Abstract
The surface of lipid droplets (LDs) in various cell types is coated with perilipin proteins encoded by the Plin genes. Perilipins regulate LD metabolism by selectively recruiting lipases and other proteins to LDs. We have studied the expression of perilipins in mouse muscle. The glycolytic fiber-enriched gastrocnemius muscle expresses predominantly Plin2-4. The oxidative fiber-enriched soleus muscle expresses Plin2-5. Expression of Plin2 and Plin4-5 is elevated in gastrocnemius and soleus muscles from mice fed a high-fat diet. This effect is preserved in peroxisome proliferator-activated receptor (PPAR)α-deficient mice. Mouse muscle derived C2C12 cells differentiated into glycolytic fibers increase transcription of these Plins when exposed to various long chain fatty acids (FAs). To understand how FAs regulate Plin genes, we used specific activators and antagonists against PPARs, Plin promoter reporter assays, chromatin immunoprecipitation, siRNA, and animal models. Our analyses demonstrate that FAs require PPARδ to induce transcription of Plin4 and Plin5. We further identify a functional PPAR binding site in the Plin5 gene and establish Plin5 as a novel direct PPARδ target in muscle. Our study reveals that muscle cells respond to elevated FAs by increasing transcription of several perilipin LD-coating proteins. This induction renders the muscle better equipped to sequester incoming FAs into cytosolic LDs.
Keywords: perilipin, Plin, Plin2, Plin3, Plin4, Plin5, S3-12, Lsdp5, TIP47, peroxisome proliferator-activated receptor δ, peroxisome proliferator-activated receptor response element, promoter, oleic acid
The main function of the muscle is to perform work. Energy to drive contraction is primarily obtained by metabolizing glucose or fatty acids (FAs), which the muscle stores as glycogen or in triacylglycerol (TAG)-containing lipid droplets (LDs), respectively. These energy-stores may impact cellular signaling when they exceed the need for storage. High intracellular content of myocellular LDs is well-known to correlate with insulin resistance (1, 2). This is, however, not an absolute phenomenon, as myocellular LD content increases in response to exercise and may even be higher in athletes than obese insulin-resistant individuals (3). Some beneficial effects of exercise are believed linked to increased capacity to oxidize LDs to prevent accumulation of lipid metabolites.
Metabolism of muscular LDs is poorly described. Similar to other cells, LDs in muscle consist of a protein coat, a single monolayer of phospholipids, and an inner core of neutral lipids, such as TAG or cholesteryl esters (CEs) (4–6). Changes in the composition of proteins embedded into the LD surface likely control the release of FAs from myocellular LDs. The perilipin proteins are particularly interesting in this context, as members of this gene family are known to regulate lipolysis. Mammalian perilipins derive from a gene family of ancient origin encoded by five Plin genes (7–9), which encode for the LD-binding perilipin1 (10, 11), perilipin2/ADRP/adipophilin (12), perilipin3/TIP47 (13), perilipin4/S3-12 (14), and perilipin5/Lsdp5/MLDP/oxPAT (7, 15, 16). These proteins are uniquely expressed. Expression of perilipin1 is confined to adipose and steroidogenic cells (17, 18). Perilipin2 and -3 are broadly expressed (12, 19), expression of perilipin4 is limited to adipose cells, brain, skeletal muscle, and heart (14, 19), whereas perilipin5 is expressed in oxidative tissues (7, 15, 16). Muscle tissues express perilipin 2-5.
Although the five perilipin proteins are likely to have unique features, they are all commonly shown to affect LD accumulation in various cell types. It is believed that the perilipins regulate lipolysis. This is best characterized for perilipin1 (4). Perilipin1 protects against adipose lipolysis when the energy status of the organism is high (postprandial), but facilitates lipolysis when stored energy needs to be released (fasted). These events are controlled by the phosphorylation status of perilipin1, which determines recruitment of lipolytic enzymes to the LD surface. This regulatory mechanism cannot be compensated for by other perilipins in the lack of perilipin1 (20–23). Distinct functional roles for the remaining perilipins are less clear. Other perilipin members similarly interact with lipases and associated factors at the LD surface (24–27). However, functional compensation precludes characterization of individual perilipins in cells and mice (28). It is clear that Plin2- and Plin5-null mice have reduced hepatic and cardiac accumulation of lipids, respectively (29, 30). Perilipin5 has a unique ability to facilitate physical linkage of LDs and mitochondria when ectopically expressed (31, 32), but the significance of this association is unclear. Depending on cell type, ectopic expression of perilipin5 either prevents (7) or enhances lipolysis (15). An obligatory role for perilipin5-mediated association of LDs and mitochondria for enhanced oxidation (31, 33) might explain the discrepancies in the initial reports. The functions of perilipin3 and perilipin4, other than binding to LDs, are poorly understood.
Depending on physiological conditions and the presence of other perilipin family members, perilipins may be cytosolic, bound to LDs, or rapidly degraded and thus nearly absent in the cell (7, 34, 35). Due to their role in lipolysis, the type of perilipins being expressed and bound to LDs are an important determinant of cellular LD metabolism. It is therefore important to identify transcription factors and pathways that control expression of the various Plin genes. Several of the Plin genes contain evolutionary conserved cis-regulatory elements occupied by peroxisome proliferator-activated receptors (PPARs). The PPARs belong to the nuclear receptor superfamily and consist of the isotypes PPARα, PPARβ/δ (hereafter referred to as PPARδ), and PPARγ. They all heterodimerize with retinoid X receptors (RXRs) onto mainly DR-1 type PPAR response elements (PPREs) in the promoter region of target genes (36, 37). Adipose expression of Plin1 and Plin4 is switched on by binding of PPARγ to their respective promoter regions (19, 38–40). Plin2 is regulated by PPARα (41–43) and PPARδ (44–46) in various cell types, whereas expression of Plin3 seems unaffected by activation of PPARs (19, 41). Regulation of Plin5 by PPARs is poorly understood. Expression of Plin5 is enhanced by activation of PPARα (7, 15, 16), but no PPRE has been identified in the Plin5 gene.
Little is known regarding transcription factors important for expression of perilipins in muscle. Given the known regulation of Plin genes by PPARs in other tissues, we analyzed FA and PPAR regulation of Plin genes. Our analyses revealed an unexpected importance for PPARδ as a FA sensor regulating the expression of the Plin4 and Plin5 genes in muscle. We further identified a conserved PPRE in the Plin5 gene. This PPRE is essential for FA-stimulated expression of perilipin5 and establishes Plin5 as a direct PPARδ target gene.
EXPERIMENTAL PROCEDURES
Materials
Restriction enzymes were purchased from New England BioLabs (Ipswich, MA). PfuTurbo® DNA polymerase was purchased from Stratagene (La Jolla, CA). The PPAR ligands, WY-14643, GW6471, GSK0660, and GW9662 were purchased from Sigma (St. Louis, MO). GW501516, rosiglitazone (Rosi), troglitazone (Tro), and GW1929 were obtained from Enzo Life Sciences (Farmingdale, NY). Reagents for quantitative real-time PCR were from Applied Biosystems (Life Technologies Corporation, Carlsbad, CA). Cell culture reagents, oligonucleotides, FAs, and other chemicals were purchased from Sigma. All other chemicals and biochemicals were of the highest quality available from commercial vendors.
Cloning of expression vectors
Full-length pENTR-4r-3r Plin1-5 vectors have been described elsewhere (47). Full-length cDNAs encoding mouse PPARα (48), PPARδ (49), Addgene plasmid 8891, PPARγ1 and -2 (3T3-L1 cDNA), and alternative translation variants of Plin2-aa123-425 and Plin5-aa16-463 were cloned into the pDONR-221 P4r-P3r vector (Life Technologies Corporation). A Kozak translation initiation site (50) was generated by inserting ACC in front of the naturally occurring AUG start codon, and stop codons were changed to TAG. To generate an expression vector without a cDNA insert, a short multi-cloning-site linker (KHSS; Start-KpnI-HindIII-SacI-SpeI: ATG- GGTACC-AAGCTT-GAGCTC-ACTAGT) was amplified for cloning into the pDONR-221 P4r-P3r vector to be exchanged with the suicidal attR4r-ccdB-chloramphenicol-attR3r- cassette. The primers used are listed in Table 1. The amplified PCR products were recombined into the pDONR-221 P4r-P3r vector using BP clonase II (Life Technologies Corporation) to produce pENTR-4r-3r-PPAR vectors. The V5-6x-His-Gly tag was amplified from a synthesized template. The PCR product was recombined into pDONR-221 P1-P4 to generate the pENTR-R1-R4-V5-6xHisG vector.
TABLE 1.
Oligos used
| Oligo Name | Site | Sequence |
| Primers to generate pcDNA3-DEST vectors | ||
| attR1-R2-cas fw | (Hind III) attB1 | ATT AAGCTT ACAAGTTTGTACAAAAAAGCTGAAC |
| attR1-R2-cas rev | (Apa I) attB2 | TAA GGGCCC ACCACTTTGTACAAGAAAGC |
| attR4r-R3r-cas fw | (Hind III) attB4r | ATT AAGCTT ACAACTTTTCTATACAAAGTTGGC |
| attR4r-R3r-cas rev | (Apa I) attB3r | TAA GGGCCC ACAACTTTATTATACAAAGTTG |
| Templates to generate pENTR tag vectors | ||
| V5-6xHis-Gly template | GGTAAGCCTATCCCTAACCCTCTCCTCGGTCTCGATTCTACGCGTACCGGTCATCATCACCATCACCATGGT | |
| KHSS template | ATGGGTACCAAGCTTGAGCTCACTAGT | |
| Primers to generate pENTR vectors | ||
| V5-6xHis-Gly-fw | attB1 | GGGGACAAGTTTGTACAAAAAAGCAGGCTTA ACCATG GGTCATCATCACCAT |
| V5-6xHis-Gly-rev | attB4 | GGGGACAACTTTGTATAGAAAAGTTGGGTGACCATGGTGATGGTGATGATGACC |
| KHSS-fw | attB4r | GGGGACAACTTTTCTATACAAAGTTGCT ACCATG GGTACCAAGCTTGAGC |
| KHSS-rev | attB3r | GGGGACAACTTTATTATACAAAGTTGTACTAGTGAGCTCAAGCTT |
| m-PPARα-fw | attB4r | GGGGACAACTTTTCTATACAAAGTTGCT ACCATG GTGGACACAGAGAGC |
| m-PPARα-rev | attB3r | GGGGACAACTTTATTATACAAAGTTGTCTAGTACATGTCTCTGTA |
| m-PPARγ1-fw | attB4r | GGGGACAACTTTTCTATACAAAGTTGCT ACCATG GTTGACACAGAGATGCC |
| m-PPARγ2-fw | attB4r | GGGGACAACTTTTCTATACAAAGTTGCT ACCATG GGTGAAACTCTGGGA |
| m-PPARγ1-rev | attB3r | GGGGACAACTTTATTATACAAAGTTGTCTAATACAAGTCCTTGTAGATCTC |
| m-PPARδ-fw | attB4r | GGGGACAACTTTTCTATACAAAGTTGCT ACCATG GAACAGCCACAGGAG |
| m-PPARδ-rev | attB3r | GGGGACAACTTTATTATACAAAGTTGTCTAGTACATGTCCTTGTA |
| m-RXRα-rw | attB4r | GGGGACAACTTTTCTATACAAAGTTGCT ACCATG GACACCAAACATTTCCTGCC |
| m-RXRα-rev | attB3r | GGGGACAACTTTATTATACAAAGTTGTCTAGGTGGCTTGATGTGG |
| m-Plin2-aa123-fw | attB4r | GGGGACAACTTTTCTATACAAAGTTGCT ACCATG GCTGGAGCCAAGGAT |
| m-Plin2-rev | attB3r | GGGGACAACTTTATTATACAAAGTTGTCTACTGAGCTTTGACCTC |
| m-Plin5-aa16-fw | attB4r | GGGGACAACTTTTCTATACAAAGTTGCT ACCATG TCCGGTGATCAGACAG |
| m-Plin5-rev | attB3r | GGGGACAACTTTATTATACAAAGTTGTCTAGAAGTCCAGCTCTGG |
| Primers to clone and mutate the mouse Plin5 LUC reporter | ||
| Plin5-promoter-fw | Hind III | TAAAGCTTGCCAGGAATGCTATTCTCGGACT |
| Plin5-promoter-rev | Hind III | TAAAGCTTTCAGGGCTCATGCCCTATGTATC |
| Plin5-PPRE-mut-fw | AGCAGGTGGCAGGAGGCTTGGGGCAGTGGGCCCGG | |
| Plin5-PPRE-mut-rev | CCGGGCCCACTGCCCCAAGCCTCCTGCCACCTGCT | |
Primers used to clone PPAR and RXR coding segments contain a Kozak initiation sequence (ACCATG; underlined , inserted into attB4r primers) or a stop codon (TAG; italic, inserted into attB3r primers). aa, amino acid; fw, forward; multi cloning site (KHSS, start-KpnI-HindIII-SacI-SpeI); rev, reverse.
The pcDNA3-DEST-R4r-R3r vector was generated by replacing the multi-cloning site of pcDNA3 (Life Technologies Corporation) with an attR4r-ccdB-chloramphenicol-attR3r- cassette (R4r-R3r). The R4r-R3r cassette was amplified with PfuTurbo® DNA polymerase (Stratagene) using pDONR-221-P4r-P3r as a template (primers see Table 1). The amplified PCR product and the pcDNA3 vector was digested with HindIII and ApaI, ligated, and transformed into ccdB SurvivalTM-T1R cells (Life Technologies Corporation) to generate the pcDNA3-DEST-R4r-R3r vector. The similar strategy was used to generate a pcDNA3-DEST-R1-R2 vector. The pcDNA3-DEST-R1-R3r vector was generated by digesting the pcDNA3-DEST-R1-R2 and pcDNA3-DEST-R4r-R3r vectors with Pst I followed by ligation of the fragment containing the R3r-att site into the cut pcDNA3-DEST-R1-R2 vector. All vectors were confirmed by sequencing (Macrogen, Korea).
The pcDNA3-DEST-R4r-R3r was recombined with the pENTR-4r-3r-PPAR vectors using LR clonase II (Life Technologies Corporation) to generate the pcDNA3-mRXRα, pcDNA3-mPPARα, pcDNA3-mPPARγ1, pcDNA3-mPPARγ2, and pcDNA3-mPPARδ expression vectors. The pcDNA3-DEST-R1-R3r was recombined with the pENTR-R1-R4-V5-6xHisG and pENTR-4r-3r-PPAR vectors to generate the pcDNA3-V5-His-PPAR expression vectors.
Cloning and mutagenesis of the Plin5 reporter
The mouse Plin2 and Plin4 LUC reporters have been described elsewhere (19, 41). The full-length mouse Plin5 promoter (−2324/+244) was amplified by a PCR strategy described previously (51), cloned into pPCR-Script (Stratagene), digested out using Hind III, and inserted into the pGL3-Basic luciferase reporter vector (Promega, Madison, WI). Site-directed mutagenesis of the DR-1 element was performed with PCR as described previously (51). Primers used are listed in Table 1.
Preparation of fatty acids
FAs were complexed to low-endotoxin FA-free BSA (Sigma, #A8806). FA (6 mM)/BSA (2.4 mM) stock solutions were generated by dissolving 6 μmol FAs in 60 μl 0.1 M NaOH, followed by FA binding to BSA at 50°C for 5 min. FA stock solutions were stored under argon at −80°C to prevent oxidization. FAs used: myristic acid (C14:0), palmitic acid (C16:0), stearic acid (C18:0), oleic acid (OA; cis-C18:1 n-9), vaccenic acid (cis-C18:1 n-7), linoleic acid (LA; C18:2 n-6), and γ-linolenic acid (C18:3n-6).
Culturing and transfection of cells
C2C12 (ATCC, #CRL-1772) and Sol8 (ATCC, #CRL-2174) cells were sub-cultured in high glucose DMEM (Sigma, #5648, supplemented with 5.958 g HEPES, 1.5 g NaHCO3, and 0.11 g sodium pyruvate/l) in the presence of penicillin (50 U/ml), streptomycin (50 μg/ml), and 20% FBS (Gibco, #26140-079, Life Technologies Corporation) at 37°C in 5% CO2. Myotubule differentiation was initiated by exchanging the 20% FCS with 2% horse serum (Diff-medium). Medium was refreshed every third day. The myoblasts decreased in number with differentiation and were replaced by a gradual increase in multi-nuclear myotubes from day 2 of differentiation (supplementary Fig. II). Spontaneous contraction was observed from day 4. The differentiation marker paired box protein 7 (Pax7) decreased, myogenic differentiation 1 (Myod1) peaked at days 2–3, whereas myosin heavy chain 2A (Myh2) increased drastically (>5,000-fold) until day 7 (result not shown). These assays confirm that both cell lines were well differentiated into contractile myotubes.
Unless otherwise indicated, C2C12 and Sol8 cells were seeded in 12-well dishes at a density of 3 × 104 cells/well. Two days later, differentiation was initiated by changing to Diff-medium. For transfection experiments, C2C12 and Sol8 cells were seeded at 6 × 104 cells/well in antibiotic-free medium. The following day, cells were given 1 ml antibiotic-free Diff-medium, prior to transfection with 2 μg DNA:4 μl Lipofectamine2000 complexed in 200 μl OPTI-MEM (Life Technologies Corporation). After 6 h, medium was replaced with Diff-medium containing antibiotics and allowed to grow for a maximum of 4 days before being harvested.
Silencing of PPARδ using siRNA
Duplexes of siRNAs (Sigma) targeting mouse PPARδ (PPARδ siRNA1: sense 5′-CCAUCAUUCUGUGUGGAGAtt-3′, antisense 5′-UCUCCACACAGAAUGAUGGtt-3′ PPARδ siRNA2: sense 5′-CCAAGUUCGAGUUUGCUGUtt-3′, antisense 5′-ACAGCAAACUCGAACUUGGtt-3′), and negative control siRNA (sense 5′-UAACGACGCGACGACGUAAtt-3′, antisense 5′-UUACGUCGUCGCGUCGUUAtt-3′) were transfected into C2C12 cells using reverse transfection. Briefly, 10 pmol RNAi duplexes (a 1:1 mix of siRNA1 and siRNA2 was used to target PPARδ) were complexed with 3 μl Lipofectamine® RNAiMAX reagent (Life Technologies Corporation) in 350 μl OPTI-MEM per well (24-well plate). Trypsinized C2C12 cells (3 × 104 cells/well) were added in 0.5 ml growth medium (containing 20% serum, but no antibiotics). Five hours after the transfection, differentiation was initiated by changing to Diff-medium. GAPDH siRNA (Life Technologies Corporation, #4390849) was used to establish transfection efficiency. The knockdown efficiency was found to be comparable 3 and 4 days after siRNA transfection.
Protein isolation and Western blotting
Cells were harvested in lysis buffer [see (41)], 1× PBS, 1% NP-40, 0.1% SDS, and complete Proteinase Inhibitor Cocktail (Roche, #4693116001), frozen, and sonicated for 2 × 2 s using Branson Sonifier 450 (Branson Ultrasonic S.A.). Frozen tissues were homogenized in lysis buffer (as for cells) using Precellys®24 (Bertin Technologies, France) for 2 × 20 s at 5,000 rpm. Protein concentrations were quantified by BC Assay (Interchim, France, #FT-40840).
Primary antibodies against mouse perilipin4/S3-12 (peptide-1 sequence MSASGDGTRVPPKSKGC) and perilipin5 (peptide-1 sequence CEAEPPRGQGKHTMMPELDF) were raised in rabbits (Affinity BioReagents). The novel antibodies were verified to not recognize any of the other mouse perilipin proteins (supplementary Fig. I).
Proteins were separated by SDS-PAGE on 4–12% NuPAGE (Life Technologies Corporation) or 10% Criterion™ Precast (Bio-Rad, Hercules, CA) gels, and transferred to a nitrocellulose membrane (GE Healthcare, UK). Membranes were incubated with the following primary antibodies: polyclonal rabbit anti-mouse perilipin1 (18, 21) (1:2,000), rabbit anti-mouse perilipin2/ADFP (Novus Biologicals, #NB110-40877, 1:400), rabbit anti-mouse perilipin3/TIP47 (52) (1:4000), rabbit anti-mouse perilipin4/S3-12 (10 μg/ml), rabbit anti-mouse perilipin5/LSDP5 (9.2 μg/ml), or His antibody (Abcam, #ab1187, 1:5,000). β-actin (Sigma, #A5441, 1:10,000) or GRP78 (BD Transduction Laboratories, #610978, 1:1,000) were used as loading controls.
Following binding of primary antibodies, membranes were incubated with species-specific horseradish peroxidase-labeled secondary antibodies (Abcam, goat to rabbit IgG-HRP, #ab6721, 1:10,000 and rabbit to mouse IgG-HRP, #524567, 1:10,000) and binding detected using ECL Plus (GE Healthcare), or alkaline phosphatase-conjugated-labeled species-specific secondary antibodies using Western Breeze® Chemiluminescent kit (Life Technologies Corporation). Chemiluminescent signals were visualized with exposure to Hyperfilm ECL (GE Healthcare). Carestream MI SE was used to quantify Western blots.
Preparation and analysis of RNA
Cells were lysed in 500 μl 1× Total RNA Lysis Solution (#4305895, Life Technologies Corporation) per well (12-well plate) and frozen at −80°C before isolation. Total RNA from cell extracts was isolated using ABI 6100 Nucleic Acid Prep-Station using the preprogrammed “RNA-Cell method” (Life Technologies Corporation).
Muscle tissue was homogenized for 2 × 30 s/5,000 rpm with zirconium dioxide ceramic beads (1.4 mm; #03961-1-103) in a Precellys®24 homogenizer (Bertin Technologies). Total RNA was subsequently isolated using the RNeasy® Mini kit (Qiagen, #74104). RNA purity and quantity were determined using Nano-Drop ND-1000 spectrophotometer (Thermo Scientific, Waltham, MA). RNA from in vivo studies was subjected to an additional quality check using Bio Analyzer prior to gene expression analysis (Agilent Technologies, Santa Clara, CA; #kit 5067-1511).
Total RNA (cells, 12 ng/μl; muscle, 50 ng/μl; and liver, 12 ng/μl) was reverse transcribed into single-stranded cDNA using high capacity cDNA reverse transcription kit (Life Technologies Corporation, #4368814). Quantitative real-time PCR amplification (1 μl cDNA reaction in 20 μl reaction volume) was performed using TaqMan® Universal PCR Master Mix on an ABI 7900HT system (Life Technologies Corporation) operating with standard settings. RNA was analyzed using predesigned TaqMan® Low Density Custom Arrays (liver and soleus) or predesigned single assays (gastrocnemius and cell cultures). Assays used: Plin1, #Mm00558672_m1; Plin2, #Mm00475794_m1; Plin3, #Mm00482206_m1; Plin4, #Mm00491061_m1; Plin5, #Mm00508852_m1; PPARα, #Mm00440939_m1; PPARδ, #Mm01305434_m1; PPARγ, #Mm01184322_m1; 36B4, #Mm00725448_s1; TBP, #Mm00446973_m1; and GAPDH, #Mm99999915_g1. Data were analyzed in RQ Manager using the ΔΔCt method. Results are presented as gene expression ± standard deviation (SD) relative to endogenous control (2−ΔΔCT).
Reporter gene expression assay
C2C12 cells (5 × 103 cells/well) were seeded in 96-well dishes in 75 μl antibiotic-free medium. The next day, 75 ng DNA (50 ng reporter, 10 ng expression vectors, and 5 ng pRL) and 0.5 μl Lipofectamie2000 were mixed in 2 × 10 μl OPTI-MEM I (Life Technologies Corporation) and added to cells incubated in 75 μl Diff-medium. After 5 h, culture medium was replaced with Diff-medium and incubated for 2 days. Fresh medium containing FAs and PPAR antagonists was added for an additional 24 h. Cells were washed in 1× PBS and lysed in 20 μl Passive Lysis buffer (#E194A, Promega). Dual luciferase activity was determined using Dual-Luciferase® Reporter Assay System (#E1910, Promega) and luciferase activity measured with a Synergy 2 Luminometer (BioTek, Winooski, VT).
ChIP experiments
Chromatin immunoprecipitation (ChIP) experiments were performed as described previously with minor modifications (53). Briefly, C2C12 cells were cross-linked with 1% formaldehyde for 10 min. Cross-linking was terminated by 10 min incubation with 0.125 M glycine. Cells were washed twice in cold 1× PBS and harvested in lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl, pH 8.0) containing complete Protease Inhibitor Cocktail (Roche, #4693116001). Lysed cells were sonicated using a Bioruptor (Diagenode, Belgium) to fragments of 300–500 bp. Chromatin was collected by centrifugation, concentration determined by A260, and diluted in RIPA buffer (0.1% SDS, 0.1% Na-deoxycholate, 1% Triton X-100, 1 mM EDTA, 0.5 mM EGTA, 140 mM NaCl, 10 mM Tris-HCl, pH 8.0) and immunoprecipitated with 2 μg antibody against RNAPII (Santa Cruz Biotechnology, CA; #sc-899) overnight at 4°C in the presence of protein A beads (GE Healthcare). Beads were washed three times in RIPA buffer and eluted in 1% SDS with 0.1 M NaHCO3. Chromatin was de-cross-linked by adding 0.2 M NaCl and incubating overnight at 65°C. DNA was purified by phenol-chloroform extraction, precipitated in ethanol with sodium acetate, and dissolved in water. DNA enrichment was quantified by real-time PCR (ABI, 7900HT) using SYBR Green Master Mix (Life Technologies Corporation). The following primers were used: Plin2 (5′-TCTGGTGCAGGACCTACCTAA, 5′-TTTGCTGTGTGGTGATCTGG), Plin3 (5′-GAGGAAACCTCCCCTACCAA, 5′-CCTCTGTCCTGTCACTCCAA), Plin4 (5′-GGTCTTCCAAACCAGCTCAC, 5′-TCTGCAGTGTCCACCAACTC), Plin5 (5′-ATCCCTACCCCACCCTCTAC, 5′-ATAAGGACGGAGGGCTGACT), and “no gene” (5′-TGGTAGCCTCAGGAGCTTGC; 5′-ATCCAAGATGGGACCAAGCTG). Primers against no gene align to a genomic region on chromosome 15 with no binding of RNAPII and served as a negative control (54).
Animal experiments
All animal use was approved and registered by the Norwegian Animal Research Authority. Mice were housed in a temperature controlled (22°C) facility with a strict 12 h light/dark cycle. Animals were euthanized by cervical dislocation; tissue samples were dissected, snap-frozen in liquid nitrogen, and stored at −80°C until further analysis.
Male backcrossed congenic PPARα−/− mice (B6.129S4-Pparatm1GonzN12; Jackson Laboratory, Bar Harbor, ME) and PPARα+/+ controls (C57BL/6J; B and K Universal Ltd., Norway) 9 weeks of age were ad libitum fed a standard chow diet [64% carbohydrate, 31.5% protein, 2% fat (fat source soya oil)] or a high-fat diet (HFD) [36% carbohydrate, 20% protein, 35.5% fat (fat source lard); #F3282, 1/2″ pellet (BioServ, Frenchtown, NJ)] for a period of 13 weeks.
Male C57BL/6N mice (Charles River, 17 weeks, ∼30 g) on a standard chow diet were given intragastric gavage of 0.5% carboxymethylcellulose (CMC) (Sigma, #C4888), 300 μl GW501516 (150 μg solved in 0.5% CMC; 5 mg/kg) or 300 μl glyceryl trioleate/triolein (Sigma #T7140). Mice were treated twice, 36 and 12 h before being euthanized (4–6 animals in each group). Mice were euthanized at the onset of the light cycle.
Statistical methods
All results are presented as means with standard error of the mean (SEM) or standard deviation (SD). One-way analysis of variance (ANOVA) followed by Tukey's multiple comparison tests or two-tailed Student's t-tests were used to assess significance (P < 0.05).
RESULTS
HFD feeding of mice increases expression of selective perilipins in muscle
Prolonged feeding of mice with a HFD increases circulating FAs and stimulates uptake and utilization of FAs as the preferred energy source in peripheral tissues. To determine if a HFD changes the expression of Plin genes in a FA-oxidative muscle type, Plin mRNA expression levels were evaluated in soleus muscle from wild-type (WT) and PPARα knock-out (KO) mice fed a chow or high-fat diet. PPARα KO mice were included in the study, because activation of PPARα is known to stimulate transcription of Plin2 and Plin5 in liver (7, 15, 16, 41–43). Plin2 mRNA expression increased 2-fold by HFD in WT but not in PPARα KO mice (Fig. 1). Plin5 mRNA expression was lower in PPARα KO mice compared with WT mice, similar to what was previously reported in liver (7). Interestingly, expression of Plin5 increased by similar magnitude in both genotypes fed a HFD (2.5-fold and 2.9-fold in PPARα KO and WT, respectively), demonstrating that regulation of Plin5 can occur independent of PPARα in muscle. In accord with the changes in gene expression, perilipin2 and perilipin5 protein increased with a HFD in WT mice (Fig. 1B). In the absence of PPARα, both perilipin2 and perilipin5 proteins were substantially lower. Perilipin2 was not detected, whereas perilipin5 levels increased by HFD also in PPARα KO mice. The mRNA and protein levels of Plin3 and Plin4 remained relatively unchanged by treatment or genotype.
Fig. 1.
Plin2 and Plin5 are increased in muscle of HFD-fed mice. Wild-type and PPARα−/− mice were fed chow or HFD for 13 weeks. A: Relative gene expression of Plin2-5 in soleus muscle normalized to 36B4. Results are presented as mean ± SEM (n = 6 for each group). Statistical differences between groups: chow versus HFD (*P < 0.05, **P < 0.01) and between genotypes with same feeding (§P < 0.05, #P < 0.01). B: Perilipin protein levels in soleus muscle. Each lane contains proteins pooled from six mice. The arrows indicate protein signals migrating at the same size as ectopically expressed perilipin2, perilipin4, and perilipin5. The identity of the smaller protein band observed for perilipin4 is unknown. C: Gene expression of Plin2-5 in gastrocnemius muscle. The results are presented as described for soleus muscle.
Next, we analyzed Plin mRNA content in gastrocnemius, which contains a mixture of glycolytic and oxidative fibers. In this tissue, Plin2 and Plin4 mRNAs increased by HFD in a PPARα-dependent manner (Fig. 1C), whereas the Plin5 mRNA increased by HFD in both WT and PPARα KO mice. Essentially the same changes in regulation were observed at the protein level (result not shown). These results implicate that a HFD alters the protein and mRNA levels of several perilipins regardless of fiber composition, and that other factors in addition to PPARα may regulate transcription of Plin5.
FAs regulate Plin genes in cultured myotubes
The increased expression of several Plin genes by the diet rich in lipids may be directly mediated by FAs or secondarily due to complex physiological changes that occur with increased levels of circulating FAs. To be able to identify molecular mechanisms inducing Plin genes in muscle tissues, we continued our investigation using cultured muscle cells. We established culturing conditions and confirmed differentiation of myoblasts into myotubes (see Experimental Procedures) in two different myotube cell cultures, C2C12 and Sol8 cells. Cells were seeded at day −2, grown to confluence, and subjected to myotube differentiation for up to 7 days. The expression of the Plin genes varied somewhat in the two cell lines, but was less affected by differentiation in C2C12 cells (see supplementary Fig. II). As only C2C12 cells expressed Plin2-5, this cell line was used in further studies.
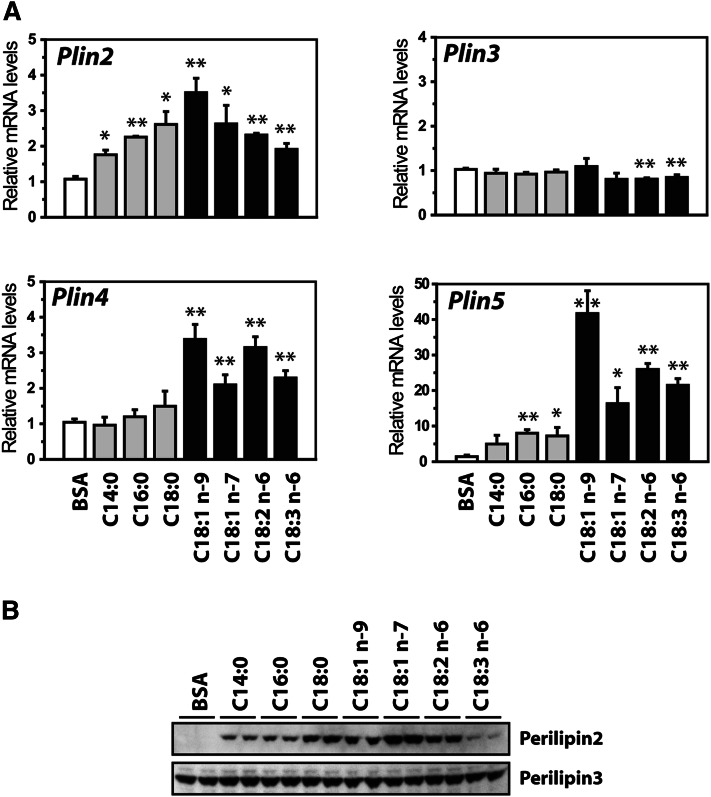
To test if the Plin genes are regulated by FAs, C2C12 cells were differentiated for 6 days and cultured for 24 h in medium supplemented with various FAs complexed to BSA. The expression of Plin2 and Plin4-5 all increased by various FAs, but with different magnitude (Fig. 2A). Plin4 and Plin5 were mainly induced by unsaturated long chain FAs, whereas Plin2 mRNA was elevated by both saturated and unsaturated long chain FAs. Similar to what we have shown in liver (41), the expression of Plin3 remained unchanged.
Fig. 2.
FAs regulate expression of Plin genes in C2C12 cells. C2C12 cells were differentiated until day 6 and stimulated with BSA alone (40 μM) or various FAs complexed to BSA (100 μM FA:40 μM BSA) for 24 h. FAs used: myristic acid (C14:0), palmitic acid (C16:0), stearic acid (C18:0), oleic acid (cis-C18:1 n-9), vaccenic acid (cis-C18:1 n-7), linoleic acid (C18:2 n-6), and γ-linolenic acid (C18:3n-6). All wells received equal amounts of BSA. A: Relative gene expression of Plin2-5 in C2C12 cells normalized to 36B4. Results are presented as mean ± SD (n = 3, *P > 0.05, **P > 0.01). B: C2C12 cells were stimulated with FA as described above. Western blots of perilipin2 and perilipin3 indicate that LDs were formed. Data are from a representative Western blot from two independent experiments.
All of the added FAs stimulated formation of small intracellular LDs (result not shown). To further verify that the added FAs were incorporated into intracellular LDs, we determined perilipin2 and perilipin3 levels (Fig. 2B). In contrast to perilipin3, perilipin2 is known to be posttranslationally regulated and rapidly degraded by the proteasome in the absence of intracellular LDs (41). The solid increase in perilipin2 and unchanged levels of perilipin3 confirms that the FAs were incorporated into intracellular LDs. No clear protein signals were observed for the perilipin4 or perilipin5 proteins, in agreement with the low mRNA expression of these Plins.
The Plin genes are differently regulated by selective activation of PPARs
Long chain FAs have previously been shown to stimulate Plin2 mRNA expression in various tested cell types (41, 46, 55), but the mechanism involved has not been clarified. FAs might activate genes by acting as physiological ligands for PPARs (36). We therefore determined the expression level of each PPAR isotype during myotube-differentiation and used the Ct values at day 0 as an indication of relative expression of each PPAR isotype. PPARδ was highly expressed with unaffected levels during differentiation in both cell lines. PPARγ was slightly downregulated during differentiation, whereas PPARα was expressed at low levels with increased expression during differentiation (supplementary Fig. III).
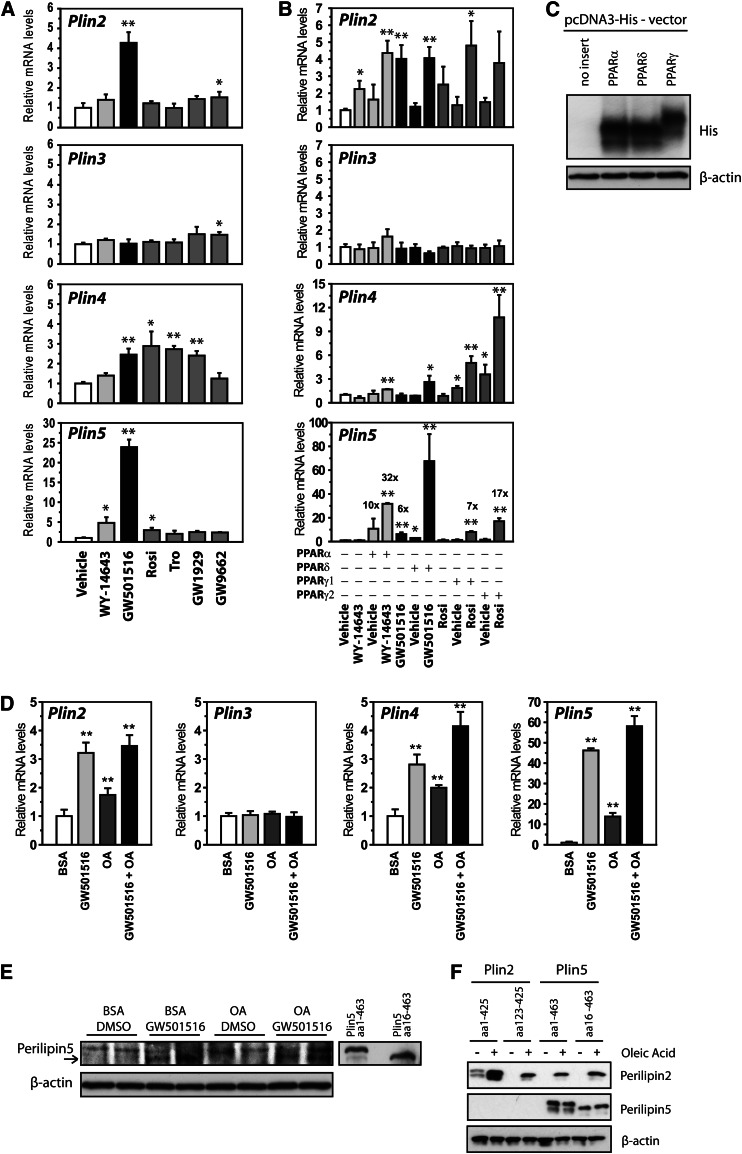
To determine if there is a PPAR isotype-dependent regulation of the Plin genes in muscle cells, cells were differentiated for 6 days and stimulated with specific PPAR activators for 24 h (Fig. 3A). Activation of PPARα (WY-14643; 10 μM) in C2C12 cells had little effect on the Plin mRNA levels, except for an increase in Plin5 mRNA (5-fold). Given the low basal expression of Plin5 in C2C12 cells (Ct = 35), such a minimal regulation is expected. Activation of PPARδ (GW501516; 0.1 μM) increased mRNA content of Plin2 (4-fold), Plin4 (5-fold), and Plin5 (46-fold), whereas Plin3 mRNA remained unchanged. Activation of PPARγ (Rosi, Tro, and GW1929; 1 μM) increased mRNA levels of Plin4 (about 3-fold) and Plin5 (2-fold). The PPARγ antagonist (GW9662; 1 μM) had no effect on the mRNA levels of the Plin genes.
Fig. 3.
PPARδ activates protein and gene expression of perilipin5 in C2C12 cells. A: C2C12 cells were differentiated until day 6 and stimulated 24 h with vehicle (0.1% DMSO) or agonists for PPARα (WY-14643; 10 μM), PPARδ (GW501516; 0.1 μM), or PPARγ (Rosi/BRL-49653, 1 μM; Tro, 1 μM; and GW1929, 1 μM), or an antagonist for PPARγ (GW9662; 1 μM). All wells received equal amounts of vehicle (0.1% DMSO). Relative gene expression of Plin2-Plin5 normalized to 36B4. B: C2C12 cells were transfected with pcDNA3 vector (control) or various pcDNA3-PPAR expression vectors at day 0, differentiated for 3 days and stimulated with selective PPAR activators for 24 h. Activators of PPARα (WY-14643; 10 μM), PPARδ (GW501516; 0.1 μM), or PPARγ (Rosi/BRL-49653; 1 μM). Relative gene expression of Plin2-5 normalized to 36B4. Results are presented as mean ± SD (n = 3, *P > 0.05, **P > 0.01). C: C2C12 cells were transfected with pcDNA3-6xHis-V5-PPARα, -PPARδ, or -PPARγ expression vectors and the relative expression level of each PPAR isoform was determined by Western blot using a His antibody. D, E: C2C12 cells were differentiated in the presence of a PPARδ activator (GW501516; 0.1 μM) and/or BSA-OA (100 μM) from day 0 until day 7. D: Relative gene expression of Plin2-5 normalized to 36B4. Results are presented as mean ± SD (n = 3). Statistical differences refer to BSA (**P < 0.01). E: The short perilipin5 (perilipin5 aa16-463) protein was increased following PPARδ agonist (GW501516) treatment. The Western blot shows two independent samples per treatment. F: C2C12 cells were transfected with pcDNA3-Plin2, -Plin2 aa125-425, -Plin5, or -Plin5 aa16-463 at day 0, differentiated for 3 days, and stimulated with BSA-OA (100 μM) for an additional 24 h. Perilipin2 and perilipin5 protein amounts were analyzed using specific perilipin antibodies. Each lane contains proteins pooled from three independent wells.
The profound response to activation of PPARδ compared with other PPARs might be explained by the relatively higher expression level of this particular PPAR in cultured C2C12 cells. To determine the efficiency for each PPAR to regulate Plin genes, C2C12 cells were transfected to ectopically express PPARα, PPARδ, or PPARγ, differentiated for 3 days, and stimulated for 24 h with PPAR isoform specific ligands (Fig. 3B). Comparable ectopic expression of the various PPARs was confirmed by expressing 6xHis-PPAR vectors in C2C12 cells and visualizing the fusion proteins with an antibody recognizing the His epitope (Fig. 3C). Ectopic expression and activation of PPARα induced Plin2 and Plin5 mRNAs. Very similar results were observed with ectopic expression and activation of PPARδ, which induced expression of Plin2, Plin4, and Plin5. Ectopic expression and activation of PPARγ1 or PPARγ2 foremost increased Plin4 mRNA. These results demonstrate that each PPAR differently regulates expression of Plin2-5 mRNAs in C2C12 cells.
To determine if addition of FAs and PPARδ alters expression of the perilipin4 and perilipin5 proteins, C2C12 cells were differentiated in the presence of BSA-OA and/or the PPARδ agonist (GW501516). The fold induction of Plin2, Plin4, and Plin5 mRNAs (Fig. 3D) were somewhat higher than overnight stimulation of differentiated myotubes (compare Fig. 2 and Fig. 3B). Stimulation with the PPARδ agonist during differentiation induced the short perilipin5 protein [amino acids 16–463, see (7)], whereas no clear signal was observed for OA treatment (Fig. 3E). A clear band indicative for perilipin4 protein was not observed. Given the low levels of perilipin5 protein, we tested if Plin5 behaves similarly to Plin2, which is posttranslationally stabilized by the cellular content of LDs (7, 34). Vectors expressing the full-length perilipin2, the shorter alternative translated perilipin2 (plin2 a123-425), perilipin5, and perilipin5 a16-463 were transfected into C2C12 cells. As expected, expression of the two perilipin2 isoforms was highly dependent on BSA-OA supplementation of the culture media (Fig. 3F). In contrast, ectopic expression of the perilipin5 isoforms was unaffected by addition of BSA-OA. Together, these results show that activation of PPARδ induces Plin5 mRNA and protein in C2C12 cells.
FAs are unable to induce Plin mRNAs in the presence of a PPARδ antagonist
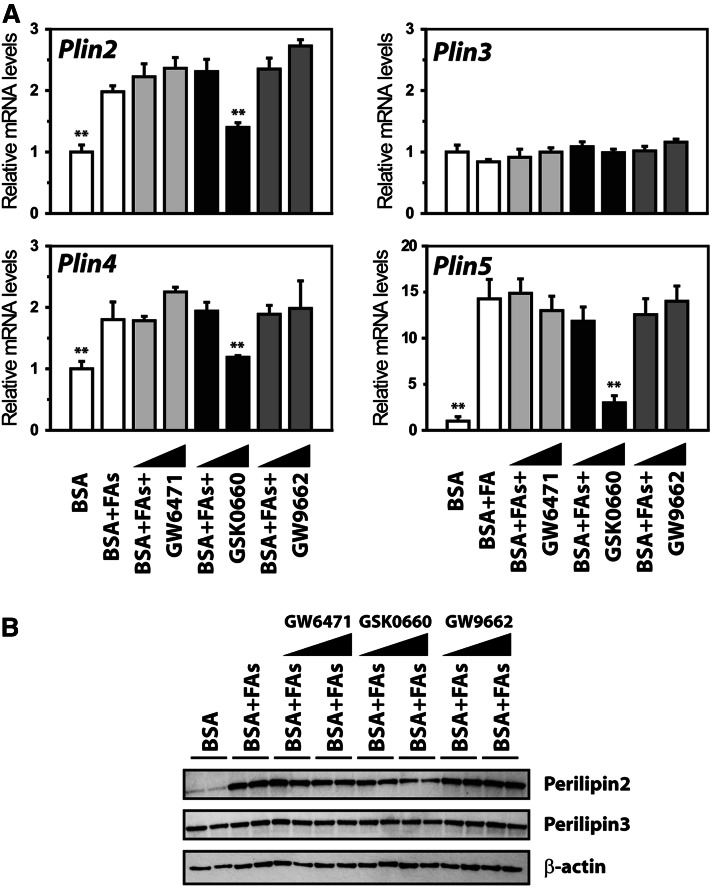
To test if the observed FA effect on Plin mRNAs depends on a particular PPAR, differentiated C2C12 cells were incubated with a combination of BSA-OA and BSA-LA (FAs; 50 μM each) in the presence of selective PPAR inhibitors GW6471 for PPARα (56), GSK0660 for PPARδ (57), and GW9662 for PPARγ (58). Incubation with FAs increased mRNA levels of Plin2, Plin4, and Plin5. Coincubation with antagonists for PPARα or PPARγ did not affect the FA-stimulating effect. In contrast, coincubation with the PPARδ antagonist blunted the effect of FAs on Plin2, Plin4, and Plin5 mRNAs (Fig. 4A). The PPARδ antagonist was also effective in preventing induction of the perilipin2 protein (Fig. 4B).
Fig. 4.
Long chain FAs fail to induce Plin5 expression in the presence of a PPARδ antagonist. C2C12 cells were differentiated until day 6 and treated for 24 h with BSA or BSA-FA (50 μM BSA-OA and 50 μM BSA-LA) in combination with antagonists for PPARα (GW6471; 0.1 or 1 μM), PPARδ (GSK0660; 0.1 or 1 μM), or PPARγ (GW9662; 0.1 or 1 μM). All wells received equal amounts of vehicle (0.1% DMSO) and BSA (40 μM). A: Relative gene expression of Plin2-5 normalized to 36B4. Results are presented as mean ± SD (n = 3). Statistical differences were evaluated against cells receiving BSA-FAs (**P > 0.01). B: C2C12 cells were stimulated as above and perilipin2 and perilipin3 proteins were detected. One representative Western blot from two independent experiments is shown.
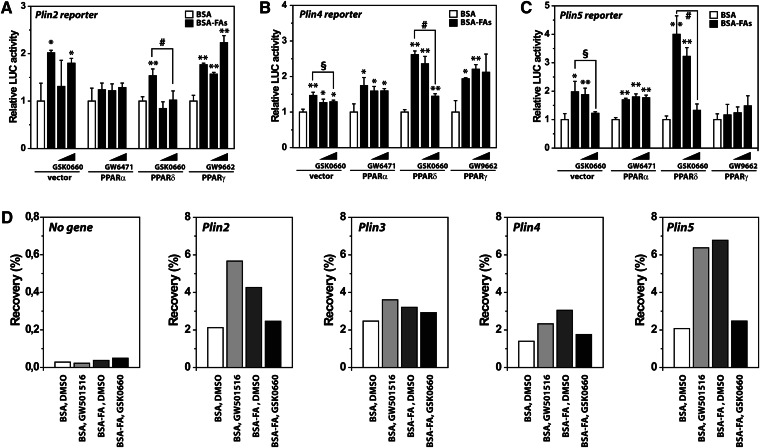
To determine if the observed increase in Plin mRNAs by FAs was due to transcriptional stimulation, Plin2 (41), Plin4 (19), and Plin5 luciferase reporters were cotransfected with PPAR expression vectors into myoblast C2C12 cells. Cells were subsequently differentiated into myotubes prior to 24 h incubation with FAs and PPAR antagonists. Ectopic expression of each PPAR stimulated basal reporter activity with different efficiency (not shown). Addition of FAs increased reporter activity further, and in this context, PPARδ had a unique effect (Fig. 5A–C). Coexpression of PPARδ resulted in a marked boost in Plin4 and Plin5 reporter activity upon activation with FAs, whereas the PPARδ antagonist reversed the effect of FAs.
Fig. 5.
PPARδ transactivates Plin genes. C2C12 cells were transfected at day 0 with Plin reporters and cotransfected with pcDNA3 vector (control) or various pcDNA3-PPAR expression vectors as illustrated. After differentiation for 72 h, cells were treated for 24 h with BSA-FA (50 μM OA and 50 μM LA) in combination with increasing concentrations of antagonists for PPARα (GW6471; 0.1 or 1 μM), PPARδ (GSK0660; 0.1 or 1 μM), or PPARγ (GW9662; 0.1 or 1 μM). A: Luciferase values for the mouse Plin2 reporter. B: Luciferase values for the mouse Plin4 reporter. C: Luciferase values for the mouse Plin5 reporter. One representative of three experiments is shown. Results are presented as mean ± SD (n = 3). Statistical differences from BSA (*P > 0.05, **P > 0.01) or between treatments as indicated in the figure (§P > 0.05, #P > 0.01). D: C2C12 cells were differentiated until day 6 and treated for 24 h with BSA, a PPARδ agonist (GW501516; 0.1 μM), or BSA-FA (50 μM OA and 50 μM LA) with or without an antagonist to PPARδ (GSK0660; 1 μM). Cells were processed for ChIP using RNA Pol II antibody. Relative recovery at the no gene loci (control, a chromosomal nontranscribed loci), and the Plin2-5 locus were determined by ChIP-qPCR.
To strengthen the observation that FAs increase Plin mRNAs by stimulating transcriptional activity, we analyzed Pol II recruitment to the Plin genes using ChIP with primers located 100–200 bp downstream of the transcriptional start sites. Stimulation with either FAs or the PPARδ agonist (GW501516) increased Pol II recruitment to the Plin2, Plin4, and Plin5 genes, whereas coincubation with the PPARδ antagonist (GSK0660) reversed the effect of FAs (Fig. 5D). Taken together, our results suggest that FAs stimulate transcription of several Plin genes in a PPARδ-dependent manner.
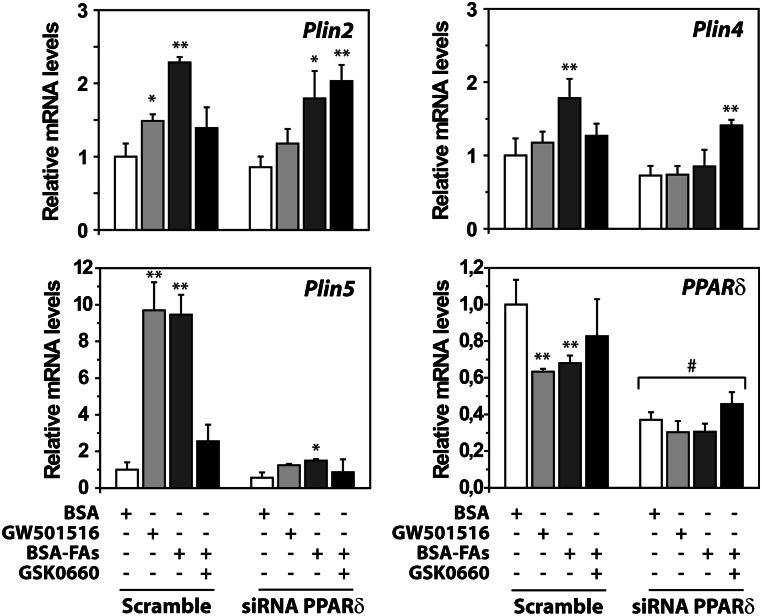
Silencing of PPARδ attenuated the effect of FAs on Plin5 and Plin4 mRNAs
To determine the effect of FAs in cells with reduced expression of PPARδ, C2C12 cells were reverse transfected with scramble siRNA or siRNAs against PPARδ and subjected for myotube differentiation. PPARδ mRNA was knocked down by 70% from day 2 to day 4 post-transfection (Fig. 6). Two days post-transfection, cells were incubated with the PPARδ activator (GW501516; 0.1 μM), FAs (100 μM) alone, or in combination with the PPARδ antagonist (GSK0660, 1 μM). Incubation with the FAs increased Plin2, Plin4, and Plin5 mRNA levels in scramble-transfected cells, with the FA effect attenuated upon addition of the PPARδ antagonist (Fig. 6). Strikingly, the stimulating effect on Plin5 mRNA by FA incubation was abolished in PPARδ siRNA-transfected cells, underlying the importance of PPARδ in Plin5 regulation. Silencing of PPARδ also blunted FA-stimulated induction of Plin4 mRNA, but had little effect on Plin2 mRNA.
Fig. 6.
Silencing of PPARδ attenuates the stimulating effect of FAs on Plin gene transcription. C2C12 cells were transfected at day 0 with scramble or siRNA against PPARδ and differentiated for 2 days prior to stimulation for 24 h with BSA, GW501516 (0.1 μM), BSA-FAs (50 μM OA and 50 μM LA), or BSA-FAs in combination with antagonist to PPARδ (GSK0660; 1 μM). All wells received equal amounts of vehicle (0.1% DMSO) and BSA (40 μM). Relative expression of various RNAs against 36B4 was evaluated using quantitative real-time PCR analysis. Relative expression of Plin2, Plin4, Plin5, and PPARδ. Results are shown as mean ± SD (n = 3). Difference from control (BSA; *P > 0.05, **P > 0.01). Difference among scramble or PPARδ siRNA-treated cells receiving similar treatment (#P > 0.01).
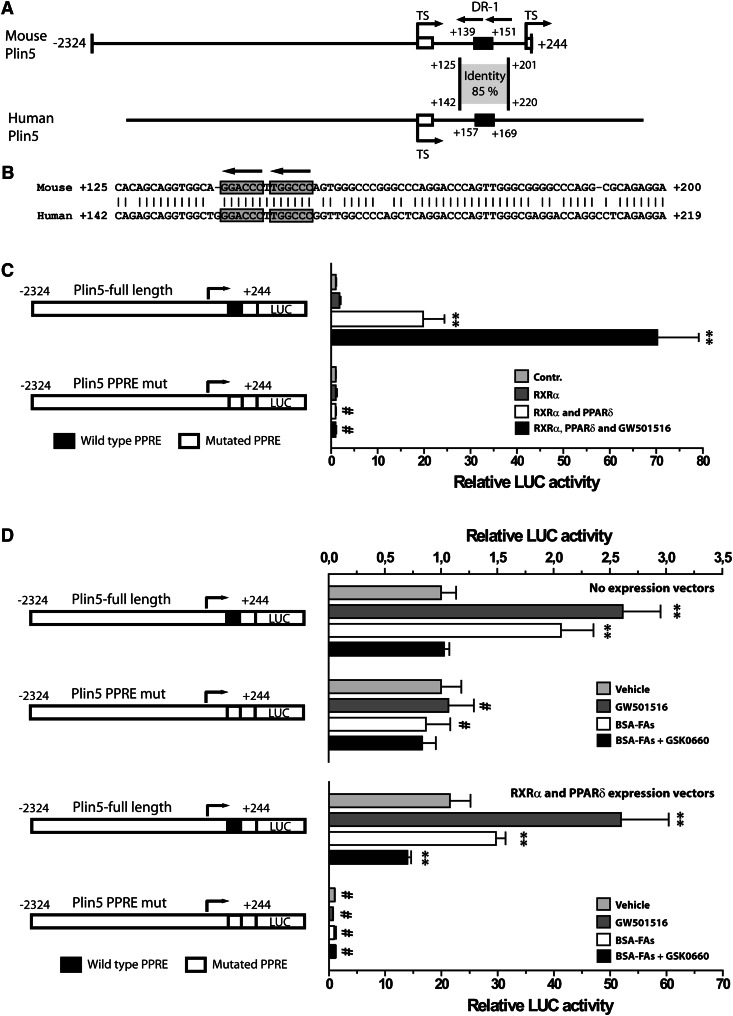
The Plin5 intron 1 contains an evolutionary conserved PPRE
The Plin1, Plin2, and Plin4 promoters all contain functionally characterized PPREs (19, 41). Our results prompted us to analyze the Plin5 gene for the presence of a functional PPRE. A promising DR-1 type element was identified in intron 1, in an area conserved to the human Plin5 gene (Fig. 7A, B). To test the functionality of this element, we mutated the DR-1 element and performed reporter assays with Plin5 WT or mutated reporter constructs. C2C12 cells were transfected with reporters and expression vectors encoding for RXRα and/or PPARδ and differentiated into myotubes for 3 days. Cells were then stimulated with vehicle or the PPARδ activator GW501516 (0.1 μM) for 24 h. Transfection with the Plin5 WT or mutated reporters had similar basal activity, demonstrating that the mutation itself did not affect basal transcriptional activity. Coexpression with RXRα and PPARδ, and stimulation with GW501516 gradually increased Plin5 WT reporter activity up to a maximal 70-fold increase (Fig. 7C). In contrast, no induction was observed with the mutated Plin5 reporter construct.
Fig. 7.
PPARδ regulates Plin5 through a conserved PPRE in intron 1. A: A schematic presentation of the cloned mouse Plin5 reporter (nucleotides −2,324 to +244), and the corresponding human Plin5 promoter. The two alternative transcriptional start sites (TS) are indicated by arrows. The localization of the conserved intron region and percentage identity to the human intronic segment is shown. The nucleotide position of the black boxes points out the localization of the PPREs. B: Sequence alignment and homology between nucleotides in the mouse and the human Plin5 intron 1 regions. The two half sites in the conserved PPRE are enclosed by boxes, and conserved nucleotides are indicated by vertical lines. C: Transient transfection of Plin5 full-length or mutated reporters into C2C12 cells. Cells were transfected, differentiated for 3 days, and stimulated with the indicated ligands for 24 h. Cells were cotransfected with pcDNA3 (control), pcDNA3-RXRα, or pcDNA3-PPARδ expression plasmids and stimulated with vehicle (DMSO) or GW501516 (0.1 μM). Results are presented as mean ± SD (n = 3). D: Transient transfection of Plin5 WT or mutated reporters in combination with no expression plasmid or RXRα and PPARδ expression plasmids. Cells were stimulated with BSA (light gray), GW501516 (0.1 μM; dark gray), BSA-FAs (50 μM OA and 50 μM LA; white), or BSA-FAs in combination with GSK0660 (1 μM; black). For (C) and (D), statistical differences from control treatment (**P < 0.01) or between Plin5 reporter constructs with same treatment (#P < 0.01).
We finally tested if the PPARδ-dependent regulation of Plin5 gene expression by FAs depends on the identified response element. Plin5 WT or mutated reporters were transfected into cells together with empty or RXRα and PPARδ expression vectors and stimulated as shown. For the Plin5 WT reporter, stimulation with GW501516 or FAs gave an expected increase in reporter activity, whereas GSK0660 attenuated the FA effect (Fig. 7D). In contrast, for the Plin5 mutated reporter construct, none of the treatments had any effect on reporter activity.
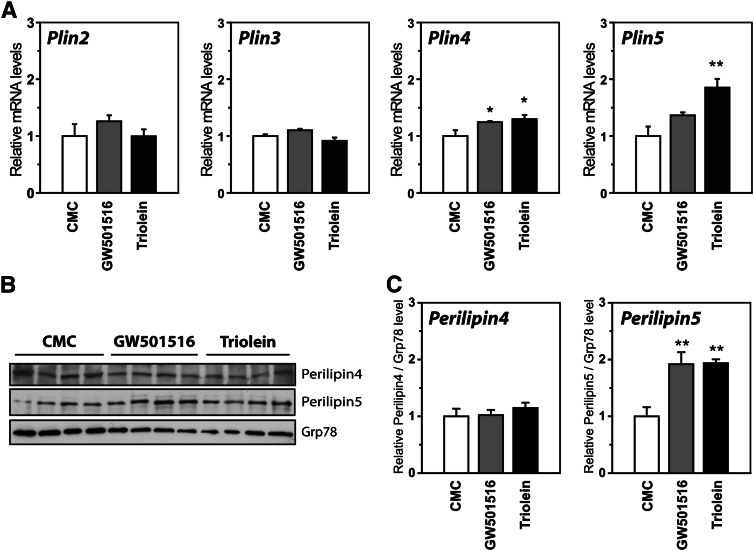
Expression of Plin5 is induced by a PPARδ agonist and oleic acid in vivo
Our results from cell studies support that FAs regulate expression of Plin4 and Plin5 in muscle by activating PPARδ. To determine if the same occurs in muscle tissue, mice were given an oral gavage of a PPARδ agonist (GW501516) or FAs in the form of triglycerides (glyceryl trioleate; triolein). Treatment with GW501516 increased Plin4 mRNA and showed a tendency for an induction of Plin5 mRNA in soleus muscle (Fig. 8A), whereas triolein treatment increased both Plin4 and Plin5 mRNAs. The same inductions occurred in gastrocnemius muscle (results not shown).The induction was stronger for Plin5, and only the perilipin5 protein was significantly elevated by these treatments (Fig. 8B, C).
Fig. 8.
Oleic acid or PPARδ agonist treatment of mice elevates Plin5 in soleus muscle. Mice were gavaged twice (36 and 12 h before being euthanized) with vehicle (0.5% CMC), GW501516 (5 mg/kg), or 400 μl glyceryl trioleate/triolein. Mice were euthanized at the onset of the light cycle. A: Relative gene expression of Plin2-5 in soleus muscle relative to TBP. Results are presented as mean ± SEM (n = 4–6 per group). Statistical differences from CMC treatment (*P < 0.05, **P < 0.01). B: Western blot of perilipin4, perilipin5, and grp78 (control) proteins in soleus muscle. Each treatment is represented by four independent mice. C: Relative levels of perilipins/grp78 (n = 4, difference from CMC treatment, **P < 0.01).
DISCUSSION
Activation of PPARα and PPARγ are known to stimulate expression of Plin genes in liver (7, 15, 16, 41–43) and adipose tissue (19, 38–40), respectively. By using PPAR isoform-specific agonists and antagonists, we demonstrate that PPARs selectively regulate Plin genes in muscle cells. PPARδ is required for FA-induced transcription of Plin5 through a conserved DR-1 element in intron 1 of the Plin5 gene. The identification of this functional response element classifies Plin5 as a novel direct PPAR target gene, similar to Plin1, Plin2, and Plin4. Plin3 seems to be an exception in the family, by not being regulated by PPARs.
The different responses to PPAR activation are likely important for the distinct Plin tissue expression profiles (7, 19). In muscle, enhanced PPARα and PPARδ activation increases Plin5 expression, whereas the adipose-enriched Plin4 is preferentially induced by enhanced PPARγ activity. In contrast, Plin2 shows no clear PPAR isoform preference, and is nonresponsive to manipulation of PPARδ expression. In addition to PPAR redundancy, the distinct regulation of Plin2 may also be influenced by other transcription factors (59, 60). Several factors may influence recruitment of PPAR isoforms to a particular PPRE and subsequent transcriptional regulation. Among these are tissue variability in chromatin packing within each Plin locus, specific coregulator recruitment, the presence of activating or repressing ligands, or the nucleotide composition of the PPREs and nearby binding sites (36). The level of endogenous ligands clearly plays a role in our cell studies. PPARδ is highly expressed in muscle tissues and C2C12, yet perilipin5 is only detected in C2C12 cells after prolonged culturing in the presence of a specific PPARδ agonist. Another factor is likely the nucleotide compositions of the various PPREs found to regulate Plin genes. These sequences are highly conserved across species (human, mouse, and rat), but vary considerably among the various Plin genes (19, 41). This suggests that the PPAR isoform regulating each Plin gene is evolutionarily conserved and that this PPAR-specific regulation is physiologically important.
Expression of Plin5 was initially described to correlate with PPARα activity (7, 15, 16), but conflicting observations provided evidence for alternative regulation of the gene. In the lack of PPARα, the fold-induction of Plin5 is preserved by fasting in liver (7). We now demonstrate that Plin5 expression is elevated in muscle by a HFD in the absence of PPARα. Fasting and a HFD can be viewed as contrary physiological conditions with low or excess energy, respectively, but have elevated circulating FAs in common, caused by hormone-stimulated lipolysis of adipose TAG or increased digestion and uptake of FAs. Increased expression of Plins has been observed in various cell types upon stimulation with FAs (41, 46, 61), but the molecular mechanisms have been unclear. Our data provide a molecular explanation for regulation of Plin5 in the absence of PPARα. Long chain unsaturated FAs may bind directly to the ligand binding pocket of PPARδ and regulate its transcriptional activity (62), which in turn stimulates transcription of the Plin5 gene. The observation that PPARδ may sense circulating free FAs and regulate a subset of hepatic genes independent of PPARα (63) supports such a mechanism. The discovery of a phospholipid as the endogenous PPARα ligand (64) also questions the role of PPARα as a direct FA sensor. The role of PPARα may rather involve the receptor's ability to stimulate transcription of genes facilitating muscular FA uptake (65). Our data suggest that PPARδ is the physiological regulator of perilipin5 in muscle.
PPARδ is nearly ubiquitously expressed and the predominate PPAR isoform expressed in rodent skeletal muscle. In contrast to PPARα (65), forced muscle-specific expression of PPARδ improves endurance in mice and promotes a shift from glycolytic to oxidative muscle fibers (66, 67). Oxidative muscle tissues also contain higher levels of perilipin5 [see (7) and Fig. 1]. The direct regulation of Plin5 by PPARδ may therefore explain the uneven distribution of Plin5 expressed in muscle fibers. Other factors found to drive formation of oxidative muscles includes PGC-1α (68), PGC-1β (69), ERRα (70), ERRγ (71), and the corepressor NCoR1 (72). Overexpression of PGC-1α in muscle may stimulate Plin5 expression (73), but the importance of the other factors have not been studied. Nevertheless, it is clear that Plin5 belongs to the pool of genes that distinguish oxidative type I from glycolytic type II fibers.
The function of genes enriched in oxidative fibers is primarily linked to FA oxidation, myosin fiber types, mitochondrial biogenesis, and increased mitochondrial oxidative capacity (66–72). The role of perilipin5 in this setting is not clear. Due to low endogenous perilipin5 expression in cultured cells, much of our molecular understanding of the protein is based on cellular studies with ectopic perilipin5 expression. When overexpressed, perilipin5 recruits Abhd5 (24), ATGL (25, 26), and HSL (27) to the surface of LDs, and promotes association of LDs and mitochondria (31). It remains to be elucidated if a portion of these effects are caused by high ectopic expression. Although it is unclear how interactions between perilipin5 and the above mentioned proteins regulate lipolysis, accumulating evidence from mice demonstrate that perilipin5 preserves LDs. Perilipin5-null mice lack LDs in the heart (30) whereas cardiac-specific perilipin5 transgenic mice accumulate LDs (74, 75). Alteration in cardiac perilipin5 primarily affects TAG storage, which would be expected based on the preferred binding of perilipin5 to LDs filled with TAG (47). The role of perilipin4 is less clear. PPARγ stimulates perilipin4 expression in adipocytes (19), which points to a role in energy storage. Perilipin4 may be recruited to nascent LDs formed in cultured adipose cells exposed to high concentrations of FAs (14). A more recent publication demonstrates that perilipin4 preferentially binds to LDs filled with CEs with an ability to enhance accumulation of such LDs when ectopically expressed (47).
Additional functional analyses are required to fully understand the function of Plin4 and Plin5 in LD metabolism and their roles in muscle physiology. The regulation of Plin5 by PPARδ may provide insight into why lipid stores in muscles are beneficial and detrimental in athletic and obese subjects, respectively. PPARδ-mediated regulation of Plin4 and Plin5 may render the muscle tissue better equipped to fine-tune LD metabolism by having increased levels of these LD binding proteins. When present at the surface of LDs, they may help to preserve LDs and increase the cellular capacity to prevent lipotoxicity.
Supplementary Material
Acknowledgments
The authors thank Sverre Holm for technical assistance with animal work, Christin Zwafink, Christina Steppeler, and Tone Lise Aarnes Hjørnevik for technical assistance, and members of the Nebb laboratory for scientific discussions.
Footnotes
Abbreviations:
- CE
- cholesteryl ester
- ChIP
- Chromatin immunoprecipitation
- CMC
- carboxymethylcellulose
- HFD
- high-fat diet
- KO
- knock-out
- LA
- linoleic acid
- LD
- lipid droplet
- OA
- oleic acid
- PPAR
- peroxisome proliferator-activated receptor
- PPRE
- peroxisome proliferator-activated receptor response element
- Rosi
- rosiglitazone
- RXR
- retinoid X receptor
- TAG
- triacylglycerol
- Tro
- troglitazone
- WT
- wild-type
This work was supported by grants from the Medical Faculty at University of Oslo, Henning and Johan Throne-Holst Foundation, Novo Nordisk Foundation, Aktieselskabet Freia Chocolade Fabriks Medical Foundation, and Anders Jahres Foundation.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of three figures.
REFERENCES
- 1.Eckardt K., Taube A., Eckel J. 2011. Obesity-associated insulin resistance in skeletal muscle: role of lipid accumulation and physical inactivity. Rev. Endocr. Metab. Disord. 12: 163–172 [DOI] [PubMed] [Google Scholar]
- 2.Samuel V. T., Shulman G. I. 2012. Mechanisms for insulin resistance: common threads and missing links. Cell. 148: 852–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amati F., Dube J. J., Alvarez-Carnero E., Edreira M. M., Chomentowski P., Coen P. M., Switzer G. E., Bickel P. E., Stefanovic-Racic M., Toledo F. G., et al. 2011. Skeletal muscle triglycerides, diacylglycerols, and ceramides in insulin resistance: another paradox in endurance-trained athletes? Diabetes. 60: 2588–2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brasaemle D. L. 2007. The perilipin family of structural lipid droplet proteins: stabilization of lipid droplets and control of lipolysis. J. Lipid Res. 48: 2547–2559 [DOI] [PubMed] [Google Scholar]
- 5.Bickel P. E., Tansey J. T., Welte M. A. 2009. PAT proteins, an ancient family of lipid droplet proteins that regulate cellular lipid stores. Biochim. Biophys. Acta. 1791: 419–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farese R. V., Jr, Walther T. C. 2009. Lipid droplets finally get a little R-E-S-P-E-C-T. Cell. 139: 855–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dalen K. T., Dahl T., Holter E., Arntsen B., Londos C., Sztalryd C., Nebb H. I. 2007. LSDP5 is a PAT protein specifically expressed in fatty acid oxidizing tissues. Biochim. Biophys. Acta. 1771: 210–227 [DOI] [PubMed] [Google Scholar]
- 8.Lu X., Gruia-Gray J., Copeland N. G., Gilbert D. J., Jenkins N. A., Londos C., Kimmel A. R. 2001. The murine perilipin gene: the lipid droplet-associated perilipins derive from tissue-specific, mRNA splice variants and define a gene family of ancient origin. Mamm. Genome. 12: 741–749 [DOI] [PubMed] [Google Scholar]
- 9.Kimmel A. R., Brasaemle D. L., McAndrews-Hill M., Sztalryd C., Londos C. 2010. Adoption of PERILIPIN as a unifying nomenclature for the mammalian PAT-family of intracellular lipid storage droplet proteins. J. Lipid Res. 51: 468–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greenberg A. S., Egan J. J., Wek S. A., Moos M. C., Jr, Londos C., Kimmel A. R. 1993. Isolation of cDNAs for perilipins A and B: sequence and expression of lipid droplet-associated proteins of adipocytes. Proc. Natl. Acad. Sci. USA. 90: 12035–12039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blanchette-Mackie E. J., Dwyer N. K., Barber T., Coxey R. A., Takeda T., Rondinone C. M., Theodorakis J. L., Greenberg A. S., Londos C. 1995. Perilipin is located on the surface layer of intracellular lipid droplets in adipocytes. J. Lipid Res. 36: 1211–1226 [PubMed] [Google Scholar]
- 12.Brasaemle D. L., Barber T., Wolins N. E., Serrero G., Blanchette-Mackie E. J., Londos C. 1997. Adipose differentiation-related protein is an ubiquitously expressed lipid storage droplet-associated protein. J. Lipid Res. 38: 2249–2263 [PubMed] [Google Scholar]
- 13.Wolins N. E., Rubin B., Brasaemle D. L. 2001. TIP47 associates with lipid droplets. J. Biol. Chem. 276: 5101–5108 [DOI] [PubMed] [Google Scholar]
- 14.Wolins N. E., Skinner J. R., Schoenfish M. J., Tzekov A., Bensch K. G., Bickel P. E. 2003. Adipocyte protein S3–12 coats nascent lipid droplets. J. Biol. Chem. 278: 37713–37721 [DOI] [PubMed] [Google Scholar]
- 15.Wolins N. E., Quaynor B. K., Skinner J. R., Tzekov A., Croce M. A., Gropler M. C., Varma V., Yao-Borengasser A., Rasouli N., Kern P. A., et al. 2006. OXPAT/PAT-1 is a PPAR-induced lipid droplet protein that promotes fatty acid utilization. Diabetes. 55: 3418–3428 [DOI] [PubMed] [Google Scholar]
- 16.Yamaguchi T., Matsushita S., Motojima K., Hirose F., Osumi T. 2006. MLDP, a novel PAT family protein localized to lipid droplets and enriched in the heart, is regulated by peroxisome proliferator-activated receptor alpha. J. Biol. Chem. 281: 14232–14240 [DOI] [PubMed] [Google Scholar]
- 17.Greenberg A. S., Egan J. J., Wek S. A., Garty N. B., Blanchette-Mackie E. J., Londos C. 1991. Perilipin, a major hormonally regulated adipocyte-specific phosphoprotein associated with the periphery of lipid storage droplets. J. Biol. Chem. 266: 11341–11346 [PubMed] [Google Scholar]
- 18.Servetnick D. A., Brasaemle D. L., Gruia-Gray J., Kimmel A. R., Wolff J., Londos C. 1995. Perilipins are associated with cholesteryl ester droplets in steroidogenic adrenal cortical and Leydig cells. J. Biol. Chem. 270: 16970–16973 [DOI] [PubMed] [Google Scholar]
- 19.Dalen K. T., Schoonjans K., Ulven S. M., Weedon-Fekjaer M. S., Bentzen T. G., Koutnikova H., Auwerx J., Nebb H. I. 2004. Adipose tissue expression of the lipid droplet-associating proteins S3–12 and perilipin is controlled by peroxisome proliferator-activated receptor-gamma. Diabetes. 53: 1243–1252 [DOI] [PubMed] [Google Scholar]
- 20.Tansey J. T., Sztalryd C., Gruia-Gray J., Roush D. L., Zee J. V., Gavrilova O., Reitman M. L., Deng C. X., Li C., Kimmel A. R., et al. 2001. Perilipin ablation results in a lean mouse with aberrant adipocyte lipolysis, enhanced leptin production, and resistance to diet-induced obesity. Proc. Natl. Acad. Sci. USA. 98: 6494–6499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sztalryd C., Xu G., Dorward H., Tansey J. T., Contreras J. A., Kimmel A. R., Londos C. 2003. Perilipin A is essential for the translocation of hormone-sensitive lipase during lipolytic activation. J. Cell Biol. 161: 1093–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martinez-Botas J., Anderson J. B., Tessier D., Lapillonne A., Chang B. H., Quast M. J., Gorenstein D., Chen K. H., Chan L. 2000. Absence of perilipin results in leanness and reverses obesity in Lepr(db/db) mice. Nat. Genet. 26: 474–479 [DOI] [PubMed] [Google Scholar]
- 23.Miyoshi H., Souza S. C., Zhang H. H., Strissel K. J., Christoffolete M. A., Kovsan J., Rudich A., Kraemer F. B., Bianco A. C., Obin M. S., et al. 2006. Perilipin promotes hormone-sensitive lipase-mediated adipocyte lipolysis via phosphorylation-dependent and -independent mechanisms. J. Biol. Chem. 281: 15837–15844 [DOI] [PubMed] [Google Scholar]
- 24.Granneman J. G., Moore H. P., Mottillo E. P., Zhu Z. 2009. Functional interactions between Mldp (LSDP5) and Abhd5 in the control of intracellular lipid accumulation. J. Biol. Chem. 284: 3049–3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang H., Bell M., Sreenevasan U., Hu H., Liu J., Dalen K., Londos C., Yamaguchi T., Rizzo M. A., Coleman R., et al. 2011. Unique regulation of adipose triglyceride lipase (ATGL) by perilipin 5, a lipid droplet-associated protein. J. Biol. Chem. 286: 15707–15715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Granneman J. G., Moore H. P., Mottillo E. P., Zhu Z., Zhou L. 2011. Interactions of perilipin-5 (Plin5) with adipose triglyceride lipase. J. Biol. Chem. 286: 5126–5135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang H., Hu L., Dalen K., Dorward H., Marcinkiewicz A., Russell D., Gong D., Londos C., Yamaguchi T., Holm C., et al. 2009. Activation of hormone-sensitive lipase requires two steps, protein phosphorylation and binding to the PAT-1 domain of lipid droplet coat proteins. J. Biol. Chem. 284: 32116–32125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sztalryd C., Bell M., Lu X., Mertz P., Hickenbottom S., Chang B. H., Chan L., Kimmel A. R., Londos C. 2006. Functional compensation for adipose differentiation-related protein (ADFP) by Tip47 in an ADFP null embryonic cell line. J. Biol. Chem. 281: 34341–34348 [DOI] [PubMed] [Google Scholar]
- 29.Chang B. H., Li L., Paul A., Taniguchi S., Nannegari V., Heird W. C., Chan L. 2006. Protection against fatty liver but normal adipogenesis in mice lacking adipose differentiation-related protein. Mol. Cell. Biol. 26: 1063–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuramoto K., Okamura T., Yamaguchi T., Nakamura T. Y., Wakabayashi S., Morinaga H., Nomura M., Yanase T., Otsu K., Usuda N., et al. 2012. Perilipin 5, a lipid droplet-binding protein, protects heart from oxidative burden by sequestering fatty acid from excessive oxidation. J. Biol. Chem. 287: 23852–23863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang H., Sreenevasan U., Hu H., Saladino A., Polster B. M., Lund L. M., Gong D. W., Stanley W. C., Sztalryd C. 2011. Perilipin 5, a lipid droplet-associated protein, provides physical and metabolic linkage to mitochondria. J. Lipid Res. 52: 2159–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang H., Sztalryd C. 2011. Oxidative tissue: perilipin 5 links storage with the furnace. Trends Endocrinol. Metab. 22: 197–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bosma M., Minnaard R., Sparks L. M., Schaart G., Losen M., de Baets M. H., Duimel H., Kersten S., Bickel P. E., Schrauwen P., et al. 2012. The lipid droplet coat protein perilipin 5 also localizes to muscle mitochondria. Histochem. Cell Biol. 137: 205–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu G., Sztalryd C., Lu X., Tansey J. T., Gan J., Dorward H., Kimmel A. R., Londos C. 2005. Post-translational regulation of adipose differentiation-related protein by the ubiquitin/proteasome pathway. J. Biol. Chem. 280: 42841–42847 [DOI] [PubMed] [Google Scholar]
- 35.Xu G., Sztalryd C., Londos C. 2006. Degradation of perilipin is mediated through ubiquitination-proteasome pathway. Biochim. Biophys. Acta. 1761: 83–90 [DOI] [PubMed] [Google Scholar]
- 36.Poulsen L., Siersbaek M., Mandrup S. 2012. PPARs: fatty acid sensors controlling metabolism. Semin. Cell Dev. Biol. 23: 631–639 [DOI] [PubMed] [Google Scholar]
- 37.Mandard S., Muller M., Kersten S. 2004. Peroxisome proliferator-activated receptor alpha target genes. Cell. Mol. Life Sci. 61: 393–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arimura N., Horiba T., Imagawa M., Shimizu M., Sato R. 2004. The peroxisome proliferator-activated receptor gamma regulates expression of the perilipin gene in adipocytes. J. Biol. Chem. 279: 10070–10076 [DOI] [PubMed] [Google Scholar]
- 39.Nagai S., Shimizu C., Umetsu M., Taniguchi S., Endo M., Miyoshi H., Yoshioka N., Kubo M., Koike T. 2004. Identification of a functional peroxisome proliferator-activated receptor responsive element within the murine perilipin gene. Endocrinology. 145: 2346–2356 [DOI] [PubMed] [Google Scholar]
- 40.Shimizu M., Takeshita A., Tsukamoto T., Gonzalez F. J., Osumi T. 2004. Tissue-selective, bidirectional regulation of PEX11 alpha and perilipin genes through a common peroxisome proliferator response element. Mol. Cell. Biol. 24: 1313–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dalen K. T., Ulven S. M., Arntsen B. M., Solaas K., Nebb H. I. 2006. PPAR{alpha} activators and fasting induce the expression of adipose differentiation-related protein in liver. J. Lipid Res. 47: 931–943 [DOI] [PubMed] [Google Scholar]
- 42.Edvardsson U., Ljungberg A., Linden D., William-Olsson L., Peilot-Sjogren H., Ahnmark A., Oscarsson J. 2006. PPARalpha activation increases triglyceride mass and adipose differentiation-related protein in hepatocytes. J. Lipid Res. 47: 329–340 [DOI] [PubMed] [Google Scholar]
- 43.Targett-Adams P., McElwee M. J., Ehrenborg E., Gustafsson M. C., Palmer C. N., McLauchlan J. 2005. A PPAR response element regulates transcription of the gene for human adipose differentiation-related protein. Biochim. Biophys. Acta. 1728: 95–104 [DOI] [PubMed] [Google Scholar]
- 44.Chawla A., Lee C. H., Barak Y., He W., Rosenfeld J., Liao D., Han J., Kang H., Evans R. M. 2003. PPARdelta is a very low-density lipoprotein sensor in macrophages. Proc. Natl. Acad. Sci. USA. 100: 1268–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmuth M., Haqq C. M., Cairns W. J., Holder J. C., Dorsam S., Chang S., Lau P., Fowler A. J., Chuang G., Moser A. H., et al. 2004. Peroxisome proliferator-activated receptor (PPAR)-beta/delta stimulates differentiation and lipid accumulation in keratinocytes. J. Invest. Dermatol. 122: 971–983 [DOI] [PubMed] [Google Scholar]
- 46.Tobin K. A., Harsem N. K., Dalen K. T., Staff A. C., Nebb H. I., Duttaroy A. K. 2006. Regulation of ADRP expression by long-chain polyunsaturated fatty acids in BeWo cells, a human placental choriocarcinoma cell line. J. Lipid Res. 47: 815–823 [DOI] [PubMed] [Google Scholar]
- 47.Hsieh K., Lee Y. K., Londos C., Raaka B. M., Dalen K. T., Kimmel A. R. 2012. Perilipin family members preferentially sequester to either triacylglycerol-specific or cholesteryl-ester-specific intracellular lipid storage droplets. J. Cell Sci. 125: 4067–4076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Issemann I., Green S. 1990. Activation of a member of the steroid hormone receptor superfamily by peroxisome proliferators. Nature. 347: 645–650 [DOI] [PubMed] [Google Scholar]
- 49.Brun R. P., Tontonoz P., Forman B. M., Ellis R., Chen J., Evans R. M., Spiegelman B. M. 1996. Differential activation of adipogenesis by multiple PPAR isoforms. Genes Dev. 10: 974–984 [DOI] [PubMed] [Google Scholar]
- 50.Kozak M. 1987. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 15: 8125–8148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dalen K. T., Ulven S. M., Bamberg K., Gustafsson J-Å., Nebb H. I. 2003. Expression of the insulin responsive glucose transporter GLUT4 in adipocytes is dependent on liver X receptor alpha. J. Biol. Chem. 278: 48283–48291 [DOI] [PubMed] [Google Scholar]
- 52.Miura S., Gan J. W., Brzostowski J., Parisi M. J., Schultz C. J., Londos C., Oliver B., Kimmel A. R. 2002. Functional conservation for lipid storage droplet association among Perilipin, ADRP, and TIP47 (PAT)-related proteins in mammals, Drosophila, and Dictyostelium. J. Biol. Chem. 277: 32253–32257 [DOI] [PubMed] [Google Scholar]
- 53.Håkelien A. M., Delbarre E., Gaustad K. G., Buendia B., Collas P. 2008. Expression of the myodystrophic R453W mutation of lamin A in C2C12 myoblasts causes promoter-specific and global epigenetic defects. Exp. Cell Res. 314: 1869–1880 [DOI] [PubMed] [Google Scholar]
- 54.Boergesen M., Pedersen T. A., Gross B., van Heeringen S. J., Hagenbeek D., Bindesboll C., Caron S., Lalloyer F., Steffensen K. R., Nebb H. I., et al. 2012. Genome-wide profiling of liver X receptor, retinoid X receptor, and peroxisome proliferator-activated receptor alpha in mouse liver reveals extensive sharing of binding sites. Mol. Cell. Biol. 32: 852–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gao J., Ye H., Serrero G. 2000. Stimulation of adipose differentiation related protein (ADRP) expression in adipocyte precursors by long-chain fatty acids. J. Cell. Physiol. 182: 297–302 [DOI] [PubMed] [Google Scholar]
- 56.Xu H. E., Stanley T. B., Montana V. G., Lambert M. H., Shearer B. G., Cobb J. E., McKee D. D., Galardi C. M., Plunket K. D., Nolte R. T., et al. 2002. Structural basis for antagonist-mediated recruitment of nuclear co-repressors by PPARalpha. Nature. 415: 813–817 [DOI] [PubMed] [Google Scholar]
- 57.Shearer B. G., Steger D. J., Way J. M., Stanley T. B., Lobe D. C., Grillot D. A., Iannone M. A., Lazar M. A., Willson T. M., Billin A. N. 2008. Identification and characterization of a selective peroxisome proliferator-activated receptor beta/delta (NR1C2) antagonist. Mol. Endocrinol. 22: 523–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Davies S. S., Pontsler A. V., Marathe G. K., Harrison K. A., Murphy R. C., Hinshaw J. C., Prestwich G. D., Hilaire A. S., Prescott S. M., Zimmerman G. A., et al. 2001. Oxidized alkyl phospholipids are specific, high affinity peroxisome proliferator-activated receptor gamma ligands and agonists. J. Biol. Chem. 276: 16015–16023 [DOI] [PubMed] [Google Scholar]
- 59.Wei P., Taniguchi S., Sakai Y., Imamura M., Inoguchi T., Nawata H., Oda S., Nakabeppu Y., Nishimura J., Ikuyama S. 2005. Expression of adipose differentiation-related protein (ADRP) is conjointly regulated by PU.1 and AP-1 in macrophages. J. Biochem. 138: 399–412 [DOI] [PubMed] [Google Scholar]
- 60.Gu J. Q., Ikuyama S., Wei P., Fan B., Oyama J., Inoguchi T., Nishimura J. 2008. Pycnogenol, an extract from French maritime pine, suppresses toll-like receptor 4-mediated expression of adipose differentiation-related protein in macrophages. Am. J. Physiol. Endocrinol. Metab. 295: E1390–E1400 [DOI] [PubMed] [Google Scholar]
- 61.Lockridge J. B., Sailors M. L., Durgan D. J., Egbejimi O., Jeong W. J., Bray M. S., Stanley W. C., Young M. E. 2008. Bioinformatic profiling of the transcriptional response of adult rat cardiomyocytes to distinct fatty acids. J. Lipid Res. 49: 1395–1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu H. E., Lambert M. H., Montana V. G., Parks D. J., Blanchard S. G., Brown P. J., Sternbach D. D., Lehmann J. M., Wisely G. B., Willson T. M., et al. 1999. Molecular recognition of fatty acids by peroxisome proliferator-activated receptors. Mol. Cell. 3: 397–403 [DOI] [PubMed] [Google Scholar]
- 63.Sanderson L. M., Degenhardt T., Koppen A., Kalkhoven E., Desvergne B., Muller M., Kersten S. 2009. Peroxisome proliferator-activated receptor beta/delta (PPARbeta/delta) but not PPARalpha serves as a plasma free fatty acid sensor in liver. Mol. Cell. Biol. 29: 6257–6267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chakravarthy M. V., Lodhi I. J., Yin L., Malapaka R. R., Xu H. E., Turk J., Semenkovich C. F. 2009. Identification of a physiologically relevant endogenous ligand for PPARalpha in liver. Cell. 138: 476–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Finck B. N., Bernal-Mizrachi C., Han D. H., Coleman T., Sambandam N., LaRiviere L. L., Holloszy J. O., Semenkovich C. F., Kelly D. P. 2005. A potential link between muscle peroxisome proliferator- activated receptor-alpha signaling and obesity-related diabetes. Cell Metab. 1: 133–144 [DOI] [PubMed] [Google Scholar]
- 66.Luquet S., Lopez-Soriano J., Holst D., Fredenrich A., Melki J., Rassoulzadegan M., Grimaldi P. A. 2003. Peroxisome proliferator-activated receptor delta controls muscle development and oxidative capability. FASEB J. 17: 2299–2301 [DOI] [PubMed] [Google Scholar]
- 67.Gan Z., Burkart-Hartman E. M., Han D. H., Finck B., Leone T. C., Smith E. Y., Ayala J. E., Holloszy J., Kelly D. P. 2011. The nuclear receptor PPARbeta/delta programs muscle glucose metabolism in cooperation with AMPK and MEF2. Genes Dev. 25: 2619–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lin J., Wu H., Tarr P. T., Zhang C. Y., Wu Z., Boss O., Michael L. F., Puigserver P., Isotani E., Olson E. N., et al. 2002. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 418: 797–801 [DOI] [PubMed] [Google Scholar]
- 69.Kamei Y., Ohizumi H., Fujitani Y., Nemoto T., Tanaka T., Takahashi N., Kawada T., Miyoshi M., Ezaki O., Kakizuka A. 2003. PPARgamma coactivator 1beta/ERR ligand 1 is an ERR protein ligand, whose expression induces a high-energy expenditure and antagonizes obesity. Proc. Natl. Acad. Sci. USA. 100: 12378–12383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huss J. M., Torra I. P., Staels B., Giguere V., Kelly D. P. 2004. Estrogen-related receptor alpha directs peroxisome proliferator-activated receptor alpha signaling in the transcriptional control of energy metabolism in cardiac and skeletal muscle. Mol. Cell. Biol. 24: 9079–9091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rangwala S. M., Wang X., Calvo J. A., Lindsley L., Zhang Y., Deyneko G., Beaulieu V., Gao J., Turner G., Markovits J. 2010. Estrogen-related receptor gamma is a key regulator of muscle mitochondrial activity and oxidative capacity. J. Biol. Chem. 285: 22619–22629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yamamoto H., Williams E. G., Mouchiroud L., Canto C., Fan W., Downes M., Heligon C., Barish G. D., Desvergne B., Evans R. M., et al. 2011. NCoR1 is a conserved physiological modulator of muscle mass and oxidative function. Cell. 147: 827–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Koves T. R., Sparks L. M., Kovalik J. P., Mosedale M., Arumugam R., DeBalsi K. L., Everingham K., Thorne L., Phielix E., Meex R. C., et al. 2013. PPARgamma coactivator-1alpha contributes to exercise-induced regulation of intramuscular lipid droplet programming in mice and humans. J. Lipid Res. 54: 522–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pollak N. M., Schweiger M., Jaeger D., Kolb D., Kumari M., Schreiber R., Kolleritsch S., Markolin P., Grabner G. F., Heier C., et al. 2013. Cardiac-specific overexpression of perilipin 5 provokes severe cardiac steatosis via the formation of a lipolytic barrier. J. Lipid Res. 54: 1092–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang H., Sreenivasan U., Gong D. W., O'Connell K. A., Dabkowski E. R., Hecker P. A., Ionica N., Konig M., Mahurkar A., Sun Y., et al. 2013. Cardiomyocyte-specific perilipin 5 overexpression leads to myocardial steatosis and modest cardiac dysfunction. J. Lipid Res. 54: 953–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.