Abstract
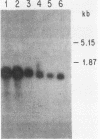
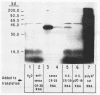
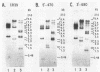
Differential hybridization and molecular cloning have been used to isolate CR39, a cDNA which hybridizes to a 1.2-kilobase (kb) mRNA in rat liver. The level of CR39 mRNA was increased seven- to ninefold over normal levels by dietary cholestyramine and mevinolin and decreased about fourfold compared with normal levels by cholesterol feeding or administration of mevalonate. Similar changes in the mRNA levels of 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reductase and HMG-CoA synthase were observed under the various conditions. In vitro translation of either CR39 hybrid selected RNA or 1.2-kb CR39 RNA generated by an SP6 in vitro transcription system produced a polypeptide of 39,000 daltons. As deduced from the nucleotide sequence of a full-length CR39 cDNA, the rat CR39 polypeptide contained 344 amino acids and had a molecular weight of 39,615. The predicted amino acid composition and submit molecular weight of the rat CR39 were very similar to those of prenyltransferases isolated from chicken, pig, and human. The sequence of amino acid residues 173 through 203 in the rat CR39 polypeptide showed that 17 out of 30 matched an active-site peptide of avian liver prenyltransferase. Thus, alterations in the rate of cholesterogenesis resulted in the coordinate regulation of three mRNAs encoding HMG-CoA reductase, HMG-CoA synthase, and CR39, the latter being tentatively identified as prenyltransferase.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barnard G. F., Langton B., Popják G. Pseudo-isoenzyme forms of liver prenyl transferase. Biochem Biophys Res Commun. 1978 Dec 14;85(3):1097–1103. doi: 10.1016/0006-291x(78)90655-1. [DOI] [PubMed] [Google Scholar]
- Barnard G. F., Popják G. Characterization of liver prenyl transferase and its inactivation by phenylglyoxal. Biochim Biophys Acta. 1980 Feb 22;617(2):169–182. doi: 10.1016/0005-2760(80)90160-5. [DOI] [PubMed] [Google Scholar]
- Barnard G. F., Popják G. Human liver prenyltransferase and its characterization. Biochim Biophys Acta. 1981 Sep 15;661(1):87–99. doi: 10.1016/0005-2744(81)90086-3. [DOI] [PubMed] [Google Scholar]
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Bergstrom J. D., Wong G. A., Edwards P. A., Edmond J. The regulation of acetoacetyl-CoA synthetase activity by modulators of cholesterol synthesis in vivo and the utilization of acetoacetate for cholesterogenesis. J Biol Chem. 1984 Dec 10;259(23):14548–14553. [PubMed] [Google Scholar]
- Blin N., Stafford D. W. A general method for isolation of high molecular weight DNA from eukaryotes. Nucleic Acids Res. 1976 Sep;3(9):2303–2308. doi: 10.1093/nar/3.9.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brems D. N., Bruenger E., Rilling H. C. Isolation and characterization of a photoaffinity-labeled peptide from the catalytic site of prenyltransferase. Biochemistry. 1981 Jun 23;20(13):3711–3718. doi: 10.1021/bi00516a007. [DOI] [PubMed] [Google Scholar]
- Brems D. N., Rilling H. C. Photoaffinity labeling of the catalytic site of prenyltransferase. Biochemistry. 1979 Mar 6;18(5):860–864. doi: 10.1021/bi00572a019. [DOI] [PubMed] [Google Scholar]
- Brown M. S., Goldstein J. L. Multivalent feedback regulation of HMG CoA reductase, a control mechanism coordinating isoprenoid synthesis and cell growth. J Lipid Res. 1980 Jul;21(5):505–517. [PubMed] [Google Scholar]
- Chan Y. L., Gutell R., Noller H. F., Wool I. G. The nucleotide sequence of a rat 18 S ribosomal ribonucleic acid gene and a proposal for the secondary structure of 18 S ribosomal ribonucleic acid. J Biol Chem. 1984 Jan 10;259(1):224–230. [PubMed] [Google Scholar]
- Chang T. Y., Chang C. C. Revertants of a Chinese hamster ovary cell mutant resistant to suppression by an analogue of cholesterol: isolation and partial biochemical characterization. Biochemistry. 1982 Oct 12;21(21):5316–5323. doi: 10.1021/bi00264a030. [DOI] [PubMed] [Google Scholar]
- Chang T. Y., Limanek J. S. Regulation of cytosolic acetoacetyl coenzyme A thiolase, 3-hydroxy-3-methylglutaryl coenzyme A synthase, 3-hydroxy-3-methylglutaryl coenzyme A reductase, and mevalonate kinase by low density lipoprotein and by 25-hydroxycholesterol in Chinese hamster ovary cells. J Biol Chem. 1980 Aug 25;255(16):7787–7795. [PubMed] [Google Scholar]
- Chin D. J., Luskey K. L., Faust J. R., MacDonald R. J., Brown M. S., Goldstein J. L. Molecular cloning of 3-hydroxy-3-methylglutaryl coenzyme a reductase and evidence for regulation of its mRNA. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7704–7708. doi: 10.1073/pnas.79.24.7704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke C. F., Edwards P. A., Lan S. F., Tanaka R. D., Fogelman A. M. Regulation of 3-hydroxy-3-methylglutaryl-coenzyme A reductase mRNA levels in rat liver. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3305–3308. doi: 10.1073/pnas.80.11.3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke C. F., Fogelman A. M., Edwards P. A. Diurnal rhythm of rat liver mRNAs encoding 3-hydroxy-3-methylglutaryl coenzyme A reductase. Correlation of functional and total mRNA levels with enzyme activity and protein. J Biol Chem. 1984 Aug 25;259(16):10439–10447. [PubMed] [Google Scholar]
- Clarke C. F., Fogelman A. M., Edwards P. A. Transcriptional regulation of the 3-hydroxy-3-methylglutaryl coenzyme A reductase gene in rat liver. J Biol Chem. 1985 Nov 15;260(26):14363–14367. [PubMed] [Google Scholar]
- Cochran B. H., Reffel A. C., Stiles C. D. Molecular cloning of gene sequences regulated by platelet-derived growth factor. Cell. 1983 Jul;33(3):939–947. doi: 10.1016/0092-8674(83)90037-5. [DOI] [PubMed] [Google Scholar]
- Collins F. S., Weissman S. M. The molecular genetics of human hemoglobin. Prog Nucleic Acid Res Mol Biol. 1984;31:315–462. doi: 10.1016/s0079-6603(08)60382-7. [DOI] [PubMed] [Google Scholar]
- Deininger P. L. Approaches to rapid DNA sequence analysis. Anal Biochem. 1983 Dec;135(2):247–263. doi: 10.1016/0003-2697(83)90680-2. [DOI] [PubMed] [Google Scholar]
- Dihanich M. E., Najarian D., Clark R., Gillman E. C., Martin N. C., Hopper A. K. Isolation and characterization of MOD5, a gene required for isopentenylation of cytoplasmic and mitochondrial tRNAs of Saccharomyces cerevisiae. Mol Cell Biol. 1987 Jan;7(1):177–184. doi: 10.1128/mcb.7.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards P. A., Lan S. F., Fogelman A. M. Alterations in the rates of synthesis and degradation of rat liver 3-hydroxy-3-methylglutaryl coenzyme A reductase produced by cholestyramine and mevinolin. J Biol Chem. 1983 Sep 10;258(17):10219–10222. [PubMed] [Google Scholar]
- Edwards P. A., Lan S. F., Tanaka R. D., Fogelman A. M. Mevalonolactone inhibits the rate of synthesis and enhances the rate of degradation of 3-hydroxy-3-methylglutaryl coenzyme A reductase in rat hepatocytes. J Biol Chem. 1983 Jun 25;258(12):7272–7275. [PubMed] [Google Scholar]
- Faust J. R., Brown M. S., Goldstein J. L. Synthesis of delta 2-isopentenyl tRNA from mevalonate in cultured human fibroblasts. J Biol Chem. 1980 Jul 25;255(14):6546–6548. [PubMed] [Google Scholar]
- Faust J. R., Goldstein J. L., Brown M. S. Synthesis of ubiquinone and cholesterol in human fibroblasts: regulation of a branched pathway. Arch Biochem Biophys. 1979 Jan;192(1):86–99. doi: 10.1016/0003-9861(79)90074-2. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Gil G., Brown M. S., Goldstein J. L. Cytoplasmic 3-hydroxy-3-methylglutaryl coenzyme A synthase from the hamster. II. Isolation of the gene and characterization of the 5' flanking region. J Biol Chem. 1986 Mar 15;261(8):3717–3724. [PubMed] [Google Scholar]
- Gil G., Goldstein J. L., Slaughter C. A., Brown M. S. Cytoplasmic 3-hydroxy-3-methylglutaryl coenzyme A synthase from the hamster. I. Isolation and sequencing of a full-length cDNA. J Biol Chem. 1986 Mar 15;261(8):3710–3716. [PubMed] [Google Scholar]
- Gould R. G., Swyryd E. A. Sites of control of hepatic cholesterol biosynthesis. J Lipid Res. 1966 Sep;7(5):698–707. [PubMed] [Google Scholar]
- Gubler U., Hoffman B. J. A simple and very efficient method for generating cDNA libraries. Gene. 1983 Nov;25(2-3):263–269. doi: 10.1016/0378-1119(83)90230-5. [DOI] [PubMed] [Google Scholar]
- Hall R. H. N6-(delta 2-isopentenyl)adenosine: chemical reactions, biosynthesis, metabolism, and significance to the structure and function of tRNA. Prog Nucleic Acid Res Mol Biol. 1970;10:57–86. doi: 10.1016/s0079-6603(08)60561-9. [DOI] [PubMed] [Google Scholar]
- Itakura K., Rossi J. J., Wallace R. B. Synthesis and use of synthetic oligonucleotides. Annu Rev Biochem. 1984;53:323–356. doi: 10.1146/annurev.bi.53.070184.001543. [DOI] [PubMed] [Google Scholar]
- Kersten H. On the biological significance of modified nucleosides in tRNA. Prog Nucleic Acid Res Mol Biol. 1984;31:59–114. doi: 10.1016/s0079-6603(08)60375-x. [DOI] [PubMed] [Google Scholar]
- Lee K. L., Isham K. R., Stringfellow L., Rothrock R., Kenney F. T. Molecular cloning of cDNAs cognate to genes sensitive to hormonal control in rat liver. J Biol Chem. 1985 Dec 25;260(30):16433–16438. [PubMed] [Google Scholar]
- Lehrach H., Diamond D., Wozney J. M., Boedtker H. RNA molecular weight determinations by gel electrophoresis under denaturing conditions, a critical reexamination. Biochemistry. 1977 Oct 18;16(21):4743–4751. doi: 10.1021/bi00640a033. [DOI] [PubMed] [Google Scholar]
- Liscum L., Luskey K. L., Chin D. J., Ho Y. K., Goldstein J. L., Brown M. S. Regulation of 3-hydroxy-3-methylglutaryl coenzyme A reductase and its mRNA in rat liver as studied with a monoclonal antibody and a cDNA probe. J Biol Chem. 1983 Jul 10;258(13):8450–8455. [PubMed] [Google Scholar]
- Luskey K. L., Chin D. J., MacDonald R. J., Liscum L., Goldstein J. L., Brown M. S. Identification of a cholesterol-regulated 53,000-dalton cytosolic protein in UT-1 cells and cloning of its cDNA. Proc Natl Acad Sci U S A. 1982 Oct;79(20):6210–6214. doi: 10.1073/pnas.79.20.6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxson R., Cohn R., Kedes L., Mohun T. Expression and organization of histone genes. Annu Rev Genet. 1983;17:239–277. doi: 10.1146/annurev.ge.17.120183.001323. [DOI] [PubMed] [Google Scholar]
- McClanahan T., McEntee K. Specific transcripts are elevated in Saccharomyces cerevisiae in response to DNA damage. Mol Cell Biol. 1984 Nov;4(11):2356–2363. doi: 10.1128/mcb.4.11.2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrabian M., Callaway K. A., Clarke C. F., Tanaka R. D., Greenspan M., Lusis A. J., Sparkes R. S., Mohandas T., Edmond J., Fogelman A. M. Regulation of rat liver 3-hydroxy-3-methylglutaryl coenzyme A synthase and the chromosomal localization of the human gene. J Biol Chem. 1986 Dec 5;261(34):16249–16255. [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Osborne T. F., Goldstein J. L., Brown M. S. 5' end of HMG CoA reductase gene contains sequences responsible for cholesterol-mediated inhibition of transcription. Cell. 1985 Aug;42(1):203–212. doi: 10.1016/s0092-8674(85)80116-1. [DOI] [PubMed] [Google Scholar]
- Parnes J. R., Velan B., Felsenfeld A., Ramanathan L., Ferrini U., Appella E., Seidman J. G. Mouse beta 2-microglobulin cDNA clones: a screening procedure for cDNA clones corresponding to rare mRNAs. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2253–2257. doi: 10.1073/pnas.78.4.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J., Sonenberg N. Insertion mutagenesis to increase secondary structure within the 5' noncoding region of a eukaryotic mRNA reduces translational efficiency. Cell. 1985 Mar;40(3):515–526. doi: 10.1016/0092-8674(85)90200-4. [DOI] [PubMed] [Google Scholar]
- Reed B. C., Rilling H. C. Substrate Binding of avian liver prenyltransferase. Biochemistry. 1976 Aug 24;15(17):3739–3745. doi: 10.1021/bi00662a015. [DOI] [PubMed] [Google Scholar]
- Reynolds G. A., Basu S. K., Osborne T. F., Chin D. J., Gil G., Brown M. S., Goldstein J. L., Luskey K. L. HMG CoA reductase: a negatively regulated gene with unusual promoter and 5' untranslated regions. Cell. 1984 Aug;38(1):275–285. doi: 10.1016/0092-8674(84)90549-x. [DOI] [PubMed] [Google Scholar]
- Reynolds G. A., Goldstein J. L., Brown M. S. Multiple mRNAs for 3-hydroxy-3-methylglutaryl coenzyme A reductase determined by multiple transcription initiation sites and intron splicing sites in the 5'-untranslated region. J Biol Chem. 1985 Aug 25;260(18):10369–10377. [PubMed] [Google Scholar]
- Rilling H. C. Eukaryotic prenyltransferases. Methods Enzymol. 1985;110:145–152. doi: 10.1016/s0076-6879(85)10069-8. [DOI] [PubMed] [Google Scholar]
- Rodwell V. W., Nordstrom J. L., Mitschelen J. J. Regulation of HMG-CoA reductase. Adv Lipid Res. 1976;14:1–74. doi: 10.1016/b978-0-12-024914-5.50008-5. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalnik D. G., Brown D. A., Brown P. C., Friedman R. L., Hardeman E. C., Schimke R. T., Simoni R. D. Mechanisms of 3-hydroxy-3-methylglutaryl coenzyme A reductase overaccumulation in three compactin-resistant cell lines. J Biol Chem. 1985 Feb 25;260(4):1991–1994. [PubMed] [Google Scholar]
- Slakey L. L., Craig M. C., Beytia E., Briedis A., Feldbruegge D. H., Dugan R. E., Qureshi A. A., Subbarayan C., Porter J. W. The effects of fasting, refeeding, and time of day on the levels of enzymes effecting the conversion of -hydroxy- -methylglutaryl-coenzyme A to squalene. J Biol Chem. 1972 May 25;247(10):3014–3022. [PubMed] [Google Scholar]
- St John T. P., Davis R. W. Isolation of galactose-inducible DNA sequences from Saccharomyces cerevisiae by differential plaque filter hybridization. Cell. 1979 Feb;16(2):443–452. doi: 10.1016/0092-8674(79)90020-5. [DOI] [PubMed] [Google Scholar]
- Weinberg R. A., Penman S. Processing of 45 s nucleolar RNA. J Mol Biol. 1970 Jan 28;47(2):169–178. doi: 10.1016/0022-2836(70)90337-2. [DOI] [PubMed] [Google Scholar]
- White L. W., Rudney H. Regulation of 3-hydroxy-3-methylglutarate and mevalonate biosynthesis by rat liver homogenates. Effects of fasting, cholesterol feeding, and triton administration. Biochemistry. 1970 Jun 23;9(13):2725–2731. doi: 10.1021/bi00815a021. [DOI] [PubMed] [Google Scholar]
- Young R. A., Davis R. W. Efficient isolation of genes by using antibody probes. Proc Natl Acad Sci U S A. 1983 Mar;80(5):1194–1198. doi: 10.1073/pnas.80.5.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R. A., Davis R. W. Yeast RNA polymerase II genes: isolation with antibody probes. Science. 1983 Nov 18;222(4625):778–782. doi: 10.1126/science.6356359. [DOI] [PubMed] [Google Scholar]