Abstract
Growth hormone (GH) acutely stimulates lipolysis and fat oxidation, a process that operates postabsorptively and involves activation of the JAK-STAT pathway in the target tissue; no in vivo data exist regarding subsequent GH-regulated gene transcription. We obtained serum samples and muscle biopsies in human subjects before and 2 h after administration of a GH bolus. A significant (∼75%) elevation in serum FFA levels was recorded post GH. Microarray identified 79 GH-regulated genes in muscle. With qRT-PCR, we then examined the expression of selected genes in the presence and absence of glucose-induced suppression of lipolysis. Four genes involved in the JAK-STAT5 signaling pathway were regulated by GH, including SOCS1–3 and CISH, in addition to three genes associated with insulin action: NFκB1A, PIK3C2B, and PRKAG2. The gene encoding ANGPTL4, a protein involved in lipolysis and suppression of LPL activity, exhibited the most pronounced upregulation (5.6-fold) after GH, which was abrogated by concomitant suppression of lipolysis. Therefore, the GH-induced stimulation of ANGPTL4 gene expression seems secondary to induction of lipolysis. This new concept implies that abundant supply of circulating FFA decreases the need for alternative triglyceride-derived FFA through distinct inhibition of LPL mediated by increased ANGPTL4 gene expression in human muscle.
Keywords: angiopoietin-related protein-like 4, growth hormone, human gene array, lipid metabolism
Growth hormone (GH) is an important regulator of lipid metabolism in skeletal muscle (1). The secretion of GH is pulsatile in a pattern opposite to insulin with suppressed postprandial levels and increased activity during fasting and exercise. Following exposure to a physiological intravenous GH bolus, distinctive temporal changes in substrate fluxes are recorded in skeletal muscle, characterized by reversible suppression of glucose uptake that peaks after 2 h together with increased uptake of lipids (2). Triglycerides (TG) in lipoproteins are hydrolyzed to FFA by lipoprotein lipase (LPL) before entering the muscle cell (3). Interestingly, GH treatment downregulates LPL activity (4), and hypersecretion of GH as seen in acromegaly is associated with hypertriglyceridemia (5). It has been shown in humans that GH stimulates turnover and oxidation of free fatty acids (FFA) together with unaltered VLDL-TG kinetics (6). This finding suggests that circulating FFA rather than VLDL-TG constitutes the major source for increased lipid metabolism in muscle following GH stimulation, but the mechanisms that regulate these processes are unknown.
Experimental data in human subjects suggest that these insulin-antagonistic effects of GH, i.e., suppression of glucose uptake and stimulation of FFA, are causally linked, as pharmacological antilipolysis resulting in suppressed FFA levels abrogates GH-induced insulin resistance during a glucose clamp (7). In animal models and cell cultures, this is associated with inhibition of the IRS1-Akt pathway in muscle (8) and fat (9). However, in human subjects, GH-induced insulin resistance does not translate into suppression of pertinent insulin signaling pathways as assessed by western blotting and activity assays (10–13). This indicates that additional mechanisms apart from the IRS1-Akt signaling pathway mediate the effects of GH on skeletal muscle metabolism.
It has also been recorded in human subjects that systemic exposure to GH of endogenous as well as exogenous origin acutely translates into detectable in vivo GH receptor signaling in muscle in terms of tyrosine-phosphorylation of the transcription factor STAT5 (11, 14–16). Moreover, this is followed by increased insulin-like growth factor 1 (IGF-1) and suppressor of cytokine signaling (SOCS) gene expression, both of which are known STAT5 targets (10, 11). Because regulation of target genes mediates numerous actions of GH (17), it is likely that identification of genes regulated by GH in human skeletal muscle may provide novel insight into the metabolic responses described above. Indeed, this approach has been used in murine skeletal muscle cell lines in vitro with GH stimulation alone (18) and in combination with insulin (19). However, the acute response under physiological conditions in humans is unknown. The DNA microarray technology measures the expression levels of large numbers of genes simultaneously, which provides a means to disentangle the transcriptional mechanisms underlying the acute effects of GH.
The aim of this study was to identify the impact of a single GH pulse on gene expression in human skeletal muscle in vivo. To this end, eight healthy male subjects underwent muscle biopsies before and 2 h after administration of an intravenous GH bolus. Microarray technology was used, including functional classification of GH-responsive genes and subsequent qRT-PCR verification. To test for direct and indirect effects of GH, expression of target genes was investigated when GH was given in combination with insulin-induced suppression of lipolysis.
RESEARCH DESIGN AND METHODS
This study presents data obtained from two clinical studies from which data of a different nature have been published previously (10, 13).
Group 1 consisted of eight healthy men ages 24.6 ± 1.8 years (mean ± SE) with a mean body mass index (BMI) of 24.2 ± 1.2 kg*m−2 who were studied in a randomized, crossover design on three occasions (13): 1) after an intravenous GH bolus (0.5 mg Genotropin, Miniquick, Pfizer, New York, NY), 2) after an intravenous GH bolus in combination with an oral glucose load (OGTT) (75 g), and 3) after an OGTT alone. Muscle biopsies from vastus lateralis were collected at t = 0 min and t = 120 min. Blood samples were drawn at least every 30 min.
Group 2 consisted of eight patients with GH deficiency (GHD) ages 57.0 ± 2.5 years (mean ± SE) with a BMI of 30.6 ± 1.4 kg*m−2, who were treated with stable doses of GH and other pituitary substitution therapy for at least three months. The study design is described in detail in a previous publication (10). We used skeletal muscle biopsies from these patients taken on two separate examination days, separated by at least four weeks. On one study day, a muscle biopsy was taken after discontinuation of GH therapy. On a separate day, a muscle biopsy was taken during a low-dose GH infusion commenced 6.5 h earlier. On each examination day, i.e., when the biopsies were taken, patients also underwent a 6 h hyperinsulinemic-euglycemic clamp with insulin infused at 0.5 mU/kg/min (10). Blood samples were drawn at least every 2 h.
Both study protocols were approved by the Regional Scientific Ethics Committee of Denmark (M-20070052; M-20070176), and all participants gave oral and written informed consent to participate. The study was conducted in accordance to the Helsinki Declaration.
Study design
We performed Human Gene 1.0 ST Array (Affymetrix Inc., Santa Clara, CA) analysis on muscle biopsies from group 1 obtained before and 120 min after an intravenous GH bolus to test for acute effects of GH stimulation in human skeletal muscle (study day 1). Regulated genes were identified as a gene displaying a fold change of ≤ 0.7 or ≥ 1.4, with a P-value < 0.05 in response to GH treatment. These criteria are similar to those published for similar studies (20–22). Expression of regulated genes was examined on the mRNA level in samples from group 1 and on the mRNA and protein levels from GHD patients from group 2.
RNA isolation
Skeletal muscle powder samples were homogenized in TriZol reagent (Life Technologies Inc., Life Technologies, Roskilde, Denmark), and total RNA was extracted following the manufacturer's protocol. RNA was quantified by measuring absorbance at 260 and 280 nm using a NanoDrop 8000 (NanoDrop Products, Wilmington, DE); the inclusion criterion was a ratio of at least 1.8. The integrity of the RNA was determined using Bioanalyzer 2100 (Agilent, Santa Clara, CA). A RIN score > 7 was obtained for all samples.
Human Gene 1.0 ST Array labeling data analysis
Total RNA (100 ng) was labeled with the Ambion WT Expression Kit (Ambion, Carlsbad, CA) according to the manufacturer's instructions. Samples were hybridized overnight to the GeneChip Human Gene 1.0 ST Array (Affymetrix) and scanned using an Affymetrix GCS 3000 7G scanner.
Data analysis was performed in the GeneSpring GX11.5 software (Agilent). Cel files were imported and quantile normalized with the iterPLIER16 algorithm followed by baseline transformation with median scaling to the median of all arrays. Differential gene expression analysis was performed on transcript values using paired t-test.
Hierarchical clustering was performed vertically on genes and horizontally on samples by Euclidian distance and centroid linkage (default in GeneSpring). A DAVID (Database for Annotation, Visualization and Integrated Discovery) analysis was performed on the regulated genes using the available online tool (DAVID Bioinformatics Resources v6.7).
Cell culture
C2C12 cells (ATCC, Manassas, VA) were cultured in DMEM (1 g/l glucose), 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin. When cells reached confluency, the growth medium was substituted with differentiation medium consisting of DMEM (1 g/l glucose), 2% horse serum, 100 U/ml penicillin, and 100 μg/ml streptomycin. Differentiation medium was changed every second day for a total of seven days. Cells were starved in serum-free medium for 2 h prior to a 2 h incubation with either palmitate [0.5 mM, conjugated to 1% (w/v) FFA-free BSA (Sigma)) and/or GH (0.5 µg/ml, Genotropin], after which cells were harvested and collected by scraping in cold PBS. All experiments were conducted in duplicate.
Real-time RT-PCR
cDNA was constructed using random hexamer primers as described by the manufacturer (Verso cDNA kit; Abgene, Epsom, UK). Then KAPA SYBR FAST qPCR Master Mix (Kapa Biosystems Inc., Woburn, MA) and the following primer pairs were added: SOCS-1, 5′-ACACGCACTTCCGCACATTC-3′ and 5′-CGAGGCCATCTTCACGCTAAG-3′; SOCS-2, 5′-GGTCGAGGCGATCAGTG-3′ and 5′-TCCTTGAAGTCAGTGCGAATC-3′; SOCS-3, 5′-GCCCTTTGCGCCCTTT-3′ and 5′-CGGCCACCTGGACTCCTATGA-3′; IGF-1, 5′-GACAGGGGCTTTATTTCAAC-3′ and 5′-CTCCAGCCTCCTTAGATCAC-3′; cytokine inducible SH2-containing protein (CISH), 5′-GCCCTGAGCCCTGGTAGTCC-3′ and 3′-GACACATTCACAGACGGGTGG-5′; angiopoietin-related protein-like 4 (ANGPTL4), 5′- TAGTCCACTCTGCCTCTCCC-3′ and 3′-GAGATGGCCCAGCCAGTT-5′; β2-microglobulin, 5′-AATGTCGGATGGATGAAACC-3′ and 5′-TCTCTCTTTCTGGCCTGGAG-3′; β-actin, 5′-ACGGGGTCACCCACACTGTGC-3′ and 3′CTAGAAGCATTTGCGGTGGACGATG-5′.
The PCR reactions were performed in duplicate using KAPA SYBR® FAST qPCR Kit (Kapa Biosystems Inc., Woburn, MA) in a LightCycler 480 (Roche Applied Science, Penzberg, Germany) using the following protocol: one step at 95°C for 3 min., then 95°C for 10 s, 60°C for 20 s, and 72°C for 10 s The increase in fluorescence was measured in real time during the extension step. The relative gene expression was estimated using the default “Advanced Relative Quantification” mode of the software version LCS 480 1.5.0.39 (Roche Applied Science, Penzberg, Germany). All samples were amplified in duplicate. A similar setup was used for negative controls, except that the reverse transcriptase was omitted and no PCR products were detected under these conditions.
Protein lysate preparation and Western blotting
Using a Precellys 24 homogenizer (Bertin Technologies, Montigny-le-Bretonneux, France), the muscle biopsies were homogenized in an ice-cold buffer containing 20 mM Tris, 50 mM NaCl, 50 mM NaF, 5 mM Na4P2O7, 250 mM sucrose, 1% (v/v) Triton X-100, 2 mM DTT, 0.1 mM benzamidine, 0.5 mM PMSF, 50 µg/ml soybean trypsin inhibitor, and 4 µg/ml leupeptin (pH 7.4). Samples were rotated for 20 min at 4°C. Insoluble materials were removed by centrifugation at 14,000 g for 20 min at 4°C. Total protein concentrations were determined using Bradford protein assay (Bio-Rad), and protein samples were resolved by SDS-PAGE on precast StainFree 4–15% gels (Bio-Rad), transferred onto PVDF membranes, blocked for 2 h in 0.3% iBlock, incubated in anti-CISH antibodies (ab88383, Abcam, Cambridge, MA), incubated for 1 h in appropriate HRP-conjugated secondary AB, and then proteins of interest were visualized by ECL using a ChemiDoc XRS system (Bio-Rad). Specific protein signals were quantified using Image Lab (v4.0.1, Bio-Rad). Total protein was quantified by the Bio-Rad StainFree technique using Image Lab.
Serum ANGPTL4 and metabolites
Serum ANGPTL4 was determined in duplicate by an ELISA assay (RayBiotech, Norcross, GA) with an intra- and interassay CV of 6% and 10%, respectively. The assay was performed as described in the manufacturer's protocol. Qualitative readings were done on a Victor3 1420 Multilabel Counter (PerkinElmer, Waltham, MA).
Total cholesterol and triglycerides in serum were measured using standard enzymatic kits.
Statistics
Data are presented as means ± SE when normally distributed, and significance is assumed when P ≤ 0.05. Statistical evaluation of differences between normally distributed data was assessed by paired t-test. When testing for fold changes in mRNA, statistical analysis was performed on log-transformed data. Two-way repeated measures ANOVA was used on ANGPTL4 serum protein data. Posthoc tests were performed using Student-Newman-Keuls test. Correlation analysis was performed by Person's Product Moment correlation analysis.
RESULTS
Circulating GH and lipids
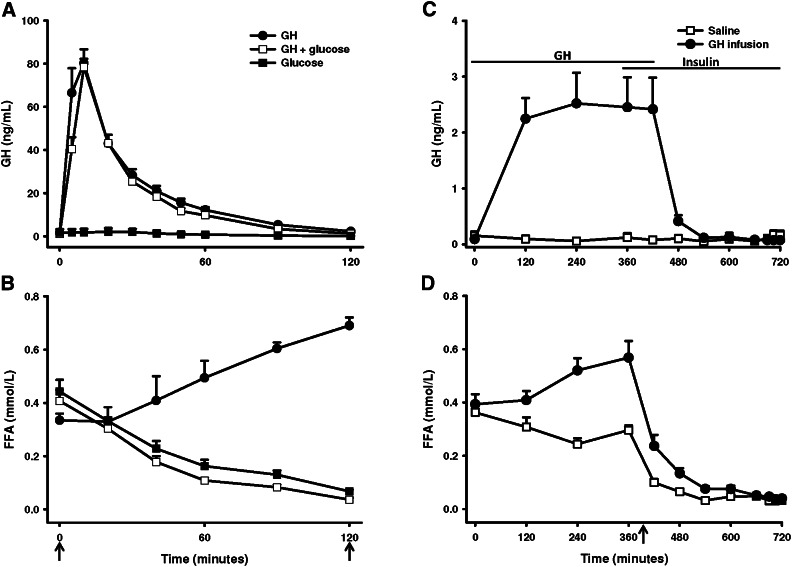
Data on circulating hormones (GH, insulin), glucose, and FFA have previously been published (13). In short, the GH bolus in group 1 induced a serum GH peak of 80.3 ± 6.4 µg/l (mean ± SE, P < 0.001), which had returned to basal levels of ∼1.5 µg/l at t = 120 min. The same was observed for GH administered during an OGTT (GHt=10 = 78.5 ± 3.8 µg/l), whereas OGTT alone was not associated with detectable GH excursions (Fig. 1A). In group 2, GH infusion caused serum GH levels of 0.09 ± 0.03 µg/l at t = 0 to increase to a mean value of 2.41 ± 0.24 µg/l (mean ± SE, P < 0.001) in the time interval t = 120–420 min. During saline infusion, no change in serum GH was recorded (Fig. 1B). In both the GH and saline infusion, insulin increased 5-fold during the hyperinsulinemic clamp (13).
Fig. 1.
Circulating GH and FFA. (A) Serum GH and (B) serum FFA in group 1 after GH, OGTT, or GH+OGTT. GH and/or glucose was initiated at t = 0. (C) Serum GH and (D) serum FFA in group 2 after saline or GH infusion and hyperinsulinemic euglycemic clamp. Arrows indicate time of biopsy. Data are presented as means ± SE.
In group 1, GH induced a ∼75% increase in serum FFA levels after 120 min (P < 0.001). This lipolytic effect of GH was suppressed by the concomitant OGTT, as characterized by a ∼85% suppression after 120 min (P < 0.001). OGTT alone induced a decrease in FFA of ∼90% (P < 0.001). The degree of FFA suppression (AUCFFA) was similar when comparing OGTT with OGTT+GH (P = 0.083). In group 2, infusion of GH induced a ∼45% increase in FFA (P = 0.04), as opposed to a ∼18% suppression during saline infusion (P = 0.01) (Fig. 1C). In both situations, FFA was almost completely suppressed by the hyperinsulinemic clamp (Fig. 1D).
In group 1, triglyceride levels decreased after OGTT both alone and during concomitant GH stimulation (Table 1). Total cholesterol levels increased ∼4% after both GH and OGTT but not after the combined OGTT and GH stimulation. In group 2, cholesterol levels were slightly increased 360 min after initiation of GH or saline infusion, and this effect was independent of GH stimulation.
TABLE 1.
Serum triglycerides and cholesterol
| Triglycerides |
Cholesterol |
|||
| Treatment | Before | After | Before | After |
| Group 1 | ||||
| Glucose | 0.89 ± 0.18 | 0.79 ± 0.19 * | 3.50 ± 0.37 | 3.61 ± 0.41* |
| GH | 0.83 ± 0.44 | 0.84 ± 0.23 | 3.65 ± 0.67 | 3.81 ± 0.66* |
| GH + glucose | 0.80 ± 0.15 | 0.66 ± 0.14 * | 3.66 ± 0.41 | 3.69 ± 0.40 |
| Group 2 | ||||
| Saline | 1.03 ± 0.30 | 1.09 ± 0.38 | 3.91 ± 0.85 | 4.25 ± 0.91* |
| GH | 1.21 ± 0.44 | 1.28 ± 0.32 | 3.90 ± 0.94 | 4.24 ± 0.77* |
Triglyceride and cholesterol measured in serum from group 1 before (t = 0) and after (t = 120) and group 2 before (t = 0) and after (t = 360) indicated treatment. Data presented as means ± SD [mM].
*P < 0.05 compared with serum levels before treatment.
Acute GH-responsive genes in muscle
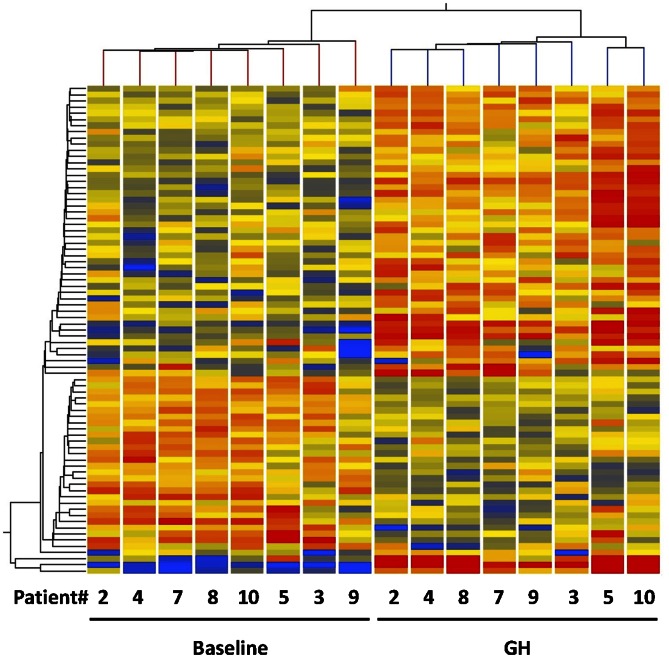
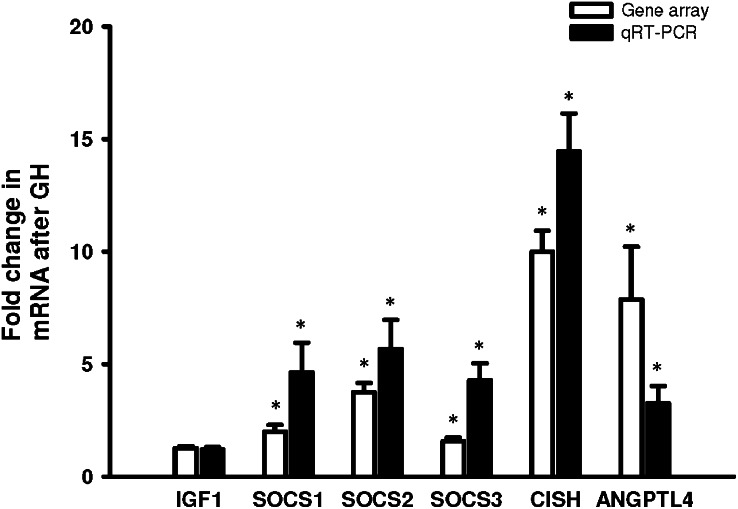
Genes that were differentially expressed in muscle tissue before and 120 min after GH were identified in a gene array analysis. After GH stimulation, 79 genes satisfied the 0.7- or 1.4-fold changes in expression and P < 0.05. Hierarchical clustering was performed, and the samples fell into two distinct clusters, one containing baseline biopsies and the other biopsies taken after GH treatment (Fig. 2). To verify the microarray data, mRNA levels of six genes (IGF-1, SOCS1–3, CISH, and ANGPTL4) were confirmed by qRT-PCR (Fig. 3). Regulated genes were analyzed by the Ingenuity Pathway Analysis tool to group the genes into biological functions. The result is depicted in Table 2, in which the main biological functions identified are growth, proliferation, differentiation, gene expression, movement, and amino acid metabolism. Table 3 lists the genes involved in amino acid metabolism, carbohydrate metabolism, lipid metabolism, cellular growth and proliferation, and gene expression. See supplementary Tables I and II for a complete list of gene array data and a DAVID analysis supporting the findings by the Ingenuity Pathway Analysis. Ingenuity analysis of the genes revealed that four genes, SOCS1–3 and CISH, involved in the JAK-STAT5 signaling pathway were regulated, whereas IGF-1 was not found to be regulated within the 2 h window. Six genes that have been shown to be induced during a state of insulin resistance and type-2 diabetes mellitus (T2DM) were identified: SOCS1–3, NFκB1A, PIK3C2B, and PRKAG2. In addition, ANGPTL4 and TXNIP, putative regulators of substrate metabolism, were regulated after GH stimulation.
Fig. 2.
Hierarchical cluster analysis on genes regulated by GH in human skeletal muscle. Numbers at the bottom of the figure indicate patient identities in either baseline or GH situation. Upregulated genes are red and downregulated genes are blue. Analysis was performed with GeneSpring.
Fig. 3.
Microarray validation by qRT-PCR. Microarray (open bars) and qRT-PCR verification (filled bars) of selected genes. Data presented as means ± SE of fold changes in expression of GH-treated samples relative to baseline samples. By paired t-test, *P ≤ 0.05 versus baseline.
TABLE 2.
Top five molecular and cellular functions
| Function | P | Number of Molecules |
| Cellular development | 4.89E-06 – 4.95E-02 | 27 |
| Cellular growth and proliferation | 4.89E-06 – 4.95E-02 | 28 |
| Gene expression | 5.89E-06 – 3.98E-02 | 21 |
| Cellular movement | 3.71E-04 – 4.77E-02 | 15 |
| Amino acid metabolism | 5.21E-04 – 5.21E-04 | 2 |
Functions sorted by P value along with the number of associated genes. Analysis performed with Ingenuity Pathway Analysis tool.
TABLE 3.
GH-responsive genes by function
| Gene | Gene Symbol | Direction | P | Fold Change |
| Amino acid metabolism | ||||
| Adrenoceptor β 2, surface | ADRB2 | Up | < 0.001 | 1.61 |
| Bone morphogenetic protein 4 | BMP4 | Up | < 0.001 | 1.63 |
| Carbohydrate metabolism | ||||
| Adrenoceptor β 2, surface | ADRB2 | Up | < 0.001 | 1.61 |
| Chemokine (C-X3-C motif) ligand 1 | CX3CL1 | Down | 0.032 | 1.49 |
| Dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 | DYRK2 | Down | < 0.001 | 1.50 |
| Phosphoinositide-3-kinase, class 2, β polypeptide | PIK3C2B | Down | < 0.001 | 1.45 |
| 5′-AMP-activated protein kinase, subunit γ-2 | PRKAG2 | Up | 0.031 | 1.46 |
| Transforming growth factor, β 3 | TGFB3 | Up | 0.022 | 1.46 |
| Tumor necrosis factor (ligand) superfamily, member 10 | TNFSF10 | Up | < 0.001 | 1.64 |
| Lipid metabolism | ||||
| Adrenoceptor β 2, surface | ADRB2 | Up | < 0.001 | 1.61 |
| Angiopoietin-related protein 4 | ANGPTL4 | Up | 0.001 | 5.56 |
| Bone morphogenetic protein 4 | BMP4 | Up | < 0.001 | 1.63 |
| FK506 binding protein 5 | FKBP5 | Up | < 0.001 | 1.59 |
| Low density lipoprotein receptor | LDLR | Down | 0.003 | 1.46 |
| Phosphoinositide-3-kinase, class 2, β polypeptide | PIK3C2B | Down | < 0.001 | 1.45 |
| 5′-AMP-activated protein kinase, subunit γ-2 | PRKAG2 | Up | 0.031 | 1.46 |
| Tumor necrosis factor (ligand) superfamily, member 10 | TNFSF10 | Up | < 0.001 | 1.64 |
| Cellular growth and proliferation | ||||
| Angiopoietin-related protein 4 | ANGPTL4a | Up | 0.001 | 5.56 |
| Bone morphogenetic protein 4 | BMP4 | Up | < 0.001 | 1.63 |
| Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 | CITED2 | Up | < 0.001 | 1.68 |
| Chemokine (C-X3-C motif) ligand 1 | CX3CL1 | Down | 0.032 | 1.49 |
| Discoidin domain receptor family, member 1 | DDR1 | Up | < 0.001 | 1.62 |
| FK506 binding protein 5 | FKBP5 | Up | < 0.001 | 1.59 |
| Frizzled-7 | FZD7 | Down | < 0.001 | 1.74 |
| Immediate-early response-3 | IER3 | Up | < 0.001 | 4.06 |
| Krueppel-like factor 10 | KLF10 | Up | 0.001 | 1.76 |
| microRNA 1-1 | mir-1 | Down | 0.012 | 1.66 |
| microRNA 133a-1 | mir-133 | Down | 0.003 | 1.91 |
| microRNA 29a | mir-29 | Down | < 0.001 | 1.41 |
| microRNA 95 | mir-95 | Up | 0.044 | 1.71 |
| Myostatin | MSTN | Down | 0.001 | 1.41 |
| v-Myc myelocytomatosis viral oncogene homolog | MYC | Up | 0.019 | 1.56 |
| Enhancer of filamentation 1 | NEDD9 | Down | 0.002 | 2.10 |
| Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, α | NFKB1A | Down | 0.001 | 1.67 |
| Pyruvate dehydrogenase kinase, isozyme 4 | PDK4a | Up | 0.009 | 1.48 |
| 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 | PFKFB3 | Up | 0.041 | 1.58 |
| Protein phosphatase 1 regulatory subunit 15A | PPP1R15A | Down | 0.003 | 1.44 |
| S100 calcium binding protein B | S100B | Up | 0.028 | 1.43 |
| Serum/glucocorticoid-regulated kinase 1 | SGK1 | Up | < 0.001 | 1.55 |
| Suppressor of cytokine signaling 1 | SOCS1 | Up | 0.004 | 1.86 |
| Suppressor of cytokine signaling 3 | SOCS3 | Up | 0.007 | 1.50 |
| Transforming growth factor, β 3 | TGFB3 | Up | 0.022 | 1.46 |
| Tumor necrosis factor (ligand) superfamily, member 10 | TNFSF10 | Up | < 0.001 | 1.64 |
| Thioredoxin-interacting protein | TXNIP | Up | < 0.001 | 1.52 |
| Wingless-type MMTV integration site family, member 9A | WNT9A | Up | 0.003 | 1.92 |
| Gene expression | ||||
| AT rich interactive domain 5B (MRF1-like) | ARID5B | Down | < 0.001 | 1.52 |
| Bone morphogenetic protein 4 | BMP4 | Up | < 0.001 | 1.63 |
| Cytokine inducible SH2-containing protein | CISH | Up | < 0.001 | 9.74 |
| Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 | CITED2 | Up | < 0.001 | 1.68 |
| FOS-like antigen 2 | FOSL2 | Up | 0.003 | 1.82 |
| Frizzled family receptor 5 | FZD5 | Up | < 0.001 | 2.97 |
| Frizzled-7 | FZD7 | Down | < 0.001 | 1.74 |
| Immediate-early response-3 | IER3 | Up | < 0.001 | 4.06 |
| Krueppel-like factor 10 | KLF10 | Up | 0.001 | 1.76 |
| Myostatin | MSTN | Down | 0.001 | 1.41 |
| v-Myc myelocytomatosis viral oncogene homolog | MYC | Up | 0.019 | 1.56 |
| Myogenic differentiation 1 | MYOD1 | Up | 0.049 | 1.43 |
| Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, α | NFKB1A | Down | 0.001 | 1.67 |
| Nuclear receptor subfamily 1, group D, member 1 | NR1D1 | Down | < 0.001 | 1.71 |
| Polo-like kinase 2 | PLK2 | Up | 0.008 | 1.47 |
| Suppressor of cytokine signaling 1 | SOCS1 | Up | 0.004 | 1.86 |
| Suppressor of cytokine signaling 2 | SOCS2 | Up | < 0.001 | 3.58 |
| Suppressor of cytokine signaling 3 | SOCS3 | Up | 0.007 | 1.50 |
| Transcription factor 21 | TCF21 | Down | 0.012 | 1.43 |
| Transforming growth factor, β 3 | TGFB3 | Up | 0.022 | 1.46 |
| Tumor necrosis factor (ligand) superfamily, member 10 | TNFSF10 | Up | < 0.001 | 1.64 |
Genes in human skeletal muscle with an absolute fold change (FC) above 1.4 120 min after GH that are involved in either amino acid metabolism, lipid metabolism, carbohydrate metabolism, cellular growth and proliferation, or gene expression are presented with FC and corresponding P value, by paired t-test.
Gene that has previously been shown to be regulated by FFA alone (47).
GH regulation of target genes in skeletal muscle
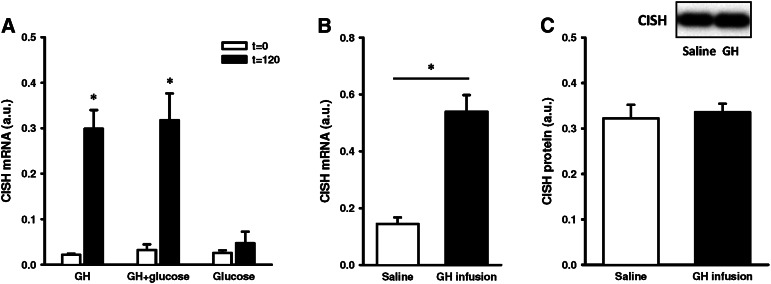
The gene array and qRT-PCR both confirmed acute GH regulation of the STAT5 target gene CISH. To further characterize GH regulation of CISH, gene expression was assessed in muscle from healthy subjects subjected to either GH alone, OGTT, or GH + OGTT. As depicted in Fig. 4A, CISH gene expression increased 18-fold after GH irrespective of concomitant OGTT, while OGTT alone induced no change in CISH mRNA. In addition, infusion of GH to GHD patients increased CISH gene expression 4-fold (Fig. 4B). However, as shown in Fig. 4C, 7 h GH infusion did not increase skeletal muscle CISH protein content.
Fig. 4.
CISH expression. (A) CISH mRNA in skeletal muscle from healthy men, before and 120 min after GH, OGTT, and GH/OGTT. (B) CISH mRNA and (C) CISH protein (CISH protein/total protein) in skeletal muscle from GHD patients under a 6 h saline or GH infusion. Data presented as means ± SE. By paired t-test, *P ≤ 0.05.
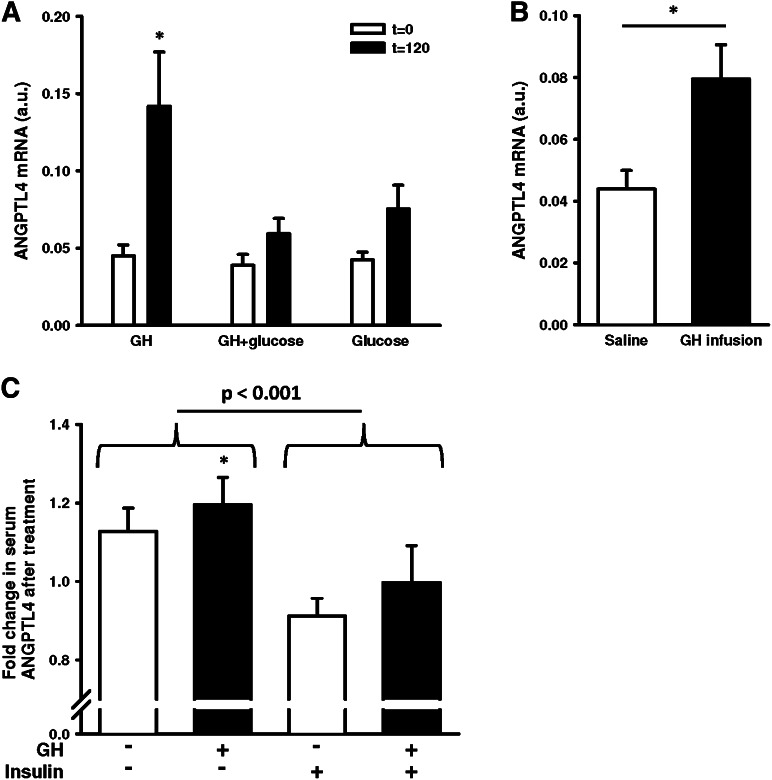
Similar to CISH, the gene array and qRT-PCR both showed acute induction of ANGPTL4. We therefore examined ANGPTL4 expression in muscle stimulated with GH during an OGTT and with OGTT alone. OGTT in combination with GH abrogated the increase in ANGPTL4 gene expression observed with GH alone (Fig. 5A).
Fig. 5.
ANGPTL4 expression. (A) ANGPTL4 mRNA in skeletal muscle from healthy men, before and 120 min after GH, OGTT, and GH/OGTT. (B) ANGPTL4 mRNA in skeletal muscle from GHD patients under a 6 h saline or GH infusion. (C) Fold changes in circulating ANGPTL4 in GHD patients measured by ELISA. Fold changes calculated as ANGPTL4 protein levels after indicated treatment relative to the protein levels before treatment. GH: 6 h infusion; saline: 6 h infusion; insulin: 6 h hyperinsulinemic-euglycemic clamp. Data presented as means ± SE. Statistics by paired t-test and two-way repeated measurements ANOVA, *P ≤ 0.05.
To further investigate the relationship between GH and ANGPTL4, we investigated skeletal muscle biopsies from group 2 and found a 2-fold increase in ANGPTL4 expression when GHD patients were infused with GH (Fig. 5B). ANGPTL4 encodes a protein that is secreted from most tissues into the circulation (23). We therefore measured ANGPTL4 levels in serum from GHD patients. As shown in Fig. 5C, ANGPTL4 levels increased 1.2-fold after GH infusion, while GH in combination with insulin induced no change. The hyperinsulinemic clamp decreased circulating ANGPTL4 by ∼20%. In group 1, there was no correlation between serum GH and ANGPTL4 mRNA (Pearson's r = 0.33, P = 0.14). However, if the data from the combined GH and glucose treatment were left out, a correlation between ANGPTL4 mRNA and serum GH was present (Pearson's r = 0.56, P = 0.04). This finding, combined with a described association between serum FFA and ANGPTL4 levels (24), led us to perform correlation analyses between the elevations in FFA and ANGPTL4 mRNA. We found a correlation between serum FFA and ANGPTL4 mRNA in group 1 (Pearson's r = 0.54, P = 0.01) and in group 2 (Pearson's r = 0.7, P = 0.004). In group 1, CISH was positively correlated to GH (Pearson's r = 0.65, P < 0.001), whereas no correlation between FFA and CISH was found.
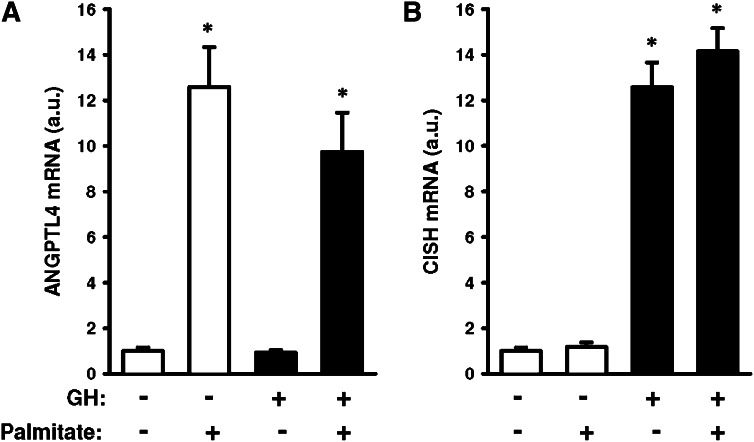
Finally, we incubated C2C12 murine muscle cells to isolate the direct effects of GH and FFA (palmitate) on ANGPTL4 and CISH gene expression. As evident from Fig. 6, we found that palmitate increased ANGPTL4 gene expression ∼12-fold (P < 0.001), whereas palmitate alone had no effect on CISH expression. GH induced a ∼13-fold increase in CISH gene expression (P < 0.001), while no direct effect of GH was observed on ANGPTL4 gene expression.
Fig. 6.
ANGPTL4 and CISH expression in C2C12. (A) ANGPTL4 mRNA. (B) CISH mRNA. Cells were incubated for 2 h in palmitate (0.5 mM) and/or GH (0.5 µg/ml). Data normalized to control situation and presented as means ± SE. Statistics by Student t-test, *P ≤ 0.05.
DISCUSSION
GH is a potent regulator of skeletal muscle metabolism in vivo, and the underlying mechanisms include regulation of gene expression (1). The aims of the present study were to identify genes in skeletal muscle that are acutely regulated after a bolus of GH and to investigate whether these genes are involved in lipid metabolism. The in vivo design integrated both direct effects of stimulation of the GH receptor in skeletal muscle mediated through JAK-STAT signaling, as well as indirect effects, such as elevated FFA levels, due to GH-stimulated lipolysis in adipose tissue. Our results show that several genes encoding regulators of signal transduction, lipid metabolism, and local growth factors were increased in human skeletal muscle 120 min after a single bolus of GH.
This study is the first to report GH-mediated regulation of ANGPTL4 gene expression in human tissue in vivo. ANGPTL4 has recently been identified as an important regulator of plasma lipoprotein levels in humans (25). Overexpression of ANGPTL4 in transgenic animals causes hypertriglyceridemia (26), and human subjects that carry an inactive genetic variant of ANGPTL4 have ∼25% lower triglyceride levels than subjects with functional ANGPTL4 (27). ANGPTL4 has been demonstrated to inhibit LPL activity by converting active LPL dimers into inactive monomers (25). It could therefore be speculated that preference for circulating FFA rather than VLDL-TG as substrates for oxidation during GH stimulation (6) is mediated by inhibition of LPL by ANGPTL4. ANGPTL4 has also been shown to directly stimulate lipolysis in 3T3-L1 adipocytes (28), and increased circulating ANGPTL4 may therefore enhance GH-stimulated FFA release from adipose tissue, ensuring substrate availability for lipid oxidation in skeletal muscle. To pursue this hypothesis, it could be of relevance to study the effect of GH exposure on lipolysis and LPL activity in the 2–3% of Caucasians who carry the E40K ANGPTL4 gene variant that results in expression of inactive ANGPTL4 protein (27).
ANGPTL4 was originally identified as a gene regulated by the peroxisome proliferator-activated receptors (PPAR)α, PPARδ, and PPARγ that function as transcription factors (29, 30). FFA is a natural ligand of the PPARs (31), and incubation of both murine and human myotubes with FFA in vitro has repeatedly showed increased ANGPTL4 mRNA expression in several microarray studies and was confirmed by incubation of C2C12 cells in this study (32–34). Both saturated (palmitate), monounsaturated (oleate), biunsaturated (linoleic acid), and polyunsaturated (eicosapentaenoic acid) fatty acids induce ANGPTL4 gene expression, showing that the effect of FFA on ANGPTL4 gene induction is independent of saturation status (33). The regulation of ANGPTL4 by FAA may be dependent on PPARδ in skeletal muscle cells (35). In our study ANGPTL4 mRNA increased in response to GH stimulation only when this was associated with an increase in FFA levels. We did not see regulation of ANGPTL4 gene expression when GH was given during an OGTT that suppressed FFA levels, suggesting that ANGPTL4 in skeletal muscle is not a direct target of signaling through the GH receptor/STAT5 pathway. This is further supported by our finding that stimulation of C2C12 cells with GH alone had no effect on ANGPPTL4 expression, whereas palmitate potently induced expression of ANGPTL4, which is in accordance with a previously published gene array study on the effect of GH in C2C12 cells (36). In vitro studies show that GH stimulation of preadipocytes that express STAT5, but not the PPARγ receptor, is associated with decreased ANGPTL4 gene expression (37). Previous work identifying STAT5A and STAT5B binding sites did not find STAT5 to bind in the vicinity of the ANGPTL4 gene (38). The observed increase in ANGPTL4 gene expression in human skeletal muscle after GH stimulation in vivo is therefore most likely mediated by stimulation of lipolysis in adipose tissue and a subsequent increase in circulating FFA levels. This concept implies that abundant supply of circulating FFA decreases the need for alternative triglyceride-derived FFA by a distinct inhibition of LPL by increased ANGPTL4 gene expression. This argument could be mechanistically tested in humans in future studies by directly increasing FFA levels through infusion of intralipids or by inhibiting lipolysis with acipimox during GH stimulation.
It has been firmly established that STAT5 plays a prominent role in mediating the signal from the GH receptor to gene transcription (13, 14, 39), and in accordance with this, we recorded increased expression of four known STAT5 targets. CISH, which was the most potently induced gene in our array, has not previously been shown to be regulated by GH in tissues from human subjects. Correlation analyses in the present study support the notion that CISH gene expression is directly regulated by GH, not as a secondary effect by GH-mediated rises in FFA. This is in accordance with earlier findings in which CISH was increased by the largest magnitude in a microarray analysis of murine C2C12 myotubes stimulated with GH (36). CISH, which is also known as cytokine-induced STAT (CIS) inhibitor, is structurally related to the other regulated STAT5 targets, SOCS1–3 (40), and like the SOCS genes, a STAT5 binding sequence is found in the CISH promoter region (41). CISH inhibits GH signaling through STAT5 by binding to the GH receptor (42). Little is known about the role of CISH in human skeletal muscle, but it is likely that the protein participates in a negative signaling feedback loop (40). In the present study, IGF-1 was not regulated 120 min after GH exposure. IGF-1 is a well-established target of STAT5 transcriptional activity in a wide array of tissues (43), and we previously detected increased IGF-1 mRNA in human skeletal muscle after 240 min of GH infusion (11) and after 6 h GH infusion to patients with GHD (10). It has recently been shown that GH stimulation is associated with a rapid loss of the transcriptional repressor Bcl6 at the SOCS and CISH promoters but not at the IGF-1 promoter (44). Sustained Bcl6 binding to the IGF-1 promoter could provide a mechanistic explanation for relative delay in IGF-1 transcription in skeletal muscle, but this remains to be experimentally proven.
GH stimulation was associated with decreased transcription of the PIK3C2B gene, which encodes a catalytic subunit of the phosphatidylinositol-4-phosphate 3-kinase (PI3k) enzyme complex. PI3k is involved in a plethora of intracellular signaling processes, including insulin receptor signaling through insulin receptor substrate 1 (IRS1), and impaired signaling through this signaling pathway has been suggested as a mechanism by which GH induces insulin resistance. However, the PIK3C2B gene encodes a class II PI3k subunit, not the class I of PI3k that is involved in insulin signaling. This is in accordance with previous findings of intact insulin-stimulated PI3k activity in human skeletal muscle (12). Only a little is known about the function of class II PI3k, although it has been shown that the C2β subunit, which is encoded by the PIK3C2B gene, is directly activated by growth factors in vitro (45). It is therefore possible that the observed downregulation of PIK3C2B is a result of direct GH signaling through the GH receptor.
The present data demonstrate that changes in gene expression after a single GH bolus is markedly different from those observed after prolonged GH treatment (20). In fact, only three genes (PDK4, SGK, and NFκB1A) were found to exhibit comparable changes in expression when comparing the present data with those reported by Sjögren et al., who examined muscle biopsies in hypopituitary men after two weeks of GH replacement (20). This could in part be due to effects from GH- induced hepatic and peripheral production of IGF-1 after more prolonged treatment. However, two weeks of GH treatment also induced expression of genes encoding enzymes involved in lipid oxidation in skeletal muscle, including fatty acid-binding protein, acyl-CoA thioesterase, and acyl-CoA synthetase (20). Similarly, in human adipose tissue, prolonged GH treatment increased genes involved in lipid metabolism, including PNPLA3, which encodes a protein with lipase activity (46). Together, the data from both the acute and prolonged GH regulation of gene expression underscore the importance of GH as a regulator of lipid metabolism.
In conclusion, the present study demonstrates that a single bout of GH induces gene expression of regulators of substrate metabolism and cellular growth in human skeletal muscle in vivo. While CISH gene expression seems to be directly induced by GH, our data suggest that the GH-induced increase in ANGPTL4 gene expression is linked to the concomitant increase in serum FFA levels, which sheds new light on the regulation and action of both ANGPTL4 and GH. This should encourage future mechanistic in vivo studies on the impact of GH on lipid metabolism in general and the link between GH, FFA, and ANGPTL4 in particular.
Supplementary Material
Acknowledgments
The authors acknowledge Hanne Steen for excellent technical assistance with the microarray analysis, and K. N. Rasmussen, H. F. Petersen, and E. S. Hornemann for excellent technical assistance with the human experiments. We thank Dr. Eduardo N. Chini, Mayo Clinic Rochester, for generously letting us conduct the cell cultural experiments in his laboratory.
Footnotes
Abbreviations:
- ANGPTL4
- angiopoietin-related protein-like 4
- CISH
- cytokine inducible SH2-containing protein
- GH
- growth hormone
- GHD
- GH deficiency
- IGF-1
- insulin-like growth factor 1
- NFκB1A
- nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor α
- OGTT
- oral glucose tolerance test
- PDK4
- pyruvate dehydrogenase kinase, isozyme 4
- PIK3C2B
- phosphoinositide-3-kinase, class 2, β polypeptide
- PRKAG2
- 5′-AMP-activated protein kinase, subunit γ-2
- SGK1
- serum/glucocorticoid-regulated kinase 1
- SOCS1–3
- suppressor of cytokine signaling 1–3
- TG
- triglyceride
This work was supported by Novo Nordisk via an unrestricted grant. J.O.L.J. has received unrestricted research grant and lecture fees from Pfizer Inc.
REFERENCES
- 1.Møller N., Jørgensen J. O. 2009. Effects of growth hormone on glucose, lipid, and protein metabolism in human subjects. Endocr. Rev. 30: 152–177 [DOI] [PubMed] [Google Scholar]
- 2.Møller N., Jørgensen J. O., Schmitz O., Møller J., Christiansen J., Alberti K. G., Ørskov H. 1990. Effects of a growth hormone pulse on total and forearm substrate fluxes in humans. Am. J. Physiol. 258: E86–E91 [DOI] [PubMed] [Google Scholar]
- 3.Merkel M., Eckel R. H., Goldberg I. J. 2002. Lipoprotein lipase: genetics, lipid uptake, and regulation. J. Lipid Res. 43: 1997–2006 [DOI] [PubMed] [Google Scholar]
- 4.Richelsen B., Pedersen S. B., Kristensen K., Børglum J. D., Nørrelund H., Christiansen J. S., Jørgensen J. O. 2000. Regulation of lipoprotein lipase and hormone-sensitive lipase activity and gene expression in adipose and muscle tissue by growth hormone treatment during weight loss in obese patients. Metabolism. 49: 906–911 [DOI] [PubMed] [Google Scholar]
- 5.Nikkila E. A., Pelkonen R. 1975. Serum lipids in acromegaly. Metabolism. 24: 829–838 [DOI] [PubMed] [Google Scholar]
- 6.Krag M. B., Gormsen L. C., Guo Z., Christiansen J. S., Jensen M. D., Nielsen S., Jørgensen J. O. 2007. Growth hormone-induced insulin resistance is associated with increased intramyocellular triglyceride content but unaltered VLDL-triglyceride kinetics. Am. J. Physiol. Endocrinol. Metab. 292: E920–E927 [DOI] [PubMed] [Google Scholar]
- 7.Nielsen S., Møller N., Christiansen J. S., Jørgensen J. O. 2001. Pharmacological antilipolysis restores insulin sensitivity during growth hormone exposure. Diabetes. 50: 2301–2308 [DOI] [PubMed] [Google Scholar]
- 8.Dominici F. P., Argentino D. P., Munoz M. C., Miquet J. G., Sotelo A. I., Turyn D. 2005. Influence of the crosstalk between growth hormone and insulin signalling on the modulation of insulin sensitivity. Growth Horm. IGF Res. 15: 324–336 [DOI] [PubMed] [Google Scholar]
- 9.del Rincon J. P., Iida K., Gaylinn B. D., McCurdy C. E., Leitner J. W., Barbour L. A., Kopchick J. J., Friedman J. E., Draznin B., Thorner M. O. 2007. Growth hormone regulation of p85alpha expression and phosphoinositide 3-kinase activity in adipose tissue: mechanism for growth hormone-mediated insulin resistance. Diabetes. 56: 1638–1646 [DOI] [PubMed] [Google Scholar]
- 10.Krusenstjerna-Hafstrøm T., Clasen B. F., Møller N., Jessen N., Pedersen S. B., Christiansen J. S., Jørgensen J. O. 2011. Growth hormone (GH)-induced insulin resistance is rapidly reversible: an experimental study in GH-deficient adults. J. Clin. Endocrinol. Metab. 96: 2548–2557 [DOI] [PubMed] [Google Scholar]
- 11.Nielsen C., Gormsen L. C., Jessen N., Pedersen S. B., Møller N., Lund S., Jørgensen J. O. 2008. Growth hormone signaling in vivo in human muscle and adipose tissue: impact of insulin, substrate background, and growth hormone receptor blockade. J. Clin. Endocrinol. Metab. 93: 2842–2850 [DOI] [PubMed] [Google Scholar]
- 12.Jessen N., Djurhuus C. B., Jørgensen J. O., Jensen L. S., Møller N., Lund S., Schmitz O. 2005. Evidence against a role for insulin-signaling proteins PI 3-kinase and Akt in insulin resistance in human skeletal muscle induced by short-term GH infusion. Am. J. Physiol. Endocrinol. Metab. 288: E194–E199 [DOI] [PubMed] [Google Scholar]
- 13.Krusenstjerna-Hafstrøm T., Madsen M., Vendelbo M. H., Pedersen S. B., Christiansen J. S., Møller N., Jessen N., Jørgensen J. O. 2011. Insulin and GH signaling in human skeletal muscle in vivo following exogenous GH exposure: impact of an oral glucose load. PLoS ONE. 6: e19392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jørgensen J. O., Jessen N., Pedersen S. B., Vestergaard E., Gormsen L., Lund S. A., Billestrup N. 2006. GH receptor signaling in skeletal muscle and adipose tissue in human subjects following exposure to an intravenous GH bolus. Am. J. Physiol. Endocrinol. Metab. 291: E899–E905 [DOI] [PubMed] [Google Scholar]
- 15.Vestergaard E. T., Gormsen L. C., Jessen N., Lund S., Hansen T. K., Møller N., Jørgensen J. O. 2008. Ghrelin infusion in humans induces acute insulin resistance and lipolysis independent of growth hormone signaling. Diabetes. 57: 3205–3210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vendelbo M. H., Jørgensen J. O., Pedersen S. B., Gormsen L. C., Lund S., Schmitz O., Jessen N., Møller N. 2010. Exercise and fasting activate growth hormone-dependent myocellular signal transducer and activator of transcription-5b phosphorylation and insulin-like growth factor-I messenger ribonucleic acid expression in humans. J. Clin. Endocrinol. Metab. 95: E64–E68 [DOI] [PubMed] [Google Scholar]
- 17.Cesena T. I., Cui T. X., Piwien-Pilipuk G., Kaplani J., Calinescu A. A., Huo J. S., Iniguez-Lluhi J. A., Kwok R., Schwartz J. 2007. Multiple mechanisms of growth hormone-regulated gene transcription. Mol. Genet. Metab. 90: 126–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sadowski C. L., Wheeler T. T., Wang L. H., Sadowski H. B. 2001. GH regulation of IGF-I and suppressor of cytokine signaling gene expression in C2C12 skeletal muscle cells. Endocrinology. 142: 3890–3900 [DOI] [PubMed] [Google Scholar]
- 19.Sadowski C. L., Choi T. S., Le M., Wheeler T. T., Wang L. H., Sadowski H. B. 2001. Insulin induction of SOCS-2 and SOCS-3 mRNA expression in C2C12 skeletal muscle cells is mediated by Stat5*. J. Biol. Chem. 276: 20703–20710 [DOI] [PubMed] [Google Scholar]
- 20.Sjögren K., Leung K. C., Kaplan W., Gardiner-Garden M., Gibney J., Ho K. K. 2007. Growth hormone regulation of metabolic gene expression in muscle: a microarray study in hypopituitary men. Am. J. Physiol. Endocrinol. Metab. 293: E364–E371 [DOI] [PubMed] [Google Scholar]
- 21.Flores-Morales A., Stahlberg N., Tollet-Egnell P., Lundeberg J., Malek R. L., Quackenbush J., Lee N. H., Norstedt G. 2001. Microarray analysis of the in vivo effects of hypophysectomy and growth hormone treatment on gene expression in the rat. Endocrinology. 142: 3163–3176 [DOI] [PubMed] [Google Scholar]
- 22.Viguerie N., Clement K., Barbe P., Courtine M., Benis A., Larrouy D., Hanczar B., Pelloux V., Poitou C., Khalfallah Y., et al. 2004. In vivo epinephrine-mediated regulation of gene expression in human skeletal muscle. J. Clin. Endocrinol. Metab. 89: 2000–2014 [DOI] [PubMed] [Google Scholar]
- 23.Ge H., Yang G., Huang L., Motola D. L., Pourbahrami T., Li C. 2004. Oligomerization and regulated proteolytic processing of angiopoietin-like protein 4. J. Biol. Chem. 279: 2038–2045 [DOI] [PubMed] [Google Scholar]
- 24.Kersten S., Lichtenstein L., Steenbergen E., Mudde K., Hendriks H. F., Hesselink M. K., Schrauwen P., Muller M. 2009. Caloric restriction and exercise increase plasma ANGPTL4 levels in humans via elevated free fatty acids. Arterioscler. Thromb. Vasc. Biol. 29: 969–974 [DOI] [PubMed] [Google Scholar]
- 25.Lichtenstein L., Kersten S. 2010. Modulation of plasma TG lipolysis by Angiopoietin-like proteins and GPIHBP1. Biochim. Biophys. Acta. 1801: 415–420 [DOI] [PubMed] [Google Scholar]
- 26.Adachi H., Kondo T., Koh G. Y., Nagy A., Oike Y., Araki E. 2011. Angptl4 deficiency decreases serum triglyceride levels in low-density lipoprotein receptor knockout mice and streptozotocin-induced diabetic mice. Biochem. Biophys. Res. Commun. 409: 177–180 [DOI] [PubMed] [Google Scholar]
- 27.Romeo S., Pennacchio L. A., Fu Y., Boerwinkle E., Tybjaerg-Hansen A., Hobbs H. H., Cohen J. C. 2007. Population-based resequencing of ANGPTL4 uncovers variations that reduce triglycerides and increase HDL. Nat. Genet. 39: 513–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanderson L. M., Degenhardt T., Koppen A., Kalkhoven E., Desvergne B., Muller M., Kersten S. 2009. Peroxisome proliferator-activated receptor beta/delta (PPARbeta/delta) but not PPARalpha serves as a plasma free fatty acid sensor in liver. Mol. Cell. Biol. 29: 6257–6267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoon J. C., Chickering T. W., Rosen E. D., Dussault B., Qin Y., Soukas A., Friedman J. M., Holmes W. E., Spiegelman B. M. 2000. Peroxisome proliferator-activated receptor gamma target gene encoding a novel angiopoietin-related protein associated with adipose differentiation. Mol. Cell. Biol. 20: 5343–5349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kersten S., Mandard S., Tan N. S., Escher P., Metzger D., Chambon P., Gonzalez F. J., Desvergne B., Wahli W. 2000. Characterization of the fasting-induced adipose factor FIAF, a novel peroxisome proliferator-activated receptor target gene. J. Biol. Chem. 275: 28488–28493 [DOI] [PubMed] [Google Scholar]
- 31.Kliewer S. A., Sundseth S. S., Jones S. A., Brown P. J., Wisely G. B., Koble C. S., Devchand P., Wahli W., Willson T. M., Lenhard J. M., et al. 1997. Fatty acids and eicosanoids regulate gene expression through direct interactions with peroxisome proliferator-activated receptors alpha and gamma. Proc. Natl. Acad. Sci. USA. 94: 4318–4323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Staiger H., Haas C., Machann J., Werner R., Weisser M., Schick F., Machicao F., Stefan N., Fritsche A., Haring H. U. 2009. Muscle-derived angiopoietin-like protein 4 is induced by fatty acids via peroxisome proliferator-activated receptor (PPAR)-delta and is of metabolic relevance in humans. Diabetes. 58: 579–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hessvik N. P., Bakke S. S., Fredriksson K., Boekschoten M. V., Fjorkenstad A., Koster G., Hesselink M. K., Kersten S., Kase E. T., Rustan A. C., et al. 2010. Metabolic switching of human myotubes is improved by n-3 fatty acids. J. Lipid Res. 51: 2090–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crunkhorn S., Dearie F., Mantzoros C., Gami H., da Silva W. S., Espinoza D., Faucette R., Barry K., Bianco A. C., Patti M. E. 2007. Peroxisome proliferator activator receptor gamma coactivator-1 expression is reduced in obesity: potential pathogenic role of saturated fatty acids and p38 mitogen-activated protein kinase activation. J. Biol. Chem. 282: 15439–15450 [DOI] [PubMed] [Google Scholar]
- 35.Robciuc M. R., Skrobuk P., Anisimov A., Olkkonen V. M., Alitalo K., Eckel R. H., Koistinen H. A., Jauhiainen M., Ehnholm C. 2012. Angiopoietin-like 4 mediates PPAR delta effect on lipoprotein lipase-dependent fatty acid uptake but not on beta-oxidation in myotubes. PLoS ONE. 7: e46212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Resmini E., Morte B., Sorianello E., Gallardo E., de Luna N., Illa I., Zorzano A., Bernal J., Webb S. M. 2011. Identification of novel GH-regulated genes in C2C12 cells. Horm. Metab. Res. 43: 919–930 [DOI] [PubMed] [Google Scholar]
- 37.Kawai M., Namba N., Mushiake S., Etani Y., Nishimura R., Makishima M., Ozono K. 2007. Growth hormone stimulates adipogenesis of 3T3-L1 cells through activation of the Stat5A/5B-PPARgamma pathway. J. Mol. Endocrinol. 38: 19–34 [DOI] [PubMed] [Google Scholar]
- 38.Basham B., Sathe M., Grein J., McClanahan T., D'Andrea A., Lees E., Rascle A. 2008. In vivo identification of novel STAT5 target genes. Nucleic Acids Res. 36: 3802–3818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Herrington J., Smit L. S., Schwartz J., Carter-Su C. 2000. The role of STAT proteins in growth hormone signaling. Oncogene. 19: 2585–2597 [DOI] [PubMed] [Google Scholar]
- 40.Flores-Morales A., Greenhalgh C. J., Norstedt G., Rico-Bautista E. 2006. Negative regulation of growth hormone receptor signaling. Mol. Endocrinol. 20: 241–253 [DOI] [PubMed] [Google Scholar]
- 41.Verdier F., Rabionet R., Gouilleux F., Beisenherz-Huss C., Varlet P., Muller O., Mayeux P., Lacombe C., Gisselbrecht S., Chretien S. 1998. A sequence of the CIS gene promoter interacts preferentially with two associated STAT5A dimers: a distinct biochemical difference between STAT5A and STAT5B. Mol. Cell. Biol. 18: 5852–5860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ram P. A., Waxman D. J. 1999. SOCS/CIS protein inhibition of growth hormone-stimulated STAT5 signaling by multiple mechanisms. J. Biol. Chem. 274: 35553–35561 [DOI] [PubMed] [Google Scholar]
- 43.Mathews L. S., Norstedt G., Palmiter R. D. 1986. Regulation of insulin-like growth factor I gene expression by growth hormone. Proc. Natl. Acad. Sci. USA. 83: 9343–9347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chia D. J., Rotwein P. 2010. Defining the epigenetic actions of growth hormone: acute chromatin changes accompany GH-activated gene transcription. Mol. Endocrinol. 24: 2038–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Falasca M., Maffucci T. 2012. Regulation and cellular functions of class II phosphoinositide 3-kinases. Biochem. J. 443: 587–601 [DOI] [PubMed] [Google Scholar]
- 46.Zhao J. T., Cowley M. J., Lee P., Birzniece V., Kaplan W., Ho K. K. 2011. Identification of novel GH-regulated pathway of lipid metabolism in adipose tissue: a gene expression study in hypopituitary men. J. Clin. Endocrinol. Metab. 96: E1188–E1196 [DOI] [PubMed] [Google Scholar]
- 47.Abbot E. L., McCormack J. G., Reynet C., Hassall D. G., Buchan K. W., Yeaman S. J. 2005. Diverging regulation of pyruvate dehydrogenase kinase isoform gene expression in cultured human muscle cells. FEBS J. 272: 3004–3014 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.