Abstract
Peroxisome proliferator-activated receptor (PPAR)α is a nuclear receptor that coordinates liver metabolism during fasting. Fatty acid synthase (FAS) is an enzyme that stores excess calories as fat during feeding, but it also activates hepatic PPARα by promoting synthesis of an endogenous ligand. Here we show that the mechanism underlying this paradoxical relationship involves the differential regulation of FAS in at least two distinct subcellular pools: cytoplasmic and membrane-associated. In mouse liver and cultured hepatoma cells, the ratio of cytoplasmic to membrane FAS-specific activity was increased with fasting, indicating higher cytoplasmic FAS activity under conditions associated with PPARα activation. This effect was due to a nutrient-dependent and compartment-selective covalent modification of FAS. Cytoplasmic FAS was preferentially phosphorylated during feeding or insulin treatment at Thr-1029 and Thr-1033, which flank a dehydratase domain catalytic residue. Mutating these sites to alanines promoted PPARα target gene expression. Rapamycin-induced inhibition of mammalian/mechanistic target of rapamycin complex 1 (mTORC1), a mediator of the feeding/insulin signal to induce lipogenesis, reduced FAS phosphorylation, increased cytoplasmic FAS enzyme activity, and increased PPARα target gene expression. Rapamycin-mediated induction of the same gene was abrogated with FAS knockdown. These findings suggest that hepatic FAS channels lipid synthesis through specific subcellular compartments that allow differential gene expression based on nutritional status.
Keywords: de novo lipogenesis, starvation, peroxisome proliferator-activated receptor α
Peroxisome proliferator-activated receptor (PPAR)α, one of three known members of a nuclear receptor family targeted to treat lipid disorders, diabetes, and obesity, is highly expressed in the liver. Its induction by fasting promotes lipid uptake, fatty acid β-oxidation, ketogenesis, and gluconeogenesis (1, 2). Ligand binding to PPARα causes it to heterodimerize with retinoid X receptor (RXR)α, allowing activation of gene transcription at peroxisome proliferator response elements (PPRE) (3, 4). Synthetic PPARα ligands, such as fibrates, used for human lipid disorders (5) have been known for decades, but potential endogenous ligands were identified only recently (6, 7). Mice with liver-specific deletion of the lipogenic enzyme fatty acid synthase (FAS) have impaired PPARα activity (8), and FAS activates PPARα by producing an endogenous phospholipid ligand (6). FAS also activates PPARα in brain and macrophages (9, 10).
Mammalian FAS synthesizes long-chain fatty acids, primarily palmitate, through the activities of seven functional domains: acyl carrier, acyl transferase, β-ketoacyl synthase, β-ketoacyl reductase, β-hydroxyacyl dehydratase, enoyl reductase, and thioesterase (11). Like PPARα, FAS is highly expressed in liver (12). In times of nutrient excess, hepatic FAS converts carbohydrate to lipid that is stored in lipid droplets or secreted in the form of VLDL (13). Nutrient excess is associated with elevated levels of insulin, known to induce FAS expression.
These accepted physiological roles for PPARα and FAS appear to conflict with the observation that inactivation of FAS impairs PPARα activation. How might FAS activate a process stimulated by feeding such as insulin-responsive lipogenesis and also activate a process stimulated by fasting such as the induction of PPARα-dependent gene expression?
We hypothesized that distinct subcellular pools of FAS mediate these disparate effects. Compartmentalization would permit regulation of an FAS pool generating lipids for signaling that would be distinct from an FAS pool generating lipids for energy storage. In support of this hypothesis, we demonstrate that FAS at two separate subcellular locations is differentially regulated by nutrients and insulin, that this regulation involves preferential dehydratase domain phosphorylation for the FAS pool that regulates PPARα, and that the effects of the kinase mammalian/mechanistic target of rapamycin complex 1 (mTORC1) on PPARα activity require FAS.
MATERIALS AND METHODS
Animals
Male C57BL/6J mice at eight weeks of age were provided ad libitum access to chow diet (Purina #5053) or fasted for 18 h. All mice were kept on Aspen bedding and had free access to water. Protocols were approved by the Washington University Animal Studies Committee.
FAS enzyme activity assay
Using a modification of a previously described assay (14), 20 µl of sample at 1 µg protein/µl was added to 70 µl of assay buffer [0.14 M potassium phosphate buffer (pH 7.0), 1.4 mM EDTA (pH 8.0), 1.4 mM DTT, 0.24 mM NADPH, 0.1 mM acetyl-CoA]. The rate of NADPH oxidation was monitored at 340 nm at baseline and again after adding 10 µl of 0.85 mg/ml malonyl-CoA (Sigma). The substrate-dependent rate was determined by subtracting the baseline NADPH oxidation rate from the rate after addition of malonyl-CoA. The rate of NADPH oxidation was normalized to FAS protein levels as determined by western blotting and densitometry to determine specific activity.
Subcellular fractionation
Perfused liver from C57BL/6J mice was homogenized in 20 mM HEPES buffer (pH 7.4) and centrifuged at 100 g for 30 min, and then the pellet was discarded. The supernatant was centrifuged at 500 g for 60 min; 1,200 g for 20 min; 10,000 g for 20 min; 20,000 g for 30 min; 40,000 g for 30 min; 70,000 g for 30 min; 100,000 g for 60 min; and 179,000 g for 75 min. After each spin, the pellet was washed and resuspended, while the supernatant was centrifuged again. All spins were done at 4°C. To obtain crude membrane and cytoplasmic fractions from mouse liver, freshly isolated perfused liver was homogenized in HEPES buffer and centrifuged at 10,000 g for 45 min at 4°C. The resulting pellet was discarded, and the supernatant centrifuged at 179,000 g for 180 min at 4°C. The supernatant (cytoplasm) and pellet (crude membrane) were collected, and the pellet was washed and resuspended in HEPES buffer. To obtain membrane and cytoplasmic extracts from Hepa1-6 cells, a Subcellular Protein Fractionation Kit for Cultured Cells (78840) from Thermo Fisher Scientific was used according to the manufacturer's protocol.
Antibodies
Rabbit polyclonal antibodies against FAS (ab22759), PMP70 (ab3421), and phosphothreonine (ab9337) were from Abcam. Mouse monoclonal antibody against α-tubulin (sc-5286) and rabbit polyclonal antibodies against Cav1 (sc-894) and β-tubulin (sc-9104, used to control for loading in western blotting experiments) were from Santa Cruz Biotechnology. Rabbit polyclonal antibodies against PDI (226), GM130 (2296), Na+/K+ ATPase (3010), Akt (9272), phospho-Akt (S473) (9271), S6 ribosomal protein (2217), and phospho-S6 ribosomal protein (Ser235/236) (2F9/4856), and rabbit monoclonal antibodies against p70 S6 kinase (2708) and CoxIV (4850) were from Cell Signaling Technology.
FAS solubility
Solubility assays were performed as previously described (15) with minor modifications. Membranes were isolated from mouse liver by ultracentrifugation and resuspended in buffer containing 20 mM HEPES buffer (pH 7.4), 1 mM EDTA, and 255 mM sucrose. The membrane fraction was subjected to treatment with various solvents (1 M NaCl, 0.1 M Na2CO3 at pH 11.5, 1% SDS or 1% Triton X-100) and then centrifuged once more (4°C, 180,000 g, 30 min). The resulting pellets and supernatants were analyzed by western blotting.
Cell culture
Hepa1-6 and Hek293T cells were maintained in DMEM + 10% FBS. Prior to insulin treatment for FAS activity assays, Hepa1-6 cells were cultured in DMEM + 0.5% FBS for 6 h. All insulin treatments were performed in DMEM + 10% FBS.
Pulse-chase study
Confluent Hepa1-6 cells in 6 cm dishes were incubated in methionine-free media for 30 min. The cells were then pulsed with 500 µCi of 35S-methionine per dish. After 1 h, cells for the “0” time point were harvested. For subsequent time points, cells were washed with PBS, chased with nonradioactive complete media, and incubated for an additional 45, 90, or 180 min before harvesting. Cells were fractionated into cytoplasm and membrane as described above. FAS was immunoprecipitated from each fraction, samples were subjected to SDS-PAGE, the gel was transferred onto PVDF membrane, and the bands corresponding to labeled FAS were visualized by autoradiography. Autoradiograms were then analyzed by densitometry.
RT-PCR
Total RNA was extracted with TRIzol reagent (Invitrogen) and reverse transcribed using an iScript™ cDNA synthesis kit (Invitrogen). Semiquantitative RT-PCR was performed using SYBR® Green reagent (Applied Biosystems) with an ABI Prism 7700 PCR instrument.
Mutagenesis and plasmid construction
A retroviral plasmid, pBABE-Puro, containing human FAS (16) generated by Max Loda (Dana Farber) was utilized to generate FAS phosphosite mutants. A 3.4 kb fragment of FAS/pBABE-Puro, including the two putative phosphorylation sites (hFAS S1028 and T1032) and two flanking BsrGI sites, was amplified by PCR and subcloned into an intermediate Topo vector. Site-directed mutagenesis of the Topo-FAS plasmid changed the codons corresponding to S1028 and T1032 to alanines, yielding two single mutants. The S1028A/T1032A double mutant was made by sequential mutagenesis, using the S1028A mutant as a template. Mutated FAS fragments were then excised and cloned back into pBabe-Puro using the two BsrGI sites to generate mutant, full-length FAS cDNAs. Mutations as well as correct orientation of the reinserted FAS fragments were verified by DNA sequencing.
Green fluorescent protein (GFP)-tagged FAS was generated by amplifying the cDNA encoding FAS from pBabe-Puro-FAS by RT-PCR, adding restriction sites for XhoI and EcoRI on the 5′ and 3′ ends, respectively. The amplified product was cloned into pEGFP-C3 using the XhoI and EcoRI sites, yielding an N-terminal GFP-tagged FAS construct.
Lentiviral shRNA-mediated knockdown and human FAS expression
A plasmid encoding a mouse FAS shRNA (TRCN0000075703) was obtained from Open Biosystems. The packaging vector psPAX2 (12260) and envelope vector pMD2.G (12259) were obtained from Addgene. Hek293T cells at 70% confluence in a 15 cm dish were transfected using Lipofectamine 2000 with 8 µg psPAX2, 2.25 µg pMD2.G, and 9 µg shRNA. After 48 h, media was collected and filtered through 0.45 µm syringe filters. Polybrene was added and the media used to treat 50–70% confluent Hepa1-6 cells. After 24 h, the media was aspirated and replaced with media containing retroviral particles encoding human FAS (see below). Forty-eight hours after addition of the retroviral media, cells were selected with puromycin. After another 48 h, cells were harvested and knockdown of mouse FAS as well as expression of human FAS were assessed.
To generate retroviral particles encoding human FAS, Hek293T cells in 10 cm dishes were transfected using Lipofectamine 2000 with 3 µg FAS plasmid and 3 µg ψA helper plasmid. After 48 h, media were collected, filtered using 0.45 µM syringe filters, then polybrene was added, and the media was used to treat 50–70% confluent Hepa1-6 cells. After 48 h, 2 µg/ml puromycin was added, and after an additional 48 h, cells were harvested.
In experiments assessing PPARα target gene expression in cells expressing mutant FAS, the endogenous murine FAS of Hepa1-6 cells was knocked down prior to retroviral expression of human FAS as described above.
PPRE-luciferase reporter assay
Media containing lentiviral particles encoding shRNA for murine FAS and media containing retroviral particles encoding wild-type or S1028A/T1032A double-mutant human FAS were prepared as described above. Seventy percent confluent Hepa1-6 cells in 10 cm dishes were treated with retroviral media for either wild-type or S1028A/T1032A FAS for 24 h, after which the media was aspirated and replaced with lentiviral media. After another 24 h, the media was again aspirated and replaced with fresh media containing puromycin.
After two days of puromycin selection, the media was aspirated, replaced with charcoal-stripped media, and incubated for 1 h. Charcoal-stripped media was also used for subsequent steps. Hepa1-6 cells were transfected with plasmids encoding 3× PPRE-luciferase and Renilla luciferase by electroporation. The electroporation for each 10 cm dish of cells was done as follows: 5 µg of PPRE-luciferase plasmid and 5 µg of Renilla luciferase plasmid were added to the bottom of a cuvette. Cells were harvested by trypsinization and spun after adding media. The media was aspirated, and cells were washed once with PBS. The PBS was aspirated, and cells were resuspended in 0.5 ml PBS and transferred to the cuvette followed by electroporation at 360 V and 250 µF (time constant of 4.5–5 s−1). One milliliter of media was added to the cuvette, cells were transferred to a 15 ml tube, and media containing puromycin was added up to 6 ml. Cells were allowed to recover for 10 min, then plated.
One day following transfection, cells were harvested by scraping, washed with room-temperature PBS three times, resuspended in PBS, and plated on a 96-well plate. Luminescence from firefly luciferase and Renilla luciferase was then measured using the Dual-Glo Luciferase Assay System (Promega) according to the manufacturer's instructions. PPRE-luciferase activity was calculated as the ratio of firefly luciferase to Renilla luciferase luminescence.
Mass spectrometry
To identify posttranslational modifications in hepatic FAS, perfused C57BL/6J mouse livers were homogenized in lysis buffer containing 1% Triton X-100. The lysate was spun at 10,000 g for 45 min, and the pellet was discarded. FAS was immunoprecipitated from 10 mg of the lysate by overnight incubation using a polyclonal rabbit anti-FAS antibody. IP beads were washed, boiled in sample buffer, and subjected to SDS-PAGE. The gel was stained with Coomassie, the gel segment corresponding to FAS was excised and further cut into small pieces (1 mm2), destained with 50% CH3CN containing 25 mM NH4HCO3, dehydrated, reduced with 20 mM DTT for 1 h at 55°C, washed and dehydrated, alkylated with 100 mM iodoacetamide for 1 h in the dark at room temperature, then subjected to cycles of washing and dehydration followed by drying in a centrifugal evaporator. In-gel digestion was performed with 0.02 mg/ml trypsin overnight or 0.02 mg/ml chymotrypsin for 6 h at 37°C. Peptides were extracted from the gel pieces using 5% TFA in 50% CH3CN and reconstituted in 0.1% FA in 3% CH3CN.
Immobilized metal ion affinity chromatography (IMAC) was used to enrich the sample for phosphopeptides. The sample was incubated with IMAC beads for 1 h at room temperature. Peptides were eluted from the beads in IMAC buffer, and the sample was diluted with 0.1% FA in 3% CH3CN. Samples were then analyzed by LC-MS/MS with a NanoLC-LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific) in data-dependent mode. Acquired spectra were searched against Swiss-Prot database through Mascot server to identify the protein and its posttranslational modifications. Nonenriched samples were also run to allow a universal search for protein modifications as well as to search for acetyl modifications.
To identify FAS modifications specific to membrane-associated FAS and cytoplasmic FAS, membrane and cytoplasmic fractions were isolated from C57BL/6J mice as described above. FAS was immunoprecipitated from equal amounts of membrane and cytoplasmic lysates (1–10 mg/each) by overnight incubation using a polyclonal rabbit anti-FAS antibody. The samples were then subjected to SDS-PAGE and analyzed as described above.
Statistics
Data are presented as mean ± standard error of the mean. Comparisons between two groups were performed using an unpaired, two-tailed t-test. ANOVA was used for comparisons involving more than two groups.
RESULTS
Hepatic FAS is present in subcellular compartments
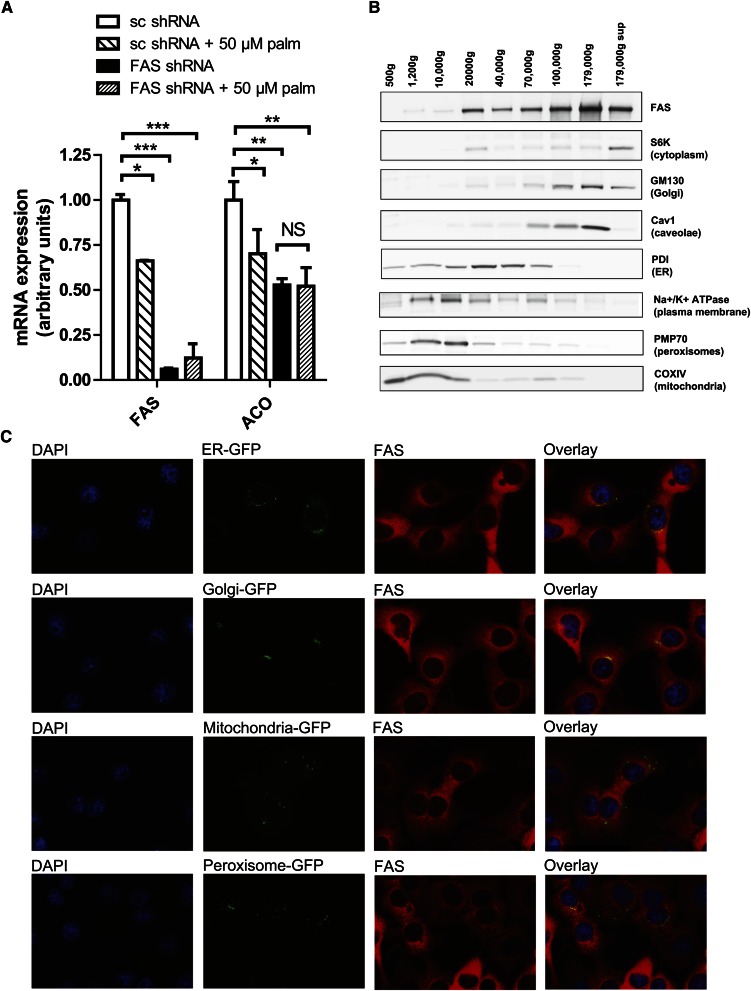
FAS synthesizes palmitate, and FAS deficiency in liver decreases PPARα target genes. If the effect of FAS deficiency on PPARα simply reflects palmitate availability, then exogenous palmitate should rescue the effect. It did not. Treatment of Hepa1-6 cells with 50 µM palmitate failed to rescue expression of the PPARα target gene ACO following FAS knockdown (Fig. 1A). Higher concentrations of palmitate (125–500 µM) were toxic (data not shown).
Fig. 1.
Hepatic FAS is not exclusively cytoplasmic. (A) Expression levels of FAS (left) and the PPARα-dependent gene ACO (right) in Hepa1-6 cells. Cells were treated with a control (scrambled, sc) shRNA or an FAS shRNA in the presence of exogenous BSA-conjugated palmitate or vehicle (BSA alone) for 8 h. *P ≤ 0.05, **P ≤ 0.005, ***P ≤ 0.0005. NS, not significant. (B) Subcellular distribution of FAS protein in mouse liver by differential centrifugation followed by western blotting. Organelle markers: S6K = P70/S6 kinase (cytoplasmic marker), GM130 = Golgi Matrix protein 130 (Golgi marker), Cav1 = Caveolin1 (caveolae marker), PDI = protein disulfide isomerase (endoplasmic reticulum marker), Na+/K+ ATPase (plasma membrane marker), PMP70 = peroxisomal membrane protein 70 (peroxisomal marker), and COXIV = cytochrome C oxidase IV (mitochondrial marker). (C) Immunofluorescent staining of FAS and expression of GFP-tagged organelle markers in murine Hepa1-6 cells. Nuclei stained with DAPI are presented on the far left, GFP images are presented second from left, FAS images are presented second from right, and merged GFP/FAS images are presented on the far right.
Since the FAS knockdown effect was not rescued with exogenous palmitate, it is plausible that not only the product of the FAS reaction but also the location of its synthesis mediates downstream effects. Dogma holds that FAS is a cytoplasmic enzyme. To determine whether FAS is also present at other sites, we fractioned mouse liver FAS by ultracentrifugation (Fig. 1B). FAS cofractionated with the cytoplasmic marker S6K but also with markers for several organelles. Immunofluorescent staining for FAS in murine Hepa1-6 liver cells demonstrated colocalization of FAS with endoplasmic reticulum (ER) and Golgi markers but not peroxisomal or mitochondrial markers (Fig. 1C). FAS did not appear in the nucleus (Fig. 1C).
Membrane-associated and cytoplasmic FAS are differentially regulated
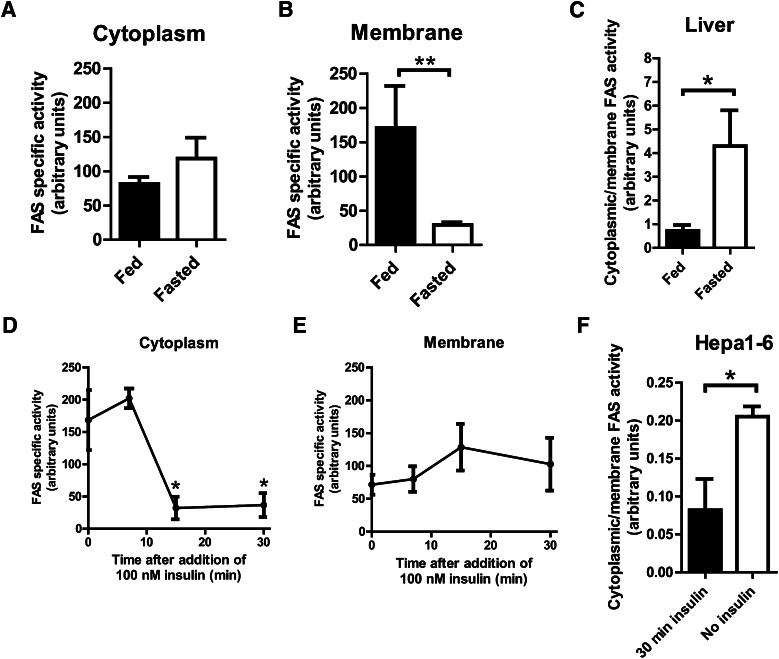
FAS is induced by insulin and nutrients (12). Surprisingly, the specific activity of mouse liver cytoplasmic FAS was not increased in the fed state when insulin levels are high (Fig. 2A). Membrane-associated, FAS-specific activity was increased with feeding (Fig. 2B). The cytoplasmic/membrane activity ratio in liver was increased with fasting, when PPARα is activated (Fig. 2C). In Hepa1-6 cells, a transformed liver cell line, insulin significantly decreased cytoplasmic FAS activity (Fig. 2D), an effect that was not seen in the membrane fraction (Fig. 2E). As with mouse liver, the cytoplasmic/membrane activity ratio in Hepa1-6 cells was increased in the absence of added insulin (Fig. 2F), a mimic of fasting.
Fig. 2.
Differential regulation of the activities of membrane-associated FAS and cytoplasmic FAS. (A) Specific activity of FAS in the cytoplasmic fraction of mouse liver. Mice were fed ad lib (fed) or fasted for 18 h (fasted). Activity was normalized to FAS protein levels as measured by western blotting. N = 9/group. (B) Specific activity of FAS in the membrane (Golgi/ER) fraction of mouse liver. Mice were fed ad lib (fed) or fasted for 18 h (fasted). Activity was normalized to FAS protein levels as measured by western blotting. N = 5/group. **P ≤ 0.005. (C) FAS-specific activities in (A) and (B) expressed as the ratio of FAS-specific activity in cytoplasm to FAS-specific activity in membrane. *P ≤ 0.05. (D) Specific activity of FAS in the cytoplasmic fraction of Hepa1-6 cells. Cells were treated with 100 nM insulin for indicated times. Activity was normalized to FAS protein levels as measured by western blotting. N = 3/group. *P ≤ 0.05. (E) Specific activity of FAS in the membrane (Golgi/ER) fraction of Hepa1-6 cells. Cells were treated with 100 nM insulin for indicated times. Activity was normalized to FAS protein levels as measured by western blotting. N = 3/group. (F) FAS-specific activities in (D) and (E) expressed as the ratio of FAS-specific activity in cytoplasm to FAS-specific activity in membrane. *P ≤ 0.05.
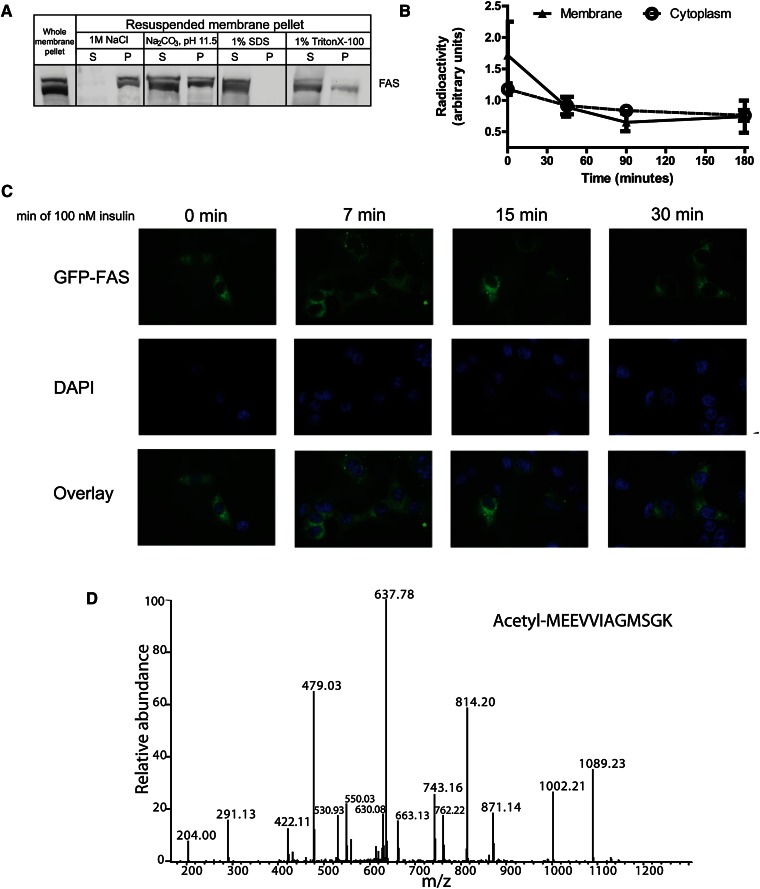
To begin to address the possibility that membrane-associated FAS is an artifact of preparation, we treated isolated fractions with different solvents. Membrane-associated FAS resisted solubilization by 1 M NaCl, remaining in the pelleted fraction, but it was largely solubilized by 0.1 M Na2CO3 (Fig. 3A). Treatment with detergent (1% SDS or 1% Triton X-100) solubilized most FAS protein (Fig. 3A). These results suggest (17–19) that FAS manifests a strong peripheral membrane interaction.
Fig. 3.
Distinct characteristics of membrane and cytoplasmic FAS. (A) Detection of FAS protein by western blotting in pellets and supernatants of membrane fractions following high-salt, carbonate, and detergent treatments. Mouse liver homogenate was fractionated by differential centrifugation into cytoplasm (not shown) and membrane pellet (lane 1). The pellet was resuspended, exposed to solvents as indicated, and again centrifuged to separate pellet (P) from the new supernatant (S). (B) Pulse-chase analysis of FAS protein in membrane and cytoplasm of Hepa1-6 cells. Cells were pulsed with 35S-labeled methionine for 1 h, then chased with media containing nonlabeled methionine for the indicated times. (C) Expression of GFP-tagged human FAS in Hepa1-6 cells treated with insulin for the indicated times. Images demonstrate no detectable shifts of FAS between cytoplasmic and membrane sites with insulin treatment. (D) Representative spectrum of N-terminally acetylated peptide of FAS. N-terminal acetylation effectively marks the initial amino acid of the protein, precluding the existence of additional expressed N-terminal exons that might constitute distinct FAS isoforms.
A pulse-chase study showed that radiolabeled FAS decreased over time in the membrane-associated and cytoplasmic compartments (Fig. 3B), suggesting that there was no ordered flux of protein from one compartment to another over the time course of this experiment. There was no discernible change in the distribution of FAS between membrane and cytoplasm when cells were treated with insulin (Fig. 3C).
Given the presence of a putative open reading frame (with a potential alternative start codon) 5′ to the published first exon of both mouse and human FAS, we considered the possibility that compartmentalized FAS represented differential splicing leading to nonidentical protein isoforms, only one of which is membrane-targeted. However, mass spectrometric analysis of FAS in membrane and cytoplasm failed to detect the predicted alternative amino acids at the N-terminus, and it identified the published FAS protein sequence as being N-terminally acetylated (Fig. 3D). This modification, which marks the N-terminus of most eukaryotic proteins (20), was present in membrane and cytoplasmic fractions of FAS, precluding the existence of an additional N-terminal sequence. All regions of the FAS protein were similarly represented in each fraction, decreasing the possibility that compartment location was determined by altered protein sequence due to a process such as exon exclusion (data not shown).
Collectively, these results suggest that the enzyme activities of cytoplasmic and membrane-associated FAS are differentially regulated, a phenomenon that does not appear to be due to intracellular trafficking of the protein or differences in its primary structure.
Cytoplasmic FAS is preferentially phosphorylated
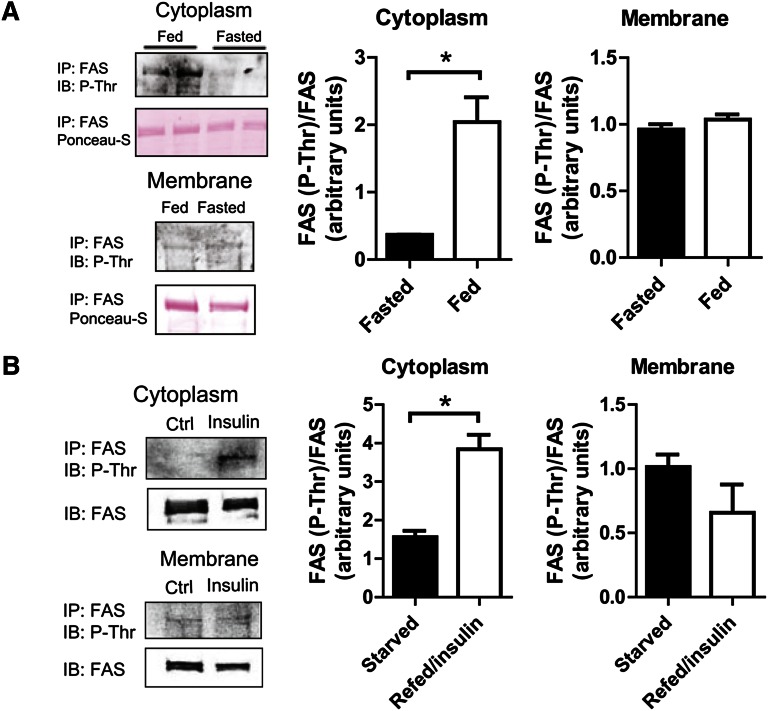
To address the possibility that differential regulation of cytoplasmic and membrane-associated FAS is caused by a covalent modification, we immunoprecipitated hepatic FAS from fasting and fed mice, and then tested different fractions for the presence of phosphothreonine by western blotting. Cytoplasmic FAS in fed mice was strongly threonine phosphorylated, a modification that was almost undetectable in fasted mice (Fig. 4A). Phosphorylation of membrane-associated FAS was low under both conditions (Fig. 4A). In Hepa1-6 cells, insulin treatment (a mimic of feeding) stimulated threonine phosphorylation of cytoplasmic but not membrane-associated FAS (Fig. 4B).
Fig. 4.
Cytoplasmic FAS is threonine phosphorylated with feeding or insulin treatment. (A) FAS threonine phosphorylation in response to feeding in mouse liver. FAS was immunoprecipitated from cytoplasmic and membrane fractions and analyzed for phosphothreonine by western blotting. Mice were fed ad lib (fed) or fasted for 18 h (fasted). Representative blots are shown. Data are averages of two independent experiments. *P ≤ 0.05. (B) FAS threonine phosphorylation in response to insulin in Hepa1-6 cells. FAS was immunoprecipitated from Hepa1-6 cytoplasmic and membrane fractions and analyzed for phosphothreonine by western blotting. Cells were cultured in 0.5% FBS media for 4 h prior to harvest (starved) or in 0.5% FBS media for 4 h, then treated with 1 nM insulin in 10% FBS media for 15 min (refed/insulin). Representative blots are shown. Data are averages of two independent experiments. *P ≤ 0.05.
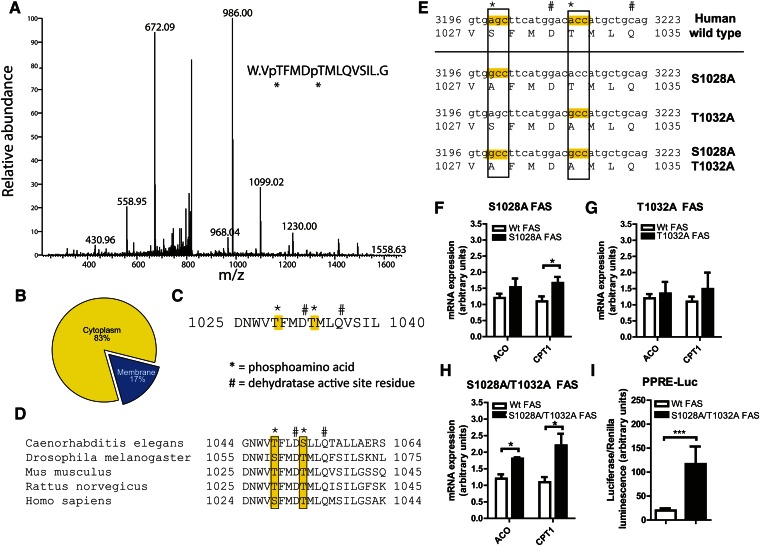
Analysis of FAS protein from unfractionated mouse liver by mass spectrometry revealed only a single peptide that was threonine phosphorylated. This modification was detected at two residues, Thr-1029 and Thr-1033 (a representative spectrum is shown in Fig. 5A). When liver FAS was separated into cytoplasmic and membrane-associated fractions and subjected to the same analysis, the phosphorylated peptide was found predominantly in the cytoplasm (Fig. 5B) despite similar total amounts of the peptide in both fractions (data not shown). These results suggest that the phosphorylated FAS species detected in the cytoplasm with feeding or insulin (Fig. 4A, B) is modified at Thr-1029 and Thr-1033.
Fig. 5.
Phosphorylation of cytoplasmic FAS at the dehydratase domain catalytic site controls downstream PPARα target gene expression. (A) Representative spectrum of the FAS P-T1029/P-T1033 phosphopeptide from wild-type mouse liver. (B) Distribution of P-T1029/P-T1033 phosphopeptides identified by mass spectrometry in cytoplasm and membrane fractions of mouse liver. Although the proportion of phosphorylation differed based on fraction, peptide abundances (phosphorylated + nonphosphorylated) were similar for the membrane and cytoplasm fractions (not shown). (C) Position of P-T1029 and P-T1033 amino acid residues in relation to the FAS dehydratase domain active site residues. D1032 is one of two dehydratase domain catalytic residues in FAS. (D) Sequence alignment of the FAS putative phospho-amino acids and dehydratase domain active sites in several species. (E) FAS phosphosite mutants in human FAS. (F) RT-PCR analyses of PPARα target gene expression in Hepa1-6 cells expressing wild-type or S1028A mutant FAS. Endogenous FAS was knocked down using lentiviral shRNA for murine FAS. Wild-type or mutant human FAS was expressed using retroviruses. Data are averages of three independent experiments. *P ≤ 0.05. (G) RT-PCR analyses of PPARα target gene expression in Hepa1-6 cells expressing wild-type or T1032A mutant FAS. Assay performed as in (F). Data are averages of three independent experiments. (H) RT-PCR analyses of PPARα target gene expression in Hepa1-6 cells expressing wild-type or S1028A/T1032A mutant FAS. Assay performed as in (F). Data are averages of three independent experiments. *P ≤ 0.05. (I) PPRE-luciferase activity in Hepa1-6 cells expressing wild-type or S1028A/T1032A mutant FAS. Wild-type or mutant human FAS was expressed using retroviruses. Endogenous FAS was knocked down using lentiviral shRNA for murine FAS. Cells were cotransfected with plasmids encoding 3× PPRE-firefly luciferase and Renilla luciferase. PPRE-luciferase activity is reported as the ratio of firefly/Renilla luciferase luminescence. N = 3–6/group. ***P ≤ 0.0005.
These residues are in the dehydratase domain of FAS. The function of this domain requires two catalytic residues, His-878 and Asp-1032, and a third residue, Gln-1036, that maintains the orientation of the catalytic residues (21). The phosphorylated residues we identified (denoted by * in Fig. 5C) are in close proximity to the catalytic residue D1032 and the structural residue Q1036 (denoted by # in Fig. 5C). Sequence alignment of the dehydratase regions from different species revealed that in addition to strict conservation of the active site residues D1032 and Q1036 (denoted by #), the phosphoresidues we identified are also conserved as either serines or threonines in humans, mice, rats, D. melanogaster, and C. elegans (boxes in Fig. 5D).
Since the evolutionary conservation of these phosphorylation sites suggests involvement in FAS function, we mutated S1028 and T1032 in human FAS (corresponding to the T1029 and T1033 in murine FAS) to alanines, generating two single mutants (S1028A and T1032A) and one double mutant (S1028A/T1032A) (Fig. 5E, mutated sites are indicated by boxes and the active site residues by #). Wild-type or mutant human FAS was then expressed in Hepa1-6 cells following knockdown of endogenous mouse FAS. Compared with cells expressing wild-type human FAS, cells expressing the S1028A mutation had increased levels of the PPARα target gene CPT1 (Fig. 5F), whereas cells expressing the T1032A mutation did not show changes in PPARα target genes (Fig. 5G). However, expression of the double-mutant S1028A/T1032A was associated with increased levels of both ACO and CPT1 (Fig. 5H). To implicate PPAR transcriptional activity in this effect, we performed a PPRE-luciferase reporter assay. After expression of wild-type or S1028A/T1032A double-mutant FAS and knockdown of endogenous mouse FAS, cells were transfected with a plasmid encoding three tandem PPREs fused to a firefly luciferase reporter gene. Luciferase activity was increased in cells expressing the S1028A/T1032A double-mutant FAS compared with wild-type FAS (Fig. 5I), suggesting that effects of the FAS mutant on PPARα target genes are mediated by PPARα transcriptional activity. One interpretation of these data is that the inability to phosphorylate FAS disinhibits FAS enzyme activity to promote PPARα transcription.
mTORC1 phosphorylates and inactivates FAS and inhibits PPARα activity
mTORC1 was recently identified as a physiologically important negative regulator of hepatic PPARα (22). mTOR, the kinase component of mTORC1, is a serine/threonine kinase that preferentially phosphorylates sites with hydrophobic residues at the +1 position (23). Since the phosphorylated residues we identified have the highly hydrophobic phenylalanine (F1030) and methionine (M1034) at the +1 positions, we addressed a role for mTORC1 in FAS phosphorylation. Treating Hepa1-6 cells with the mTORC1 inhibitor rapamycin for 30 min abolished the insulin-induced increase in cytoplasmic FAS threonine phosphorylation (Fig. 6A) and was associated with an increase in cytoplasmic FAS-specific activity (Fig. 6B). Treatment of these cells with Torin 1 at 250 nM also abolished insulin-induced FAS phosphorylation (data not shown). Treating Hepa1-6 cells with rapamycin for 24 h (a sufficient time to reach a new steady state for mRNA levels) increased expression of the PPARα target gene CPT1 (Fig. 6C). These findings confirm those made in a different system (22) and extend that work by implicating FAS in the mTORC1-PPARα axis.
Fig. 6.
FAS phosphorylation is inhibited by rapamycin and impacts CPT1 expression. (A) Cytoplasmic FAS phosphorylation in response to rapamycin in Hepa1-6 cells. Hepa1-6 cells were treated with vehicle, 100 nM insulin, or 100 nM insulin + 100 nM rapamycin for 30 min. The cytoplasmic fractions were isolated, and then FAS was immunoprecipitated and analyzed for phosphothreonine by western blotting. *P ≤ 0.05. (B) Cytoplasmic FAS activity in response to rapamycin in Hepa1-6 cells. Cells were treated with 100 nM insulin and vehicle (DMSO) or 100 nM insulin + 100 nM rapamycin for 30 min, and then FAS enzyme activity was assayed. Activity was normalized to FAS protein levels as measured by western blotting. Data are averages of two independent experiments. *P ≤ 0.05. (C) CPT1 expression levels in response to rapamycin in Hepa1-6 cells. Cells were treated with vehicle (DMSO) or 100 nM rapamycin for 24 h. Data are averages of two independent experiments. *P ≤ 0.05. (D) FAS expression levels following FAS knockdown in Hepa1-6 cells. N = 3–5/group. ***P ≤ 0.0005. (E) CPT1 expression levels in response to rapamycin following FAS knockdown in Hepa1-6 cells. N = 3–5/group. *P ≤ 0.05. NS, not significant.
To better define the interaction between mTORC1, FAS, and PPARα, FAS was knocked down in Hepa1-6 cells followed by rapamycin treatment. FAS knockdown, confirmed in the presence of rapamycin (Fig. 6D), decreased CPT1 expression (Fig. 6E). The induction of CPT1 levels with rapamycin occurring with FAS expression (Fig. 6C) was lost with FAS knockdown (Fig. 6E, solid bar). These results suggest that in this cell line under these conditions, the induction of the PPARα target gene CPT1 caused by inhibition of mTORC1 is FAS-dependent.
DISCUSSION
FAS synthesizes lipid for energy storage and participates in the generation of a lipid ligand involved in the activation of fatty acid oxidation. Energy storage occurs with feeding, and activation of fatty acid oxidation occurs with fasting. To clarify how the same enzyme mediates both processes, we pursued the possibility that distinct pools of FAS are differentially regulated in the liver.
We found FAS in the cytosol, but we also localized FAS to organelles (Fig. 1) through a strong peripheral membrane interaction (Fig. 3A). FAS-specific activity was relatively higher with feeding/insulin in membranes and relatively higher with fasting in the cytosol (Fig. 2). This effect did not appear to involve movement of FAS between compartments or primary sequence differences between these pools of FAS. Instead, this activity difference was associated with preferential phosphorylation of cytoplasmic (but not membrane) FAS with feeding (Fig. 4) at conserved sites within a catalytic domain (Fig. 5). Mutation of these sites increased endogenous PPARα target gene expression as well as activity of a PPRE-dependent reporter gene (Fig. 5), consistent with disinhibition of FAS in the absence of phosphorylation. Inhibition of mTORC1 with rapamycin decreased FAS phosphorylation, increased cytosolic FAS enzyme activity, and increased expression of the PPARα target gene CPT1, an effect that was FAS-dependent (Fig. 6). One interpretation of these findings is that hepatic FAS exists in at least two differentially regulated subcellular pools, cytoplasmic and membrane-associated (Fig. 7). Cytoplasmic FAS is phosphorylated with feeding to limit PPARα activation, and it is dephosphorylated with fasting to promote PPARα activation.
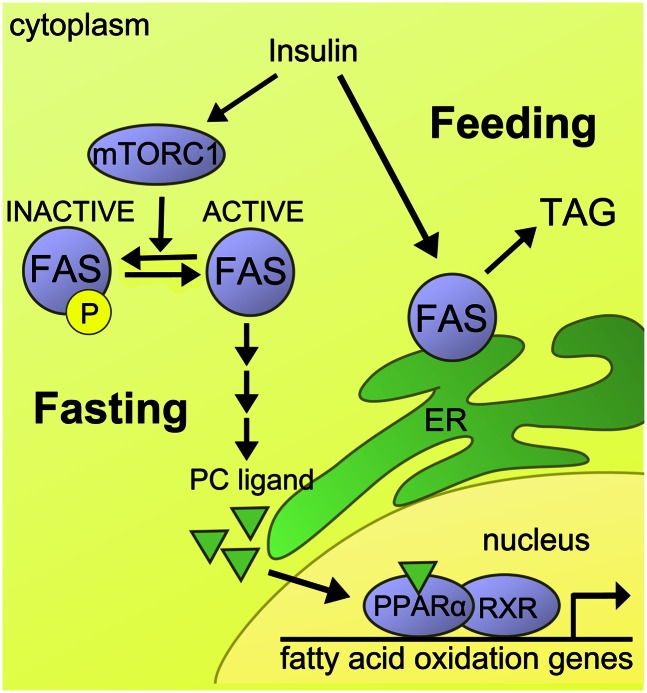
Fig. 7.
Schematic depiction of insulin/feeding-regulated FAS phosphorylation and FAS-mediated PPARα activation. In the fed state, mTORC1 promotes phosphorylation of FAS, thus limiting downstream generation of a phosphatidylcholine ligand that activates PPARα-dependent gene expression. In the fasting state, dephosphorylated FAS in the cytoplasm is permissive for the generation of the ligand activating PPARα-dependent gene expression. PC, phosphatidylcholine; TAG, triacylglycerol.
Our findings provide molecular definition and physiological context to an observation made nearly four decades ago in birds. Using pigeon liver as a model and exclusively studying FAS in the cytoplasm, Qureshi and colleagues found that feeding induced 32P incorporation into FAS, which was associated with a loss of enzyme activity (24). In vitro treatment with phosphatases dephosphorylated FAS and restored enzyme activity. The authors of this study did not identify a physiological role for this covalent modification, and it is not known whether the phosphosites we found are conserved in pigeon FAS due to the unavailability of sequence data for this species. Regardless, our work suggests that the phosphorylation state of cytoplasmic FAS may channel lipid flow to impact phospholipids inducing gene expression in the nucleus.
Physiological, mass spectrometric, and crystal structure data indicate that phospholipids interact with nuclear receptors (6, 25–29). FAS appears to be linked to PPARα through phosphatidylcholine synthesis mediated by the Kennedy pathway (6). Viewed with previous studies showing that phosphorylation regulates the CDP-choline branch of the Kennedy pathway (30, 31), our identification of functionally relevant FAS phosphorylation sites raises the possibility that phosphorylation at several nodes within a cascade of lipid signaling from the cytoplasm to the nucleus coordinates FAS-mediated PPARα activation.
Palmitate is the direct product of the FAS reaction. If the mere availability of palmitate were required to activate PPARα, exogenous palmitate would correct FAS deficiency. However, the addition of palmitate to liver cells with FAS deficiency does not restore defects in PPARα-dependent genes (Fig. 1), and elevated serum palmitate levels that accompany inactivation of liver FAS in mice does not rescue impaired activation of PPARα-dependent genes (8). Thus, palmitate produced by FAS appears to be compartmentalized, a notion supported by our finding of preferential phosphorylation depending on cellular location and nutritional state.
There is precedent for compartmentalization in metabolism. Exogenous administration of T3, the active form of thyroid hormone that can be produced locally from its precursor T4, does not rescue gene expression defects in the setting of hypothyroidism. But administration of T4, which is metabolized to generate T3 locally, restores downstream effects (32). There is also precedent for compartmentalization in lipid signaling. Phosphatidic acid derived from glycerolipid synthesis has effects on mTORC2 that are opposite from those induced by phosphatidic acid derived from membrane lipolysis (33). These observations are consistent with our model (Fig. 7). In the fed state, cytoplasmic FAS is phosphorylated to limit lipid production resulting in PPARα activation, while membrane FAS, less susceptible to phosphorylation, likely produces lipids for energy storage or export. Given the rapid demands of lipid synthesis prompted by transition from the fasting to the fed state, the induction of membrane FAS may be predominantly substrate-driven through allosteric activation by the glycolytic intermediate fructose-1,6-bisphosphate (34).
mTORC1 may control the reciprocal activity of FAS in different compartments. mTORC1 is activated by insulin and nutrients, prefers substrates like those we identified in the dehydratase domain, and is known to suppress PPARα in the liver (22). FAS and mTORC1 appear to interact in the central nervous system where the physiological effects of FAS inhibition are blunted by rapamycin (35), consistent with our model suggesting that mTORC1 inhibition would increase FAS activity.
Our work provides evidence that hepatic FAS is in the cytoplasm as well as peripherally associated with membranes. These two pools are differentially regulated by nutrients and insulin, and they are differentially susceptible to phosphorylation, thus providing a conceptual framework for understanding how FAS-mediated PPARα activation is linked to the fasting state. These observations could have clinical implications. Selective pharmacological targeting of FAS to achieve inhibition of lipid storage without impairing PPARα activation could treat fatty liver and other disorders associated with nutrient excess.
Footnotes
Abbreviations:
- ER
- endoplasmic reticulum
- GFP
- green fluorescent protein
- mTORC1
- mammalian/mechanistic target of rapamycin complex 1
- PPAR
- peroxisome proliferator-activated receptor
- PPRE
- peroxisome proliferator response element
- RXR
- retinoid X receptor
This work was supported by National Institutes of Health Grants DK-076729, DK-088083, DK-20579, DK-56341, F32-DK-083895, and T32-DK-07120, and by an American Heart Association predoctoral fellowship award.
REFERENCES
- 1.Aoyama T., Peters J. M., Iritani N., Nakajima T., Furihata K., Hashimoto T., Gonzalez F. J. 1998. Altered constitutive expression of fatty acid-metabolizing enzymes in mice lacking the peroxisome proliferator-activated receptor alpha (PPARalpha). J. Biol. Chem. 273: 5678–5684 [DOI] [PubMed] [Google Scholar]
- 2.Kersten S., Seydoux J., Peters J. M., Gonzalez F. J., Desvergne B., Wahli W. 1999. Peroxisome proliferator-activated receptor alpha mediates the adaptive response to fasting. J. Clin. Invest. 103: 1489–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kliewer S. A., Umesono K., Noonan D. J., Heyman R. A., Evans R. M. 1992. Convergence of 9-cis retinoic acid and peroxisome proliferator signalling pathways through heterodimer formation of their receptors. Nature. 358: 771–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Palmer C. N., Hsu M. H., Griffin H. J., Johnson E. F. 1995. Novel sequence determinants in peroxisome proliferator signaling. J. Biol. Chem. 270: 16114–16121 [DOI] [PubMed] [Google Scholar]
- 5.Lalloyer F., Staels B. 2010. Fibrates, glitazones, and peroxisome proliferator-activated receptors. Arterioscler. Thromb. Vasc. Biol. 30: 894–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chakravarthy M. V., Lodhi I. J., Yin L., Malapaka R. R., Xu H. E., Turk J., Semenkovich C. F. 2009. Identification of a physiologically relevant endogenous ligand for PPARalpha in liver. Cell. 138: 476–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Narala V. R., Adapala R. K., Suresh M. V., Brock T. G., Peters-Golden M., Reddy R. C. 2010. Leukotriene B4 is a physiologically relevant endogenous peroxisome proliferator-activated receptor-alpha agonist. J. Biol. Chem. 285: 22067–22074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chakravarthy M. V., Pan Z., Zhu Y., Tordjman K., Schneider J. G., Coleman T., Turk J., Semenkovich C. F. 2005. “New” hepatic fat activates PPARalpha to maintain glucose, lipid, and cholesterol homeostasis. Cell Metab. 1: 309–322 [DOI] [PubMed] [Google Scholar]
- 9.Chakravarthy M. V., Zhu Y., Lopez M., Yin L., Wozniak D. F., Coleman T., Hu Z., Wolfgang M., Vidal-Puig A., Lane M. D., et al. 2007. Brain fatty acid synthase activates PPARalpha to maintain energy homeostasis. J. Clin. Invest. 117: 2539–2552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schneider J. G., Yang Z., Chakravarthy M. V., Lodhi I. J., Wei X., Turk J., Semenkovich C. F. 2010. Macrophage fatty-acid synthase deficiency decreases diet-induced atherosclerosis. J. Biol. Chem. 285: 23398–23409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stoops J. K., Ross P., Arslanian M. J., Aune K. C., Wakil S. J., Oliver R. M. 1979. Physicochemical studies of the rat liver and adipose fatty acid synthetases. J. Biol. Chem. 254: 7418–7426 [PubMed] [Google Scholar]
- 12.Semenkovich C. F. 1997. Regulation of fatty acid synthase (FAS). Prog. Lipid Res. 36: 43–53 [DOI] [PubMed] [Google Scholar]
- 13.Jensen-Urstad A. P., Semenkovich C. F. 2012. Fatty acid synthase and liver triglyceride metabolism: Housekeeper or messenger? Biochim. Biophys. Acta. 1821: 747–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ullman A. H., White H. B., 3rd 1981. Assay of fatty acid synthase using a bicyclic dione as substrate. Methods Enzymol. 72: 303–306 [DOI] [PubMed] [Google Scholar]
- 15.Luo X., Feng L., Jiang X., Xiao F., Wang Z., Feng G. S., Chen Y. 2008. Characterization of the topology and functional domains of RKTG. Biochem. J. 414: 399–406 [DOI] [PubMed] [Google Scholar]
- 16.Fiorentino M., Zadra G., Palescandolo E., Fedele G., Bailey D., Fiore C., Nguyen P. L., Migita T., Zamponi R., Di Vizio D., et al. 2008. Overexpression of fatty acid synthase is associated with palmitoylation of Wnt1 and cytoplasmic stabilization of beta-catenin in prostate cancer. Lab. Invest. 88: 1340–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakamura N., Rabouille C., Watson R., Nilsson T., Hui N., Slusarewicz P., Kreis T. E., Warren G. 1995. Characterization of a cis-Golgi matrix protein, GM130. J. Cell Biol. 131: 1715–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackson D. D., Stevens T. H. 1997. VMA12 encodes a yeast endoplasmic reticulum protein required for vacuolar H+-ATPase assembly. J. Biol. Chem. 272: 25928–25934 [DOI] [PubMed] [Google Scholar]
- 19.Fujiki Y., Hubbard A. L., Fowler S., Lazarow P. B. 1982. Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J. Cell Biol. 93: 97–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jornvall H. 1975. Acetylation of Protein N-terminal amino groups structural observations on alpha-amino acetylated proteins. J. Theor. Biol. 55: 1–12 [DOI] [PubMed] [Google Scholar]
- 21.Pasta S., Witkowski A., Joshi A. K., Smith S. 2007. Catalytic residues are shared between two pseudosubunits of the dehydratase domain of the animal fatty acid synthase. Chem. Biol. 14: 1377–1385 [DOI] [PubMed] [Google Scholar]
- 22.Sengupta S., Peterson T. R., Laplante M., Oh S., Sabatini D. M. 2010. mTORC1 controls fasting-induced ketogenesis and its modulation by ageing. Nature. 468: 1100–1104 [DOI] [PubMed] [Google Scholar]
- 23.Hsu P. P., Kang S. A., Rameseder J., Zhang Y., Ottina K. A., Lim D., Peterson T. R., Choi Y., Gray N. S., Yaffe M. B., et al. 2011. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science. 332: 1317–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qureshi A. A., Jenik R. A., Kim M., Lornitzo F. A., Porter J. W. 1975. Separation of two active forms (holo-a and holo-b) of pigeon liver fatty acid synthetase and their interconversion by phosphorylation and dephosphorylation. Biochem. Biophys. Res. Commun. 66: 344–351 [DOI] [PubMed] [Google Scholar]
- 25.Lodhi I. J., Yin L., Jensen-Urstad A. P., Funai K., Coleman T., Baird J. H., El Ramahi M. K., Razani B., Song H., Fu-Hsu F., et al. 2012. Inhibiting adipose tissue lipogenesis reprograms thermogenesis and PPARgamma activation to decrease diet-induced obesity. Cell Metab. 16: 189–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davies S. S., Pontsler A. V., Marathe G. K., Harrison K. A., Murphy R. C., Hinshaw J. C., Prestwich G. D., Hilaire A. S., Prescott S. M., Zimmerman G. A., et al. 2001. Oxidized alkyl phospholipids are specific, high affinity peroxisome proliferator-activated receptor gamma ligands and agonists. J. Biol. Chem. 276: 16015–16023 [DOI] [PubMed] [Google Scholar]
- 27.Billas I. M., Moulinier L., Rochel N., Moras D. 2001. Crystal structure of the ligand-binding domain of the ultraspiracle protein USP, the ortholog of retinoid X receptors in insects. J. Biol. Chem. 276: 7465–7474 [DOI] [PubMed] [Google Scholar]
- 28.Li Y., Choi M., Cavey G., Daugherty J., Suino K., Kovach A., Bingham N. C., Kliewer S. A., Xu H. E. 2005. Crystallographic identification and functional characterization of phospholipids as ligands for the orphan nuclear receptor steroidogenic factor-1. Mol. Cell. 17: 491–502 [DOI] [PubMed] [Google Scholar]
- 29.Ortlund E. A., Lee Y., Solomon I. H., Hager J. M., Safi R., Choi Y., Guan Z., Tripathy A., Raetz C. R., McDonnell D. P., et al. 2005. Modulation of human nuclear receptor LRH-1 activity by phospholipids and SHP. Nat. Struct. Mol. Biol. 12: 357–363 [DOI] [PubMed] [Google Scholar]
- 30.Choi M. G., Park T. S., Carman G. M. 2003. Phosphorylation of Saccharomyces cerevisiae CTP synthetase at Ser424 by protein kinases A and C regulates phosphatidylcholine synthesis by the CDP-choline pathway. J. Biol. Chem. 278: 23610–23616 [DOI] [PubMed] [Google Scholar]
- 31.Park T. S., O'Brien D. J., Carman G. M. 2003. Phosphorylation of CTP synthetase on Ser36, Ser330, Ser354, and Ser454 regulates the levels of CTP and phosphatidylcholine synthesis in Saccharomyces cerevisiae. J. Biol. Chem. 278: 20785–20794 [DOI] [PubMed] [Google Scholar]
- 32.Grijota-Martinez C., Diez D., Morreale de Escobar G., Bernal J., Morte B. 2011. Lack of action of exogenously administered T3 on the fetal rat brain despite expression of the monocarboxylate transporter 8. Endocrinology. 152: 1713–1721 [DOI] [PubMed] [Google Scholar]
- 33.Zhang C., Wendel A. A., Keogh M. R., Harris T. E., Chen J., Coleman R. A. 2012. Glycerolipid signals alter mTOR complex 2 (mTORC2) to diminish insulin signaling. Proc. Natl. Acad. Sci. USA. 109: 1667–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Plate C. A., Joshi V. C., Sedgwick B., Wakil S. J. 1968. Studies on the mechanism of fatty acid synthesis. XXI. The role of fructose 1,6-diphosphate in the stimulation of the fatty acid synthetase from pigeon liver. J. Biol. Chem. 243: 5439–5445 [PubMed] [Google Scholar]
- 35.Proulx K., Cota D., Woods S. C., Seeley R. J. 2008. Fatty acid synthase inhibitors modulate energy balance via mammalian target of rapamycin complex 1 signaling in the central nervous system. Diabetes. 57: 3231–3238 [DOI] [PMC free article] [PubMed] [Google Scholar]