Abstract
Many of the apolipoproteins in HDL can elicit cholesterol efflux via ABCA1, a critical initial step in HDL formation. Recent work has indicated that omnipresent amphipathic helices play a critical role, and these have been studied intensively in the most common HDL protein, apolipoprotein (apo)A-I. However, little information exists about helical domain arrangement in other apolipoproteins. We studied two of the smallest apolipoproteins known to interact with ABCA1, human apoA-II and apoC-I, in terms of ability to reorganize phospholipid (PL) bilayers and to promote ABCA1-mediated cholesterol. We found that both proteins contained helical domains that were fast and slow with respect to solubilizing PL. ABCA1-medated efflux required a minimum of a bihelical polypeptide comprised of at least one each of a slow and fast lipid reorganizing domain. In both proteins, the fast helix was located at the C terminus preceded by a slow helix. Helical placement in apoC-I was not critical for ABCA1 activity, but helix swaps in apoA-II dramatically disrupted cholesterol efflux, indicating that the tertiary structure of the longer apolipoprotein is important for the pathway. This work has implications for a more complete molecular understanding of apolipoprotein-mediated cholesterol efflux.
Keywords: amphipathic helix, apolipoprotein-mediated cholesterol efflux, tertiary structure
High-density lipoproteins (HDL) are a family of plasma particles composed primarily of phospholipids, cholesterol, and proteins known as apolipoproteins. For over half a century, it has been repeatedly demonstrated that high levels of plasma HDL-cholesterol (HDL-C) are associated with a reduced risk of cardiovascular disease (CVD), at least in large human populations (1, 2). However, therapies that substantially raise the level of plasma HDL-C, particularly those that work via inhibition of its catabolism (3, 4) have been disappointing with regard to CVD protection. This has focused attention on a better understanding of the process of HDL biogenesis, which may prove to be a more productive therapeutic target. The first step is the assembly of phospholipids and cholesterol with lipid-poor apolipoproteins produced in the liver and intestine. This is dependent on the plasma membrane protein ATP-binding cassette transporter A1 (ABCA1, recently reviewed in Ref. 5). Although abundant in tissues that produce apoA-I, ABCA1 also plays important roles in reverse cholesterol transport from the periphery to the liver, especially in cells, such as macrophages, with excess intracellular cholesterol. When this transporter is dysfunctional, such as in Tangier disease, the removal of excess cholesterol from sites like the arterial wall is impaired (6–9). Genetic ablation of ABCA1 in peripheral macrophages results in increased atherosclerosis in rodent models (10).
A variety of exchangeable apolipoproteins can mediate ABCA1-dependent cholesterol efflux, including apolipoprotein (apo)A-I, A-II, A-IV, C-I, C-II, C-III, and E, though they must be largely lipid-unassociated prior to the interaction (11). However, they lack a clear consensus sequence like those that mediate traditional receptor:ligand interactions, indicating that a more generalized structural feature is at play. Current consensus holds that this feature is the amphipathic α-helix, a motif common to all exchangeable apolipoproteins. Indeed, a multitude of small synthetic helical peptides have been shown to promote ABCA1-specific cholesterol efflux from a variety of cell types, and many of these have been explored as potential therapeutics (12, 13). In addition, a significant amount of work has shown that helical features such as mean hydrophobicity, size of the hydrophobic face, and amino acids at helical junctions are important for this function (14). Other groups suggested that a linear array of acidic amino acids aligned along the junction of the hydrophobic and hydrophilic faces of two amphipathic α-helices is a critical element for this process (15). However, peptides designed around cholesterol efflux have not yet fulfilled the promise of producing a robust therapeutic for CVD (16, 17). One reason for this could be that small synthetic peptides lack elements present in the full-length apolipoproteins that are required for subsequent HDL particle maturation and metabolism. Unfortunately, our understanding of the tertiary arrangement of these helical domains in the larger context of full-length apolipoproteins remains incomplete, with the most information limited to apoA-I. Given the large number of apolipoproteins that can access cellular lipid via ABCA1, it should be possible to identify common tertiary structural features within this family that mediate not only the lipid accumulation steps but also subsequent particle maturation. Such knowledge would add to our understanding of the physiological mechanism of HDL biogenesis, and may open new ways to modulate HDL levels at the biosynthesis end of its lifecycle.
Here, we began to address this problem by studying two of the smallest apolipoproteins known to interact with ABCA1, apoC-I and apoA-II. Their small size allowed the facile synthesis of significant domains within each protein, with the aim of identifying minimal structural features that mediate the reorganization of phospholipid surfaces and functional interaction with ABCA1. This led to the identification of a common bihelical structural motif and a more complete understanding of how these elements are arranged in the primary structure of human apolipoproteins.
MATERIALS AND METHODS
Reagents and cell lines
Dulbecco's Modified Eagle Medium (DMEM) and fetal bovine serum (FBS) were acquired from Invitrogen (Carlsbad, CA). [1,2-3H(N)]cholesterol was supplied by Amersham Biosciences (Piscataway, NJ). 8-bromoadenosine 3′,5′-cyclic monophosphate sodium salt (cAMP) and gentamicin were purchased from Sigma (St. Louis, MO). The RAW264.7 macrophages used in the cholesterol efflux assay were obtained from the American Type Culture Collection (Manassas, VA) and were maintained in DMEM supplemented with 10% FBS and 10 μg/ml gentamicin. Radiolabeling and efflux measurements were performed in DMEM supplemented with 0.2% fatty-acid free BSA (Calbiochem, Gibbstown, NJ). 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) was purchased from Avanti Polar Lipids (Birmingham, AL). Human plasma apoC-I was purchased from Athens Research and Technology (Athens, GA). The apoC-I pMaI-c2 vector was a generous gift from the Howlett Laboratory of the University of Melbourne, Victoria, Australia. All other reagents were the highest quality available.
Expression of apoC-I point mutants
Human mature apoC-I was generated in an E. coli expression system similar to those we have used to produce other recombinant apolipoproteins (18) using the pMal-c2 expression vector as described by Atcliffe et al. (19). Briefly, the apoC-I was expressed with an N-terminal maltose-binding protein tag, which was used for batch purification on an amylose affinity resin and eluted with free maltose. The affinity tag was cleaved using factor Xa protease. Separation of tag from protein was accomplished by separation on a Superdex 75 (GE Healthcare) gel filtration column in 3.5 M guanidine HCl to prevent aggregation of the protein. The recombinant apoC-I was over 95% pure as determined by Tris/tricine SDS-PAGE analysis. Circular dichroism spectroscopy measurements showed that the spectra of our recombinant apoC-I and plasma-isolated apoC-I are superimposable, with two minima at 208 and 222 nm, indicating that the secondary structure of the recombinant protein is predominantly α-helical proteins and similar to the plasma form (data not shown).
Peptide synthesis
ApoC-I- and apoA-II-based peptides were synthesized by GenScript Corp. (Scotch Plains, NJ), 21st Century Biochemicals (Marlboro, MA), or Peptide 2.0 (Chantilly, VA) and were acetylated on the N terminus and amidated on the C terminus. All peptides were greater than 95% pure as determined by high performance liquid chromatography. Peptides were stored lyophilized at −80°C until use. When needed, peptides were solubilized in ultrapure water ± 3M guanidine, then dialyzed into Standard Tris Buffer (STB, 10 mM Tris HCl, 150 mM NaCl, 1 mM EDTA, 0.02% sodium azide, pH 7.4), and stored under N2 until use. Secondary structure content was estimated by circular dichroism spectroscopy on a Jasco J-715 spectropolarimeter. Samples were dialyzed into 20 mM phosphate buffer (pH 7.4) and diluted to a concentration of 100 μg/ml. A background scan of phosphate buffer was subtracted from each sample scan. Plasma apoA-II from normolipidemic patients was isolated and purified as previously described for apoA-I (20).
DMPC liposome clearance assay
Peptides or plasma proteins were added to DMPC liposomes at the specified mole lipid:mole α-helix ratio, and the absorbance of the solution at 325 nm was recorded every 30 s for 20 min. Samples were measured in triplicate, and absorbances were averaged. The data were expressed as a normalized optical density (OD), which was calculated by dividing the sample OD by the initial OD (OD0). Two trials were measured on different days using fresh preparations of liposomes for each set of peptides. All measurements were made on an Amersham Biosciences Ultraspec 4000 UV/Visible spectrophotometer within a temperature-controlled cuvette at 24.5°C.
Free cholesterol efflux assay
RAW 264.7 macrophages were grown to 80% confluency, then incubated with media containing 1.0 μCi/ml [1,2-3H(N)]cholesterol for labeling ± 0.3 mM cAMP. In these cells, the presence of cAMP leads to expression of ABCA1 on the cell surface (21). After washing away any radiolabel not internalized by the cells, media containing the peptide or plasma protein of interest ± 0.3 mM cAMP were incubated with the cells for 8 h. At the beginning of this incubation, a t0 control plate was washed with PBS, and the labeled cells in each well were dissolved in isopropanol. This solution was then dried under air, resolubilized in toluene, mixed with scintillation fluid, and counted on a 1900CA Packard liquid scintillation analyzer to determine the total amount of labeled cholesterol internalized by the cells at the beginning of the efflux experiment. After 8 h, a sample of media was filtered to remove floating cells and debris and then counted to determine how much labeled cholesterol was transferred to the peptide or plasma protein in the media during the incubation. Each protein or peptide was tested in triplicate, and the percentage efflux calculated was the average. Percentage efflux is the counts in the media divided by the total internalized counts per well calculated using the t0 control plate, with background efflux to STB subtracted. The percentage efflux of each peptide was compared with baseline using a two-tailed Student t-test, with P < 0.05 indicating an ability of the peptide to stimulate free cholesterol (FC) efflux. For each set of peptides, at least two independent assays were performed using cells prepared on separate days.
RESULTS
Role of a linear array of acidic residues in apoC-I in the reorganization of phospholipid liposomes and ABCA1-mediated cholesterol efflux
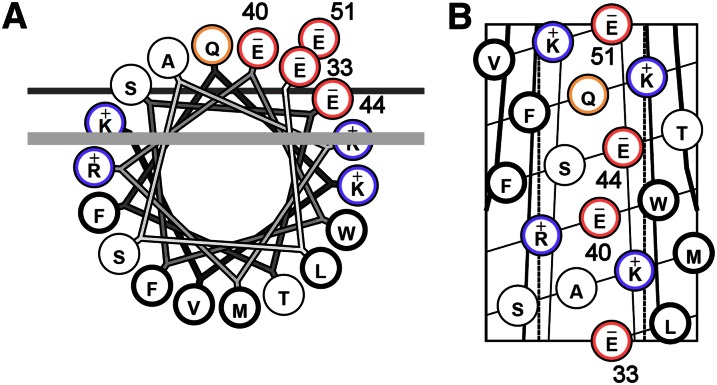
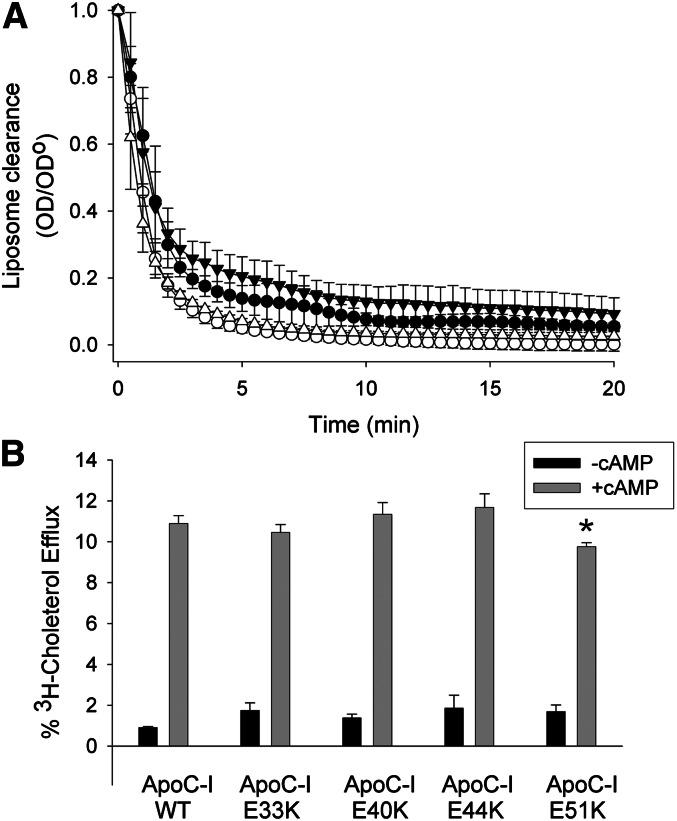
Natarajan et al. (15) identified an evolutionarily conserved linear array of three acidic amino acid residues spanning helices 9 and 10 of human apoA-I that appeared to be required for promotion of ABCA1-mediated cholesterol efflux, at least in peptides derived from apoA-I. Our analysis of the primary structure of apoC-I (Fig. 1) identified a similar array of four glutamic acid residues (residues 33, 40, 44, and 51) spanning both putative helical domains of the protein (see Fig. 3A for the sequence of apoC-I) that were in a similar orientation relative to the nonpolar face as the peptides studied by Natarajan et al. To determine the importance of this array in the context of apoC-I, we systematically reversed the charge at each position by replacing the resident Glu with a basic Lys residue using our E. coli-based expression system. Because the ability to promote ABCA1-mediated cholesterol efflux has been linked to the ability to solubilize phospholipid bilayers (22, 23) (see Discussion), we first tested the ability of these mutants to spontaneously reorganize DMPC liposomes. Preliminary experiments demonstrated that apoC-I isolated from human plasma and the recombinant form generated in the bacterial system performed comparably in this assay (supplementary Fig. IA). In this analysis, the protein or peptide of interest is added to turbid DMPC liposomes at the lipid gel-to-liquid crystal transition temperature, and the binding and emulsification of the lipid by the protein is indicated by a decrease in the solution turbidity as the peptides reorganize the liposomes into small lipoproteins. Fig. 2A shows that WT apoC-I was efficient at reorganizing the liposomes under these conditions, clearing the liposomes completely in about 10 min. ApoC-I E33K and apoC-I E51K were slightly more effective than WT, while apoC-I K40K was slightly less effective. In all cases, the differences from the WT protein were small compared with other apolipoprotein mutants we have studied in the past (24) and probably do not represent biologically relevant differences.
Fig. 1.
Helical wheel projection (A) and helical net (B) representation of apoC-I helix 2 (residues 33–51). (A) Each circle represents an amino acid. The four glutamate residues highlighted in red form a linear acidic array on the polar face of the helix that has been proposed to be required for ABCA1-mediated cholesterol efflux (15). The thick line represents the outside limit of the lipid bilayer in front, and the thin line represents the same in back. (B) Helical net with glutamates numbered. Basic residues are colored blue and acidic residues are colored red. For more information on these helical projections, see Ref. 53.
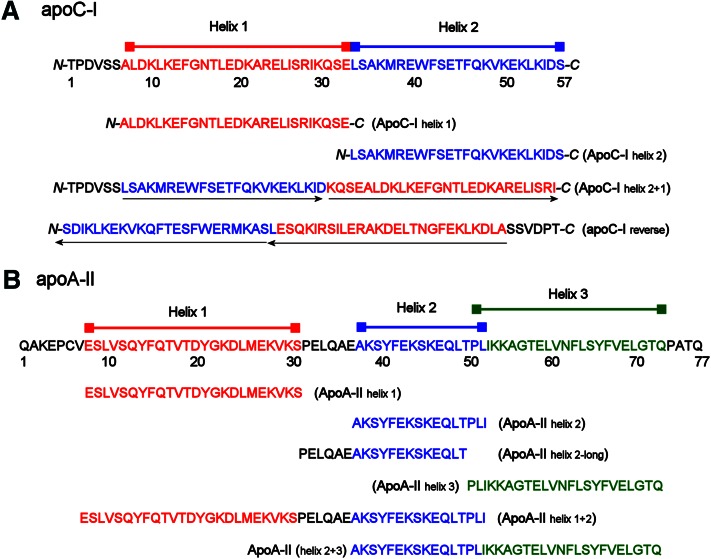
Fig. 3.
Synthetic peptides reflecting individual helical domains of apoC-I and apoA-II. (A) Full-length sequence of human mature apoC-I. Consensus helical domains based on sequence analyses are indicated, as are two bihelical constructs. The first simply reverses the order of the helices in the WT sequence (maintaining a predicted turn sequence KQSE between the helices), while the second completely reverses the amino acid order of the WT protein. The orientations of the helices in these two mutants with respect to the WT protein are shown by the black arrows under the sequences. (B) Full-length sequence of human mature (monomeric) apoA-II. Consensus helical domains based on several sequence analyses (see text) are shown in red, blue, and green, with correspondingly colored bars above. Individual peptides designed to mimic each individual helix (including two possibilities for helix 2) are aligned under the sequence. The sequence of two bihelical constructs is also shown.
Fig. 2.
Ability of apoC-I and its charge reversion point mutants to clear DMPC liposomes and promote ABCA1-mediated cholesterol efflux. (A) ApoC-I was added to DMPC liposomes at a lipid:protein mass ratio of 2.5:1 at the DMPC gel to liquid-crystal phase transition at temperature of 24.5°C. As the apolipoprotein binds to and reorganizes the lipid, the liposomes are solubilized to lipoprotein discs and therefore scatter less light over time. For each sample, the light scattering at time 0 min was normalized to 1.0, and each subsequent reading was expressed as a fraction (OD divided by OD0). Filled circles, WT recombinant apoC-I; open circles, apoC-I E33K; filled triangles, apoC-I E40K; open triangles, apoC-I E51K. The experiments were repeated three times on separate days and the average values are plotted ± 1 sample SD. (B) RAW mouse macrophages were exchange-labeled with tritiated cholesterol and then treated with or without 8-Br-cAMP to upregulate ABCA1 expression as described in Methods. Each apolipoprotein (5 µg/ml) was then incubated with the cells for 8 h with or without 8-Br-cAMP, and the radioactivity appearing in the media were assessed by scintillation counting. The data are expressed as the percentage of label present in the cells immediately before the incubation period (T = 0 h). Black bars show cholesterol efflux in the absence of cAMP (ABCA1 independent), while the shaded bars show cholesterol efflux in the presence of cAMP (ABCA1 component). The experiment was run in triplicate and expressed as averages ± 1 sample SD. *P < 0.05 compared with WT apoC-I (two-tailed Student t-test).
We next evaluated the point mutants for their ability to promote ABCA1-mediated cholesterol efflux. Again, we verified that the human plasma and recombinant forms of apoC-I performed comparably in the assay (supplementary Fig. IB), and concentration experiments showed that cholesterol efflux saturates above ∼15 µg/ml under these conditions (not shown). Therefore, we tested the mutants at a concentration of 5 µg/ml to avoid obscuring any differences due to efflux saturation. The cholesterol efflux results are shown in Fig. 2B. In the absence of cAMP, the efflux of radiolabeled cholesterol to WT apoC-I was minimal, but this increased to about 11% in the presence of cAMP, reflecting the activity of induced ABCA1 (21). The apoC-I mutants E33K, E40K, and E44K were similar to WT. ApoC-I E51K exhibited a statistically lower degree of cholesterol efflux, but this effect was again small. Further mutations that introduced dual charge reversions at these sites also failed to produce significant effects on either DMPC clearance or ABCA1-mediated cholesterol efflux (not shown). Thus, the data suggest that this linear array of acidic amino acids is not a critical motif for mediating ABCA1 lipid efflux, at least in apoC-I. We were unable to detect a similar acidic array in apoA-II, further arguing that it is not a universal feature mediating ABCA1-mediated lipid efflux.
Design of peptides modeling the individual α-helices of apoC-I and apoA-II
To further examine human apoC-I and apoA-II for important structural elements with respect to ABCA1-mediated efflux, we synthesized short peptides representing each individual α-helix within these apolipoproteins. The determinations of the beginning and end of each α-helix in apoC-I are based on the analyses of Gursky et al. (25), who determined helices of lipid-free apoC-I with proline scanning and circular dichroism measurements, Rozek et al., who used NMR on lipid-bound apoC-I to estimate helices (26), and Segrest et al. (27), who used sequence analysis to delineate helical regions within lipid-free apoC-I. Putative helical breakpoints were favored if they resulted in a peptide with strong amphipathic character and were 22 amino acids in length as observed in a wide range of exchangeable apolipoproteins (28).The sequences of the apoC-I peptides are shown in Fig. 3A. In a similar manner, by observing the proline punctuation within apoA-II, modeling the protein with the PredictProtein program (29), and combining these findings with the sequence analysis of lipid-free apoA-II by Segrest et al. (27), designations for apoA-II helical peptides were obtained (Fig. 3B). Due to some ambiguity in determining the boundaries of apoA-II helix 2, we produced a short form (residues 38–53, called apoA-II helix 2) and a long form (residues 32–50, apoA-IIhelix 2-long) to ensure that any functional differences were not due to inappropriate truncation. We found that the long form was functionally identical to the shortened peptide in both PL reorganization and cholesterol efflux (data not shown), and therefore, we refer to the short form throughout this article. All peptides were N-terminally acylated and C-terminally amidated to aid in helical formation in solution, as previously described (30).
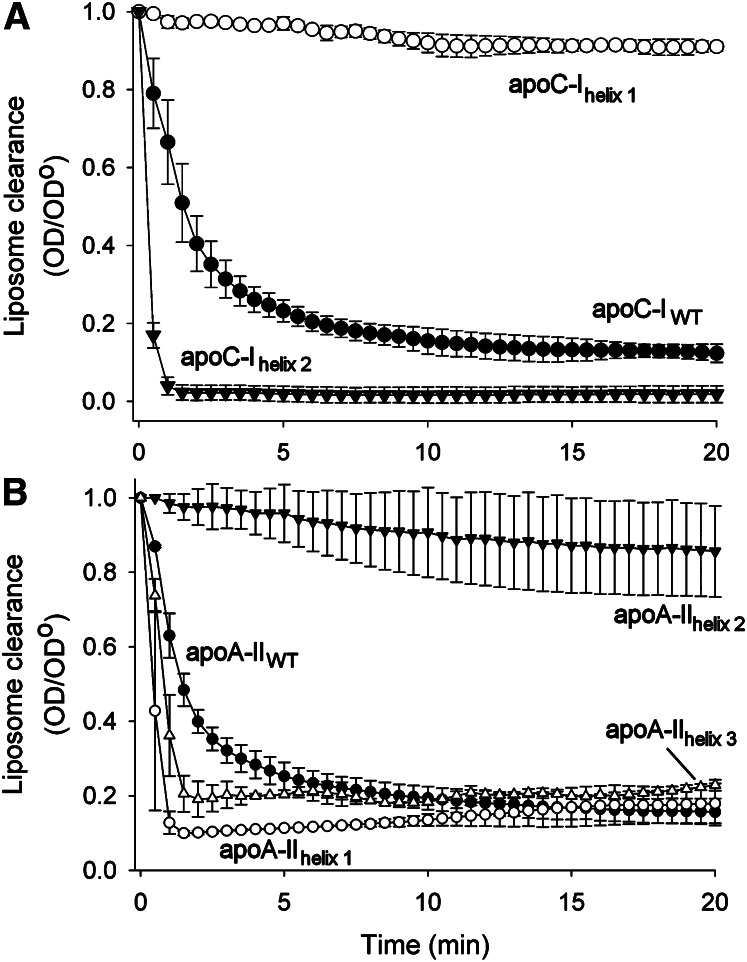
Ability to reorganize phospholipid surfaces
For these experiments, each helix of apoC-I was compared with human plasma-isolated apoC-I on an equimolar helical basis (i.e., 2:1 molar ratio; the same number of putative α-helical domains was present in each sample solution). Similarly, each helix of apoA-II was compared with human plasma-isolated apoA-II on an equimolar helical basis. Fig. 4A confirms the results shown in Fig. 2A, indicating that apoC-I rapidly cleared DMPC liposomes within about 10 min under these conditions. By contrast, apoC-Ihelix 1 failed to significantly modify liposome light scattering, exhibiting a similar clearance profile to DMPC liposomes incubated with buffer alone (not shown). This indicates that this helix either i) failed to bind to the lipids or ii) was able to bind the liposomes but was unable to rearrange the bilayer structure to smaller particles. ApoC-Ihelix 2 however, exhibited markedly increased clearance, with the reaction being effectively complete by the second time point (1 min). These data suggest that the majority of the ability of intact apoC-I to bind liposomes lies in its C-terminal helix. Fig. 4B shows a similar distribution of lipid-binding abilities among the helices of apoA-II. Whereas intact apoA-II was effective, apoA-II helix 2 was largely ineffective (as was the long form). However apoA-II helix 1 and apoA-II helix 3 were both more effective than the WT protein. These data indicate that i) both apolipoproteins contain helices which are capable of “fast” lipid clearance and those which are incapable or “slow”; ii) both apolipoproteins have a fast lipid-binding helix present at the C terminus of the mature protein; and iii) the N-terminal helix in each protein can be either fast or slow. Gel filtration analysis showed that the particles produced by the individual peptides of both apoC-I and apoA-II generated peptide/lipid complexes that were generally consistent in size with those generated by the native apolipoprotein, although the size distributions were much broader in the case of the peptides (not shown).
Fig. 4.
Ability of individual helices within apoC-I and apoA-II to clear DMPC liposomes. (A) Peptides modeling the individual α-helices of apoC-I were added to DMPC liposomes at a 23 mol lipid:1 mol α-helix ratio. The solution's turbidity over time is plotted. Conditions and data treatment were as indicated in Fig. 1. (B) Peptides modeling the individual α-helices of apoA-II were added to DMPC liposomes at a 10 mol lipid:1 mol α-helix ratio. Conditions and data treatment were as indicated in Fig. 1. All points are the means of three replicates, and bars represent mean ± 1 sample SD.
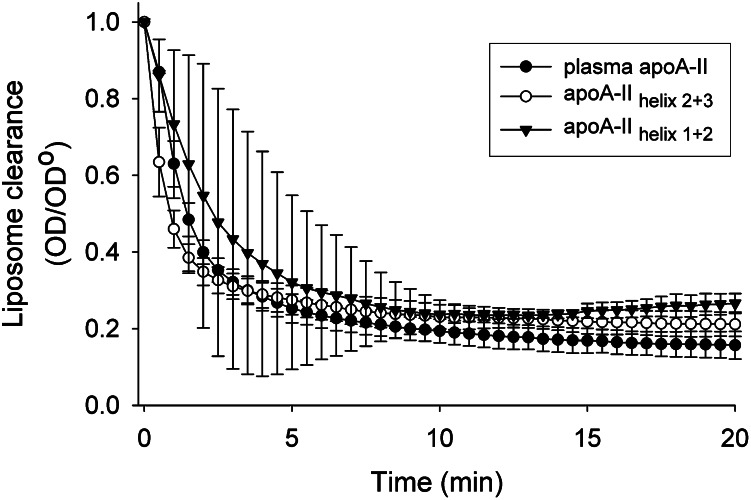
We noted that the isolated fast lipid-binding helices studied above were all significantly more effective than the intact protein when tested at an equimolar helical concentration. In the case of apoC-I, it appears that the presence of at least one slow helix in combination with a fast helix may act as a brake on lipid binding/reorganization. To determine whether this is the case with the helical domains of apoA-II, two bihelical peptides were synthesized representing two naturally occurring pairs of fast emulsifying and nonemulsifying helices in apoA-II, apoA-II helix 1+2 (which spanned residues 8–53 of the mature sequence), and apoA-II helix 2+3 (which spanned residues 38–73) (see Fig. 3). Fig. 5 shows that, despite some experimental variation, apoA-II helix 1+2 cleared lipid at a rate approximately similar to WT apoA-II, but much more slowly than apoA-II helix 1 by itself (Fig. 4B). Similarly, apoA-II helix 2+3 cleared lipid like WT apoA-II, but it was not as extreme as helix 3 in isolation. This indicates that the presence of a slow lipid-binding helix can attenuate the lipid reorganizing properties of a fast helix. We also determined whether this is a cooperative phenomenon by comparing the effect of incubating a mixture of the individual helices. We compared the clearance rates of apoC-I to an equimolar mixture of apoC-I helix 1 and apoC-I helix 2 (mixed as separate peptides). We also compared mixtures of appropriate helices in isolation to the bihelical peptides shown in Fig. 5. In all cases, the peptide mixtures cleared lipid faster than the corresponding bihelical peptide (i.e., similar to that for the fast helix in isolation) (not shown). This indicates that helices must be connected in order for the slow helix to attenuate the action of the fast helix.
Fig. 5.
Ability of bihelical peptides derived from the sequence of apoA-II to clear DMPC liposomes. Bihelical peptides modeling both potential helical pairings within apoA-II were added to DMPC liposomes at a 10 mol lipid:1 mol α-helix ratio, with the peptides added at an equimolar helical concentration as apoA-II. Conditions and data treatment for both panels were as indicated in Fig. 1. All points are the mean of three replicates, and bars represent mean ±1 sample SD.
ABCA1-mediated FC efflux
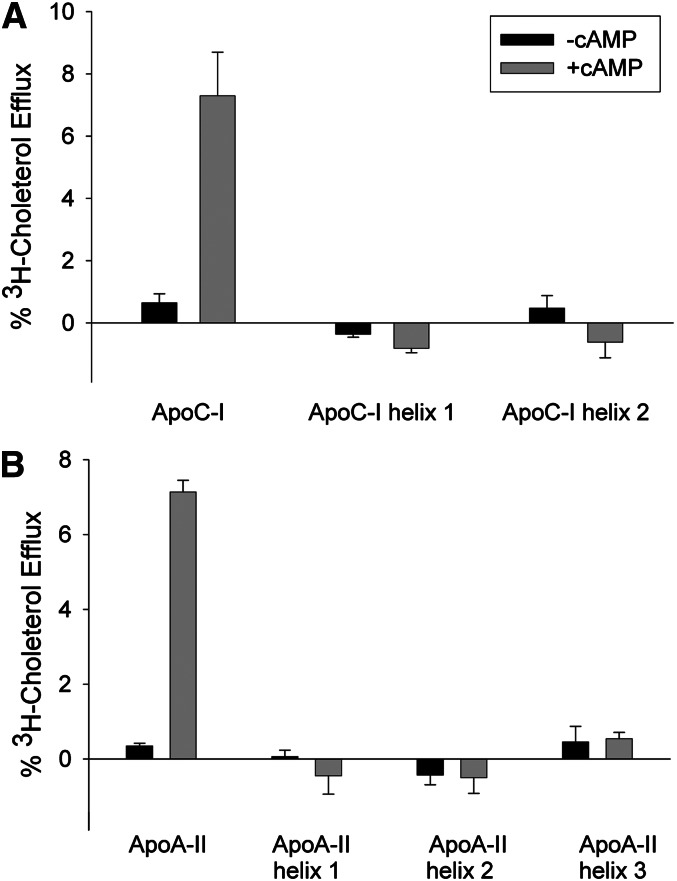
We next evaluated the ability of each helix in apoC-I and apoA-II to promote ABCA1-mediated cholesterol efflux. Again, individual helices were compared with plasma proteins on an equimolar α-helical basis at subsaturating concentrations. Fig. 6A demonstrates that neither apoC-I helix can serve as an efficient acceptor of cholesterol from ABCA1, although the intact protein was functional. Fig. 6B shows that the same is true for the individual helices of apoA-II, with the intact protein also exhibiting activity. This suggests that neither a fast lipid-emulsifying helix nor a nonlipid-emulsifying helix is sufficient for cholesterol efflux via ABCA1 when present in isolation.
Fig. 6.
ABCA1-dependent efflux of cholesterol to lipid-free plasma proteins and individual helical peptides. (A) Cholesterol efflux to lipid-free human plasma apoC-I and individual C-I helical peptides. The experiment was performed as in Fig. 1, except that the human plasma apoC-I was at 5 µg per ml and the individual helices were present at the same molar helix concentration. (B) Cholesterol efflux to lipid-free human plasma apoA-II and individual A-II helical peptides. The human plasma apoA-II was present at 5 µg per ml and the individual helical peptides were present at the same molar helix concentration. Bars represent mean ± 1 sample SD of three replicates. Black bars represent the absence of cAMP, and gray bars represent the presence of cAMP.
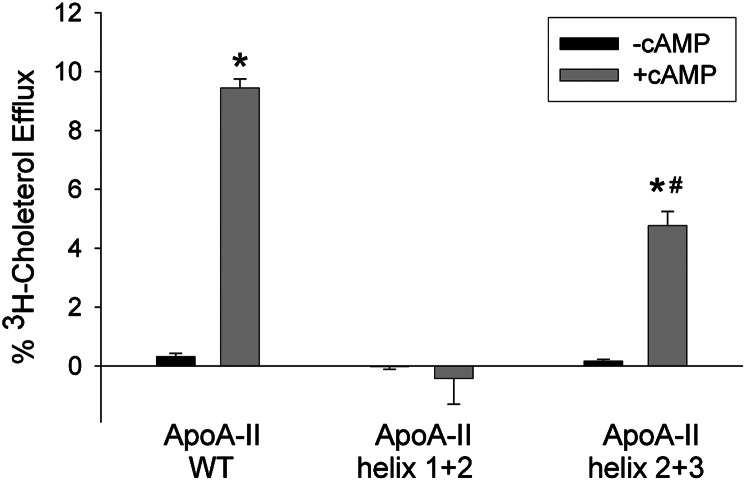
In previous studies using synthetic apoA-I mimetic peptides, Sethi et al. (31) postulated that the minimal structural element for ABCA1 cholesterol acceptors is a pair of helices, with one being a fast emulsifier and one slow. This is consistent with our observation that intact apoC-I (essentially a bihelical peptide) was functional while its individual fast or slow helices were not. To test this in the context of apoA-II, we evaluated the apoA-II bihelical peptides studied in Fig. 5. Full-length apoA-II was functional as expected; however, the apoA-II helix 1+2, which contained both a fast and poor lipid-emulsifying helix (in order N to C), failed to promote significant cholesterol efflux (Fig. 7). Interestingly, apoA-I helix 2+3, which has a fast lipid-binding helix at its C terminus, could promote ABCA1-mediated cholesterol efflux, although not as effectively as full-length apoA-II (Fig. 7). This supports the idea that at least two helices are required for specific cholesterol efflux via ABCA1, with a poor and fast lipid-emulsifying helix arranged N to C, respectively.
Fig. 7.
ABCA1-dependent efflux of cholesterol to lipid-free human plasma apoA-II and its bihelical peptides. Experimental conditions and presentation are exactly as described for Fig. 6B. *P < 0.05 presence of cAMP (gray bar) versus absence of cAMP (black bar) in protein sample (two-tailed Student t-test). #P < 0.05 presence of cAMP (gray bar) versus absence of cAMP (black bar) in WT apoA-II.
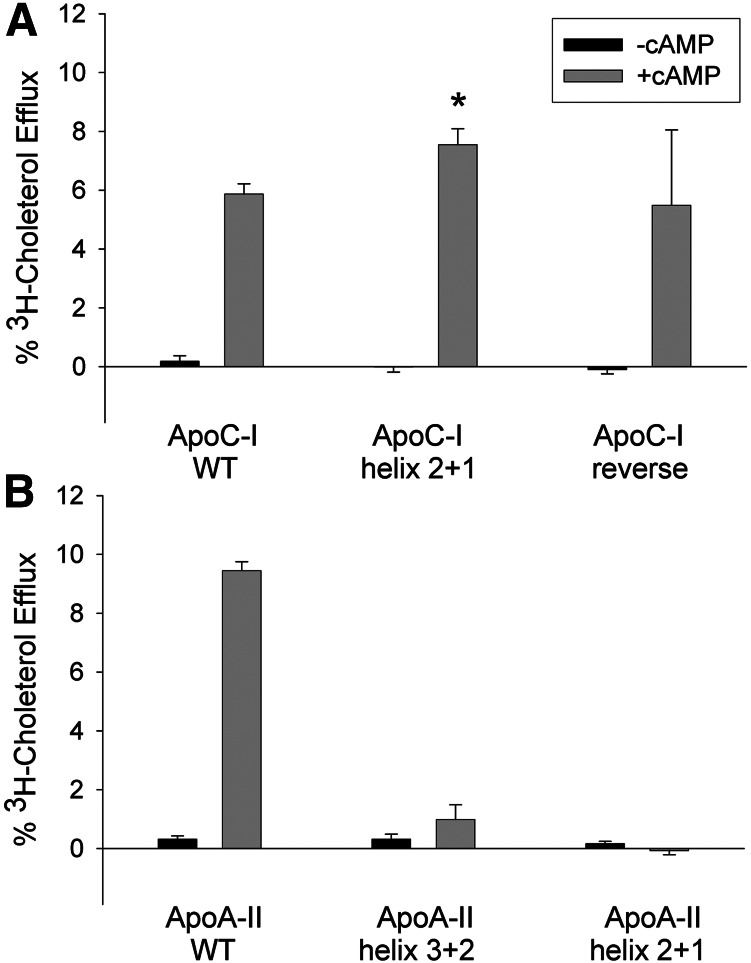
To further test the requirement of an N to C distribution of the slow and fast helices, we generated a series of helical swap constructs. In apoC-I, we reversed the position of helices 1 and 2 within the context of the full-length protein (ApoC-I helix 2+1, Fig. 3). This placed the fast helix on the N-terminal side and the slow helix at the C-terminal side, but kept each helix in otherwise the same orientation with respect to the dipole moment of the molecule. Fig. 8A shows that ApoC-I helix 2+1 was actually slightly more effective than WT apoC-I at promoting cholesterol efflux. We also tested a variant called ApoC-I reverse (essentially WT apoC-I synthesized backward). This has the effect of placing helix 1 at the C terminus and helix 2 at the N terminus like ApoC-I helix 2+1, but the sequence is reversed in relation to each helical dipole. This peptide also promoted cholesterol efflux with an efficiency comparable to WT apoC-I. Thus, the N- to C-terminal orientation of the fast and slow helices did not appear to be a major factor in determining ABCA1 cholesterol efflux in apoC-I.
Fig. 8.
Effect of helical rearrangement on ABCA1-dependent efflux of cholesterol to lipid-free apoC-I (A) and apoA-II (B). Experimental conditions and presentation are exactly as described for Fig. 6B. *P < 0.05 compared with WT apoC-I (two-tailed Student t-test).
Unlike in apoC-I, reversing the helical order in the apoA-II bihelical peptides completely abolished ABCA1-mediated cholesterol efflux. Fig. 7A shows that ApoA-II helix 2+3 exhibited partial efflux activity compared with WT apoA-II. However, ApoA- II helix 3+2, which has its fast and slow helices arranged N to C, was nonfunctional. ApoA-II helix 2+1, like ApoA-II helix 1+2, was also nonfunctional.
DISCUSSION
A popular model for the apolipoprotein-mediated removal of cellular lipids (32) holds that lipid-free (or lipid-poor) apolipoproteins interact with ABCA1 and stabilize its presence at the cell surface. This is consistent with prior studies showing that apoA-I can be cross-linked to ABCA1 and that apoA-I binding can prolong the residence of ABCA1 at the cell surface (33–35). Sustained lipid floppase activity by ABCA1 promotes the binding of apolipoproteins, via their amphipathic helices, to specific lipid sites on the plasma membrane. Nascent HDL particles are then formed by microsolubilization of membrane lipids. There is no specific apolipoprotein consensus sequence that mediates this process, and our experiments show that even relatively specific features, such as the linear array of acidic residues identified by Natarajan et al. (15), were not critical for either DMPC reorganization or ABCA1-mediated cholesterol efflux, at least in the case of apoC-I (Fig. 2). At this point, it can be stated that apolipoproteins that work through ABCA1 share the following features: i) their sequences contain 11 and 22 mer amphipathic helices that can have a variety of charge distributions on the polar face (e.g., class A, Y, or G helices) (36); ii) within a given protein, these helical domains can have varying hydrophobic moments (see supplementary Table I for the physical properties of these peptides) that result in different abilities to associate with lipid (37, 38); and iii) they can exist in a soluble lipid-free conformation or as a lipid-bound particle. Beyond this, the specific properties that confer activity with ABCA1, at least within the context of naturally occurring apolipoproteins, are poorly understood.
One possibility is that the order of these helical domains in the peptide sequence may be a key factor in modulating ABCA1-mediated cholesterol efflux. The results of the current study indicate that helix 2 of apoC-I and helix 3 from apoA-II avidly reorganized DMPC liposomes when present in isolation. In both cases, these fast lipid-binding helices are located at the extreme C-terminal end of the sequence. Similarly, numerous studies have shown that the extreme C-terminal amphipathic helix of apoA-I (helix 10, having the highest hydrophobic moment of any helix in apoA-I) is required for optimal lipid binding (38) and ABCA1-mediated cholesterol efflux (24, 39, 40). In fact, this C-terminal helical domain can even be added to a truncated form of apoA-I and still be effective in both processes, as long as the helix is located at the C terminus (24). ApoE appears to have a similar architecture. Vedhachalam et al. showed that the C-terminal lipid binding region (residues 222–299) of apoE is required for efficient ABCA1-mediated cholesterol efflux (41). A 40 mer peptide representing the extreme C-terminal residues 260–299 cleared DMPC liposomes rapidly, indicating that this domain likely represents a fast helix, similar to that located at apoA-I's C terminus. This occurrence in four different proteins suggests that a C-terminal avid lipid-binding helix is a common theme among the exchangeable apolipoproteins. Most structural studies indicate that the C-terminal domains of apolipoproteins are structurally dynamic in the lipid-free form and that these domains are driven to interact with lipid in order to stabilize the helical structure (42–44). Once this domain has taken hold, the remainder of the protein can then unfold from a four-helical bundle to an extended lipid-bound form (41, 45–47). Interestingly, our helical swap mutations indicated that the C-terminal location of the fast lipid-binding helix is not an absolute requirement for ABCA1-mediated cholesterol efflux. In apoC-I, essentially a bihelical peptide, the helical domains can be switched within the sequence or even completely reversed with respect to the helical dipole moment without significant effects on cholesterol efflux. Similar findings have been documented in studies using synthetic peptides based on apoA-I (15). In apoA-II, the situation was more complicated. A bihelical peptide that contained a fast helix at the C terminus (apoA-II helix 2+3) could partially promote cholesterol efflux. However, apoA-II helix 1+2, which had a fast-binding helix at the N terminus failed to promote cholesterol efflux, despite its ability to clear DMPC liposomes. We confirmed this finding by producing another version, apoA-II helix 3+2, which also could clear DMPC liposomes effectively but was unable to work with ABCA1 to promote cholesterol efflux (not shown). Therefore, we conclude that small apolipoproteins, such as apoC-I, which may be limited in the extent of their tertiary structure, do not have a strong dependence on the N or C placement of the fast lipid-binding helix. However, apolipoproteins that contain more than two helical domains, such as apoA-II and apoA-I, may require the fast lipid-binding helix to be located in a dynamic C-terminal location in order to bind lipid and promote ABCA1-mediated cholesterol efflux. This may be because the more extensive tertiary structure of these longer proteins is critical at some stage of the pathway and is disrupted by rearranging the helices.
One protein that does not follow this pattern is apoA-IV. This apolipoprotein contains a highly unique charged domain at its C terminus that likely does not form an amphipathic helix (48). Furthermore, our studies have shown that the critical apoA-IV lipid-binding elements occur near its N terminus, in sharp contrast to the others discussed above. Interestingly, our recent crystal structural analysis of apoA-IV (49) and comparisons with the crystal structure of the highly related apoA-I (50) provide intriguing hints that apoA-IV may have undergone a gene reversion event sometime in its evolution from a common ancestor apolipoprotein (51). Indeed, many structural features that appear at the N terminus of apoA-I appear in the C-terminal aspects of apoA-IV. Thus, the unique tertiary architecture of apoA-IV is likely permissive to a lipid-binding helical domain present near the N terminus.
Another outcome of this study is that apolipoproteins that successfully promote ABCA1-mediated cholesterol efflux contain mixture of at least one each of a fast and a slow lipid-binding helix. ApoC-I could only promote cholesterol efflux when both slow and fast helices were present and only when those two helices were physically connected in the same peptide. Similarly, in apoA-II, the individual helical domains were not able to promote ABCA1-mediated lipid efflux despite avid DMPC clearance. However, helices 2 (slow) and 3 (fast) were active when present in the same bihelical peptide (Fig. 7). The observation that all ABCA1-competent constructs contained at least one poor and one fast lipid-binding helix suggests that the combination is important during the pathway. The addition of helix 9 (an 11 mer with low lipid affinity) to the strong lipid-binding helix 10 in apoA-I increases the specificity of the peptide for ABCA1-mediated cholesterol efflux (i.e., it reduces the ability of helix 10 alone to nonspecifically solubilize membrane lipids, leading to cellular toxicity) (15). Sethi et al. (31) clearly demonstrated that asymmetry with respect to lipid binding among the helices in synthetic bihelical peptides improves the specificity of ABCA1-mediated lipid efflux. They suggested that the poor lipid-binding helix acts as a check on the lipid affinity of the fast helix, thus preventing nonspecific or detergent-like removal of membrane lipids. In such case, ABCA1 becomes necessary to overcome energy barriers to allow the apolipoprotein to accumulate lipid. In our study, we did not notice an overtly toxic effect of our individual helical peptides on the RAW macrophages. For example, apoA-II helix 3 and apoC-I helix 2 in isolation were highly effective phospholipid “detergents” (Fig. 4), but they did not promote detergent-like lipid removal in our system, as evidenced by the minimal levels of cholesterol efflux in control cells that were not treated with cAMP (Fig. 6). It may be that two fast PL-solubilizing domains are required for the cytotoxic effect (31). The poor lipid-binding helix may play a role in the ABCA1 pathway in addition to preventing cell toxicity, perhaps mediating intermolecular interactions among the apolipoprotein monomers as the particle is being assembled. Alternatively, the poor lipid-binding helix may physically tether the pair close to the surface of the lipid bilayer. This controlled submersion of the lipid-binding helix could facilitate the microsolubilization process. Of note, Sviridov et al. (52) recently showed that the presence of high volume hydrophobic residues (tryptophan and phenylalanine) near the hinge between two helical domains was important for lipid solubilization and ABCA1-mediated cholesterol efflux in the synthetic peptide 5A. However, inspection of Fig. 3 shows that the helical connection regions in apoC-I and A-II are devoid of these residues, suggesting that other factors are at play in these native apolipoproteins.
In summary, this article adds two apolipoproteins to our knowledge base in terms of the individual contributions of their amphipathic helical domains to the process of phospholipid reorganization/microsolubilization and ABCA1-mediated cholesterol efflux. The evidence shows a common organizational theme among most exchangeable apolipoproteins that includes the presence of an avid lipid-binding domain situated at a usually conformationally dynamic C terminus. The relationship of this avid domain to other lower affinity domains appears to dictate whether the protein can engage the ABCA1 pathway for lipid efflux. Further studies on the role of these domains in other steps of the pathway, including stabilization of cell surface ABCA1 and HDL particle maturation, will be helpful in delineating the molecular details of ABCA1-mediated cholesterol efflux and HDL biogenesis.
Supplementary Material
Acknowledgments
The authors thank Dr. Apryll Stalcup, Department of Chemistry, University of Cincinnati, for generous donation of time on her spectropolarimeter.
Footnotes
Abbreviations:
- CVD
- cardiovascular disease
- DMPC
- dimyristoyl-phospatidylcholine
- DPPC
- 1,2-dipalmitoyl-sn-glycero-3-phosphocholine
- DPPE
- 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine
- FC
- free cholesterol
- HDL-C
- HDL-cholesterol
- NBD-OLPC
- nitrobenzoxadiazole 1-oleoyl-2-[12-[(7-nitro-2-1,3-benzoxadiazol-4-yl) amino] dodecanoyl]-sn-glycero-3-phosphocholine
- OD
- optical density
- OD0
- initial optical density
- PL
- phospholipid
This work was supported by National Institutes of Health Grant R01 HL-67093 (W.S.D.) and by an American Heart Association predoctoral fellowship (L.E.S.).
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of one figure and one table.
REFERENCES
- 1.Gordon T., Castelli W. P., Hjortland M. C., Kannel W. B., Dawber T. R. 1977. High density lipoprotein as a protective factor against coronary heart disease. The Framingham Study. Am. J. Med. 62: 707–714 [DOI] [PubMed] [Google Scholar]
- 2.Assmann G., Schulte H., vonEckardstein A., Huang Y. D. 1996. High-density lipoprotein cholesterol as a predictor of coronary heart disease risk. The PROCAM experience and pathophysiological implications for reverse cholesterol transport. Atherosclerosis. 124: S11–S20 [DOI] [PubMed] [Google Scholar]
- 3.Barter P. J., Caulfield M., Eriksson M., Grundy S. M., Kastelein J. J., Komajda M., Lopez-Sendon J., Mosca L., Tardif J. C., Waters D. D., et al. 2007. Effects of torcetrapib in patients at high risk for coronary events. N. Engl. J. Med. 357: 2109–2122 [DOI] [PubMed] [Google Scholar]
- 4.Schwartz G. G., Olsson A. G., Abt M., Ballantyne C. M., Barter P. J., Brumm J., Chaitman B. R., Holme I. M., Kallend D., Leiter L. A., et al. 2012. Effects of dalcetrapib in patients with a recent acute coronary syndrome. N. Engl. J. Med. 367: 2089–2099 [DOI] [PubMed] [Google Scholar]
- 5.Yvan-Charvet L., Wang N., Tall A. R. 2010. Role of HDL, ABCA1, and ABCG1 transporters in cholesterol efflux and immune responses. Arterioscler. Thromb. Vasc. Biol. 30: 139–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rust S., Rosier M., Funke H., Real J., Amoura Z., Piette J. C., Deleuze J. F., Brewer H. B., Duverger N., Denefle P., et al. 1999. Tangier disease is caused by mutations in the gene encoding ATP-binding cassette transporter 1. Nat. Genet. 22: 352–355 [DOI] [PubMed] [Google Scholar]
- 7.Bodzioch M., Orso E., Klucken J., Langmann T., Bottcher A., Diederich W., Drobnik W., Barlage S., Buchler C., Porsch-Ozcurumez M., et al. 1999. The gene encoding ATP-binding cassette transporter 1 is mutated in Tangier disease. Nat. Genet. 22: 347–351 [DOI] [PubMed] [Google Scholar]
- 8.Brooks-Wilson A., Marcil M., Clee S. M., Zhang L. H., Roomp K., van Dam M., Yu L., Brewer C., Collins J. A., Molhuizen H. O., et al. 1999. Mutations in ABC1 in Tangier disease and familial high-density lipoprotein deficiency. Nat. Genet. 22: 336–345 [DOI] [PubMed] [Google Scholar]
- 9.Lawn R. M., Wade D. P., Garvin M. R., Wang X., Schwartz K., Porter J. G., Seilhamer J. J., Vaughan A. M., Oram J. F. 1999. The Tangier disease gene product ABC1 controls the cellular apolipoprotein-mediated lipid removal pathway. J. Clin. Invest. 104: R25–R31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aiello R. J., Brees D., Bourassa P. A., Royer L., Lindsey S., Coskran T., Haghpassand M., Francone O. L. 2002. Increased atherosclerosis in hyperlipidemic mice with inactivation of ABCA1 in macrophages. Arterioscler. Thromb. Vasc. Biol. 22: 630–637 [DOI] [PubMed] [Google Scholar]
- 11.Remaley A. T., Stonik J. A., Demosky S. J., Neufeld E. B., Bocharov A. V., Vishnyakova T. G., Eggerman T. L., Patterson A. P., Duverger N. J., Santamarina-Fojo S., et al. 2001. Apolipoprotein specificity for lipid efflux by the human ABCAI transporter. Biochem. Biophys. Res. Commun. 280: 818–823 [DOI] [PubMed] [Google Scholar]
- 12.Remaley A. T., Thomas F., Stonik J. A., Demosky S. J., Bark S. E., Neufeld E. B., Bocharov A. V., Vishnyakova T. G., Patterson A. P., Eggerman T. L., et al. 2003. Synthetic amphipathic helical peptides promote lipid efflux from cells by an ABCA1-dependent and an ABCA1-independent pathway. J. Lipid Res. 44: 828–836 [DOI] [PubMed] [Google Scholar]
- 13.Wool G. D., Reardon C. A., Getz G. S. 2008. Apolipoprotein A-I mimetic peptide helix number and helix linker influence potentially anti-atherogenic properties. J. Lipid Res. 49: 1268–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D'Souza W., Stonik J. A., Murphy A., Demosky S. J., Sethi A. A., Moore X. L., Chin-Dusting J., Remaley A. T., Sviridov D. 2010. Structure/function relationships of apolipoprotein a-I mimetic peptides: implications for antiatherogenic activities of high-density lipoprotein. Circ. Res. 107: 217–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Natarajan P., Forte T. M., Chu B., Phillips M. C., Oram J. F., Bielicki J. K. 2004. Identification of an apolipoprotein A-I structural element that mediates cellular cholesterol efflux and stabilizes ATP binding cassette transporter A1. J. Biol. Chem. 279: 24044–24052 [DOI] [PubMed] [Google Scholar]
- 16.Watson C. E., Weissbach N., Kjems L., Ayalasomayajula S., Zhang Y., Chang I., Navab M., Hama S., Hough G., Reddy S. T., et al. 2011. Treatment of patients with cardiovascular disease with L-4F, an apo-A1 mimetic, did not improve select biomarkers of HDL function. J. Lipid Res. 52: 361–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bloedon L. T., Dunbar R., Duffy D., Pinell-Salles P., Norris R., DeGroot B. J., Movva R., Navab M., Fogelman A. M., Rader D. J. 2008. Safety, pharmacokinetics, and pharmacodynamics of oral apoA-I mimetic peptide D-4F in high-risk cardiovascular patients. J. Lipid Res. 49: 1344–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tubb M. R., Smith L. E., Davidson W. S. 2009. Purification of recombinant apolipoproteins A-I and A-IV and efficient affinity tag cleavage by tobacco etch virus protease. J. Lipid Res. 50: 1497–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Atcliffe B. W., MacRaild C. A., Gooley P. R., Howlett G. J. 2001. The interaction of human apolipoprotein C–I with sub-micellar phospholipid. Eur. J. Biochem. 268: 2838–2846 [DOI] [PubMed] [Google Scholar]
- 20.Durbin D. M., Jonas A. 1997. The effect of apolipoprotein A-II on the structure and function of apolipoprotein A-I in a homogeneous reconstituted high density lipoprotein particle. J. Biol. Chem. 272: 31333–31339 [DOI] [PubMed] [Google Scholar]
- 21.Oram J. F., Lawn R. M., Garvin M. R., Wade D. P. 2000. ABCA1 is the cAMP-inducible apolipoprotein receptor that mediates cholesterol secretion from macrophages. J. Biol. Chem. 275: 34508–34511 [DOI] [PubMed] [Google Scholar]
- 22.Vedhachalam C., Liu L., Nickel M., Dhanasekaran P., Anantharamaiah G. M., Lund-Katz S., Rothblat G. H., Phillips M. C. 2004. Influence of ApoA-I structure on the ABCA1-mediated efflux of cellular lipids. J. Biol. Chem. 279: 49931–49939 [DOI] [PubMed] [Google Scholar]
- 23.Gillotte K. L., Zaiou M., Lund-Katz S., Anantharamaiah G. M., Holvoet P., Dhoest A., Palgunachari M. N., Segrest J. P., Weisgraber K. H., Rothblat G. H., et al. 1999. Apolipoprotein-mediated plasma membrane microsolubilization. Role of lipid affinity and membrane penetration in the efflux of cellular cholesterol and phospholipid. J. Biol. Chem. 274: 2021–2028 [DOI] [PubMed] [Google Scholar]
- 24.Panagotopulos S. E., Witting S. R., Horace E. M., Hui D. Y., Maiorano J. N., Davidson W. S. 2002. The role of apolipoprotein A-I helix 10 in apolipoprotein-mediated cholesterol efflux via the ATP-binding cassette transporter ABCA1. J. Biol. Chem. 277: 39477–39484 [DOI] [PubMed] [Google Scholar]
- 25.Gursky O. 2001. Solution conformation of human apolipoprotein C-1 inferred from proline mutagenesis: far- and near-UV CD study. Biochemistry. 40: 12178–12185 [DOI] [PubMed] [Google Scholar]
- 26.Rozek A., Sparrow J. T., Weisgraber K. H., Cushley R. J. 1999. Conformation of human apolipoprotein C-I in a lipid-mimetic environment determined by CD and NMR spectroscopy. Biochemistry. 38: 14475–14484 [DOI] [PubMed] [Google Scholar]
- 27.Segrest J. P., Jones M. K., De Loof H., Brouillette C. G., Venkatachalapathi Y. V., Anantharamaiah G. M. 1992. The amphipathic helix in the exchangeable apolipoproteins: a review of secondary structure and function. J. Lipid Res. 33: 141–166 [PubMed] [Google Scholar]
- 28.Segrest J. P., De Loof H., Dohlman J. G., Brouillette C. G., Anantharamaiah G. M. 1990. Amphipathic helix motif: classes and properties. Proteins. 8: 103–117 [DOI] [PubMed] [Google Scholar]
- 29.Rost B., Yachdav G., Liu J. 2004. The PredictProtein server. Nucleic Acids Res. 32(Web Server issue): W321–W326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Venkatachalapathi Y. V., Phillips M. C., Epand R. M., Epand R. F., Tytler E. M., Segrest J. P., Anantharamaiah G. M. 1993. Effect of end group blockage on the properties of a class A amphipathic helical peptide. Proteins. 15: 349–359 [DOI] [PubMed] [Google Scholar]
- 31.Sethi A. A., Stonik J. A., Thomas F., Demosky S. J., Amar M., Neufeld E., Brewer H. B., Davidson W. S., D'Souza W., Sviridov D., et al. 2008. Asymmetry in the lipid affinity of bihelical amphipathic peptides. A structural determinant for the specificity of ABCA1-dependent cholesterol efflux by peptides. J. Biol. Chem. 283: 32273–32282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vedhachalam C., Ghering A. B., Davidson W. S., Lund-Katz S., Rothblat G. H., Phillips M. C. 2007. ABCA1-induced cell surface binding sites for ApoA-I. Arterioscler. Thromb. Vasc. Biol. 27: 1603–1609 [DOI] [PubMed] [Google Scholar]
- 33.Chroni A., Liu T., Fitzgerald M. L., Freeman M. W., Zannis V. I. 2004. Cross-linking and lipid efflux properties of ApoA-I mutants suggest direct association between ApoA-I helices and ABCA1. Biochemistry. 43: 2126–2139 [DOI] [PubMed] [Google Scholar]
- 34.Martinez L. O., Agerholm-Larsen B., Wang N., Chen W., Tall A. R. 2003. Phosphorylation of a PEST sequence in ABCA1 promotes calpain degradation and is reversed by ApoA-I. J. Biol. Chem. 278: 37368–37374 [DOI] [PubMed] [Google Scholar]
- 35.Wang N., Chen W., Linsel-Nitschke P., Martinez L. O., Agerholm-Larsen B., Silver D. L., Tall A. R. 2003. A PEST sequence in ABCA1 regulates degradation by calpain protease and stabilization of ABCA1 by apoA-I. J. Clin. Invest. 111: 99–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Segrest J. P., Garber D. W., Brouillette C. G., Harvey S. C., Anantharamaiah G. M. 1994. The amphipathic alpha helix: a multifunctional structural motif in plasma apolipoproteins. Adv. Protein Chem. 45: 303–369 [DOI] [PubMed] [Google Scholar]
- 37.Mishra V. K., Palgunachari M. N., Datta G., Phillips M. C., Lund-Katz S., Adeyeye S. O., Segrest J. P., Ananthanarayanan M. 1998. Studies of synthetic peptides of human apolipoprotein A-I containing tandem amphipathic alpha-helixes. Biochemistry. 37: 10313–10324 [DOI] [PubMed] [Google Scholar]
- 38.Palgunachari M. N., Mishra V. K., Lund-Katz S., Phillips M. C., Adeyeye S. O., Alluri S., Anantharamaiah G. M., Segrest J. P. 1996. Only the two end helixes of eight tandem amphipathic helical domains of human apo A-I have significant lipid affinity. Implications for HDL assembly. Arterioscler. Thromb. Vasc. Biol. 16: 328–338 [DOI] [PubMed] [Google Scholar]
- 39.Kono M., Tanaka T., Tanaka M., Vedhachalam C., Chetty P. S., Nguyen D., Dhanasekaran P., Lund-Katz S., Phillips M. C., Saito H. 2010. Disruption of the C-terminal helix by single amino acid deletion is directly responsible for impaired cholesterol efflux ability of apolipoprotein A-I Nichinan. J. Lipid Res. 51: 809–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saito H., Dhanasekaran P., Nguyen D., Deridder E., Holvoet P., Lund-Katz S., Phillips M. C. 2004. Alpha-helix formation is required for high affinity binding of human apolipoprotein A-I to lipids. J. Biol. Chem. 279: 20974–20981 [DOI] [PubMed] [Google Scholar]
- 41.Vedhachalam C., Narayanaswami V., Neto N., Forte T. M., Phillips M. C., Lund-Katz S., Bielicki J. K. 2007. The C-terminal lipid-binding domain of apolipoprotein E is a highly efficient mediator of ABCA1-dependent cholesterol efflux that promotes the assembly of high-density lipoproteins. Biochemistry. 46: 2583–2593 [DOI] [PubMed] [Google Scholar]
- 42.Choy N., Raussens V., Narayanaswami V. 2003. Inter-molecular coiled-coil formation in human apolipoprotein E C-terminal domain. J. Mol. Biol. 334: 527–539 [DOI] [PubMed] [Google Scholar]
- 43.Fan D., Li Q., Korando L., Jerome W. G., Wang J. 2004. A monomeric human apolipoprotein E carboxyl-terminal domain. Biochemistry. 43: 5055–5064 [DOI] [PubMed] [Google Scholar]
- 44.Tanaka M., Koyama M., Dhanasekaran P., Nguyen D., Nickel M., Lund-Katz S., Saito H., Phillips M. C. 2008. Influence of tertiary structure domain properties on the functionality of apolipoprotein A-I. Biochemistry. 47: 2172–2180 [DOI] [PubMed] [Google Scholar]
- 45.Tanaka M., Dhanasekaran P., Nguyen D., Ohta S., Lund-Katz S., Phillips M. C., Saito H. 2006. Contributions of the N- and C-terminal helical segments to the lipid-free structure and lipid interaction of apolipoprotein A-I. Biochemistry. 45: 10351–10358 [DOI] [PubMed] [Google Scholar]
- 46.Davidson W. S., Hazlett T., Mantulin W. W., Jonas A. 1996. The role of apolipoprotein AI domains in lipid binding. Proc. Natl. Acad. Sci. USA. 93: 13605–13610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Westerlund J. A., Weisgraber K. H. 1993. Discrete carboxyl-terminal segments of apolipoprotein E mediate lipoprotein association and protein oligomerization. J. Biol. Chem. 268: 15745–15750 [PubMed] [Google Scholar]
- 48.Weinberg R. B. 1994. Identification of functional domains in the plasma apolipoproteins by analysis of inter-species sequence variability. J. Lipid Res. 35: 2212–2222 [PubMed] [Google Scholar]
- 49.Deng X., Morris J., Dressmen J., Tubb M. R., Tso P., Jerome W. G., Davidson W. S., Thompson T. B. 2012. The structure of dimeric apolipoprotein A-IV and its mechanism of self-association. Structure. 20: 767–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mei X., Atkinson D. 2011. Crystal structure of C-terminal truncated apolipoprotein A-I reveals the assembly of high density lipoprotein (HDL) by dimerization. J. Biol. Chem. 286: 38570–38582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karathanasis S. K., Oettgen P., Haddad I. A., Antonarakis S. E. 1986. Structure, evolution, and polymorphisms of the human apolipoprotein A4 gene (APOA4). Proc. Natl. Acad. Sci. USA. 83: 8457–8461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sviridov D. O., Andrianov A. M., Anishchenko I. V., Stonik J. A., Amar M. J., Turner S., Remaley A. T. 2013. Hydrophobic amino acids in the hinge region of the 5A apolipoprotein mimetic peptide are essential for promoting cholesterol efflux by the ABCA1 transporter. J. Pharmacol. Exp. Ther. 344: 50–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Segrest J. P., Jones M. K., Mishra V. K., Ananatharamaiah G. M. 2002. Experimental and computational studies of the interactions of amphipathic peptides with lipid surfaces. In Peptide-Lipid Interactions. Vol. 52. S. A. Simon and T. J. McIntosh, editors. 391–429. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.