Summary
Cdc42-interacting protein 4 (CIP4), a member of the F-BAR family of proteins, plays important roles in a variety of cellular events by regulating both membrane and actin dynamics. In many cell types, CIP4 functions in vesicle formation, endocytosis and membrane tubulation. However, recent data indicate that CIP4 is also involved in protrusion in some cell types, including cancer cells (lamellipodia and invadopodia) and neurons (ribbed lamellipodia and veils). In neurons, CIP4 localizes specifically to extending protrusions and functions to limit neurite outgrowth early in development. The mechanism by which CIP4 localizes to the protruding edge membrane and induces lamellipodial/veil protrusion and actin rib formation is not known. Here, we show that CIP4 localization to the protruding edge of neurons is dependent on both the phospholipid content of the plasma membrane and the underlying organization of actin filaments. Inhibiting phosphatidylinositol (3,4,5)-trisphosphate (PIP3) production decreases CIP4 at the membrane. CIP4 localization to the protruding edge is also dependent on Rac1/WAVE1, rather than Cdc42/N-WASP. Capping actin filaments with low concentrations of cytochalasin D or by overexpressing capping protein dramatically decreases CIP4 at the protruding edge, whereas inactivating Arp2/3 drives CIP4 to the protruding edge. We also demonstrate that CIP4 dynamically colocalizes with Ena/VASP and DAAM1, two proteins known to induce unbranched actin filament arrays and play important roles in neuronal development. Together, this is the first study to show that the localization of an F-BAR protein depends on both actin filament architecture and phospholipids at the protruding edge of developing neurons.
Key words: CIP4, F-BAR, Lamellipodia, Actin polymerization
Introduction
Lamellipodia and filopodia are dynamic actin-based structures that are crucial for important biological events in all cells (Ridley, 2011; Saarikangas et al., 2010). In neurons, these structures are required for a variety of processes during neuronal development including neuronal migration, neurite formation, neuronal polarization, axonal guidance and branching (Dent et al., 2011). Actin filaments are major structural components of both filopodia and lamellipodia. Therefore, molecules that regulate actin organization and dynamics also play essential roles in neuronal development.
Actin filaments in lamellipodia and filopodia of all cells are organized with the bulk of their barbed ends oriented toward the protruding edge membrane (Pollard and Borisy, 2003). The polymerization of actin filaments is regulated by coordination between actin capping and anti-capping proteins. Capping proteins limit addition of new actin monomers to existing actin filaments, favoring a dendritic array that leads to lamellipodia formation (Cooper and Sept, 2008). Conversely, anti-capping (pro-elongating) proteins allow the polymerization of several actin filaments in long, unbranched arrays, favoring filopodia formation (Bear and Gertler, 2009; Mejillano et al., 2004; Schafer, 2004). The interplay of these and other proteins determines the actin structure within cells and consequently their mode of outgrowth and migration.
Cdc42-interacting protein 4 (CIP4), a member of the F-BAR superfamily, is an adaptor protein that interacts directly with negatively-charged membrane phospholipids through its F-BAR domain and regulates actin polymerization indirectly by binding active Cdc42 via its HR1 domain and various actin-associated proteins through its SH3 domain (Aspenström, 2009; Heath and Insall, 2008; Roberts-Galbraith and Gould, 2010). Although the most characterized function of F-BAR proteins is their role in endocytosis, many F-BAR proteins including CIP4 have also been shown to play a role in filopodial and lamellipodial protrusion (Carlson et al., 2011; Guerrier et al., 2009; Lee et al., 2010; Saengsawang et al., 2012; Zaidel-Bar et al., 2010). In non-neuronal cells, CIP4 induces polymerization associated with actin comet tails, endocytosis, and tubulation through its interaction with Cdc42, N-WASP and dynamin (Hartig et al., 2009). Recently, we showed that in cortical neurons CIP4 localizes to the protruding edges of immature neurons and the growth cones of polarized neurons (Saengsawang et al., 2012). We also showed that CIP4 induces lamellipodia and veil formation and increased the density of long, thin actin filaments (defined as actin ribs) around neuronal cell bodies. Notably, this excess formation of actin ribs and veils resulted in inhibition of filopodia and neurite formation. However, the mechanism by which CIP4 localizes to the peripheral edge of neurons and induces lamellipodia/veil/actin rib formation is not known, and more broadly, little is known about the cytoskeletal mechanisms that drive veil and actin rib formation in neurons (Cohan et al., 2001; Grabham et al., 2003; Mongiu et al., 2007; Yang et al., 2012). Here we report that both the lipid composition of the membrane and the underlying actin filament architecture determine CIP4 localization in neurons, where it functions to inhibit neurite outgrowth by forming ribbed and veiled protrusions. We show that CIP4 is key to forming ribs/veils and tubulovesicular structures in neurons, but is not required for dendritic and bundled actin filament networks that form lamellipodia and filopodia protrusions, respectively.
Results
Throughout this study we measured the levels of CIP4-EGFP/tdTomato at the protruding edge of living cortical neurons expressing low to moderate levels of the fusion protein via total internal reflection fluorescence microscopy (TIRFM). It is important to stress that CIP4-EGFP/tdTomato concentrates at protruding membranes and sometimes at non-protruding membranes where actin polymerization is balanced with retrograde actin flow. However it is not targeted to retracting or stable regions of the plasma membrane (Saengsawang et al., 2012). Live-cell TIRFM allowed us to compare the effects of acute pharmacological treatments on the dynamics of CIP4 directly over time, in single cells. Although this methodology does not allow us to measure changes in endogenous CIP4, all antibodies to CIP4 tested to date label CIP4 knockout neurons via immunocytochemistry (Saengsawang et al., 2012), making it impossible to collect reliable data on the localization of endogenous CIP4 in cortical neurons.
F-BAR and SH3 domains are required for CIP4 localization to the membrane
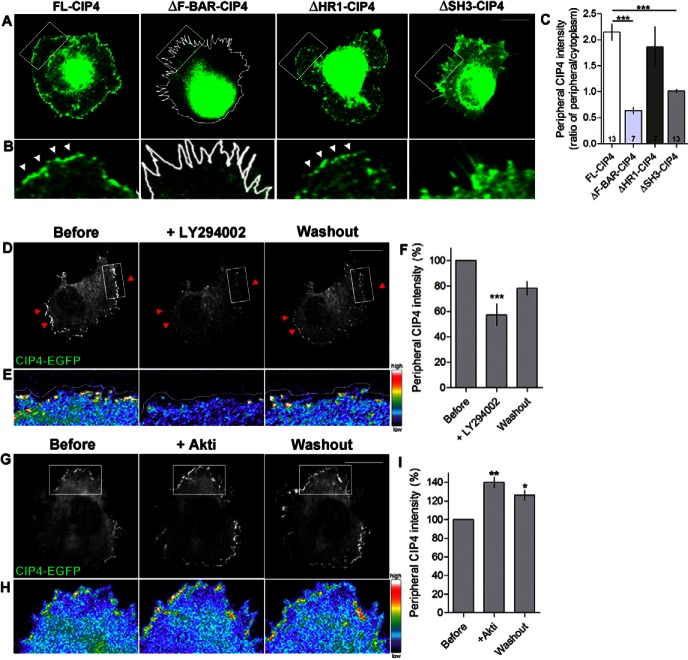
We have previously shown through analysis of domain mutants that the F-BAR and SH3 domains of CIP4 are instrumental in forming ribs and veils and thus inhibiting filopodia formation in stage 1 (pre-neurite outgrowth) cortical neurons (Saengsawang et al., 2012). To determine if this directly translated into the amount of CIP4 at protruding plasma membranes we used several of the same myc-labeled domain mutants and quantified the ratio of myc at the plasma membrane to the levels in the cytoplasm. Results indicate that deletion of either the F-BAR (binds acidic phospholipids) or SH3 (binds actin-associated proteins) domains of CIP4 dramatically inhibit the amount of CIP4 at the periphery of stage 1 cortical neurons (Fig. 1A–C). However, deletion of the HR1 domain (binds active Cdc42) had little affect on the localization of CIP4 to protruding membranes. These data suggest that both the phospholipid content and actin regulatory proteins play important roles in targeting CIP4 to protruding membranes. To determine if this was the case we first studied the effects of phospholipids on CIP4 localization.
Fig. 1.
Domains of CIP4 and lipids required for CIP4 localization at the edge of protruding membranes in cortical neurons. (A) Images of stage 1 cortical neurons transfected with full-length myc-CIP4, myc-ΔF-BAR-CIP4, myc-ΔHR1-CIP4 or myc-ΔSH3-CIP4 and labeled with an antibody to myc. (B) Images magnified from boxes in A. White arrowheads indicate CIP4 at the peripheral membrane. (C) Bar graphs of the average intensity ratio of CIP4, ΔF-BAR-CIP4, ΔHR1-CIP4 and ΔSH3-CIP4 at the peripheral/central region of the neurons. Numbers in the bar graphs indicate number of cells quantified from at least two independent experiments. (D,G) Images of stage 1 cortical neurons transfected with CIP4-EGFP before, during and after LY294002 (10 µM) (D) or Akti (5 µM) (G) treatments. (E,H) Images magnified from boxes in D and G showing the intensity of CIP4 at the peripheral area in pseudocolor. White lines in A, B and E indicate the edge of the neurons. (F,I) Bar graphs of the average intensity of CIP4 at the peripheral membrane in CIP4-transfected neurons before treatment, after 20 minutes of LY294002 treatment (F) or 10 minutes in Akti (I), and after washout. Data are expressed as mean ± s.e.m. *P<0.05 **P<0.01 and ***P<0.001 compared with before treatment (one-way ANOVA with Dunnett's post-test comparison); n = 6 and 3 cells for LY and Akti, respectively, from three independent preparations. Scale bars: 10 µm.
PIP3 is required for CIP4 localization at the protruding edge of neurons
CIP4 localizes to tubulovesicular structures in several different cell lines (Frost et al., 2008; Kamioka et al., 2004; Tian et al., 2000), but concentrates primarily at the edge of protruding membranes of developing cortical neurons (Saengsawang et al., 2012). This difference in localization may be due to distinct lipid or protein content in the differing cell types. Thus, we asked whether the level of certain negatively charged phospholipids would affect CIP4 localization to the protruding edge membrane of stage 1 cortical neurons. F-BAR proteins are known to interact with two negatively charged phospholipids, phosphatidylinositol (4,5)-bisphosphate (PIP2) and phosphatidylinositol (3,4,5)-trisphosphate (PIP3), through their F-BAR domain (Tsujita et al., 2006). Since CIP4 localizes specifically to the protruding edge(s) of neurons and PIP3 concentrates at these same regions (Henle et al., 2011), we hypothesized that CIP4 may be associating with PIP3.
To test this hypothesis we treated stage 1 cortical neurons with LY294002, a PI(3)K inhibitor, which decreases the level of PIP3 synthesis, and measured the level of CIP4 at the protruding edge of living neurons before, during and after washout of the drug. In the presence of LY294002, the intensity of CIP4 at protruding membranes decreased to 57.0±8.8% of pretreatment levels (Fig. 1D–F), indicating that a portion of CIP4 localization to the protruding regions requires PIP3. Subsequent washout of the drug returned CIP4 to levels that were not significantly different from pretreatment levels (Fig. 1F), suggesting that loss of CIP4 was largely reversible and was not likely due to general toxicity of the drug. To verify the effect of LY294002 on the level of PIP3 and PIP2, we transfected neurons with GFP-Akt-PH (an indicator of PIP3) or GFP-PLC-PH (an indicator of PIP2), and treated the neurons with LY294002. The level of GFP-Akt-PH decreased, while the level of GFP-PLC-PH increased in the presence of LY294002 (supplementary material Fig. S1), indicating that the drug decreases PIP3 but also appears to increase PIP2 levels at the membrane.
Because a major target of PIP3 at the cell membrane is the kinase Akt, we determined if inhibiting Akt would increase the availability of PIP3, to which CIP4 could then bind. Consistent with this hypothesis, treatment with Akti, a PH domain antagonist (not a kinase domain antagonist), increased CIP4 levels at the edge of protruding membranes by 40±5.7% (Fig. 1G–I). Washout of Akti was partially reversible (Fig. 1I). These results suggest that CIP4 localization depends directly on the levels of PIP3 at the membrane and that CIP4 may compete with other proteins (Manning and Cantley, 2007) to bind the available PIP3 at protruding membranes in cortical neurons.
Active Rac1 regulates CIP4 localization and function in neurons
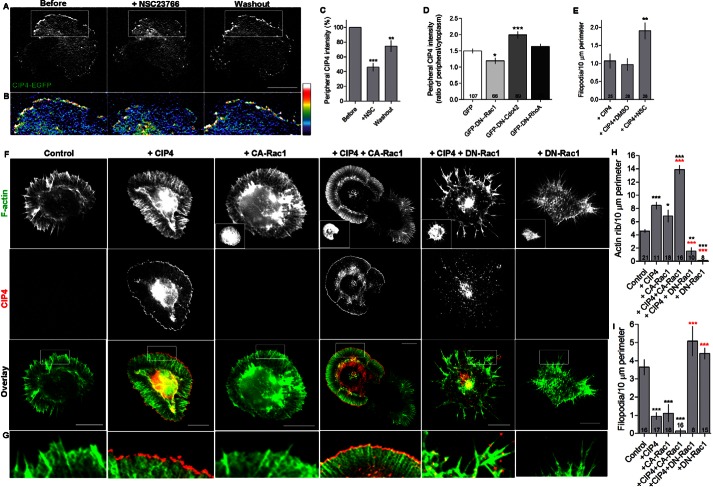
We next examined whether Rho GTPases could influence the localization of CIP4 at the protruding edge. Active Rho-GTPases localize to the inner leaflet of the plasma membrane in regions of membrane dynamics. We reasoned that the Rho-GTPase Rac1 may play a role in CIP4 localization and function at the plasma membrane in neurons because PIP3 has been shown to activate Rac1 (Welch et al., 1998) and induce lamellipodial formation (Weiner et al., 2002). In addition, active Rac1 has been shown to affect CIP4 distribution in Swiss 3T3 cells (Aspenström, 1997). We treated stage 1 cortical neurons expressing CIP4-EGFP with NSC23766, a specific inhibitor for Rac1 (Gao et al., 2004), and observed the intensity of CIP4 before, during and after washout of the drug (Fig. 2A,B). In the presence of the Rac1 inhibitor the level of CIP4 at the protruding edge decreased to 46±5.6% of pre-treatment levels (Fig. 2A–C). Upon washout, CIP4 levels partially recovered, but remained significantly less than pre-treatment levels (Fig. 2C).
Fig. 2.
Active Rac1 is required for CIP4 localization and function at the peripheral edge in neurons. (A) Image of a stage 1 cortical neuron transfected with CIP4-EGFP before, during and after NSC23766 (100 µM) treatment. (B) Pseudocolored image magnified from boxes in A showing the intensity of CIP4 at the peripheral area. (C) Bar graph of average intensity of CIP4 at the peripheral membrane of CIP4-transfected neurons before treatment, after 10 minutes in NSC23766 and after washout (n = 8 cells). (D) Bar graph showing average ratios of CIP4 at the peripheral membrane to the cytoplasm in stage 1 neurons after overnight transfection with GFP (n = 107), GFP-DN-Rac1 (n = 66), GFP-DN-Cdc42 (n = 89) and GFP-DN-RhoA (n = 53) from two independent preparations. (E) Bar graph showing filopodial number of untreated CIP4-expressing stage 1 neurons and neurons treated with DMSO or NSC23766 for 30 minutes. Number of neurons are indicated in the bar graphs. (F) Images of stage 1 cortical neurons transfected with CIP4-tdTomato (shown in red) and GFP-CA-Rac1 or GFP-DN-Rac1 (shown in insets) then stained with phalloidin 670 (shown in green). (G) Images magnified from boxes in F showing the intensity of CIP4 at the peripheral areas. (H,I) Bar graphs showing actin ribs (H) and filopodial number (I) of control neurons and neurons transfected with different constructs. Data for all graphs (C, D and E) are expressed as mean ± s.e.m. with *P<0.05 **P<0.01 and ***P<0.001 compared with before treatment (one-way ANOVA with Dunnett's post-test comparison). For graphs H and I, black asterisks are in comparison to controls and red asterisks are in comparison to CIP4 (one-way ANOVA with Bonferroni's post-test comparison). Scale bars: 10 µm.
The dependence of CIP4 localization on active Rac1 was curious given that CIP4 was discovered as a specific Cdc42 interacting protein (Aspenström, 1997). Therefore, we tested if inhibition of other Rho-GTPases also affected CIP4 localization. We transfected cortical neurons with CIP4-tdTomato and either GFP, GFP-N17-Rac1 (DN-Rac1), GFP-N17-Cdc42 (DN-Cdc42) or GFP-N17-RhoA (DN-RhoA) and fixed the cultures one day later. We discovered that DN-Rac1 significantly decreased the peripheral CIP4 intensity, while DN-Cdc42 significantly increased peripheral CIP4 intensity (Fig. 2D). DN-RhoA did not affect the intensity of CIP4 at the periphery. We confirmed the specificity of Rac1 for CIP4 localization to the protruding edge by expressing constitutively active and dominant negative Rac1 constructs. Cortical neurons were co-transfected with CIP4-tdTomato and GFP-V12-Rac1 (GFP-CA-Rac1) or GFP-N17-Rac1 (DN-Rac1), followed by fixation and labeling of actin filaments with phalloidin. CIP4 was concentrated at the protruding edge of neurons coexpressing CA-Rac1, similar to CIP4-transfected neurons (Fig. 2F,G). Conversely, CIP4 was dispersed in a punctate fashion throughout the cytoplasm and did not localize to the periphery of neurons expressing CIP4 and DN-Rac1 (Fig. 2F,G). Together, these results suggest that CIP4 localization to the periphery of neurons is positively regulated by Rac1 and negatively regulated by Cdc42.
Previously we showed that CIP4 overexpression reduced the number of filopodia in stage 1 neurons by causing the extension of lamellipodia/veils (Saengsawang et al., 2012). Additionally, CIP4 transfected neurons developed many more actin ribs, defined as thin actin bundles within the lamellipodium that do not protrude more than a micron past the periphery, but fewer filopodia compared to controls (Saengsawang et al., 2012). Here we confirmed that result and show that CA-Rac1 also increased the number of ribs and decreased the number of filopodia in stage 1 cortical neurons (Fig. 2H,I). CIP4 and CA-Rac1 co-expression resulted in an additive increase in actin ribs and markedly fewer filopodia (Fig. 2H,I). In contrast, when neurons were transfected with CIP4 and DN-Rac1, they developed fewer actin ribs but more filopodia compared to CIP4 alone (Fig. 2H,I). The increased number of filopodia was recapitulated, albeit to a lesser extent, with a 30-minute treatment with the Rac1 inhibitor NSC23766 (Fig. 2E). These results indicate that active Rac1 functions synergistically with CIP4 at the protruding edge to limit filopodia numbers, while increasing the number of actin ribs in stage 1 cortical neurons.
CIP4 colocalizes with WAVE1 at the protruding edge of neurons
To determine if Rac1 signals through a known or novel pathway by interacting with CIP4 we explored whether a candidate Rho-GTPase interacting protein was dynamically colocalized with CIP4 at the protruding edge of living cortical neurons. We focused on WAVE1 (Wiskott-Aldrich syndrome protein VErprolin homologous 1) which is a known scaffolding protein that regulates actin polymerization and organization downstream of Rac1 (Soderling and Scott, 2006). WAVE1 is expressed specifically in the brain at high levels, where it has been shown to continuously distribute along the protruding edge of growth cones, similar to CIP4 (Nozumi et al., 2003; Saengsawang et al., 2012; Soderling et al., 2007). In addition, dCIP4/TOCA1 has been shown to colocalize with WAVE1 on tubulovesicular structures in Drosophila S2 cells (Fricke et al., 2009) and CIP4 was shown to co-immunoprecipitate with WAVE1 in 293T cell lysates (Roignot et al., 2010).
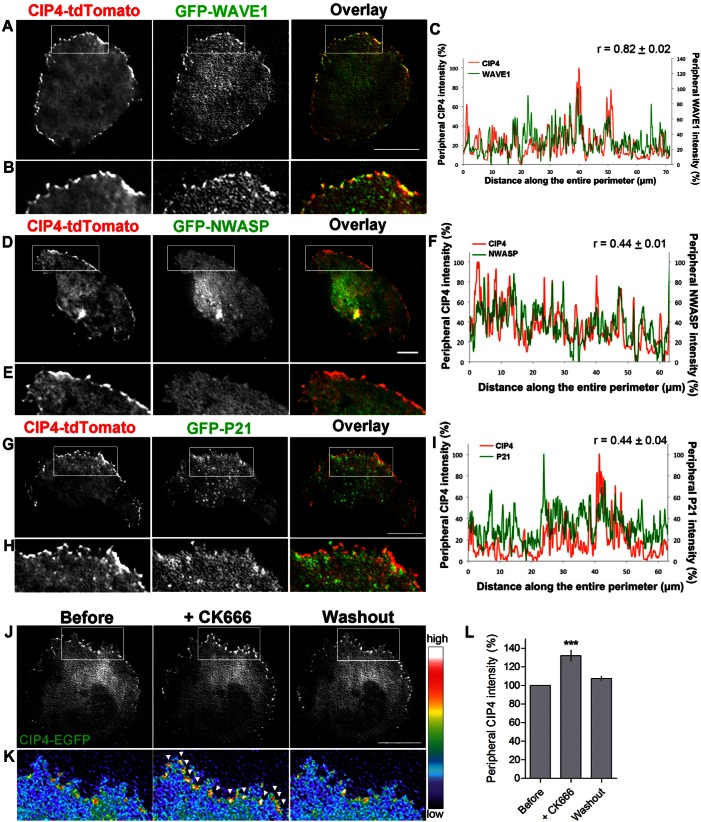
To test whether CIP4 colocalizes with WAVE1 at the protruding edge of living neurons, we co-transfected CIP4-tdTomato and GFP-WAVE1 into the cortical neurons and imaged stage 1 neurons after 1 day in vitro (DIV). Time-lapse TIRFM imaging showed that CIP4 and WAVE1 dynamically colocalized at the protruding edge of the neurons (Fig. 3A–C; supplementary material Movie 1). Line scans of the intensity of both CIP4 and WAVE1 around the periphery of the neuron showed similar patterns of distribution. To quantitatively determine the amount of colocalization around the periphery of living neurons we computed the Pearson's correlation coefficient (r), which ranges from 0 (no colocalization) to 1 (entirely colocalized), from peripheral traces of several living neurons and confirmed a high degree of colocalization (r = 0.82±0.02, n = 4 neurons) between CIP4 and WAVE1. To confirm these results, we determined if CIP4 and endogenous WAVE1 colocalized at the protruding edge of neurons. Because we do not have a suitable antibody for immunocytochemistry of CIP4 we expressed myc-CIP4, fixed and labeled stage 1 cortical neurons with antibodies to myc and WAVE1. Imaging these neurons showed that myc-CIP4 and endogenous WAVE1 colocalized (r = 0.70±0.02, n = 19) at the protruding edge of stage 1 cortical neurons (supplementary material Fig. S2A–C). In contrast, there was little colocalization between CIP4-tdTomato and GFP-N-WASP (r = 0.44±0.01, n = 4) (Fig. 3D–F) or between myc-CIP4 and endogenous N-WASP (r = 0.20±0.01, n = 6) (supplementary material Fig. S2D–F). N-WASP is another activator of actin polymerization that has been reported to directly interact with dCIP4/TOCA1 downstream of Cdc42 in non-neuronal cells (Fricke et al., 2009). Together these results demonstrate that in stage 1 cortical neurons CIP4 associates with WAVE1, but not N-WASP at the protruding edge.
Fig. 3.
CIP4 colocalizes with WAVE1 but not with N-WASP or Arp2/3 in cortical neurons. (A,D,G) Images of stage 1 cortical neurons transfected with CIP4-tdTomato and GFP-WAVE1, GFP-N-WASP or EGFP-P21 (part of the Arp2/3 complex). (B,E,H) Images magnified from boxes in A, D and G showing that CIP4 colocalizes with WAVE1 (B) but not with N-WASP (E) or P21 (G), respectively. (C,F,I) Representative graphs showing the normalized intensity of CIP4 (red) and WAVE1 (C), N-WASP (F) or P21 (I) (green) from lines scans at the peripheral membrane of neurons shown in A, D and G, respectively. Numbers above the graphs indicate the average Pearson's correlation coefficient (r) (n = 4, 4 and 6 neurons for C, F and I, respectively). (J) Image of a stage 1 cortical neuron transfected with CIP4-EGFP and tdTomato-F-tractin before treatment, after 5 minutes of Arp2/3 inhibitor treatment (CK666, 10 µM), and after washout. (K) Images magnified from boxes in J showing the intensity of CIP4 at the peripheral area in pseudocolor. (L) Bar graphs of average intensity of CIP4 at the peripheral membrane of the cells in CIP4-transfected neurons before, 2–5 minutes after CK666 treatment and after washout. Data are expressed as mean ± s.e.m. ***P<0.001 compared with before treatment (one-way ANOVA with Dunnett's post-test comparison); n = 5 treatments. Scale bars: 10 µm.
CIP4 does not function through Arp2/3
Since Arp2/3 is a well-known downstream target of WAVE1 and plays a major role in lamellipodia formation in various cell types, we determined whether CIP4 colocalizes with P21, a protein in the Arp2/3 complex. We co-transfected CIP4-tdTomato and GFP-P21 into cortical neurons and imaged stage 1 neurons after 1DIV. However, we detected little colocalization of CIP4 with P21 (r = 0.44±0.04, n = 6) at the protruding edge of living neurons (Fig. 3G–I). We confirmed these results by determining whether CIP4 colocalized with endogenous Arp2/3 complex by using an Arp3 antibody. We still found little overlap between myc-CIP4 and endogenous Arp3 (r = 0.40±0.03, n = 7) at the protruding edge of stage 1 neurons (supplementary material Fig. S2G–I). Additionally, we determined whether pharmacological inhibition of Arp2/3 affects CIP4 localization in neurons. We treated cortical neurons expressing CIP4-EGFP with CK666, a specific Arp2/3 inhibitor (Nolen et al., 2009; Wu et al., 2012), and observed CIP4 distribution before, during and after washout of the drug. Arp2/3 inhibition with CK666 treatment increased CIP4 localization at the protruding edge by 32±6.0%, which decreased to the pre-treatment level after drug washout (Fig. 3J–L). These results suggest that Arp2/3 activation and the formation of dendritic actin networks are incompatible with the actin ribbed network that results from expression of CIP4 protein at the protruding edge of neurons. Rather, inhibition of Arp2/3 would favor unbranched actin filament growth at the protruding edge, which would result in more actin rib formation via CIP4.
Barbed end capping of actin filaments limits CIP4 at the protruding edge
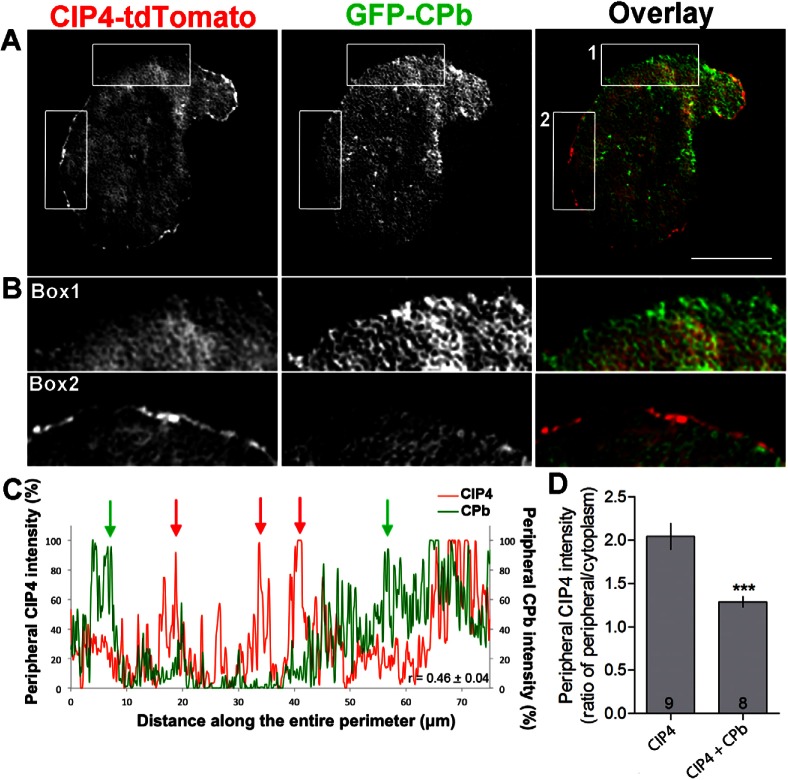
One consequence of forming a dendritic actin network in lamellipodia is an increased demand for capping of branched actin filaments, so they remain short enough to provide sufficient protrusive forces (Pollard and Borisy, 2003). Capping proteins (alpha and beta) are responsible for capping actin filament barbed ends in cells (Cooper and Sept, 2008). To determine if CIP4 and capping protein had an antagonistic relationship, similar to Arp2/3, at the protruding edge membrane of newly plated neurons, we overexpressed GFP-capping protein-beta (GFP-CPb) with CIP4-TdTomato and determined their dynamic distribution in cortical neurons (Fig. 4A). Images of living stage 1 neurons showed that regions of GFP-CPb and CIP4-tdTomato were largely non-overlapping (Fig. 4B). A line scan of CIP4 (red) and CPb (green) showed that they formed an alternating pattern along the peripheral edge of the neuron (Fig. 4C). Quantification of colocalization of CIP4 and CPb in several cells resulted in a low Pearson's correlation coefficient (r = 0.46±0.04, n = 6). In addition, the intensity of CIP4 at the periphery was lower in neurons overexpressing capping protein b (Fig. 4D). These results further support the idea that CIP4 preferably localizes to growing, rather than capped, actin barbed ends at the protruding edge of cortical neurons.
Fig. 4.
Capping protein β decreases CIP4 level at the protruding edge. (A) Image of a stage 1 cortical neuron transfected with CIP4-tdTomato and GFP-capping protein β. (B) Images magnified from boxes in A showing that CIP4 does not colocalize with capping protein β. (C) Representative graph showing the normalized intensity of CIP4 (red) and capping protein β (green) at the peripheral membrane of neuron shown in A. Green and red arrows indicate regions with a high intensity of capping protein β or CIP4, respectively. Value of r shown in the graph indicates the average Pearson's correlation coefficient (n = 6 neurons). (D) Bar graph of average intensity ratio of CIP4 at the peripheral/cytoplasm in CIP4-transfected neurons with and without capping protein β. Data are expressed as mean ± s.e.m. ***P<0.001 (two-tailed unpaired t-test with Welch's correction). Numbers in the bar graph indicate number of cells quantified from at least two independent experiments. Scale bar: 10 µm.
To determine whether uncapped barbed ends of actin filaments are required for CIP4 localization to the protruding edge membrane we transfected cortical neurons with CIP4-EGFP and mRuby-lifeact (to label F-actin). We then imaged these neurons while treating them with nanomolar concentrations of cytochalasin D (CyD), a drug that specifically caps actin filaments at low concentrations (Bear et al., 2002; Dent et al., 2007; Urbanik and Ware, 1989). Before CyD treatment, CIP4 dynamically localized to the protruding edge of the growth cone as we have shown previously (Saengsawang et al., 2012) (Fig. 5A). After CyD treatment CIP4 decreased to 57.8±8.0% of pretreatment levels at the protruding edge of the growth cone. Surprisingly, coincident with the disappearance of CIP4 at the protruding edge, CIP4-positive puncta appeared in the central region of the growth cone (Fig. 5A,B; supplementary material Movie 2). After formation, these CIP4 puncta were transported away from the protruding edge with actin retrograde flow (Fig. 5A,E). The retrogradely moving puncta do not colocalize with Rab5 (supplementary material Fig. S3A), Clathrin (supplementary material Fig. S3B), transferrin or FM4-64 labeled vesicles (data not shown) indicating they are not likely to be clathrin-coated vesicles. Both the decrease in CIP4 at the protruding edge and the increase in the number of CIP4-positive puncta in the central region of the growth cone returned to pretreatment levels after removal of CyD (Fig. 5C,D). Interestingly, upon drug washout CIP4 puncta in the central region of the growth cone formed actin comet tails, which appeared to propel CIP4 puncta forward, throughout the growth cone (Fig. 5A,F). These CIP4 puncta, propelled by actin comet tails, are similar to those that have been described in Drosophila S2 cells transfected with dCIP4/TOCA1 and are likely to represent CIP4-labeled endocytic vesicles (Fricke et al., 2009). Notably, in the presence of latrunculin, a drug that depolymerizes actin filaments, CIP4 did not induce tubulation (supplementary material Fig. S4) as described in non-neuronal cells treated with latrunculin (Itoh 2005); a further indicator that CIP4 functions differently in neurons and non-neuronal cells. Together, these results suggest that uncapped barbed ends of actin filaments are important for CIP4 localization at the protruding edge and decreasing these barbed ends through CyD-induced actin capping results in CIP4 redistribution to retrogradely transported vesicle-like structures that, when actin polymerization resumes (CyD washout), undergo actin-based propulsion.
Fig. 5.
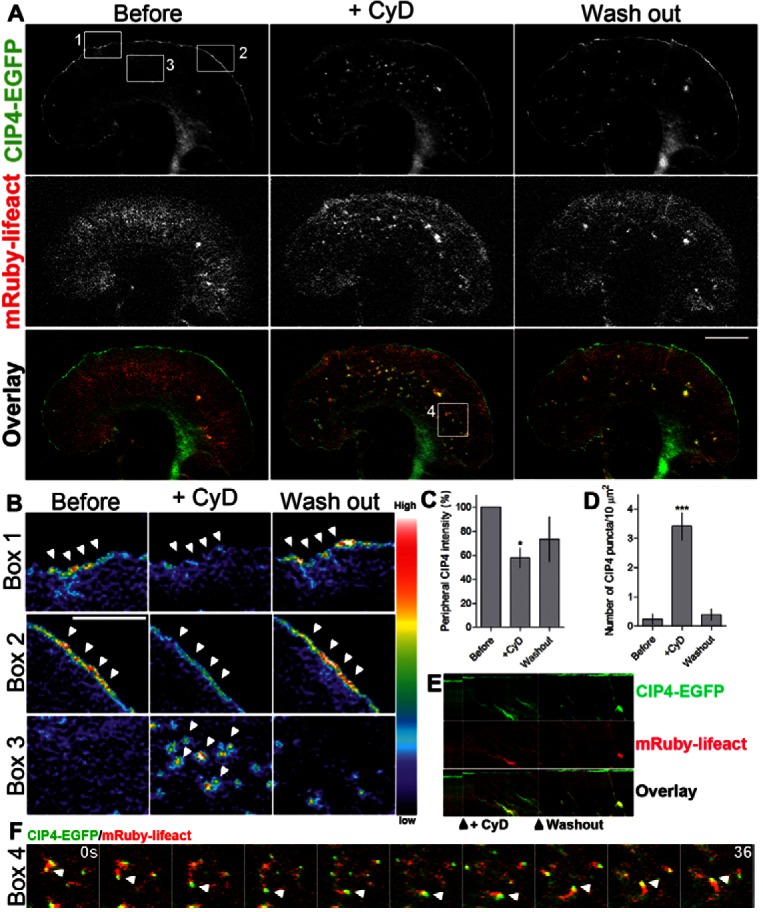
CIP4 localization at the protruding membrane depends on barbed ends of actin filaments. (A) Image of a growth cone from a cortical neuron transfected with CIP4-EGFP and mRuby-lifeact before and after cytochalasin D (CyD) treatment (100 nM). (B) Pseudocolored images magnified from boxes 1, 2 and 3 in A, showing the intensity of CIP4 in the peripheral area (box 1 and 2) and internal area (box 3) of the growth cone before and after CyD treatment. (C,D) Bar graphs of the average intensity of CIP4 at the peripheral area of the cells (C) and number of CIP4 puncta in the internal area (D) before, 4 minutes after CyD treatment and after washout. Data are expressed as mean ± s.e.m. *P<0.05 and ***P<0.001 compared with before treatment (one-way ANOVA with Dunnett's post-test comparison); n = 7 treatments. (E) Kymograph showing CIP4 and F-actin dynamics before CyD treatment, during and after CyD was washed out. (F) Selected frames from box 4 in A, showing time-lapse images of CIP4 and actin comet tails after CyD washout. Scale bars: 10 µm (A), 5 µm (B).
Because capping actin filaments with CyD and inhibiting PI3K with LY294002 decreased CIP4 at the protruding edge to similar levels (∼57%) and the fact that PIP3 has been shown to inhibit capping proteins, although with lesser potency than PIP2 (Schafer et al., 1996), we sought to determine if the effects of treatment with LY294002 on CIP4 localization were only due to actin filament capping. To test this we treated neurons with both LY294002 and Cytochalasin D and observed CIP4 localization at the protruding edge of living neurons. After only one minute treatment with both drugs CIP4 intensity at the protruding edge decreased to 42.1±8.0% (supplementary material Fig. S5A–C) of pretreatment levels. This decrease in CIP4 was substantially greater than LY294002 (Fig. 1A–C) or CyD alone (Fig. 5C), both of which decreased the level of CIP4 to 57% after 20 minutes or 4 minutes, respectively. In addition, one minute incubation with either LY294002 or CyD decreased CIP4 levels at the protruding edge to only 88.7±7.7% and 91.5±10.0% (supplementary material Fig. S5D) of pretreatment levels, respectively. Together, these results suggest that inhibition of PIP3 production and actin filament capping have synergistic effects on levels of CIP4 at the membrane and are likely to be acting by separate mechanisms that converge on CIP4 and affect its distribution in neurons.
CIP4 functions with DAAM1 and Ena/VASP proteins at the protruding edge
The previous data suggest that CIP4 is functioning through a PIP3/Rac1/WAVE1 pathway, but not through an Arp2/3-rich dendritic actin network, in cortical neurons. Rather, CIP4 appears to concentrate at the protruding edge of these neurons when uncapped actin filaments are concentrated at the membrane. Two families of actin-associated proteins that play important roles in unbranched actin filament polymerization are the formin and Ena/VASP families of proteins. Previous work has shown that at least one of the diaphanous related formins (mDia2) is not present in embryonic cortical neurons (Dent et al., 2007). Therefore, we focused on a formin that is present in prenatal cortex, DAAM1 (Kida et al., 2004; Nakaya et al., 2004). The formin DAAM1 (Dishevelled-associated activators of morphogenesis-1) is part of a class of proteins known to promote actin nucleation and elongation of unbranched actin filaments (Goode and Eck, 2007; Paul and Pollard, 2009; Prokop et al., 2011). DAAM1 has also been shown to interact with CIP4 and to regulate cell morphology and actin dynamics in COS7 cells (Aspenström et al., 2006). Although DAAM1 and other formins are commonly known to be activated by Rho, DAAM1 has been shown to function downstream of Rac1 in developing neurons of the Drosophila nervous system (Matusek et al., 2008).
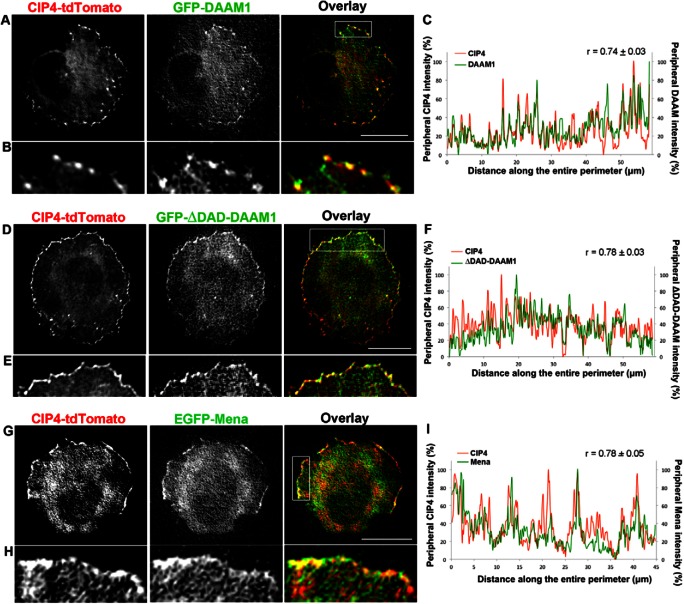
Since we recently showed that CIP4 is expressed early during CNS development (Saengsawang et al., 2012) and functions at the protruding edge, downstream of Rac1 (Fig. 2), we asked whether CIP4 and DAAM1 dynamically colocalized to the protruding edge by co-expressing CIP4-tdTomato and GFP-DAAM1. Time-lapse TIRF imaging showed that DAAM1 colocalized (r = 0.74±0.03, n = 6) with CIP4 at the protruding edge of living cortical neurons (Fig. 6A–C; supplementary material Movie 3). Additionally, ΔDAD-DAAM1, a constitutively active form of DAAM1 (Liu et al., 2008), also colocalized (r = 0.78±0.05, n = 5) with CIP4 at the protruding edge (Fig. 6D–F). Although we have not shown that DAAM1 and CIP4 directly bind one another these results identify DAAM1 as an actin elongating protein that coordinates with CIP4 to function at the protruding edge of neurons and may contribute to the polymerization of long, thin actin filaments bundles resulting in ribbed lamellipodial structures.
Fig. 6.
CIP4 colocalizes with DAAM1 and Mena at the protruding membrane of cortical neurons. (A,D,G) Images of stage 1 cortical neurons transfected with CIP4-tdTomato and GFP-DAAM1 (A) or GFP-ΔDAD-DAAM1 (D) or EGFP-Mena (G). (B,E,H) Images magnified from boxes in A, D and G, showing the colocalization of CIP4 and GFP-DAAM1, GFP-ΔDAD-DAAM1 or GFP-Mena at the peripheral area, respectively. (C,F,I) Representative graphs showing the normalized intensity of CIP4 (red) and DAAM1, ΔDAD-DAAM1 or Mena (green) at the peripheral membrane of neurons shown in A, D and G. Value of r shown in the graph indicates the average Pearson's correlation coefficient; n = 6, 5 and 6 neurons for C, F and I, respectively. Scale bars: 10 µm.
Another important actin-associated protein family whose activity results in formation of unbranched actin filaments, and is present in embryonic cortical neurons, is the Ena/VASP family of proteins. Here we focused on Mena, as it is highly expressed in prenatal cortex (Lanier et al., 1999). To determine if CIP4 and Mena dynamically colocalized we transfected cortical neurons with CIP4-tdTomato and EGFP-Mena and imaged stage 1 neurons in time-lapse. CIP4 and Mena clearly colocalized (r = 0.78±0.05, n = 6) at the protruding edge of living neurons (Fig. 6G–I; supplementary material Movie 4). Because EGFP-Mena was also present at fairly high levels in the cytoplasm, we determined if endogenous Mena and CIP4 colocalize at the protruding edge. We expressed myc-CIP4, fixed and labeled stage 1 cortical neurons with an antibody to Mena. Imaging these neurons showed that myc-CIP4 and endogenous Mena colocalized (r = 0.70±0.02, n = 6) at the protruding edge of stage 1 cortical neurons (supplementary material Fig. S2J–L). These data indicate that Mena and CIP4 may have coordinated functions at the protruding edge of cortical neurons.
To determine if Mena was necessary for CIP4 localization at the protruding edge, we transfected neurons with CIP4-EGFP and mCherry-FP4-Mito, a construct that targets all three Ena/VASP family members (Mena/VASP/EVL) to the mitochondrial surface, effectively resulting in a functional Ena/VASP knockout cell (Bear et al., 2000; Dent et al., 2007). Importantly, the control for these experiments is transfection with a construct that only varies in one amino acid, termed mCherry-AP4-Mito. Ena/VASP inactivation with mCherry-FP4-Mito did not result in an apparent change of CIP4 at the protruding edge, as evidenced by similar CIP4-EGFP label in both AP4-Mito and FP4-Mito transfected neurons (Fig. 7A,C). This result indicates Ena/VASP is not likely targeting CIP4 to the membrane. However, CIP4 may maintain peripheral localization during the relatively slow sequestration (compared to the drug treatments used earlier) of Ena/VASP proteins to the mitochondrial surface over the period of a day.
Fig. 7.
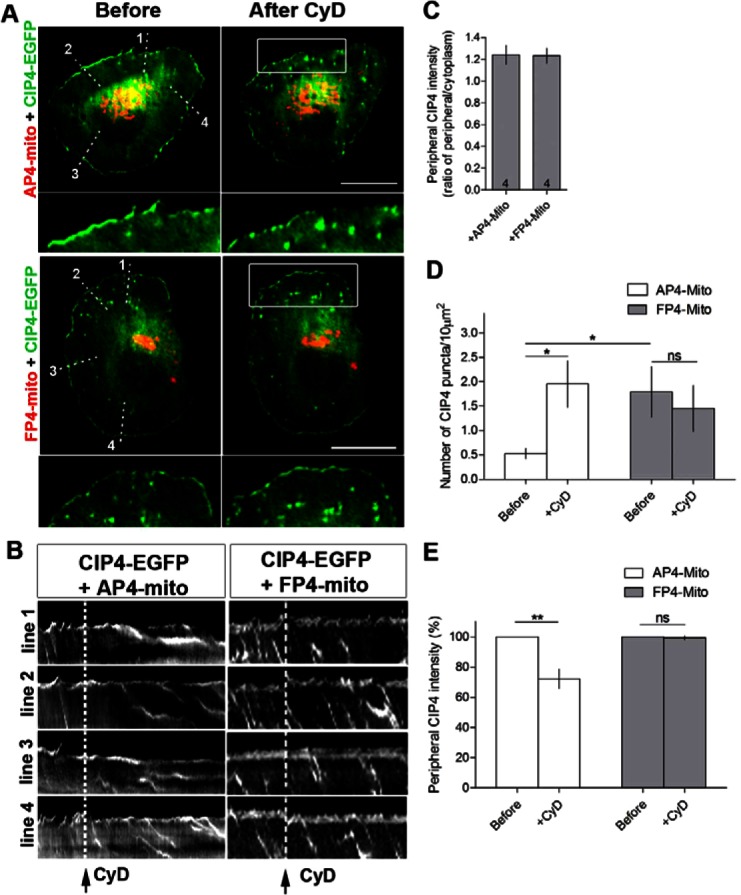
Functional inactivation of Ena/VASP proteins increases internal CIP4 puncta and renders neurons refractory to cytochalasin D treatment. (A) Image of stage 1 cortical neurons transfected with CIP4-EGFP and mCherry-FP4-mito (which sequesters all Ena/VASP proteins to the mitochondria) or mCherry-AP4-mito (targets to mitochondria but does not sequester Ena/VASP proteins) before and after cytochalasin D (CyD) treatment. (B) Kymograph from time-lapse series of CIP4-EGFP from lines drawn in A. Dashed lines indicate the time point where cytochalasin D was added. (C) Bar graph of peripheral CIP4 intensity ratio in AP4-mito or FP4-mito transfected neurons. Numbers in the bar graph indicate number of cells quantified from at least two independent experiments. (D) Bar graph of average number of CIP4 puncta in the internal area of the cells or (E) peripheral CIP4 intensity in AP4-mito or FP4-mito transfected neurons before and after CyD treatment. Data are expressed as mean ± s.e.m. *P<0.05 and **P<0.01 compared with before treatment (paired t-test); n = 6 and 4 cells for AP4 and FP4-mito, respectively, from at least three independent experiments. Scale bars: 10 µm.
While no apparent effect was observed at the protruding edge, mCherry-FP4-Mito transfection did increase the number of CIP4 puncta in the cytoplasm of stage 1 neurons (Fig. 7A,D), reminiscent of cytochalasin D treatment (Fig. 5). These CIP4-EGFP-positive puncta also moved rearward with actin retrograde flow (Fig. 7B), similar to those induced by CyD (Fig. 5E). To determine if CyD was still capable of reducing CIP4 at the membrane and inducing CIP4-positive puncta, we imaged both FP4-mito and AP4-Mito transfected neurons during treatment with CyD. The AP4-Mito transfected neurons showed a marked increase in CIP4-positive puncta moving retrogradely, however no increase in the already high level of puncta was observed in the FP4-Mito-transfected neurons (Fig. 7B,D). Furthermore, CyD treatment was not able to decrease the amount of CIP4 at the protruding edge of the FP4-Mito expressing cells like it did with the AP4-Mito expressing cells (Fig. 7B,E). These results suggest that the slow sequestration of Ena/VASP proteins to the mitochondrial surface allows for changes in CIP4 at the protruding edge that make it more resistant to actin capping. Together, these data are consistent with the hypothesis that Ena/VASP proteins act as actin filament anti-capping factors (Bear et al., 2002) and that CIP4 localization at the protruding edge of neurons is dependent on uncapped actin filaments.
Discussion
Here we demonstrate that the localization and function of CIP4 at the protruding edge of neurons depends on both lipid composition and the uncapped barbed ends of actin filaments. Increasing PIP3 content in the membrane resulted in more CIP4, while decreasing PIP3 resulted in less CIP4, at the protruding edge membrane. Increasing actin filament barbed-end capping, by addition of a low concentration of Cytochalasin D, by expression of capping protein or by inhibition of Ena/VASP proteins, disrupted CIP4 localization at the protruding edge of stage 1 cortical neurons. Furthermore, we show that Rac1 is the predominant Rho family GTPase that functions with CIP4 in neurons through WAVE1, resulting in a ribbed- and veil-rich actin network. Although Arp2/3 functions downstream of Rac in many cell types, we found it was not part of the pathway linking CIP4 with actin filaments. Rather, DAAM1 and Ena/VASP proteins coordinate with CIP4 at the membrane. Finally, an acute increase in barbed end actin capping by addition of cytochalasin D or chronic inactivation of Ena/VASP family anti-capping proteins, partially converts the localization of CIP4 from the protruding edge to retrogradely flowing CIP4 puncta that resemble tubulovesicular structures seen in other cell types that overexpress CIP4 (Frost et al., 2008; Kamioka et al., 2004; Saengsawang et al., 2012; Tian et al., 2000). Together these data suggest a model (Fig. 8) in which CIP4 plays an important role in the formation of ribs/veils and tubulovesicular structures in neurons but does not directly function in pathways resulting in formation of lamellipodia and filopodia.
Fig. 8.
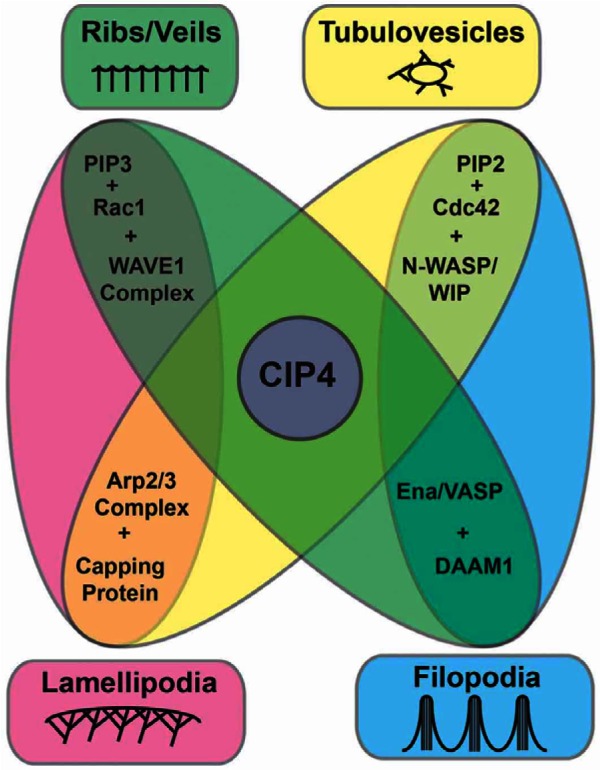
Model of CIP4 localization and function at the protruding edge of stage 1 cortical neurons. CIP4 requires PIP3 and active Rac1 to localize at the protruding edge of neurons, where it functions with WAVE1 and DAAM1 and/or Ena/VASP proteins to promote actin polymerization into long thin filaments (actin ribs). These actin ribs (green box and oval), which are thinner than filopodia and only extend slightly beyond the protruding edge, are connected by veils and result in a ‘lamellipodia-like’ structure in the light microscope. However, they differ from classic lamellipodia (pink box and oval) in that they function with Ena/VASP and DAAM1 instead of with the Arp 2/3 complex and capping proteins. However, when the PIP2/Cdc42/N-WASP/WIP pathway is activated it can signal, through CIP4, the Arp2/3 complex and capping protein to produce tubulovesicular structures (yellow box and oval). If the Ena/VASP and DAAM1 pathways are activated downstream of PIP2/Cdc42/N-WASP, but CIP4 is not present, then filopodia are formed (blue box and oval).
PIP2 and PIP3 are important regulators of actin cytoskeletal dynamics in all cell types. In many cells, PIP3 accumulates at the leading edge of cells, coincident with increases in actin nucleation, in response to cell stimulation (Insall and Weiner, 2001). This asymmetric distribution of PIP3 was also observed at the protruding edge of growth cones, in response to neurotrophin stimulation (Henle et al., 2011). Interestingly we found that PIP3 is required for CIP4 localization. Inhibiting PIP3 synthesis with LY294002 decreased the level of CIP4 at the protruding edge, which was independent of Akt activation, since Akt inhibition did not produce the same effect. In contrast, inhibition of Akt increased CIP4 localization at the protruding edge. This result may be due to an increase in the available PIP3, leading to increased Rac1 activity. Nevertheless, inhibition of PI3K can also inhibit the formation of PIP2 and the AKT-PH domain probe that we used can recognize both PIP2 and PIP3 (Oikawa et al., 2008). Therefore, we cannot rule out the possibility that PIP2 might also play a role in CIP4 localization to the protruding edge of neurons. Further study with other types of PH domains are required to test this hypothesis.
We found that Rac1 is required for CIP4 localization in neurons. Inhibition of Rac1 with a pharmacological inhibitor (NSC23766) or transfection with a dominant negative Rac1 construct decreased CIP4 at the protruding edge. In contrast, constitutively active Rac1 increased CIP4 at the protruding edge. CIP4 is known to interact with Cdc42 in other cell types and takes its name from the GTPase (Aspenström, 1997). However, Cdc42 and RhoA do not appear to be necessary for the distribution of CIP4 at the protruding edge of neurons and may even inhibit such localization. We show that transfection of neurons with DN-Cdc42 increased the levels of CIP4 at the protruding membrane, while DN-RhoA had no effect. Additionally, we detected little colocalization between CIP4 and N-WASP in neurons, a protein that has been shown to interact with CIP4 in non-neuronal cells, downstream of Cdc42. Indeed we found that CIP4 colocalized with WAVE1, another actin associated protein that functions downstream of Rac1, at the protruding edge of stage 1 cortical neurons.
We then sought to identify proteins that directly interact with actin and function downstream of CIP4 at the protruding edge. Arp2/3 and formins are two major proteins that induce nucleation of actin filaments. Arp2/3 activation results in the formation of short branched actin filaments, while formins assemble long, unbranched actin filaments (Pollard, 2007). The balance between the presence and activity of these two actin nucleators is therefore crucial for the regulation of actin organization. Activation of Arp2/3 favors lamellipodia formation, while activation of formins favors filopodia formation (Small et al., 2002; Block et al., 2008). Surprisingly, inhibition of the Arp2/3 complex promotes CIP4 localization at the protruding edge. Since Arp2/3 functions downstream of active Rac1, inhibiting Arp2/3 might increase Rac1 availability and therefore allow Rac1 to recruit more CIP4 to the membrane.
The level of Arp2/3 is very low in neurons compared to other cell types (Strasser et al., 2004). It is therefore possible that the actin nucleating formin proteins might play a major role in nucleating actin in neurons. DAAM1 has been shown to function downstream of Rac1 in the developing neurons in the Drosophila nervous system (Matusek et al., 2008) and we show that CIP4 colocalizes with DAAM1 at the protruding edge of stage 1 neurons. Moreover, although Ena/VASP proteins do not nucleate actin filaments directly (Barzik et al., 2005), they play a key role in developing cortical neurons by enhancing unbranched actin polymerization that results in the formation of filopodia (Dent et al., 2007). We show here that Mena, one of three Ena/VASP family proteins expressed in cortical neurons (Lanier et al., 1999), colocalizes with CIP4 at the protruding edge of stage 1 neurons. Moreover, functional inactivation of Ena/VASP proteins results in the formation of CIP4 puncta that resemble tubulovesicular structures in other cell types. Thus, DAAM1 and Ena/VASP proteins are likely to link CIP4 function to actin polymerization resulting in lamellar regions enriched in actin ribs and veils.
In our previous study we suggested that CIP4 inhibits neurite formation by forming lamellipodial structures in early differentiated neurons (Saengsawang et al., 2012). The present work suggests that it is not the formation of lamellipodia per se, but rather a ribbed/veiled actin network that gives rise to a lamellipodial-like structure in neurons. This ribbed and veiled lamellar structure is composed of actin filaments of a different molecular and structural makeup compared with lamellipodia in other cell types. The classic lamellipodial structure in non-neuronal cells is highly dependent on activation of PIP3-Rac1-WAVE1 and an Arp2/3-capping protein dendritic actin network (Fig. 8, pink oval). Although the ribbed/veiled lamellar network shown here is also dependent on PIP3-Rac1-WAVE1, it is not activated by Arp2/3-capping protein, but rather by CIP4, Ena/VASP or DAAM1 (Fig. 8, green oval). The ribbed/veiled actin network can be converted into actin-associated tubulovesicular structures by favoring PIP2-Cdc42-N-WASP and Arp2/3-capping protein (Fig. 8, yellow oval). Finally, activation of PIP2-Cdc42-N-WASP and Ena/VASP or DAAM1, in the absence of CIP4, results in filopodia formation (Fig. 8, blue oval). We suggest that CIP4 is not required for classic lamellipodia or filopodia, but is essential for a ribbed/veiled lamellar actin network and actin-associated tubulovesicular structures. These results suggest that CIP4 may play a different role than another CIP4 family member TOCA-1, because TOCA-1 has been shown to induce formation of filopodia and endocytic tubules in neuronal cell lines in an N-WASP and Cdc42-dependent fashion (Bu et al., 2009; Bu et al., 2010). Thus, according to the model in Fig. 8, TOCA-1 may be functioning at the nexus of the yellow and blue ovals to induce filopodia or tubulovesicles, depending on the presence and activation state of downstream proteins.
This model (Fig. 8) is consistent with the data presented here and suggests why CIP4 needs to be downregulated in late embryonic cortical development (Saengsawang et al., 2012). It is known that filopodia serve a key function in cortical neurite initiation (Dent et al., 2007). Thus, CIP4 would have to be downregulated to efficiently produce filopodia, which themselves develop into neurites (Dent et al., 2007). If CIP4 is maintained at high levels it would favor the formation of actin ribs/veils and tubulovesicular structures, which may play important roles in neuronal migration, as has been shown for the F-BAR protein SrGAP2 (Guerrier et al., 2009), but inhibit the formation of lamellipodia and filopodia, which are essential for productive axon and dendrite outgrowth.
In conclusion, our findings support the following model for the induction of rib/veil-rich lamellar structures in cortical neurons. First, PIP3 production recruits active Rac1 and the WAVE1 complex at the protruding edge. Concurrently, activation of anti-capping proteins (Ena/VASP) or actin elongating proteins (DAAM1) occurs in the same region of membrane. Both the activation of PIP3-Rac-WAVE1 and Ena/VASP or DAAM1 in a temporally and spatially specific manner may be needed to recruit CIP4 proteins, since both the F-BAR (membrane binding) and SH3 (DAAM1 binding) domains are essential for localization to protruding membranes (Fig. 1). Furthermore, the polymerization of CIP4 proteins is required for their localization to the protruding edge (Saengsawang et al., 2012). Thus, CIP4 may be acting as a type of coincidence detector that only localizes to protruding membranes when both membrane binding and barbed-end actin-associated proteins are activated in localized regions of the membrane. The number of CIP4 molecules may dictate the organization of actin filaments. If there is excess CIP4 present along the protruding edge, thin actin ribs are likely to form in a lamellar region, whereas small, dispersed regions of CIP4 may favor filopodia formation. Indeed, CIP4 is concentrated at the tips of both extending actin ribs and filopodia in stage 1 neurons (Saengsawang et al., 2012). Here we report a series of proteins that regulate CIP4 localization and function and coordinate with CIP4 to induce actin polymerization and form ribbed/veiled protrusions at the protruding edge of neurons. This model does not exclude the possibility that the activity of CIP4 could also be regulated by post-translational modifications such as phosphorylation. Further studies will be required to test this model.
Materials and Methods
Plasmids and reagents
Full-length CIP4-EGFP was a gift from Dr Andrew Craig (Queen's University, Kingston, Ontario, Canada). Full-length CIP4 was cloned into ptdTomato-N1 vector (Clontech) using CIP4-EGFP plasmid as a template to create CIP4-tdTomato. mRuby-lifeact was a gift from Dr Roland Wedlich-Soldner (Max Planck Institute, Martinsried, Germany) and was subcloned into pCAGG vector. Full-length tdTomato-F-tractin and EGFP-F-tractin were used to label F-actin in live imaging experiments (Johnson and Schell, 2009) and were gifts from Dr Michael J. Schell (Uniformed Services University, Bethesda, MD). GFP-V12-Rac1, GFP-N17-Rac1, GFP-N17-Cdc42, GFP-N17-Rho, GFP-PLC-PH and GFP-Akt-PH have been validated in previous studies (Myers et al., 2012; Robles et al., 2005; Woo and Gomez, 2006). GFP-WAVE1 was a gift from Dr Scott Soderling (Duke University, NC) (Soderling et al., 2007). GFP-DAAM1 and GFP-ΔDAD-DAAM1 were gifts from Dr Raymond Habas (Temple University, Philadelphia) (Liu et al., 2008). GFP-N-WASP and GFP-P21 were gifts from Dr Lorene Lanier (University of Minnesota) (Strasser et al., 2004). mCherry-Rab5 and DsRed-Clathrin were gifts from Dr Anjon Audhya (University of Wisconsin-Madison). GFP-Capping protein beta, rabbit anti-Mena antibody, mCh-FP4-Mito and mCh-AP4-Mito were from Dr Frank Gertler (Dent et al., 2007). Anti-Mena antibody was used at 1∶1000. Rabbit anti-WAVE1 and rabbit anti-N-WASP antibody (Abcam) were used at 1∶500 and 1∶1000, respectively. Rabbit anti-Arp3 antibody (Millipore) was used at 1∶100. Mouse anti-myc antibody (Invitrogen) was used at 1∶1000. HRP-conjugated secondary antibodies (Jackson IR) were used at 1∶10,000. Actin-stain 670 phalloidin (Cytoskeleton, Inc.) was used at 1∶50. NSC23766 and Akt inhibitor VIII were from Santa Cruz Biotechnology. LY294002, CK666 and Cytochalasin D were from Sigma.
Cortical neuron culture and transfection
All mouse procedures were approved by the University of Wisconsin Committee on Animal Care and were in accordance with NIH guidelines. Cortical (E15.5) neuron cultures were prepared from Swiss Webster mice (Taconic) essentially as described (Viesselmann et al., 2011). Briefly, cortices were dissected, trypsinized and dissociated. Dissociated cortical neurons were resuspended in Nucleofector solution (Mouse Neuron Kit, Lonza, Cologne, Germany) and transfected with an Lonza Nucleofector according to the manufacturer's directions. Transfected neurons were plated on poly-D-lysine (Sigma)-coated glass coverslips adhered to the bottom of 35 mm plastic culture dishes that had a 15 mm hole drilled through the bottom of the chamber. Neurons were plated in plating medium (PM) (Neurobasal medium with 5% FBS (Hyclone), B27 supplement, 2 mM glutamine, 37.5 mM NaCl and 0.3% glucose). After 1 hour, this medium was replaced with serum-free medium (SFM), which was PM without FBS. Neurons were then imaged or fixed after 1 day in vitro (1DIV).
Immunocytochemistry and imaging
For wide-field imaging cortical neurons were fixed in 4% paraformaldehyde/KREBs/sucrose at 37°C. Cultures were rinsed in PBS and blocked with 10% BSA/PBS, permeabilized in 0.2% Triton X-100/PBS and labeled with primary and secondary antibodies. Actin-stain 670 phalloidin (Cytoskeleton, Inc.) was used to label actin filaments (1∶50). For wide-field imaging neurons were fixed, labeled and imaged on a Nikon TE300 inverted microscope as described previously (Saengsawang et al., 2012). All fluorescence live-cell imaging was performed using a Nikon TIRFM microscope as described (Hu et al., 2008). During time-lapse microscopy neurons were kept at 37°C. For live-cell imaging the culture dish was closed with a glass ring, coverslip and silicone grease. All images were collected, measured and analyzed in MetaMorph imaging software (Molecular Devices). Pearson's correlation coefficient was calculated using ImageJ software with JACoP plugin (Bolte and Cordelières, 2006) where a coefficient close to 1.0 indicates colocalization, and 0 indicates low probability for colocalization. Figures were compiled in Photoshop (Adobe).
Statistical analysis
All statistical tests were performed with GraphPad Prism 4.0c. For each data set, two-tailed unpaired t-test with Welch's correction, one-way or two-way ANOVA with Bonferroni or Dunnett’s post-test comparison and two-way repeated measures ANOVA were performed, depending on the type of data sets.
Supplementary Material
Acknowledgments
We thank all the members of the Dent lab (especially Thomas Fothergill) for helpful discussions and critical comments on the manuscript. We thank Fiona Ukken for the help with plasmid cloning. We thank Katherine Kalil for the use of the electroporator and the developmental neurobiology group at UW for critical advice during the project. We thank Frank Gertler for his generous sharing of Ena/VASP-related reagents.
Footnotes
Author contributions
W.S. and E.W.D. conceived the project. W.S. performed most of the experiments and wrote the manuscript with E.W.D. K.L.T. performed experiments in Figure 2. K.L.T., D.C.L., K.M., A.P. and L.P. helped with cortical neurons preparation, experiments and data analysis. T.M.G. provided plasmids, reagents and advice for revising the manuscript. E.W.D. provided advice, overall direction and supervised the execution of the project. All authors read and edited the manuscript.
Funding
This work was supported by grants from the National Institutes of Health [grant number R01-NS064014 to E.W.D.]; and the UW Graduate School (Henry Vilas Associates Award) to E.W.D. Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.117473/-/DC1
References
- Aspenström P. (1997). A Cdc42 target protein with homology to the non-kinase domain of FER has a potential role in regulating the actin cytoskeleton. Curr. Biol. 7, 479–487 10.1016/S0960-9822(06)00219-3 [DOI] [PubMed] [Google Scholar]
- Aspenström P. (2009). Roles of F-BAR/PCH proteins in the regulation of membrane dynamics and actin reorganization. Int. Rev. Cell Mol. Biol 272, 1–31 10.1016/S1937-6448(08)01601-8 [DOI] [PubMed] [Google Scholar]
- Aspenström P., Richnau N., Johansson A. S. (2006). The diaphanous-related formin DAAM1 collaborates with the Rho GTPases RhoA and Cdc42, CIP4 and Src in regulating cell morphogenesis and actin dynamics. Exp. Cell Res. 312, 2180–2194 10.1016/j.yexcr.2006.03.013 [DOI] [PubMed] [Google Scholar]
- Barzik M., Kotova T. I., Higgs H. N., Hazelwood L., Hanein D., Gertler F. B., Schafer D. A. (2005). Ena/VASP proteins enhance actin polymerization in the presence of barbed end capping proteins. J. Biol. Chem. 280, 28653–28662 10.1074/jbc.M503957200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear J. E., Gertler F. B. (2009). Ena/VASP: towards resolving a pointed controversy at the barbed end. J. Cell Sci. 122, 1947–1953 10.1242/jcs.038125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear J. E., Loureiro J. J., Libova I., Fässler R., Wehland J., Gertler F. B. (2000). Negative regulation of fibroblast motility by Ena/VASP proteins. Cell 101, 717–728 10.1016/S0092-8674(00)80884-3 [DOI] [PubMed] [Google Scholar]
- Bear J. E., Svitkina T. M., Krause M., Schafer D. A., Loureiro J. J., Strasser G. A., Maly I. V., Chaga O. Y., Cooper J. A., Borisy G. G. et al. (2002). Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell 109, 509–521 10.1016/S0092-8674(02)00731-6 [DOI] [PubMed] [Google Scholar]
- Block J., Stradal T. E., Hänisch J., Geffers R., Köstler S. A., Urban E., Small J. V., Rottner K., Faix J. (2008). Filopodia formation induced by active mDia2/Drf3. J. Microsc. 231, 506–517 10.1111/j.1365-2818.2008.02063.x [DOI] [PubMed] [Google Scholar]
- Bolte S., Cordelières F. P. (2006). A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224, 213–232 10.1111/j.1365-2818.2006.01706.x [DOI] [PubMed] [Google Scholar]
- Bu W., Chou A. M., Lim K. B., Sudhaharan T., Ahmed S. (2009). The Toca-1-N-WASP complex links filopodial formation to endocytosis. J. Biol. Chem. 284, 11622–11636 10.1074/jbc.M805940200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu W., Lim K. B., Yu Y. H., Chou A. M., Sudhaharan T., Ahmed S. (2010). Cdc42 interaction with N-WASP and Toca-1 regulates membrane tubulation, vesicle formation and vesicle motility: implications for endocytosis. PLoS ONE 5, e12153 10.1371/journal.pone.0012153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson B. R., Lloyd K. E., Kruszewski A., Kim I. H., Rodriguiz R. M., Heindel C., Faytell M., Dudek S. M., Wetsel W. C., Soderling S. H. (2011). WRP/srGAP3 facilitates the initiation of spine development by an inverse F-BAR domain, and its loss impairs long-term memory. J. Neurosci. 31, 2447–2460 10.1523/JNEUROSCI.4433-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohan C. S., Welnhofer E. A., Zhao L., Matsumura F., Yamashiro S. (2001). Role of the actin bundling protein fascin in growth cone morphogenesis: localization in filopodia and lamellipodia. Cell Motil. Cytoskeleton 48, 109–120 [DOI] [PubMed] [Google Scholar]
- Cooper J. A., Sept D. (2008). New insights into mechanism and regulation of actin capping protein. Int. Rev. Cell Mol. Biol 267, 183–206 10.1016/S1937-6448(08)00604-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent E. W., Kwiatkowski A. V., Mebane L. M., Philippar U., Barzik M., Rubinson D. A., Gupton S., Van Veen J. E., Furman C., Zhang J. et al. (2007). Filopodia are required for cortical neurite initiation. Nat. Cell Biol. 9, 1347–1359 10.1038/ncb1654 [DOI] [PubMed] [Google Scholar]
- Dent E. W., Gupton S. L., Gertler F. B. (2011). The growth cone cytoskeleton in axon outgrowth and guidance. Cold Spring Harb. Perspect. Biol. 3, a001800 10.1101/cshperspect.a001800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricke R., Gohl C., Dharmalingam E., Grevelhörster A., Zahedi B., Harden N., Kessels M., Qualmann B., Bogdan S. (2009). Drosophila Cip4/Toca-1 integrates membrane trafficking and actin dynamics through WASP and SCAR/WAVE. Curr. Biol. 19, 1429–1437 10.1016/j.cub.2009.07.058 [DOI] [PubMed] [Google Scholar]
- Frost A., Perera R., Roux A., Spasov K., Destaing O., Egelman E. H., De Camilli P., Unger V. M. (2008). Structural basis of membrane invagination by F-BAR domains. Cell 132, 807–817 10.1016/j.cell.2007.12.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Dickerson J. B., Guo F., Zheng J., Zheng Y. (2004). Rational design and characterization of a Rac GTPase-specific small molecule inhibitor. Proc. Natl. Acad. Sci. USA 101, 7618–7623 10.1073/pnas.0307512101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode B. L., Eck M. J. (2007). Mechanism and function of formins in the control of actin assembly. Annu. Rev. Biochem. 76, 593–627 10.1146/annurev.biochem.75.103004.142647 [DOI] [PubMed] [Google Scholar]
- Grabham P. W., Reznik B., Goldberg D. J. (2003). Microtubule and Rac 1-dependent F-actin in growth cones. J. Cell Sci. 116, 3739–3748 10.1242/jcs.00686 [DOI] [PubMed] [Google Scholar]
- Guerrier S., Coutinho-Budd J., Sassa T., Gresset A., Jordan N. V., Chen K., Jin W. L., Frost A., Polleux F. (2009). The F-BAR domain of srGAP2 induces membrane protrusions required for neuronal migration and morphogenesis. Cell 138, 990–1004 10.1016/j.cell.2009.06.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartig S. M., Ishikura S., Hicklen R. S., Feng Y., Blanchard E. G., Voelker K. A., Pichot C. S., Grange R. W., Raphael R. M., Klip A. et al. (2009). The F-BAR protein CIP4 promotes GLUT4 endocytosis through bidirectional interactions with N-WASp and Dynamin-2. J. Cell Sci. 122, 2283–2291 10.1242/jcs.041343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath R. J., Insall R. H. (2008). F-BAR domains: multifunctional regulators of membrane curvature. J. Cell Sci. 121, 1951–1954 10.1242/jcs.023895 [DOI] [PubMed] [Google Scholar]
- Henle S. J., Wang G., Liang E., Wu M., Poo M. M., Henley J. R. (2011). Asymmetric PI(3,4,5)P3 and Akt signaling mediates chemotaxis of axonal growth cones. J. Neurosci. 31, 7016–7027 10.1523/JNEUROSCI.0216-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X., Viesselmann C., Nam S., Merriam E., Dent E. W. (2008). Activity-dependent dynamic microtubule invasion of dendritic spines. J. Neurosci. 28, 13094–13105 10.1523/JNEUROSCI.3074-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insall R. H., Weiner O. D. (2001). PIP3, PIP2, and cell movement—similar messages, different meanings? Dev. Cell 1, 743–747 10.1016/S1534-5807(01)00086-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T., Erdmann K. S., Roux A., Habermann B., Werner H., De Camilli P. (2005). Dynamin and the actin cytoskeleton cooperatively regulate plasma membrane invagination by BAR and F-BAR proteins. Dev Cell. 9, 791–804 10.1016/j.devcel.2005.11.005 [DOI] [PubMed] [Google Scholar]
- Johnson H. W., Schell M. J. (2009). Neuronal IP3 3-kinase is an F-actin-bundling protein: role in dendritic targeting and regulation of spine morphology. Mol. Biol. Cell 20, 5166–5180 10.1091/mbc.E09-01-0083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamioka Y., Fukuhara S., Sawa H., Nagashima K., Masuda M., Matsuda M., Mochizuki N. (2004). A novel dynamin-associating molecule, formin-binding protein 17, induces tubular membrane invaginations and participates in endocytosis. J. Biol. Chem. 279, 40091–40099 10.1074/jbc.M404899200 [DOI] [PubMed] [Google Scholar]
- Kida Y., Shiraishi T., Ogura T. (2004). Identification of chick and mouse Daam1 and Daam2 genes and their expression patterns in the central nervous system. Brain Res. Dev. Brain Res. 153, 143–150 10.1016/j.devbrainres.2004.07.014 [DOI] [PubMed] [Google Scholar]
- Lanier L. M., Gates M. A., Witke W., Menzies A. S., Wehman A. M., Macklis J. D., Kwiatkowski D., Soriano P., Gertler F. B. (1999). Mena is required for neurulation and commissure formation. Neuron 22, 313–325 10.1016/S0896-6273(00)81092-2 [DOI] [PubMed] [Google Scholar]
- Lee K., Gallop J. L., Rambani K., Kirschner M. W. (2010). Self-assembly of filopodia-like structures on supported lipid bilayers. Science 329, 1341–1345 10.1126/science.1191710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Sato A., Khadka D., Bharti R., Diaz H., Runnels L. W., Habas R. (2008). Mechanism of activation of the Formin protein Daam1. Proc. Natl. Acad. Sci. USA 105, 210–215 10.1073/pnas.0707277105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning B. D., Cantley L. C. (2007). AKT/PKB signaling: navigating downstream. Cell 129, 1261–1274 10.1016/j.cell.2007.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matusek T., Gombos R., Szécsényi A., Sánchez-Soriano N., Czibula A., Pataki C., Gedai A., Prokop A., Raskó I., Mihály J. (2008). Formin proteins of the DAAM subfamily play a role during axon growth. J. Neurosci. 28, 13310–13319 10.1523/JNEUROSCI.2727-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejillano M. R., Kojima S., Applewhite D. A., Gertler F. B., Svitkina T. M., Borisy G. G. (2004). Lamellipodial versus filopodial mode of the actin nanomachinery: pivotal role of the filament barbed end. Cell 118, 363–373 10.1016/j.cell.2004.07.019 [DOI] [PubMed] [Google Scholar]
- Mongiu A. K., Weitzke E. L., Chaga O. Y., Borisy G. G. (2007). Kinetic-structural analysis of neuronal growth cone veil motility. J. Cell Sci. 120, 1113–1125 10.1242/jcs.03384 [DOI] [PubMed] [Google Scholar]
- Myers J. P., Robles E., Ducharme-Smith A., Gomez T. M. (2012). Focal adhesion kinase modulates Cdc42 activity downstream of positive and negative axon guidance cues. J. Cell Sci. 125, 2918–2929 10.1242/jcs.100107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaya M. A., Habas R., Biris K., Dunty W. C., Jr, Kato Y., He X., Yamaguchi T. P. (2004). Identification and comparative expression analyses of Daam genes in mouse and Xenopus. Gene Expr. Patterns 5, 97–105 10.1016/j.modgep.2004.06.001 [DOI] [PubMed] [Google Scholar]
- Nolen B. J., Tomasevic N., Russell A., Pierce D. W., Jia Z., McCormick C. D., Hartman J., Sakowicz R., Pollard T. D. (2009). Characterization of two classes of small molecule inhibitors of Arp2/3 complex. Nature 460, 1031–1034 10.1038/nature08231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozumi M., Nakagawa H., Miki H., Takenawa T., Miyamoto S. (2003). Differential localization of WAVE isoforms in filopodia and lamellipodia of the neuronal growth cone. J. Cell Sci. 116, 239–246 10.1242/jcs.00233 [DOI] [PubMed] [Google Scholar]
- Oikawa T., Itoh T., Takenawa T. (2008). Sequential signals toward podosome formation in NIH-src cells. J. Cell Biol. 182, 157–169 10.1083/jcb.200801042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul A. S., Pollard T. D. (2009). Review of the mechanism of processive actin filament elongation by formins. Cell Motil. Cytoskeleton 66, 606–617 10.1002/cm.20379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard T. D. (2007). Regulation of actin filament assembly by Arp2/3 complex and formins. Annu. Rev. Biophys. Biomol. Struct. 36, 451–477 10.1146/annurev.biophys.35.040405.101936 [DOI] [PubMed] [Google Scholar]
- Pollard T. D., Borisy G. G. (2003). Cellular motility driven by assembly and disassembly of actin filaments. Cell 112, 453–465 10.1016/S0092-8674(03)00120-X [DOI] [PubMed] [Google Scholar]
- Prokop A., Sánchez-Soriano N., Gonçalves-Pimentel C., Molnár I., Kalmár T., Mihály J. (2011). DAAM family members leading a novel path into formin research. Commun. Integr. Biol. 4, 538–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley A. J. (2011). Life at the leading edge. Cell 145, 1012–1022 10.1016/j.cell.2011.06.010 [DOI] [PubMed] [Google Scholar]
- Roberts-Galbraith R. H., Gould K. L. (2010). Setting the F-BAR: functions and regulation of the F-BAR protein family. Cell Cycle 9, 4091–4097 10.4161/cc.9.20.13587 [DOI] [PubMed] [Google Scholar]
- Robles E., Woo S., Gomez T. M. (2005). Src-dependent tyrosine phosphorylation at the tips of growth cone filopodia promotes extension. J. Neurosci. 25, 7669–7681 10.1523/JNEUROSCI.2680-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roignot J., Taïeb D., Suliman M., Dusetti N. J., Iovanna J. L., Soubeyran P. (2010). CIP4 is a new ArgBP2 interacting protein that modulates the ArgBP2 mediated control of WAVE1 phosphorylation and cancer cell migration. Cancer Lett. 288, 116–123 10.1016/j.canlet.2009.06.030 [DOI] [PubMed] [Google Scholar]
- Saarikangas J., Zhao H., Lappalainen P. (2010). Regulation of the actin cytoskeleton-plasma membrane interplay by phosphoinositides. Physiol. Rev. 90, 259–289 10.1152/physrev.00036.2009 [DOI] [PubMed] [Google Scholar]
- Saengsawang W., Mitok K., Viesselmann C., Pietila L., Lumbard D. C., Corey S. J., Dent E. W. (2012). The F-BAR protein CIP4 inhibits neurite formation by producing lamellipodial protrusions. Curr. Biol. 22, 494–501 10.1016/j.cub.2012.01.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer D. A. (2004). Cell biology: barbed ends rule. Nature 430, 734–735 10.1038/430734a [DOI] [PubMed] [Google Scholar]
- Schafer D. A., Jennings P. B., Cooper J. A. (1996). Dynamics of capping protein and actin assembly in vitro: uncapping barbed ends by polyphosphoinositides. J. Cell Biol. 135, 169–179 10.1083/jcb.135.1.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small J. V., Stradal T., Vignal E., Rottner K. (2002). The lamellipodium: where motility begins. Trends Cell Biol. 12, 112–120 10.1016/S0962-8924(01)02237-1 [DOI] [PubMed] [Google Scholar]
- Soderling S. H., Scott J. D. (2006). WAVE signalling: from biochemistry to biology. Biochem. Soc. Trans. 34, 73–76 10.1042/BST0340073 [DOI] [PubMed] [Google Scholar]
- Soderling S. H., Guire E. S., Kaech S., White J., Zhang F., Schutz K., Langeberg L. K., Banker G., Raber J., Scott J. D. (2007). A WAVE-1 and WRP signaling complex regulates spine density, synaptic plasticity, and memory. J. Neurosci. 27, 355–365 10.1523/JNEUROSCI.3209-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser G. A., Rahim N. A., VanderWaal K. E., Gertler F. B., Lanier L. M. (2004). Arp2/3 is a negative regulator of growth cone translocation. Neuron 43, 81–94 10.1016/j.neuron.2004.05.015 [DOI] [PubMed] [Google Scholar]
- Tian L., Nelson D. L., Stewart D. M. (2000). Cdc42-interacting protein 4 mediates binding of the Wiskott-Aldrich syndrome protein to microtubules. J. Biol. Chem. 275, 7854–7861 10.1074/jbc.275.11.7854 [DOI] [PubMed] [Google Scholar]
- Tsujita K., Suetsugu S., Sasaki N., Furutani M., Oikawa T., Takenawa T. (2006). Coordination between the actin cytoskeleton and membrane deformation by a novel membrane tubulation domain of PCH proteins is involved in endocytosis. J. Cell Biol. 172, 269–279 10.1083/jcb.200508091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanik E., Ware B. R. (1989). Actin filament capping and cleaving activity of cytochalasins B, D, E, and H. Arch. Biochem. Biophys. 269, 181–187 10.1016/0003-9861(89)90098-2 [DOI] [PubMed] [Google Scholar]
- Viesselmann C., Ballweg J., Lumbard D., Dent E. W. (2011). Nucleofection and primary culture of embryonic mouse hippocampal and cortical neurons. J. Vis. Exp. 47, e2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner O. D., Neilsen P. O., Prestwich G. D., Kirschner M. W., Cantley L. C., Bourne H. R. (2002). A PtdInsP(3)- and Rho GTPase-mediated positive feedback loop regulates neutrophil polarity. Nat. Cell Biol. 4, 509–513 10.1038/ncb811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch H., Eguinoa A., Stephens L. R., Hawkins P. T. (1998). Protein kinase B and rac are activated in parallel within a phosphatidylinositide 3OH-kinase-controlled signaling pathway. J. Biol. Chem. 273, 11248–11256 10.1074/jbc.273.18.11248 [DOI] [PubMed] [Google Scholar]
- Woo S., Gomez T. M. (2006). Rac1 and RhoA promote neurite outgrowth through formation and stabilization of growth cone point contacts. J. Neurosci. 26, 1418–1428 10.1523/JNEUROSCI.4209-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Asokan S. B., Berginski M. E., Haynes E. M., Sharpless N. E., Griffith J. D., Gomez S. M., Bear J. E. (2012). Arp2/3 is critical for lamellipodia and response to extracellular matrix cues but is dispensable for chemotaxis. Cell 148, 973–987 10.1016/j.cell.2011.12.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Zhang X. F., Pollard T. D., Forscher P. (2012). Arp2/3 complex-dependent actin networks constrain myosin II function in driving retrograde actin flow. J. Cell Biol. 197, 939–956 10.1083/jcb.201111052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidel-Bar R., Joyce M. J., Lynch A. M., Witte K., Audhya A., Hardin J. (2010). The F-BAR domain of SRGP-1 facilitates cell-cell adhesion during C. elegans morphogenesis. J. Cell Biol. 191, 761–769 10.1083/jcb.201005082 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.