Abstract
Impairment in the elimination of misfolded proteins generates cellular toxicity and leads to various late-onset neurodegenerative diseases. However, the mechanisms by which cells recognize abnormal cellular proteins for selective clearance remain unknown. Lack of the mahogunin ring finger-1 (MGRN1) E3 ubiquitin ligase in mice causes the development of age-dependent spongiform neurodegeneration. Here, we report for the first time that the MGRN1 E3 ubiquitin ligase interacts and nicely co-localizes with the cytosolic molecular chaperone Hsp70. The expression of MGRN1 increased following exposure to a variety of stressors. The inhibition of autophagy not only elevated endogenous MGRN1 levels but also caused MGRN1 to be recruited to cytosolic ubiquitin-positive inclusion bodies. Finally, we showed that the overexpression of MGRN1 protects against cell death mediated by oxidative and endoplasmic reticulum stress. These data suggest that MGRN1 selectively targets misfolded proteins for degradation and may exhibit viable therapeutic potential for the treatment of spongiform neurodegeneration.
In healthy cells, the replacement of old or damaged proteins via the regular renewal of new polypeptides is an essential cellular function. To achieve a functional proteome and normal cellular homeostasis, cells maintain a delicate balance between protein synthesis and degradation1. Numerous studies have shown that failure in the removal of abnormal proteins results in the accumulation of misfolded proteins and, eventually, cell death2,3. The presence of accumulated proteinaceous species intracellularly is a major cytopathological hallmark of various neurodegenerative diseases4,5. To eliminate and prevent the intracellular accumulation of misfolded protein aggregates, cells employ protein quality control mechanisms through the ubiquitin proteasome system and alternatively, the autophagy-lysosomal pathway, which also work at multiple levels6. Genetic mutations, conditions of environmental stress, and aberrant changes in protein complexes lead to the massive accumulation of oligomeric or aggregated misfolded proteins, which cannot be degraded and digested merely through proteasomes. The deposition of protein aggregates globally impairs the ubiquitin proteasome system and further adds to the aggregation of misfolded proteins and formation of inclusions7. To survive under such threatening conditions, cells degrade misfolded proteinaceous inclusions through autophagy, which may integrate selective chaperone-mediated autophagy (CMA) with specific E3 ubiquitin ligases8,9.
An overburden of misfolded protein aggregates impairs cellular quality control mechanisms. Eventually, failure in the clearance of damaged proteins results in the progression of various human protein conformation disorders and neurodegenerative diseases10,11,12. Recent studies have uncovered the role of dysregulated autophagy in neurodegeneration6,13. CMA specifically promotes the degradation of proteins with KFERQ motifs, which constitute 30% of all cytosolic proteins that retain biochemically related motifs14. In this mechanism, the Hsc70 chaperone binds to the critical substrate protein via the KFERQ motifs. In addition, the lysosomal membrane receptor lysosome-associated membrane protein 2a (Lamp2a) helps in their subsequent degradation15. Recently, it was shown that CMA regulates the activity of the myocyte enhancer factor 2D (MEF2D) transcription factor, which is required for neuronal survival and contributes to the pathobiology of Parkinson's disease16.
It has been clearly established that E3 ubiquitin ligases recognize and polyubiquitinate misfolded proteins and promote their degradation, thereby eliminating abnormal protein deposits17,18,19,20. Emerging studies suggest that various E3 ubiquitin ligases (e.g., CHIP, E6-AP, Gp78, and Parkin–PINK1) and the novel DJ-1 E3 complex are directly involved in cellular quality control mechanisms and aberrant protein degradation21,22,23,24,25. Loss of function of the Mahogunin ring finger-1 (MGRN1) E3 ubiquitin ligase leads to late-onset spongiform neurodegeneration26. MGRN1 promotes proteasome-independent ubiquitylation of TSG101 and regulates endosomal trafficking27. Co-sequestration of MGRN1 with cytosolic-exposed prion protein aggregates leads to altered lysosomal morphology. Its depletion may also be implicated in the biology of neuronal dysfunction and disease28. It is clear that CMA plays a significant role in neurodegenerative diseases and aging29. Still, we are far from understanding how CMA specifically targets misfolded proteins for degradation and which functional interactions between chaperones and E3 ubiquitin ligases determine this unique function in the dense pool of cytosolic proteins.
In the current study, we found that endogenous MGRN1 levels are increased after exposure to various cellular stressors. MGRN1 co-immunoprecipitates with the Hsp70 chaperone and nicely co-localizes with cytosolic Hsp70-positive heat-denatured misfolded luciferase inclusion bodies. Moreover, MGRN1 is involved in the degradation and clearance of Hsp70-associated misfolded proteins via the selective autophagy pathway. Notably, we found one highly related KFERQ motif in the MGRN1 amino acid sequence and a HSF-1 binding sequence in the 5′-untranslated region of the MGRN1 gene, which also supports the current findings. Partial knockdown of MGRN1 exacerbated the cytotoxic effects generated by various stress-inducing agents, while the overexpression of MGRN1 mitigated the proteotoxicity induced by various stressors. The current study suggests a cytoprotective role of the MGRN1 ubiquitin ligase in protein quality control mechanisms through the elimination of misfolded protein-mediated toxicity in cells.
Results
MGRN1 is induced under various cellular stress conditions and interacts with Hsp70
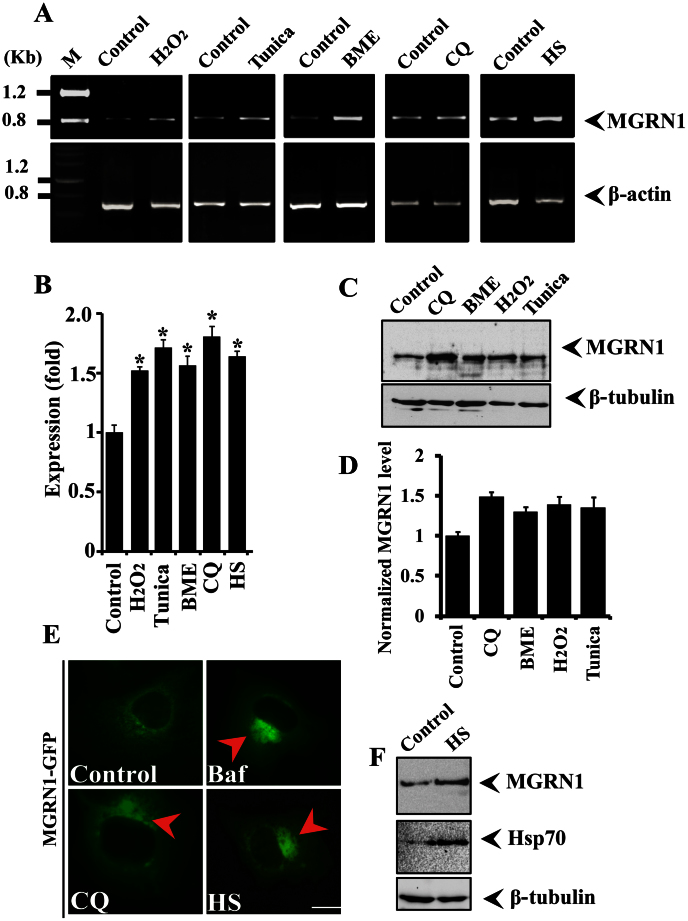
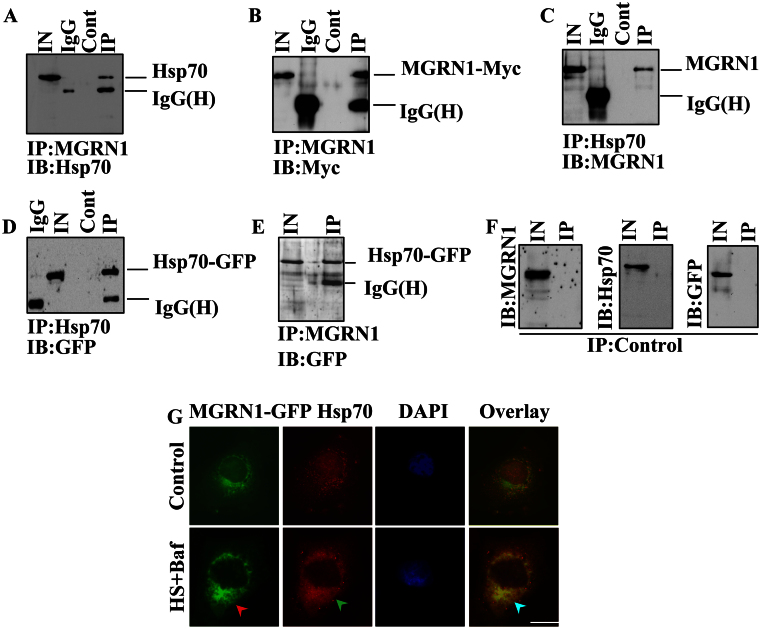
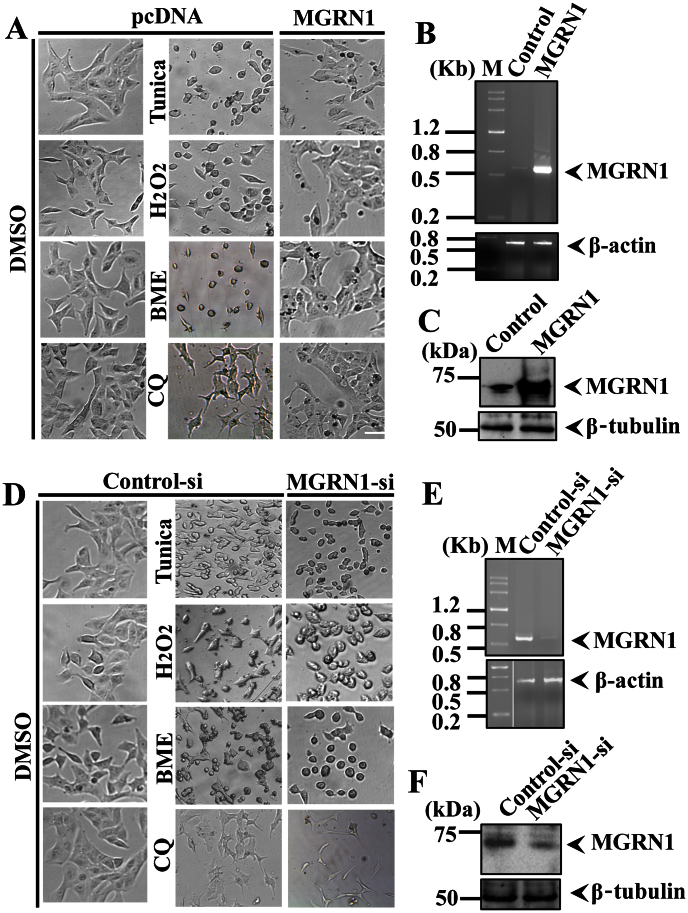
It has been shown that MGRN1 interacts with the aggregates of cytosolic prion proteins (PrPs) that are associated with neurodegeneration28. However, the mechanisms through which the loss of MGRN1 function impairs cellular protein quality control mechanisms and causes spongiform neurodegeneration is not known. Mitochondrial dysfunction and high levels of oxidative stress have also been observed in MGRN1 mutant mice30. Therefore, we set out to explore the possible role of MGRN1 in cellular protein quality control mechanisms under various types of stress conditions. We exposed A549 cells to oxidative (H2O2), ER (Tunicamycin and β-mercaptoethanol), and HS (heat stress), as well as to autophagy dysfunction (Chloroquine) stress, and then examined MGRN1 mRNA and protein levels. We noticed that MGRN1 mRNA levels were significantly increased under various stress conditions (Fig. 1A). We have also noticed approximately 0.5 (Oxidative stress), 0.70 (ER), 0.80 (autophagy dysfunction) and 0.65 (Heat stress) fold increase in MGRN1 mRNA levels under various stress conditions as compared with control cells (Fig. 1B). Therefore, we thought it was also important to check MGRN1 protein levels after treatment with different types of stress-inducing agents. MGRN1 protein levels were also elevated after exposure to the various cellular insults generated by the stressor agents (Fig. 1C and D). To our surprise, we did not find very high levels of MGRN1 proteins compared to mRNA levels after exposure with various stress inducing agents. We thought that the decrease in the protein levels of MGRN1 in the stressed cells could be due to its strong association with misfolded protein inclusions. To confirm this hypothesis, we overexpressed MGRN1-GFP construct in cells and treated with various stress inducing agents. We have observed that under various stress conditions MGRN1 protein nicely accumulated in periphery nuclear region and make clear big inclusions like structures (Fig. 1E). Most likely these inclusions are non-SDS-soluble in nature and therefore it is hard to observe their soluble forms. It is known that heat shock proteins levels are elevated after HS and that Hsp70 is involved in the protection of cells against various types of stress31,32. In the current study, we found that the response of MGRN1 to HS was very similar to that of Hsp70 (Fig. 1F). Because the expression profile of MGRN1 induced by the variety of cellular insults was similar to that of Hsp70, we hypothesized that MGRN1 interacts with the Hsp70 chaperone and that this cooperative function may enhance the clearance of misfolded proteins. Unexpectedly, we found that MGRN1 contains one biochemically related KFERQ motif, which is important for CMA (Supplementary Fig. 1B). To strengthen this finding, we next analyzed the 5′-untranslated region of the MGRN1 gene and observed that it contains a consensus heat shock factor 1 (HSF1) binding sequence that is 90% homologous with the yeast HSF-1 binding sequence. This preliminary result indicated that this gene can respond to heat shock (Supplementary Fig. 1A). We used co-immunoprecipitation to explore the possibility of an interaction between MGRN1 and Hsp70. An immunoprecipitation experiment using the lysates of Cos-7 cells that overexpressed MGRN1-Myc and Hsp70-EGFP revealed that Hsp70 interacts with MGRN1 (Fig. 2A). To further confirm the observed interaction, we next performed a more detailed immunoprecipitation study; the same co-transfected cell lysates pull down with MGRN1 antibody were subjected to immunoblot analysis using anti-Myc (Fig. 2B). It was important to check reverse interaction between Hsp70 and MGRN1; therefore, in the next immunoprecipitation experiment, we pulled down co-transfected cell lysates using an Hsp70 antibody and developed the blot using MGRN1 antibody (Fig. 2C). Next, we performed experiments using different controls; the same cell lysates were processed for pull down by anti-Hsp70 and the blot probed with GFP antibody (Fig. 2D). MGRN1 interaction was further analyzed with Hsp70 chaperone, we used MGRN1 antibody in immunoprecipitation experiment and blot was developed with GFP antibody (Fig. 2E). In control pulled down experiments we used only beads and blots were incubated with anti-MGRN1, anti-Hsp70 and anti-GFP antibodies (Fig. 2F). To further confirm the novel interaction of Hsp70 and MGRN1, we treated MGRN1-GFP transfected cells with Bafilomycin and heat stress (Baf + HS) and observed that GFP positive MGRN1 inclusions like structures were colocalized with induced aggregated Hsp70 chaperone (Fig. 2G). These results suggest that MGRN1 confers tolerance to the expression of a stress response and interacts with Hsp70.
Figure 1. MGRN1 expression is induced in response to various stressors.
(A) A549 cells were plated into six-well tissue culture plates and treated with 0.2 mM H2O2 for 2 h, 5 μg/ml tunicamycin for 5 h, 5 mM β-mercaptoethanol for 2 h, or 20 μM chloroquine (CQ) for 08 h. Alternatively, for heat stress (HS) exposure, cells were exposed to 43°C for 30 minutes and then returned to 37°C for 1 h. After treatment, cells were collected, and total cellular RNA was isolated and subjected to reverse transcription polymerase chain reaction (RT-PCR) using primers for MGRN1 and β-actin. (B) Quantitation of MGRN1 mRNA levels using quantitative real time RT-PCR in the experiment as described in A. (C) Representative immunoblot showing MGRN1 protein levels in cells treated with different stress-inducing agents, as described in section A. Blots were probed with MGRN1 and β-tubulin antibodies. (D) Quantification of the MGRN1 band intensities are shown in C, which were collected from three independent experiments. Data are expressed as a ratio of MGRN1 to β-tubulin. Values are the mean ± SD of three independent experiments. (E) Cells were transiently transfected with MGRN1-GFP construct and after 48 hrs of transfection treated with 50 nM Bafilomycin (Baf), 30 μM chloroquine (CQ) or HS exposure 43°C for 30 minutes. Treated cells were directly subjected to fluorescence microscopy analysis. (F) Cells were exposed to HS at 43°C for 30 minutes and then processed for immunoblotting. Blots were probed with MGRN1, Hsp70, and β-tubulin antibodies. Values are the means ± S.D. of three independent experiments.*, p < 0.05 compared with the control. Scale bar, 20 μm.
Figure 2. MGRN1 interacts with Hsp70.
(A–B) Transfection was used to overexpress MGRN1-Myc and Hsp70-EGFP in Cos-7 cells. Cells were collected, lysed, and processed for immunoprecipitation (IP) with an MGRN1 antibody 24 hrs post-transfection. Blots were probed with an Hsp70 (A) antibody and Myc (B) antibody. (C–D) As described in A, the samples were processed for IP using an Hsp70 antibody and the blots were probed with anti-MGRN1 (C) and anti-GFP (D). (E) Hsp70-EGFP transfected cell lysate were pulled down by MGRN1 antibody and blot was developed with GFP antibody. (F) As described in section A-E; same samples were pulled down by beads only (control) and blots were obtained with anti-MGRN1, anti-Hsp70 and anti-GFP antibodies. (G) The cells were transfected as described in A with MGRN1-GFP plasmid and 36 hrs of post transfection some cells pretreated with 50 nM Bafilomycin (Baf) and exposed to 43°C for 30 minutes heat shock (HS). Immediately after heat shock treatment cells were processed to immunofluorescence staining using Hsp70 antibody. Rhodamine-conjugated secondary antibody was used to stain Hsp70 protein. Arrows indicate the recruitment of MGRN1 to the induced Hsp70 inclusions.
Recruitment of MGRN1 to components of inclusion bodies following inhibition of autophagy
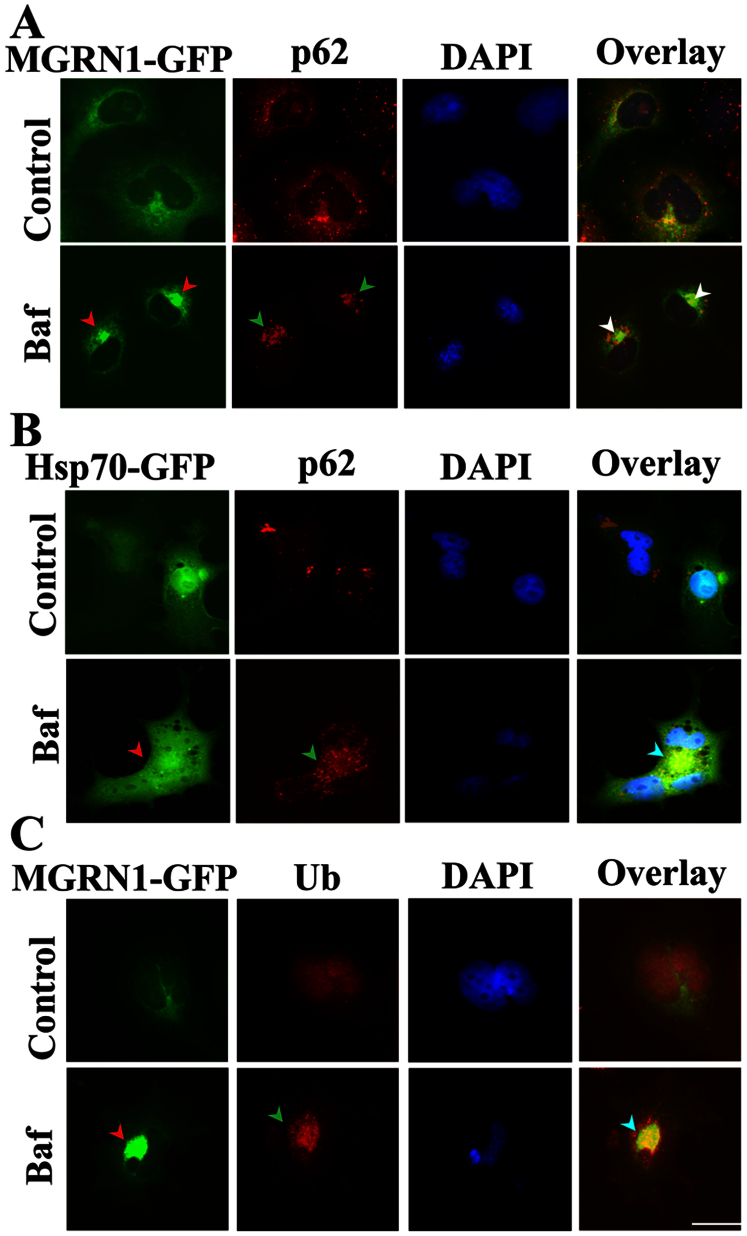
Inclusion bodies (IBs) are generally positive for p62 and ubiquitin33,34. The key observation of the interaction between MGRN1 and Hsp70 raised the critical question of whether there are cumulative effects of MGRN1 and Hsp70 in the selective autophagy pathway. To assess whether MGRN1 is recruited to the general components of IBs, we next performed immunofluorescence staining of p62 in MGRN1-GFP overexpressing cells after treatment with Bafilomycin. Treatment with Bafilomycin resulted in the formation of distinct IBs that were co-localized with p62. MGRN1 was also clearly recruited towards these IBs (Fig. 3A). In Baf-treated cells, these IBs were also positive for both Hsp70 and p62 (Fig. 3B). Next, we observed that Baf treatment caused the recruitment of MGRN1 primarily toward perinuclear ubiquitin-positive IB structures (Fig. 3C). Overall, these results clearly indicate that MGRN1 is predominantly redistributed in perinuclear regions and recruited to p62-, Hsp70-, and ubiquitin-positive IBs.
Figure 3. Co-localization of MGRN1 with cytoplasmic p62 bodies, and ubiquitin (Ub) following autophagy dysfunction.
(A) Cos-7 cells were plated on two-chamber slides. On the following day, the cells were transiently transfected with the MGRN1-GFP construct. After 36 h of transfection, the cells were treated with 50 nM Bafilomycin (Baf) for 12 h. Post-treatment, the cells were fixed and subjected to immunofluorescence staining using an p62 antibody. A rhodamine-conjugated secondary antibody was used to label the p62 antibody. Nuclei were stained with 4′,6- diamidino-2-phenylindole (DAPI). Arrows indicate the recruitment of MGRN1 to p62 aggregates. (B) Cells were transfected with Hsp70-EGFP and treated with Baf as described in A. Cells were observed using a fluorescence microscope and their nuclei were stained using DAPI. Arrows indicate the co-localization of Hsp70 with p62 aggregates. (C) Cells were transiently transfected with the MGRN1-GFP construct and treated with Baf as described in A; after treatment, the cells were processed for immunofluorescence staining using ubiquitin (Ub) antibody. Rhodamine-conjugated secondary antibody was used to label ubiquitin. Nuclei were stained with DAPI. Arrows indicate the redistribution of MGRN1 with Ub-positive aggregates. Scale bar, 20 μm.
MGRN1 is co-localized with Hsp70-anchored misfolded luciferase inclusion formations
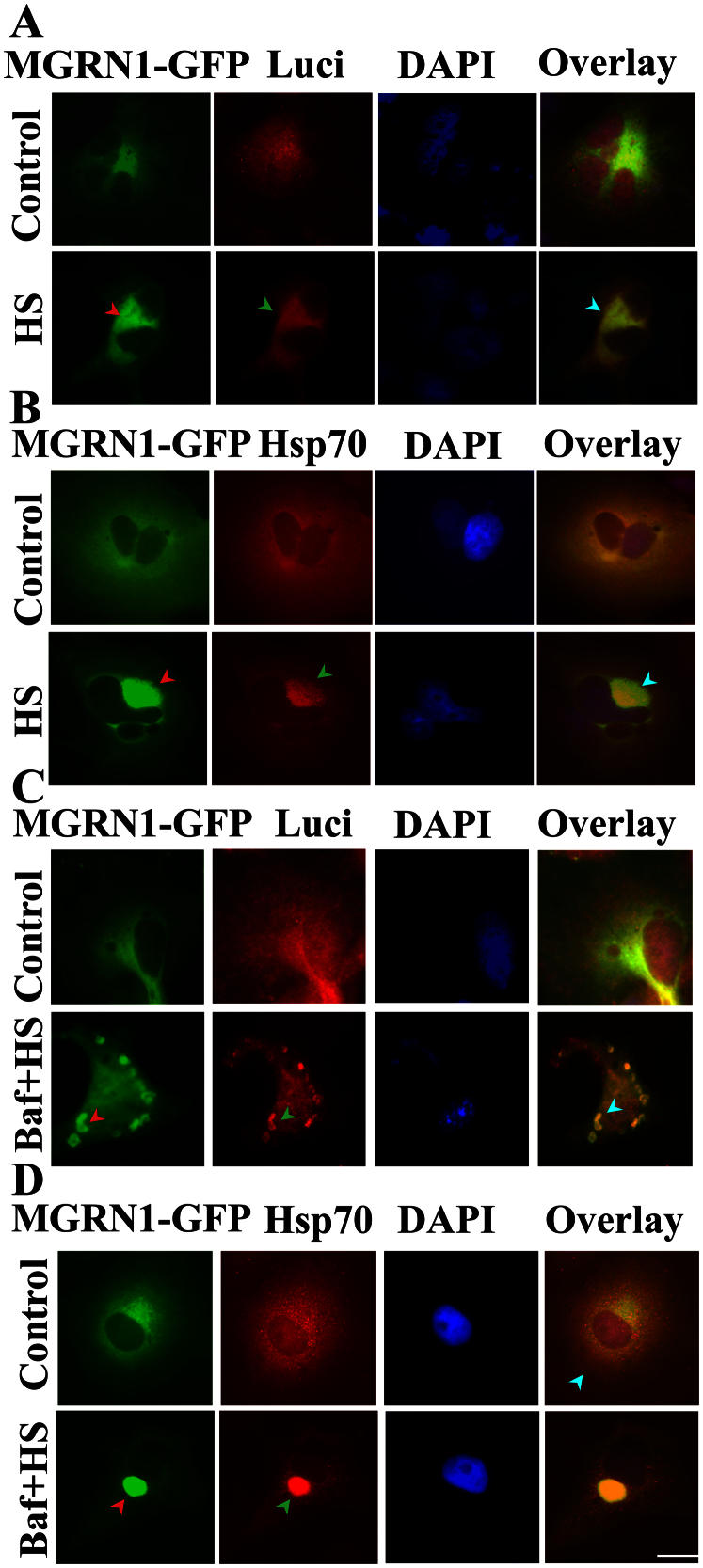
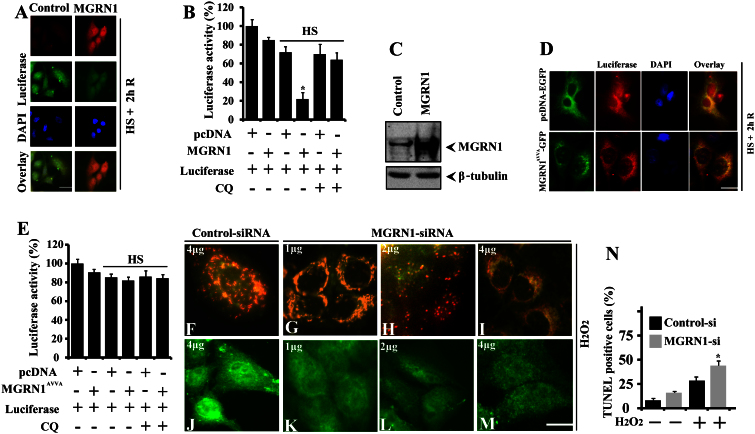
We assessed the interaction, recruitment, and co-localization of MGRN1 with the pre-formed general components of IBs, which are induced in response to impaired autophagy. Based on the surprising results, we decided to further confirm and explore the interaction of MGRN1 with heat-denatured luciferase protein, which is misfolded upon thermal stress. MGRN1-GFP was overexpressed in cells. A portion of the cells were co-transfected with MGRN1 and luciferase constructs. Immediately after the heat shock response, we noticed that MGRN1 levels were increased and that MGRN1 recruited towards perinuclear regions with denatured luciferase (Fig. 4A) and the Hsp70 chaperone (Fig. 4B). To more clearly confirm that this perinuclear distribution was specifically associated with the selective autophagy pathway, we used Baf-pretreated cells for heat shock. In this experiment, we observed that denatured luciferase aggregates were dispersed throughout the cytoplasm and strongly co-localized with both MGRN1 (Fig. 4C) and the Hsp70 (Fig. 4D) chaperone. These results suggest that MGRN1 may target misfolded proteins through the Hsp70 chaperone and promote their clearance through autophagy.
Figure 4. Co-localization of MGRN1 with luciferase and Hsp70 in heat-stressed cells following autophagy inhibition.
(A) Cos-7 cells were plated on two-chamber slides and co-transfected with MGRN1-GFP and luciferase expression constructs. Thirty-six hours later, the cells were exposed to HS at 43°C for 1 h and then processed for immunofluorescence staining using luciferase antibody. A rhodamine-conjugated secondary antibody was used to label luciferase. Arrows indicate the co-localization of MGRN1 with luciferase following HS treatment. (B) The cells were transfected with MGRN1-GFP construct. The cells were exposed to HS as described in A, and fixed cells were subjected to immunofluorescence staining using an Hsp70 antibody. Arrows indicate the redistribution of MGRN1 with Hsp70 following HS exposure. A rhodamine-conjugated secondary antibody was used to stain Hsp70. A fluorescence microscope was used to acquire images. (C) Cos-7 cells were grown on two-chamber slides and co-transfected with luciferase and MGRN1-GFP expression constructs. Thirty-six hours later, a portion of the samples were treated with 50 nM Bafilomycin (Baf) for 12 h. Cells were heat stressed via exposure to 43°C for 1 h and then subjected to immunofluorescence staining with anti-luciferase antibody. A rhodamine-conjugated secondary antibody to label luciferase. Overlay images include DAPI nuclear staining (blue color). Arrows indicate cytosolic aggregation and co-localization of MGRN1 with luciferase following treatment with Baf + HS. (D) The cells were transfected with MGRN1-GFP plasmid. Thirty-six hours post-transfection, the cells were treated with Baf and HS as described in A and subjected to immunofluorescence staining using an Hsp70 antibody. A rhodamine-conjugated secondary antibody was used to label Hsp70. Nuclei were stained with DAPI. Arrows indicate the cytosolic aggregation and co-localization of Hsp70 with MGRN1 following Baf + HS treatment. Images were obtained using a fluorescence microscope. Scale bar, 20 μm.
MGRN1 alleviates cellular insults generated by various stress-inducing agents
Thus far, our results indicated that MGRN1 may play a significant role in preventing the accumulation of misfolded proteins. However, the mechanism by which MGRN1 defends against the various cellular insults is not known. To explore this question in light of our current observations, we hypothesized that MGRN1 targets the misfolded proteinaceous species associated with the Hsp70 chaperone through autophagy. To confirm this hypothesis and simultaneously investigate the role of MGRN1 in protecting against various stresses, we treated control and MGRN1 overexpressing cells with 10 μg/ml tunicamycin for 12 h or 5 mM βmercaptoethanol for 3 h to induce ER stress, 0.5 mM H2O2 for 5 h to induce oxidative stress, and 50 μM CQ for 12 h to induce autophagy dysfunction. After treatment with the stress inducers, cells were visualized using bright field microscopy (Fig. 5A). Overexpression of MGRN1 was confirmed using reverse transcription polymerase chain reaction (RT-PCR) (Fig. 5B) and immunoblot analysis with an MGRN1 antibody (Fig. 5C). To further confirm the protective effect of MGRN1 against various cellular insults, we used siRNA oligonucleotides against MGRN1 to partially knockdown endogenous MGRN1. Scrambled siRNA was used as a control. We achieved approximately 70–80% knockdown of endogenous MGRN1 levels. After transfection, cells were treated with various stress-inducing agents and processed for bright field microscopy image analysis as shown in Fig. 5D. MGRN1 knockdown was confirmed by both reverse transcription polymerase chain reaction (RT-PCR) (Fig. 5E) and immunoblot analysis using an MGRN1 antibody (Fig. 5F). These results further substantiate that MGRN1 participates in cellular defense mechanisms in response to the proteotoxic effects caused by various stress-inducing agents.
Figure 5. MGRN1 protects cells against the effects of various stress-inducing agents.
(A–C) A549 cells were transiently transfected with pcDNA (control) and MGRN1-Myc constructs (transfected with a 24-well tissue culture plate). After 48 h, cells were treated with 10 μg/ml tunicamycin (Tunica) for 12 h, 5 mM β-mercaptoethanol (BME) for 3 h, 0.5 mM H2O2 for 5 h and 50 μM CQ for 12 h. Cells treated with the various stressors were visualized using a bright field microscope as indicated in A. After treatment, cells were collected, and total cellular RNA was isolated and subjected to RT-PCR using primers for MGRN1 and β-actin (B). Some of the transfected cells were processed for immunoblot analysis using MGRN1 and β-tubulin antibodies (C). (D–F) The cells were transiently transfected with control (scrambled siRNA) and MGRN1 siRNA oligonucleotides and treated with the various stress-inducing agents such as 05 μg/ml tunicamycin (Tunica) for 08 h, 2.5 mM β-mercaptoethanol (BME) for 1.5 h, 0.1 mM H2O2 for 4 h and 25 μM CQ for 10 h. The cells treated with the various stressors were visualized using bright field microscopy as indicated in D. Post-treatment, the transfected cells were used to isolate total cellular RNA and processed for RT-PCR using primers for MGRN1 and β-actin (E). Some cells were used to make cell lysates, which were subjected to immunoblot analysis with anti-MGRN1 and β-tubulin antibodies (F).
MGRN1 overexpression induces the degradation of misfolded luciferase protein, and knockdown leads to mitochondrial membrane depolarization and cytochrome c release
The MGRN1 E3 ubiquitin ligase regulates endosomal trafficking through the proteasomal-independent ubiquitylation pathway27. In the current study, we unexpectedly found that MGRN1 interacts with the Hsp70 chaperone. MGRN1 also co-localizes with both perinuclear IBs and misfolded proteins following the inhibition of autophagy. These results prompted us to further investigate the functional role of MGRN1 in the degradation of misfolded proteins. To directly demonstrate the role of MGRN1 in the degradation of denatured luciferase protein, we overexpressed MGRN1 along with a luciferase construct in Cos-7 cells. Transfected cells were exposed to 43°C for 30 minutes and then returned to 37°C for 2 h of recovery. In some experiments, cells were treated with 100 μM CQ before the HS exposure. Cells were then processed for immunofluorescence staining (Fig. 6A) and a luciferase activity assay (Fig. 6B). As shown in Fig. 6A and B, MGRN1 overexpression led to the efficient degradation of heat-denatured luciferase protein, which was prevented by the CQ-mediated inhibition of autophagy (Fig. 6B). Overexpression of MGRN1 was confirmed by performing an immunoblot using an MGRN1 antibody (Fig. 6C). To further confirm this result we cotransfected catalytically inactive form of MGRN1 (MGRN1AVVA-GFP) along with luciferase constructs. Transfected cells were exposed to heat stress treatment with CQ. Cells were then subjected to immunofluorescence staining (Fig. 6D) and a luciferase activity assay (Fig. 6E). We noticed that overexpression of catalytically inactive form of MGRN1 did not degrade heat denatured luciferase. Taken together, our results suggest that MGRN1 overexpression significantly reduced the levels of heat-denatured, misfolded luciferase protein through autophagy. We noticed that MGRN1 defends against oxidative stress in cells and generates cytoprotective effects. To further explore this finding, we next investigated the effect of MGRN1 on mitochondrial membrane depolarization and cytochrome c release. Cells were transiently transfected with control (scrambled siRNA) or MGRN1-specific siRNA oligonucleotides in a concentration-dependent manner. Following 48 h of transfection, the same cells were subjected to mild oxidative stress (0.05 mM H2O2 for 10 minutes) and then processed for JC-1 staining to examine changes in mitochondrial membrane depolarization. In addition, some cells were used for immunofluorescence staining of cytochrome c. The voltage sensitive fluorescence dye JC-1 stains polarized mitochondria red. Conversely, green fluorescence indicates a low or depolarized mitochondrial membrane potential. Compared with the treatment with control siRNA, partial knockdown of MGRN1 in a concentration-dependent manner made cells highly sensitive to mild oxidative insults, as determined by the signal detected with fluorescence microscopy (Fig. 6F–I). Partial knockdown of MGRN1 released cytochrome c from the mitochondria (Fig. 6J–M). Mitochondrial dysfunction in MGRN1 mutant mice has also been reported30. We also observed a significant increase in TUNEL-positive cells in the MGRN1 miRNA-transfected cells treated with oxidative stress inducing agent (Fig. 6N). In the current study, we also noticed that knockdown of MGRN1 made cells highly sensitive to oxidative insults. Taken together, our results suggest that a loss of MGRN1 function plays a critical role in mitochondrial dysfunction and the associated oxidative stress.
Figure 6. MGRN1 promotes the degradation of misfolded heat-denatured luciferase, and knockdown results the release of cytochrome c in responses to oxidative stress.
(A–C) Cos-7 cells were co-transfected with an MGRN1-Myc plasmid along with a firefly luciferase expression construct. Cells were processed for immunofluorescence staining using anti-luciferase and MGRN1 antibodies as shown in section A. The same set of cells was then subjected to a luciferase activity assay (B) and immunoblot analysis using (C) MGRN1 and β-tubulin antibodies. Values are presented as the mean ± SD of three independent experiments. *, p < 0.05 compared with the control group that was heat-stressed but transfected with an empty pcDNA construct. (D–E) Cells were cotransfected with pcDNA-EGFP, MGRN1AVVA-GFP mutant and luciferase plasmids and after heat treatment and recovery, cells were processed for immunofluorescence staining (D) using anti-luciferase antibody and luciferase assay (E) as described in “experimental procedure”. Values are presented as the mean ± SD of three independent experiments. *, p < 0.05 compared with the control group that was heat-stressed but transfected with an empty pcDNA construct. (F–M) A549 cells were transiently transfected with control (scrambled) siRNA (F, J) or with different concentrations of MGRN1 siRNA oligonucleotides as indicated and post transfected cells were treated with 0.05 mM H2O2 for 10 minutes and then subjected to either 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolcarbocyanine iodide (JC-1) staining to detect the changes in mitochondrial membrane potential. A functional coupled mitochondrial activity as red and the depolarized membrane becomes green (F–I) or immunostaining using anti-cytochrome c (J–M). (N) Percentage of TUNEL positive control (scrambled) siRNA and MGRN1-siRNA transfected cells treated with H2O2. Values are presented as the mean ± SD of three independent experiments. *, p < 0.05 compared with the control-si cells treated with H2O2. Scale bar, 20 μm.
MGRN1 protects against cell death mediated by ER and oxidative stress
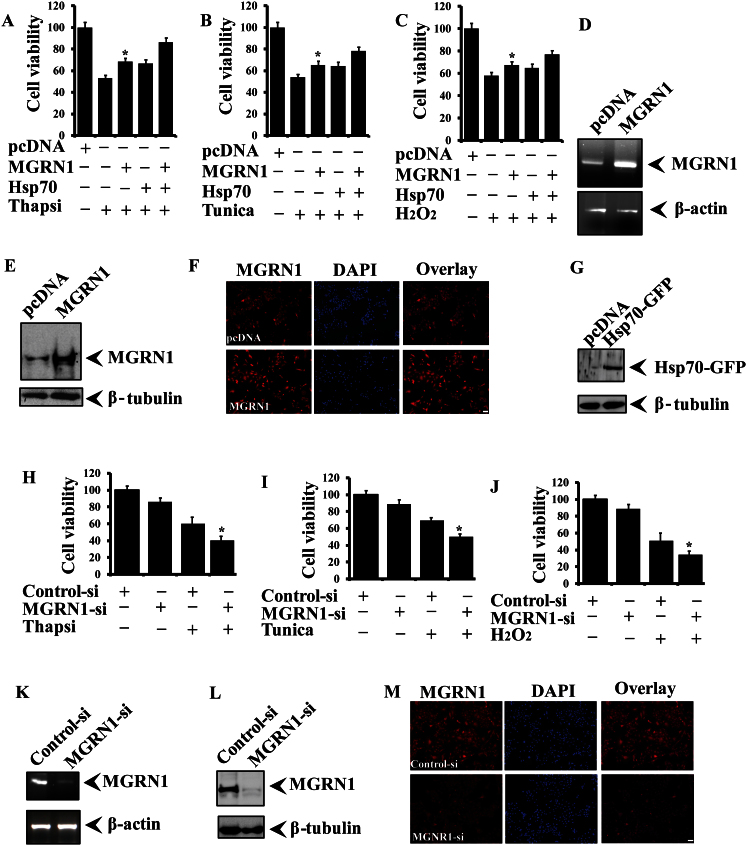
In the current report, we noticed that endogenous levels of MGRN1 are increased after exposure to different stressors and promote the degradation of misfolded proteins. These results all suggest that MGRN1 may contribute to a critical protective survival response under stressed conditions. To evaluate this possibility, we overexpressed MGRN1 with or without co-transfection of Hsp70. After transfection, cells were treated with different stress-inducing agents. ER (thapsigargin and tunicamycin) and oxidative stress (H2O2) cause a decline in cell viability. The overexpression of MGRN1 alleviated cytotoxicity and protected against the cell death induced by ER (Fig. 7A and B) and oxidative stress (Fig. 7C). Overexpression of Hsp70 along with MGRN1 generated an additional protective effect against cytotoxic insult. MGRN1 overexpression was confirmed using RT-PCR (Fig. 7D), immunoblot analysis (Fig. 7E), and immunofluorescence staining (Fig. 7F) with an MGRN1 antibody. Overexpression of GFP-tagged Hsp70 was confirmed by immunoblotting using a GFP antibody (Fig. 7G). To further confirm the cytoprotective nature of MGRN1, we knocked down endogenous levels of MGRN1 with specific siRNA oligonucleotides and exposed the cells to ER and oxidative stress as described earlier. We observed that cells were highly sensitive to the stress; cell viability was significantly reduced after the partial knockdown of MGRN1 in cells under ER stress (Fig. 7H and I) and oxidative stress (Fig. 7J). Knockdown of MGRN1 was confirmed using RT-PCR (Fig. 7K), immunoblotting (Fig. 7L), and immunofluorescence staining (Fig. 7M).
Figure 7. MGRN1 contributes to the cytoprotective potential and alleviates cell death induced by various stressors.
(A–C) A549 cells were transiently transfected with the MGRN1-Myc plasmid and some of the cells co-transfected with the Hsp70-EGFP construct. After 48 hours of transfection, cells were treated with 5 μg/ml thapsigargin for 8 h (A), 10 μg/ml tunicamycin for 10 h (B), or 0.5 mM H2O2 for 5 h (C) and cell viability was measured using an MTT assay. Values are presented as the mean ± S.D. of three independent experiments, each performed in duplicate. *, p < 0.05 compared with the empty pcDNA-transfected (stress inducing treated) experiment. (D–G) Some transfected cells were processed for RT-PCR (D), immunoblot (E) and for immunofluorescence staining (F). Overexpressed Hsp70 was detected by using GFP antibody (G). (H–J) As described in above sections (A–C), cells were transiently transfected with control (scrambled siRNA) or MGRN1 siRNA oligonucleotides and treated with thapsigargin (H), tunicamycin (I) and H2O2 (J). Cell viability was measured using an MTT assay. Values are the mean ± S.D. of three independent experiments, each performed in duplicate. *, p < 0.05 compared with the control (scrambled siRNA-stress inducing treated) experiment. (K–M) As described in H, the cells were collected and processed for RT-PCR (K), immunoblotting (L) and immunofluorescence staining (M) with an MGRN1 antibody. For nuclear staining, fixed cells were incubated with DAPI for 10 minutes.
Discussion
Here, we present the first report that mahogunin ring finger-1 (MGRN1), a putative E3 ubiquitin ligase, interacts with Hsp70-anchored misfolded protein inclusions and promotes their degradation via autophagy. The end product of the MGRN1 gene is a member of the “really interesting new gene” (RING) domain family, which contains the E3 ubiquitin ligases35. Under proteotoxic insult, cells generate defense mechanisms that maintain a delicate balance between protein folding and degradation with the help of molecular chaperones and E3 ubiquitin ligases. A recent study showed that some newly synthesized and pre-existing proteins sequester with amyloidogenic aggregates. Furthermore, the lack of function of those critical proteins may generate toxicity in cells36. Nearly 30% of newly synthesized proteins are not folded properly37. Under cellular stress conditions, this imbalance in proteostasis may be aggravated by an overabundance of misfolded protein aggregates or an insufficient chaperone capacity.
It has been shown that the massive aggregation of misfolded proteins impairs the ubiquitin proteasome system7. How cells survive after proteasomal inhibition and which other pathways are adopted for the clearance of misfolded proteins remains a valid and open question. Numerous reports suggest that the existence of CMA also reduces the overabundance of misfolded proteins9. However, how CMA recognizes and specifically targets misfolded proteins for degradation through autophagy is not fully understood.
The possibility of an interaction between lysosomes, chaperones, and E3 ubiquitin ligases remains unresolved. The pentapeptide consensus motif retaining cytosolic proteins that are recognized by chaperones for autophagy; this degradation pathway does not require vesicle trafficking29. Neural precursor cells that express both developmentally down-regulated protein 4 (NEDD4) and atrophin-interacting Protein 4 (AIP4)/Itch-E3 ubiquitin ligases and have been implicated in the degradation of melanocytic transmembrane proteins (i.e., Melan-A/MART-1) by lysosomes in pigmented cells. This study suggests that E3 ubiquitin ligases mediate the ubiquitylation process that controls lysosomal sorting38. In mouse mutation in MGRN1 ubiquitin ligase cause embryonic lethality and generates spongiform neurodegeneration26. MGRN1 mono-ubiquitinates tumor susceptibility gene 101 (TSG101) and under in vitro and in vivo conditions27. MGRN1 physically interacts with melanocortin receptors (MCRs), and the overexpression of Gαs eliminates the inhibitory effect of MGRN1 on MC1R signaling39. Previous reports support the current findings that suggest that MGRN1-E3 ubiquitin ligases may have a potential role in the degradation of misfolded proteins via autophagy. For the first time, we showed that MGRN1 mRNA and protein levels were substantially increased after treatment with various stress-inducing agents. Other E3 ligases, including E6-AP, CHIP, and Parkin, are also increased after exposure to various stressors and play a cytoprotective role against stress-mediated cell death21,40,41.
Recently, it has been reported that the molecular chaperone Hsp70 interacts with bis (monoacylglycero) phosphate (BMP) protein, and this interaction stabilizes the lysosomal membrane and thus reduces lysosomal pathology in Niemann-Pick disease in fibroblasts42. Because we noticed that MGRN1 follows the same induced expression profile as Hsp70 following heat shock stress, we hypothesized that MGRN1 might recognize Hsp70-associated misfolded proteins and promote their degradation through autophagy. We noticed that MGRN1 interacts with the Hsp70 chaperone. Notably, when we continued our work, we found that the 5′-untranslated region of MGRN1 retained a heat shock factor 1 consensus binding site and one CMA motif, which further supports the current findings. It has been reported previously that Hsp70 interacts with E3 ubiquitin ligases and regulates a delicate balance between protein folding and the degradation of misfolded proteins43. Because of our previous results, it was important to demonstrate the functional significance of MGRN1 and Hsp70 under conditions of cellular stress. To answer this question, we performed a series of immunofluorescence experiments. We observed that following the inhibition of autophagy, MGRN1 was nicely co-localized with p62, Hsp70, and ubiquitin-positive cytoplasmic IBs. It has been shown previously that p62 binds to ubiquitinated proteins and co-localizes with ubiquitin-positive aggregates after Baf treatment44. We also found co-localization of MGRN1 with both Hsp70 protein and denatured luciferase protein after HS treatment. The immunofluorescence staining analysis demonstratesd that MGRN1 is recruited with Hsp70-associated heat-denatured luciferase protein after the inhibition of autophagy. The recruitment of MGRN1 with heat-denatured luciferase inclusion-like structures convinced us to further investigate the functional role of MGRN1 in the degradation of misfolded cytoplasmic proteins. Next, we showed that MGRN1 overexpression promotes the degradation of heat-denatured luciferase protein through autophagy. Autophagy inhibits the aggregation of non-native proteins and cell organelles and subsequently supports cell survival under stressed conditions45,46. These studies all support our current finding showing that Hsp70-associated misfolded proteins are preferentially targeted by MGRN1 for selective autophagy.
Exposure to various cellular stressors causes protein misfolding and aggregation and may be a causal factor for known neurodegenerative diseases. After stress exposure, cells can respond in different manners; our current results demonstrate that MGRN1 actively participates in cell survival under various proteotoxic insults. Next, we further confirmed the cytoprotective role of MGRN1 during aberrant cellular stress responses. We noticed that MGRN1 overexpression counteracts the effects of various cellular stressors and promotes cell survival. Partial knockdown of MGRN1 induced cytotoxicity and consequently suppressed the cell growth mediated by various stress-inducing agents. Mitochondrial malfunction and oxidative stress have been observed in Parkinson's disease (PD), Alzheimer's disease (AD), and Huntington's disease (HD)47. It has been established that mitochondrial dysfunction and cellular stress are causally associated with neurodegeneration. MGRN1 null mutant mice have lower expression levels of mitochondrial proteins, which leads to mitochondrial dysfunction at an early age30. In the current study, we demonstrated that down-regulation of MGRN1 dramatically altered mitochondrial membrane depolarization and cytochrome c release. Previous studies and our current results clearly indicate that MGRN1 generates an oxidative stress response, and loss of its function most likely leads to mitochondrial defects in cells. Growing evidence suggests that the overexpression of molecular chaperones and E3 ubiquitin ligases alleviates the toxic effects of misfolded proteins in cells and plays a significant role in the biology of protein misfolding and neurodegenerative diseases48,49,50,51.
Finally, we showed that knockdown of MGRN1 under conditions of stress induces cell death and overexpression of MGRN1 protects against cell death induced by various stressors; this cytoprotective effect is more prominent with congruent Hsp70 overexpression. Altogether, our study provides evidences that MGRN1 is recruited to Hsp70-associated misfolded protein inclusions and promotes their degradation via autophagy. In the future, a detailed understanding of the molecular pathomechanism underlying the role of MGRN1 in protein-folding diseases will provide greater knowledge of how misfolded proteins lead to cellular damage and death.
Methods
Materials
TRIzol reagent, 3-(4,5-dimethylthiazol-2-yl)-2,5 diphenyltetrazolium bromide (MTT), N6,2′-O-dibutyryladenosine-3′,5′-cyclic monophosphate (dbcAMP), Thapsigargin, Tunicamycin, Chloroquine, Bafilomycin, 2-Mercaptoethanol and all cell culture reagents were obtained from Sigma. Lipofectamine® 2000, optiMEM, reverse transcription-PCR kits and JC-1 (5,5′, 6,6′-tetrachloro-1,1′,3,3′ tetraethylbenzimidazolocarbocyanine iodide) were purchased from Life Technologies and Molecular Probes respectively. Dual luciferase reporter gene assay kit was purchased from Promega. Protein G-agarose beads, NBT (nitroblue tetrazolium salt) and BCIP (5-bromo-4-chloro-3-indolyl phosphate, toluidinium salt) from Roche Applied Science. Monoclonal anti-Hsp70, polyclonal anit-GFP, polyclonal anti-MGRN1, monoclonal anti-luciferase, monoclonal anti-ubiquitin, monoclonal anti-β-tubulin and MGRN1 specific siRNA oligonucleotides was purchased from Santa Cruz Biotechnology. Polyclonal anti-MGRN1, monoclonal anti-p62/SQSTM1 and polyclonal-anti-cytochrome-c was purchased from Sigma. Alkaline phosphatase-conjugated anti-mouse, anti-rabbit, anti-goat IgG, Goat anti-mouse IgG- fluorescein isothiocyanate, IgG-Rhodamine, Goat anti-rabbit IgG-fluorescein isothiocyanate and IgG-Rhodamine were purchased from Vector Laboratories. Horseradish peroxidase-conjugated anti-mouse and anti-rabbit IgG were from Amersham Biosciences. Human Mahogunin (MGRN1) construct was gifted by Dr. C. Olivares Sánchez from university of Murcia, Espinardo (SPAIN) as described elsewhere39. Plasmid pcDNA™ 3.1 was purchased from Life Technologies. Plasmids pEGFP-Hsp70 (Addgene plasmid 15215), Plasmid pcDNA3-EGFP (Addgene 13031), pcDNA3-cmyc (Addgene 16011) and Luciferase-pcDNA3 (Addgene plasmid 18964) were purchased from Addgene.
Cell culture, transfection, cell viability and reporter gene assay
Cos-7 cells and A549 cell lines were maintained in Dulbecco's modified Eagle's medium (Sigma-Aldrich). All cells were grown at 37°C in 5% CO2 and supplemented with 10% heat-inactivated fetal bovine serum, 100 U/ml penicillin and 100 μg/ml streptomycin. Cells were plated into 6-well, 24 well and 96 well tissue culture plates before one day of transfection at a subconfluent density for various experiments. Cells were transiently transfected with expression vectors using the Lipofectamine® 2000 reagent according to the manufacturer's instructions. The transfection efficiency was about 70–80%. For cell viability assays, cells were plated into 96-well plates (5 × 103 cells/well) and transiently transfected with different expression plasmids and MGRN1 specific siRNA oligonucleotides; 48 h later, cells were exposed to a variety of stress agents for different time periods. Cell viability was measured by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay as described previously52. For reporter gene assay Cos-7 cells were transiently transfected with luciferase expression plasmid together with empty pcDNA3.1, MGRN1 and the MGRN1AVVA plasmids for 36 hours. Cells were either maintained in a normal CO2 incubator or heat-stressed at 43°C for 30 min and then returned to the normal CO2 incubator for 2 h. In some experiments, cells were treated with 100 μM chloroquine (CQ) before heat stress (HS). Cells were then collected and assayed for luciferase activity according to the manufacturer's protocol from Promega (Dual luciferase reporter gene assay kit).
Co-immunoprecipitation experiment and immunoblotting technique
Post transfected cells were washed with cold phosphate-buffered saline, scraped, pelleted by centrifugation, and lysed on ice for 30 min with Nonidet P-40 lysis buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1% Nonidet P-40, complete protease inhibitor mixture). Prepared cell lysates were briefly sonicated, centrifuged for 10 min at 15,000 × g at 4°C, and the supernatants (total soluble extract) were used for immunoprecipitation as described earlier53. For each immunoprecipitation experiment, about 250 μg protein in 0.2 ml NP40 lysis buffer was incubated with 4 μg of primary antibody. Protein samples were separated through SDS-polyacrylamide gel electrophoresis and processed for immunoblotting using nitrocellulose membrane. After incubation in blocking buffer (5% skim milk prepared in TBST), blot was probed with primary antibody overnight at 4°C, diluted in TBST. The membrane was washed with TBST buffer for 10 minutes, 3 times and incubated with secondary antibody for 90 minutes. Blot detection was carried out with ECL substrate (Amersham) and some membranes were developed in GB3 buffer (1 M NaCl, 0.1mM MgCl2, 10 M tris) and NBT (nitroblue tetrazolium)/BCIP (5-bromo-4-chloro-3-indoxyl phosphate substrate for alkaline phosphatase detection (10 ml GB3, 68 μl NBT, 34 μl BCIP). All primary antibodies were used in 1:1000 dilutions for immunoblotting.
Immunofluorescence technique, TUNEL assay and statistical analysis
Cells grown in 2 well chamber slide and 6-well tissue-cultured plate, on the following day, they were transiently transfected with the appropriate plasmids. Post transfected cells were twice washed with phosphate-buffered saline, fixed with 4% paraformaldehyde in phosphate-buffered saline for 20 minutes, permeabilized with 0.5% Triton X-100 in phosphate-buffered saline for 5 min, washed 4 times extensively and then blocked with 5% nonfat dried milk in TBST (Tris-buffered saline with Tween 20) for 90 minutes. Appropriate primary antibody (1:1000) incubation was used for overnight at 4°C. After incubation cells were washed with TBST several times and incubated with fluorescein isothiocyanate-conjugated and rhodamine-conjugated secondary antibody (1:500 dilutions) for 90 minutes, washed several times, and mounted in antifade solution. Nuclear staining was carried out with 5 minutes incubation in 10 mM 4,6-diamidino-2-phenylindole (DAPI) for subsequent to secondary antibody incubation and images were obtained using a fluorescence microscope. For TUNEL assay, A549 cells were plated into four well chamber slides and on the next day cells were transfected with control-siRNA and MGRN1-siRNA oligonucleotides. Post transfected cells treated with H2O2 (1.5 mM for 4 h). After treatment cells were processed for TUNEL staining according to manufacturer's instructions52. Statistical differences values are presented as mean ± SD. Statistical analysis between groups and inter group comparisons were determined by using the Student's t test, and p < 0.05 indicated statistical significance.
Reverse transcriptase PCR analysis, quantitative real time RT-PCR analysis, RNAi experiments and JC-1 staining
Cells were treated with various stress inducers. Post treated cells were used for total RNA isolation by using TRIzol reagent and then semi-quantitative RT-PCR was carried out with a RT-PCR kit (Life Technologies). The cycle number for MGRN1 34 and 21 and for β-actin was 23. MGRN1 and β-actin PCR conditions were the same: 30 min at 50°C for reverse transcription of RNA, an initial denaturation step at 94°C for 5 min and further cycling through 94°C for 30 s denaturation, 55°C for 45 s annealing, 72°C for 1 min, final extension step at 72°C for 5 min and 5 min at 4°C to cool down the reaction. For quantitative real time PCR for MGRN1 was subjected with iQ SYBR green super mix (BioRad) after cDNA synthesis from total RNA isolated from culture cells using TRIzol reagent. The real time PCRs were processed by using an iCycler iQ real-time Thermocycler Detection System (Bio-Rad). All reactions were normalized with ribosomal 18 S RNA as internal control. In reactions negative controls were prepared with the reaction lacking the template DNA. All reactions were carried out in triplicates. The sequences of the various primers were: MGRN1F, 5′-ATGGGCTCCATTCTCAGC-3′; MGRN1R, 5′- GTTGCTGTTGTCGCTGTTCT-3′; MGRN1F1, 5′- CGGGCCCTCCTGCAGATC-3′; MGRN1R1, 5′-CTGGCAGGTAGATGTCAGCA-3′; ActinF, 5′-TGACGGGGTCACCCACACTGTGCCCATCTA-3′; ActinR, 5′-CTAGAAGCATTTGCGGTGGACGATGGAGGG-3′. For RNA interference experiment, cells were plated into 6-well tissue culture plates, and on the next day the cells were transiently transfected with either MGRN1-siRNA or Control-siRNA oligonucleotides. After 48 hours, cells were collected for total RNA isolation and processed for RT-PCR, some knockdown cells were used for immunoblot analysis. To measure mitochondrial membrane potential, cells were plated into 2 well chamber slides at subconfluent density. After 24 hours, some cells were transiently transfected with either MGRN1-siRNA or Control-siRNA oligonucleotides. Post transfected cells treated with H2O2 (0.05 mM for 10 minutes) were probed with 5 μM JC-1 fluorescence dye for 45 min in the CO2 incubator and extensively washed with prewarmed PBS at 37°C. JC-1 stained cells were qualitatively observed under a fluorescence microscope using 568-nm filter.
Author Contributions
D.C. and A.M. executed the experiments. A.M. designed the experiments and wrote the manuscript. All authors reviewed the manuscript.
Supplementary Material
Supplementary information
Acknowledgments
This work was supported by the Department of Biotechnology, Government of India. AM was supported by Ramalinganswami Fellowship and Innovative Young Biotechnologist Award (IYBA) scheme from the Department of Biotechnology, Government of India. The authors would like to thank Mr. Bharat Pareek for his technical assistance and entire lab management during the manuscript preparation. We also thank to all for gifted plasmids: Dr. C. Olivares Sánchez and Dr. J.C. García-Borrón (University of Murcia, Campus de Espinardo, Murcia, Spain) for pcDNA3-MGRN1 L (−)-myc plasmid, Dr. Teresa M. Gunn (McLaughlin Research Institute Great Falls, MT) for the MGRN1-GFP and MGRN1AVVA-GFP constructs, Dr. Lois Greene (Laboratory of Cell Biology, NHLBI, NIH, Bethesda, MD) for the pEGFP hsp70 construct, Dr. Douglas T Golenbock (University of Massachusetts Medical School, Worcester MA) for the pcDNA3-EGFP plasmid, Dr. Wafik S. El-Deiry (Penn State Hershey Cancer Institute, University Drive Hershey, PA) for the pcDNA3-cmyc and Dr. William Kaelin from Dana Farber Cancer Institute and the Howard Hughes Medical Institute for luciferase-pcDNA3 plasmid.
References
- Morimoto R. & Cuervo A. Protein homeostasis and aging: taking care of proteins from the cradle to the grave. The journals of gerontology. Series A, Biological sciences and medical sciences 64, 167–170 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M. et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441, 880–884 (2006). [DOI] [PubMed] [Google Scholar]
- Berke S. & Paulson H. Protein aggregation and the ubiquitin proteasome pathway: gaining the UPPer hand on neurodegeneration. Current opinion in genetics & development 13, 253–261 (2003). [DOI] [PubMed] [Google Scholar]
- Ross C. & Poirier M. Protein aggregation and neurodegenerative disease. Nature medicine 10 Suppl, 7 (2004). [DOI] [PubMed] [Google Scholar]
- Taylor J., Hardy J. & Fischbeck K. Toxic proteins in neurodegenerative disease. Science (New York, N.Y.) 296, 1991–1995 (2002). [DOI] [PubMed] [Google Scholar]
- Rubinsztein D. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature 443, 780–786 (2006). [DOI] [PubMed] [Google Scholar]
- Bennett E., Bence N., Jayakumar R. & Kopito R. Global impairment of the ubiquitin-proteasome system by nuclear or cytoplasmic protein aggregates precedes inclusion body formation. Molecular cell 17, 351–365 (2005). [DOI] [PubMed] [Google Scholar]
- Yao T.-P. The role of ubiquitin in autophagy-dependent protein aggregate processing. Genes & cancer 1, 779–786 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejarano E. & Cuervo A. Chaperone-mediated autophagy. Proceedings of the American Thoracic Society 7, 29–39 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopito R. Aggresomes, inclusion bodies and protein aggregation. Trends in cell biology 10, 524–530 (2000). [DOI] [PubMed] [Google Scholar]
- Morimoto R. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes & development 22, 1427–1438 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhangani D. & Mishra A. Protein Quality Control System in Neurodegeneration: A Healing Company Hard to Beat but Failure is Fatal. Mol Neurobiol. [DOI] [PubMed] [Google Scholar]
- Nixon R. Autophagy in neurodegenerative disease: friend, foe or turncoat? Trends in neurosciences 29, 528–535 (2006). [DOI] [PubMed] [Google Scholar]
- Chiang H. & Dice J. Peptide sequences that target proteins for enhanced degradation during serum withdrawal. J Biol Chem 263, 6797–6805 (1988). [PubMed] [Google Scholar]
- Chiang H., Terlecky S., Plant C. & Dice J. A role for a 70-kilodalton heat shock protein in lysosomal degradation of intracellular proteins. Science (New York, N.Y.) 246, 382–385 (1989). [DOI] [PubMed] [Google Scholar]
- Yang Q. et al. Regulation of neuronal survival factor MEF2D by chaperone-mediated autophagy. Science (New York, N.Y.) 323, 124–127 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhangani D., Jana N. R. & Mishra A. Misfolded Proteins Recognition Strategies of E3 Ubiquitin Ligases and Neurodegenerative Diseases. Mol Neurobiol (2012). [DOI] [PubMed] [Google Scholar]
- Cyr D., Höhfeld J. & Patterson C. Protein quality control: U-box-containing E3 ubiquitin ligases join the fold. Trends in biochemical sciences 27, 368–375 (2002). [DOI] [PubMed] [Google Scholar]
- Goldberg A. Protein degradation and protection against misfolded or damaged proteins. Nature 426, 895–899 (2003). [DOI] [PubMed] [Google Scholar]
- Mishra A. et al. E6-AP promotes misfolded polyglutamine proteins for proteasomal degradation and suppresses polyglutamine protein aggregation and toxicity. J Biol Chem 283, 7648–7656 (2008). [DOI] [PubMed] [Google Scholar]
- Mishra A., Godavarthi S. K., Maheshwari M., Goswami A. & Jana N. R. The ubiquitin ligase E6-AP is induced and recruited to aggresomes in response to proteasome inhibition and may be involved in the ubiquitination of Hsp70-bound misfolded proteins. J Biol Chem 284, 10537–10545 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata S., Minami Y., Minami M., Chiba T. & Tanaka K. CHIP is a chaperone-dependent E3 ligase that ubiquitylates unfolded protein. EMBO reports 2, 1133–1138 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhangani D., Joshi A. P. & Mishra A. E3 ubiquitin ligases in protein quality control mechanism. Mol Neurobiol 45, 571–585 (2012). [DOI] [PubMed] [Google Scholar]
- Xiong H. et al. Parkin, PINK1, and DJ-1 form a ubiquitin E3 ligase complex promoting unfolded protein degradation. The Journal of clinical investigation 119, 650–660 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Z. et al. Gp78, an ER associated E3, promotes SOD1 and ataxin-3 degradation. Human molecular genetics 18, 4268–4281 (2009). [DOI] [PubMed] [Google Scholar]
- He L. et al. Spongiform degeneration in mahoganoid mutant mice. Science (New York, N.Y.) 299, 710–712 (2003). [DOI] [PubMed] [Google Scholar]
- Kim B., Olzmann J., Barsh G., Chin L.-S. & Li L. Spongiform neurodegeneration-associated E3 ligase Mahogunin ubiquitylates TSG101 and regulates endosomal trafficking. Molecular biology of the cell 18, 1129–1142 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti O. & Hegde R. Functional depletion of mahogunin by cytosolically exposed prion protein contributes to neurodegeneration. Cell 137, 1136–1147 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey A., Zhang C. & Cuervo A. Chaperone-mediated autophagy in aging and disease. Current topics in developmental biology 73, 205–235 (2006). [DOI] [PubMed] [Google Scholar]
- Sun K., Johnson B. S. & Gunn T. M. Mitochondrial dysfunction precedes neurodegeneration in mahogunin (Mgrn1) mutant mice. Neurobiology of aging 28, 1840–1852 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry J., Chrétien P., Lambert H., Hickey E. & Weber L. Heat shock resistance conferred by expression of the human HSP27 gene in rodent cells. The Journal of cell biology 109, 7–15 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestril R., Chi S. H., Sayen M. R., O'Reilly K. & Dillmann W. H. Expression of inducible stress protein 70 in rat heart myogenic cells confers protection against simulated ischemia-induced injury. J Clin Invest 93, 759–767 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zatloukal K. et al. p62 Is a common component of cytoplasmic inclusions in protein aggregation diseases. Am J Pathol 160, 255–263 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe J. et al. Ubiquitin is a common factor in intermediate filament inclusion bodies of diverse type in man, including those of Parkinson's disease, Pick's disease, and Alzheimer's disease, as well as Rosenthal fibres in cerebellar astrocytomas, cytoplasmic bodies in muscle, and mallory bodies in alcoholic liver disease. J Pathol 155, 9–15 (1988). [DOI] [PubMed] [Google Scholar]
- He L., Eldridge A., Jackson P., Gunn T. & Barsh G. Accessory proteins for melanocortin signaling: attractin and mahogunin. Annals of the New York Academy of Sciences 994, 288–298 (2003). [DOI] [PubMed] [Google Scholar]
- Olzscha H. et al. Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell 144, 67–78. [DOI] [PubMed] [Google Scholar]
- Schubert U. et al. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature 404, 770–774 (2000). [DOI] [PubMed] [Google Scholar]
- Levy F. et al. Ubiquitylation of a melanosomal protein by HECT-E3 ligases serves as sorting signal for lysosomal degradation. Mol Biol Cell 16, 1777–1787 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Oliva A. B., Olivares C., Jimenez-Cervantes C. & Garcia-Borron J. C. Mahogunin ring finger-1 (MGRN1) E3 ubiquitin ligase inhibits signaling from melanocortin receptor by competition with Galphas. J Biol Chem 284, 31714–31725 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikshit P. & Jana N. R. The co-chaperone CHIP is induced in various stresses and confers protection to cells. Biochem Biophys Res Commun 357, 761–765 (2007). [DOI] [PubMed] [Google Scholar]
- Imai Y. et al. CHIP is associated with Parkin, a gene responsible for familial Parkinson's disease, and enhances its ubiquitin ligase activity. Mol Cell 10, 55–67 (2002). [DOI] [PubMed] [Google Scholar]
- Kirkegaard T. et al. Hsp70 stabilizes lysosomes and reverts Niemann-Pick disease-associated lysosomal pathology. Nature 463, 549–602. [DOI] [PubMed] [Google Scholar]
- McClellan A. J., Tam S., Kaganovich D. & Frydman J. Protein quality control: chaperones culling corrupt conformations. Nat Cell Biol 7, 736–741 (2005). [DOI] [PubMed] [Google Scholar]
- Korolchuk V., Mansilla A., Menzies F. & Rubinsztein D. Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Molecular cell 33, 517–527 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B. & Kroemer G. Autophagy in the pathogenesis of disease. Cell 132, 27–42 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Levine B., Cuervo A. & Klionsky D. Autophagy fights disease through cellular self-digestion. Nature 451, 1069–1075 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beal M. Mitochondria take center stage in aging and neurodegeneration. Annals of neurology 58, 495–505 (2005). [DOI] [PubMed] [Google Scholar]
- Mishra A. et al. E6-AP association promotes SOD1 aggresomes degradation and suppresses toxicity. Neurobiol Aging (2012). [DOI] [PubMed] [Google Scholar]
- Cummings C. et al. Over-expression of inducible HSP70 chaperone suppresses neuropathology and improves motor function in SCA1 mice. Human molecular genetics 10, 1511–1518 (2001). [DOI] [PubMed] [Google Scholar]
- Tsai Y., Fishman P., Thakor N. & Oyler G. Parkin facilitates the elimination of expanded polyglutamine proteins and leads to preservation of proteasome function. J Biol Chem 278, 22044–22055 (2003). [DOI] [PubMed] [Google Scholar]
- Wyttenbach A. et al. Heat shock protein 27 prevents cellular polyglutamine toxicity and suppresses the increase of reactive oxygen species caused by huntingtin. Human molecular genetics 11, 1137–1151 (2002). [DOI] [PubMed] [Google Scholar]
- Mishra A. & Jana N. R. Regulation of turnover of tumor suppressor p53 and cell growth by E6-AP, a ubiquitin protein ligase mutated in Angelman mental retardation syndrome. Cell Mol Life Sci 65, 656–666 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra A., Godavarthi S. K. & Jana N. R. UBE3A/E6-AP regulates cell proliferation by promoting proteasomal degradation of p27. Neurobiol Dis 36, 26–34 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information