Abstract
Malignant breast tissue contains a rare population of multi-potent cells with the capacity to self-renew; these cells are known as cancer stem-like cells (CSCs) or tumor-initiating cells. Primitive mammary CSCs/progenitor cells can be propagated in culture as floating spherical colonies termed ‘mammospheres'. We show here that the expression of the autophagy protein Beclin 1 is higher in mammospheres established from human breast cancers or breast cancer cell lines (MCF-7 and BT474) than in the parental adherent cells. As a result, autophagic flux is more robust in mammospheres. We observed that basal and starvation-induced autophagy flux is also higher in aldehyde dehydrogenase 1-positive (ALDH1+) population derived from mammospheres than in the bulk population. Beclin 1 is critical for CSC maintenance and tumor development in nude mice, whereas its expression limits the development of tumors not enriched with breast CSCs/progenitor cells. We found that decreased survival in autophagy-deficient cells (MCF-7 Atg7 knockdown cells) during detachment does not contribute to an ultimate deficiency in mammosphere formation. This study demonstrates that a prosurvival autophagic pathway is critical for CSC maintenance, and that Beclin 1 plays a dual role in tumor development.
Keywords: breast cancer, autophagy, cancer stem-like/progenitor cell, Beclin 1
Introduction
Cancer stem cells are an exciting avenue for cancer studies, and offer a novel target for drug discovery. The theory of cancer stem-like cells (CSCs) or tumor-initiating cells posits that tumors are organized in a cellular hierarchy that is maintained by a subpopulation of cells displaying stem cell properties.1 These properties include self-renewal, which drives tumorigenesis and tumor growth, as well as differentiation, which generates the tumor bulk and contributes to cellular heterogeneity.2 Recent findings have shown that more decentralized tumors with cancer cells are able to switch between these different types.3 Accumulating evidence suggests that CSCs enhance tumorigenesis and drug resistance.4, 5, 6 CSCs were first identified in the hematopoietic system,7 but have subsequently been identified in many different types of solid tumors, including breast cancer.8, 9, 10 These initial studies show that cell-surface marker profiles, such as CD44high/CD24−/low ESA+, can be used to isolate subpopulations in which CSCs/progenitor cells have been enriched. Another candidate marker for the stem/progenitor cell population in both the normal mammary gland and mammary carcinomas is ALDH1 activity.11, 12 However, it is unreliable if only surface markers are used for CSC isolation. The abilities to regenerate xenograft tumors in the mammary fat pad of immunodeficient mice and to form mammospheres in serum-free medium were strongly suggested to characterize breast CSCs.13, 14
Macroautophagy (for convenience referred to simply as ‘autophagy' below) is an evolutionarily conserved lysosomal degradation pathway in which the cell ‘self-digests' its intracellular materials, and recycles its cytoplasmic components.15 Basal autophagy is a homeostatic process that allows cells to break down long-lived proteins, and is responsible for regulating organelle turnover. This function is essential for cell fitness, and limits the accumulation of reactive oxygen species. Under conditions of metabolic stress, including nutrient deprivation, autophagy is stimulated to provide the nutrients and energy the cell requires for survival.16 Moreover, selective autophagy helps the cell get rid of damaged organelles, toxic aggregation-prone proteins and invading microorganisms.17
Autophagy is induced by an initial membrane nucleation step that requires the ULK1 complex, and a class III phosphoinositide 3-kinase complex that includes Beclin 1.18 The isolation membrane selects its cargo and elongates until its edges fuse to form a double-membraned structure known as the autophagosome. Two ubiquitin-like conjugation systems forming Atg5–Atg12 and microtubule-associated protein 1 light chain 3-phosphatidylethanolamine (LC3 II), respectively, are necessary for this elongation of the isolation membrane to occur.19, 20 The autophagosome matures by fusing with endosomes and lysosomes, finally forming the autolysosome where cargo degradation occurs.21, 22 The mammalian target of rapamycin (mTOR) constitutes a central checkpoint that downregulates autophagy.23
Autophagy is upregulated by several tumor suppressor genes, and its impairment is associated with increased tumorigenesis.17, 24, 25 Beclin 1 is a haploinsufficient tumor-suppressor gene that can be either monoallelically deleted or display reduced expression in various cancers, including breast cancer. Heterozygous deletion of Becn1 in mice increases the incidence of spontaneous tumors.26, 27 Two possible mechanisms are currently proposed to explain how a defective survival mechanism can promote tumorigenesis. In normal cells, the coordinated induction of autophagy degrades damaged proteins, lipids and organelles, preventing a toxic buildup of cellular garbage in a p62-dependent manner, thus protecting the cell against changes in cell signaling, oxidative stress and DNA damage, thereby preventing chromosomal instability, genetic change and tumorigenesis.28 In the absence of autophagy, inflammation resulting from enhanced necrosis can stimulate tumorigenesis.29, 30 Paradoxically, autophagy may also promote the survival of starved tumor cells in regions of the tumor with a poor blood supply.29 Autophagy may also play a cytoprotective role during anticancer therapy.31
Here we analyze the autophagy capacities of breast CSCs in three-dimensional cultures known as mammospheres. We found that the autophagic flux was greater in CSC-enriched breast tumor cells than in non-CSC-enriched adherent ones, which correlated with high Beclin 1 expression in mammospheres. We also found that the maintenance and tumorigenicity of breast CSCs is dependent on Beclin 1 and autophagy. The roles of Beclin 1 and autophagy differ in CSCs/progenitor cells and in bulk cancer cells.
Results
Mammospheres display greater autophagic flux
To elucidate the basal and starvation-stimulated autophagic capacities of breast CSCs/progenitor cells, we used an in vitro system in which primitive mammary CSCs/progenitor cells can be propagated in culture as floating spherical colonies termed ‘mammospheres'. A single-cell suspension was obtained from either fresh human breast tumors of eight breast cancer patients (Table 1) or various breast cancer cell lines, including MCF-7, HMLER CD24low/− (HMLER CD24−) and BT474. These cells were cultured under adherent and mammosphere conditions (Supplementary Figure S1a, left). We subsequently analyzed the frequency of mammosphere generation using a well-established assay.32 Our data (Supplementary Figure S1a, right) indicate that mammospheres contain both the CSCs capable of self-renewal and progenitor cells, which is consistent with findings reported by Ponti et al.14 Limiting-dilution transplantation is a well-established assay used to characterize CSC cells.1 We therefore performed this assay, and compared the tumor-forming abilities of adherent cells and mammospheres in nude mice (Supplementary Figure S1b). Data indicated that mammosphere cells were at least 100 times more tumorigenic than adherent cells. The volume of the tumors produced by mammospheric cells was greater than that of tumors produced by adherent cells (Supplementary Figure S1c), indicating that the mammospheres contained more tumorigenic breast CSCs/progenitor cells than adherent cells. From these findings, we concluded that the mammosphere is an adequate model in which are enriched mammary CSCs/progenitor cells. We therefore used mammospheres to elucidate the autophagic capacities of mammary CSCs/progenitor cells.
Table 1. Patients and tumor characteristics.
| Case no. | Age | Diagnosis | Tumor size (cm2) |
Receptor status |
HER2 | Grade | Lymp node status | Clinical stage | Mammospheres per 1000 cells | |
|---|---|---|---|---|---|---|---|---|---|---|
| ER | PR | |||||||||
| 1 | 48 | IDC | 3.5 × 2.0 | (+) | (+) | (−) | III | N1 | II | 59 |
| 2 | 34 | IDC | 3.8 × 2.5 | (+) | (−) | (+) | II | N2 | III | 178 |
| 3 | 44 | IDC | 4.3 × 2.3 | (−) | (−) | (−) | III | N1 | II | 228 |
| 4 | 45 | IDC | 2.9 × 2.0 | (−) | (−) | (+) | III | N1 | II | 178 |
| 5 | 52 | IDC | 3.3 × 1.8 | (+) | (+) | (−) | III | N2 | III | 61 |
| 6 | 43 | IDC | 4.2 × 1.9 | (+) | (+) | (−) | III | N2 | III | 107 |
| 7 | 55 | IDC | 3.0 × 1.6 | (+) | (−) | (−) | III | N1 | II | 175 |
| 8 | 50 | IDC | 3.9 × 2.1 | (+) | (+) | (+) | III | N1 | II | 128 |
Abbreviations: ER, estrogen receptor; IDC, invasive ductal breast cancer; PR, progesterone receptor.
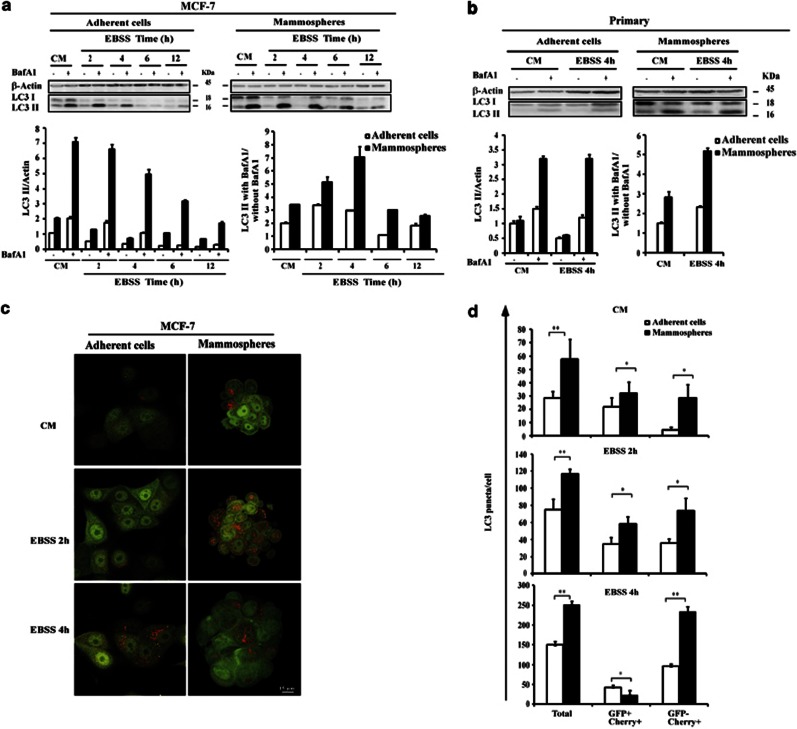
We analyzed the accumulation of LC3 II, the lipidated form of LC3 associated with the autophagosome membrane. The amount of LC3 II reflects the number of autophagosomes. Because the steady-state levels of autophagosomes are determined by synthesis and consumption in the lysosomal compartment, the effects on autophagy are best determined after clamping autophagosome consumption with bafilomycin A1 (Baf A1), an inhibitor of vacuolar-type H+-ATPase.33 The differences in the amount of LC3 II in the samples in the presence and absence of Baf A1 correspond to the amounts of LC3 II delivered to the lysosomes for degradation (that is, the autophagic flux) (Figure 1a). The amount of LC3 II markedly increased when MCF-7 mammospheres were used. We also determined the amount of autophagy cargo p62 that was delivered to lysosomes for degradation. We observed a higher rate of p62 degradation in mammospheres than in adherent MCF-7 cells (Supplementary Figure S2). Taken together, these data support the hypothesis that autophagosome synthesis is greater in mammospheres than in adherent cells, both at the basal level and following Earle's balanced salt solution (EBSS) treatment. The robust autophagic flux under basal conditions and during starvation was also observed in primary mammospheres (Figure 1b).
Figure 1.
Autophagic flux in mammospheres and adherent cells. (a) Immunoblot analysis of LC3 I and LC3 II levels in MCF-7 adherent cells (left) and mammospheres (right). Cells were incubated in complete medium (CM) or with EBSS for the time indicated in the presence or absence of Baf A1 (final concentration: 100 nℳ) for 2 h. (b) Immunoblot analysis of LC3 I and LC3 II levels in primary adherent cells (left) and primary mammospheres (right). Cells were incubated in CM or with EBSS for 4 h in the presence or absence of Baf A1 (final concentration: 100 nℳ) for 2 h. Actin immunoblotting was used as a loading control. The LC3 II/actin ratio was determined using the Bio1D software. Results are representative of three independent experiments. Control was set as 1 in adherent untreated cells. The autophagic flux determined as the ratio between the LC3 II levels with Baf A1 and without Baf A1 in (a) and (b) (bottom) is expressed in arbitrary units. (c) MCF-7-mCherry-GFP-LC3 adherent cells and mammospheres were incubated in CM or with EBSS for the times indicated, fixed and then visualized by confocal microscopy. Bars: 15 μm. (d) The number of GFP+/mCherry+(yellow) and GFP−/mCherry-LC3+ (red) dots was scored on ∼50 to 100 cells. The data are presented as means±s.d. from three independent experiments and analyzed using Student's t-test (*P<0.05, **P<0.01).
To confirm western blot analysis, changes in the cellular localization of the autophagosome marker LC3 were evaluated in cells stably transfected with GFP-LC3. At basal level, GFP-LC3 assays revealed diffuse staining in untreated MCF-7-GFP LC3 cells. A marked increase in the number of GFP-LC3 dots was displayed when MCF-7-GFP LC3 mammospheres were used. It should be noted that mammospheres were incubated overnight in the same medium as adherent cells before treatment. To block the lysosomal degradation of LC3 II, experiments were also performed in the presence of Baf A1. The accumulation of GFP-LC3 dots was considerably enhanced in mammospheres in the presence of Baf A1 (Supplementary Figure S2c). Statistical analysis indicates that at the basal level and under starvation conditions, autophagic flux was significantly higher in the mammospheres than in the adherent cells confirming the western blotting analysis (Supplementary Figure S2d).
To avoid using autophagic flux calculations based on Baf A1, we used MCF-7 cells stably transfected with mCherry-GFP-LC3 plasmid. We observed that mammospheres had more autophagosomes and more intense autophagic flux than adherent cells by analyzing the distribution of the mCherry-EGFP-LC3 fusion protein (Figure 1c). This fusion protein can be used to distinguish between autophagosomes (GFP+/mCherry+; yellow dots) and autolysosomes (GFP−/mCherry+; red dots), in which GFP fluorescence was quenched inside the acidic compartments.34 Statistical analysis indicates that, whatever medium is considered, the numbers of total LC3 puncta and autolysosomes were higher (two- and two- to six-fold, respectively) in the mammospheres than in the adherent cells (Figure 1d). Taken together, these data demonstrate that both basal autophagy and starvation-stimulated autophagy are robustly active in mammospheres.
Robust autophagic flux in mammospheres is correlated with CSC/progenitor phenotype
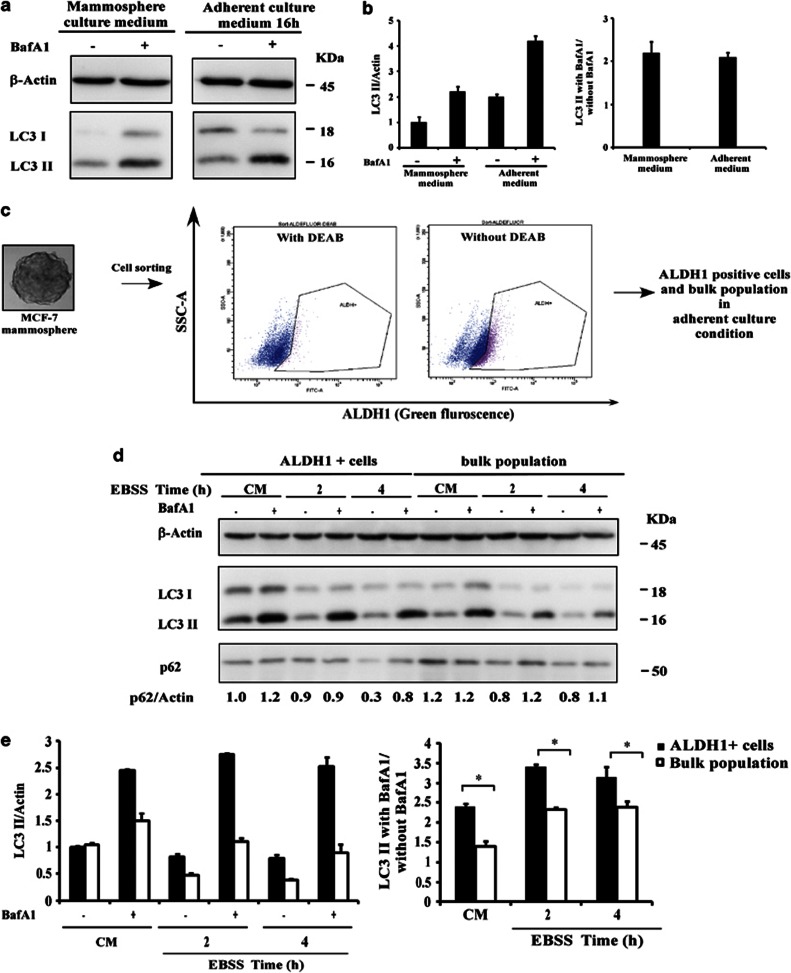
To find out whether the culture conditions were responsible for the upregulation of autophagy observed in mammospheres, we firstly tested the basal autophagic flux in mammospheres cultured under different conditions. Our data indicated that serum-free mammosphere medium did not affect the basal autophagic flux of the mammospheres, suggesting that the CSC/progenitor phenotype is responsible for the high level of autophagy (Figures 2a and b).
Figure 2.
Robust autophagic flux in mammospheres is correlated with the CSC/progenitor phenotype. (a) Immunoblot analysis of LC3 I and LC3 II levels in MCF-7 mammospheres. Cells were incubated in complete mammosphere medium or adherent medium for 16 h in the presence or absence of Baf A1 (final concentration: 100 nℳ) for 2 h. (b) The LC3 II/actin ratio and autophagy flux were determined as described in Figure 1c. ALDH1+ cells were isolated by fluorescence-activated cell sorting. ALDH1 activity in an MCF-7 mammosphere was measured by flow cytometry using the ALDEFLUOR reagent in the presence or absence of the ALDH1 inhibitor DEAB. Cells expressing ALDH1 were sorted using the ALDH1+ gate based on cells treated with the ALDH1 inhibitor, DEAB. ALDH1+ cells are collected, and then incubated in complete adherent medium. (d) Immunoblot analysis of p62, LC3 I and LC3 II levels in ALDH1+ cells (left) and bulk population mammospheres (right) were incubated in CM or with EBSS for the times indicated in the presence or absence of Baf A1 (final concentration: 100 nℳ) for 2 h. (e) The LC3 II/actin ratio and autophagy flux were determined as described in Figure 1.
Breast CSC cells were found to display properties including ALDH1 activity, which is used to isolate CSC/progenitor cells from malignant tissues and cell lines. We therefore used ALDEFLUOR assay to isolate CSC/progenitor cell population derived from MCF-7 mammospheres (Figure 2c). Western blot analysis of LC3 showed that in the presence of Baf A1, the amount of LC3 II accumulation was markedly increased when ALDH1+ cells were used (Figures 2d). We also determined the amount of the autophagy cargo p62 that was delivered to lysosomes for degradation. We observed a higher rate of p62 degradation in ALDH1+ cells than in the bulk population (Figure 2d). Taken together, these findings indicate that autophagosome synthesis is greater in ALDH1+ cells than in the bulk population, both at the basal level and following EBSS treatment.
From these findings, taken together, we conclude that a high level of autophagy is a hallmark of breast CSCs/progenitor cells.
Beclin 1 expression is upregulated in mammospheres
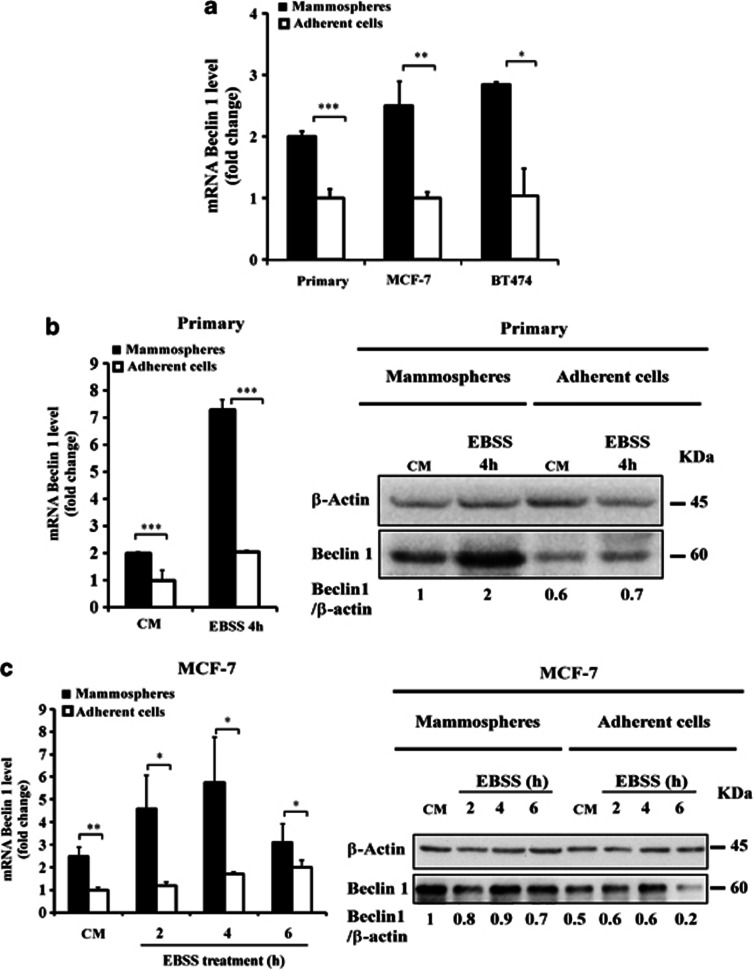
Autophagy is tightly regulated by the activity of the Beclin 1 complex in initiating the formation of autophagosomes.16 We therefore compared Beclin 1 expression in mammospheres and in adherent cells from primary breast cancer or MCF-7 and BT474 cell lines. Real-time quantitative polymerase chain reaction (RT–qPCR) analysis indicated that at the basal level, Beclin 1 mRNA was upregulated to a greater extent in primary mammospheres than in adherent cells (Figure 3a). Similar data were obtained for the MCF-7 and BT474 cell lines cultured under adherent and mammosphere conditions (Figure 3a). Beclin 1 mRNA level was also higher in primary mammospheres than in primary adherent cells under starvation conditions (Figure 3b, left). Similar data were obtained for the MCF-7 cell lines cultured under adherent and mammosphere conditions (Figure 3c, left). Western blot analysis indicated that at basal levels Beclin 1 protein expression was also upregulated to a greater extent in primary mammospheres than in adherent cells (Figure 3b, right). Beclin 1 protein level expression was still higher in primary mammospheres than in primary adherent cells under starvation conditions. Similar data were obtained for the MCF-7 cell lines (Figure 3c, right). From this finding, we concluded that mRNA and protein expression of Beclin 1 are both upregulated in breast CSCs/progenitor cells.
Figure 3.
Beclin 1 expression is upregulated in mammospheres.(a) The single-cell suspension from either fresh breast cancer samples or various different breast cancer cell lines was cultured under adherent or mammosphere conditions. The expression of the BECN1 gene in mammospheres and adherent cells was investigated using RT–qPCR. Results are expressed as the mean±s.d. for three independent determinations and analyzed using Student's t-test (*P<0.05, **P<0.01, ***P<0.001). The fold change was calculated relative to control adherent cells. (b) Primary adherent cells and primary mammospheres were incubated in complete medium (control) or with EBSS for 4 h, and then the expression level of the BECN1 gene was determined as in (a) using quantitative RT–qPCR (left). Results are expressed as described in (a). Immunoblot analysis of Beclin 1 levels in primary mammospheres and adherent cells (right). Control was set as in 1 in untreated cells. (c) MCF-7 mammospheres and adherent cells were incubated in complete medium or with EBSS for the times indicated, and then the expression of BECN1 gene was determined as in (a, left). Results are expressed as described in (a). Immunoblot analysis of Beclin 1 levels in MCF-7 mammospheres and adherent cells (right).
Starvation-induced autophagy in breast CSCs/progenitor cells is Beclin 1 dependent
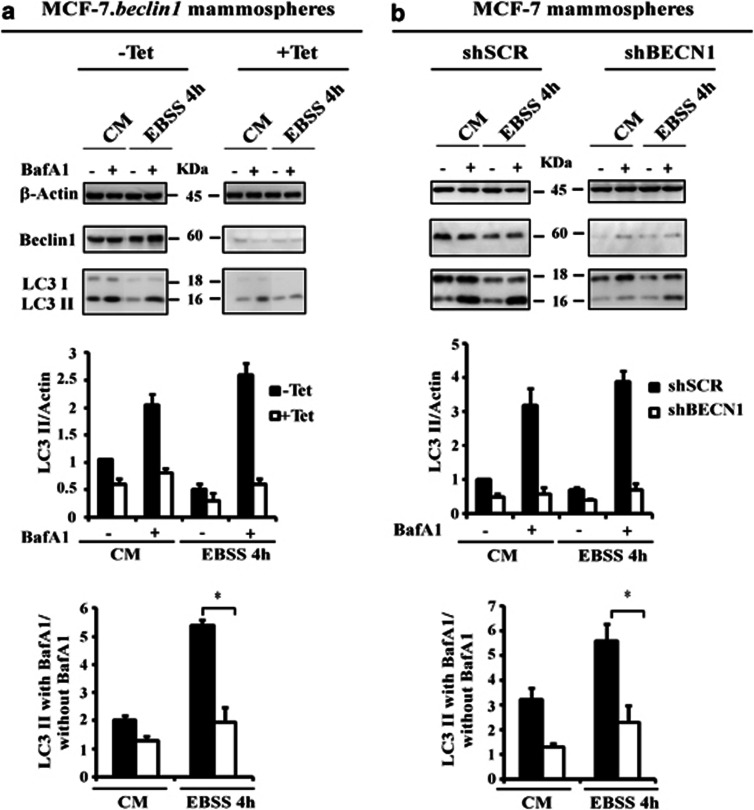
We next investigated the involvement of Beclin 1 in the regulation of autophagy in mammospheres. We first used MCF-7.beclin 1tet-off cells that express detectable endogenous levels of Beclin 1 to investigate the effect of Beclin 1 on starvation-induced autophagy in mammospheres. As expected, the autophagic flux was decreased (three fold) by downregulation of Beclin 1 expression (Figure 4a), indicating that Beclin 1 is required for autophagy to be stimulated by EBSS in mammospheres. We then compared the increase in the autophagic flux caused by EBSS in control mammospheres (control or pMKO.1-puro scrambled control short hairpin RNA (shSCR)) to that in mammospheres incubated in complete medium or with EBSS following short hairpin RNA (shRNA)-mediated silencing of Beclin 1. As expected, autophagic flux was lower (three fold) when Beclin 1 expression was reduced (Figure 4b).
Figure 4.
Starvation-induced autophagy in mammospheres is Beclin 1-dependent. (a) MCF-7.beclin 1tet-off adherent cells were cultured in the presence or absence of 2 μg/ml of tetracycline for 1 week. The single-cell suspension of cells from each condition was cultured under mammosphere conditions in the presence or absence of 2 μg/ml of tetracycline. Mammospheres were incubated in complete medium (CM) or with EBSS for indicated time in the presence or absence of Baf A1 (final concentration: 100 nℳ) for 2 h. Lysates were separated on sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) gels, and membranes were immunoblotted with the antibodies indicated (upper). The LC3 II/actin ratio and autophagic flux were determined as described in Figure 1 (bottom). (b) MCF-7 adherent cells were stably transfected with shRNA-mediated scramble (shSCR) or shRNA-mediated silencing of Beclin 1 (shBECN1). The single-cell suspension from each condition was cultured under mammosphere conditions and analyzed as indicated in Figure 4a. *P<0.05.
The degree of autophagy inhibition was also measured in the adherent cells and during the mammosphere assay (Supplementary Figure S3).Western blot analysis of LC3II accumulation shows that autophagic flux was lower (three fold) when Beclin 1 was reduced, whatever the test conditions.
Beclin 1 is critical for maintaining the proliferation of breast CSCs and progenitor cells
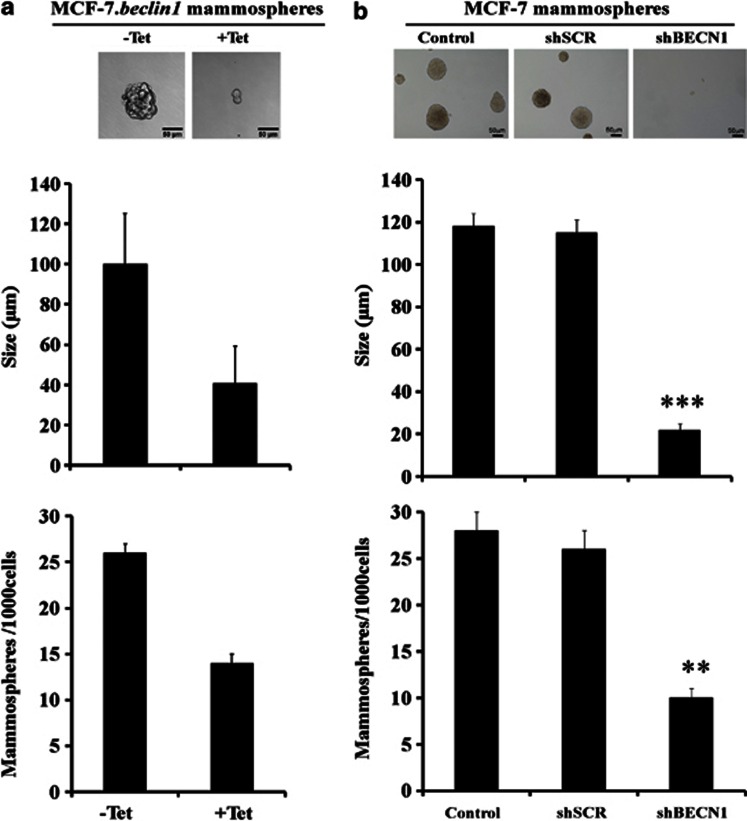
The number of mammospheres formed after serial passages at clonal density reflects the self-renewal of primitive CSC, whereas the size of the mammospheres reflects progenitor cell proliferation. To investigate the function of autophagy in the self-renewal potential of breast CSCs and progenitor cell proliferation, we first compared primary and secondary mammosphere formation in the presence and absence of tetracycline (Figure 5a). Surprisingly, we found that both the size (40 versus 100 μm) and number (15 versus 25) of mammospheres decreased when Beclin 1 expression was reduced.
Figure 5.
Beclin 1 is critical for the maintenance of breast CSCs and progenitor cells proliferation. (a) MCF-7.beclin 1tet-off adherent cells were cultured as in Figure 4a. Mammospheres from each condition were seeded at a single cell per well in a 96-well plate. Primary mammospheres were enzymatically dissociated with trypsin to obtain a single-cell suspension. Cells were then seeded at one single cell per well in a 96-well plate, and secondary mammospheres formed after 11 days. Representative phase-contrast photomicrographs of mammospheres formed after 11 days (upper). The number and the size of the mammospheres were quantified (bottom). Results are representative of three independent experiments. (b) MCF-7 adherent cells were stably transfected with empty pMKO.1 vector (control) or shRNA-mediated scramble (shSCR) or shRNA-mediated silencing of Beclin 1 (shBECN1). The single-cell suspension of cells from each condition was cultured under mammosphere conditions and analyzed as indicated in Figure 5a. **P<0.01; ***P<0.001.
We then investigated primary and secondary MCF-7 mammosphere formation following shRNA-mediated silencing of Beclin 1. Once again, we found that the size (20 versus 120 μm) and number of mammospheres (10 versus 27) fell when Beclin 1 expression was lower (pMKO.1-puro Beclin 1 shRNA (shBECN1); Figure 5b), suggesting that Beclin 1 is critical for maintaining the proliferation of breast CSCs and progenitor cells. To exclude an MCF-7-specific phenotype, in vitro self-renewal assays following shRNA-mediated silencing of Beclin 1 were carried out in two additional breast cancer cell lines, SKBR3 and SK-3rd (Supplementary Figure S4). The data obtained indicate that, whatever cell lines are used, Beclin 1 is always critical for maintaining the proliferation of breast CSCs and progenitor cells. It was worth noting that the mammospheres formed by SK-3rd cells are more numerous and larger than those formed by SKBR3 cells, confirming our previous findings as described in Ginestier et al.35
Beclin 1 is critical for breast CSC/progenitor cell tumorigenicity
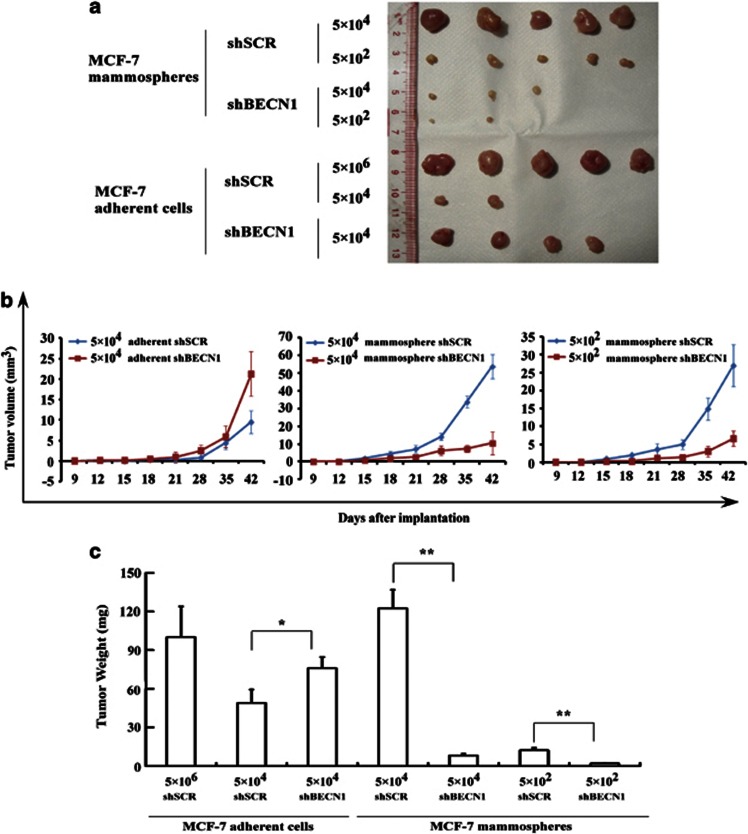
To evaluate the effect of Beclin 1 on the efficiency of xenograft formation of breast CSCs/progenitor cells, we analyzed the in vivo tumorigenicity of MCF-7 adherent cells in nude mice following the shRNA-mediated silencing of Beclin 1 (Figure 6). As a positive control, 100% of mice inoculated with 5 × 106 adherent MCF-7 cells (shSCR) generated tumors (Figure 6a). As expected, tumor volume (Figure 6b) and tumor weight (Figure 6c) were highest with adherent MCF-7 cells when Beclin 1 expression was lower (shSCR versus shBECN1, P=0.014; χ2-test). However, the incidence of tumor formation was lower when adherent shSCR MCF-7 cells were injected into nude mice (2/5 versus 4/5) (Table 2) than when adherent shBECN1 MCF-7 cells were injected, which is consistent with findings reported by Linag et al.36
Figure 6.
Beclin 1 is critical for mammosphere tumorigenicity. (a) MCF-7 adherent cells were stably transfected with shRNA-mediated scramble (shSCR) or shRNA-mediated silencing of Beclin 1 (shBECN1). The single-cell suspension corresponding to each condition was cultured under mammosphere conditions. Incidence of tumors induced by injecting the indicated number of adherent and mammospheric cells into nude mice. (b) Tumor volume and (c) tumor weight was determined as described in Materials and methods section. Tumor volume and weight from mammospheric MCF-7 shSCR cells were higher than in mammospheric MCF-7 shBECN1 cells. In contrast, adherent MCF-7 Beclin 1+ cells were less tumorigenic than MCF-7 beclin 1−/low cells. *P<0.05; **P<0.01.
Table 2. Incidence of tumors by MCF-7 mammosphere and adherent cells in nude mice following the shRNA-mediated silencing of Beclin 1.
| Number of cells inoculated | 5 × 106 | 5 × 104 | 5 × 102 |
|---|---|---|---|
| Mammosphere MCF-7 shSCR | 5/5 | 5/5 | |
| Mammosphere MCF-7 shBECN1 | 3/5 | 2/5 | |
| Adherent MCF-7 shSCR | 5/5 | 2/5 | |
| Adherent MCF-7 shBECN1 | 4/5 |
Abbreviations: shBECN1, Beclin 1 shRNA; shSCR, scrambled control shRNA; shRNA, short hairpin RNA.
We next analyzed the in vivo tumorigenicity of MCF-7 mammospheres in nude mice following the shRNA-mediated silencing of Beclin 1 (Figure 6). Consistent with the findings of the in vitro assay of self-renewal potential (Figure 5), tumor volume (Figure 6b) and tumor weight (Figure 6c) were higher with mammospheric MCF-7 cells when Beclin 1 expression was higher (shSCR versus shBECN1, P<0.01; χ2-test). The incidence of tumor formation was higher when mammospheric shSCR MCF-7 cells were injected into nude mice (5/5 versus 2/5) (Table 2) than when mammospheric shBECN1 MCF-7 cells were injected.
We also analyzed in vivo tumorigenicity of SK-3rd and SKBR3 adherent cells and mammospheres in nude mice following the shRNA-mediated silencing of Beclin 1. Tumor volume (Supplementary Figures S5a, b, d and e) was highest for both adherent SK-3rd and SKBR3 cells when Beclin 1 expression was low. The incidence of tumor formation was lower when both adherent SK-3rd- and SKBR3-shSCR cells were injected into nude mice (2/5 versus 3/5; 1/5 versus 2/5, respectively) (Supplementary Figures S5c and f) than when adherent shBECN1 cells were injected, which is consistent with the findings for MCF-7 cells.
In line with the tumorigenicity of MCF-7 cells, tumor volume (Supplementary Figures S5a, b, d and e) was higher with both mammospheric SK-3rd and SKBR3 cells when Beclin 1 expression was higher (shSCR versus shBECN1). The incidence of tumor formation when mammospheric shSCR cells were injected into nude mice was higher (Supplementary Figures S5c and f) than when mammospheric shBECN1 cells were injected. The data from these experiments indicate that whatever cell lines were used in our animal model, Beclin 1 was always critical for the tumorigenicity of breast CSCs/progenitor cells.
Autophagy is critical for the maintenance of breast CSCs
The interconnection between autophagy and maintenance/proliferation of breast CSCs/progenitor cells was further investigated using chemical inhibitors of autophagy (Supplementary Figure S6) and shRNA-dependent suppression of the expression of essential autophagy genes, such as ATG7 (Supplementary Figure S7a).
The Vps34 class III phosphatidylinositol 3-kinase complex plays an essential role in autophagosome formation. We found that the mammospheres were smaller (30 versus 120 μm) when they had been treated with 3-MA, a Vps34 class III phosphatidylinositol 3-kinase inhibitor, whereas the number of mammospheres formed remained significantly similar (Supplementary Figure S6), indicating that progenitor cells proliferation was dramatically inhibited by 3-MA, but that this agent did not affect the self-renewal of breast CSCs. 3-MA, which is used at very high concentrations to inhibit autophagy (usually 10 mℳ), has potential off-target effects that might complicate the interpretation of the results.37
Although phosphatidylinositol 3-kinase inhibitors block the formation of autophagosomes, the other major pharmacological inhibitors in experimental use block later stages of autophagy.38 Vinblastine, a microtubule-disrupting agent, inhibits autophagosome–lysosome fusion, a step in the pathway that requires microtubules. Baf A1 and chloroquine inhibit the final degradation of the autophagic cargo inside autolysosomes. We found that secondary mammosphere formation was totally abolished when the mammospheres were treated with vinblastine, Baf A1 and chloroquine (Supplementary Figure S6).
As both chemical inhibitors of autophagy and depletion of Beclin 1 could inhibit the maintenance/proliferation of breast CSCs/progenitor cells by mechanisms independent of their effects on autophagy, we knocked down the expression of another essential autophagy gene, ATG7, in MCF-7 cells, using specific shRNAs, before analyzing mammosphere formation. We found that both the number (20 versus 41) and the size of the mammospheres decreased significantly (88 versus 53 μm, P<0.001) when Atg7 expression was reduced (Supplementary Figure S7b and c). The degree of autophagy inhibition was also measured in the adherent cells and during the mammosphere assay (Supplementary Figure S7d and e). Western blot analysis of LC3 II accumulation shows that autophagy flux was always lower (2.5-fold) when Atg7 was reduced, whatever the test conditions.
Collectively, these findings indicate that Atg7 is critical for maintaining breast CSCs and progenitor cell proliferation.
Interestingly, it has been demonstrated that substratum detachment promotes autophagy as a survival mechanism.39 The interconnection between autophagy, an early time point following detachment and the number of breast CSCs/progenitor cells was further investigated using ALDEFLUOR assay. The effect of decreased survival during detachment could contribute to the ultimate deficiency observed in mammosphere formation, and so we analyzed the percentage of ALDH1+ cells among the MCF-7 shSCR and MCF-7 shATG7 cells before and after detaching the cells for 24 h. The data from these experiments show that there was no change in the percentage of ALDH1+ after detachment in either cell population (Supplementary Figure S8). These data support that maintaining CSCs by autophagy promotes the stem cell phenotype.
Discussion
Autophagy is upregulated by several tumor suppressor genes, and impaired autophagy is associated with increased tumorigenesis.17, 24, 25 Recent data suggest that suppressing autophagy inhibits the genesis and progression of mammary tumors in animals with an intact immune system.40 Hence, the precise role of autophagy during cancer progression and treatment is both tissue- and context-dependent. Breast CSCs promote both tumorigenesis and drug resistance. An appropriate CSC model is critical for analyzing autophagy and its involvement in CSC biology. In this study, we characterized mammospheres from patients and human breast cancer cell lines. Our data indicate that mammosphere cultures of breast cancer cells have higher stemness marker expression and also greater tumorigenicity than adherent cells in nude mice, indicating that the mammospheres contained more CSCs/progenitor cells than the parental adherent cultures, which is consistent with other studies of the growth of breast CSCs as mammospheres in three-dimensional cultures.13, 14 Although the ‘whole' mouse mammary and human breast tumors probably arise from a combination of ‘adherent' and ‘mammosphere'-like cells, CSCs play a critical role in ‘total' tumor progression. Statistical analysis indicates that both at basal level and under starvation conditions, autophagic flux was significantly higher in the mammospheres than in the adherent cells. Mammospheres are formed by growing breast cancer cells under low attachment conditions; hence, extracellular matrix detachment can induce robust autophagy in all cells during mammosphere formation. However, we observed that basal and starvation-induced autophagy flux was also higher in the ALDH1+ population derived from mammospheres than in the bulk population. These findings suggest that CSC/progenitor phenotype displays higher autophagic flux than the non-CSC/progenitor phenotype. Utilizing the side population from MCF-7 cells, Zhou et al.41 suggested that the PTEN/mTOR/STAT3 pathway is required for the maintenance of breast cancer stem cells.4 Activation of mTOR downregulates autophagy, and leads to the phosphorylation of eukaryotic initiation factor 4E binding protein 1, and of the 70-kDa polypeptide 1 ribosomal protein S6 kinase-1 (p70S6K), two proteins involved in protein synthesis. We monitored the activity of mTOR by immunodetecting the levels of phosphorylation of its substrates, 4E binding protein 1 and p70S6K, at Thr37/46 and Thr389, respectively (data not shown). The phosphorylation of 4E binding protein 1 and pS706K in the mammospheres was no lower than that in the adherent cells, indicating that the upregulation of the autophagic flux in the mammosphere was not due to mTOR inhibition. Our data are consistent with the absence of any evidence for mTOR regulation of normal or malignant mammary stem/progenitor cells as reported by Yue et al.42
We found that Beclin 1 expression was upregulated at the protein and mRNA levels in breast CSCs/progenitor cells. Beclin 1 is a potential target for miRNA miR-30a, and this miRNA could downregulate Beclin 1 expression, resulting in lower autophagic activity.43 We used RT–qPCR to compare miRNA miR-30a levels in the mammospheres and in the adherent cells, and did not observe any correlation between the upregulation of Beclin 1 expression and the downregulation of miR-30a in breast CSCs (data not shown).
Surprisingly, suppression of autophagy in breast CSCs/progenitor cells by a chemical inhibitor or Beclin 1/Atg7 depletion reduces both the size and formation efficiency of mammospheres. These data indicate that autophagy is required for CSCs to be maintained, which would be consistent with an essential regulator function of Atg7 in hematopoietic stem cell maintenance.44 However, an alternative explanation is that ATG knockdown cells exhibit increased anoikis when first introduced into the mammosphere assay, which ultimately results in mammosphere-deficiency phenotypes over extended periods. We did not observe any change in the percentage of ALDH1+ in either cell population after detachment. These findings rule out this hypothetical explanation, and suggest that autophagy is indeed required for stem cell maintenance and expansion.
Consistent with the tumor-suppressing function of Beclin 1 detected in studies using tumor cell lines in monolayer cultures and mouse models,26, 27, 36 Beclin 1 depletion in monolayer cultures was found to increase tumorigenesis. Despite these early data implicating Beclin 1 in the suppression of breast cancer, our data using mammospheres and nude mice models indicate that Beclin 1 depletion did not lead to tumor development. Furthermore, the suppression of autophagy in breast CSCs/progenitor cells by Beclin 1 depletion inhibited tumorigenesis. These findings suggest that autophagy may be protumorigenic in breast CSCs/progenitor cells. Our data are consistent with those reported by Wei et al.,45 indicating that the suppression of autophagy by the deletion of FIP200 (200-kDa FAK family interacting protein), an important regulator of autophagy in mammalian cells, inhibits mammary tumorigenesis.
Materials and methods
Antibodies and reagents
Anti-actin antibody and rabbit polyclonal anti-LC3B and anti-Atg7 antibodies were supplied by Sigma-Aldrich (St Louis, MO, USA). Anti-Beclin 1 antibody was from BD Biosciences PharMingen (San Jose, CA, USA). Baf A1 was from Sigma. EBSS and fetal bovine serum were from Invitrogen (Carlsbad, CA, USA). The ECLTM western blotting detection kit and donkey anti-rabbit antibody were purchased from Amersham Bioscience (Buckinghamshire, UK). Goat anti-mouse and swine anti-goat antibodies were obtained from Bio-Rad (Richmond, CA, USA) and Caltag (Burlingame, CA, USA).
Tumor specimens
Fresh tumor tissues were obtained from eight female breast cancer patients via an ultrasound-guided vacuum-assisted Vacora biopsy system (Bard Biopsy System, Tempe, AZ, USA), as approved by the Research Ethics Board at the Breast Tumor Center, Sun Yat-Sen Memorial Hospital. The patients were fully informed and consented to the collection of clinical samples (Table 1). All samples reached the laboratory within 20 min, and were immediately mechanically disaggregated and digested with collagenase as described elsewhere.8 Single-cell suspensions were obtained by filtering through a 40-μm filter.
Cell culture
Primary cells were cultured both as adhering cells in Dulbecco's modified Eagle's medium supplemented with 20% fetal bovine serum, and suspended in serum-free Dulbecco's modified Eagle's medium/F12 supplemented with B27 (1:50; Invitrogen), 20 ng/ml epidermal growth factor (BD Biosciences), 0.4% bovine serum albumin (Sigma) and 4 μg/ml insulin (Sigma), as previously described.14, 35, 46
Human breast cancer cell line MCF-7 and MCF-7-GFP/LC3 cells were kindly provided by M Jaattela (Institute of Cancer Biology, Copenhagen, Denmark), and cultured in RPMI supplemented with 2 mℳℒ-glutamine and 10% fetal bovine serum. MCF-7/tet-off cells stably transfected with beclin 1 (MCF-7.beclin 1tet-off ) were kindly provided by B Levine (Department of Microbiology, University of Texas Southwestern Medical Center, Dallas, TX, USA), and cultured in Dulbecco's modified Eagle's medium with 200 μg/ml hygromycin B. Beclin 1 expression was repressed by treatment for 3 days with 2 μg/ml tetracycline.
The human mammary epithelial cell line infected with a retrovirus carrying hTERT, SV40 large T and the oncogenic allele H-RasV12 (referred to as HMLER) was a generous gift from RA Weinberg Whitehead Institute, Massachusetts Institute of Technology. Epithelial–mesenchymal transition can promote stem cell phenotype in mammary epithelial cells.47 HMLER CD24−/low (CD24−) is a subclone that was established as described.48 This model is known to be rich for the ‘stemness' phenotype. HMLER CD24− cells were cultured as recommended by Lonza (Tebu, Le Perray en Yvelines, France). BT474 cells were obtained from ATCC (Manassas, VA, USA), and cultured and maintained according to ATCC instructions.
Mammosphere formation and frequency assay
Single cells were plated at 103 cells/ml in ultra-low attachment culture dishes in serum-free Dulbecco's modified Eagle's medium/F12 supplemented with B27 (1:50; Invitrogen), 20 ng/ml epidermal growth factor (BD Biosciences), 0.4% bovine serum albumin (Sigma) and 4 μg/ml insulin (Sigma), as described previously.14, 35, 46 To propagate mammospheres in vitro, primary mammospheres were enzymatically dissociated with 0.05% trypsin for 15 min at 37 °C to obtain a single-cell suspension, and then cultured in suspension to produce the next generation of mammospheres. Sphere formation was assessed 11 days after seeding the cells, and the percentage of mammospheres per well and the size of the mammospheres were analyzed. HMLER CD24low/− mammosphere culture was performed in ultra-low attachment plates in serum-free MEBM-2 (Lonza).
Xenograft tumor formation
All the parental adherent cells (MCF-7, SKBR3, SK-3rd and stably knocked down clones) were firstly enzymatically dissociated with trypsin to get single cells. These single cells were then cultured under adherent or mammosphere culture condition to generate adherent daughter or mammospheric cells, respectively. Adherent cells and mammosphere cultures were collected, enzymatically dissociated, washed in phosphate-buffered saline and resuspended in phosphate-buffered saline/Matrigel mixture (1:1 volume). A measure of 0.1 ml of this mixture was then implanted in the mammary fat pad of 5-week-old female Athymic Nude-Fox1nu mice (Harlan, France). The mice were maintained in individually ventilated cages (Tecniplast, Lyon, France) under constant temperature and humidity; all experiments were performed under laminar flow (Tecniplast). The mice received estradiol supplementation (0.4 mg/kg) every 7 days for 42 days after cell injection, and were observed and palpated for tumor appearance. Tumor growth was measured weekly using calipers. Tumor volume was determined using the standard formula: L × W2 × 0.52, where L and W are the longest and shortest diameters, respectively. All animal work was carried out according to the Guidelines of the United Kingdom Co-ordinating Committee on Cancer Research.
Detection of ALDH1+ cells
ALDH1 enzyme activity was detected using the ALDEFLUOR kit (Aldagen, Durham, NC, USA) according to the manufacturer's protocol. Briefly, cells (1 × 106/ml) were loaded with the enzyme substrate either alone or in the presence of the specific enzyme inhibitor diethylamino-benzyaldehyde (DEAB). After 30 min at 37 °C, the cells were analyzed using BD LSR Fortessa flow cytometry with 488-nm blue laser and 530/30 bandpass filter (green fluorescence). ALDH1+ cells were identifiable by having greater fluorescence than cells in which enzyme activity was inhibited by DEAB.
Isolation of ALDH1+ cells by fluorescence-activated cell sorting
Single MCF-7 cells were plated at 103 cells/ml in ultra-low attachment culture dishes in the mammosphere medium. After 7 days, ALDH activity in MCF-7 mammosphere (107 cells) was measured by flow cytometry using the ALDEFLUOR reagent in the presence or absence of DEAB. Cells expressing ALDH1 (ALDH1+ gate) were sorted on FACS Aria I flow cytometer with 488-nm blue laser and 530/30 bandpass filter. ALDH1+ cells were collected, and then plated in complete adherent medium. Cells were then analyzed as indicated in the legend of Figures 2c–e.
Western blot analysis
Western blot analysis was carried out as described previously.49 All experiments were performed at least three times. Representative autoradiograms are shown. The antibody dilutions were as follows: anti-LC3, 1:5000; anti-Beclin 1, 1:2500; and anti-β-actin, 1:20 000.
Generation of Beclin 1 shRNA-expressing retroviruses and transfection of breast cancer cells
The retroviral vector pMKO.1-puro was obtained from Invitrogen. The following phosphorylated and annealed small interfering RNA oligos against scrambled control shRNA or Beclin 1 shRNA were inserted into the AgeI and EcoRI sites of pMKO.1-puro: Beclin 1 shRNA1, forward, 5′-CCGGCTCAGGAGAGGAGCCATTTCTCGAGAAATGGCTCCTCTCCTGAGTTTTTG-3′ and reverse, 5′-AATTCAAAAACTCAGGAGAGGAGCCATTTCTCGAGAAATGGCTCCTCTCCTGAG-3′ scrambled control shRNA, forward 5′-CCGGGTTCTCCGAACGTGTCACGTCTCGAGACGTGACACGTTCGGAGAATTTTTG-3′ and reverse, 5′-AATTCAAAAAGTTCTCCGAACGTGTCACGTCTCGAGACGTGACACGTTCGGAGAA-3′.
The resultant vectors were designated as shSCR and shBECN1, respectively. A total of 2 × 106 HEK 293T cells were co-transfected with 1 μg of empty pMKO.1-puro, shSCR or shBECN1 and 1 μg of pCL-Ampho (Imgenex, San Diego, CA, USA), a vector encoding a replication-defective helper virus, using FuGENE 6 (Roche Diagnostics, Meylan, France). At 24 h after transfection, the supernatants, containing amphotropic retroviruses, were collected, filtered (0.45 μm), supplemented with Sequa-brene (final concentration, 8 μg/ml) and used for infection. A total of 1 × 106 MCF-7 cells were infected with the amphotropic retroviruses, and 24 h later, selection of the infected cells was started in the medium containing 500 ng/ml puromycin. The selected cell populations were subjected to western blotting with an anti-Beclin 1 antibody to investigate the silencing efficiency of the Beclin 1 shRNA.
Atg7 shRNA and noncoding shRNA lentiviral particles (Santa Cruz, Santa Cruz, CA, USA) were used to infect directly immortalized MCF-7 cells, and stable clones were selected using puromycin as described previously.50 The selected cell populations were subjected to western blotting with an anti-Atg7 antibody to investigate the silencing efficiency of the Atg7-shRNA.
GFP-LC3 or mCherry-GFP-LC3 assay
The assay was performed in MCF-7-GFP-LC3 cells (kindly provided by M Jaattela, Institute of Cancer Biology, Copenhagen, Denmark) or mCherry-GFP-LC3 cells (kindly provided by N Brady, German Cancer Research Center, Heidelberg, Germany). Autophagy was then measured by confocal counting of cells with GFP-LC3 puncta or GFP+/mCherry+ (yellow) and GFP−/mCherry-LC3+ (red) puncta.
Immunofluorescence and microscopy
Adherent cells were grown to 50% confluence on a coverslip. Mammospheres were cultured overnight in the same medium, and adherent cells and the corresponding mammospheres were then cultured in complete or starvation medium for the times indicated. The cells were washed twice with phosphate-buffered saline, and then fixed with freshly prepared 3.7% formaldehyde at 4 °C for 20 min. Cells were then washed in 1 × phosphate-buffered saline, and slides were mounted with a coverslip using the DAKO Mounting Medium. Cell images were obtained using an LSM 510 META confocal microscope (Zeiss, LE PECQ, France) equipped with an argon laser (wavelength excitation 488 nm) and a Plan-Apochromat × 63/1.4 oil objective lens. GFP fluorescence was collected with a 505 nm long-pass emission filter. The pinhole diameter was set at 1.0 Airy Unit, giving a 0.8 μm optical slice thickness. Stacks of images were collected every 0.37 μm along the z axis.
Since the spheres are three-dimensional specimens, optical sectioning was carried out along the XZ plane to get a Z stack for each specimen. Images were then imported, and further analyzed.
Quantitative evaluation and statistical analysis of data
The three-dimensional reconstructions of mammospheres captured by confocal microscopy were performed using the Imaris software package (Bitplane Scientific Software, St Paul, MN, USA).
To determine the number of GFP-LC3 vacuoles in three dimension, we performed surface rendering using the Imaris MeasurementPro module. After identifying the objects from their intensity and their size, the data was exported to MS Excel and further analyzed. At least 50–100 cells per sample were counted in duplicate samples per condition per experiment.
RT–qPCR analysis
Total RNA was extracted using the TriZOL reagent, according to the manufacturer's instructions (Invitrogen). RNA quality was checked by capillary electrophoresis using the Bioanalyzer 2100 device (Agilent Technologies, Massy, France). One microgram of total RNA was reverse-transcribed using oligo(dT) and random hexamer primers, using the iScript cDNA synthesis kit according to the manufacturer's recommendations (Bio-Rad). Quantitative real-time PCR was performed using 4 μl of 50-fold diluted cDNA in a final volume of 10 μl using the SsoFast EvaGreen supermix according to the manufacturer's instructions (Bio-Rad) in a CFX96 thermal cycler (Bio-Rad). Previously described PCR primers for the BECN1 gene target were used.51 The relative expression of the transcripts was normalized on the endogenous control gene ACTB using the delta-delta Cq calculation method.
Statistical analysis
Statistical analysis of the differences between the groups was performed using Student's t-test. P<0.05 was considered statistically significant.
Acknowledgments
We thank B Levine for providing us with the MCF-7.beclin 1 cell line, M Jäättelä and N Brady for providing the MCF-7 GFP-LC3 and the mCherry-GFP-LC3 cell lines, respectively. We thank A Hamaï for his careful reading of our manuscript. We thank N Modjtahedi for helpful discussions. This work was supported by grants from INSERM, Université Paris-Sud 11, PHC-PFCC programme français de cooperation avec la Chine, INCa, ANR, the National Natural Science Foundation of China (30830110, 30921140312, 30801376, 30972785, 81172524), 973 Projects from Ministry of Science and Technology of China (2010CB912800, 2011CB504203 , 2009CB521706), Development of Important New Drugs from Ministry of Health of China (2011ZX09102-010-02), Clinical Key Project of Public Health Administration of China, Sun Yat-sen University Training Project (11ykpy28,11ykzd12) and Key Laboratory of malignant tumor gene regulation and target therapy of Guangdong Higher Education Institutes, Sun Yat-sen University (KLB09001). SG was supported with Parker B Francis Fellowship in Respiratory Disease.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
Supplementary Material
References
- Clarke MF, Dick JE, Dirks PB, Eaves CJ, Jamieson CH, Jones DL, et al. Cancer stem cells—perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res. 2006;66:9339–9344. doi: 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- Clevers H. The cancer stem cell: premises, promises and challenges. Nat Med. 2011;17:313–319. doi: 10.1038/nm.2304. [DOI] [PubMed] [Google Scholar]
- Gupta PB, Fillmore CM, Jiang G, Shapira SD, Tao K, Kuperwasser C, et al. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell. 2011;146:633–644. doi: 10.1016/j.cell.2011.07.026. [DOI] [PubMed] [Google Scholar]
- Pardal R, Clarke MF, Morrison SJ. Applying the principles of stem-cell biology to cancer. Nat Rev Cancer. 2003;3:895–902. doi: 10.1038/nrc1232. [DOI] [PubMed] [Google Scholar]
- Clement V, Sanchez P, de Tribolet N, Radovanovic I, Ruiz i Altaba A. HEDGEHOG-GLI1 signaling regulates human glioma growth, cancer stem cell self-renewal, and tumorigenicity. Curr Biol. 2007;17:165–172. doi: 10.1016/j.cub.2006.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangemi R, Paleari L, Orengo AM, Cesario A, Chessa L, Ferrini S, et al. Cancer stem cells: a new paradigm for understanding tumor growth and progression and drug resistance. Curr Med Chem. 2009;16:1688–1703. doi: 10.2174/092986709788186147. [DOI] [PubMed] [Google Scholar]
- Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- Kim CF, Jackson EL, Woolfenden AE, Lawrence S, Babar I, Vogel S, et al. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell. 2005;121:823–835. doi: 10.1016/j.cell.2005.03.032. [DOI] [PubMed] [Google Scholar]
- Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charafe-Jauffret E, Ginestier C, Iovino F, Tarpin C, Diebel M, Esterni B, et al. Aldehyde dehydrogenase 1-positive cancer stem cells mediate metastasis and poor clinical outcome in inflammatory breast cancer. Clin Cancer Res. 2010;16:45–55. doi: 10.1158/1078-0432.CCR-09-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dontu G, Al-Hajj M, Abdallah WM, Clarke MF, Wicha MS. Stem cells in normal breast development and breast cancer. Cell Prolif. 2003;36 (Suppl 1:59–72. doi: 10.1046/j.1365-2184.36.s.1.6.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponti D, Costa A, Zaffaroni N, Pratesi G, Petrangolini G, Coradini D, et al. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res. 2005;65:5506–5511. doi: 10.1158/0008-5472.CAN-05-0626. [DOI] [PubMed] [Google Scholar]
- Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- Mehrpour M, Esclatine A, Beau I, Codogno P. Overview of macroautophagy regulation in mammalian cells. Cell Res. 2010;20:748–762. doi: 10.1038/cr.2010.82. [DOI] [PubMed] [Google Scholar]
- Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nat Rev Cancer. 2007;7:961–967. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- Mizushima N. Autophagy in protein and organelle turnover. Cold Spring Harb Symp Quant Biol. 2011;LXXVI:3. doi: 10.1101/sqb.2011.76.011023. [DOI] [PubMed] [Google Scholar]
- Weidberg H, Shvets E, Elazar Z. Biogenesis and cargo selectivity of autophagosomes. Annu Rev Biochem. 2011;80:125–156. doi: 10.1146/annurev-biochem-052709-094552. [DOI] [PubMed] [Google Scholar]
- Kroemer G, Jaattela M. Lysosomes and autophagy in cell death control. Nat Rev Cancer. 2005;5:886–897. doi: 10.1038/nrc1738. [DOI] [PubMed] [Google Scholar]
- Yorimitsu T, Klionsky DJ. Autophagy: molecular machinery for self-eating. Cell Death Differ. 2005;12 (Suppl 2:1542–1552. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codogno P, Meijer AJ. Autophagy and signaling: their role in cell survival and cell death. Cell Death Differ. 2005;12 (Suppl 2:1509–1518. doi: 10.1038/sj.cdd.4401751. [DOI] [PubMed] [Google Scholar]
- Hippert MM, O'Toole PS, Thorburn A. Autophagy in cancer: good, bad, or both. Cancer Res. 2006;66:9349–9351. doi: 10.1158/0008-5472.CAN-06-1597. [DOI] [PubMed] [Google Scholar]
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci USA. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikic I, Johansen T, Kirkin V. Selective autophagy in cancer development and therapy. Cancer Res. 2010;70:3431–3434. doi: 10.1158/0008-5472.CAN-09-4027. [DOI] [PubMed] [Google Scholar]
- Mathew R, White E. Autophagy in tumorigenesis and energy metabolism: friend by day, foe by night. Curr Opin Genet Dev. 2011;21:113–119. doi: 10.1016/j.gde.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E, DiPaola RS. The double-edged sword of autophagy modulation in cancer. Clin Cancer Res. 2009;15:5308–5316. doi: 10.1158/1078-0432.CCR-07-5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau K, Luo S, Rubinsztein DC. Cytoprotective roles for autophagy. Curr Opin Cell Biol. 2010;22:206–211. doi: 10.1016/j.ceb.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds BA, Weiss S. Clonal and population analyses demonstrate that an EGF-responsive mammalian embryonic CNS precursor is a stem cell. Dev Biol. 1996;175:1–13. doi: 10.1006/dbio.1996.0090. [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC, Cuervo AM, Ravikumar B, Sarkar S, Korolchuk V, Kaushik S, et al. In search of an ‘autophagomometer'. Autophagy. 2009;5:585–589. doi: 10.4161/auto.5.5.8823. [DOI] [PubMed] [Google Scholar]
- Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, et al. Let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- Wu YT, Tan HL, Shui G, Bauvy C, Huang Q, Wenk MR, et al. Dual role of 3-methyladenine in modulation of autophagy via different temporal patterns of inhibition on class I and III phosphoinositide 3-kinase. J Biol Chem. 2010;285:10850–10861. doi: 10.1074/jbc.M109.080796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung C, Lock R, Gao S, Salas E, Debnath J. Induction of autophagy during extracellular matrix detachment promotes cell survival. Mol Biol Cell. 2008;19:797–806. doi: 10.1091/mbc.E07-10-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H, Wei S, Gan B, Peng X, Zou W, Guan J-L. Suppression of autophagy by FIP200 deletion inhibits mammary tumorigenesis. Genes Dev. 2011;25:1510–1527. doi: 10.1101/gad.2051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wulfkuhle J, Zhang H, Gu P, Yang Y, Deng J, et al. Activation of the PTEN/mTOR/STAT3 pathway in breast cancer stem-like cells is required for viability and maintenance. Proc Natl Acad Sci USA. 2007;104:16158–16163. doi: 10.1073/pnas.0702596104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkaya H, Paulson A, Charafe-Jauffret E, Ginestier C, Brown M, Dutcher J, et al. Regulation of mammary stem/progenitor cells by PTEN/Akt/beta-catenin signaling. PLoS Biol. 2009;7:e1000121. doi: 10.1371/journal.pbio.1000121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Wu H, Liu X, Li B, Chen Y, Ren X, et al. Regulation of autophagy by a beclin 1-targeted microRNA, miR-30a, in cancer cells. Autophagy. 2009;5:816–823. doi: 10.4161/auto.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen M, Soilleux EJ, Djordjevic G, Tripp R, Lutteropp M, Sadighi-Akha E, et al. The autophagy protein Atg7 is essential for hematopoietic stem cell maintenance. J Exp Med. 2011;208:455–467. doi: 10.1084/jem.20101145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H, Wei S, Gan B, Peng X, Zou W, Guan JL. Suppression of autophagy by FIP200 deletion inhibits mammary tumorigenesis. Genes Dev. 2011;25:1510–1527. doi: 10.1101/gad.2051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel AP, Lievre M, Thomas C, Hinkal G, Ansieau S, Puisieux A. Generation of breast cancer stem cells through epithelial–mesenchymal transition. PLoS One. 2008;3:e2888. doi: 10.1371/journal.pone.0002888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Bauvy C, Souquere S, Tonelli G, Liu L, Zhu Y, et al. The Bcl-2 homology domain 3 mimetic gossypol induces both Beclin 1-dependent and Beclin 1-independent cytoprotective autophagy in cancer cells. J Biol Chem. 2010;285:25570–25581. doi: 10.1074/jbc.M110.118125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghavami S, Mutawe MM, Schaafsma D, Yeganeh B, Unruh H, Klonisch T, et al. Geranylgeranyl transferase 1 modulates autophagy and apoptosis in human airway smooth muscle. Am J Physiol Lung Cell Mol Physiol. 2012;302:L420–L428. doi: 10.1152/ajplung.00312.2011. [DOI] [PubMed] [Google Scholar]
- Djavaheri-Mergny M, Amelotti M, Mathieu J, Besancon F, Bauvy C, Souquere S, et al. NF-kappaB activation represses tumor necrosis factor-alpha-induced autophagy. J Biol Chem. 2006;281:30373–30382. doi: 10.1074/jbc.M602097200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.