Abstract
Recent genome-wide association studies (GWAS) have identified 35 loci that significantly associate with coronary artery disease (CAD) susceptibility. The majority of the genes represented in these loci have not previously been studied in the context of atherosclerosis. To characterize the roles of these candidate genes in the vessel wall, we determined their expression levels in endothelial, smooth muscle, and macrophage cells isolated from healthy, prelesioned, and lesioned mouse aortas. We also performed expression quantitative locus (eQTL) mapping of these genes in human endothelial cells under control and proatherogenic conditions. Of the 57 genes studied, 31 were differentially expressed in one or more cell types in disease state in mice, and the expression levels of 8 were significantly associated with the CAD SNPs in human cells, 7 of which were also differentially expressed in mice. By integrating human and mouse results, we predict that PPAP2B, GALNT4, MAPKAPK5, TCTN1, SRR, SNF8, and ICAM1 play a causal role in the susceptibility to atherosclerosis through a role in the vasculature. Additionally, we highlight the genetic complexity of a subset of CAD loci through the differential expression of multiple candidate genes per locus and the involvement of genes that lie outside linkage disequilibrium blocks.
Keywords: coronary artery disease, genome-wide association study, atherosclerosis, genetics, endothelium, smooth muscle cells, macrophages
Inheritance plays an important role in the pathogenesis of coronary artery disease (CAD) (1, 2). Recent advances in genome-wide association studies (GWAS) have provided insight into the many different genetic factors that contribute to the disease (3–7). Using data acquired from thousands of patients and healthy controls, these studies have collectively identified 35 genetic loci associated with CAD (8, 9). Although 10 of the recently identified CAD risk loci work through known risk factors, such as lipids and blood pressure, this is not the case for the majority of loci (3), implying that key pathways leading to coronary atherosclerosis are yet to be discovered. Given their firm association with disease risk, novel CAD loci provide a solid foundation to unravel disease networks.
As GWAS results do not provide functional information on the loci identified, additional studies are needed to determine the candidate genes and their role in disease. The immediate challenges associated with the validation of GWAS candidate genes include identifying the likely cell types in which the risk variants and genes function, determining which of the multiple candidate genes represented by each CAD locus contribute to disease, and defining the functions of poorly annotated candidate genes. Expression analyses of candidate genes under defined conditions in model organisms and their associations with risk variants in human samples provide a powerful way to address these issues and predict the causal candidate genes.
It is likely that at least some of the novel genes (that is, those not affecting known risk factors, such as plasma lipids or blood pressure) are perturbing vessel wall or inflammatory cell functions. Endothelial cells (EC) play a critical role in the initiation and progression of atherosclerosis. During atherogenesis, low density lipoproteins (LDL) accumulate in the intimal region of the vessel wall and over time become oxidized and otherwise modified to generate proinflammatory species capable of activating ECs and inducing the surface expression of monocyte adhesion molecules (10). Certain oxidized phospholipid species in the modified LDL, such as oxidized 1-palmitoyl-2-arachidonyl-sn-glycero-3-phosphocholine (oxPAPC), are likely to contribute to the inflammatory response (10). This leads to the recruitment of blood monocytes and lymphocytes to the vessel wall. The monocytes differentiate to macrophages and then take up the modified lipoproteins to give rise to the cholesterol-loaded “foam cells.” In response to inflammation, vascular smooth muscle cells (SMC) migrate from the media into the intima, forming a “fibrous cap.” The intima of vessels with advanced aortic lesions is a heterogeneous collection consisting predominantly of ECs, macrophages, and SMCs. We therefore hypothesized that some of the CAD candidate genes would be involved with the disease process in these cell types present in the vascular wall.
In this study, we report our results from a two-pronged approach in understanding the contribution of candidate CAD GWAS genes to disease pathogenesis. First, we utilized a mouse model to examine the expression of these genes with disease progression in the three major cell types found in atherosclerotic lesions. Each candidate gene is characterized by its transcription in vascular cells in different stages of atherosclerosis. Second, we used an expression quantitative trait locus (eQTL) mapping approach to understand how the CAD risk variants correlate with gene expression in ECs from healthy aortas of 147 human donors under control and proatherogenic conditions. Together, these data provide firm directions for predicting the causal candidate genes in CAD risk loci and elucidating their roles in the disease pathology.
MATERIALS AND METHODS
Animals
Unless otherwise indicated, C57BL/6J (BL6) male mice fed a chow diet were used for experiments. BL6 mice were obtained from the Jackson Laboratory or bred in our colony; BL6 ApoetmUnc mice (Apoe−/−) were maintained in a colony in the University of California Los Angeles (UCLA) vivarium. Mice at both 4 and 24 weeks of age were used for the study; BL6 wild-type (wt) mice were used in samples representing healthy vascular cells, and BL6 Apoe−/− mice were used for prelesioned vascular cells. The aortas from BL6 Apoe−/− mice fed a chow diet at 4 weeks of age were considered prelesioned, and BL6 Apoe−/− mice at 24 weeks of age exhibited clear atherosclerosis. For macrophage studies, Ldlr−/− mice on a BALB/cJ background were either maintained on a chow diet for isolation of macrophages or placed on a Western diet (Open Source D12079B) at 8 weeks of age for 16 weeks for isolation of lipid-loaded macrophages, commonly referred to as foam cells. Cells were collected from four to six mice per condition. Mice were euthanized using isoflurane in accordance with UCLA Animal Resource Committee policies. All protocols involving mice were approved by UCLA Institutional Review Board.
Microscopy/Oil Red O staining
Dissected aortas were frozen in the OCT compound. Sections of aortic arch were fixed onto Superfrost slides and stained with Oil Red O dye. Microscopy photos were taken at 20× magnification.
Mouse endothelial cell isolation
The cell isolation protocol was adapted from a previously published method (11) (supplementary Fig. I). After a mouse was euthanized, the chest cavity was opened, and lungs, trachea, and esophagus were removed, and the aortic arch was excised under a dissecting microscope. Care was taken to maintain consistency in dissection of the arch region. No perfusion of the vasculature was performed. After rinsing in cold PBS, the vessel was placed on a glass slide, the surrounding connective tissue was removed, and the aorta was opened en face. To visualize the endothelial layer, the opened aorta was stained with 30 μl hematoxylin for 3 min. The stain was rinsed off with cold PBS. The collagenase liberase blendzyme 2 (Roche) was diluted 1:100 with PBS, and 25 μl was added to the top of the aorta and incubated at 37°C for 8 min. The slide with collagenase-treated aorta was then placed under a dissecting microscope, and the ECs were gently pried off using a 26 gauge needle. This process continued until all ECs were removed, as determined by the lack of hematoxylin-dyed nuclei on the surface of the sample. The liquid containing the ECs was then pipetted with a thin pipet tip into RNA extraction buffer.
Macrophage/foam cell isolation
Ldlr−/− mice at 16 weeks of age either on chow or Western diet were injected intraperitoneally with 4% thioglycollate (Brewer Thioglycollate Medium, BD#211716). On the fourth day after injection, macrophages were collected through lavage of the peritoneal cavity using PBS buffer. Collected cells were treated with ACK lysis buffer to remove red blood cells and were then cultured overnight in DMEM media containing 20% FBS. The next morning the cell culture dishes were rinsed with PBS, and RNA from adherent cells was collected using buffer RLT from the Qiagen RNeasy kit.
Human cell culture and treatment
Human aortic endothelial cells (HAEC) were isolated from aortic explants of 147 heart transplant donors in the UCLA transplant program and grown to confluence in 100 mm dishes as described previously (12). All protocols involving humans were approved by UCLA Institutional Review Board. Cell purity was 95% as indicated by positive staining for platelet endothelial cell adhesion molecule 1 as well as factor 8 and by acetylated LDL take up assay. At 100% confluence, cells were treated in duplicate with either M199 media (Mediatech, Manassas, VA) containing 1% FBS (HyClone; Thermo Scientific, Logan, UT) or additionally with 40 μg/ml oxPAPC for 4 h.
Total RNA extraction
Total RNA from mouse aortic endothelial cells (MAEC) was extracted using the Ambion RNAqueous®-Micro Kit following the manufacturer's instructions. The buffer and collagenase solution containing scraped ECs was pipetted directly into 100 μl of Lysis buffer in a 200 μl PCR tube, vortexed, and then incubated at 42°C for 30 min prior to isolation. Total RNA from mouse SMCs was extracted from aortas with the intimal layer removed using procedure described above. The tissue was homogenized in Qiazol for 30 s, and RNA was isolated with the Qiagen RNeasy kit. The complete removal of cells present in the intimal layer was observed by lack of cells stained with hematoxylin. Total RNA from macrophage and HAECs was isolated using Qiagen RNeasy Kit. RNA quantity and quality was checked by the Nanodrop and the Agilent 2100 Bioanalyzer, respectively. Only RNA samples with RNA integrity numbers (RIN) of 7.0 or higher were used for subsequent processing. For mouse studies, RNA expression profiles from four to six mice per condition and for human studies RNA expression profiles from four samples per donor (two control-treated and two oxPAPC-treated) meeting quality control conditions were analyzed.
mRNA amplification
Since the total amount of RNA obtained from MAECs was around 100 picograms, insufficient for global expression analyses, we amplified the RNA using the NuGEN WT-Ovation One-Direct RNA amplification system. The procedure followed manufacturer's protocol; preamplification work was conducted in a sterile hood treated with RNase ZAP and DNA-OFF and using laboratory equipment exclusive to preamplification to eliminate contamination.
cDNA synthesis and RT-qPCR
As the output of the NuGEN WT-Ovation One-Direct RNA amplification system is cDNA, no separate cDNA synthesis was required for amplified RNA for MAECs. Macrophage and SMC cDNA was synthesized with Applied Biosystems High-Capacity cDNA Reverse Transcription Kit. Roche and KAPA Biosystems SYBR green reagents were used for RT-qPCR, and reactions were processed on 384 well plates on the Roche LightCycler 480. Of the 55 genes we determined to be represented by published GWAS loci (8), mouse gene homologs were identified for 52 genes and primers were designed for the mouse genes. Sequences for RT-qPCR primers are presented in supplementary Table I. Mouse gene 9530008L14Rik is the homolog of human gene C6orf105, and mouse gene Zfp259 is the homolog of human gene ZNF259. All other mouse genes share the names of their human homologs. Genes with Cp values less than 35 were considered to be expressed. Final expression values were calculated by normalizing the raw values for each transcript by the appropriate housekeeping gene (B2M for MAECs, lesions, and SMCs; 36B4 for macrophages), averaging the technical replicates, and then averaging these values for the four to six mice per condition.
Gene expression profiling and analysis
Gene expression profiles of the human cells were determined using the Affymetrix HT HG-U133A microarray, which contains 18,630 probes. Intensity values were normalized with the robust multiarray average normalization method implemented in the affy package in Bioconductor (13). Expression data are available in Gene Expression Omnibus accession GSE30169. Differential expression of mouse genes was assessed using Mann-Whitney U test and corrected for multiple comparisons using the qvalue package in R (14). Multivariate analysis across cell types was assessed by Hotelling's T2 test. Genes with false discovery rate (FDR) less than 10% were considered differentially expressed.
Genotyping and association analysis
HAEC genomic DNA was isolated using the Qiagen DNeasy kit. SNP genotyping was performed using the Affymetrix SNP 6.0 microarray platform as described previously (15). Microarray images were processed using the Affymetrix Genotyping Console 4.1 to make the SNP calls. SNPs were filtered out based on the following criteria: 296,063 SNPs with minor allele frequency less than 10%; 18,030 SNPs with less than 95% genotyping rate among the donors; 13,213 SNPs were set to missing because they were detected as heterozygous in haploid genotypes; and 3,890 SNPs were excluded because they failed the Hardy-Weinberg equilibrium test (P < 10−4). Principal component analysis with 11 HapMap3 populations indicated the presence of multiple ethnicities among the HAEC donors (supplementary Fig. II); therefore, a mixed-model approach to account for the population structure as implemented in the EMMAX program was used to perform the association analysis (16).
We applied the linear mixed model:
where μ = mean, x = SNP, β = SNP effect, and u = random effects due to genetic relatedness, with Var(u) = σg2K and Var(e) = σe2, where K = IBS (identity-by-state) matrix across all genotypes in the panel. We computed a restricted maximum likelihood estimate for σg2 and σe2, and we performed association based on the estimated variance component with an F test to test that β does not equal 0. Expression values of the 18,630 probes in control and oxPAPC-treated conditions were considered as quantitative traits, and the association analysis of SNPs was carried out using the 574,391 informative SNPs that passed the filtering criteria. In the donor population, 108 males and 39 females were present; therefore, sex was considered as a covariate in the mixed model for association analysis. For cis (local) associations, we considered the SNPs within the ±1 Mb window on each side of the probe locations. We considered the remaining SNPs for trans (distal) associations. For cis and trans associations, 35,186,928 and 10,665,717,400 P-values, respectively, were calculated. FDRs were calculated using the Benjamini and Hochberg (17) approach for the cis and trans association P-values in the control and oxPAPC dataset separately. P-value cut-offs of 1.16 × 10−4 and 1.12 × 10−4, which corresponded to 5% FDR, were used for the control and oxPAPC datasets, respectively, for cis associations. P-value cut-offs of 1.90 × 10−8 and 1.87 × 10−8, which corresponded to 5% FDR, were used for the control and oxPAPC datasets, respectively, for trans associations. The results of the association analysis are available for user-friendly querying at systems.genetics.ucla.edu. This resource can be used to determine how EC gene expression is regulated by additional CAD-associated SNPs that may be discovered as a result of future GWAS and meta-analyses, as well as with diseases whose pathology may involve the vascular endothelium.
RESULTS
A majority of GWAS candidate genes are expressed in at least one of the cell types present in mouse atherosclerotic lesions
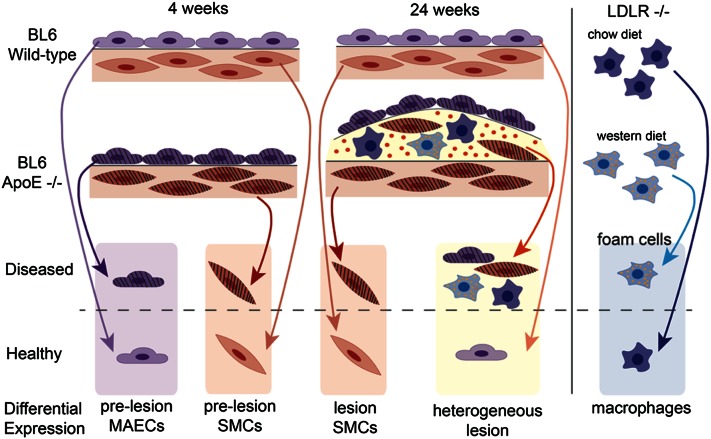
We determined the transcriptional profiles of ECs, SMCs, and macrophage/foam cells representing the three predominant cell types present in atherosclerotic lesions from healthy and diseased arteries, as well as lesions that represent a mixture of the three cell types (Fig. 1). While BL6 wt mice do not develop atherosclerosis on a chow diet at 4 or 24 weeks, BL6 Apoe−/−mice fed the same diet are hyperlipidemic and their arteries are “prelesioned” at 4 weeks and have atherosclerotic lesions at 24 weeks (supplementary Fig. I). Therefore, in the context of CAD, we considered vascular cells taken from BL6 wt mice to be “healthy” and those taken from BL6 Apoe−/− mice to be “diseased.” As BL6 Apoe−/− mice have not yet accumulated lipid deposits or macrophages at 4 weeks of age, the intimal cells taken from these mice are prelesioned endothelial cells, compared with healthy endothelial cells taken from their wt counterparts. The intimal samples taken from diseased mice at 24 weeks of age represent the heterogeneous cell composition present in advanced atherosclerotic lesions. Medial SMCs were collected from both healthy and diseased aortas at both prelesioned and lesioned stages. This set of atherosclerotic lesion cell types was completed by assaying the transcriptional profile of macrophages compared with lipid-loaded foam cells resembling those found in lesions. We used Ldlr−/− mice fed a chow diet or Western diet for these experiments, as this background is required for peritoneal macrophages to form foam cells (18).
Fig. 1.
Experimental outline of cell collection for gene expression studies of GWAS loci genes. MAECs isolated from 4-week-old BL6 wt aortas represent healthy MAECs, whereas MAECs isolated from Apoe−/− aortas are from prelesioned vessels and considered diseased. Aortas isolated from 24-week-old Apoe−/− mice had developed atherosclerotic lesions and their intimal cells were diseased, whereas the intima from wt mice at 24-weeks of age remained free of lipid deposits and were thus termed healthy. After the 4- and 24-week-old vessels had been stripped of their intimal cells, the remainder was composed primarily of SMCs and was used for the appropriate SMC samples. Macrophages isolated from Ldlr−/− mice had the same characteristics of the foam cells found in lesions and were considered diseased, while those collected from chow-fed Ldlr−/− mice did not have lipids present and served as the healthy control. Differential expression was determined as the fold-change from the expression level of a gene at the baseline condition of healthy to the expression level of the gene in the diseased condition.
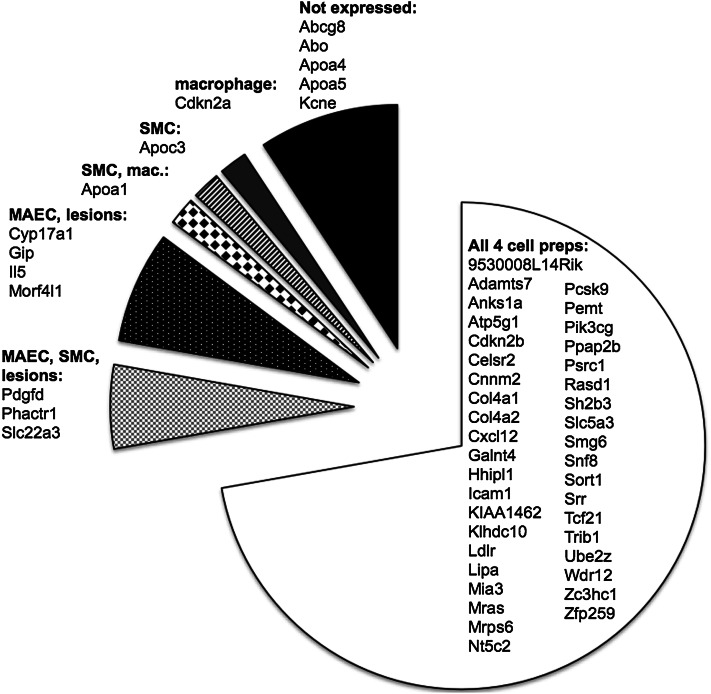
The 35 published GWAS loci for CAD represent 55 candidate genes reported in the original studies (3). We utilized RT-qPCR to assess the presence of the mouse homologs for 51 of these transcripts among the three sets of mouse cell types as well as among advanced lesions. Although 46 of the GWAS candidate genes were expressed in at least one of the cell preparations, there were 5 transcripts whose expression was below the detection threshold in all samples. We did not detect the expression of Abcg8, Abo, Apoa4, Apoa5, or Kcne, suggesting that the involvement of these genes in the pathogenesis of atherosclerosis is not in the vascular wall (Fig. 2). We did not identify any transcripts that were undetectable under the healthy condition of a cell type and detectable in the diseased condition, or vice versa. Thirty-six of the candidate genes were expressed in all three cell types. Pdgfd, Phactr1, and Slc22a3 were only expressed in MAECs, SMCs, and lesions, while Cyp17a1, Gip, Il5, and Morf4l1 were only expressed in MAECs and advanced lesions.
Fig. 2.
Expression of GWAS CAD candidate gene homologs in vascular cell types present during atherosclerosis. Transcripts were detected for 54 of the GWAS candidate genes in at least one of the cell types assayed, as shown by the different shades in the chart: MAECs, intimal lesions, SMCs, and macrophages (mac). The 6 genes with no detectable gene transcript are in the black section. Three genes (Icam1, Klhdc10, and Galnt4) that are not in GWAS loci but show significant association with CAD SNPs in human endothelial cells were also assayed.
Differential expression of GWAS candidate genes in disease conditions in mice
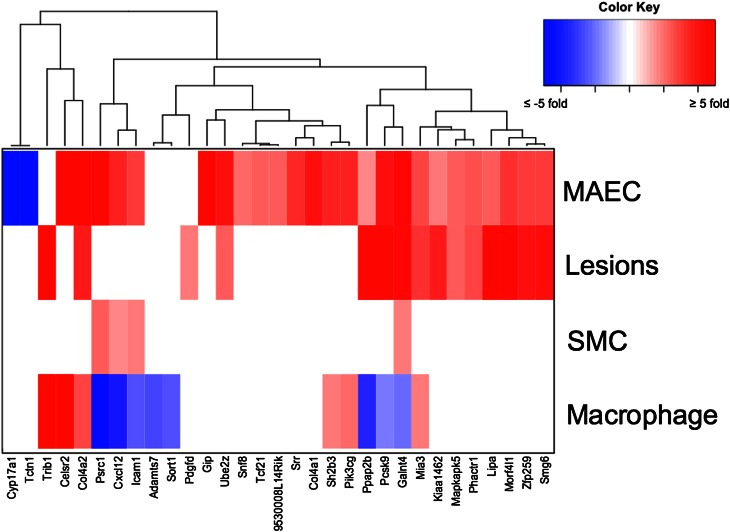
To determine whether the candidate genes were differentially expressed in diseased cells, we calculated the change in gene expression in the three cell types during lesion development using the healthy condition as the baseline (Fig. 1). Thirty-one of the 46 GWAS candidate genes that are expressed in the vessel wall showed differential expression between the diseased and healthy states in at least one of the comparisons (Fig. 3, supplementary Table II). A majority of these genes are upregulated in diseased cells, with only 9 of them showing downregulation compared with healthy cells. None of the 31 candidate genes are differentially expressed in prelesioned medial SMCs, although 4 candidate genes showed differential expression in SMCs taken from atherosclerotic lesions in mice at 24 weeks of age. Twelve candidate genes showed differential expression in only one cell type: Adamts7, Celsr2, and Sort1 in foam cells; Cyp17a1, Col4a1, Gip, Snf8, Srr, Tcf21, Tctn1, and 9530008L14Rik in prelesioned MAECs; and Pdgfd in the lesioned intima exclusively.
Fig. 3.
GWAS CAD candidate genes exhibiting differential expression from healthy to diseased states in vascular cell types. The 31 candidate genes that show differential expression in at least one of the cell types are grouped using hierarchical clustering based on the fold-change and direction of expression. No genes were differentially expressed at the four-week medial SMC time point.
GWAS candidate genes with differential expression from healthy to disease state in more than one cell type
Galnt4 exhibited statistically significant differential expression in all four cell preparations, while seven genes showed dysregulation in the diseased condition in three of the cell types tested (Fig. 3, supplementary Table II). Col4a2, Mia3, Pcsk9, and Ppap2b were upregulated in diseased MAECs, foam cells, and atherosclerotic lesions. Cxcl12, Icam1, and Psrc1 were upregulated in diseased MAECs and medial cells, but they were downregulated in foam cells. Six of these transcripts had a greater than 10-fold induction, including Col4a2 with an 11-fold increase in diseased MAECs, Celsr2 with a 22- and 17- fold increase in endothelial cells and foam cells, respectively, and Trib1 with a 15-fold increase in atherosclerotic lesions and 10-fold increase in foam cells. Seven genes showed differential expression exclusively in the prelesioned endothelium and the lesioned intima: Kiaa1462, Lipa, Mapkapk5, Phactr1, Smg6, Ube2z, and Zfp259. Hierarchical clustering of the genes based on the level of dysregulation identified two distinct clusters: one in which CAD genes are upregulated in diseased ECs and lesion cells, and another in which they are downregulated in macrophage/foam cells (Fig. 3). To identify differential expression from healthy to diseased mice across all four cell types as a whole, we applied multivariate statistical testing for each gene and found statistical significance across tissues in six genes (supplementary Table V). These differentially expressed genes were Col4a2, Cxcl12, Icam1, Mia3, Ppap2b, and Pik3cg, all of which were also determined as differentially expressed in separate cell types.
Genetic regulation of human endothelial genes in GWAS loci
To understand how the genetic variants associated with CAD perturb gene expression and thereby contribute to disease susceptibility, we studied the genetic regulation of transcript abundance in HAECs. We cultured the cells under control conditions or conditions in which HAECs were stimulated with oxPAPC for 4 h to mimic the activation of the endothelium with mmLDL. We observed that approximately 1,500 genes had a significant cis association with nearby SNPs in control or oxPAPC-treated cells, 1,239 of which overlap. Trans associations in control and oxPAPC-treated cells were observed in 185 and 164 genes, respectively, and 22 of these overlapped between two datasets (supplementary Fig. III).
We then specifically focused on how the SNPs most significantly associated with CAD in multiple GWAS loci (8) affect nearby gene expression. Of these SNPs, 17 were genotyped in our study. We determined proxy SNPs for 11 of them, but we did not have information for 7 of the peak SNPs (supplementary Table III). In 7 of the 28 CAD loci studied, we observed significant association between the peak CAD SNP or its proxy SNP and the nearby transcript levels, suggesting that these endothelial genes play a role in disease susceptibility (Table 1). It is plausible that the CAD SNPs are functional in tissues as well as in endothelial cells; therefore, to determine whether these 7 SNPs had significant associations with nearby genes in cells other than HAECs, we queried the Phenotype-Genotype Integrator (ncbi.nlm.nih.gov/gap/PheGenI), the University of Chicago eQTL database (eqtl.uchicago.edu), and Genevar database (19) by looking up the associations of the 7 SNPs with nearby gene expression from various eQTL studies. In 4 loci, cis eQTLs were observed in monocytes obtained from 1,490 humans (20), and in adipose and skin tissues as well as lymphoblastoid cell lines obtained from 856 humans (21) (Table 1).
TABLE 1.
Association of CAD GWAS peak SNPs with nearby genes
| SNP | Locus | Proxya | GWAS P-value | Risk Allele | Reported Gene(s) | cis eQTL Gene(s) | eQTL P-value (Control, oxPAPC) | eQTLs in Other Tissuesc |
| rs11556924 | 7q32.2 | 9.00E-18 | C | ZC3HC1 | KLHDC10 | 6.72E-8, 1.16E-5 | KLHDC10 (m) | |
| rs216172 | 17p13.3 | rs7217226 (r2 = 1) | 1.00E-09 | C | SMG6, SRR | SRR | 1.77E-7, 3.42E-8 | SRR (a, l) |
| rs46522 | 17q21.32 | 2.00E-08 | T | UBE2Z, GIP, ATP5G1, SNF8 | SNF8 | 5.85E-5, 1.50E-4 | ATP5G1 (m, a, l, s), UBE2Z (m, a, l, s) | |
| rs3184504 | 12q24.12 | rs653178 (r2 = 0.873) | 6.00E-06b | T | SH2B3 | MAPKAPK5 | 1.79E-4, 4.21E-2 | |
| rs3184504 | 12q24.12 | rs653178 (r2 = 0.873) | 6.00E-06b | T | SH2B3 | TCTN1 | 8.92E-4, 1.19E-3 | |
| rs7136259 | 12q21.33 | 6.00E-10 | T | ATP2B1 | GALNT4 | 8.80E-4, 4.26E-5 | GALNT4 (m) | |
| rs17114036 | 1p32.2 | rs6588635 (r2 = 0.831) | 4.00E-19 | A | PPAP2B | PPAP2B | 2.94E-4, 3.36E-3 | |
| rs1122608 | 19p13.2 | 1.00E-09 | G | LDLR | ICAM1 | 1.22E-3, 9.85E-4 |
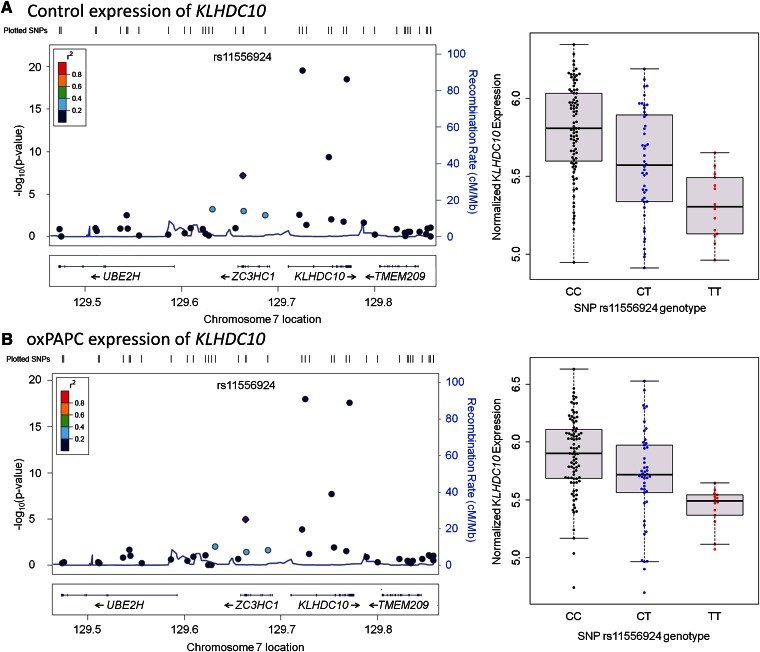
We observed that in four of the seven loci, the significantly associated gene was outside of the linkage disequilibrium (LD) block. For example, in the 7q32.2 locus, SNP rs11556924 encodes a missense mutation in the ZC3HC1 gene; however, we identified that the SNP was associated with the expression level of the nearby KLHDC10 gene, which also had a significant cis association in monocytes with the same SNP (20). The risk allele (C) was associated with higher expression of KLHDC10 (Fig. 4). In the 17q24.12 locus, SNP rs3184504 encodes a missense mutation in the SH2B3 gene; however, in our dataset, its proxy SNP rs653178 was associated with the transcript levels of two nearby genes, MAPKAPK5 and TCTN1. The risk allele (T) was associated with lower expression of MAPKAPK5 but higher expression of TCTN1 (supplementary Figs. IV and V). The association between SNP rs653178 and MAPKAPK5 was significant only in control cells but not in oxPAPC-treated cells, suggesting a differential role for this gene in various disease stages. In the 12q21.33 locus, SNP rs7136259, an intergenic SNP located between ATP2B1 and MRPLP2P1, was associated with the expression level of the nearby GALNT4 gene, and this association was replicated in monocytes (20). The risk allele (T) was associated with lower expression of GALNT4 (supplementary Fig. VI). In the 19p13.2 locus, the risk allele (G) of the SNP rs1122608, located in an intron of the SMARCA4 gene, was associated with the lower levels of the nearby ICAM1 transcript (supplementary Fig. VII).
Fig. 4.
Regional association plots of KLHDC10 transcript levels. Left panels show the regional association of KLHDC10 gene expression in (A) control and (B) oxPAPC conditions. Association of the genotyped SNPs and the transcript expression as indicated by –log10(P) values are plotted. The color of the SNPs indicates the strength of linkage disequilibrium with the peak SNP, which is in purple. Recombination rates are based on 1,000 genomes European populations (46). Right panels show the expression levels of KLHDC10 in individuals based on their SNP rs11556924 genotypes.
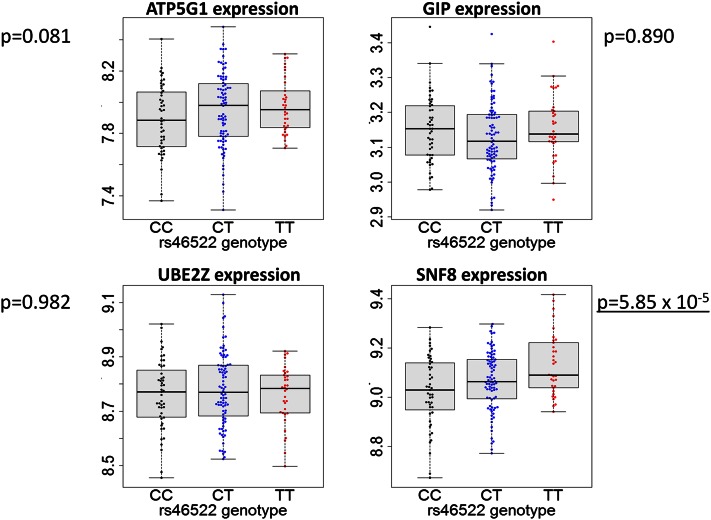
In three of the seven loci, the significantly associated gene was within the same LD block as the variant, in some cases allowing for differentiation between multiple candidate genes reported per locus. For example, in the 17q21.32 locus, the risk allele (T) of SNP rs46522, located in the intron of UBE2Z gene, was associated with higher expression level of the SNF8 gene but not the other three genes in the same locus (Fig. 5, supplementary Fig. VIII). In monocytes (20), however, the same SNP was associated with the expression levels of two other genes in the locus, ATP5G1 and UBE2Z, suggesting a different mechanism for disease susceptibility in endothelial cells and monocytes. In the 17p13.3 locus, the CAD SNP rs216172 is located in an intron of the SMG6 gene; however, we observed that the proxy SNP rs7217226 for rs216172 (r2 = 1 in 1,000 genomes of European population) was associated with the expression level of the nearby SRR gene. The risk allele (C) of the SNP rs216172 was associated with the higher expression of SRR (supplementary Fig. IX). In the 1p32.2 locus, SNP rs17114036 is located in an intron of PPAP2B gene. Its proxy SNP rs6588635 (r2 = 0.831 in 1,000 genomes of European population) was associated with the PPAP2B transcript levels (supplementary Fig. X). The risk allele (A) was associated with lower expression of PPAP2B.
Fig. 5.
Association of gene expression in the 17q21.32 locus with the CAD risk SNP. The linkage disequilibrium block in the 17q21 locus contains four genes. The expression levels of these four genes under control conditions were stratified based on the donor genotype of the CAD risk SNP rs46522. P-values of association were calculated based on the mixed model detailed in Materials and Methods.
Differential expression of predicted causal human GWAS genes in murine vascular cells in vivo
To determine how the genes associated with human GWAS SNPs change in different stages of the disease, we evaluated their expression in ECs, lesions, SMCs, and macrophages in our mouse models. Among the 8 genes with cis eQTLs, seven of them (all but Klhdc10) showed differential expression in one of the cell types in at least one of the disease conditions (Table 2). Srr was upregulated in the prelesioned MAECs and this regulation was consistent with the association of higher expression of this gene with the risk allele of the CAD SNP in HAECs. Snf8 was upregulated in the SMCs from atherosclerotic mice and its high expression was associated with the risk allele in HAECs. Three cis eQTL genes in HAECs, Galnt4, Icam1, and Ppap2b, were dysregulated in different stages of atherosclerosis in the three cell types we studied in mice, suggesting a multicellular role for these genes. Mapkapk5 and Tctn1 dysregulation were in the opposite direction of genetic regulation by the risk SNP suggesting that these two genes play different roles in disease susceptibility and progression. Klhdc10 expression did not change with disease despite a significant association with the CAD risk SNP. Taken together, these results suggest that the majority of the cis eQTL genes identified in HAECs play a role in cells present in the vasculature during disease progression.
TABLE 2.
Regulation of cis eQTL gene expression in mouse and humans
| Gene | Fold-change: Mice | Direction with Respect to Healthy Cells | Mouse Cell Type | HAEC eQTL Risk Allele Association |
| SRR | 2.9** | Up | MAEC | High |
| SNF8 | 1.4* | Up | SMC | High |
| GALNT4 | 4.3* | Up | MAEC | Low |
| 2.6* | Up | Lesion | ||
| 1.7** | Down | Macrophage/foam | ||
| ICAM1 | 2.0** | Up | Lesion | Low |
| 1.5** | Up | SMC | ||
| 2.1** | Up | Macrophage/foam | ||
| PPAP2B | 8.6** | Up | Lesion | Low |
| 2.6** | Down | Macrophage/foam | ||
| MAPKAPK5 | 2.1* | Up | Lesion | Low |
| TCTN1 | 6.3* | Down | MAEC | High |
| KLHDC10 | – | – | – | High |
*P < 0.05, **P < 0.01 based on Student t-test.
DISCUSSION
Epidemiological and family studies predict the genetic component of CAD risk to be 40% to 60% (8). While GWAS have identified 35 genomic loci associated with a 7% to 37% increase in CAD risk, the majority of the identified loci are not associated with known risk factors and as such represent novel pathways involved in disease etiology. CAD is a complex disease that involves a variety of cell types, tissues, and pathways. To study the role of these genes in the vessel wall specifically, we assembled a comprehensive dataset using an in vivo mouse model of atherosclerosis and in vitro aortic endothelial gene expression from approximately 150 human donors. Discrete samples consisting of three major cell types that contribute to atherosclerosis, ECs, SMCs, and macrophages, along with the heterogeneous mixture of cells that constitute atherosclerotic lesions in the intima, were assessed for changes in gene expression that correlate with the progression of disease. This dataset allowed for the thorough assessment of GWAS candidate gene expression in healthy and diseased vascular walls. Our data address two important challenges: the first is the identification of causal genes at GWAS loci, and the second is their expression patterns in disease-relevant cell types through which they contribute to disease.
Our mouse GWAS gene expression dataset provides new information for nearly all CAD candidate genes. There are many candidate genes for which no additional information linking them to the disease is available, such as the differentially expressed genes shown in Fig. 6: 9530008L14Rik, Adamts7, Cyp17a1, Kiaa1462, Mia3, Morf4l1, Phactr1, Ppap2b, Sh2b3, Smg6, Srr, Tcf21, Ube2z, and Zfp259. Of the differentially expressed genes that have prior association with atherosclerosis, Celsr2, Pcsk9, Pik3cg, Psrc1, and Srr, the cell type differentially expressing these genes had not been previously observed. It is probable that the six genes with undetectable transcripts in the vascular cell types are not directly active and do not affect atherosclerosis through action in the vasculature. The seven genes that are differentially expressed in both the prelesioned endothelium and the lesioned intima but with no differential expression in smooth muscle or macrophage cell type (Fig. 3) may be induced in the endothelium early during atherogenesis and continue their dysregulation throughout the development of the atheroma. The seven genes that show differential expression in three cell types may be candidates for more widespread involvement in CAD. We can also use this dataset to gain further insight into the functions of well-studied candidate genes, such as PCSK9. The gene product of PCSK9 promotes LDLR degradation and has a direct proatherogenic effect (22, 23), and it has emerged as a new pharmacological target for hypercholesterolemia with various PCSK9 inhibitors now being evaluated in clinical trials (24, 25). Although this protein has been reported as present in human atherosclerotic plaques (26), no previous studies describe expression in preatherosclerotic endothelial cells. Our studies suggest a role for PCSK9 in the arterial wall and provide information suggesting an additional role for this potential therapeutic target.
Fig. 6.
Overview of GWAS differential gene expression in the cells of the murine vessel wall.
Utilizing both in vivo mouse and in vitro human gene expression analysis, we can draw some preliminary conclusions for GWAS loci that represent more than one gene, as summarized in Fig. 6 and supplementary Table IV. The involvement of some loci appears to be relatively straightforward, such as the 21q22 locus. The genes SLC5A3, MRPS6, and KCNE2 are reported as candidates for this region, and our data indicate that only one gene, MRPS6, is differentially expressed in only one cell type, SMCs, suggesting that this gene and cell type are responsible. Other loci, such as 17q21.32, seem to be more complex. For example, four candidate genes are reported for this locus: UBE2Z, GIP, ATP5G1, and SNF8 (3). In mice, we find that Atp5g is only differentially expressed in foam cells, Ube2z and Gip are differentially expressed exclusively in the intimal layer, and Snf8 is upregulated specifically in the medial SMCs. On the other hand, the CAD risk SNP was associated with SNF8 expression in HAECs but with ATP5G and UBE2Z in monocytes (20), adipose, and skin tissues, as well as in lymphoblastoid cell lines (21). Since all four of the genes in this locus are expressed in different cell types and at different stages of disease progression, these results suggest that this locus plays a complex role in contributing to CAD. The locus 11q23.3 also represents multiple genes: ZNF259 and APOA5-A4-C3-A1. Although apolipoproteins have been well-established as contributors to heart disease through lipid metabolism, we found that another gene at this locus, ZNF259, was upregulated in the prelesioned endothelium and intimal lesions, indicating the need for follow-up studies to identify the function of this gene. Another complex locus is 10q24.32, which harbors the genes CYP17A1, CNNM2, and NT5C2 and has been associated with CAD risk and blood pressure in humans (27). Our results show that only one gene, CYP17A1, is differentially expressed in the prelesioned endothelium, suggesting that the CYP17A1 gene plays a role in increasing CAD susceptibility.
An additional facet of this GWAS locus complexity is that the candidate genes they represent may not necessarily be defined by LD blocks. Our eQTL results based on HAEC gene expression demonstrate that genes outside of the LD block of a given locus may also be candidates for involvement in disease susceptibility. Four of the seven CAD SNPs have significant associations with nearby genes that are not in the same LD block as the SNP, perhaps signifying that these SNPs may tag regulatory variants rather than protein-coding regions. This is consistent with recent evidence from the 1,000 Genomes Project that showed only 19% of GWAS signals resulted in nonsynonymous changes in protein coding genes, suggesting that the majority of the GWAS signals are associated with the regulation of gene expression (28).
The integration of these gene expression data from two independent experimental systems allowed us to confidently predict that a subset of GWAS candidate genes contribute to CAD through a role in the artery wall. Some of the candidate genes we have identified have already been shown to play a role in disease progression. For example, GALNT4 expression is associated with a SNP that is located between ATP2B1 and MRPLP2P1 in the 12q21.33 locus in both HAECs and monocytes. The GALNT4 gene encodes the N-acetyl galactosaminyl transferase 4 enzyme and is thought to play an important role in endothelial-platelet interactions by O-glycosylating the threonine residues of the P-selectin glycoprotein ligand (PSGL-1) (29). Coding polymorphisms in this gene have been associated with incidence of myocardial infarction in an Irish population (30), and PSGL-1 interacts with P-selectin expressed on the surface of ECs and regulates tethering and rolling of monocytes on these cells. The CAD risk allele was associated with lower expression of GALNT4 in HAECs, and the transcript was downregulated in macrophages but increased in prelesioned MAECs, lesioned intima, and medial smooth muscle cells. Taken together, our results and reports from previous studies suggest that GALNT4 confers susceptibility to CAD via structural variants that affect its protein structure and regulatory variants in the 12q21.33 locus that affect its expression, thereby mediating platelet-endothelial interactions.
Another relatively well-studied locus is 19p13.2. Although we predict the causal gene with respect to vascular activity in this region to be ICAM1, this locus also contains LDLR, a gene with a well-established role in cholesterol homeostasis and atherosclerosis. One possibility is that the CAD SNP is in LD with other SNPs that encode missense mutations in the LDLR protein, thereby affecting its function. On the other hand, ICAM1 has also been shown to play a critical role in the initiation of atherosclerosis (31), and we have observed upregulation in MAECs, SMCs from lesioned aortas, and macrophage foam cells (Table 2). In the context of these findings, it is probable that the CAD-associated variants in this locus have a multitude of regulatory functions that affect the structure of LDLR and expression of ICAM1 in different cell types and that affect atherosclerosis.
In the 1p32.2 locus, we predict PPAP2B to be the causal gene. PPAP2B encodes the lipid phosphate phosphohydrolase 3 (LPP3) enzyme, which belongs to the LPP family of cell-surface-associated lipid phosphatases that hydrolyze and inactivate lysophosphatidic acid (LPA) and the related lysophospholipid sphingosine-1-phoshate. PPAP2B also functions as a cell-associated integrin ligand (32, 33). We observed that lower expression of PPAP2B was associated with the risk allele in HAECs, whereas its expression was upregulated in lesions and MAECs. This was consistent with different functions that have been attributed for PPAP2B in ECs and SMCs. In ECs, PPAP2B localizes to adherens junctions and regulates β-catenin signaling, thereby affecting cell migration (34). In SMCs, it regulates lysophospholipid signaling responses and affects SMC phenotypic plasticity (35). Our results indicate that PPAP2B may be involved in susceptibility to atherosclerosis via its dual function in vascular wall cells.
In the 12q24.12 loci, the significant association between the CAD SNP and nearby gene expression was outside the LD region. Based on our eQTL and in vivo vascular expression data, we predict that MAPKAPK5 and TCTN1 are causal genes for CAD risk in the 12q24.12 locus. These genes have not been previously studied in the context of atherosclerosis. MAPKAPK5 is a protein kinase that functions downstream of p38 in the MAPK pathway, a mediator of inflammatory response (36). Studies have shown that it is active in endothelial cells, where it mediates EC migration and angiogenesis (37). Little is known regarding a role for TCTN1 in cardiovascular disease. It has a role in hedgehog signaling during development (38), and as it plays a role in ciliogenesis, mutations in this gene cause Joubert syndrome (39), a ciliopathy that affects the brain. There is evidence for ciliated endothelial cells in atherogenic, disturbed-flow regions of the vessel wall (40), and it is possible this gene functions in atherosclerosis through affecting ciliogenesis.
In the 7q32.2 region, based on HAEC and monocyte eQTL results, we predict KLHDC10 to be the causal gene. This gene is not well characterized, but it has recently been shown to regulate oxidative stress-induced apoptosis in multiple cell types (41). Because the expression levels of this gene were not regulated in the different vascular cell types with disease progression in vivo, it is probable that the SNP may play a regulatory role for the expression levels of this gene without consequences for increased CAD risk.
In the 17p13.3 and the 17q21.32 loci, we suggest that the human genes SRR and SNF8 are candidates for further study in vascular cells, based on corresponding expression data between the two different experimental approaches reported in this study. SRR codes for serine racemase, which synthesizes D-serine from L-serine. SRR variants have been associated with schizophrenia (42), and they are thought to be primarily expressed in the brain (43). SNF8 codes for a subunit of the endosomal sorting complex required for transport II (ESCRT-II) (44) and has been shown to mediate sorting of ubiquitinated proteins for lysosomal degradation (45). These genes have not been shown to play a role in the etiology of atherosclerosis previously; however, our results suggest a role for endothelial involvement of these genes in atherosclerosis.
As additional GWAS studies are completed and new loci identified, it is important to recognize that simply identifying a genetic variant will not reveal the extent of involvement of these loci in the disease process. In this study, we provide comprehensive expression data for 58 candidate genes in different cell types and disease conditions of the vasculature. We believe this resource will be valuable for researchers planning focused functional follow-up studies on these loci. Furthermore, as we have shown here, in many cases the significantly associated loci play complex roles; therefore, in identifying genetic regions for follow-up studies, it will be useful to consider areas that may not be within the defined LD block and potential regulatory regions in addition to the candidate genes reported. Further, it is possible that the genomic regions marked by these loci contribute to disease through more than one pathway and in multiple cell types. Detailed biochemistry studies are required to understand the function of candidate genes in the vascular cells in relation to CAD susceptibility.
Supplementary Material
Footnotes
Abbreviations:
- CAD
- coronary artery disease
- EC
- endothelial cell
- eQTL
- expression quantitative trait locus
- GWAS
- genome-wide association study
- HAEC
- human aortic endothelial cell
- LD
- linkage disequilibrium
- MAEC
- mouse aortic endothelial cell
- oxPAPC
- oxidized 1-palmitoyl-2-arachidonoyl-sn-glycero-3-phosphocholine
- SMC
- smooth muscle cell
- SNP
- single nucleotide polymorphism
- wt
- wild-type
This work was supported by National Institutes of Health Grants HL-30568 and HL-28481 (A.J.L. and J.A.B.), Leducq Transatlantic Networks of Excellence in Cardiovascular Research Program Award (A.J.L.), American Heart Association predoctoral fellowship 10PRE3420032 and Ruth L. Kirschstein National Research Service Award GM-07185 (A.E.), American Heart Association postdoctoral fellowship 10POST3660048, and Ruth L. Kirschstein National Research Service Award T32 HL-69766 (M.C.).
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of 10 figures and 5 tables.
REFERENCES
- 1.Stylianou I. M., Bauer R. C., Reilly M. P., Rader D. J. 2012. Genetic basis of atherosclerosis: insights from mice and humans. Circ. Res. 110: 337–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kathiresan S., Srivastava D. 2012. Genetics of human cardiovascular disease. Cell. 148: 1242–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schunkert H., Konig I. R., Kathiresan S., Reilly M. P., Assimes T. L., Holm H., Preuss M., Stewart A. F., Barbalic M., Gieger C., et al. 2011. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat. Genet. 43: 333–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.IBC 50K CAD Consortium 2011. Large-scale gene-centric analysis identifies novel variants for coronary artery disease. PLoS Genet. 7: e1002260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clarke R., Peden J. F., Hopewell J. C., Kyriakou T., Goel A., Heath S. C., Parish S., Barlera S., Franzosi M. G., Rust S., et al. 2009. Genetic variants associated with Lp(a) lipoprotein level and coronary disease. N. Engl. J. Med. 361: 2518–2528 [DOI] [PubMed] [Google Scholar]
- 6.Lu X., Wang L., Chen S., He L., Yang X., Shi Y., Cheng J., Zhang L., Gu C. C., Huang J., et al. 2012. Genome-wide association study in Han Chinese identifies four new susceptibility loci for coronary artery disease. Nat. Genet. 44: 890–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soranzo N., Spector T. D., Mangino M., Kuhnel B., Rendon A., Teumer A., Willenborg C., Wright B., Chen L., Li M., et al. 2009. A genome-wide meta-analysis identifies 22 loci associated with eight hematological parameters in the HaemGen consortium. Nat. Genet. 41: 1182–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peden J. F., Farrall M. 2011. Thirty-five common variants for coronary artery disease: the fruits of much collaborative labour. Hum. Mol. Genet. 20: R198–R205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roberts R., Stewart A. F. 2012. Genes and coronary artery disease: where are we? J. Am. Coll. Cardiol. 60: 1715–1721 [DOI] [PubMed] [Google Scholar]
- 10.Lee S., Birukov K. G., Romanoski C. E., Springstead J. R., Lusis A. J., Berliner J. A. 2012. Role of phospholipid oxidation products in atherosclerosis. Circ. Res. 111: 778–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jongstra-Bilen J., Haidari M., Zhu S. N., Chen M., Guha D., Cybulsky M. I. 2006. Low-grade chronic inflammation in regions of the normal mouse arterial intima predisposed to atherosclerosis. J. Exp. Med. 203: 2073–2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Navab M., Hough G. P., Stevenson L. W., Drinkwater D. C., Laks H., Fogelman A. M. 1988. Monocyte migration into the subendothelial space of a coculture of adult human aortic endothelial and smooth muscle cells. J. Clin. Invest. 82: 1853–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gautier L., Cope L., Bolstad B. M., Irizarry R. A. 2004. Affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 20: 307–315 [DOI] [PubMed] [Google Scholar]
- 14.Storey J. D., Tibshirani R. 2003. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA. 100: 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Romanoski C. E., Lee S., Kim M. J., Ingram-Drake L., Plaisier C. L., Yordanova R., Tilford C., Guan B., He A., Gargalovic P. S., et al. 2010. Systems genetics analysis of gene-by-environment interactions in human cells. Am. J. Hum. Genet. 86: 399–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang H. M., Sul J. H., Service S. K., Zaitlen N. A., Kong S. Y., Freimer N. B., Sabatti C., Eskin E. 2010. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 42: 348–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benjamini Y., Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B. 57: 289–300 [Google Scholar]
- 18.Li A. C., Binder C. J., Gutierrez A., Brown K. K., Plotkin C. R., Pattison J. W., Valledor A. F., Davis R. A., Willson T. M., Witztum J. L., et al. 2004. Differential inhibition of macrophage foam-cell formation and atherosclerosis in mice by PPARalpha, beta/delta, and gamma. J. Clin. Invest. 114: 1564–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang T. P., Beazley C., Montgomery S. B., Dimas A. S., Gutierrez-Arcelus M., Stranger B. E., Deloukas P., Dermitzakis E. T. 2010. Genevar: a database and Java application for the analysis and visualization of SNP-gene associations in eQTL studies. Bioinformatics. 26: 2474–2476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeller T., Wild P., Szymczak S., Rotival M., Schillert A., Castagne R., Maouche S., Germain M., Lackner K., Rossmann H., et al. 2010. Genetics and beyond--the transcriptome of human monocytes and disease susceptibility. PLoS ONE. 5: e10693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grundberg E., Small K. S., Hedman A. K., Nica A. C., Buil A., Keildson S., Bell J. T., Yang T. P., Meduri E., Barrett A., et al. 2012. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat. Genet. 44: 1084–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farnier M. 2011. The role of proprotein convertase subtilisin/kexin type 9 in hyperlipidemia: focus on therapeutic implications. Am. J. Cardiovasc. Drugs. 11: 145–152 [DOI] [PubMed] [Google Scholar]
- 23.Denis M., Marcinkiewicz J., Zaid A., Gauthier D., Poirier S., Lazure C., Seidah N. G., Prat A. 2012. Gene inactivation of proprotein convertase subtilisin/kexin type 9 reduces atherosclerosis in mice. Circulation. 125: 894–901 [DOI] [PubMed] [Google Scholar]
- 24.Stein E. A., Mellis S., Yancopoulos G. D., Stahl N., Logan D., Smith W. B., Lisbon E., Gutierrez M., Webb C., Wu R., et al. 2012. Effect of a monoclonal antibody to PCSK9 on LDL cholesterol. N. Engl. J. Med. 366: 1108–1118 [DOI] [PubMed] [Google Scholar]
- 25.Marian A. J. 2010. PCSK9 as a therapeutic target in atherosclerosis. Curr. Atheroscler. Rep. 12: 151–154 [DOI] [PubMed] [Google Scholar]
- 26.Turpeinen H., Raitoharju E., Oksanen A., Oksala N., Levula M., Lyytikainen L. P., Jarvinen O., Creemers J. W., Kahonen M., Laaksonen R., et al. 2011. Proprotein convertases in human atherosclerotic plaques: the overexpression of FURIN and its substrate cytokines BAFF and APRIL. Atherosclerosis. 219: 799–806 [DOI] [PubMed] [Google Scholar]
- 27.Ehret G. B., Munroe P. B., Rice K. M., Bochud M., Johnson A. D., Chasman D. I., Smith A. V., Tobin M. D., Verwoert G. C., Hwang S. J., et al. 2011. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 478: 103–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abecasis G. R., Auton A., Brooks L. D., DePristo M. A., Durbin R. M., Handsaker R. E., Kang H. M., Marth G. T., McVean G. A. 2012. An integrated map of genetic variation from 1,092 human genomes. Nature. 491: 56–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bennett E. P., Hassan H., Mandel U., Mirgorodskaya E., Roepstorff P., Burchell J., Taylor-Papadimitriou J., Hollingsworth M. A., Merkx G., van Kessel A. G., et al. 1998. Cloning of a human UDP-N-acetyl-alpha-D-Galactosamine:polypeptide N-acetylgalactosaminyltransferase that complements other GalNAc-transferases in complete O-glycosylation of the MUC1 tandem repeat. J. Biol. Chem. 273: 30472–30481 [DOI] [PubMed] [Google Scholar]
- 30.O'Halloran A. M., Patterson C. C., Horan P., Maree A., Curtin R., Stanton A., McKeown P. P., Shields D. C. 2009. Genetic polymorphisms in platelet-related proteins and coronary artery disease: investigation of candidate genes, including N-acetylgalactosaminyltransferase 4 (GALNT4) and sulphotransferase 1A1/2 (SULT1A1/2). J. Thromb. Thrombolysis. 27: 175–184 [DOI] [PubMed] [Google Scholar]
- 31.Cybulsky M. I., Gimbrone M. A., Jr 1991. Endothelial expression of a mononuclear leukocyte adhesion molecule during atherogenesis. Science. 251: 788–791 [DOI] [PubMed] [Google Scholar]
- 32.Humtsoe J. O., Bowling R. A., Jr, Feng S., Wary K. K. 2005. Murine lipid phosphate phosphohydrolase-3 acts as a cell-associated integrin ligand. Biochem. Biophys. Res. Commun. 335: 906–919 [DOI] [PubMed] [Google Scholar]
- 33.Humtsoe J. O., Feng S., Thakker G. D., Yang J., Hong J., Wary K. K. 2003. Regulation of cell-cell interactions by phosphatidic acid phosphatase 2b/VCIP. EMBO J. 22: 1539–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Humtsoe J. O., Liu M., Malik A. B., Wary K. K. 2010. Lipid phosphate phosphatase 3 stabilization of beta-catenin induces endothelial cell migration and formation of branching point structures. Mol. Cell. Biol. 30: 1593–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Panchatcharam M., Miriyala S., Salous A., Wheeler J., Dong A., Mueller P., Sunkara M., Escalante-Alcalde D., Morris A. J., Smyth S. S. 2013. Lipid phosphate phosphatase 3 negatively regulates smooth muscle cell phenotypic modulation to limit intimal hyperplasia. Arterioscler. Thromb. Vasc. Biol. 33: 52–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ono K., Han J. 2000. The p38 signal transduction pathway: activation and function. Cell. Signal. 12: 1–13 [DOI] [PubMed] [Google Scholar]
- 37.Yoshizuka N., Chen R. M., Xu Z., Liao R., Hong L., Hu W. Y., Yu G., Han J., Chen L., Sun P. 2012. A novel function of p38-regulated/activated kinase in endothelial cell migration and tumor angiogenesis. Mol. Cell. Biol. 32: 606–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reiter J. F., Skarnes W. C. 2006. Tectonic, a novel regulator of the Hedgehog pathway required for both activation and inhibition. Genes Dev. 20: 22–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garcia-Gonzalo F. R., Corbit K. C., Sirerol-Piquer M. S., Ramaswami G., Otto E. A., Noriega T. R., Seol A. D., Robinson J. F., Bennett C. L., Josifova D. J., et al. 2011. A transition zone complex regulates mammalian ciliogenesis and ciliary membrane composition. Nat. Genet. 43: 776–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Egorova A. D., van der Heiden K., Poelmann R. E., Hierck B. P. 2012. Primary cilia as biomechanical sensors in regulating endothelial function. Differentiation. 83: S56–S61 [DOI] [PubMed] [Google Scholar]
- 41.Sekine Y., Hatanaka R., Watanabe T., Sono N., Iemura S. I., Natsume T., Kuranaga E., Miura M., Takeda K., Ichijo H. 2012 doi: 10.1016/j.molcel.2012.09.018. The Kelch repeat protein KLHDC10 regulates oxidative stress-induced ASK1 activation by suppressing PP5. Mol. Cell. 48: 692–704. [DOI] [PubMed] [Google Scholar]
- 42.Labrie V., Fukumura R., Rastogi A., Fick L. J., Wang W., Boutros P. C., Kennedy J. L., Semeralul M. O., Lee F. H., Baker G. B., et al. 2009. Serine racemase is associated with schizophrenia susceptibility in humans and in a mouse model. Hum. Mol. Genet. 18: 3227–3243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miya K., Inoue R., Takata Y., Abe M., Natsume R., Sakimura K., Hongou K., Miyawaki T., Mori H. 2008. Serine racemase is predominantly localized in neurons in mouse brain. J. Comp. Neurol. 510: 641–654 [DOI] [PubMed] [Google Scholar]
- 44.Schmidt A. E., Miller T., Schmidt S. L., Shiekhattar R., Shilatifard A. 1999. Cloning and characterization of the EAP30 subunit of the ELL complex that confers derepression of transcription by RNA polymerase II. J. Biol. Chem. 274: 21981–21985 [DOI] [PubMed] [Google Scholar]
- 45.Malerod L., Stuffers S., Brech A., Stenmark H. 2007. Vps22/EAP30 in ESCRT-II mediates endosomal sorting of growth factor and chemokine receptors destined for lysosomal degradation. Traffic. 8: 1617–1629 [DOI] [PubMed] [Google Scholar]
- 46.Pruim R. J., Welch R. P., Sanna S., Teslovich T. M., Chines P. S., Gliedt T. P., Boehnke M., Abecasis G. R., Willer C. J. 2010. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 26: 2336–2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.