Abstract
Multiple reports have demonstrated a role for ceramide kinase (CERK) in the production of eicosanoids. To examine the effects of the genetic ablation of CERK on eicosanoid synthesis, primary mouse embryonic fibroblasts (MEFs) and macrophages were isolated from CERK−/− and CERK+/+ mice, and the ceramide-1-phosphate (C1P) and eicosanoid profiles were investigated. Significant decreases were observed in multiple C1P subspecies in CERK−/− cells as compared to CERK+/+ cells with overall 24% and 48% decreases in total C1P. In baseline experiments, the levels of multiple eicosanoids were significantly lower in the CERK−/− cells compared with wild-type cells. Importantly, induction of eicosanoid synthesis by calcium ionophore was significantly reduced in the CERK−/− MEFs. Our studies also demonstrate that the CERK−/− mouse has adapted to loss of CERK in regards to airway hyper-responsiveness as compared with CERK siRNA treatment. Overall, we demonstrate that there are significant differences in eicosanoid levels in ex vivo CERK−/− cells compared with wild-type counterparts, but the effect of the genetic ablation of CERK on eicosanoid synthesis and the serum levels of C1P was not apparent in vivo.
Keywords: eicosanoids, ceramide-1-phosphate, cytosolic phospholipase A2, inflammation
Eicosanoids are a class of inflammatory signaling molecules derived from a common precursor, arachidonic acid (AA). There are over 100 different eicosanoids, which can be further classified into four categories: prostaglandins, leukotrienes, prostacyclins, and thromboxanes. Eicosanoids have both pro- and anti-inflammatory functions, and therefore these lipids have roles in numerous disease states such as cancer, diabetes, rheumatoid arthritis, asthma, and cardiovascular disease (1–9).
The biosynthesis of eicosanoids is triggered in response to a variety of inflammatory signals (e.g., cytokines, growth factors, mechanical trauma, etc.) in which AA is liberated from the sn-2 position of membrane phospholipids by a phospholipase A2 enzyme. In most cases, this initial rate-limiting step in eicosanoid biosynthesis is started by the activation of group IVA phospholipase A2 (cPLA2α) (10). This activation of cPLA2α in cells requires the association of the enzyme with intracellular membranes in a Ca2+-dependent manner, which is mediated by the N-terminal C2 domain of the enzyme (11–14). Previously, our group has demonstrated that cPLA2α is activated by direct binding of the C2 domain to ceramide-1-phosphate (C1P) (15–18). This direct binding of the C2 domain to C1P increases the residence time of cPLA2α on the cellular membranes, thereby increasing the catalytic ability of the enzyme. The C1P binding site on the enzyme consists of at least three cationic amino acids, R57, K58, and R59, which are present in the β-groove of cPLA2α (19, 20). Mutation of these amino acids to alanine has no effect on basal activity or the binding to other phospholipids, but the association of the enzyme with C1P is lost in vitro. In cells, this mutant of cPLA2α is not activated in response to inflammatory agonists demonstrating the requirement of the cPLA2α/C1P interaction in eicosanoid biosynthesis (15, 20).
C1P is produced by phosphorylation of ceramide by ceramide kinase (CERK), and as the C1P/cPLA2α interaction is required for eicosanoid production, a role for CERK was hypothesized in this biosynthesis cascade. In this regard, our laboratory previously demonstrated that downregulation of CERK by siRNA inhibited eicosanoid production induced by a variety of agonists (15, 21, 22). However, genetic ablation of CERK in mice has produced mixed results, both ex vivo and in vivo. For example, Graf et al. (23) found that the CERK−/− animals were sensitive to both antigen (Ag)-induced and serum transfer-induced arthritis in contrast to the cPLA2α knockout suggesting that cPLA2α pathways are fully functional in the CERK−/− animals. In contrast, this same laboratory group reported that basal prostaglandin E2 (PGE2) synthesis was reduced in the bronchoalveolar (BAL) fluid of CERK−/− mice (24). Importantly, these studies showed an appreciable amount of D-e-C18:1/16:0 C1P was still present in the CERK−/− cells, and other C1P subspecies were not characterized. Igarashi and coworkers also generated a CERK knockout mouse and demonstrated a minor effect on total C1P levels (25). Hence, these reports provided evidence that there is at least one alternative pathway for the synthesis of C1P in addition to CERK (23). This alternative pathway of C1P production is still unknown, and developmental adaptation via this uncharacterized pathway was possible as the total C1P levels were only minorly affected as reported by both Graf et al. (23) and Igarashi and coworkers (25). Furthermore, only a few eicosanoids have been characterized for CERK−/− cells opening the possibility of CERK regulation of uncharacterized eicosanoids.
In this regard, we examined the effect of genetic ablation of CERK on both C1P subspecies production and eicosanoid synthesis by HPLC-ESI-MS/MS. A decrease in several subspecies of C1P was observed for an overall moderate but significant decrease in total C1P in ex vivo cells from the CERK−/− animal. Furthermore, tissue culture conditions had significant effects on compensating for the loss of C1P induced by the genetic ablation of CERK. Interestingly, novel C1P subspecies were observed in the CERK−/− cells in comparison to wild-type cells corroborating the hypothesis of developmental adaptation via upregulation of a separate anabolic pathway. We also demonstrate that ablation of CERK produces a dysfunction in basal eicosanoid synthesis as well as in the eicosanoid response to the calcium ionophore, A23187. Lastly, we demonstrate that the CERK−/− mouse is not dysregulated in eicosanoid production induced by airway hyper-responsiveness (AHR) in stark contrast to mice treated with CERK siRNA. Overall, the results of this study demonstrate that the CERK−/− mouse model has partially adapted when examining eicosanoid synthesis ex vivo and in vivo.
MATERIALS AND METHODS
Cell culture
Primary mouse embryonic fibroblasts (MEFs) were isolated from 13 or 14 days pregnant mice as previously described (26) and maintained for three or less passages in high glucose Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) supplemented with 20% fetal bovine serum (FBS) (Invitrogen) and 2% penicillin/streptomycin (BioWhittaker) at standard incubation conditions. For immortalized MEFs, the cells were passaged every 3 days, and after 20 serial passages of the primary MEFs, immortalized MEFs were obtained. Immortalized MEFs were grown in high glucose DMEM (Invitrogen) supplemented with 10% FBS (Invitrogen) and 2% penicillin/streptomycin (BioWhittaker) at 5% CO2 at 37°C and passaged every 2 days. Bone marrow-derived macrophages (BMDMs) were isolated as previously described (27). Briefly, BALB/c mice were sacrificed, and the femoral and tibial marrows were flushed with sterile PBS using a 27-gauge needle. Red blood cells were then removed by osmotic shock. The cells were resuspended in culture medium supplemented with 15% FBS and 10 ng/ml M-CSF (hereafter BMDM medium), seeded at a density of 2.0 × 105 cells per cm2, and incubated at 37°C in a humidified incubator with 5% CO2. The following day adherent cells were discarded, while nonadherent cells were centrifuged at 1,000 g for 5 min. Cell pellet was resuspended in fresh BMDM medium, and allowed to further differentiate. On day 3, fresh BMDM medium was added to the culture dish. On day 6, the nonadherent cells were discarded and fresh BMDM medium was added.
Baseline eicosanoid experiments
Short-term (4 h).
MEFs (2 × 106) were plated in 10 cm dishes in the appropriate medium and grown under standard incubator conditions overnight. The next morning the medium was removed and replaced with high glucose DMEM supplemented with 2% FBS for 2 h. At the end of the 2 h time point the medium was removed and fresh medium (high glucose DMEM, no FBS supplement) was added to the cells. The cells were then incubated for an additional 4 h. At the end of the 4 h treatment the medium was collected and the cells were harvested for analysis. For 10% FBS supplemented medium, the cells were rinsed with fresh 10% serum and then fresh medium (high glucose DMEM, 10% FBS supplement) was added to the cells. The cells were also incubated for 4 h, medium was collected, and cells were harvested for analysis.
Long-term (24 h).
BMDM cells were plated in 10 cm dishes in the appropriate medium and grown under standard conditions overnight. The next day the medium was removed and replaced with medium supplemented with 2% FBS for 2 h. After 2 h the medium was removed and fresh medium with 10% FBS supplement was added to the cells. The cells were incubated for 24 h. At the end of the 24 h treatment the medium was collected and cells were harvested for analysis.
A23187 treatment experiments
MEFs (2 × 106) were plated on 10 cm plates in the appropriate medium and grown under standard incubator conditions overnight. The next day the medium was removed and replaced with high glucose DMEM with no FBS supplement for 2 h. At the end of the 2 h time point the medium was removed and cells were treated in high glucose DMEM (no FBS supplement) with A23187 (5 μM) (Sigma-Aldrich), DMSO (1:5000) (Sigma-Aldrich), or medium for 5 min. At the end of the 5 min treatment the medium was collected and the cells were harvested for analysis.
AHR
Wild-type BALB/c and CERK−/− mice (BALB/c background, littermates) were obtained from Dr. Frederic Bornancin of Novartis International and utilized for these studies (23). Mice were ip injected on day 1 and day 5 with ovalbumin (OVA) (50 μg) while control mice received saline sham injections at the same time points. For in vivo downregulation of CERK, wild-type BALB/c mice were ip injected with control or CERK siRNA on day 13 (1 μg/g body weight) as described (28–30) and briefly below. To induce the phenotype, mice were challenged with aerosolized OVA (1% in PBS for 60 min) on days 14–20. On day 21 (10 h postchallenge), mice were sacrificed and BAL fluid was collected. BAL fluid was evaluated for eicosanoid production via enzyme-linked immunosorbent assays (ELISAs). Cell populations were evaluated via H and E staining. For in vivo downregulation of CERK, mice were ip injected with control or CERK siRNA (OnTarget siStable, Dharmacon) on day 13 (1 μg/g body weight). Prior to administration, siRNA was bound to siPORT Amine transfection reagent according to instructions from the manufacturer. Briefly, 45 μL siPORT Amine was combined with 110 μl saline and was incubated at room temperature for 30 min. siRNA (45 μl) was then incubated with the siPORT Amine solution for 30 min. The siPORT Amine siRNA solution was administered in a total volume of 200 μl.
Analysis of C1P levels in mouse plasma
Blood was collected from mice via heart puncture into a syringe containing EDTA (100 μl) to prevent clotting. Whole blood was transferred to a glass tube and then centrifuged at 1,000 rpm for 10 min to separate plasma. Plasma was collected and transferred to a clean glass tube. Plasma (50 μl) was extracted according to the C1P/sphingolipid analysis protocol described below, and previously described by our laboratory (20).
Eicosanoid detection
MEFs (2 × 106) were plated on 10 cm plates in the appropriate medium and grown under standard incubator conditions overnight. The next day cells were subjected to the relevant treatment. After treatment, the plates were placed on ice and the medium was collected for eicosanoid analysis. Eicosanoids were extracted from the collected medium using a solid phase extraction method and analyzed as described by Blaho et al. (31) with slight modifications as reported by our laboratory (32). Briefly, to 4 ml of the medium, 100 μl of internal standard containing the following deuterated eicosanoids was added (100 pg/μl, 10 ng total): (d4) 6-keto prostaglandin F1α (6-keto PGF1α), (d4) prostaglandin F2α (PGF2α), (d4) PGE2, (d4) prostaglandin D2 (PGD2), (d8) 5-hydroxyeicosatetranoic acid (HETE), and (d8) AA. Four hundred microliters of 10% methanol and 20 μl of glacial acetic acid were also added to the samples. Strata-X SPE columns (Phenomenex) were washed with 2 ml methanol and then 2 ml of dH2O. The samples were applied, and then the sample vials were rinsed with 2 ml of 5% methanol, which was then applied to the columns. The eicosanoids were eluted with 2 ml isopropanol. The eluent was dried under vacuum and the samples were reconstituted in 100 μl of 50:50 ethanol:dH2O for LC/MS/MS analysis.
The reconstituted eicosanoids were analyzed via HPLC ESI-MS/MS. A 30 min reversed-phase LC method utilizing a Kinetex C18 column (100 × 2.1 mm, 2.6 μm) was used to separate the eicosanoids at a flow rate of 200 μl/min at 50°C. The column was equilibrated with 100% Solvent A [acetonitrile:water:formic acid (40:60:0.02, v/v/v)] for 5 min and then 10 μl of sample was injected. 100% Solvent A was used for the first minute of elution. Solvent B [acetonitrile:isopropanol (50:50, v/v)] was increased in a linear gradient to 25% Solvent B to 3 min, to 45% until 11 min, to 60% until 13 min, to 75% until 18 min, and to 100% until 20 min. 100% Solvent B was held until 25 min, then was decreased to 0% in a linear gradient until 26 min, and then held until 30 min. The eicosanoids were then analyzed using a tandem quadrupole mass spectrometer (ABI 4000 Q-Trap®, Applied Biosystems) via multiple reaction monitoring (MRM) in negative-ion mode. Eicosanoids were monitored using precursor → product MRM pairs, which can be found in supplementary Table II. The mass spectrometer parameters used were: curtain gas, 30; CAD, high; ion spray voltage, −3,500 V; temperature, 500°C; gas 1, 40; gas 2, 60; declustering potential, collision energy, and cell exit potential vary per transition. MRM transitions utilized for the eicosanoids can be found in supplementary Table I.
C1P/sphingolipid analysis
MEFs (2 × 106) or BMDMs were plated on 10 cm plates in the appropriate medium and grown under standard incubator conditions overnight. The next day cells were subjected to the relevant treatment. After treatment the plates were placed on ice, cells were washed twice with ice-cold PBS, and cells were harvested by scraping in 200 μl of PBS followed by sonication to obtain a homogenous mixture. Lipids were extracted from the remaining cells using a modified Bligh and Dyer method and analyzed as described by Wijesinghe et al. (21). Briefly, to 200 μl of the cells in PBS, 1.5 ml of 2:1 methanol:chloroform was added. The samples were spiked with 500 pmol of d18:1/12:0 C1P, sphingomyelin, ceramide, and monohexosylceramide as the internal standard (Avanti). The mixture was sonicated to disperse the cell clumps and incubated for 6 h at 48°C. The samples were then sonicated, followed by centrifugation to separate particulates. The extracts were then dried down and reconstituted in methanol. The reconstituted samples were then sonicated, incubated at 48°C for 15 min, vortexed, and then incubated for an additional 15 min at 48°C. The samples were then centrifuged to separate particulates and used for analysis of C1P, ceramide, sphingomyelin, and monohexosylceramide. The lipids were separated using a Kinetix C18 column (50 × 2.1 mm, 2.6 μm) (Phenomenex) on a Prominence HPLC system (Shimadzu) and eluted using a linear gradient (Solvent A, 58:41:1 CH3OH/water/HCOOH, 5 mM ammonium formate; Solvent B, 99:1 CH3OH/HCOOH, 5 mM ammonium formate, 20–100% B in 3.5 min and at 100% B for 4.5 min at a flow rate of 0.4 ml/min at 60°C). Electrospray ionization with tandem mass spectroscopy using an API 4000 QTRAP instrument (Applied Biosystems, MDS Sciex) was used to detect C1P, ceramide, sphingomyelin, and monohexosylceramide under positive ionization. MRM transitions utilized for C1P, ceramide, sphingomyelin, and monohexosylceramide can be found in supplementary Table II.
3H-AA labeling experiments
Pulse labeling (4 h).
MEFs (0.5 × 105) were plated in 24-well plates in the appropriate medium and were grown under standard incubation conditions overnight. The next day the medium was replaced and 0.25 μCi [3H]AA was added for an incubation period of 4 h. Then the medium was collected and the cells were harvested. The radioactivity was determined in both the supernatants and the cells using a scintillation counter as previously described by our laboratory (22, 33, 34).
Steady-state labeling (A23187).
MEFs (0.5 × 105) were plated in 24-well plates in the appropriate medium supplemented with 0.25 μCi [3H]AA per well and grown under standard incubation conditions overnight. The next day the cells were washed with PBS (Invitrogen) and then incubated with fresh medium containing 0% FBS for 2 h. After 2 h the 0% serum medium was removed and the cells were treated with A23187 (5 μM in 0% FBS medium), DMSO (1:5000 in 0% FBS medium), or medium for 5 min. After 5 min the medium was collected and centrifuged, and the cells were harvested. The radioactivity was determined in both the supernatants and the cells using a scintillation counter as previously described by our laboratory (22, 33–35).
Statistical analysis
Data from experiments involving the comparison of only two groups are plotted as mean ± standard deviation. Significance testing was performed using a two-tailed independent sample t-test to compare means. For all other studies involving multiple experimental groups, measured values are presented as mean ± standard error. One-way ANOVA and Tukey's post hoc analysis were used for pair-wise comparison of experimental groups. Analysis was performed on statistical software (IBM SPSS statistics 19.0) with P < 0.05 being considered significant.
Ethical considerations
Breeding pairs of the CERK−/− mice (BALB/c background) and wild-type BALB/c counterparts were obtained from Dr. Frederick Bornancin of Novartis International (23) and were bred and kept in the animal care facility at Virginia Commonwealth University (VCU). The mouse studies were undertaken under the supervision and approval of the VCU IACUC (protocol number AM10089) following standards set by the Federal and State government. The animal assurance number for VCU is A2381-01.
RESULTS
Genetic ablation of CERK reduces basal C1P levels
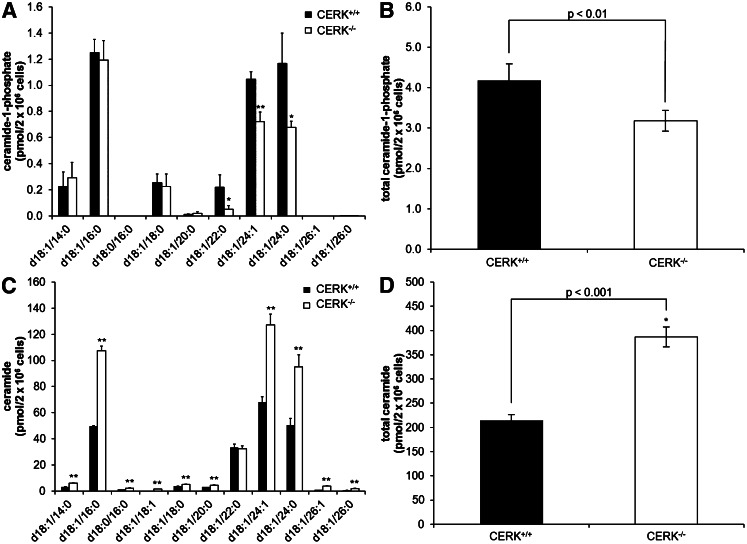
Previously, we confirmed that knockdown of CERK by siRNA technology reduced C1P levels in A549 cells (15) demonstrating that CERK was an anabolic enzyme for C1P formation in these cells. Additionally, Graf et al. (23) demonstrated the production of C18:1/16:0 C1P was decreased in the genetic ablation model of CERK, but only this form was examined. For this study, we examined the effect of genetic ablation of CERK on the levels of various C1P subspecies using immortalized CERK−/− MEFs in comparison to CERK+/+ MEFs. Using HPLC-ESI-MS/MS, the levels of C18:1/22:0 C1P, C18:1/24:1 C1P, and C18:1/24:0 C1P in the CERK−/− MEFs were significantly lower than the levels observed in the CERK+/+ MEFs (Fig. 1A). For example, C18:1/22:0 C1P was downregulated approximately 75% in the CERK−/− MEFs, C18:1/24:1 C1P was downregulated approximately 30%, and C18:1/24:0 C1P was downregulated approximately 40%. Overall, the total amount of C1P was downregulated approximately 24% (Fig. 1B). Interestingly, we did not observe a significant difference in the amount of C18:1/16:0 C1P when the CERK+/+ and CERK−/− MEFs were compared in contrast to previous reports in ex vivo cells from the CERK−/− mouse (23). To ensure that the C1P downregulation observed in the CERK−/− MEFs was not due to a reduction in ceramide, ceramide levels were also analyzed (Fig. 1C). All chain lengths except C18:1/22:0 C1P showed an increase in ceramide (Fig. 1D).
Fig. 1.
Genetic ablation of CERK reduces basal C1P levels. Immortalized MEFs (2 × 106) were plated in 10 cm dishes and incubated under standard conditions overnight. The next morning the medium was replaced with 2% serum medium for 2 h and was then changed to 0% serum medium for an additional 4 h. Cells were harvested, extracted, and analyzed via HPLC-ESI MS/MS as described in the Materials and Methods section to evaluate the levels of C1P. C1P levels by individual chain length (A), total C1P (B), ceramide levels by individual chain length (C), and total ceramide (D). (* indicates a statistical significance of P < 0.01, ** indicates a statistical significance of P < 0.001.) Data are representative of n = 6 on at least three separate occasions.
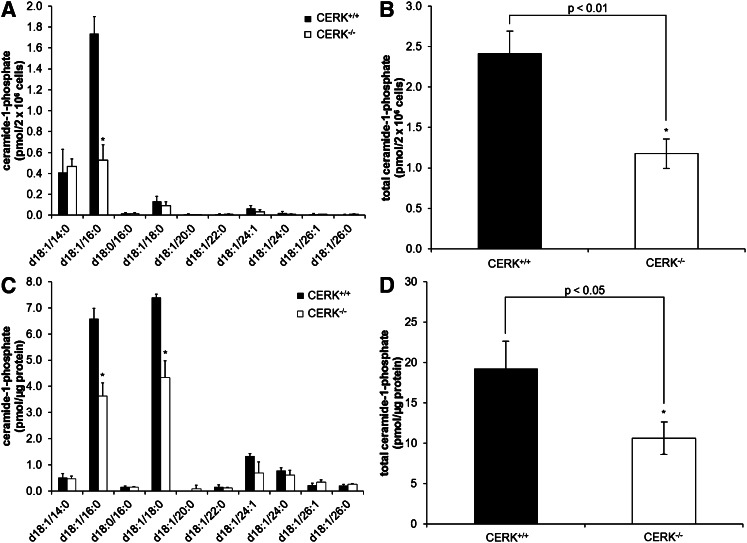
To further characterize the levels of C1P subspecies in the CERK ablation model, and determine whether immortalization affected the type of C1P produced, the C1P profile was determined in primary MEFs as well as in primary BMDMs for CERK−/− and CERK+/+ mice. A decrease in C1P was also observed in the primary MEFs, but interestingly, the C18:1/16:0 subspecies was the only chain length of C1P that was decreased in the CERK−/− cells analogous to the findings of Graf and coworkers (23, 36) (Fig. 2A). Overall, a 51% decrease of C1P was observed in the CERK−/− MEFs (Fig. 2B). In BMDMs, both C18:1/16:0 C1P and C18:1/18:0 C1P were significantly reduced in the CERK−/− cells (Fig. 2C) with a total reduction in C1P similar to that of primary MEFs (e.g., an approximately 45% reduction in total C1P in the CERK−/− cells) (Fig. 2D). These data demonstrate that CERK is a major regulator of C1P biosynthesis, but the specific chain lengths of C1P subspecies produced by CERK vary in response to immortalization and between cell types.
Fig. 2.
Genetic ablation of CERK reduces basal C1P levels in primary cells. Primary MEFs (2 × 106) were plated in 10 cm dishes and incubated under standard conditions overnight. The next morning the medium was replaced with 2% serum medium for 2 h and was then changed to 0% serum medium and incubated for 4 h. Cells were harvested, extracted, and analyzed via HPLC-ESI MS/MS as described in the Materials and Methods section to evaluate the levels of C1P. C1P levels by individual chain length (A) and total C1P (B) (* indicates a statistical significance of P < 0.05). Data are representative of n = 4 on at least three separate occasions. BMDMs (2 × 106) were plated in 10 cm dishes and incubated under standard conditions overnight. The next morning the medium was replaced with 2% serum medium for 2 h and was then changed to fresh 10% serum medium and incubated for 24 h. Cells were harvested, extracted, and analyzed via HPLC-ESI MS/MS as described in the Materials and Methods section to evaluate the levels of C1P. C1P levels by individual chain length (C) and total C1P (D) (* indicates a statistical significance of P < 0.001). Data are representative of n = 3 on at least two separate occasions.
Genetic ablation of CERK affects basal eicosanoid synthesis
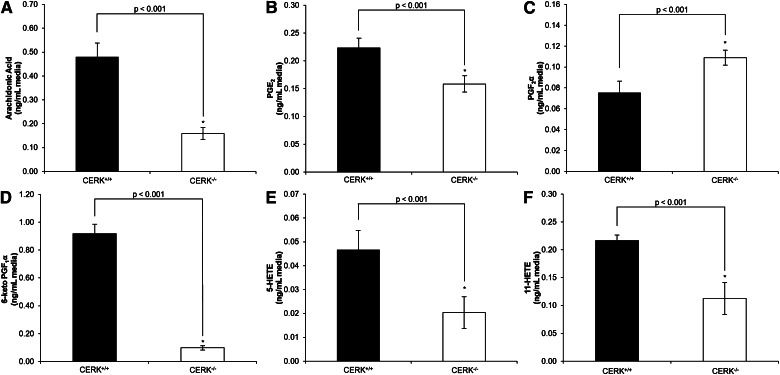
Previously, we have shown that the association of CERK-derived C1P with cPLA2α is required for the synthesis of eicosanoids in response to inflammatory agonists (15). Therefore, we chose to characterize CERK ablation models for eicosanoid production. First, we examined the basal eicosanoid production produced by immortalized MEFs from CERK+/+ mice using HPLC-ESI-MS/MS. Of the 15 eicosanoids examined, six were detected in quantifiable amounts (Table 1). Specifically, AA, PGE2, PGF2α, 6-keto PGF1α, 5-HETE, and 11-HETE were basally produced in appreciable amounts by CERK+/+ MEFs. Importantly, the production these eicosanoids were significantly reduced in CERK−/− cells with the exception of PGF2α, which was surprisingly increased in the CERK−/− MEFs (Fig. 3, Table 1). For example, the levels of both 5-HETE and 11-HETE were reduced by approximately 50% in the media from the CERK−/− MEFs compared with CERK+/+ MEFs, while PGE2 was reduced approximately 30%. AA and 6-keto PGF1α demonstrated more dramatic decreases of approximately 67% and 90%, respectively. PGF2α presented with an upregulation of approximately 45% higher concentration in the media for the CERK−/− MEFs compared with the wild-type MEFs. As it is possible that genetic ablation of CERK and therefore reduced C1P levels could affect secretion and release of eicosanoids, cellular levels of eicosanoids were examined. Levels of cellular eicosanoids were either unchanged between the CERK+/+ and CERK−/− MEFs or the cellular levels corresponded with the media results (data not shown).
TABLE 1.
Eicosanoids produced by immortalized MEFs in 0% serum
| Eicosanoid | CERK+/+ (ng/ml media) | CERK−/− (ng/ml media) | Control (CERK+/+) (%) |
| AA | 0.48 | 0.16 | 33 |
| PGE2 | 0.22 | 0.16 | 71 |
| PGD2 | — | — | ND |
| PGF2α | 0.08 | 0.11 | 145 |
| 6-keto PGF1α | 0.92 | 0.10 | 11 |
| PGJ2 | — | — | ND |
| PGI2 | — | — | ND |
| 5-HETE | 0.05 | 0.02 | 44 |
| 8-HETE | — | — | ND |
| 11-HETE | 0.22 | 0.11 | 52 |
| 12-HETE | — | — | ND |
| 15-HETE | — | — | ND |
| 20-HETE | — | — | ND |
| LTB4 | — | — | ND |
| LTE4 | — | — | ND |
ND, not detected; PGI2, prostacyclin; LTB4, leukotriene B4; LTE4, leukotriene E4.
Fig. 3.
Genetic ablation of CERK induces a loss in the production of basal eicosanoids. Immortalized MEFs (2 × 106) were plated in 10 cm dishes and incubated under standard conditions overnight. The next morning the medium was replaced with 2% serum medium for 2 h and was then switched to 0% serum medium and incubated for 4 h. Medium was then collected and analyzed via HPLC-ESI MS/MS as described in the Materials and Methods section to evaluate the levels of (A) AA, (B) PGE2, (C) PGF2α, (D) 6-keto PGF1α, (E) 5-HETE, and (F) 11-HETE. * indicates a statistical significance of P < 0.001. Data are representative of n = 3 on at least three separate occasions.
The dysfunction in eicosanoid synthesis was also observed in the primary CERK−/− MEFs, although the reduction in basal eicosanoid synthesis was of a lesser extent. For example, AA and 6-keto PGF1α demonstrated the most robust decrease in the immortalized MEFs (67% and 89%, respectively), but AA and 6-keto PGF1α demonstrated only approximately 41% and 20% reduction, respectively, in the primary CERK−/− MEFs. 5-HETE and 11-HETE were downregulated approximately 40%, and both PGE2 and PGD2 were downregulated approximately 25% (supplementary Fig. I, Table 2). Interestingly, PGF2α levels were not shown to be increased in primary CERK−/− cells, but the PGF2α levels did not demonstrate a significant difference between wild-type CERK and CERK−/− cells. These results demonstrate that there is an inherent basal deficiency in eicosanoid production in the CERK−/− cells, but immortalization significantly alters the extent of the deficiency.
TABLE 2.
Eicosanoids produced by primary MEFs in 0% serum
| Eicosanoid | CERK+/+ (ng/ml media) | CERK−/− (ng/ml media) | Control (CERK+/+) (%) |
| AA | 2.69 | 1.59 | 59 |
| PGE2 | 1.14 | 0.86 | 75 |
| PGD2 | 0.05 | 0.03 | 73 |
| PGF2α | 0.20 | 0.19 | 94 |
| 6-keto PGF1α | 1.10 | 0.88 | 80 |
| PGJ2 | — | — | ND |
| PGI2 | — | — | ND |
| 5-HETE | 0.10 | 0.06 | 57 |
| 8-HETE | — | — | ND |
| 11-HETE | 0.86 | 0.67 | 61 |
| 12-HETE | — | — | ND |
| 15-HETE | — | — | ND |
| 20-HETE | — | — | ND |
| LTB4 | — | — | ND |
| LTE4 | — | — | ND |
ND, not detected; PGI2, prostacyclin; LTB4, leukotriene B4; LTE4, leukotriene E4.
After observing the differences in the MEFs, we next determined if these effects were cell-type specific. Therefore, we again utilized primary BMDMs obtained from both CERK+/+ and CERK−/− mice (supplementary Fig. II, Table 3). All eight eicosanoids produced at detectable levels in the media demonstrated significant reductions in levels in the CERK−/− BMDMs. AA, 5-HETE, 12-HETE, and 15-HETE demonstrated the most significant reductions of approximately 80%. PGE2 demonstrated an approximately 55% reduction in the CERK−/− cells, and both PGF2α and 11-HETE demonstrated approximately 30% reductions. These results suggest that the overall deficiency in eicosanoid production induced by the loss of CERK translates to multiple cell types.
TABLE 3.
Eicosanoids produced by primary bone marrow-derived macrophages
| Eicosanoid | CERK+/+ (ng/μg protein) | CERK−/− (ng/μg protein) | Control (CERK+/+) (%) |
| AA | 16.86 | 3.63 | 22 |
| PGE2 | 0.33 | 0.15 | 45 |
| PGD2 | — | — | ND |
| PGF2α | 1.04 | 0.72 | 70 |
| 6-keto PGF1α | — | — | ND |
| PGJ2 | — | — | ND |
| PGI2 | — | — | ND |
| 5-HETE | 7.21 | 1.26 | 18 |
| 8-HETE | 0.65 | 0.19 | 29 |
| 11-HETE | 34.47 | 10.27 | 30 |
| 12-HETE | 2.02 | 0.52 | 26 |
| 15-HETE | 1.80 | 0.40 | 22 |
| 20-HETE | — | — | ND |
| LTB4 | — | — | ND |
| LTE4 | — | — | ND |
ND, not detected; PGI2, prostacyclin; LTB4, leukotriene B4; LTE4, leukotriene E4.
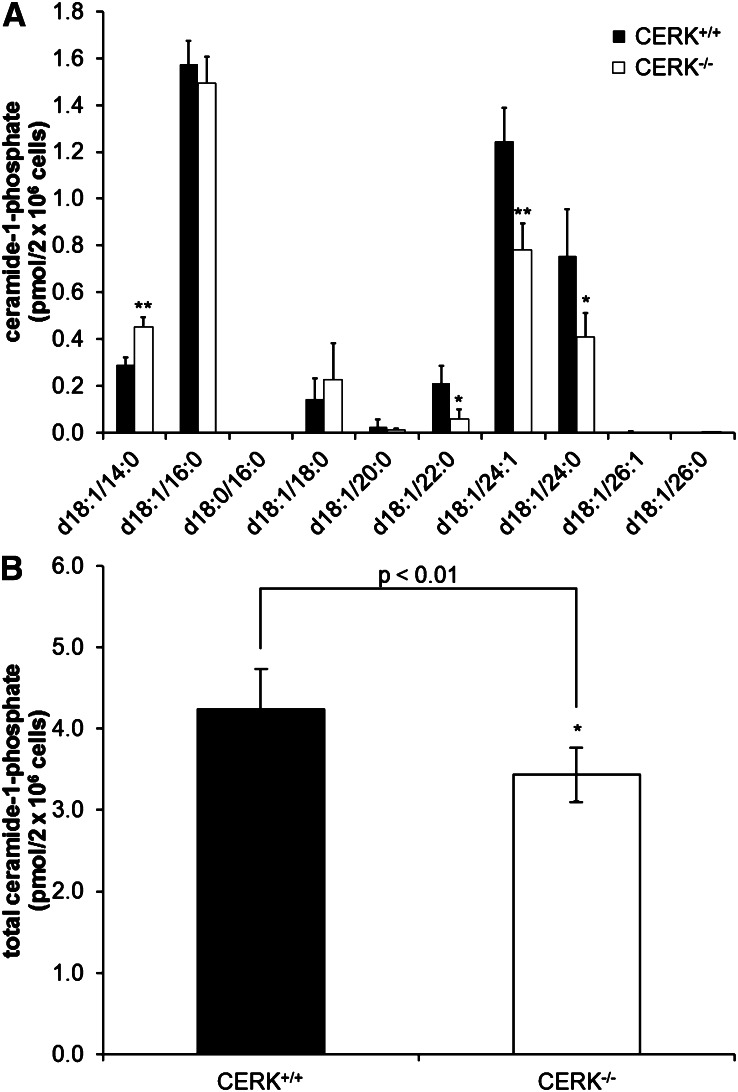
Next, the effects of serum in the tissue culture media were examined as previous reported studies with CERK−/− cells utilized 10% serum in the culture media (5). We surmised that the C1P present in the FBS used to supplement tissue culture media could affect both C1P and eicosanoid levels. In this regard, the quantities of the various chain lengths of C1P were again examined in the wild-type and the CERK−/− MEFs cultured in 10% serum (Fig. 4). Similarly to the C1P levels found when the MEFs were incubated with media containing no serum, the levels of C18:1/22:0 C1P, C18:1/24:1 C1P, and C18:1/24:0 C1P were downregulated in the CERK−/− MEFs. However, with the inclusion of 10% FBS in the media, an approximately 2-fold upregulation of C18:1/14:0 C1P was observed in the CERK−/− MEFs when compared with the CERK+/+ MEFs. When the levels of C1P are compared between the no serum treatment and the 10% serum treatment, C18:1/16:0 C1P appears to have higher levels in 10% serum. In contrast, the addition of serum in the media reduced the C18:1/24:0 levels found in both cell types. Overall, these data illustrating the differences in C1P levels based upon the amount of serum in the media suggest that MEFs have the ability to partially adapt to C1P deficiency.
Fig. 4.
Tissue culture conditions modulate the adaption of C1P levels in CERK−/− MEFs. MEFs (2 × 106) were plated in 10 cm dishes and incubated under standard conditions overnight. Cells were then switched to fresh 10% serum medium and incubated for 4 h. Cells were then collected and analyzed via HPLC-ESI MS/MS as described in the Materials and Methods section to evaluate the levels of C1P. C1P levels by individual chain length (A) and total C1P (B) (* indicates a statistical significance of P < 0.01, ** indicates a statistical significance of P < 0.001). Data are representative of n = 6 on at least three separate occasions.
To determine if the increase of C1P induced by serum correlated with an effect on eicosanoid biosynthesis, basal eicosanoid production was again examined. Of the 15 eicosanoids examined, 11 were produced in quantifiable amounts when immortalized MEFs were treated with 10% serum in contrast to the six detected when treated with 0% serum (supplementary Fig. III, Table 4). Specifically, AA, PGE2, PGD2, PGF2α, 6-keto PGF1α, prostaglandin J2 (PGJ2), 5-HETE, 8-HETE, 11-HETE, 12-HETE, and 15-HETE were basally produced in appreciable amounts by CERK+/+ MEFs. These 11 eicosanoids were also produced in quantifiable amounts in the CERK−/− MEFs. The levels of these eicosanoids were again reduced in the CERK−/− cells with the exception of PGD2 and PGF2α being produced at higher levels in the CERK−/− MEFs. The overall levels of eicosanoids were significantly upregulated in both cell types with respect to the levels produced for basal eicosanoids without FBS supplementation. Interestingly, the levels of AA, PGF2α, and 5-HETE were greatly increased when comparing the percent control values for CERK−/− MEFs between the 0% serum treatment and the 10% serum treatment (Table 4), while PGE2 levels were decreased when comparing the percent control values (Table 4). These results further illustrate the inherent difference of tissue culture conditions on basal eicosanoid production in the CERK−/− MEFs when compared with wild-type counterparts.
TABLE 4.
Eicosanoids produced by immortalized MEFs in 10% serum
| Eicosanoid | CERK+/+ (ng/ml media) | CERK−/− (ng/ml media) | Control (CERK+/+) (%) |
| AA | 7.55 | 6.06 | 80 |
| PGE2 | 17.35 | 4.38 | 25 |
| PGD2 | 0.57 | 1.05 | 185 |
| PGF2α | 0.50 | 0.90 | 180 |
| 6-keto PGF1α | 37.62 | 1.53 | 4 |
| PGJ2 | 0.47 | 0.30 | 64 |
| PGI2 | — | — | ND |
| 5-HETE | 0.24 | 0.22 | 91 |
| 8-HETE | 0.06 | 0.05 | 92 |
| 11-HETE | 9.93 | 4.66 | 47 |
| 12-HETE | 0.64 | 0.60 | 94a |
| 15-HETE | 0.17 | 0.09 | 51 |
| 20-HETE | — | — | ND |
| LTB4 | — | — | ND |
| LTE4 | — | — | ND |
ND, not detected; PGI2, prostacyclin; LTB4, leukotriene B4; LTE4, leukotriene E4.
Not significant.
After observing the differences in basal eicosanoid synthesis between the CERK+/+ and CERK−/− MEFs, we questioned whether the differences in eicosanoid synthesis were due to a dysfunction in the uptake of AA. To test this possibility, pulse labeling experiments with [3H]AA were undertaken, and no differences in the uptake of AA into the cells (supplementary Fig. IV) were observed when comparing the CERK+/+ and the CERK−/− MEFs. These results demonstrate that the dysfunction in eicosanoid synthesis observed in the CERK−/− cells is not due to an issue with AA uptake.
Genetic ablation of CERK leads to dysfunction of AA release and eicosanoid synthesis in response to calcium ionophore
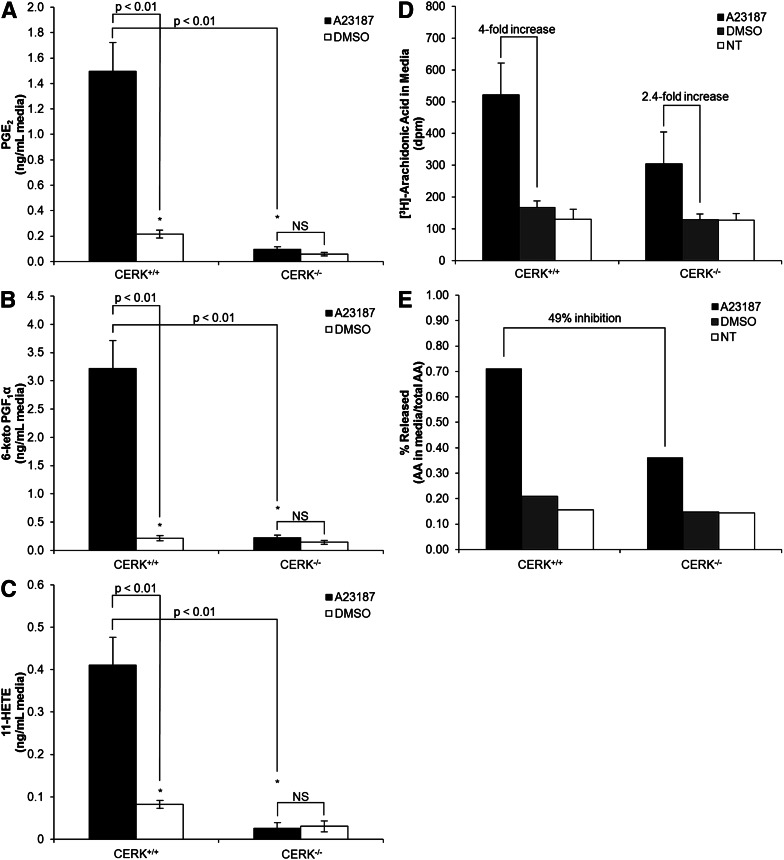
Previously, our laboratory demonstrated that the interaction between C1P and cPLA2α is required for the activation/translocation of cPLA2α in response to inflammatory agonists (e.g., the calcium ionophore, A23187). Due to this requirement of C1P, we hypothesized that the CERK−/− cells would be dysfunctional in eicosanoid synthesis in response to A23187. Wild-type MEFs demonstrated a dramatic increase in three of the quantifiable eicosanoids, PGE2, 6-keto PGF1α, and 11-HETE in response to A23187. However, CERK−/− MEFs demonstrated a startling reduction in these same eicosanoids with the fold-stimulation of the A23187 response significantly inhibited (Fig. 5 A–C). No significant differences were observed between the no treatment and the DMSO control samples. These data demonstrate that the production of eicosanoids in response to calcium ionophore is dysfunctional in CERK−/− cells.
Fig. 5.
Genetic ablation of CERK induces a reduction in A23187-induced eicosanoid production. MEFs (2 × 106) were plated in 10 cm dishes and incubated under standard conditions overnight. The next morning the medium was replaced with 0% serum medium for 2 h and was then dosed with 5 μM A23187, 1:5000 DMSO, or 0% serum sham for 5 min. Medium was then collected and analyzed via HPLC-ESI MS/MS using the method described in the Materials and Methods section to evaluate the levels of eicosanoids: PGE2 (A), 6-keto PGF1α (B), and 11-HETE (C). Data are combined results from n = 3 performed on three separate occasions. [3H]AA uptake and release is shown in medium (D) with percentage of release shown in (E). * indicates a statistical significance of P < 0.01. Data are representative of n = 3 performed on two separate occasions.
This effect of genetic ablation of CERK on eicosanoid synthesis was confirmed at the level of AA release using steady-state labeling with [3H]AA. Both CERK+/+ and CERK−/− cells showed a significant increase in [3H]AA release upon stimulation with A23187; but as predicted, the amount of AA released from CERK−/− cells was significantly less than the amount released from the CERK+/+ cells (Fig. 5D, E). No significant differences were observed between the no treatment and the DMSO control samples. Therefore, the genetic loss of CERK leads to dysregulation in both basal and induced eicosanoid synthesis.
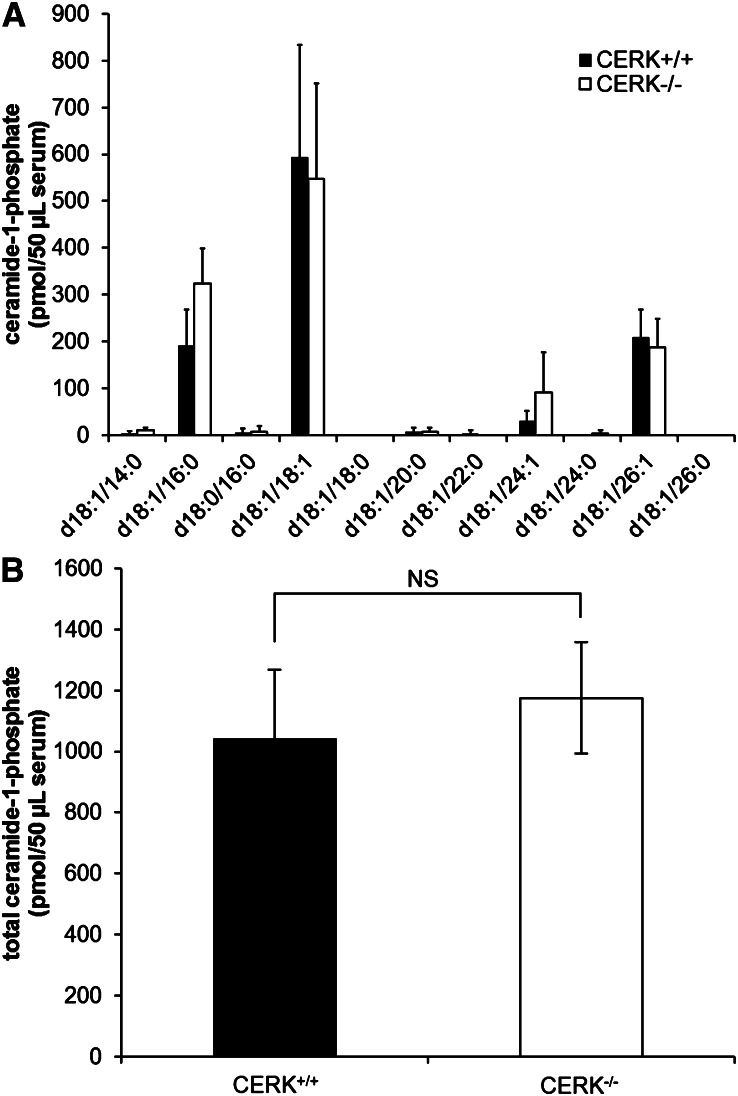
C1P levels are not affected in the plasma of CERK−/− mice
There have been reports of few phenotypes in the CERK−/− mice (23–25) in comparison to phenotypes found in the cPLA2α knockout mice (37–48), which is unexpected given the importance of the binding of C1P and cPLA2α in the activation of the enzyme and resulting eicosanoid synthesis (15). This lack of phenotypes suggests the possibility that the CERK−/− mice may have at least partially adapted to the loss of CERK. As our ex vivo cellular experiments demonstrated that tissue culture conditions modulate the levels of both C1P and eicosanoids, we analyzed the C1P profile in plasma from both the CERK+/+ and CERK−/− mice. Interestingly, no significant difference was observed in specific chain lengths of C1P (Fig. 6A) or total C1P levels (Fig. 6B) when comparing plasma from the CERK+/+ and CERK−/− mice. These data indicate that despite ex vivo cells showing reductions in C1P levels with loss of CERK, plasma levels of the CERK−/− mice are unaffected.
Fig. 6.
C1P levels are unchanged in the plasma of CERK−/− mice. Whole blood was collected via heart puncture from both wild-type and CERK knockout mice. Whole blood was immediately centrifuged to separate plasma. Plasma was then extracted as described in the Materials and Methods section to evaluate the levels of C1P. C1P levels are shown for (A) specific chain lengths and (B) total C1P. Data are representative of at least n = 4 performed on two separate occasions.
The CERK−/− mouse has adapted in regards to AHR
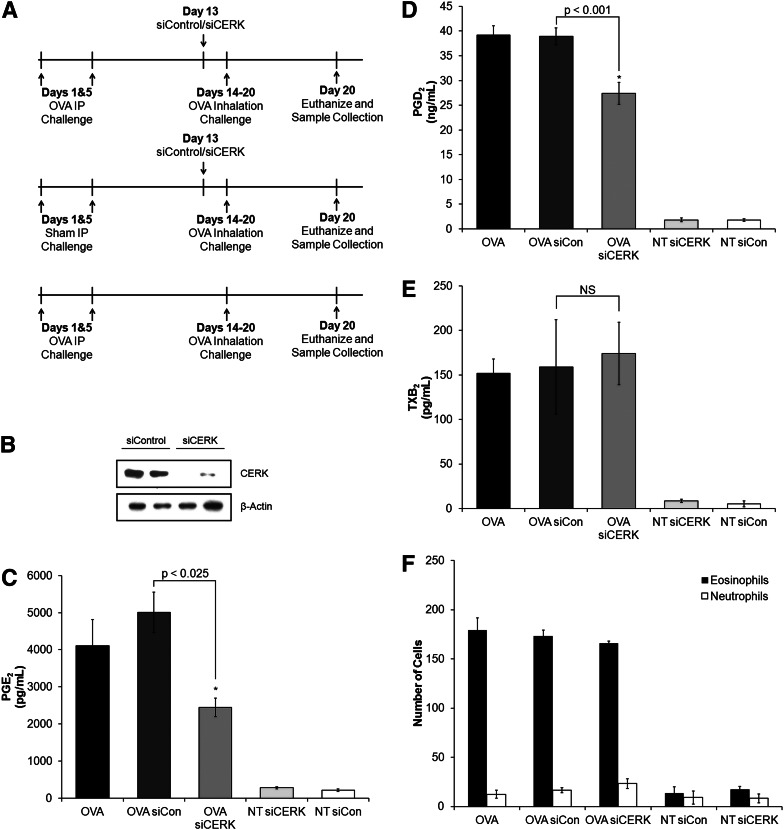
As we now hypothesized that the CERK−/− mouse has partially adapted to loss of CERK by producing C1P via a separate anabolic pathway, the genetic loss of CERK was compared with acute induced loss of CERK via siRNA in an inflammatory phenotype. AHR was evaluated in relation to the loss of CERK as the cPLA2α knockout mouse demonstrated reduced eicosanoid production upon OVA challenge/rechallenge (37). OVA was ip injected on day 1 and day 5, then these mice were rechallenged by aerosolized OVA on days 14–20 (1 h/day). siRNA was given to mice via ip injection on day 13 (Fig. 7A), and mice were sacrificed on day 21 (10 h postchallenge) with the lungs and BAL fluid collected. In our study, siRNA was utilized to knock down CERK levels in the lungs of wild-type mice, and these mice were evaluated for AHR. The CERK siRNA introduced to the mice successfully infiltrated the lungs and downregulated the production of CERK (Fig. 7B). BAL fluid was evaluated for eicosanoid production via ELISA, and three eicosanoids, PGE2, PGD2, and thromboxane B2 (TXB2) were upregulated upon challenge with OVA (Fig. 7C–E). In addition, the production of PGE2 and PGD2 was significantly decreased with the acute downregulation of CERK when mice were challenged with OVA (Fig. 7C, D), while the downregulation of CERK had no effect on TXB2 production (Fig. 7E). Cell infiltration in the BAL fluid was also characterized, and no significant differences were observed between control siRNA- and CERK siRNA-treated mice demonstrating that the observed differences were not due to loss of eosinophil infiltration (Fig. 7F). This study was then repeated with CERK+/+ and CERK−/− mice, and no difference was observed when comparing the eicosanoid production of the CERK+/+ and CERK−/− mice in accord with a previous report by Graf and coworkers (23) (data not shown). These data demonstrate the genetic ablation of CERK produces different phenotypes in comparison to acute downregulation via siRNA treatment.
Fig. 7.
CERK−/− mice have adapted in regards to AHR. Mice were ip injected on day 1 and day 5 with OVA (50 μg) and control mice received saline sham injections. To knockdown CERK, mice were injected with CERK siRNA on day 13 (1 μg/g body weight). To induce the phenotype, the mice were challenged with aerosolized OVA (1% in PBS for 60 min) on days 14–20. After the day 20 challenge, mice were sacrificed with BAL fluid and lungs collected. A: Schematic of AHR experimental design. B: Western blot depicting CERK knockdown in lungs. BAL fluid was evaluated for eicosanoid production via enzyme-linked immunosorbent assays (ELISAs) for (C) PGE2, (D) PGD2, and (E) TXB2. F: Cells contained in BAL fluid were characterized by H and E staining. * indicates statistical significance with a P value as indicated in the figure. Data are representative of n = 8 on at least three separate occasions.
DISCUSSION
In this study, we have demonstrated that CERK-derived C1P is instrumental in the production of eicosanoids, corroborating our previous studies illustrating that siRNA downregulation of CERK blocks the release AA by inhibiting the production of CERK-derived C1P (15, 22). As we have shown here, cells derived from the CERK−/− mice and tested under reduced serum conditions are dysfunctional in eicosanoid synthesis, both basally and in response to calcium ionophore. This further illustrates the effects of CERK-derived C1P on eicosanoid production, and therefore implicates these mediators as players in the inflammatory response. Of note, these findings are partially in opposition to the reports by Bornancin and coworkers (23, 24, 49). Specifically, this group explored how cPLA2α-dependent pathways were affected by the genetic ablation of CERK and this laboratory observed similar responses of cells derived from both wild-type and knockout animals to PMA/ionomycin treatment as well as the susceptibility of the whole animals to arthritis induction by both Ag and KRN serum. As cPLA2α function is imperative in these responses, Bornancin and coworkers concluded that cPLA2α-dependent pathways were functional. However, after this study was released, our laboratory reported the C1P binding site on the cPLA2α protein and showed that both reduction in C1P and mutation of this binding site reduced the activity of cPLA2α (19). These contradictory results gave rise to the current study using cells from the same genetic model, and this study demonstrates several differences in approaches that may explain the contrasting findings. For example, most of our cellular MEF results demonstrating dysregulation in eicosanoids utilized either no serum or low serum environments in comparison to the Bornancin study. Indeed, as we increase serum levels, the effect on both total C1P levels and the basal generation of eicosanoids becomes partially “rescued”. Furthermore, a novel chain length of C1P is increased in the CERK−/− cells, which may account for the minor phenotype on eicosanoids observed in a high serum environment. Regardless, MEFs from the CERK−/− mice presented with reduced levels of C1P, and the role of C1P in regulating cPLA2α responses may be cell type specific. For example, the C1P levels in BMDMs are several-fold higher than the fibroblasts regardless of serum concentration utilized for MEFs, and the effect on basal eicosanoids is also more dramatic in BMDMs from CERK−/− cells. Hence, differential effects on eicosanoids in regards to C1P may vary greatly from cell type to cell type. Indeed, the reports by Bornancin and coworkers also used different cell types, specifically primary kidney fibroblasts and peritoneal macrophages (23, 36). Unfortunately, we were unable to obtain peritoneal macrophages using thioglycolate in the CERK−/− animals in contrast to CERK+/+. Hence, we could not undertake the same studies, and relied on BMDMs for comparison. We attribute the lack of macrophage infiltration in response to thioglycolate to the reported neutropenia as neutrophils are strongly linked to macrophage infiltration (23).
Another difference between the studies is the use of PMA/ionomycin versus our use of only calcium ionophore in regards to AA release. PMA drives activation of protein kinase C, which has been strongly linked to cPLA2α activation (50–53). This activation may or may not be dependent on C1P, and unpublished findings from our laboratory show that a DAG lipase inhibitor is a dramatic stimulator of eicosanoid synthesis without the requirement of C1P. Regardless, this study and the Graf studies are not dramatically divergent with technical differences explaining many of the contrasting findings for the cell studies.
As culture conditions, technical details, and differences in cell types may explain the incongruent findings between our laboratory and the reports by Bornancin and coworkers, the only remaining conundrums in regards to CERK and eicosanoid synthesis is the report by Bornancin and coworkers as to the relative lack of phenotypes in the CERK−/− animals for eicosanoid biosynthesis as well as the more recent report using a specific CERK inhibitor (NVP-231) in vitro (49). In regards to the CERK−/− mice, this genetic ablation model does not demonstrate the phenotypes observed in the cPLA2α knockout mice (e.g., resistance to arthritis induction by both Ag and KRN serum and OVA-induced AHR) with the exception of one report by Bornanicin and coworkers showing a reduction in the levels of basal PGE2 in the BAL fluid of the CERK−/− mice (23, 24). As a decade of research from our laboratory and others has shown cPLA2α to possess a specific C1P interaction site, we hypothesized that the CERK−/− mice were able to demonstrate intact cPLA2α-dependent pathways due to an adaptation of the whole mouse to the loss of CERK-derived C1P (15–20, 22, 33–35). For example, no differences in total C1P levels were observed in the serum of the mice, which correlates well with C1P data obtained from primary hepatocytes (J. A. Mietla et al., unpublished observations). Thus, not all cells produce C1P via CERK, and as our cell studies showed, other cells can at least partially adapt to loss of CERK via upregulating novel chain lengths of C1P (J. A. Mietla et al., unpublished observations). This effect was also observed by genetic ablation of CERK in HeLa cells via zinc-finger nuclease technology (J. A. Mietla et al., unpublished observations). Indeed, the levels of C1P were increased upon ablation of CERK in these cells via an unknown anabolic pathway ( J. A. Mietla et al., unpublished observations). This type of adaptation could explain the lack of phenotypes observed in the CERK knockout mouse. The hypothesis of the adaption of the CERK−/− mice to loss of C1P is supported by our AHR studies presented in this manuscript. An effect on eicosanoid production induced by AHR was observed when CERK was downregulated via siRNA, but the same effect was not seen in the evaluation of the CERK−/− mice. These types of differences have also been observed in some phenotypes for the genetic ablation model of cPLA2α in comparison to the acute inhibition/downregulation of the enzyme by utilization of cPLA2α inhibitors and siRNA (54–56). Overall, our studies highlight the difficulty of examining complex and important pathways in mammalian physiology due to compensatory pathways, which highlight the difficulty in understanding the role of cPLA2α, in general, as well as via the activation of lipid cofactors. Thus, for examining the role of a protein-lipid interaction, it may be necessary to identify the binding site and ablate this site genetically in a mouse to determine function. This may be one alternative route to determining the role of a particular bioactive lipid in a particular biology to overcome compensatory mechanisms when targeting the biosynthetic pathways.
As stated previously, there is still the question of the CERK inhibitor (NVP-231) developed by Novartis (49). Bornancin and coworkers reported that treatment of cells with this inhibitor had no effect on the stimulation of eicosanoid synthesis by various inflammatory agonists (49). Our laboratory has only sparingly utilized this inhibitor, but concur that the inhibitor dramatically reduces total C1P levels with 1 h of treatment in primary MEFs (supplementary Fig. V). Interestingly, the inhibitor potently and dramatically lowers the synthesis of all chain lengths of C1P, with the exception of C18:1/14:0 C1P, in contrast to CERK siRNA (15, 21, 22) and CERK ablation (23, 25). Thus, the CERK inhibitor may also block the currently unknown anabolic pathways of C1P biosynthesis as well as CERK. Therefore, this inhibitor may be an excellent “tool” to examine C1P biology as the entirety of the C1P anabolic system is possibly blocked. What additional targets the inhibitor may be affecting is unknown, but the lack of effect on the levels of ceramide suggests that the unknown pathways are not CERKs. On the other hand, ceramide may be quickly metabolized to hexosylceramides in MEFs masking any increases in ceramide induced by CERK inhibition. As the levels of C1P in comparison to ceramide and phosphatidic acid (PA) are very low, a DAG kinase (DAGK) with low activity toward ceramide could account for some of the C1P formed in cells, and examination of DAG and PA levels in the presence of this inhibitor may be warranted. Furthermore, DAGKθ has high homology to CERK (57), and Bornancin and coworkers only examined DAGKα for efficacy (49). Overall, future studies will also include the use of this CERK inhibitor and examining a number of cell types and inflammatory agonists under various culture conditions to determine whether C1P plays a role in AA release and eicosanoid synthesis. The broad use of this inhibitor coupled to findings using a mouse model with the C1P interaction site ablated from cPLA2α may finally answer the question as to the role of C1P in regulating cPLA2α activation. Furthermore, investigations as to whether this inhibitor will reduce the levels of C1P in cells from the CERK−/− mouse as well as the CERK−/− HeLa cells will confirm the inhibition of a separate anabolic pathway for C1P formation. Indeed, these cell models may prove useful in defining these unknown anabolic pathways for C1P as well as investigate their roles in biological mechanisms.
In conclusion, this study utilizing ex vivo cells supports previous results from our laboratory for the role of CERK in the activation of cPLA2α and regulation of eicosanoid synthesis. This study demonstrates that cells derived from the CERK−/− mice are dysfunctional in eicosanoid synthesis, both basally and through induction by calcium ionophore. C1P levels appear to be partially rescued via addition of serum to the culture medium, which mimics the possible adaptation of the CERK−/− mice. This adaptation of the CERK−/− mice was also demonstrated via the AHR study in comparison to acute downregulation. As inflammation plays a critical role in many diseases, understanding how these interactions modulate eicosanoid levels could lead to potential new drug targets for disease therapies. These new treatments could be vast improvements over current therapies which shunt eicosanoid production from one set of enzymes to another, creating additional health issues as a result of treatment.
Supplementary Material
Acknowledgments
We thank Dr. Frederick Bornancin of Novartis International for providing breeding pairs of the CERK−/− mouse along with wild-type counterparts.
Footnotes
Abbreviations:
- AA
- arachidonic acid
- Ag
- antigen
- AHR
- airway hyper-responsiveness
- BAL
- bronchoalveolar
- BMDM
- bone marrow-derived macrophage
- C1P
- ceramide-1-phosphate
- CERK
- ceramide kinase
- cPLA2α
- group IVA cytosolic phospholipase A2
- DAGK
- diacylglycerol kinase
- HETE
- hydroxyeicosatetraenoic acid
- 6-keto PGF1α
- 6-keto prostaglandin F1α
- MEF
- mouse embryonic fibroblast
- MRM
- multiple reaction monitoring
- OVA
- ovalbumin
- PA
- phosphatidic acid
- PGD2
- prostaglandin D2
- PGE2
- prostaglandin E2
- PGF2α
- prostaglandin F2α
- PGJ2
- prostaglandin J2
- TXB2
- thromboxane B2
- VCU
- Virginia Commonwealth University
This work was supported by research grants from the Veteran's Administration (VA Merit Review I to C.E.C. and a Research Career Scientist Award to C.E.C.); National Institutes of Health Grants HL-072925 (C.E.C.), CA-154314 (C.E.C.), and NH1C06-RR-17393 (to Virginia Commonwealth University for renovation); a National Research Service Award-T32 Post-Doctoral Fellowship in Wound Healing [GM008695 (D.S.W.)]; a Career Development Award (CDA1) from the Department of Veterans Affairs (D.S.W.); National Research Service Award-T31 Pre-Doctoral Fellowship in Functional Lipidomics in Cardiovascular and Respiratory Diseases [HL094290 (J.A.M.)] and by grants from the Aubery Sage MacFarlane Endowment for Acute Lung Injury Research.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of five figures and two tables.
REFERENCES
- 1.Coussens L. M., Werb Z. 2002. Inflammation and cancer. Nature. 420: 860–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grivennikov S. I., Greten F. R., Karin M. 2010. Immunity, inflammation, and cancer. Cell. 140: 883–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Z., Nakayama T. 2010. Inflammation, a link between obesity and cardiovascular disease. Mediators Inflamm. 2010: 535918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Black P. H., Garbutt L. D. 2002. Stress, inflammation and cardiovascular disease. J. Psychosom. Res. 52: 1–23 [DOI] [PubMed] [Google Scholar]
- 5.Choy E. H., Panayi G. S. 2001. Cytokine pathways and joint inflammation in rheumatoid arthritis. N. Engl. J. Med. 344: 907–916 [DOI] [PubMed] [Google Scholar]
- 6.Wellen K. E., Hotamisligil G. S. 2005. Inflammation, stress, and diabetes. J. Clin. Invest. 115: 1111–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dandona P., Aljada A., Bandyopadhyay A. 2004. Inflammation: the link between insulin resistance, obesity and diabetes. Trends Immunol. 25: 4–7 [DOI] [PubMed] [Google Scholar]
- 8.Yang M., Kumar R. K., Hansbro P. M., Foster P. S. 2012. Emerging roles of pulmonary macrophages in driving the development of severe asthma. J. Leukoc. Biol. 91: 557–569 [DOI] [PubMed] [Google Scholar]
- 9.Woodruff P. G., Modrek B., Choy D. F., Jia G., Abbas A. R., Ellwanger A., Koth L. L., Arron J. R., Fahy J. V. 2009. T-helper type 2-driven inflammation defines major subphenotypes of asthma. Am. J. Respir. Crit. Care Med. 180: 388–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leslie C. C. 1997. Properties and regulation of cytosolic phospholipase A2. J. Biol. Chem. 272: 16709–16712 [DOI] [PubMed] [Google Scholar]
- 11.Kramer R. M., Sharp J. D. 1997. Structure, function and regulation of Ca2+-sensitive cytosolic phospholipase A2 (cPLA2). FEBS Lett. 410: 49–53 [DOI] [PubMed] [Google Scholar]
- 12.Nalefski E. A., Sultzman L. A., Martin D. M., Kriz R. W., Towler P. S., Knopf J. L., Clark J. D. 1994. Delineation of two functionally distinct domains of cytosolic phospholipase A2, a regulatory Ca(2+)-dependent lipid-binding domain and a Ca(2+)-independent catalytic domain. J. Biol. Chem. 269: 18239–18249 [PubMed] [Google Scholar]
- 13.Sharp J. D., Pickard R. T., Chiou X. G., Manetta J. V., Kovacevic S., Miller J. R., Varshavsky A. D., Roberts E. F., Strifler B. A., Brems D. N., et al. 1994. Serine 228 is essential for catalytic activities of 85-kDa cytosolic phospholipase A2. J. Biol. Chem. 269: 23250–23254 [PubMed] [Google Scholar]
- 14.Huang Z., Payette P., Abdullah K., Cromlish W. A., Kennedy B. P. 1996. Functional identification of the active site nucleophile of the human 85-kDa cytosolic phospholipase A2. Biochemistry. 35: 3712–3721 [DOI] [PubMed] [Google Scholar]
- 15.Lamour N. F., Subramanian P., Wijesinghe D. S., Stahelin R. V., Bonventre J. V., Chalfant C. E. 2009. Ceramide 1-phosphate is required for the translocation of group IVA cytosolic phospholipase A2 and prostaglandin synthesis. J. Biol. Chem. 284: 26897–26907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Subramanian P., Vora M., Gentile L. B., Stahelin R. V., Chalfant C. E. 2007. Anionic lipids activate group IVA cytosolic phospholipase A2 via distinct and separate mechanisms. J. Lipid Res. 48: 2701–2708 [DOI] [PubMed] [Google Scholar]
- 17.Lamour N. F., Stahelin R. V., Wijesinghe D. S., Maceyka M., Wang E., Allegood J. C., Merrill A. H., Jr, Cho W., Chalfant C. E. 2007. Ceramide kinase uses ceramide provided by ceramide transport protein: localization to organelles of eicosanoid synthesis. J. Lipid Res. 48: 1293–1304 [DOI] [PubMed] [Google Scholar]
- 18.Subramanian P., Stahelin R. V., Szulc Z., Bielawska A., Cho W., Chalfant C. E. 2005. Ceramide 1-phosphate acts as a positive allosteric activator of group IVA cytosolic phospholipase A2 alpha and enhances the interaction of the enzyme with phosphatidylcholine. J. Biol. Chem. 280: 17601–17607 [DOI] [PubMed] [Google Scholar]
- 19.Stahelin R. V., Subramanian P., Vora M., Cho W., Chalfant C. E. 2007. Ceramide-1-phosphate binds group IVA cytosolic phospholipase a2 via a novel site in the C2 domain. J. Biol. Chem. 282: 20467–20474 [DOI] [PubMed] [Google Scholar]
- 20.Ward K. E., Bhardwaj N., Vora M., Chalfant C. E., Lu H., Stahelin R. V. 2013. The molecular basis of ceramide-1-phosphate recognition by C2 domains. J. Lipid Res. 54: 636–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wijesinghe D. S., Allegood J. C., Gentile L. B., Fox T. E., Kester M., Chalfant C. E. 2010. Use of high performance liquid chromatography-electrospray ionization-tandem mass spectrometry for the analysis of ceramide-1-phosphate levels. J. Lipid Res. 51: 641–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pettus B. J., Bielawska A., Spiegel S., Roddy P., Hannun Y. A., Chalfant C. E. 2003. Ceramide kinase mediates cytokine- and calcium ionophore-induced arachidonic acid release. J. Biol. Chem. 278: 38206–38213 [DOI] [PubMed] [Google Scholar]
- 23.Graf C., Zemann B., Rovina P., Urtz N., Schanzer A., Reuschel R., Mechtcheriakova D., Müller M., Fischer E., Reichel C., et al. 2008. Neutropenia with impaired immune response to Streptococcus pneumoniae in ceramide kinase-deficient mice. J. Immunol. 180: 3457–3466 [DOI] [PubMed] [Google Scholar]
- 24.Niwa S., Urtz N., Baumruker T., Billich A., Bornancin F. 2010. Ovalbumin-induced plasma interleukin-4 levels are reduced in ceramide kinase-deficient DO11.10 RAG1-/- mice. Lipids Health Dis. 9: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitsutake S., Yokose U., Kato M., Matsuoka I., Yoo J. M., Kim T. J., Yoo H. S., Fujimoto K., Ando Y., Sugiura M., et al. 2007. The generation and behavorial analysis of ceramide kinase-null mice, indicating a function in cerebellar Purkinje cells. Biochem. Biophys. Res. Commun. 363: 519–524 [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y., Zhao L., Wang C., Lei B. 2003. Isolation and culture of mouse embryonic fibroblast. Sichuan Da Xue Xue Bao Yi Xue Ban. 34: 344–346 [PubMed] [Google Scholar]
- 27.Lamour N. F., Wijesinghe D. S., Mietla J. A., Ward K. E., Stahelin R. V., Chalfant C. E. 2011. Ceramide kinase regulates the production of tumor necrosis factor a (TNFα) via inhibition of TNFα-converting enzyme. J. Biol. Chem. 286: 42808–42817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yin C., Xi L., Wang X., Eapen M., Kukreja R. C. 2005. Silencing heat shock factor 1 by small interfering RNA abrogates heat shock-induced cardioprotection against ischemia-reperfusion injury in mice. J. Mol. Cell. Cardiol. 39: 681–689 [DOI] [PubMed] [Google Scholar]
- 29.Natarajan R., Salloum F. N., Fisher B. J., Kukreja R. C., Fowler A. A., 3rd 2006. Hypoxia inducible factor-1 activation by prolyl 4-hydroxylase-2 gene silencing attenuates myocardial ischemia reperfusion injury. Circ. Res. 98: 133–140 [DOI] [PubMed] [Google Scholar]
- 30.Natarajan R., Salloum F. N., Fisher B. J., Smithson L., Almenara J., Fowler A. A., 3rd 2009. Prolyl hydroxylase inhibition attenuates post-ischemic cardiac injury via induction of endoplasmic reticulum stress genes. Vascul. Pharmacol. 51: 110–118 [DOI] [PubMed] [Google Scholar]
- 31.Blaho V. A., Buczynski M. W., Brown C. R., Dennis E. A. 2009. Lipidomic analysis of dynamic eicosanoid responses during the induction and resolution of Lyme arthritis. J. Biol. Chem. 284: 21599–21612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Norton S. K., Wijesinghe D. S., Dellinger A., Sturgill J., Zhou Z., Barbour S., Chalfant C., Conrad D. H., Kepley C. L. 2012. Epoxyeicosatrienoic acids are involved in the C(70) fullerene derivative-induced control of allergic asthma. J. Allergy Clin. Immunol. 130: 761–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pettus B. J., Bielawska A., Subramanian P., Wijesinghe D. S., Maceyka M., Leslie C. C., Evans J. H., Freiberg J., Roddy P., Hannun Y. A., et al. 2004. Ceramide 1-phosphate is a direct activator of cytosolic phospholipase A2. J. Biol. Chem. 279: 11320–11326 [DOI] [PubMed] [Google Scholar]
- 34.Pettus B. J., Kitatani K., Chalfant C. E., Taha T. A., Kawamori T., Bielawski J., Obeid L. M., Hannun Y. A. 2005. The coordination of prostaglandin E2 production by sphingosine-1-phosphate and ceramide-1-phosphate. Mol. Pharmacol. 68: 330–335 [DOI] [PubMed] [Google Scholar]
- 35.Wijesinghe D. S., Subramanian P., Lamour N. F., Gentile L. B., Granado M. H., Bielawska A., Szulc Z., Gomez-Munoz A., Chalfant C. E. 2009. Chain length specificity for activation of cPLA2alpha by C1P: use of the dodecane delivery system to determine lipid-specific effects. J. Lipid Res. 50: 1986–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boath A., Graf C., Lidome E., Ullrich T., Nussbaumer P., Bornancin F. 2008. Regulation and traffic of ceramide 1-phosphate produced by ceramide kinase: comparative analysis to glucosylceramide and sphingomyelin. J. Biol. Chem. 283: 8517–8526 [DOI] [PubMed] [Google Scholar]
- 37.Uozumi N., Kume K., Nagase T., Nakatani N., Ishii S., Tashiro F., Komagata Y., Maki K., Ikuta K., Ouchi Y., et al. 1997. Role of cytosolic phospholipase A2 in allergic response and parturition. Nature. 390: 618–622 [DOI] [PubMed] [Google Scholar]
- 38.Bonventre J. V., Huang Z., Taheri M. R., O'Leary E., Li E., Moskowitz M. A., Sapirstein A. 1997. Reduced fertility and postischaemic brain injury in mice deficient in cytosolic phospholipase A2. Nature. 390: 622–625 [DOI] [PubMed] [Google Scholar]
- 39.Klivenyi P., Beal M. F., Ferrante R. J., Andreassen O. A., Wermer M., Chin M. R., Bonventre J. V. 1998. Mice deficient in group IV cytosolic phospholipase A2 are resistant to MPTP neurotoxicity. J. Neurochem. 71: 2634–2637 [DOI] [PubMed] [Google Scholar]
- 40.Bonventre J. V. 1999. The 85-kD cytosolic phospholipase A2 knockout mouse: a new tool for physiology and cell biology. J. Am. Soc. Nephrol. 10: 404–412 [DOI] [PubMed] [Google Scholar]
- 41.Fujishima H., Sanchez Mejia R. O., Bingham C. O., 3rd, Lam B. K., Sapirstein A., Bonventre J. V., Austen K. F., Arm J. P. 1999. Cytosolic phospholipase A2 is essential for both the immediate and the delayed phases of eicosanoid generation in mouse bone marrow-derived mast cells. Proc. Natl. Acad. Sci. USA. 96: 4803–4807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakatani N., Uozumi N., Kume K., Murakami M., Kudo I., Shimizu T. 2000. Role of cytosolic phospholipase A2 in the production of lipid mediators and histamine release in mouse bone-marrow-derived mast cells. Biochem. J. 352: 311–317 [PMC free article] [PubMed] [Google Scholar]
- 43.Nagase T., Uozumi N., Ishii S., Kume K., Izumi T., Ouchi Y., Shimizu T. 2000. Acute lung injury by sepsis and acid aspiration: a key role for cytosolic phospholipase A2. Nat. Immunol. 1: 42–46 [DOI] [PubMed] [Google Scholar]
- 44.Hong K. H., Bonventre J. C., O'Leary E., Bonventre J. V., Lander E. S. 2001. Deletion of cytosolic phospholipase A(2) suppresses Apc(Min)-induced tumorigenesis. Proc. Natl. Acad. Sci. USA. 98: 3935–3939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nagase T., Uozumi N., Ishii S., Kita Y., Yamamoto H., Ohga E., Ouchi Y., Shimizu T. 2002. A pivotal role of cytosolic phospholipase A(2) in bleomycin-induced pulmonary fibrosis. Nat. Med. 8: 480–484 [DOI] [PubMed] [Google Scholar]
- 46.Haq S., Kilter H., Michael A., Tao J., O'Leary E., Sun X. M., Walters B., Bhattacharya K., Chen X., Cui L., et al. 2003. Deletion of cytosolic phospholipase A2 promotes striated muscle growth. Nat. Med. 9: 944–951 [DOI] [PubMed] [Google Scholar]
- 47.Hegen M., Sun L., Uozumi N., Kume K., Goad M. E., Nickerson-Nutter C. L., Shimizu T., Clark J. D. 2003. Cytosolic phospholipase A2alpha-deficient mice are resistant to collagen-induced arthritis. J. Exp. Med. 197: 1297–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miyaura C., Inada M., Matsumoto C., Ohshiba T., Uozumi N., Shimizu T., Ito A. 2003. An essential role of cytosolic phospholipase A2alpha in prostaglandin E2-mediated bone resorption associated with inflammation. J. Exp. Med. 197: 1303–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Graf C., Klumpp M., Habig M., Rovina P., Billich A., Baumruker T., Oberhauser B., Bornancin F. 2008. Targeting ceramide metabolism with a potent and specific ceramide kinase inhibitor. Mol. Pharmacol. 74: 925–932 [DOI] [PubMed] [Google Scholar]
- 50.Kishimoto A., Takai Y., Mori T., Kikkawa U., Nishizuka Y. 1980. Activation of calcium and phospholipid-dependent protein kinase by diacylglycerol, its possible relation to phosphatidylinositol turnover. J. Biol. Chem. 255: 2273–2276 [PubMed] [Google Scholar]
- 51.Nishizuka Y. 1984. The role of protein kinase C in cell surface signal transduction and tumour promotion. Nature. 308: 693–698 [DOI] [PubMed] [Google Scholar]
- 52.Castagna M., Takai Y., Kaibuchi K., Sano K., Kikkawa U., Nishizuka Y. 1982. Direct activation of calcium-activated, phospholipid-dependent protein kinase by tumor-promoting phorbol esters. J. Biol. Chem. 257: 7847–7851 [PubMed] [Google Scholar]
- 53.Bonventre J. V., Swidler M. 1988. Calcium dependency of prostaglandin E2 production in rat glomerular mesangial cells. Evidence that protein kinase C modulates the Ca2+-dependent activation of phospholipase A2. J. Clin. Invest. 82: 168–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gijón M. A., Spencer D. M., Siddiqi A. R., Bonventre J. V., Leslie C. C. 2000. Cytosolic phospholipase A2 is required for macrophage arachidonic acid release by agonists that do and do not mobilize calcium. Novel role of mitogen-activated protein kinase pathways in cytosolic phospholipase A2 regulation. J. Biol. Chem. 275: 20146–20156 [DOI] [PubMed] [Google Scholar]
- 55.Dana R., Leto T. L., Malech H. L., Levy R. 1998. Essential requirement of cytosolic phospholipase A2 for activation of the phagocyte NADPH oxidase. J. Biol. Chem. 273: 441–445 [DOI] [PubMed] [Google Scholar]
- 56.Kim J. H., Lee B. D., Kim Y., Lee S. D., Suh P. G., Ryu S. H. 1999. Cytosolic phospholipase A2-mediated regulation of phospholipase D2 in leukocyte cell lines. J. Immunol. 163: 5462–5470 [PubMed] [Google Scholar]
- 57.Sugiura M., Kono K., Liu H., Shimizugawa T., Minekura H., Spiegel S., Kohama T. 2002. Ceramide kinase, a novel lipid kinase. Molecular cloning and functional characterization. J. Biol. Chem. 277: 23294–23300 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.