Abstract
Recent investigations have demonstrated a complex interrelationship between autophagy and cell death. A common mechanism of cell death in liver injury is tumor necrosis factor (TNF) cytotoxicity. To better delineate the in vivo function of autophagy in cell death, we examined the role of autophagy in TNF-induced hepatic injury. Atg7Δhep mice with a hepatocyte-specific knockout of the autophagy gene atg7 were generated and cotreated with D-galactosamine (GalN) and lipopolysaccharide (LPS). GalN/LPS-treated Atg7Δhep mice had increased serum alanine aminotransferase levels, histological injury, numbers of TUNEL (terminal deoxynucleotide transferase-mediated deoxyuridine triphosphate nick end-labeling)-positive cells and mortality as compared with littermate controls. Loss of hepatocyte autophagy similarly sensitized to GalN/TNF liver injury. GalN/LPS injury in knockout animals did not result from altered production of TNF or other cytokines. Atg7Δhep mice had accelerated activation of the mitochondrial death pathway and caspase-3 and -7 cleavage. Increased cell death did not occur from direct mitochondrial toxicity or a lack of mitophagy, but rather from increased activation of initiator caspase-8 causing Bid cleavage. GalN blocked LPS induction of hepatic autophagy, and increased autophagy from beclin 1 overexpression prevented GalN/LPS injury. Autophagy, therefore, mediates cellular resistance to TNF toxicity in vivo by blocking activation of caspase-8 and the mitochondrial death pathway, suggesting that autophagy is a therapeutic target in TNF-dependent tissue injury.
Keywords: apoptosis, autophagy, caspase-8, lipopolysaccharide, tumor necrosis factor
The relationship between the lysosomal, degradative pathway of macroautophagy and cell death remains unclear. Recent studies increasingly support the concept that autophagy functions to prevent rather than promote cell death.1 Autophagy increases in cultured cells in response to cellular stressors, including death-inducing stimuli. However, it is often unclear whether the increase in autophagy promotes cell death, or is an ineffective or partially protective survival response. Autophagy has been implicated as a mechanism of cell death from a variety of agents ranging from radiation to viruses.2, 3 In contrast, macroautophagy has been demonstrated to mediate survival from nutrient deprivation, ischemia/reperfusion injury and endoplasmic reticulum stress.4, 5, 6 Many of these investigations have been conducted in cultured cells, and the effects of autophagy on cell death in vivo have not been delineated. In the liver, a dramatic increase in hepatocyte autophagy occurs with starvation, but little is known about how hepatic levels of autophagy are altered in response to injurious stimuli or modulate hepatocyte cell death in vivo.7
The tumor necrosis factor (TNF) death pathway is a common mechanism of cell death in a variety of forms of tissue injury, including in the liver.8 In certain pathophysiological settings, such as toxin-induced liver injury, TNF death pathway activation occurs during which initiator caspase-8 is activated and cleaves the pro-apoptotic Bcl-2 family member Bid into truncated or tBid. Mitochondrial translocation of tBid triggers the mitochondrial death pathway with release of cytochrome c and other factors that activate downstream effector caspases to induce hepatocyte apoptosis.8 Critical hepatocyte events in this death cascade are the inhibition of nuclear factor-κB (NF-κB) signaling that leads to c-Jun N-terminal kinase (JNK) and caspase-8 overactivation.9, 10, 11 The ability of autophagy to selectively target damaged mitochondria for degradation in cultured hepatocytes12 suggests a potential role for autophagy in resisting hepatocellular death induced by TNF mitochondrial death pathway activation in vivo.
A well-established in vivo model of TNF-dependent liver injury is that resulting from 𝒟-galactosamine (GalN) and lipopolysaccharide (LPS) cotreatment.13 Subtoxic doses of the hepatocyte-specific toxin GalN sensitize hepatocytes to death from LPS-induced TNF. To determine the role of autophagy in TNF-mediated hepatocyte death in vivo, we generated an inducible, hepatocyte-specific mouse knockout of the critical autophagy gene atg7, and examined the effects of a loss of autophagy on GalN/LPS liver injury. Inhibition of autophagy increased liver injury and hepatocyte apoptosis by amplifying caspase-8 and mitochondrial death pathway activation. GalN also blocked the normal hepatic induction of autophagy in response to LPS, and a genetic increase in autophagy blocked GalN/LPS injury. These findings demonstrate a critical function for autophagy in hepatocyte resistance to TNF-mediated cell death in vivo, and indicate that levels of autophagy may be an important determinant of the extent of tissue injury from TNF toxicity.
Results
Generation of an inducible hepatocyte-specific atg7 knockout mouse
A global knockout of the autophagy gene atg5 or atg7 results in post-natal lethality.14, 15 Therefore, the in vivo effects of a loss of autophagy must be investigated in Cre-lox knockout mice. Hepatocyte atg7 has been knocked out shortly after birth by a constitutively expressed albumin promoter-driven Cre.16 However, spontaneous hepatomegaly and concomitant hepatitis develop rapidly after birth in these mice, making this model unsuitable for investigations of liver injury. Other studies have employed an inducible atg7 knockout using the Mx1-driven Cre recombinase;15 however, this promoter is neither hepatocyte nor liver specific.17 To inhibit hepatocyte autophagy in adult mice, a tamoxifen-inducible, albumin promoter-driven knockout of atg7 was generated by crossing Atg7F/F mice15 with ERt-albumin-Cre mice18 to produce ERt-albumin-Cre-Atg7F/F or Atg7Δhep mice. Atg7 protein was present in the livers of both Cre-negative and -positive uninjected mice, and was absent in tamoxifen-injected Cre-positive, but not Cre-negative, livers by immunoblotting (Supplementary Figure 1S). Tamoxifen induced a hepatocyte-specific knockout of Atg7 as indicated by the selective loss of Atg7, its downstream conjugate Atg5/12 and microtubule-associated protein 1 light chain 3 (LC3)-II along with a compensatory increase in LC3-I in the liver, but not in the kidney (Supplementary Figure 1S) or other organs (data not shown). The livers of Atg7Δhep mice had no evidence of liver injury for 2 weeks following tamoxifen injection, as indicated by normal serum alanine aminotransferase (ALT) levels (Figure 1a) and histology (Figure 1b).
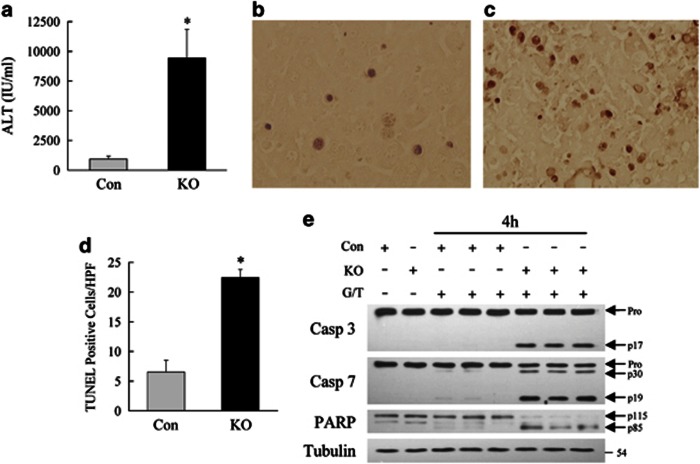
Figure 1.
Inhibition of hepatocyte autophagy increases GalN/LPS liver injury. (a) Serum ALT levels in littermate controls (Con) and Atg7Δhep (KO) mice untreated (0 h) and at 1 and 2 h after GalN/LPS administration (*P<0.001 as compared with control mice; n=9–10). (b) Histological grade of liver injury in untreated and GalN/LPS-treated mice (*P<0.03 as compared with control mice; n=5–7). (c) Serum ALT levels at 4 and 6 h (*P<0.0001 as compared with control mice; n=5–10). (d) Numbers of TUNEL-positive cells per high power field (HPF; *P<0.05; **P<0.006; n=3–8). (e) Histological grade of hepatic inflammation (*P<0.02 as compared with control mice; n=5–7). (f) Survival curve after GalN/LPS treatment (P<0.001; n=14–31)
Inhibition of hepatocyte autophagy sensitizes to TNF-dependent liver injury
To determine the function of autophagy in TNF-mediated organ injury, TNF-dependent liver injury from cotreatment with the hepatotoxin GalN and LPS was examined in Atg7Δhep mice. Littermate control and Atg7Δhep mice were tamoxifen injected and administered GalN/LPS 5 days later. GalN/LPS liver injury is normally first detectable at 4 h, becomes significant at 6 h and causes lethality after 6 h. Consistent with this time course of injury, serum ALTs in GalN/LPS-treated littermate controls were normal at 1 and 2 h (Figure 1a), minimally increased at 4 h and markedly elevated by 6 h (Figure 1c). In contrast, serum ALTs in Atg7Δhep mice were increased within 1 h of treatment and significantly elevated over those in littermate controls at all time points (Figures 1a and c). Blinded, semi-quantitative, histological grading confirmed that Atg7Δhep mice had significant liver injury within 1 h, and that their injury was greater than that in control mice at all times (Figures 1b, 2a and b). Increased injury was associated with greater cell death in knockout mouse livers. Terminal deoxynucleotide transferase-mediated deoxyuridine triphosphate nick end-labeling (TUNEL) staining revealed increased numbers of TUNEL-positive cells in the livers of Atg7Δhep mice at all times with the most pronounced effects at 4 and 6 h (Figures 1d and 2c–f). In contrast to findings for injury and death, the histological degree of inflammation was equivalent with the exception of an increase in knockouts at 1 h, a time when injury had not yet occurred in control mice (Figure 1e).
Figure 2.
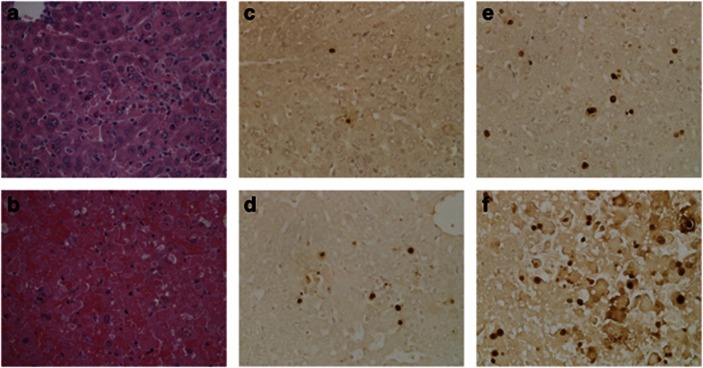
Knockout livers have increased histological liver injury and TUNEL staining. Hematoxylin and eosin stained liver sections from control (a) and knockout (b) mice 4 h after GalN/LPS administration. TUNEL staining in control (c, e) and knockout (d, f) livers at 2 h (c, d) and 4 h (e, f) after GalN/LPS. Magnifications for all, × 400
GalN/LPS-treated Atg7Δhep mice have increased mortality
GalN/LPS injury can trigger fulminant hepatic failure and death. To determine whether an inhibition of autophagy affected mortality, long-term survival after GalN/LPS was examined. Within 6 h of GalN/LPS administration, 50% of the Atg7Δhep mice had died, whereas all control mice were still alive (Figure 1f). Almost all Atg7Δhep mice were dead within 24 h, but 30% of control mice survived for 48 h (Figure 1f). An inhibition of hepatocyte autophagy, therefore, led to both increased liver injury and mortality from GalN/LPS.
Loss of autophagy did not alter macrophage cytokine production
GalN/LPS liver injury results from GalN-mediated hepatocyte sensitization to cytotoxicity from LPS-induced TNF.13 The fact that the studies were performed in a hepatocyte-specific autophagy knockout suggested that the likely mechanism of increased GalN/LPS injury was decreased hepatocyte resistance to TNF toxicity. However, to exclude the possibility of an effect on macrophage-produced TNF, hepatic cytokine induction by GalN/LPS was assayed by real-time PCR. TNF mRNA induction after GalN/LPS treatment was equivalent in control and knockout mice (Figure 3a). In addition, TNF-receptor type I protein levels were unchanged in the knockout mice (Figure 3b). LPS-induced mRNA levels of interferon-γ (Figure 3c), interleukin (IL)-6 (Figure 3d) and IL-1β (Figure 3e) were similarly unaffected by the loss of hepatocyte autophagy. Increased liver injury and mortality in Atg7Δhep mice did not result from altered LPS cytokine induction, consistent with a hepatocyte mechanism for the increase in injury.
Figure 3.
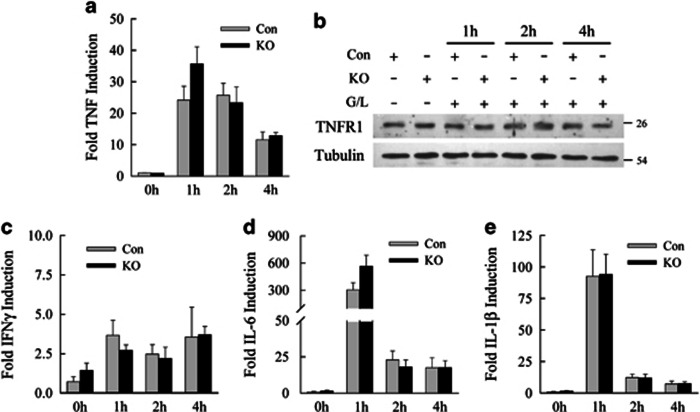
Cytokine induction by GalN/LPS is equivalent in control (Con) and knockout (KO) mice. (a) Relative TNF mRNA levels determined by real-time PCR in Con and KO mice at the indicated hours after GalN/LPS administration (n=5-6). (b) Immunoblots of total liver protein from Con and KO mice untreated or treated with GalN/LPS (G/L) for the indicated times, and probed for TNF-receptor type 1 (TNFR1) and tubulin as a loading control. Relative mRNA levels in the same mice for IFNγ (c), IL-6 (d) and IL-1β (e) (n=5–6)
Atg7Δhep mice have accelerated mitochondrial death pathway activation
Hepatic toxicity from GalN/LPS occurs through the classical TNF death cascade of caspase-8-mediated Bid cleavage, mitochondrial death pathway activation, cytochrome c release, effector caspase cleavage and apoptosis. TUNEL evidence of increased apoptosis in Atg7Δhep mice (Figure 1d) suggested that the loss of autophagy promoted death through this pathway. An examination of mitochondrial death pathway activation demonstrated accelerated activation in Atg7Δhep mice with increased levels of tBid, the active truncated form of Bid that triggers the mitochondrial death pathway in hepatocytes19 and decreased levels of cytochrome c in mitochondrial protein isolates of knockout mice 4 h after GalN/LPS (Figure 4a). Increased levels of tBid and cytochrome c were also detected at 4 h in the cytosolic fractions from knockout mice (Figure 4b).
Figure 4.
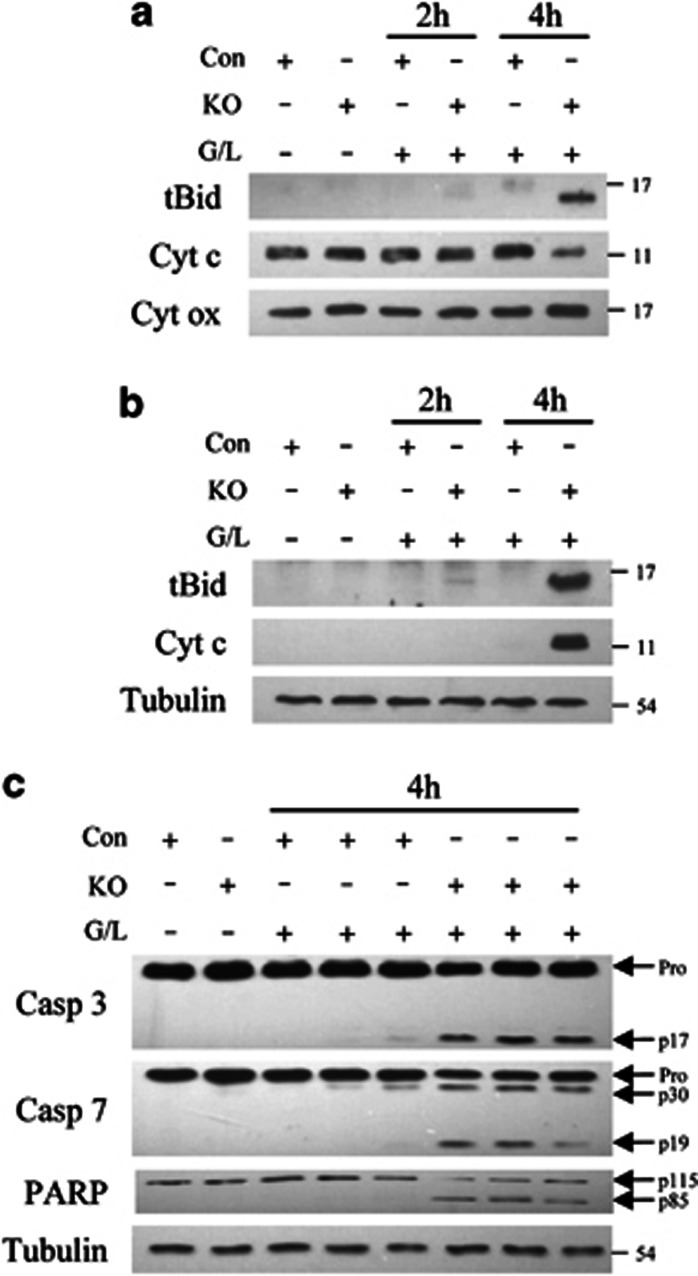
Knockout (KO) mice have increased activation of the mitochondrial death pathway. (a) Immunoblots of mitochondrial protein isolates from control (Con) and KO mice untreated or treated with GalN/LPS (G/L) for 2 or 4 h. The proteins were probed for truncated Bid (tBid), cytochrome c (Cyt c) and cytochrome oxidase (Cyt ox) as a loading control. (b) Cytosolic fractions from the same mouse livers immunoblotted for tBid, Cyt c and tubulin as a loading control. (c) Immunoblots of total hepatic protein from untreated and 4 h GalN/LPS-treated mice for caspase-3 (Casp 3), caspase-7 (Casp 7), PARP and tubulin. Arrows indicate the procaspases (Pro), the cleaved caspase-3 (p17) and -7 (p30 and p19) forms, and the intact (p115) and cleaved (p85) forms of PARP. Results are representative of three independent experiments
Mitochondrial cytochrome c release initiates cleavage of downstream effector caspases that mediate cell death. In parallel with increased mitochondrial cytochrome c release, high levels of the active, cleaved forms of effector caspases 3 and 7 were detected at 4 h in the livers of GalN/LPS-treated Atg7Δhep mice but not in littermate controls (Figure 4c). Increased caspase activity was confirmed by the finding of cleavage of the caspase substrate poly (ADP-ribose) polymerase (PARP) in the same livers (Figure 4c). Loss of hepatocyte autophagy increased apoptosis from GalN/LPS by accelerating activation of the mitochondrial death pathway and effector caspases.
Loss of hepatocyte autophagy sensitizes to liver injury from GalN/TNF
To ensure that increased liver injury from GalN/LPS represented sensitization to death from TNF, the effects of the loss of autophagy on injury from GalN/TNF were examined. GalN/TNF-treated knockout mice had a 10-fold increase in ALT levels over those in control mice at 4 h (Figure 5a). Commensurate with the elevated ALT were increases in TUNEL staining (Figure 5b–d), caspase-3 and -7 activation and PARP cleavage (Figure 5e). Consistent with findings for TNF-dependent GalN/LPS injury, hepatocytes lacking autophagy were sensitized to injury and death from TNF.
Figure 5.
Liver injury from GalN/TNF is increased in knockout (KO) mice. (a) Serum ALT levels in littermate control (Con) and KO mice 4 h after GalN/TNF treatment (*P<0.001 as compared with control mice; n=6-7). (b) TUNEL staining of control mouse liver 4 h after GalN/TNF. (c) TUNEL staining of 4 h GalN/LPS-treated KO liver. (d) Numbers of TUNEL-positive cells per high power field (HPF; *P<0.001 as compared with control mice; n=3-4). (e) Immunoblots of total hepatic protein from untreated and 4 h GalN/TNF-treated Con and KO mice for caspase-3 (Casp 3), caspase-7 (Casp 7), PARP and tubulin. Arrows indicate the procaspases (Pro), the cleaved caspase-3 (p17) and -7 (p30 and p19) forms, and the intact (p115) and cleaved (p85) forms of PARP
Mitochondrial function and number are unaffected by the loss of autophagy
Mitochondrial dysfunction can promote cell death through a loss of energy homeostasis or the generation of excessive oxidative stress. Autophagy supplies substrates for energy production,7 and our previous studies demonstrated that autophagy is critical to maintain levels of ATP required for hepatocyte resistance to oxidant stress.20 Loss of hepatocyte autophagy did not impair cellular energy stores, as ATP levels were equivalent in GalN/LPS-treated control and Atg7Δhep mice (Figure 6a). Levels of the principal hepatic antioxidant glutathione (GSH) were evaluated to assess oxidative stress. Hepatic GSH levels decreased significantly at 2–4 h after GalN/LPS administration, but the reduction was similar in control and knockout mice (Figure 6b). Mitochondrial GSH levels were unaffected by GalN/LPS treatment and equivalent in the two types of mice (Figure 6c).
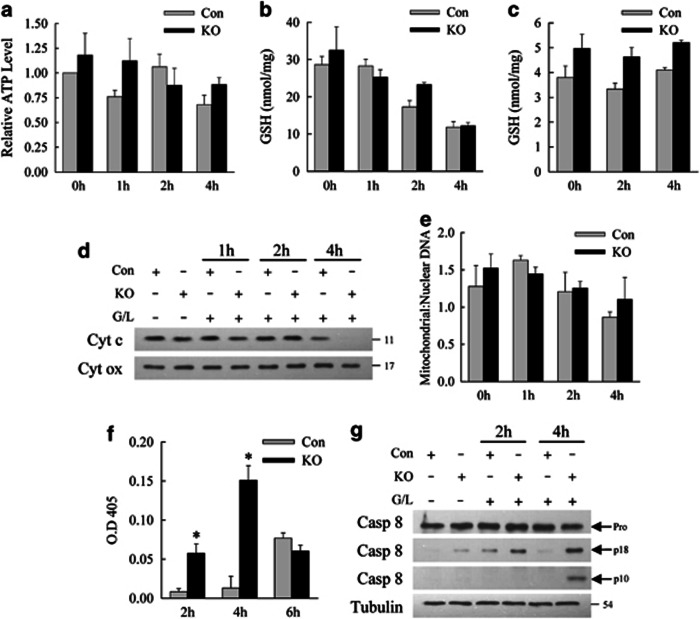
Figure 6.
Mitochondrial death pathway activation in the absence of autophagy results from increased caspase-8 activation. (a) Relative ATP levels in the livers of control (Con) and knockout (KO) mice at the indicated hours after GalN/LPS treatment (n=3-4). (b) Total hepatic GSH levels in Con and KO mice after GalN/LPS treatment (n=3). (c) Mitochondrial GSH levels in identically treated mice (n=2-3). (d) Total liver protein homogenates from untreated and GalN/LPS-treated mice immunoblotted for cytochrome c (Cyt c) and cytochrome oxidase (Cyt ox). (e) Ratio of mitochondrial to nuclear DNA in the livers of the same animals (n=3-5). (f) Relative caspase-8 activity as measured by optical density (O.D.) at 405 nm in the two types of mice treated with GalN/LPS for the indicated times (*P<0.003; n=4-5). (g) Immunoblots of hepatic cellular protein isolates from untreated and GalN/LPS-treated mice probed for caspase-8. The three images are different exposures of the same immunoblot. The procaspase (Pro) and cleaved (p18 and p10) forms of caspase-8 are indicated by arrows. Immunoblots are representative of findings from 3 independent experiments
The selective removal of damaged mitochondria by mitophagy may prevent cytochrome c release and cell death.12 To examine whether impaired mitophagy was a mechanism of TNF-sensitization in knockout mice, relative mitochondria number was determined by hepatic mitochondrial protein and DNA content. Total hepatic protein levels of cytochrome c and cytochrome oxidase were equivalent in untreated and 1 and 2 h GalN/LPS-treated control and knockout mice, consistent with equal numbers of mitochondria in the absence or presence of autophagy (Figure 6d). Levels of cytochrome c were decreased in 4 h-treated knockout mice, and to a lesser extent in control mice, due to release and degradation of this protein from mitochondrial death pathway activation as indicated by unchanged levels of cytochrome oxidase (Figure 6d). Equivalent mitochondrial number was confirmed by similar ratios of hepatic mitochondrial to nuclear DNA content in control and knockout mice (Figure 6e). The mechanism by which the loss of autophagy sensitized hepatocytes to GalN/LPS injury was not impaired energy homeostasis, increased oxidant stress or an absence of mitophagy.
Atg7Δhep mice have increased caspase-8 activation
The absence of primary mitochondrial dysfunction, together with increased Bid cleavage, suggested that loss of autophagy might promote hepatocyte caspase-8 activation to increase Bid cleavage and mitochondrial death pathway activation. Low levels of hepatic caspase-8 activity were detectable in controls at 2 and 4 h after GalN/LPS, but activity was markedly increased in knockout mouse livers at these times (Figure 6f). By 6 h, caspase-8 activity was equivalently elevated in control and knockout mice (Figure 6f). Immunoblots confirmed that the active, cleaved p18 and p10 forms of caspase-8 were increased in the livers of GalN/LPS-treated Atg7Δhep mice (Figure 6g). Loss of autophagy, therefore, led to increased caspase-8 activation that accelerated Bid cleavage and mitochondrial death pathway activation.
JNK activation is increased in Atg7Δhep mice
NF-κB and JNK signaling regulate TNF-induced death,8 and our prior studies demonstrated that inhibition of autophagy increases hepatocyte JNK activation from oxidant stress.20 NF-κB promotes cellular resistance to TNF cytotoxicity, whereas JNK overactivation sensitizes hepatocytes to death from TNF.10, 11 In GalN/LPS liver injury, JNK overactivation promotes caspase-8 cleavage.21, 22 Rapid NF-κB activation occurred in both control and Atg7Δhep mice as indicated by total degradation of the NF-κB inhibitory protein IκB within 1 h of GalN/LPS (Figure 7a). Loss of autophagy amplified JNK activation from GalN/LPS as reflected by increased levels of phosphorylated JNK and its downstream substrate c-Jun (Figure 7a). Total JNK and c-Jun levels were unaffected (Figure 7a). One mechanism by which JNK may promote caspase activation is by increasing degradation of the caspase-8 inhibitor cellular FLICE-like inhibitor protein (c-FLIP).21, 23 Levels of c-FLIPL were decreased in Atg7Δhep mice, but not until 4 h after GalN/LPS treatment (Figure 7b), a time after the initial increase in caspase-8 activity and injury. Levels of anti-apoptotic cellular inhibitor of apoptosis (cIAP) 1 and 2 were unchanged by GalN/LPS or the loss of autophagy (Figure 7b).
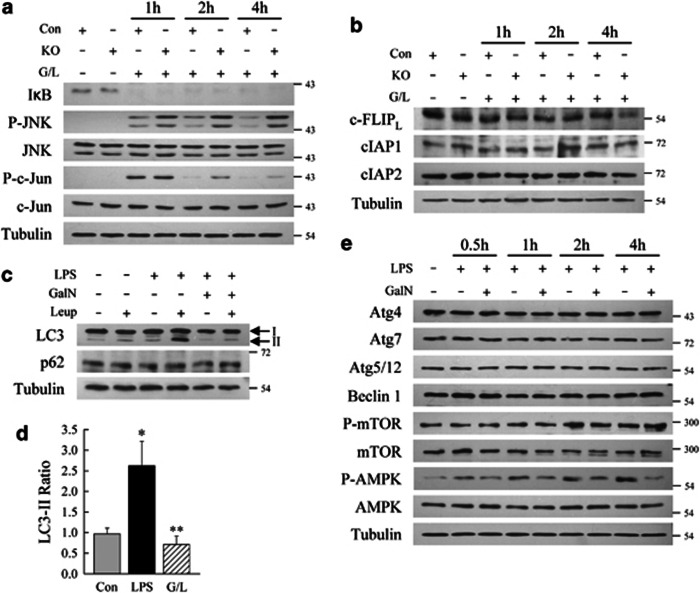
Figure 7.
Knockout (KO) mice have increased JNK activation and GalN blocks the induction of autophagy by LPS. (a) Total liver protein from control (Con) and KO mice untreated or treated with GalN/LPS (G/L) for the hours shown and immunoblotted for the total or phosphorylated (P-) forms of the indicated proteins. (b) Immunoblots of the same hepatic protein samples probed for the proteins shown. (c) Immunoblots of total liver protein from wild-type mice untreated or treated with LPS alone or GalN/LPS for 4 h. Some mice were also injected with leupeptin (Leup) 2 h before killing. Proteins were probed for LC3, p62 and tubulin, and the LC3-I and -II forms are labeled with arrows. (d) Quantification of the ratio of LC3-II in leupeptin-injected to uninjected Con, LPS-treated (LPS) and GalN/LPS-treated (G/L) mouse livers by densitometric scanning of immunoblots (*P<0.004 as compared with control; **P<0.02 as compared with LPS-injected; n=4). (e) Immunoblots of total hepatic protein from wild-type mice untreated or treated with LPS or GalN/LPS and probed for the indicated total or phosphorylated (P-) proteins. Results are representative of three independent experiments
GalN blocks the induction of autophagy by LPS
Although basal levels of autophagy regulate cell function, autophagy increases with cell stress. One stressor is LPS, which induces autophagy in various cells, including hepatocytes.24 We examined whether LPS and/or GalN altered hepatic autophagy by in vivo leupeptin injection to measure autophagic flux.25 Untreated mouse livers had low basal levels of autophagy as demonstrated by the slight increase in LC3-II protein following leupeptin injection (Figure 7c). Leupeptin induced a marked increase in LC3-II in LPS-treated mice, indicating an LPS induction of autophagy (Figure 7c). In contrast, leupeptin failed to increase LC3-II in GalN/LPS-injected mice over levels seen in control mice not injected with GalN/LPS (Figure 7c), demonstrating that GalN blocked the induction of autophagy by LPS. Quantification of autophagic flux by densitometric scanning of the LC3-II band intensities, and calculation of the ratios of LC3-II in leupeptin-injected to uninjected livers, confirmed an LPS-induced increase in autophagic flux that was blocked by GalN (Figure 7d). Basal autophagic function was maintained in GalN/LPS-treated livers, as levels of autophagic flux were only slightly decreased over those in control mice (Figure 7d), and no increase occurred in the autophagy-degraded protein p62 (Figure 7c).
To examine the mechanism by which GalN blocked the LPS-inducible increase in autophagy, the effect of GalN on the regulatory components of the autophagic pathway were determined. Levels of critical mediators of autophagy, including Atg4, Atg7, Atg5/12 and beclin 1, were unchanged by LPS or GalN/LPS treatment (Figure 7e). Similarly, the treatment neither altered the activation of mammalian target of rapamycin (mTOR), a major inhibitor of autophagy,26 as determined by levels of phospho-mTOR (Figure 7e). However, adenosine monophosphate-activated protein (AMPK), which positively regulates autophagy,27 was activated by LPS as demonstrated by increased Thr-172 phosphorylation, and this activation was blocked by GalN (Figure 7e). This inhibitory effect of GalN on AMPK may mediate the block in induction of autophagy by LPS.
Increased autophagy prevents GalN/LPS liver injury
The block in autophagy by GalN suggested that this effect may be a mechanism by which this hepatotoxin sensitizes the liver to LPS/TNF injury. To examine this possibility, we determined whether increased levels of autophagy protect against GalN/LPS injury. Studies have established that adenoviral overexpression of the autophagy genes atg7 and beclin 1 increases hepatic autophagy.28, 29 To avoid adenoviral immune effects, we generated a beclin-1-expressing recombinant adeno-associated virus (rAVV). Mice were infected with a promoterless LacZ control rAAV or the beclin-1-expressing rAAV, and 4 weeks later, were treated with GalN/LPS. The beclin 1 rAAV was effective in increasing hepatic beclin 1 protein levels (Figure 8a). Beclin 1 overexpression reduced GalN/LPS liver injury as demonstrated by decreased serum ALT levels (Figure 8b), histological evidence of liver injury (Figure 8c) and TUNEL staining (Figure 8d). Increased autophagy blocked mitochondrial death pathway activation (Figure 8e) due to inhibition of caspase-8 and Bid cleavage (Figure 8f). As a result, effector caspase and PARP cleavage was inhibited in beclin-1-expressing mice (Figure 8a). Basal levels of autophagy are therefore insufficient to protect against GalN/LPS-induced injury, indicating that GalN sensitizes the liver to TNF injury by blocking the normally protective induction of autophagy by LPS.
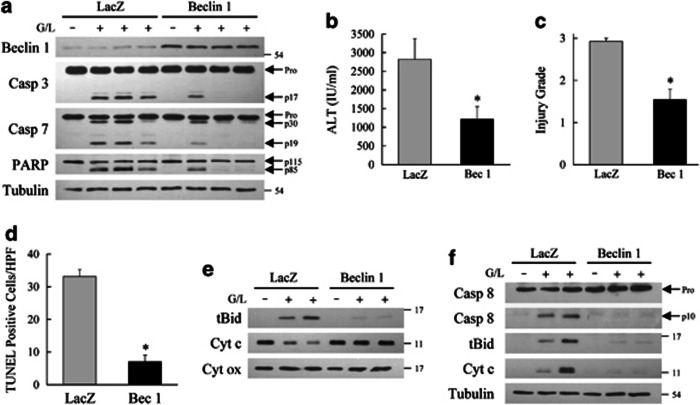
Figure 8.
Beclin-1-induced increase in autophagy prevents liver injury from GalN/LPS. (a) Mice were infected with the control LacZ or beclin-1-expresssing rAAV and left untreated or treated with GalN/LPS (G/L) for 5 h. Total liver protein was immunoblotted for beclin 1, caspase-3 (Casp 3), caspase-7 (Casp 7), PARP and tubulin. Arrows indicate the procaspases (Pro), the cleaved caspase-3 (p17) and -7 (p30 and p19) forms, and intact (p115) and cleaved (p85) PARP. (b) Serum ALT levels at 5 h after GalN/LPS treatment (*P<0.02 as compared with control mice; n=7–11). (c) Histological grade of liver injury in the same mice (*P<0.0005 as compared with control mice; n=7–11). (d) Numbers of TUNEL-positive cells per high power field (HPF; *P<0.001; n=3). (e) Mitochondrial protein isolates from rAAV-infected mice untreated or treated with GalN/LPS (G/L) for 5 h and probed for truncated Bid (tBid), cytochrome c (Cyt c) and cytochrome oxidase (Cyt ox) as a loading control. (f) Immunoblots of cytosolic protein from the same mice immunoblotted for caspase-8 and the other indicated proteins. The procaspase and cleaved caspase-8 images are different exposures of the same immunoblot. Results are representative of two independent experiments
Discussion
Delineating the mechanisms of TNF-induced cell death is critical to understanding a variety of pathophysiological conditions ranging from inflammation to cancer. Resistance to TNF cytotoxicity is particularly important in the liver where hepatocyte sensitization to TNF-induced death underlies many forms of hepatic injury.8 However, the cellular pathways that mediate TNF resistance in hepatocytes, and nontransformed cells in general, remain unclear. The present study identifies an essential function for autophagy in protection against TNF toxicity in vivo. Inhibition of hepatocyte autophagy led to increased liver injury, hepatocyte death and mortality from TNF-dependent toxic liver injury. The mechanism of injury was decreased hepatocyte resistance to TNF, as the findings were derived from a hepatocyte-specific knockout in which macrophage activation and cytokine production was unaffected. Death resulted from overactivation of caspase-8 and the mitochondrial death pathway, indicating that autophagy has a critical gatekeeper function in the extrinsic death pathway.
Recent in vitro investigations have demonstrated discordant but mainly pro-apoptotic effects of autophagy on caspase-8. The ability of autophagy to block T-cell death has been attributed to autophagic degradation of apoptotic proteins, including caspase-8.30 In contrast, studies in atg5 knockout fibroblasts have indicated that components of the autophagosome complex with caspase-8 to promote its activation.31 In transformed cells with Bcl-2 overexpression and a block in proteasomal degradation, autophagy also promoted caspase-8 activation.32 Although these studies have already suggested a role for autophagy in caspase-8 regulation, they have mainly indicated that autophagy increases caspase-8 activation and cell death.
An advantage of the present investigations is that they examined autophagic function in the TNF-dependent extrinsic death pathway in normal cells in vivo. In GalN/LPS and GalN/TNF liver injury, hepatocytes die from mitochondrial death pathway induction and caspase-dependent apoptosis. In the absence of autophagy, hepatocyte apoptosis from TNF was increased. Loss of hepatocyte autophagy amplified the TNF-dependent extrinsic death pathway as indicated by accelerated mitochondrial cytochrome c release, downstream effector caspase activation and PARP cleavage. Mitochondrial ATP content, GSH levels and number were unaffected by the loss of autophagy, indicating that primary mitochondrial damage did not initiate death. Rather, mitochondrial death pathway activation occurred following early and increased caspase-8 activation, demonstrating that autophagy modulates upstream initiator caspase activation.
Investigations by our laboratory and others have established that hepatocellular TNF resistance is dependent on NF-κB downregulation of lethal JNK overactivation.10, 11 JNK promotes caspase-8 cleavage.22 We previously reported that loss of hepatocyte autophagy promotes JNK activation from oxidant stress.20 In TNF-induced injury, the absence of autophagy similarly led to increased JNK/c-Jun signaling, which likely promoted caspase-8 activation. The caspase-8 inhibitor c-FLIPL is a JNK-regulated factor implicated in hepatocyte TNF resistance,21 and c-FLIPL levels were decreased in GalN/LPS-treated Atg7Δhep mice. Although effects on c-FLIPL may partially explain the present findings, changes in c-FLIPL were detected at 4 h, a time after which caspase-8 activation had begun. Thus, other as yet unidentified JNK-regulated factors altered by the loss of autophagy might have a role in caspase-8 overactivation.
These findings increase our understanding of the crosstalk between autophagy and cell death by identifying a critical role for autophagy in blocking apoptosis at the level of initiator caspase activation. A number of studies have shown that the two pathways modulate each other's activity, but most have demonstrated regulation of autophagy by components of the apoptotic pathway. For example, Fas-associated death domain protein and caspase-8 have been reported to regulate autophagy to promote apoptosis from IFNγ and TRAIL.33, 34 In contrast, the present study, together with other recent investigations,30, 31, 32 demonstrates that autophagy directly controls activation of components of the apoptotic pathway.
GalN blocked the induction of hepatic autophagy by LPS, suggesting that this increase may be a mechanism of hepatic resistance to TNF cytotoxicity. That GalN sensitizes to TNF-induced liver injury by blocking the protective upregulation of autophagy was proven by the ability of a beclin-1-mediated increase in autophagy to decrease GalN/LPS liver injury. The level of basal autophagic function remaining after GalN/LPS treatment is partially protective as demonstrated by the fact that a complete inhibition of autophagy in atg7 knockout mice significantly worsened injury from GalN/LPS. Hepatic autophagic function decreases in settings, such as steatosis and aging.16, 35 If basal levels of autophagy mediate hepatic resistance to TNF toxicity, then these conditions that impair autophagic function may render the liver vulnerable to TNF-induced cell death. Consistent with this possibility is that TNF mediates hepatocellular injury in steatosis.36 The mechanism of the block of an LPS induction of autophagy by GalN remains to be further defined, but may result from GalN's inhibition of AMPK, a known stimulator of autophagy.27
There has been a significant scientific emphasis on the role of autophagy in preventing chronic tissue injury, resulting from the aggregation of abnormal proteins as occurs in many neurodegenerative diseases.37 In the liver, pharmacological induction of autophagy reduces hepatic fibrosis in chronic liver injury, resulting from the cellular aggregation of mutant α1-antitrypsin.38 Our findings demonstrate an equally important in vivo function for autophagy in acute cytokine-mediated tissue injury as a critical inhibitor of mitochondrial death pathway activation. The development of new agents to acutely induce autophagy may be a valuable therapeutic approach in these diseases as well.
Materials and Methods
Animal model
Mice were maintained under 12 h light/dark cycles with unlimited access to food and water. All studies were performed in 10- to 14-week-old male mice. C57BL/6 mice were used as wild-type mice. Atg7F/F mice15 containing floxed alleles for the autophagy gene atg7 were crossed with ERt-albumin-Cre mice18 with a tamoxifen-inducible, albumin promoter-driven cre recombinase to generate ERt-albumin-Cre-Atg7F/F or Atg7Δhep mice. Both types of transgenic mice were on a C57BL/6 background. Genotypes were confirmed by PCR, with established primers. To activate Cre expression and generate mice with a hepatocyte-specific knockout of atg7, Atg7Δhep mice were injected intraperitoneally with 0.1 mg of tamoxifen (Sigma, St Louis, MO, USA) daily for 5 consecutive days, as previously described.39 Controls for all experiments with Atg7Δhep mice were littermate Atg7F/F male mice lacking the Cre transgene, which were identically injected with tamoxifen. Studies on transgenic mice were performed 5 days post-tamoxifen treatment.
Liver injury was induced by intraperitoneal injections of 100 μg/kg of LPS (E. coli 0111:B4) and 700 mg/kg of GalN (Sigma) dissolved in phosphate-buffered saline, as previously performed.40 Some mice were injected with the same dose of LPS alone. Mice were also injected intravenously with 10 μg/kg of mouse recombinant TNF (R&D Systems, Minneapolis, MN, USA) 30 min after GalN treatment.
Autophagic flux was determined by comparing hepatic LC3-II levels on immunoblots between mice injected intraperitoneally with saline or leupeptin (Fisher, Pittsburgh, PA, USA) at 40 mg/kg 2 h before killing, as previously described.25 All animal studies were approved by the Animal Care and Use Committee of the Albert Einstein College of Medicine, and followed the National Institutes of Health guidelines for animal care.
Protein isolation and western blotting
Total liver protein and mitochondrial and cytosolic protein fractions were isolated as previously described.22, 40 Protein concentrations were determined by the Bio-Rad (Hercules, CA, USA) protein assay, and western blotting preformed as previously described.22 Membranes were exposed to antibodies that recognized c-FLIP, cIAP 1 and 2, total JNK1 and JNK2, total c-Jun, PARP, TNF-receptor 1, IκB (Santa Cruz Biotechnology, Santa Cruz, CA, USA), phosphorylated JNK 1 and JNK2, phosphorylated c-Jun, LC3, Atg7, beclin 1, phosphorylated and total mTOR, phosphorylated and total AMPK, caspase-3, caspase-7, tubulin, beclin 1 (Cell Signaling, Beverly, MA, USA), caspase-8 (R&D Systems), Atg5 (Novus, Littleton, CO, USA), Atg4 (Sigma), cytochrome oxidase, β-actin (Abcam, Cambridge, MA, USA), cytochrome c (BD Biosciences, San Jose, CA, USA), p62 (Enzo, Plymouth Meeting, PA, USA) and Bid (kind gift of Xiao-Ming Yin, University of Indiana). Western blot signals were quantitated by a FluorChem densitometer (Alpha Innotech, San Leonardo, CA, USA).
ALT assay
Serum ALTs were measured by commercial kit (TECO Diagnostics, Anaheim, CA, USA).
Histology
Livers were fixed in 10% neutral formalin, stained with hematoxylin and eosin, and graded in a blinded fashion by a single pathologist for the degree of liver injury and inflammation. The percentage of hepatic parenchyma with apoptosis/necrosis or inflammation was semiquantitatively graded on a sliding scale of: 0, absent; 0.5, minimal; 1, mild; 1.5, mild-to-moderate; 2, moderate; 2.5, moderate-to-marked; and 3, marked.
TUNEL assay
TUNEL-positive cells in liver sections were detected with the commercial kit DeadEnd Colorimetric System (Promega, Madison, WI, USA). Tissue sections were deparaffinized in xylene, gradually rehydrated in decreasing concentrations of ethanol, and the assay performed according to the manufacturer's instructions. Under light microscopy, the numbers of TUNEL-positive cells in 10 randomly selected high power fields ( × 400 magnification) were counted per liver section.
Real-time reverse-transcription PCR
Total liver RNA was isolated using the commercial kit RNeasy Plus (QIAGEN, Valencia, CA, USA). Reverse transcription was carried out with 1 μg of RNA in an Eppendorf Mastercycler (Hamburg, Germany) using a high-capacity cDNA reverse transcription kit (ABI, Foster City, CA, USA). Annealing of primers was done at 25 °C for 10 min, followed by elongation at 37 °C for 2 h and inactivation of the enzyme at 85 °C for 5 min. Negative controls (no added transcriptase) were performed in parallel. PCR for TNF, interferon-γ, IL-6, IL-1β and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was performed in triplicate in a 7500 Fast Real-Time PCR System (ABI). The primer sequences in Supplementary Table S1 were purchased from Integrated DNA Technologies (Coralville, IA, USA). PCR was carried out using Power SYBR Green Master Mix (ABI). Taq polymerase was activated at 95 °C for 10 min. The cycling parameters were denaturation at 95 °C for 30 s and extension at 60 °C for 1 min (for 40 cycles). Data analysis was performed using the 2−ΔΔCT method for relative quantification. All samples were normalized to GAPDH.
ATP assay
Liver ATP content was measured with a commercial kit (Biovision, Milpitas, CA, USA), normalized to protein concentration and expressed as levels relative to untreated control samples.
GSH assay
The 5,5′-dithiobis (2-nitrobenzoic acid)–GSH disulfide recycling assay was used for the determination of total GSH in whole livers and mitochondrial GSH in isolated mitochondria, as previously described.41 Values were normalized to protein content.
Mitochondrial DNA content
Total liver DNA was isolated using the commercial kit DNeasy Blood & Tissue (QIAGEN). Real-time PCR for the cytochrome c1 and 18S rRNA genes was performed as described above, using the primers in Supplementary Figure S1. Mitochondrial DNA content was quantified by normalizing values for the cytochrome c1 gene (mitochondrial DNA) to that for the 18S rRNA gene (nuclear DNA).
Caspase-8 activity
Caspase-8 activity was measured with a colorimetric commercial kit (R&D Systems). Livers were homogenized in lysis buffer with a dounce homogenizer. Reactions were carried out on a standard amount of protein at 37 °C for 2 h, and the optical density measured on a microplate reader at a wavelength of 405 nm.
rAAV generation
The open reading frame of human beclin 1 gene (NM_003766) was inserted into pAAV expression vector driven by a CMV-enhanced chick β-actin promoter,42 confirmed by gene sequencing, and cotransfected with AAV-8 packaging plasmid and adenovirus helper plasmid into AAV-293 cells. The rAAV particles were collected and purified by cesium chloride gradient centrifugation and titered, as previously described.42 Six-week-old mice were injected intravenously via the tail vein with 3 × 1011 gene copies of the beclin 1 rAAV or a promoterless LacZ rAAV as a control. Some mice were then administered GalN/LPS at 10 weeks of age.
Statistical analysis
Numerical results are reported as means±S.E. and are derived from at least three independent experiments unless otherwise indicated. The unpaired Student's t-test was used to assess significance between control and treated groups. Survival rates were compared by the Cox proportional hazards model. Statistical significance was defined as P<0.05.
Acknowledgments
This work was supported in part by grants DK044234 (MJC) and HL059407 (GG) from the National Institutes of Health and PR2010-0002 (LF) from the Spanish Ministry of Education. We thank Pierre Chambon for the ERt-Cre mice, Masaaki Komatsu and Keiji Tanaka for the atg7F/F mice and Xiao-Ming Yin for the Bid antibody.
Glossary
- ALT
alanine aminotransferase
- AMPK
adenosine monophosphate-activated protein
- c-FLIP
cellular FLICE-like inhibitor protein
- cIAP
cellular inhibitor of apoptosis
- GalN
𝒟-galactosamine
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- GSH
glutathione
- JNK
c-Jun N-terminal kinase
- LC3
microtubule-associated protein 1 light chain 3
- LPS
lipopolysaccharide
- mTOR
mammalian target of rapamycin
- NF-κB
nuclear factor-κB
- PARP
poly (ADP-ribose) polymerase
- rAAV
recombinant adeno-associated virus
- TNF
tumor necrosis factor
- TUNEL
terminal deoxynucleotide transferase-mediated deoxyuridine triphosphate nick end-labeling
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by J Silke
Supplementary Material
References
- Kroemer G, Levine B. Autophagic cell death: the story of a misnomer. Nat Rev Mol Cell Biol. 2008;9:1004–1010. doi: 10.1038/nrm2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paglin S, Hollister T, Delohery T, Hackett N, McMahill M, Sphicas E, et al. A novel response of cancer cells to radiation involves autophagy and formation of acidic vesicles. Cancer Res. 2001;61:439–444. [PubMed] [Google Scholar]
- Espert L, Denizot M, Grimaldi M, Robert-Hebmann V, Gay B, Varbanov M, et al. Autophagy is involved in T cell death after binding of HIV-1 envelope proteins to CXCR4. J Clin Invest. 2006;116:2161–2172. doi: 10.1172/JCI26185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boya P, Gonzalez-Polo RA, Casares N, Perfettini JL, Dessen P, Larochette N, et al. Inhibition of macroautophagy triggers apoptosis. Mol Cell Biol. 2005;25:1025–1040. doi: 10.1128/MCB.25.3.1025-1040.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamacher-Brady A, Brady NR, Gottlieb RA. Enhancing macroautophagy protects against ischemia/reperfusion injury in cardiac myocytes. J Biol Chem. 2006;281:29776–29787. doi: 10.1074/jbc.M603783200. [DOI] [PubMed] [Google Scholar]
- Ding WX, Ni HM, Gao W, Hou YF, Melan MA, Chen X, et al. Differential effects of endoplasmic reticulum stress-induced autophagy on cell survival. J Biol Chem. 2007;282:4702–4710. doi: 10.1074/jbc.M609267200. [DOI] [PubMed] [Google Scholar]
- Czaja MJ. Functions of autophagy in hepatic and pancreatic physiology and disease. Gastroenterology. 2011;140:1895–1908. doi: 10.1053/j.gastro.2011.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schattenberg JM, Czaja MJ.TNF and TNF receptorsIn: Dufour JF, Clavien P-A, (eds).Signaling Pathways in Liver Diseases2nd edn.Springer-Verlag: Berlin; 2010161–179. [Google Scholar]
- Xu Y, Bialik S, Jones BE, Iimuro Y, Kitsis RN, Srinivasan A, et al. NF-κB inactivation converts a hepatocyte cell line TNF-α response from proliferation to apoptosis. Am J Physiol. 1998;275:C1058–C1066. doi: 10.1152/ajpcell.1998.275.4.C1058. [DOI] [PubMed] [Google Scholar]
- Liu H, Lo CR, Czaja MJ. NF-κB inhibition sensitizes hepatocytes to TNF-induced apoptosis through a sustained activation of JNK and c-Jun. Hepatology. 2002;35:772–778. doi: 10.1053/jhep.2002.32534. [DOI] [PubMed] [Google Scholar]
- Schwabe RF, Uchinami H, Qian T, Bennett BL, Lemasters JJ, Brenner DA. Differential requirement for c-Jun NH2-terminal kinase in TNFα- and Fas-mediated apoptosis in hepatocytes. FASEB J. 2004;18:720–722. doi: 10.1096/fj.03-0771fje. [DOI] [PubMed] [Google Scholar]
- Kim I, Lemasters JJ. Mitophagy selectively degrades individual damaged mitochondria after photoirradiation. Antioxid Redox Signal. 2011;14:1919–1928. doi: 10.1089/ars.2010.3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak M, Gaines GC, Rosenberg J, Minter R, Bahjat FR, Rectenwald J, et al. LPS-induced liver injury in D-galactosamine-sensitized mice requires secreted TNF-α and the TNF-p55 receptor. Am J Physiol Regul Integr Comp Physiol. 2000;278:R1202–R1209. doi: 10.1152/ajpregu.2000.278.5.R1202. [DOI] [PubMed] [Google Scholar]
- Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- Schuler M, Dierich A, Chambon P, Metzger D. Efficient temporally controlled targeted somatic mutagenesis in hepatocytes of the mouse. Genesis. 2004;39:167–172. doi: 10.1002/gene.20039. [DOI] [PubMed] [Google Scholar]
- Yin XM, Wang K, Gross A, Zhao Y, Zinkel S, Klocke B, et al. Bid-deficient mice are resistant to Fas-induced hepatocellular apoptosis. Nature. 1999;400:886–891. doi: 10.1038/23730. [DOI] [PubMed] [Google Scholar]
- Wang Y, Singh R, Xiang Y, Czaja MJ. Macroautophagy and chaperone-mediated autophagy are required for hepatocyte resistance to oxidant stress. Hepatology. 2010;52:266–277. doi: 10.1002/hep.23645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L, Kamata H, Solinas G, Luo JL, Maeda S, Venuprasad K, et al. The E3 ubiquitin ligase itch couples JNK activation to TNFalpha-induced cell death by inducing c-FLIPL turnover. Cell. 2006;124:601–613. doi: 10.1016/j.cell.2006.01.021. [DOI] [PubMed] [Google Scholar]
- Wang Y, Singh R, Lefkowitch JH, Rigoli RM, Czaja MJ. Tumor necrosis factor-induced toxic liver injury results from JNK2-dependent activation of caspase-8 and the mitochondrial death pathway. J Biol Chem. 2006;281:15258–15267. doi: 10.1074/jbc.M512953200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama Y, Taura K, Miura K, Schnabl B, Osawa Y, Brenner DA. Antiapoptotic effect of c-Jun N-terminal Kinase-1 through Mcl-1 stabilization in TNF-induced hepatocyte apoptosis. Gastroenterology. 2009;136:1423–1434. doi: 10.1053/j.gastro.2008.12.064. [DOI] [PubMed] [Google Scholar]
- Carchman EH, Rao J, Loughran PA, Rosengart MR, Zuckerbraun BS. Heme oxygenase-1-mediated autophagy protects against hepatocyte cell death and hepatic injury from infection/sepsis in mice. Hepatology. 2011;53:2053–2062. doi: 10.1002/hep.24324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haspel J, Shaik RS, Ifedigbo E, Nakahira K, Dolinay T, Englert JA, et al. Characterization of macroautophagic flux in vivo using a leupeptin-based assay. Autophagy. 2011;7:629–642. doi: 10.4161/auto.7.6.15100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Li P, Fu S, Calay ES, Hotamisligil GS. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab. 2010;11:467–478. doi: 10.1016/j.cmet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JH, Ahn IS, Fischer TD, Byeon JI, Dunn WA, Behrns KE, et al. Autophagy suppresses age-dependent ischemia and reperfusion injury in livers of mice. Gastroenterology. 2011;141:2188–2199. doi: 10.1053/j.gastro.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs JR, Li C, Yang Q, Li G, Garcia IG, Ju S, et al. Autophagy promotes T-cell survival through degradation of proteins of the cell death machinery. Cell Death Differ. 2012;19:144–152. doi: 10.1038/cdd.2011.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M, Takahashi Y, Khan O, Park S, Hori T, Yun J, et al. Autophagosomal membrane serves as a platform for an intracellular death-inducing signaling complex (iDISC)-mediated caspase-8 activation and apoptosis. J Biol Chem. 2012;287:12455–12468. doi: 10.1074/jbc.M111.309104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laussmann MA, Passante E, Dussmann H, Rauen JA, Wurstle ML, Delgado ME, et al. Proteasome inhibition can induce an autophagy-dependent apical activation of caspase-8. Cell Death Differ. 2011;18:1584–1597. doi: 10.1038/cdd.2011.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyo JO, Jang MH, Kwon YK, Lee HJ, Jun JI, Woo HN, et al. Essential roles of Atg5 and FADD in autophagic cell death: dissection of autophagic cell death into vacuole formation and cell death. J Biol Chem. 2005;280:20722–20729. doi: 10.1074/jbc.M413934200. [DOI] [PubMed] [Google Scholar]
- Thorburn J, Moore F, Rao A, Barclay WW, Thomas LR, Grant KW, et al. Selective inactivation of a Fas-associated death domain protein (FADD)-dependent apoptosis and autophagy pathway in immortal epithelial cells. Mol Biol Cell. 2005;16:1189–1199. doi: 10.1091/mbc.E04-10-0906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo AM, Bergamini E, Brunk UT, Droge W, Ffrench M, Terman A. Autophagy and aging: the importance of maintaining ‘clean' cells. Autophagy. 2005;1:131–140. doi: 10.4161/auto.1.3.2017. [DOI] [PubMed] [Google Scholar]
- Li Z, Yang S, Lin H, Huang J, Watkins PA, Moser AB, et al. Probiotics and antibodies to TNF inhibit inflammatory activity and improve nonalcoholic fatty liver disease. Hepatology. 2003;37:343–350. doi: 10.1053/jhep.2003.50048. [DOI] [PubMed] [Google Scholar]
- Harris H, Rubinsztein DC. Control of autophagy as a therapy for neurodegenerative disease. Nat Rev Neurol. 2012;8:108–117. doi: 10.1038/nrneurol.2011.200. [DOI] [PubMed] [Google Scholar]
- Hidvegi T, Ewing M, Hale P, Dippold C, Beckett C, Kemp C, et al. An autophagy-enhancing drug promotes degradation of mutant α1-antitrypsin Z and reduces hepatic fibrosis. Science. 2010;329:229–232. doi: 10.1126/science.1190354. [DOI] [PubMed] [Google Scholar]
- Metzger D, Li M, Chambon P. Targeted somatic mutagenesis in the mouse epidermis. Methods Mol Biol. 2005;289:329–340. doi: 10.1385/1-59259-830-7:329. [DOI] [PubMed] [Google Scholar]
- Wang Y, Schattenberg JM, Rigoli RM, Storz P, Czaja MJ. Hepatocyte resistance to oxidative stress is dependent on protein kinase C-mediated down-regulation of c-Jun/AP-1. J Biol Chem. 2004;279:31089–31097. doi: 10.1074/jbc.M404170200. [DOI] [PubMed] [Google Scholar]
- Xu Y, Jones BE, Neufeld DS, Czaja MJ. Glutathione modulates rat and mouse hepatocyte sensitivity to tumor necrosis factor toxicity. Gastroenterology. 1998;115:1229–1237. doi: 10.1016/s0016-5085(98)70095-2. [DOI] [PubMed] [Google Scholar]
- Xie J, Xie Q, Zhang H, Ameres SL, Hung JH, Su Q, et al. MicroRNA-regulated, systemically delivered rAAV9: a step closer to CNS-restricted transgene expression. Mol Ther. 2011;19:526–535. doi: 10.1038/mt.2010.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.