu Y. W., Monn M. F., Hughes H. V., Burstein S., Walker J. M.2010. Identification of endogenous acyl amino acids based on a targeted lipidomics approach. J. Lipid Res. 51: 112–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Turman M. V., Kingsley P. J., Kingsley P. J., Rouzer C. A., Cravatt B. F., Marnet L. J. 2008. Oxidative metabolism of a fatty acid amide hydrolase-regulated lipid, arachidonoyltaurine. Biochemistry. 47: 3917–3925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bisogno, T., D. Melck, L. De Petrocellis, M. Yu. Bobrov, N. M. Gretskaya, V. V. Bezuglov, N. Sitachitta, W. H. Gerwick, and V. Di Marzo 1998. Arachidonoylserotonin and other novel inhibitors of fatty acid amide hydrolase. Biochem. Biophys. Res. Commun. 248: 515–522 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.