Abstract
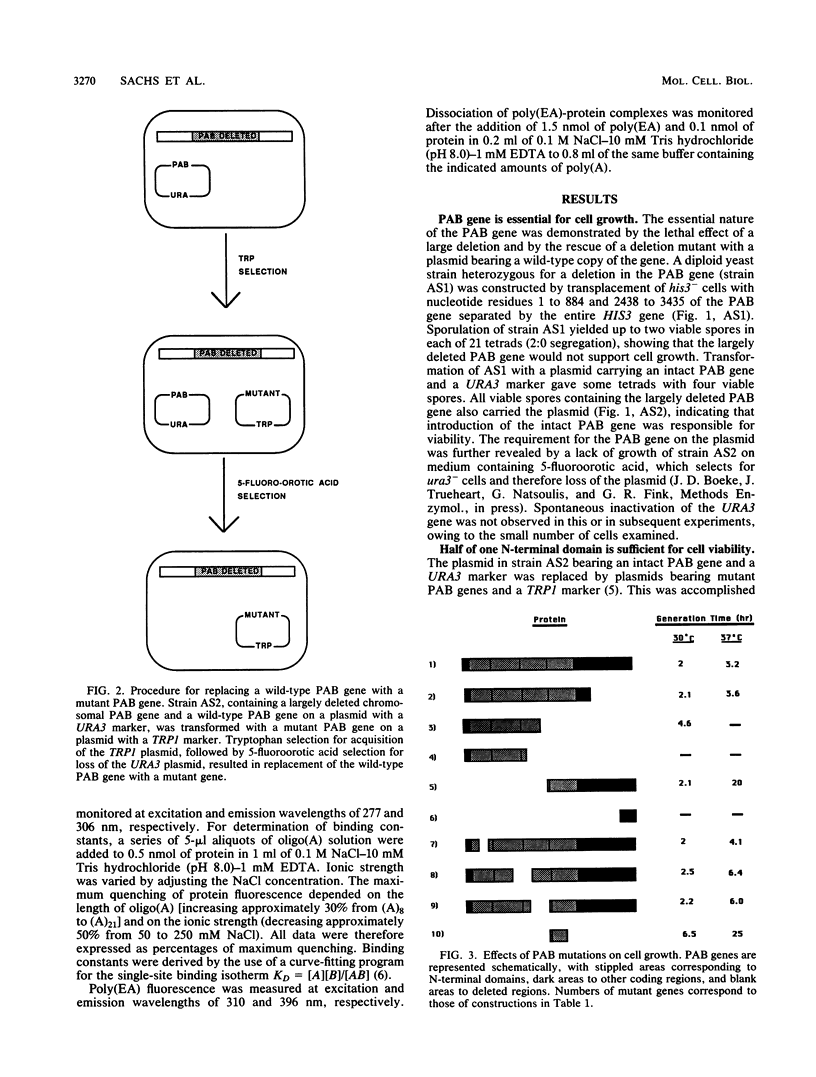
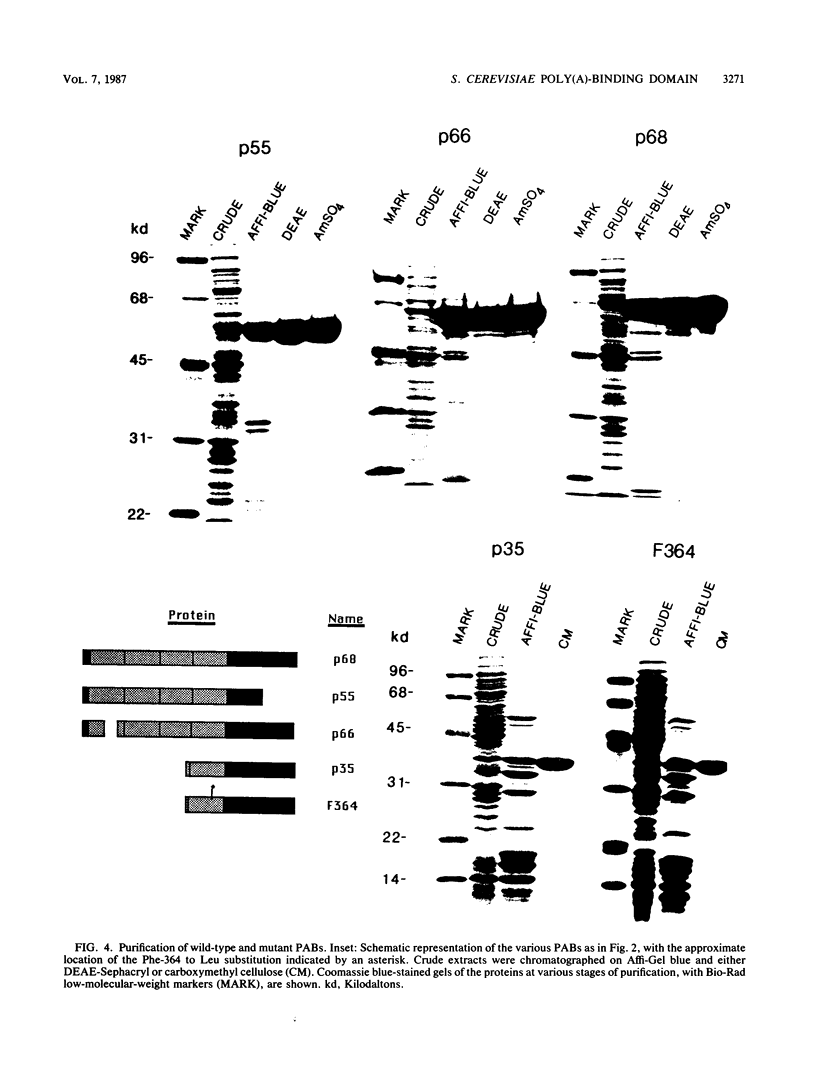
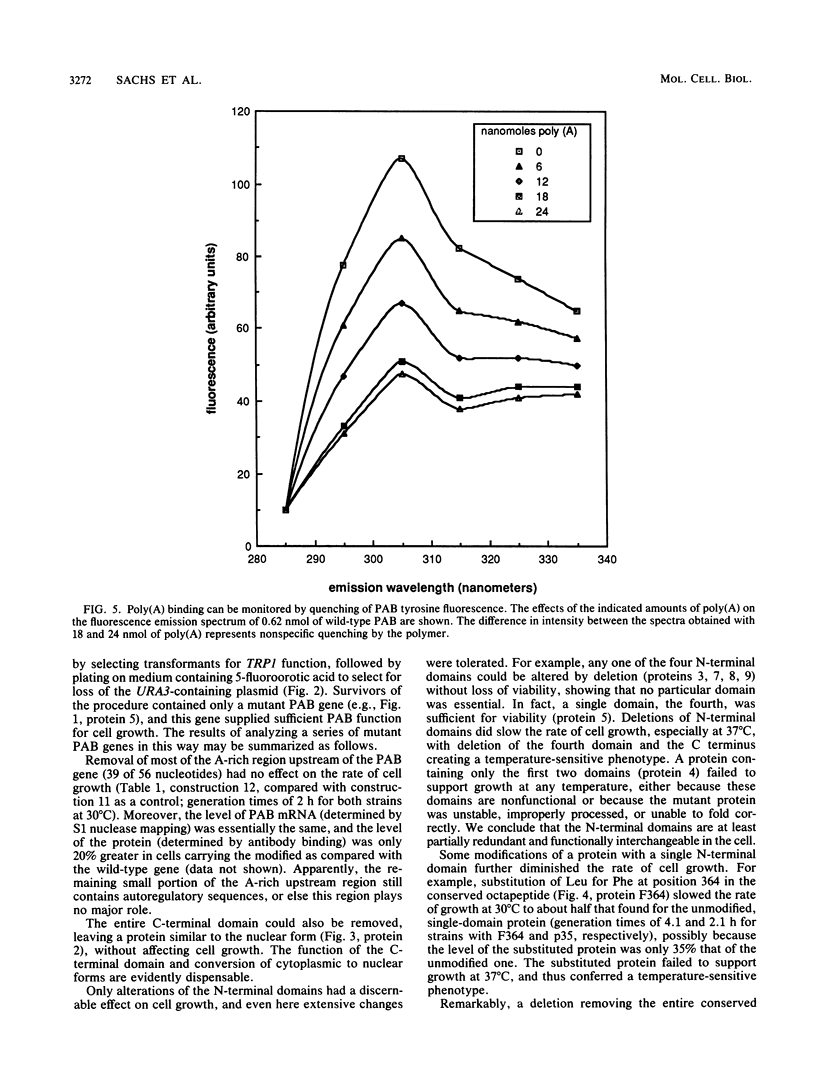
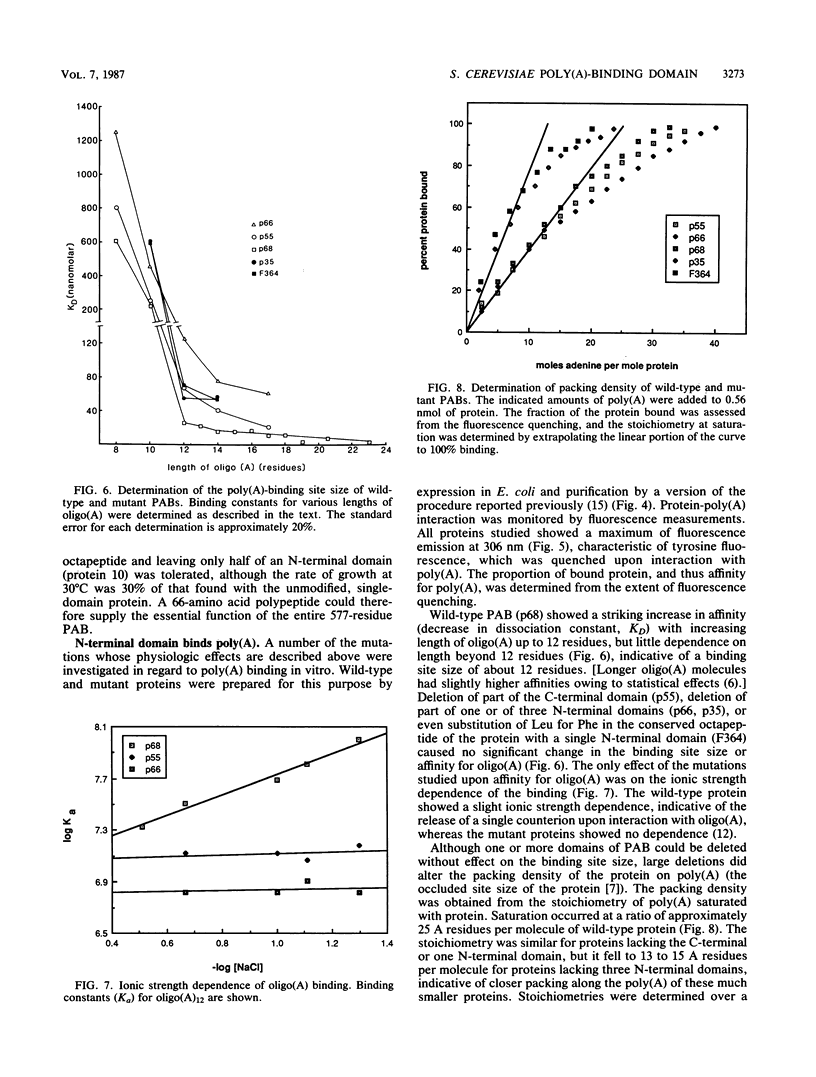
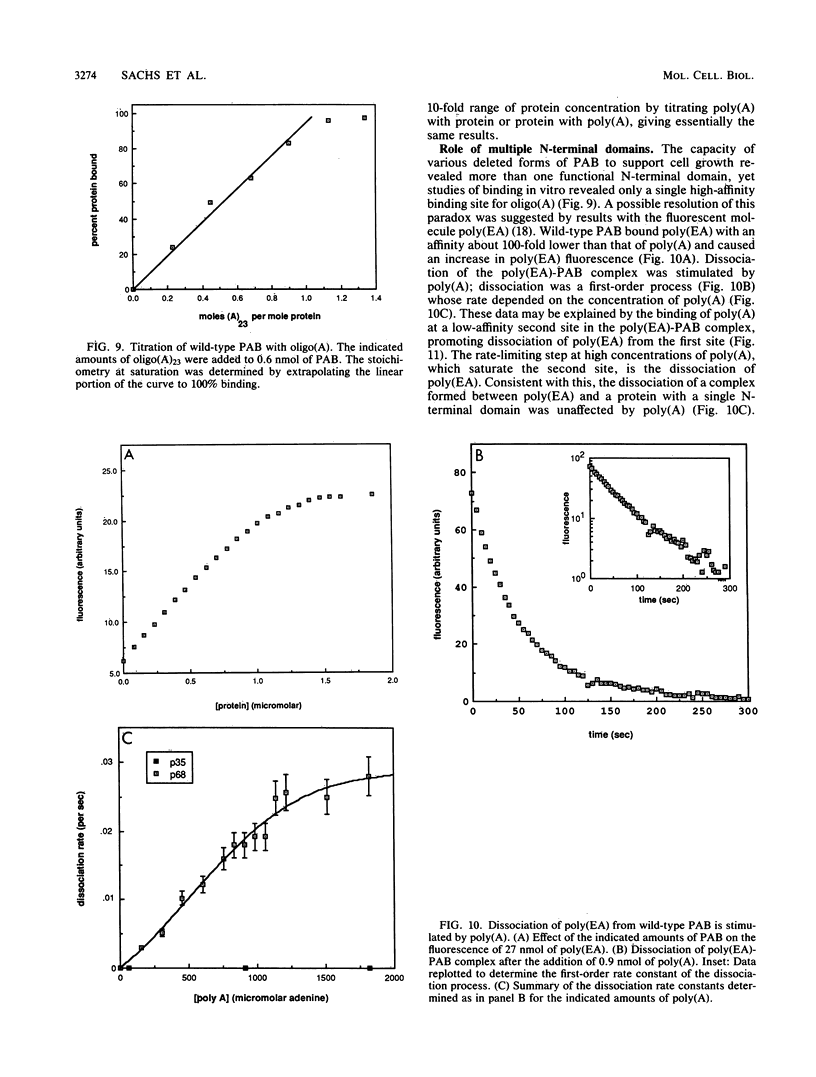
The poly(A)-binding protein (PAB) gene of Saccharomyces cerevisiae is essential for cell growth. A 66-amino acid polypeptide containing half of a repeated N-terminal domain can replace the entire protein in vivo. Neither an octapeptide sequence conserved among eucaryotic RNA-binding proteins nor the C-terminal domain of PAB is required for function in vivo. A single N-terminal domain is nearly identical to the entire protein in the number of high-affinity sites for poly(A) binding in vitro (one site with an association constant of approximately 2 X 10(7) M-1) and in the size of the binding site (12 A residues). Multiple N-terminal domains afford a mechanism of PAB transfer between poly(A) strands.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adam S. A., Nakagawa T., Swanson M. S., Woodruff T. K., Dreyfuss G. mRNA polyadenylate-binding protein: gene isolation and sequencing and identification of a ribonucleoprotein consensus sequence. Mol Cell Biol. 1986 Aug;6(8):2932–2943. doi: 10.1128/mcb.6.8.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer B. W., Kornberg R. D. Repeating structure of cytoplasmic poly(A)-ribonucleoprotein. Proc Natl Acad Sci U S A. 1980 Apr;77(4):1890–1892. doi: 10.1073/pnas.77.4.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer B. W., Kornberg R. D. The protein responsible for the repeating structure of cytoplasmic poly(A)-ribonucleoprotein. J Cell Biol. 1983 Mar;96(3):717–721. doi: 10.1083/jcb.96.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg O. G., Winter R. B., von Hippel P. H. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry. 1981 Nov 24;20(24):6929–6948. doi: 10.1021/bi00527a028. [DOI] [PubMed] [Google Scholar]
- Boeke J. D., LaCroute F., Fink G. R. A positive selection for mutants lacking orotidine-5'-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197(2):345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- Chung S. Y., Wooley J. Set of novel, conserved proteins fold pre-messenger RNA into ribonucleosomes. Proteins. 1986 Nov;1(3):195–210. doi: 10.1002/prot.340010302. [DOI] [PubMed] [Google Scholar]
- Kelly R. C., Jensen D. E., von Hippel P. H. DNA "melting" proteins. IV. Fluorescence measurements of binding parameters for bacteriophage T4 gene 32-protein to mono-, oligo-, and polynucleotides. J Biol Chem. 1976 Nov 25;251(22):7240–7250. [PubMed] [Google Scholar]
- Kowalczykowski S. C., Paul L. S., Lonberg N., Newport J. W., McSwiggen J. A., von Hippel P. H. Cooperative and noncooperative binding of protein ligands to nucleic acid lattices: experimental approaches to the determination of thermodynamic parameters. Biochemistry. 1986 Mar 25;25(6):1226–1240. doi: 10.1021/bi00354a006. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lohman T. M. Kinetics and mechanism of dissociation of cooperatively bound T4 gene 32 protein-single-stranded nucleic acid complexes. 1. Irreversible dissociation induced by sodium chloride concentration jumps. Biochemistry. 1984 Sep 25;23(20):4656–4665. doi: 10.1021/bi00315a022. [DOI] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W., Rothstein R. J. Yeast transformation: a model system for the study of recombination. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6354–6358. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROMAN H. Studies of gene mutation in Saccharomyces. Cold Spring Harb Symp Quant Biol. 1956;21:175–185. doi: 10.1101/sqb.1956.021.01.015. [DOI] [PubMed] [Google Scholar]
- Record M. T., Jr, Anderson C. F., Lohman T. M. Thermodynamic analysis of ion effects on the binding and conformational equilibria of proteins and nucleic acids: the roles of ion association or release, screening, and ion effects on water activity. Q Rev Biophys. 1978 May;11(2):103–178. doi: 10.1017/s003358350000202x. [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Ho Y. S., Shatzman A. The use of pKc30 and its derivatives for controlled expression of genes. Methods Enzymol. 1983;101:123–138. doi: 10.1016/0076-6879(83)01009-5. [DOI] [PubMed] [Google Scholar]
- Sachs A. B., Bond M. W., Kornberg R. D. A single gene from yeast for both nuclear and cytoplasmic polyadenylate-binding proteins: domain structure and expression. Cell. 1986 Jun 20;45(6):827–835. doi: 10.1016/0092-8674(86)90557-x. [DOI] [PubMed] [Google Scholar]
- Sachs A. B., Kornberg R. D. Nuclear polyadenylate-binding protein. Mol Cell Biol. 1985 Aug;5(8):1993–1996. doi: 10.1128/mcb.5.8.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver M. S., Fersht A. R. Direct observation of complexes formed between recA protein and a fluorescent single-stranded deoxyribonucleic acid derivative. Biochemistry. 1982 Nov 23;21(24):6066–6072. doi: 10.1021/bi00267a007. [DOI] [PubMed] [Google Scholar]