SUMMARY
Signals from commensal bacteria can influence immune cell development and susceptibility to infectious or inflammatory diseases. However, the mechanisms by which commensal bacteria regulate protective immunity after exposure to systemic pathogens remain poorly understood. Here, we demonstrate that antibiotic-treated (ABX) mice exhibit impaired innate and adaptive antiviral immune responses and substantially delayed viral clearance after exposure to systemic LCMV or mucosal influenza virus. Furthermore, ABX mice exhibited severe bronchiole epithelial degeneration and increased host mortality after influenza virus infection. Genome-wide transcriptional profiling of macrophages isolated from ABX mice revealed decreased expression of genes associated with antiviral immunity. Moreover, macrophages from ABX mice exhibited defective responses to type I and type II IFNs and impaired capacity to limit viral replication. Collectively, these data indicate that commensal-derived signals provide tonic immune stimulation that establishes the activation threshold of the innate immune system required for optimal antiviral immunity.
INTRODUCTION
Commensal microbial communities colonize barrier surfaces of the skin, vaginal, upper respiratory, and gastrointestinal tracts of mammals and consist of bacteria, fungi, protozoa, and viruses (Breitbart et al., 2003; Ley et al., 2006a; Scupham et al., 2006). The largest and most diverse microbial communities reside in the intestine and have beneficial properties ranging from aiding in metabolism to competing with invasive pathogens for the environmental niche (Honda and Littman, 2012; Sonnenburg et al., 2006). Studies in patients have associated alterations in bacterial communities with susceptibility to diabetes, obesity, cancer, inflammatory bowel disease (IBD), allergy, and other atopic disorders, highlighting the potential impact of hostcommensal interactions on multiple metabolic and chronic inflammatory diseases (Ley et al., 2006b; Manichanh et al., 2006; Moore and Moore, 1995; Penders et al., 2007).
Studies using gnotobiotic (germ-free), antibiotic-treated (ABX), or selectively colonized mice have demonstrated that deliberate manipulation of commensal bacterial communities results in impaired lymphoid tissue development, dysregulated immune cell homeostasis, and altered susceptibility to infectious or inflammatory diseases in the gastrointestinal tract (Abt and Artis, 2009; Littman and Pamer, 2011; Smith et al., 2007). For example, experimental colonization of mice with Clostridium spp. induced CD4+ regulatory T cells in the intestine and ameliorated intestinal inflammation in a murine model of IBD (Atarashi et al., 2011). In contrast, colonization of the intestine with segmented filamentous bacteria (SFB) is associated with increased frequencies of intestinal CD4+ T helper 17 cells and exacerbated autoimmune inflammation in murine models of arthritis, multiple sclerosis, and diabetes, demonstrating that defined commensal species can promote inflammatory diseases (Ivanov et al., 2009; Kriegel et al., 2011; Lee et al., 2011; Wu et al., 2010). Consistent with proinflammatory properties, signals from commensal bacteria can act as an adjuvant, augmenting immune responses after intestinal parasitic or bacterial infections (Benson et al., 2009; Hall et al., 2008; Ivanov et al., 2009). Conversely, commensal bacteria can increase viral infectivity in the gastrointestinal microenvironment (Kane et al., 2011; Kuss et al., 2011). Thus, commensal-derived signals are capable of limiting or exacerbating infection in the intestinal microenvironment. However, the mechanisms through which commensal-derived signals regulate innate and adaptive immunity to infection remain poorly defined.
The mammalian innate immune system has evolved diverse strategies each tailored to detect and respond to distinct pathogens. Despite this apparent specialization, crosstalk between pathways has been reported in which stimulation from one class of pathogens influences the response to another (Barton et al., 2007; Spencer et al., 1977). However, it is unclear whether commensal bacteria influence innate immune pathways in the steady-state and, if so, whether these interactions modulate responsiveness to viral pathogens. Iwasaki and colleagues reported impaired antiviral immunity in the lung after manipulation of commensal bacteria (Ichinohe et al., 2011) that was associated with defective activation of the inflammasome (Lamkanfi and Dixit, 2011). Whether depletion of commensal bacteria selectively regulates inflammasome-dependent pathways or represents broader immunological crosstalk between commensal bacteria and antiviral pathways remains to be determined.
In this report, we examine this fundamental question and demonstrate that manipulating commensal bacteria results in impaired host protective immunity after either systemic (lymphocytic choriomeningitis virus [LCMV]) or mucosal (influenza virus) infection, leading to dysregulated adaptive immune responses and underlying defects in innate antiviral pathways. Genome-wide transcriptional profiling of macrophages from naive ABX mice revealed reduced expression of genes associated with IFN activation and antiviral immunity. Moreover, macrophages from ABX mice exhibited impaired responsiveness to type I and type II IFNs and a reduced capacity to control viral replication. Restoration of IFN responsiveness in ABX mice re-established protective antiviral immunity after influenza virus infection. Taken together, these data indicate that commensal bacteria provide tonic signals that calibrate the activation threshold and sensitivity of the innate antiviral immune system.
RESULTS
Defective Immunity to LCMV Infection after ABX-Mediated Depletion of Commensal Bacteria
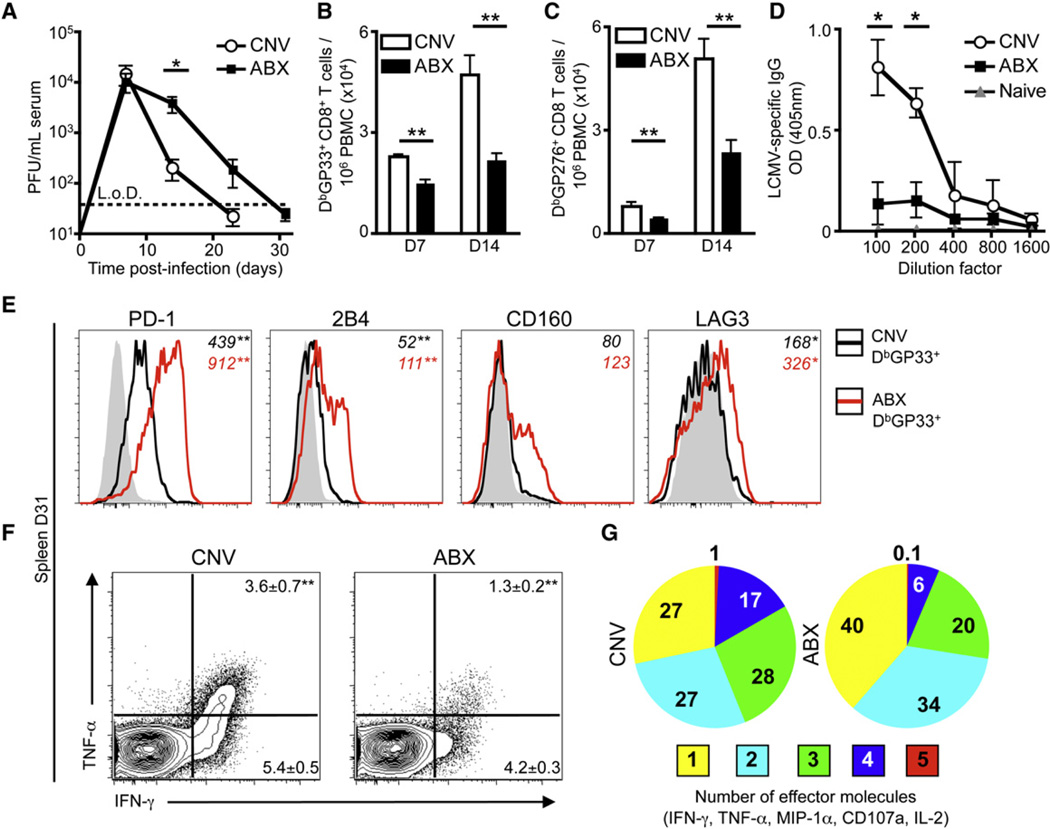
Signals from commensal bacteria regulate intestinal immune cell homeostasis in multiple settings (Hill and Artis, 2010; Round and Mazmanian, 2009); however, whether commensal-derived signals regulate immunity to pathogens that infect sites other than the gastrointestinal tract is unclear. To address this question, we administered naive C57BL/6 mice oral doses of broad-spectrum antibiotics for 2 weeks and subsequently infected them with LCMV T1b, a strain of virus that causes viremia for 1–2 weeks and that requires a robust innate and adaptive immune response for viral clearance (Blackburn et al., 2009). As reported in our earlier studies, exposure to antibiotics resulted in a reduction in intestinal commensal bacteria and dramatic reorganization of the bacterial community structure (Hill et al., 2012). After infection with LCMV, conventionally housed (CNV) mice exhibited maximal viremia at day 7 (d7) post-infection (p.i.) and successfully controlled viremia by d23 (Figure 1A). Control of infection was associated with expansion of LCMV-specific CD8+ T cells (Figures 1B and 1C) and LCMV-specific IgG in the serum (Figure 1D). In contrast, ABX mice exhibited a significant delay in clearance of circulating virus (Figure 1A, p = 0.036) and increased viral titers in the kidneys at d31 p.i. (Figure S1A available online). Impaired viral control was associated with reduced LCMV-specific CD8+ T cell responses and IgG antibody titers in the blood (Figures 1B–1D). In addition, at d31 p.i., LCMV-specific H2-DbGP33 tetramer+ CD8+ T cells isolated from ABX mice expressed increased levels of the inhibitory receptors PD-1, 2B4, CD160, and LAG-3 (Figure 1E) and were less efficient producers of multiple effector molecules (IFN-γ, TNF-α, IL-2, MIP-1α, and CD107a) (Figures 1F and 1G). Analysis of CD8+ T cell responses specific for other LCMV epitopes revealed similar impairment of the total LCMV-specific CD8 T cell response in ABX mice (Figures S1B and S1C). These results are consistent with more severe T cell exhaustion in the ABX group, a characteristic sign of impaired immunity to LCMV infection (Wherry, 2011). Therefore, deliberate manipulation of commensal-derived signals results in defective virus-specific adaptive immune responses and inefficient control of viral replication after systemic infection.
Figure 1. Systemic LCMV T1b Infection Results in Delayed Viral Clearance and an Impaired LCMV-Specific CD8+ T Cell Response in ABX Mice.
(A) Viral titer in the serum of CNV or ABX C57BL/6 mice after LCMV T1b infection (L.o.D., limit of detection).
(B and C) LCMV-specific (B) DbGP33 and (C) DbGP276 tetramer+ CD8+ T cells per 106 peripheral blood mononuclear cells (PBMC) at d7 and d14 p.i.
(D) Serial dilution of LCMV-specific IgG antibody titers in serum of CNV or ABX mice at d23 p.i. Naive serum from CNV mice used for baseline.
(E) Expression of inhibitory receptors PD-1, 2B4, CD160, LAG-3 on DbGP33 tetramer+ CD8+ T cells isolated from the spleen of CNV (black line) or ABX (red line) mice. Shaded histograms represent CD44lo CD8+ T cells. Numbers in italics represent mean fluorescence intensity (MFI).
(F and G) Splenocytes from d31 infected mice were incubated with GP33 peptide for 5 hr in the presence of BFA and assessed for production of IFN-γ, TNF-α, MIP-1α, CD107a, and IL-2. FACS plots gated on live, CD8α+ cells.
(G) Proportion of GP33 peptide responsive CD8+ T cells producing multiple effector molecules. Data representative of three independent experiments with n = 5 mice per group. Data shown are the mean ± SEM. Serum viral titer statistics determined by two-part t test for each time point. *p < 0.05, **p < 0.01. See also Figure S1.
Enhanced Susceptibility and Reduced Immunity to Influenza Virus Infection in ABX Mice
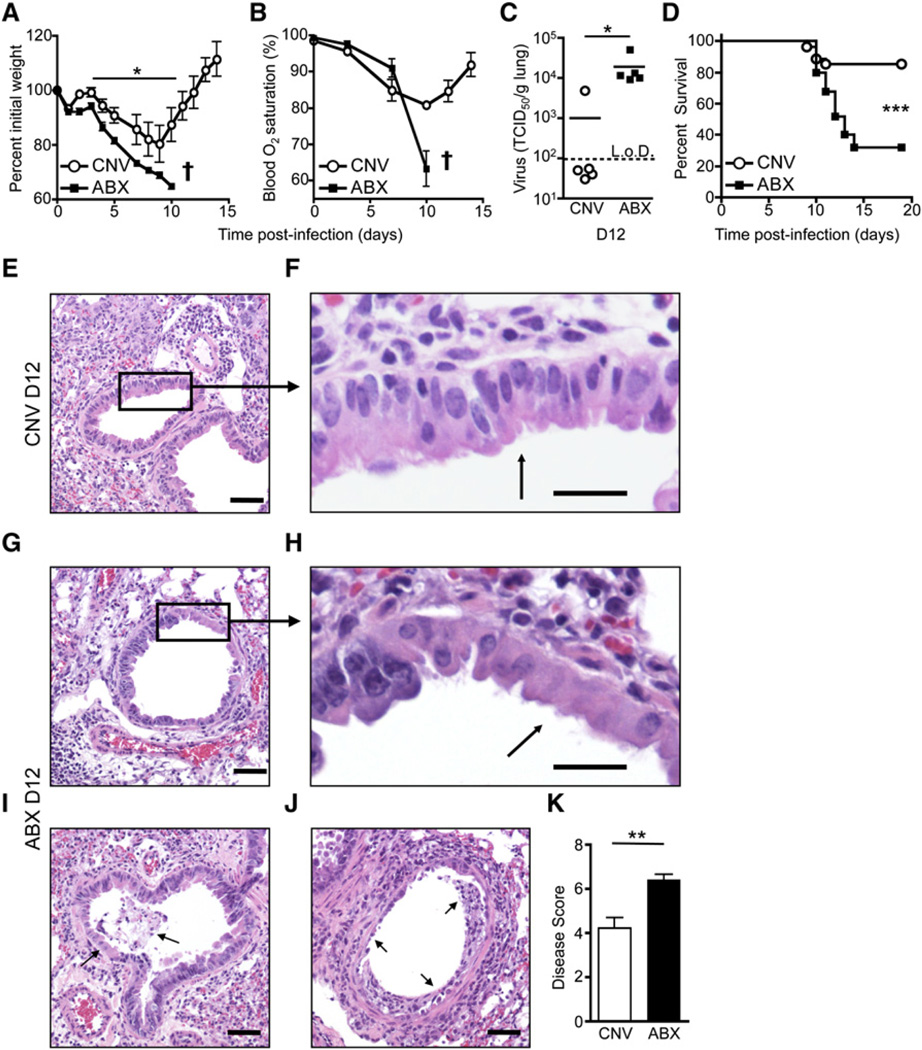
To test whether commensal bacteria might influence optimal host defense following exposure to other viral infections, we infected CNV or ABX mice with influenza virus (PR8-GP33) and analyzed immunologic, virologic, and pathologic parameters. Similar to the gastrointestinal tract (Figure S2A), there was a loss of culturable aerobic and anaerobic commensal bacteria in the upper respiratory tract of ABX mice compared to CNV mice (Figure S2B). After exposure to influenza virus infection, CNV mice lost ~20% of their original body weight (Figure 2A) and had reduced lung function as measured by blood oxygen saturation (Figure 2B). Approximately 80% of CNV mice recovered and cleared virus from the lungs by d12 p.i. (Figures 2C and 2D). Histopathological examination of lung sections from CNV mice at d12 p.i. (Figures 2E and 2F) compared to uninfected mice (Figure S2C) revealed peribronchiolar inflammation and epithelial hyperplasia, indicating ongoing tissue damage and repair (Figures 2E and 2F). In contrast, ABX mice lost significantly more weight (Figure 2A, p = 0.013), had a drastic drop in blood oxygen saturation (Figure 2B), exhibited significantly higher viral titers in the lung (Figure 2C, p = 0.016), and had increased mortality (Figure 2D, p ≤ 0.001) after influenza virus infection. Lung sections from infected ABX mice revealed more pronounced epithelial cell necrosis (Figures 2G and 2H), increased exudate and dead cells in the bronchiolar lumen (Figure 2I, arrows), and in the most severe cases, complete loss of the bronchiole epithelial layer (Figure 2J, arrows). Scoring of histological sections of the lung confirmed increased prevalence of epithelial cells with morphologic features of degeneration and necrosis in ABX mice relative to CNV mice (Figure 2K). Consistent with this, increased cell death was observed in the bronchiole alveolar lavage (BAL) fluid of ABX compared to CNV mice (Figures S2D and S2E). Similar to a previously published report (Dolowy and Muldoon, 1964), influenza virus-infected germ-free (GF) mice also exhibited increased weight loss (Figure S2F), impaired viral clearance (Figure S2G), reduced virus-specific antibody titers (Figure S2H), and more severe bronchiole epithelial degeneration compared to CNV mice (Figures S2I and S2J). In addition, CNV or ABX mice were infected with X31-GP33 virus, a less pathogenic strain of influenza virus that causes minimal morbidity and mortality in CNV mice (Bouvier and Lowen, 2010; Decman et al., 2010). Consistent with results using the PR8 strain of influenza virus, ABX mice exhibited considerably greater weight loss (Figure S3A), elevated viral titers (Figure S3B), increased lung epithelial degeneration (Figure S3C), and increased mortality (Figure S3D). Taken together, these data indicate that commensal-derived signals are critical in promoting optimal immunity against multiple viral infections at sites distinct from the gastrointestinal tract.
Figure 2. Alterations in Commensal Bacterial Communities Exacerbate Lung Pathology and Mortality to Influenza Virus.
CNV or ABX C57BL/6 mice were infected i.n. with influenza virus PR8-GP33.
(A and B) Time course of weight loss (A) and blood oxygen (B) saturation after infection (representative exp. n = 5: † signifies mice below 70% initial weight were sacrificed).
(C) Influenza virus genome copies in the lung at d12 p.i. assessed by RT-PCR and displayed as TCID50/gram of lung tissue.
(D) Survival curve after PR8-GP33 infection; CNV n = 27, ABX n = 25.
(E–J) H&E-stained lung section of CNV (E and F) or ABX (G–J) mice at d12 p.i. Black box and arrows highlight (E and F) epithelial hyperplasia, (G and H) epithelial cell necrosis, (I) cellular debris and exudate in lumen, and (J) loss of bronchiole epithelium (scale bar represents 50 µm in E, G, I, and J; 20 µm in F and H).
(K) Disease score of bronchiole epithelial degeneration at d12 p.i. Data representative of five independent experiments with n = 5–6 mice per group. Survival statistics determined by log rank test. Viral titer statistics determined by two-part t test. *p < 0.05, **p < 0.01, and ***p < 0.001. Data shown are mean ± SEM. See also Figure S2.
Diminished Generation of Influenza Virus-Specific Adaptive Immunity in ABX Mice
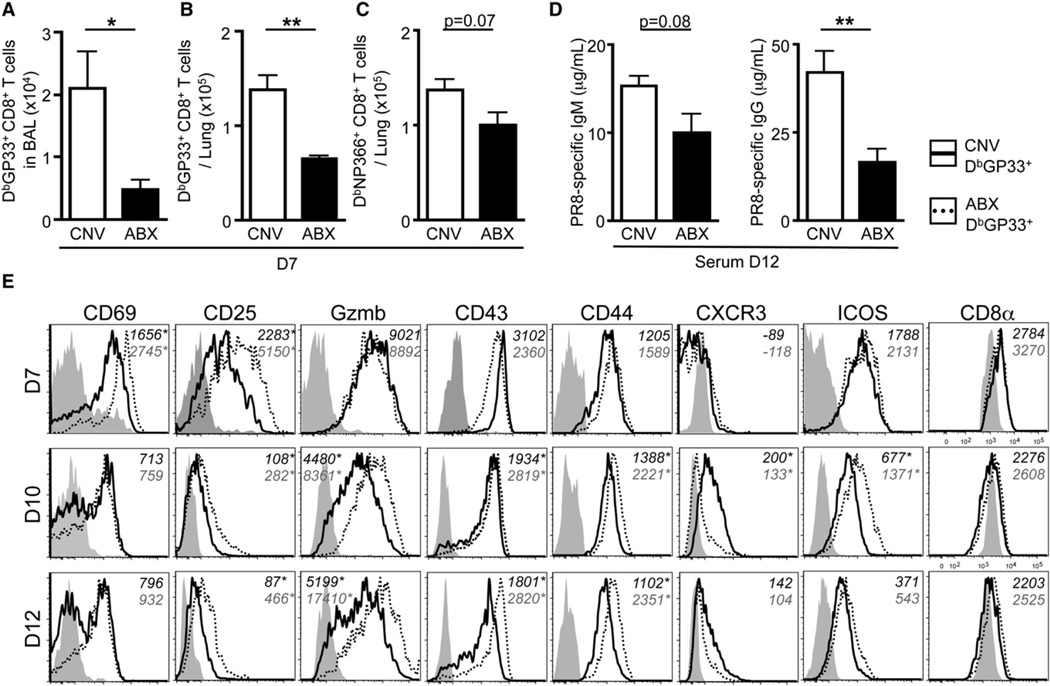
Analysis of the adaptive immune response revealed that CNV mice infected with PR8-GP33 virus generated large populations of influenza virus-specific CD8+ T cells in the BAL (Figure 3A) and lung parenchyma (Figures 3B and 3C) by d7 p.i. In contrast, the total number of influenza virus-specific CD8+ T cells present in these tissues was reduced in ABX mice (Figures 3A–3C). Consistent with these data, infection with X31-GP33 also resulted in significantly fewer tetramer+ CD8+ T cells in the BAL (Figure S3E, p = 0.034), lung parenchyma (Figure S3F, p = 0.008), mediastinal lymph nodes (Figure S3G,p = 0.05), and spleen (Figure S3H, p≤ 0.001) of ABX mice compared to CNV mice. Additionally, virus-specific CD8+ T cells in the lung of ABX mice were less capable of producing multiple effector molecules simultaneously (IFN-γ, TNF-α, IL-2, MIP-1α, and CD107a) (Figure S3I). ABX mice also exhibited lower titers of PR8-specific IgM and IgG in the serum (Figure 3D).
Figure 3. ABX Mice Have a Diminished Influenza Virus-Specific Adaptive Immune Response.
(A and B) Total number of influenza virus-specific DbGP33 tetramer+ CD8+ T cells isolated from the (A) BAL or (B) lung parenchyma at d7 p.i.
(C) DbNP366 tetramer+ CD8+ T cells isolated from the lung parenchyma at d7 p.i.
(D) PR8-specific IgM and IgG titers in the serum at d12 p.i.
(E) Phenotypic profile of DbGP33 tetramer+ CD8+ T cells isolated from the lung of CNV (solid line) or ABX (dotted line) mice at d7, d10, and d12 p.i. Gray shaded histograms are CD44lo CD8+ T cells isolated from the lung. Numbers in italics represent MFI. Data representative of three independent experiments with n = 4–5 mice per group. *p < 0.05 and **p < 0.01. Data shown are mean ± SEM. See also Figure S3.
Expression of activation markers such as CD69, CD25, and Granzyme-B is rapidly upregulated early after viral infection and then downregulated as the infection is controlled (Lawrence and Braciale, 2004; Wherry et al., 2007). Although DbGP33 tetramer+ CD8+ T cells from CNV mice downregulated early activation markers between d7 to 12 p.i., expression of CD69 and CD25 remained elevated in DbGP33 tetramer+ CD8+ T cells isolated from ABX mice (Figure 3E; Table S1). Elevated expression of these molecules, along with Granzyme-B and CD43, persisted in ABX mice at d12 p.i. These data are consistent with a delay in the adaptive immune response to influenza virus and sustained activation of virus-specific CD8+ T cells due to prolonged viral replication. Collectively, these data demonstrate that ABX-mediated alterations in commensal bacterial communities result in impaired humoral and cellular immune responses necessary for clearing systemic or mucosal viral infections.
Impaired Innate Antiviral Immune Responses in ABX Mice
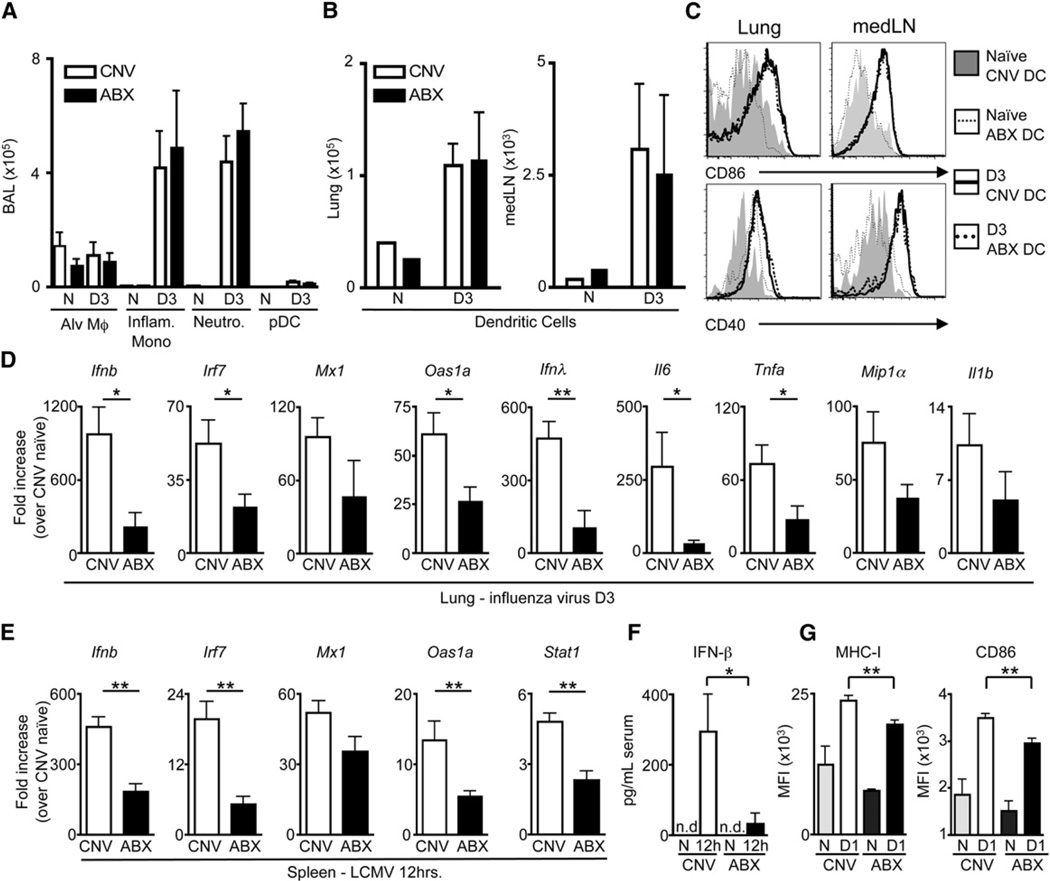
The impaired antiviral CD8+ T cell response in ABX mice after LCMV or influenza virus infection suggested a potential defect in the early innate immune response. To test this, we assessed the recruitment and activation of early responding innate immune cells after PR8-GP33 virus infection. There was a comparable influx of macrophages, inflammatory monocytes, neutrophils, plasmacytoid dendritic cells (pDC) (Figures 4A and S4A), and conventional dendritic cells (cDC) (Figure 4B) into the BAL, lung or draining lymph nodes of CNV versus ABX mice. Further, both cDC (Figure 4C) and pDC (Figure S4B) exhibited a similar activation profile in CNV and ABX mice. However, although the total number of macrophages was similar between CNV and ABX mice, there was reduced expression of macrophage-associated antiviral response genes (Ifnb, Irf7, Mx1, Oas1a, Il28b [Ifnγ], Il6, Tnfa, Ccl3 [Mip1α], and Il1b) in the lung of ABX mice compared to CNV mice (Figure 4D), indicating an impaired innate immune response. Consistent with an impaired innate immune response following influenza virus infection, as early as 12 hr post-LCMV infection there was reduced expression of mRNA encoding Ifnb, Irf7, Mx1, Oas1a, and Stat1 in the spleen (Figure 4E) and decreased IFN-β in the serum of ABX mice compared to CNV mice (Figure 4F), indicating that diminished innate antiviral immune responses are a general feature in ABX mice after either mucosal or systemic viral infection.
Figure 4. Innate Antiviral Immune Response Is Diminished in ABX Mice after Influenza Virus or LCMV Infection.
(A) Total numbers of alveolar macrophages, inflammatory monocytes, neutrophils and plasmacytoid dendritic cells isolated from BAL at d3 p.i.
(B) Total numbers of dendritic cells (DCs) isolated from the lung parenchyma and mediastinal lymph node (medLN).
(C) Expression of CD86 and CD40 on naive or d3 p.i. DCs isolated from the lung and medLN of CNV or ABX mice.
(D) Fold induction of antiviral defense gene expression in the lung at d3 post-influenza virus infection relative to lung of naive CNV mice as assessed by RT-PCR.
(E) Fold induction of antiviral defense genes in the spleen 12 hr after LCMV (T1b) infection relative to spleen of naive CNV mice as assessed by RT-PCR.
(F) IFN-β levels in the serum at 12 hr post LCMV infection as detected by ELISA.
(G) Expression of MHC-I and CD86 on peritoneal macrophages from CNV or ABX mice 24 hr post LCMV infection. Data representative of two or more independent experiments with n = 3–5 mice per group. *p < 0.05, **p < 0.01. Data shown are mean ± SEM. See also Figure S4.
The immunregulatory cytokine IL-10 has been demonstrated to influence the antiviral immune response (Brooks et al., 2006; McKinstry et al., 2009; Sun et al., 2009) and expression of this cytokine can be modulated by commensal bacteria (Amaral et al., 2008; Mazmanian et al., 2008). However, prior to infection, no differences in expression of Il10 mRNA were observed in the spleen (Figure S4C) or lung (Figure S4D) of CNV and ABX mice. Further, after influenza virus infection, a similar increase in IL-10 was detected in the BAL of CNV and ABX mice (Figure S4E), indicating that impaired antiviral immune responses in ABX mice were not associated with dysregulated IL-10 production. In contrast, proinflammatory cytokines and chemokines were reduced in the BAL after influenza virus infection (Figure S4F) or in the spleen after LCMV infection (Figure S4G) of ABX mice early after infection. Furthermore, expression of MHC-I and CD86 on macrophages from ABX, LCMV-infected mice was reduced at d1 p.i. (Figure 4G). Together, these findings indicate that ABX-mediated alterations in commensal bacterial communities result in selective dysregulation of innate immune responses after systemic or mucosal viral infection.
Defective Expression of Antiviral Defense Genes in Macrophages from ABX Mice
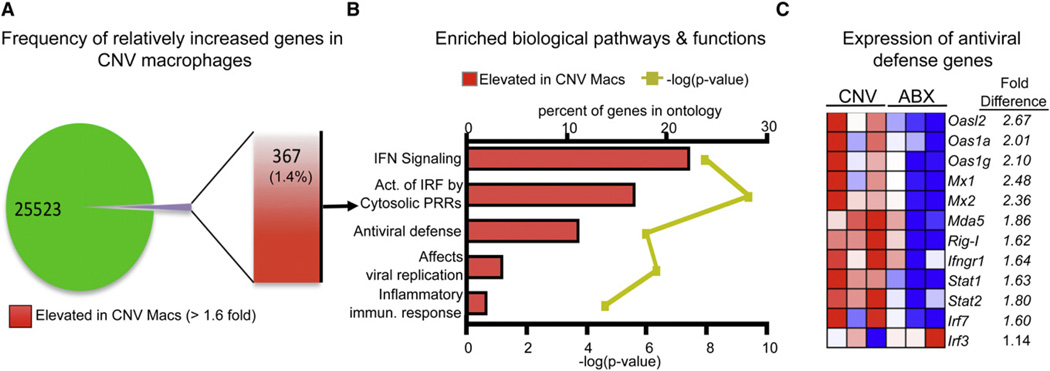
Early defects in the antiviral response in ABX mice provoke the hypothesis that commensal-derived signals regulate the activation status of innate immune cells prior to viral infection. Phenotypic characterization revealed that macrophages (Figure S5A), but not DCs (Figure S5B), isolated from the peritoneal cavity of naive ABX mice had decreased expression of IFN-γRI, MHC-I, CD40, and CD86 molecules that are critical during the early response to viral infection. To interrogate the potential mechanisms through which signals derived from commensal bacteria regulate macrophage responses and innate antiviral immunity, we employed genome-wide transcriptional profiling of macrophages isolated from CNV or ABX mice prior to viral infection. Fundamental differences in transcriptional profiles were readily apparent between macrophages isolated from CNV versus ABX mice (Figure S6A). In total, there were 367 gene transcripts upregulated by ≥1.6-fold in macrophages isolated from CNV mice relative to macrophages isolated from ABX mice (Figure 5A). Ingenuity pathways analysis identified the interferon signaling pathway as the most significantly enriched pathway in macrophages isolated from CNV mice versus ABX mice (Figure 5B, p ≤ 0.001). In addition, gene set enrichment analysis (GSEA) (Haining and Wherry, 2010; Subramanian et al., 2005) revealed that six of the top eight most enriched gene sets in macrophages from CNV mice were related to interferon responses (Figures S6B–S6D). Analysis of specific gene expression in macrophages isolated from ABX mice revealed a relative downregulation of several genes that regulate detection of virus (Ifih1 [Mda5] and Ddx58 [Rig-I]), the response to interferon signaling (Irf7, Ifngr1, Stat1, and Stat2), or inhibition of viral replication (Mx1 and Oas1a) (Figure 5C), but not genes associated with the inflammasome or TLR signaling pathways (Figures S6E and S6F), two alternative innate pathogen recognition mechanisms. Differential expression of these interferon-pathway related antiviral defense genes was independently confirmed by real-time PCR (RT-PCR) analysis (Figure S6G). Taken together, genome-wide transcriptional profiling and computational analyses suggest that signals derived from commensal bacteria calibrate the activation threshold of antiviral immune response pathways in macrophages.
Figure 5. Naive Macrophages from ABX Mice Have an Attenuated Antiviral Defense Gene Profile.
RNA was extracted from sort-purified peritoneal macrophages isolated from naive CNV or ABX mice. Extracted RNA was hybridized to an Affymetrix GeneChip microarray to assess gene expression.
(A) Frequency and total number of elevated genes in CNV macrophages compared to macrophages isolated from ABX mice.
(B) Highly enriched biological pathways and functions found within the subset of elevated genes from CNV macrophages as assessed by Ingenunity pathways analysis. Red bars indicate the percent of genes in a pathway upregulated in macrophages isolated from CNV mice. Yellow line indicates p value calculated by Fisher’s exact test.
(C) Heat map of key antiviral defense genes in macrophages isolated from CNV or ABX mice. Red, high expression; blue, low expression. See also Figures S5 and S6.
Reduced Response to IFN Stimulation or Viral Infection in Macrophages Isolated from ABX Mice
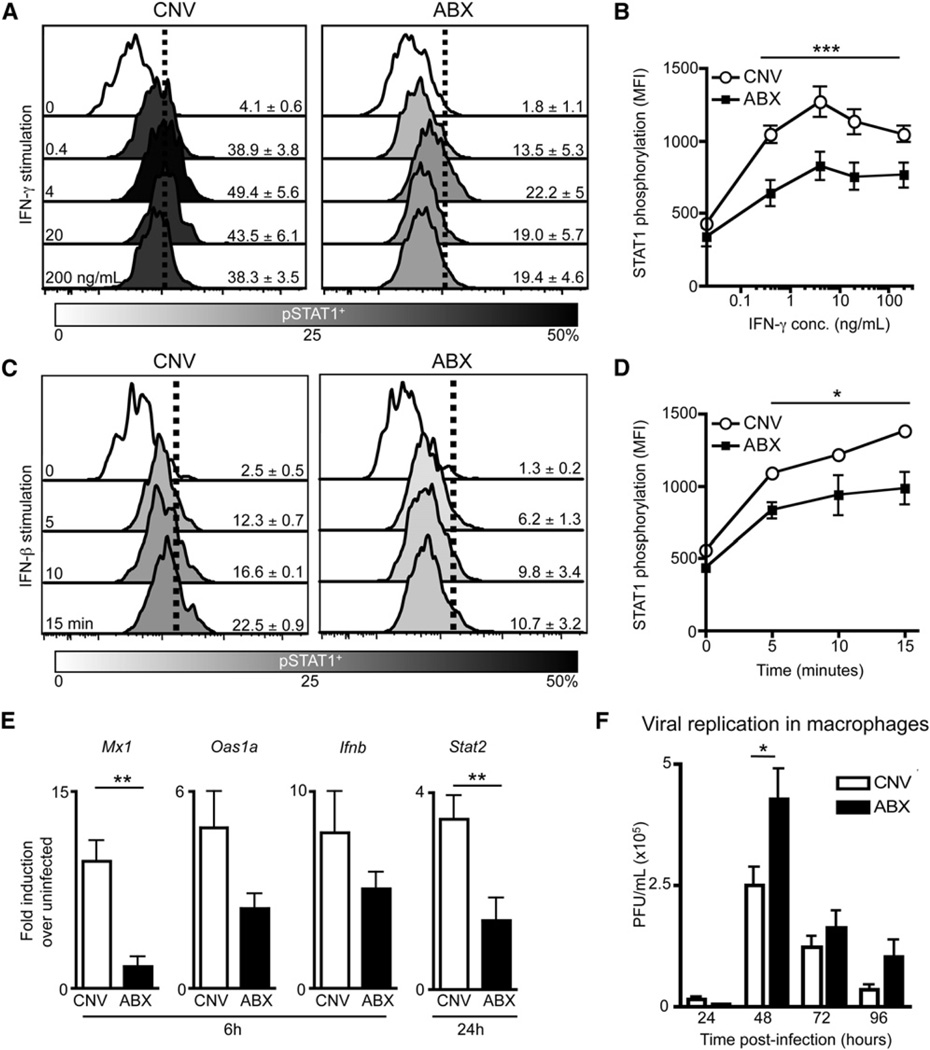
Genome-wide transcriptional profiling suggested that commensal-derived signals modulate the responsiveness of macrophages to viral infection or IFN stimulation. IFN receptor signaling results in phosphorylation of STAT1 followed by translocation into the nucleus, where STAT1 mediates transcription of interferon-responsive genes crucial to the early antiviral response (Schindler et al., 1992; Shuai et al., 1992). To functionally test IFN responsiveness, we stimulated macrophages isolated from naive CNV or ABX mice with IFN-γ or IFN-β in vitro and measured STAT1 phosphorylation. Stimulated macrophages isolated from ABX mice exhibited significantly reduced phospho-STAT1 (pSTAT1) compared to CNV macrophages after IFN-γ (Figures 6A and 6B, p ≤ 0.001) or IFN-β (Figures 6C and 6D, p = 0.025) stimulation. Similar deficiencies in STAT1 phosphorylation were observed after IFN-γ (Figures S7A and S7B) or IFN-β (Figures S7C and S7D) stimulation of macrophages isolated from GF mice. To determine whether defects in IFN responsiveness impact the response to viral infection, we infected macrophages isolated from naive CNV or ABX mice with influenza virus in vitro and examined induction of antiviral defense genes. Expression of multiple antiviral defense genes (Mx1, Oas1a, Ifnb, and Stat2) was reduced in macrophages isolated from ABX mice compared to CNV mice at 6–24 hr p.i. (Figure 6E), indicating an inherent inability to respond to viral infection. Furthermore, macrophages isolated from ABX (Figure 6F, p = 0.04) or GF (Figure S7E, p ≤ 0.01) mice supported significantly more viral replication upon in vitro infection with LCMV. These biological observations are consistent with diminished expression of antiviral defense genes identified by transcriptional profiling (Figure 5) and indicate that signals from commensal bacteria are crucial for maintaining optimal responsiveness of macrophages to IFN stimulation required to control viral replication.
Figure 6. Macrophages from Naive ABX Mice Have a Diminished Ability to Respond to IFN Stimulation and Viral Infection In Vitro.
(A–D) Peritoneal Macrophages isolated from CNV or ABX mice were stimulated with IFN-γ or IFN-β in vitro. Histograms of STAT1 phosphorylation in macrophages after (A) IFN-γ stimulation (0.4, 4, 20, 200ng/mL) or (C) IFN-β stimulation (103 units/mL for 5, 10, and 15 min). MFI of pSTAT1 in macrophages after (B) IFN-γ or (D) IFN-β stimulation.
(E and F) Peritoneal macrophages sorted from naive CNV or ABX mice were infected in vitro with (E) influenza virus (X31-GP33, MOI of5)or(F) LCMV (cl-13 strain, MOI of 0.2).
(E) Induction of antiviral defense genes in macrophages at 6 and 24 hr p.i. as assessed by RT-PCR.
(F) LCMV viral titers in supernatant at 24–96 hr p.i. Data representative of two or more independent experiments with n = 3–5 mice per group. *p < 0.05, **p < 0.01, and ***p < 0.001. Data shown are mean ± SEM. See also Figure S7.
Impaired In Vivo Antiviral Macrophage Response in ABX Mice after Influenza Virus or LCMV Infection
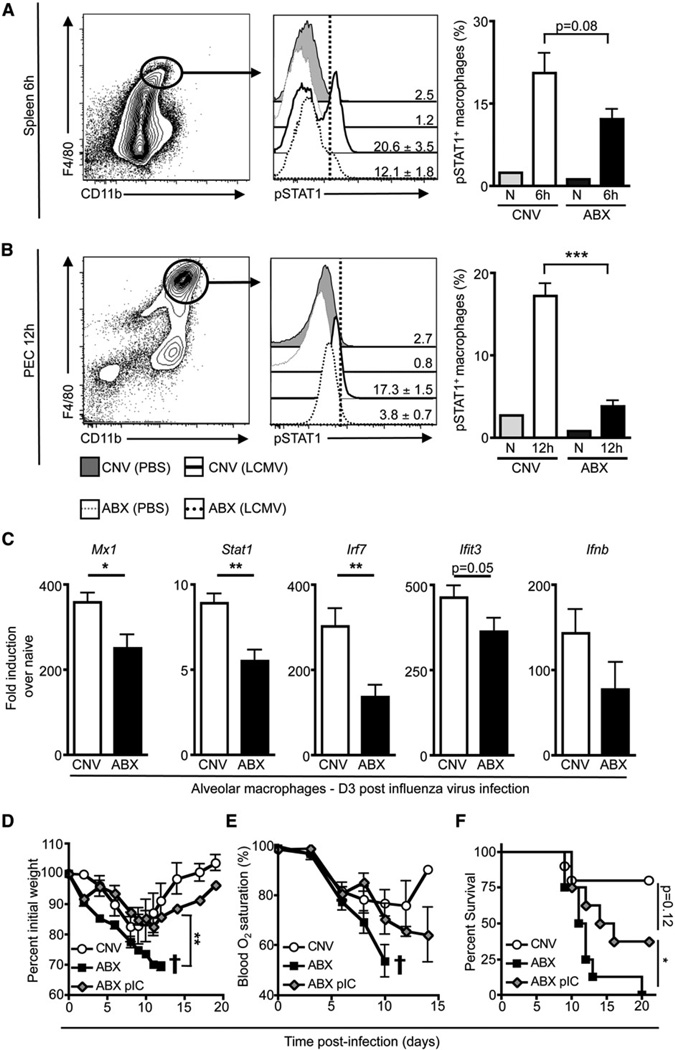
To test whether defects in the antiviral macrophage response exist in vivo after viral infection, we infected CNV or ABX mice with LCMV and assessed STAT1 phosphorylation in macrophages. As early as 6 hr after infection, macrophages from the spleen of ABX mice exhibited reduced pSTAT1 relative to splenic macrophages isolated from CNV mice (Figure 7A). By 12 hr p.i., peritoneal macrophages from ABX mice exhibited significantly decreased STAT1 phosphorylation compared to peritoneal macrophages from CNV mice (Figure 7B, p ≤ 0.001). To determine whether alveolar macrophages also exhibited a defective antiviral response after influenza virus infection, we sorted macrophages from the BAL of CNV or ABX mice at d3 post-influenza virus infection and assessed a panel of antiviral defense genes by RT-PCR. Airway macrophages from ABX mice exhibited reduced induction of antiviral defense genes including Mx1, Stat1, Irf7, Ifit3, and Ifnb (Figure 7C), indicating that the macrophage response in ABX mice was qualitatively impaired after viral infection.
Figure 7. In Vivo Antiviral Macrophage Response Is Impaired in ABX Mice after LCMV or Influenza Virus Infection.
(A and B) CNV or ABX mice were inoculated with LCMV (T1b) or PBS i.v. and (A) splenocytes at 6 hr p.i. or (B) PECs at 12 hr p.i. were immediately fixed to preserve the in vivo STAT1 phosphorylation status of macrophages.
(C) CNV or ABX mice were infected with influenza virus (PR8-GP33). At d3 p.i., alveolar macrophages were sorted from the BAL and in vivo induction of antiviral defense genes was assessed by RT-PCR. Gene expression displayed as fold induction over naive alveolar macrophages from CNV mice. Data representative of two independent experiments with n = 3–5 mice per group.
(D–F) CNV or ABX mice were infected with influenza virus (PR8-GP33). Mice received 30 µg of poly I:C (ABX+pIC group) or PBS (CNV & ABX group) i.n. at d −1 and 100 µg of poly I:C or PBS i.p. at d3. Weight loss (D) and blood oxygen (E) saturation after infection (representative exp. n = 4–6: † signifies mice below 70% initial weight were sacrificed). Weight loss statistics determined by two-way ANOVA.
(F) Survival curve after influenza virus infection. Survival curve is a combination of two independent experiments. CNV n = 10, ABX n = 8, ABX+pIC n = 12. Survival statistics determined by log rank test. *p < 0.05, **p < 0.01, and ***p < 0.001. Data shown are mean ± SEM.
To assess whether re-establishing IFN responsiveness in ABX mice could restore protective immunity to influenza virus infection, we administered poly I:C to ABX mice and then challenged them with influenza virus. ABX mice receiving PBS had drastic weight loss (Figure 7D), loss of lung function (Figure 7E), and high mortality after influenza virus infection (Figure 7F). In contrast, administration of poly I:C to influenza virus-infected ABX mice resulted in less weight loss (Figure 7D) as well as marked improvements in lung function and survival (Figures 7E and 7F), suggesting that recalibrating the immune response to IFN signaling could supplant the need for tonic commensal-derived stimulation of the innate immune system. Taken together, these observations indicate that commensal bacterial communities have a fundamental role in setting the activation threshold of the innate immune system required for optimal antiviral immune responses.
DISCUSSION
Our studies demonstrate an unexpected role for commensal bacteria in calibrating the responsiveness of antiviral immunity. ABX-mediated alterations of commensal bacteria compromised innate and adaptive immune responses after systemic or respiratory viral infection. Severe defects in the adaptive immune response to LCMV and influenza virus as well as poor control of viral replication pointed toward early defects in the initiation of antiviral responses. Indeed, genome-wide transcriptional profiling and functional assays uncovered a global defect in antiviral responsiveness of macrophages isolated from ABX mice. Upon deliberate manipulation of commensal bacteria, expression of antiviral defense genes and interferon responsive pathways were altered in the steady state. For rapidly replicating viruses, such a delay in initiating antiviral pathways and activating downstream events such as humoral and cell-mediated adaptive immune responses can have dramatic consequences leading to failure to control infection, increased host morbidity, and mortality. Together, these data suggest a model in which signals from commensal bacteria calibrate the activation threshold of innate antiviral immune responses.
Commensal bacterial communities modulate immune cell homeostasis and disease by providing either immunoregulatory or proinflammatory signals. For example, polysaccharide A, isolated from Bacteroides fragilis, can reduce the severity of intestinal inflammation in two models of IBD (Mazmanian et al., 2008). Conversely, commensal bacteria can also boost immune responses against mucosal infections (Benson et al., 2009; Hall et al., 2008; Ichinohe et al., 2011; Ivanov et al., 2009). These studies provoke the hypothesis that commensal-derived signals might influence the systemic immune response to infection. The present study demonstrates that commensal bacteria influence the activation threshold of broadly used innate antiviral response pathways such as the IFN signaling pathway. Induction of a type I IFN response is fundamental and critical for defense against the majority of viruses (Sen, 2001). Macrophages isolated from ABX mice, however, displayed major defects in expression of key interferon-stimulated response genes even prior to viral exposure compared to macrophages from CNV mice. Reduction in steady-state transcription of antiviral pathways was associated with impaired responsiveness to type I and type II IFNs or virus.
Iwasaki and colleagues observed that commensal bacteria can influence inflammasome activity, an innate signaling pathway involved in responses to bacteria, cytosolic oligomers, and a subset of viruses (Ichinohe et al., 2011; Lamkanfi and Dixit, 2011). In addition, two recent reports identified a fundamental interaction between intestinal commensal bacteria and enteric viruses in which virus can utilize bacterial products to enhance infectivity (Kane et al., 2011; Kuss et al., 2011). These reports highlight the dynamic interrelationship between viral pathogens, commensal bacteria, and the immune system. Our results reveal a previously unrecognized interplay between commensal bacteria and antiviral interferon signaling pathways in which low-level tonic signaling by commensal bacteria regulates the steady-state readiness of hard-wired antiviral pathways in macrophages.
Tonic signaling has been proposed as a mechanism to maintain optimal responsiveness of signaling pathways in other immunologic settings (Macia et al., 2009). For example, naive T cells use tonic signals from low-affinity interactions with self-MHC to regulate homeostasis and optimal dynamic responsiveness upon engagement of cognate antigen (Takeda et al., 1996; Tanchot et al., 1997). In this current study, tonic signaling was dependent on commensal-derived signals to maintain the fitness of antiviral pathways in macrophages. Although the potential impact of antibiotic treatment on the host virome is largely unexplored, the most direct interpretation of our data is that commensal bacteria calibrate the threshold of innate immune activation to viral infections and suggest steady-state innate immune crosstalk. Such crosstalk can occur in other settings. For example, latent viral infections can render mice less susceptible to bacterial challenge, an effect attributed to basal macrophage activation (Barton et al., 2007). Conversely, the bacterial species, Wolbachia, confers protection against viral infections in Drosophila (Teixeira et al., 2008). In addition to antibacterial defense genes, bacterial-derived LPS-TLR4 signaling can upregulate transcription of antiviral genes (Amit et al., 2009; Doyle et al., 2002). In the case of LPS-TLR4 signaling, antiviral gene expression is initially induced, but rapidly limited by the polycomb repressor Cbx4 (Amit et al., 2009). This latter observation suggests a potential explanation for the commensal-antiviral immune fitness axis at the transcriptional level. Induction of transcription followed by repression might maintain key antiviral genes in a state of poised transcriptional regulation, rather than a repressed or inactive state. Transcriptional poising, or the presence of both activating and repressive chromatin, enables more efficient transcriptional induction upon exposure to a true inducer of the gene of interest (Cuddapah et al., 2010). This state of transcriptional equilibrium provided by tonic commensal stimulation may enable rapid induction of antiviral defense genes upon infection. Examples of this type of regulation exist in other biological systems such as the yeast Hog1-MAPK pathway (Macia et al., 2009). Our results suggest that commensal bacteria provide such a signal to maintain antiviral innate immune pathways in a state of optimal readiness, allowing dynamic and robust responses upon challenge by viral infections.
It was remarkable that macrophages isolated from ABX mice prior to viral infection also displayed less detectable in vivo pSTAT1 compared to macrophages from naive CNV controls. Commensal-derived signals may induce tonic, low-level STAT1 activation in the steady state, which could be a key contributing factor to basal induction of antiviral defense genes prior to infection. The mechanisms through which commensal-derived signals stimulate immune cells in the periphery are poorly understood. One possibility is that peripheral immune cells are directly exposed to bacterial microbes or their products. Small numbers of live commensal bacteria can be found in the Peyer’s patches and mesenteric lymph nodes of mice in the steady state, and there is evidence that absorbed commensal products circulate throughout the host (Clarke et al., 2010; Macpherson and Uhr, 2004). Thus, direct interaction between peripheral immune cells and bacterial products is plausible. Alternatively, commensal bacteria may act indirectly on peripheral immune cells via responses evoked from epithelial or other mucosal-associated stromal cells (Artis, 2008). Defining the potential pathways involved in microbial sensing by the peripheral immune system will be crucial for understanding how microbial crosstalk influences immune cell homeostasis and host protective immunity.
Modulating commensal bacterial communities has therapeutic potential. For example, probiotic treatments can ameliorate intestinal inflammatory diseases (Sartor, 2004) and success of bacteriotherapy in cases of viral gastroenteritis demonstrates the potential use of probiotics as a treatment strategy to combat viral infection (Fang et al., 2009; Szajewska and Mrukowicz, 2005). Further, prophylactic probiotic administration can limit the duration and severity of respiratory viral infections in human subjects, suggesting that the beneficial effects of probiotics on antiviral immunity are not limited to the gastrointestinal tract (de Vrese et al., 2006). Despite many recent advances in defining the diverse and dynamic microbiome in humans and animal models of human disease, it is unclear which bacterial species or microbial products are associated with the beneficial antiviral effects of commensal bacteria observed in this study. It will be important to define the commensal bacterial species and signals that elicit these host protective effects. Such studies could lead to new approaches for therapeutically administering commensal bacteria or commensal-derived products and selectively manipulating host protective immunity.
EXPERIMENTAL PROCEDURES
Mice and Viruses
C57BL/6 mice (4 to 6 weeks old) were purchased from the National Cancer Institute (Frederick, MD). Mice were maintained in specific pathogen-free facilities at the University of Pennsylvania. GF C57BL/6 and Swiss Webster mice were maintained in plastic isolator units and fed autoclaved chow and water. The University of Pennsylvania Institutional Animal Care and Use Committee (IACUC) approved all protocols, and all experiments were performed in accordance with the guidelines of the University of Pennsylvania IACUC. Mice were inoculated intravenously (i.v.) with LCMV T1b (2 × 106 PFU), or intranasally (i.n.) with recombinant influenza viruses expressing the LCMV GP33 epitope (PR8-GP33: 368 TCID50, H3N2 X31-GP33: 1 × 105 TCID50) as described (Decman et al., 2010). LCMV viral titers were determined by plaque assay on Vero cell monolayers (Ahmed et al., 1984).
Oral Antibiotic Treatment
Mice were provided autoclaved drinking water supplemented with ampicillin (0.5 mg/mL, Sigma), gentamicin (0.5 mg/mL, Gemini Bio-Products), metronidazole (0.5 mg/mL Sigma), neomycin (0.5 mg/mL, Med-Pharmex), vancomycin (0.25 mg/mL, Novaplus), and sucralose (4mg/mL, Splenda, McNeil Nutritionals, LLC). Splenda was added to make the antibiotic cocktail more palatable. Antibiotic treatment was started 2–4 weeks prior to infection and continued for the duration of the experiment.
Pulse Oximetry
Anesthetized mice had neck and throat hair removed with a chemical depilatory agent (Nair, Church & Dwight Co.) 1 day prior to infection. MouseOx Tm Pulse-Oximeter neck sensors (Starr Life Sciences, Oakmount, PA) were placed on exposed skin and blood oxygen saturation was recorded with Starr MouseOx software v. 5.1. Pulse oximetry readings were allowed to stabilize and 15 seconds of measurements were averaged.
Isolation of Cells from the Spleen, Blood, Lymph Node, Peritoneal Cavity, Lung Tissue and Airway
Lymphocytes were isolated from the spleen, lymph node, blood, lungs, or BAL as described (Decman et al., 2010). Peritoneal exudates cells (PECs) were obtained by injecting and recovering PBS from the peritoneal cavity.
Flow Cytometry, Tetramer, and Intracellular Staining
Single-cell suspensions were stained for surface antigens with antibodies and tetramers or for intracellular cytokines (ICS) after peptide stimulation as described (Decman et al., 2010). Fluorescently conjugated antibodies used include those specific to CD3ε, CD4, CD8α, CD11c, CD19, CD25, CD43, CD45, CD69, CD119, CD160, F4/80, Ly6c, MHC-I (H-2Kb), MHC-II (I-A I-E) (eBioscience), CD5, CD44, CD80, CD86, Ly6g (clone 1A8), ICOS, LAG-3, PD-1 (clone RMP1–30), (Biolegend), CD40 (BD Biosciences), CXCR3 (R&D Systems), 2B4 (BD PharMingen), CD11b, IL-2 (eBioscience), IFN-γ (BD PharMingen), TNF-α (Biolegend), MIP-1α (R&D Systems), and granzyme B (Invitrogen). MHC class I peptide tetramers were made and used for identifying virus-specific CD8+ T cells (Altman et al., 1996). Alveolar macrophages were identified as non-T, non-B, non-NK cells (NTNBNNK), and CD11c+, F4/80+. Inflammatory monocytes were identified as NTNBNNK, CD11c−, F4/80−, Ly6g−, Ly6c+, and CD11b+. Neutrophils were identified as NTNBNNK, CD11c−, F4/80−, Ly6g+, and CD11b+. DCs were identified as NTNBNNK, F4/80−, CD11c+, and MHC-IIhi. Plasmacytoid DCs were identified as NTNBNNK, nonalveolar macrophage, noninflammatory monocyte, nonneutrophil, CD11cint, and PDCA-1+. Peritoneal and splenic macrophages were identified as NTNBNNK F4/80+, CD11b+ Samples were collected with a LSR II flow cytometer (Becton Dickinson). All flow cytometry data were analyzed by FlowJo v 8.8 (Treestar). Pie charts were created with the Pestle and SPICE programs (Mario Roederer; Vaccine Research Center, NIAID, NIH).
RNA Isolation, cDNA Preparation, and RT-PCR
RNA was isolated from cells with an RNeasy mini-kit (QIAGEN) and from lung or spleen tissue with mechanical homogenization and TRIzol isolation (Invitrogen) in accordance with the manufacturer’s instructions. PA influenza-specific primers and probes were used for determining influenza virus genome copies on the basis of a standard curve then converted to TCID50/ gram of lung tissue. Genes of interest were normalized to β-actin (or Hprt for whole spleen tissue) and displayed as fold difference relative to uninfected CNV control mice.
Histological Sections and Pathology Scoring
Lungs were inflated with 4% paraformaldehyde (PFA) and embedded in paraffin. Five micrometer sections were cut and stained with hematoxylin and eosin. Blind scoring of H&E-stained lung tissue sections by a board-certified veterinary pathologist reflect degree of luminal exudates (0–5) and degree of bronchiole epithelial degeneration and necrosis (0–5), for a maximum score of 10.
Cell Sorting and Microarray Data Analysis
Peritoneal macrophages from naive CNV or ABX mice were sorted directly into TRIzol (Invitrogen) on a BD Aria (Beckson Dickson). Test sorts were ≥95% pure. For microarray analysis, RNA was extracted from three sorted biological replicates of peritoneal macrophages from naive CNV or ABX mice. cDNA was amplified with NuGen WT Ovation Pico kit and hybridized to Affymetrix GeneChip Mouse Gene 1.0 ST microarrays at the University of Pennsylvania’s Microarray facility. Expression levels were summarized with the Robust Multichip Averaging (RMA) algorithm (Irizarry et al., 2003). The ClassNeighbors module of GenePattern (Broad Institute, Cambridge MA) was used for identifying differentially expressed genes. Gene transcripts with greater than 1.6-fold difference in expression were analyzed with Ingenuity pathway analysis software (Ingenuity Systems, www.ingenuity.com). A right-tailed Fisher’s exact test was used for calculating a p value. GSEA was performed as described (Subramanian et al., 2005).
In Vitro and In Vivo Phosflow STAT1 Staining of Macrophages
Adherent macrophages from PECs were stimulated with recombinant IFN-γ (R&D Systems) or IFN-β (PBL Interferonsource). After stimulation, media was removed and replaced with 0.05% trypsin and incubated at 37°C for 2 min. Cells were then fixed with 1.6% PFA for 10 min, permeabilized with ice cold 90% methanol, and stained for surface markers and pSTAT1 with PE-conjugated anti-STAT1 (pY701) antibody (BD biosciences). LCMV-infected CNV or ABX mice were sacrificed at 6 or 12 hr p.i. We directly fixed cells in 1.6% PFA to preserve phosphorylation status and resuspended them in 90% methanol at 4°C for 30 min as described in Krutzik et al. (2005).
In Vitro Viral Infection of Peritoneal Macrophages
Sort purified macrophages isolated from naive CNV, ABX, or GF mice were infected with either LCMV clone 13 strain (MOI of 0.2) or influenza virus X31-GP33 strain (MOI of 5) for 1 hr. At indicated time points, supernatants were collected so that viral titers could be assessed by plaque assay or macrophages were directly lysed with RLT lysis buffer (QIAGEN) and RNA was isolated as described above.
Poly I:C Administration in Influenza Virus-Infected Mice
ABX mice were administered 30 mg of polyinosinic-polycytidylic acid (poly I:C Sigma) i.n. at d −1 and 100 mg i.p. at d3 post influenza virus infection (PR8-GP33). Control CNV and ABX mice received 30 µl of PBS i.n. at d −1 and 100 µl i.p. at d3.
Statistical Analysis
Results represent means ± SEM. Statistical significance was determined by the unpaired, two tailed Student’s t test for individual time points, two-way ANOVA test for time course experiments, log rank test for survival curve, or two-part t test for comparison of groups that contained samples that were below the limit of assay detection. Statistical analyses were performed with Prism GraphPad software v4.0 (*p < 0.05; **p < 0.01; ***p < 0.001).
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge members of the Artis and Wherry laboratory for helpful discussions and critical reading of the manuscript and D. Kobuley for care of the germ-free facility. This research is supported by the US National Institutes of Health (grants AI061570, AI087990, AI074878, AI095608, AI091759, and AI095466 to D.A.; grants AI071309, AI078897, AI095608, AI083022, AI077098, and HHSN266200500030C to E.J.W.; T32-AI05528 to M.C.A.; T32-AI007532 to G.F.S and L.A.M.; T32-RR007063 and K08-DK093784 to T.A.; T32-AI007324 to M.A.P.), Irvington Institute Postdoctoral Fellowship of the Cancer Research Institute (L.C.O.), the Burroughs Wellcome Fund (D.A.), the National Institute of Diabetes and Digestive and Kidney Disease, Center for the Molecular Studies in Digestive and Liver Disease, and the Molecular Pathology and Imaging Core (DK50306). We thank N. Cohen for assistance with tracheal isolation and the Abramson Cancer Center Flow Cytometry and Cell Sorting Resource Laboratory (partially supported by NCI Comprehensive Cancer Center Support grant #2-P30 CA016520), the Wistar flow cytometry core, the UPenn Vet School Pathology Service, UPenn microarray facility, and UPenn human immunology core for invaluable technical assistance and expertise.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven figures, one table, Supplemental Experimental Procedures and can be found with this article online at doi:10.1016/j.immuni.2012.04.011.
REFERENCES
- Abt MC, Artis D. The intestinal microbiota in health and disease: the influence of microbial products on immune cell homeostasis. Curr. Opin. Gastroenterol. 2009;25:496–502. doi: 10.1097/MOG.0b013e328331b6b4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed R, Salmi A, Butler LD, Chiller JM, Oldstone MB. Selection of genetic variants of lymphocytic choriomeningitis virus in spleens of persistently infected mice. Role in suppression of cytotoxic T lymphocyte response and viral persistence. J. Exp. Med. 1984;160:521–540. doi: 10.1084/jem.160.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman JD, Moss PA, Goulder PJ, Barouch DH, McHeyzer-Williams MG, Bell JI, McMichael AJ, Davis MM. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274:94–96. [PubMed] [Google Scholar]
- Amaral FA, Sachs D, Costa VV, Fagundes CT, Cisalpino D, Cunha TM, Ferreira SH, Cunha FQ, Silva TA, Nicoli JR, et al. Commensal microbiota is fundamental for the development of inflammatory pain. Proc. Natl. Acad. Sci. USA. 2008;105:2193–2197. doi: 10.1073/pnas.0711891105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amit I, Garber M, Chevrier N, Leite AP, Donner Y, Eisenhaure T, Guttman M, Grenier JK, Li W, Zuk O, et al. Unbiased reconstruction of a mammalian transcriptional network mediating pathogen responses. Science. 2009;326:257–263. doi: 10.1126/science.1179050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artis D. Epithelial-cell recognition of commensal bacteria and maintenance of immune homeostasis in the gut. Nat. Rev. Immunol. 2008;8:411–420. doi: 10.1038/nri2316. [DOI] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton ES, White DW, Cathelyn JS, Brett-McClellan KA, Engle M, Diamond MS, Miller VL, Virgin HW., 4th Herpesvirus latency confers symbiotic protection from bacterial infection. Nature. 2007;447:326–329. doi: 10.1038/nature05762. [DOI] [PubMed] [Google Scholar]
- Benson A, Pifer R, Behrendt CL, Hooper LV, Yarovinsky F. Gut commensal bacteria direct a protective immune response against Toxoplasma gondii. Cell Host Microbe. 2009;6:187–196. doi: 10.1016/j.chom.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn SD, Shin H, Haining WN, Zou T, Workman CJ, Polley A, Betts MR, Freeman GJ, Vignali DA, Wherry EJ. Coregulation of CD8+ T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nat. Immunol. 2009;10:29–37. doi: 10.1038/ni.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier NM, Lowen AC. Animal Models for Influenza Virus Pathogenesis and Transmission. Viruses. 2010;2:1530–1563. doi: 10.3390/v20801530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart M, Hewson I, Felts B, Mahaffy JM, Nulton J, Salamon P, Rohwer F. Metagenomic analyses of an uncultured viral community from human feces. J. Bacteriol. 2003;185:6220–6223. doi: 10.1128/JB.185.20.6220-6223.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks DG, Trifilo MJ, Edelmann KH, Teyton L, McGavern DB, Oldstone MB. Interleukin-10 determines viral clearance or persistence in vivo. Nat. Med. 2006;12:1301–1309. doi: 10.1038/nm1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke TB, Davis KM, Lysenko ES, Zhou AY, Yu Y, Weiser JN. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat. Med. 2010;16:228–231. doi: 10.1038/nm.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuddapah S, Barski A, Zhao K. Epigenomics of T cell activation, differentiation, and memory. Curr. Opin. Immunol. 2010;22:341–347. doi: 10.1016/j.coi.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vrese M, Winkler P, Rautenberg P, Harder T, Noah C, Laue C, Ott S, Hampe J, Schreiber S, Heller K, Schrezenmeir J. Probiotic bacteria reduced duration and severity but not the incidence of common cold episodes in a double blind, randomized, controlled trial. Vaccine. 2006;24:6670–6674. doi: 10.1016/j.vaccine.2006.05.048. [DOI] [PubMed] [Google Scholar]
- Decman V, Laidlaw BJ, Dimenna LJ, Abdulla S, Mozdzanowska K, Erikson J, Ertl HC, Wherry EJ. Cell-intrinsic defects in the proliferative response of antiviral memory CD8 T cells in aged mice upon secondary infection. J. Immunol. 2010;184:5151–5159. doi: 10.4049/jimmunol.0902063. [DOI] [PubMed] [Google Scholar]
- Dolowy WC, Muldoon RL. Studies of Germfree Animals. I. Response of Mice to Infection with Influenza a Virus. Proc. Soc. Exp. Biol. Med. 1964;116:365–371. doi: 10.3181/00379727-116-29249. [DOI] [PubMed] [Google Scholar]
- Doyle S, Vaidya S, O’Connell R, Dadgostar H, Dempsey P, Wu T, Rao G, Sun R, Haberland M, Modlin R, Cheng G. IRF3 mediates a TLR3/TLR4-specific antiviral gene program. Immunity. 2002;17:251–263. doi: 10.1016/s1074-7613(02)00390-4. [DOI] [PubMed] [Google Scholar]
- Fang SB, Lee HC, Hu JJ, Hou SY, Liu HL, Fang HW. Dose-dependent effect of Lactobacillus rhamnosus on quantitative reduction of faecal rotavirus shedding in children. J. Trop. Pediatr. 2009;55:297–301. doi: 10.1093/tropej/fmp001. [DOI] [PubMed] [Google Scholar]
- Haining WN, Wherry EJ. Integrating genomic signatures for immunologic discovery. Immunity. 2010;32:152–161. doi: 10.1016/j.immuni.2010.02.001. [DOI] [PubMed] [Google Scholar]
- Hall JA, Bouladoux N, Sun CM, Wohlfert EA, Blank RB, Zhu Q, Grigg ME, Berzofsky JA, Belkaid Y. Commensal DNA limits regulatory T cell conversion and is a natural adjuvant of intestinal immune responses. Immunity. 2008;29:637–649. doi: 10.1016/j.immuni.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill DA, Artis D. Intestinal bacteria and the regulation of immune cell homeostasis. Annu. Rev. Immunol. 2010;28:623–667. doi: 10.1146/annurev-immunol-030409-101330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill DA, Siracusa MC, Abt MC, Kim BS, Kobuley D, Kubo M, Kambayashi T, Larosa DF, Renner ED, Orange JS, et al. Commensal bacteria-derived signals regulate basophil hematopoiesis and allergic inflammation. Nat. Med. 2012;18:538–546. doi: 10.1038/nm.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K, Littman DR. The Microbiome in Infectious Disease and Inflammation. Annu Rev Immunol. 2012;30:759–795. doi: 10.1146/annurev-immunol-020711-074937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichinohe T, Pang IK, Kumamoto Y, Peaper DR, Ho JH, Murray TS, Iwasaki A. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc. Natl. Acad. Sci. USA. 2011;108:5354–5359. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane M, Case LK, Kopaskie K, Kozlova A, MacDearmid C, Chervonsky AV, Golovkina TV. Successful transmission of a retrovirus depends on the commensal microbiota. Science. 2011;334:245–249. doi: 10.1126/science.1210718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegel MA, Sefik E, Hill JA, Wu HJ, Benoist C, Mathis D. Naturally transmitted segmented filamentous bacteria segregate with diabetes protection in nonobese diabetic mice. Proc. Natl. Acad. Sci. USA. 2011;108:11548–11553. doi: 10.1073/pnas.1108924108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krutzik PO, Hale MB, Nolan GP. Characterization of the murine immunological signaling network with phosphospecific flow cytometry. J. Immunol. 2005;175:2366–2373. doi: 10.4049/jimmunol.175.4.2366. [DOI] [PubMed] [Google Scholar]
- Kuss SK, Best GT, Etheredge CA, Pruijssers AJ, Frierson JM, Hooper LV, Dermody TS, Pfeiffer JK. Intestinal microbiota promote enteric virus replication and systemic pathogenesis. Science. 2011;334:249–252. doi: 10.1126/science.1211057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamkanfi M, Dixit VM. Modulation of inflammasome pathways by bacterial and viral pathogens. J. Immunol. 2011;187:597–602. doi: 10.4049/jimmunol.1100229. [DOI] [PubMed] [Google Scholar]
- Lawrence CW, Braciale TJ. Activation, differentiation, and migration of naive virus-specific CD8+ T cells during pulmonary influenza virus infection. J. Immunol. 2004;173:1209–1218. doi: 10.4049/jimmunol.173.2.1209. [DOI] [PubMed] [Google Scholar]
- Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc. Natl. Acad. Sci. USA. 2011;108(Suppl 1):4615–4622. doi: 10.1073/pnas.1000082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006a;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006b;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- Littman DR, Pamer EG. Role of the commensal microbiota in normal and pathogenic host immune responses. Cell Host Microbe. 2011;10:311–323. doi: 10.1016/j.chom.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macia J, Regot S, Peeters T, Conde N, Solé R, Posas F. Dynamic signaling in the Hog1 MAPK pathway relies on high basal signal transduction. Sci. Signal. 2009;2:ra13. doi: 10.1126/scisignal.2000056. [DOI] [PubMed] [Google Scholar]
- Macpherson AJ, Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science. 2004:1662–1665. doi: 10.1126/science.1091334. [DOI] [PubMed] [Google Scholar]
- Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L, Nalin R, Jarrin C, Chardon P, Marteau P, et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut. 2006;55:205–211. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- McKinstry KK, Strutt TM, Buck A, Curtis JD, Dibble JP, Huston G, Tighe M, Hamada H, Sell S, Dutton RW, Swain SL. IL-10 deficiency unleashes an influenza-specific Th17 response and enhances survival against high-dose challenge. J. Immunol. 2009;182:7353–7363. doi: 10.4049/jimmunol.0900657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore WE, Moore LH. Intestinal floras of populations that have a high risk of colon cancer. Appl. Environ. Microbiol. 1995;61:3202–3207. doi: 10.1128/aem.61.9.3202-3207.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penders J, Thijs C, van den Brandt PA, Kummeling I, Snijders B, Stelma F, Adams H, van Ree R, Stobberingh EE. Gut micro-biota composition and development of atopic manifestations in infancy: the KOALA Birth Cohort Study. Gut. 2007;56:661–667. doi: 10.1136/gut.2006.100164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartor RB. Therapeutic manipulation of the enteric microflora in inflammatory bowel diseases: antibiotics, probiotics, and prebiotics. Gastroenterology. 2004;126:1620–1633. doi: 10.1053/j.gastro.2004.03.024. [DOI] [PubMed] [Google Scholar]
- Schindler C, Shuai K, Prezioso VR, Darnell JE., Jr Interferon-dependent tyrosine phosphorylation of a latent cytoplasmic transcription factor. Science. 1992;257:809–813. doi: 10.1126/science.1496401. [DOI] [PubMed] [Google Scholar]
- Scupham AJ, Presley LL, Wei B, Bent E, Griffith N, McPherson M, Zhu F, Oluwadara O, Rao N, Braun J, Borneman J. Abundant and diverse fungal microbiota in the murine intestine. Appl. Environ. Microbiol. 2006;72:793–801. doi: 10.1128/AEM.72.1.793-801.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen GC. Viruses and interferons. Annu. Rev. Microbiol. 2001;55:255–281. doi: 10.1146/annurev.micro.55.1.255. [DOI] [PubMed] [Google Scholar]
- Shuai K, Schindler C, Prezioso VR, Darnell JE., Jr Activation of transcription by IFN-gamma: tyrosine phosphorylation of a 91-kD DNA binding protein. Science. 1992;258:1808–1812. doi: 10.1126/science.1281555. [DOI] [PubMed] [Google Scholar]
- Smith K, McCoy KD, Macpherson AJ. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin. Immunol. 2007;19:59–69. doi: 10.1016/j.smim.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Sonnenburg JL, Chen CT, Gordon JI. Genomic and metabolic studies of the impact of probiotics on a model gut symbiont and host. PLoS Biol. 2006;4:e413. doi: 10.1371/journal.pbio.0040413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer JC, Ganguly R, Waldman RH. Nonspecific protection of mice against influenza virus infection by local or systemic immunization with Bacille Calmette-Gueérin. J. Infect. Dis. 1977;136:171–175. doi: 10.1093/infdis/136.2.171. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Madan R, Karp CL, Braciale TJ. Effector T cells control lung inflammation during acute influenza virus infection by producing IL-10. Nat. Med. 2009;15:277–284. doi: 10.1038/nm.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szajewska H, Mrukowicz JZ. Use of probiotics in children with acute diarrhea. Paediatr. Drugs. 2005;7:111–122. doi: 10.2165/00148581-200507020-00004. [DOI] [PubMed] [Google Scholar]
- Takeda S, Rodewald HR, Arakawa H, Bluethmann H, Shimizu T. MHC class II molecules are not required for survival of newly generated CD4+ T cells, but affect their long-term life span. Immunity. 1996;5:217–228. doi: 10.1016/s1074-7613(00)80317-9. [DOI] [PubMed] [Google Scholar]
- Tanchot C, Lemonnier FA, Peérarnau B, Freitas AA, Rocha B. Differential requirements for survival and proliferation of CD8 naıïve or memory T cells. Science. 1997;276:2057–2062. doi: 10.1126/science.276.5321.2057. [DOI] [PubMed] [Google Scholar]
- Teixeira L, Ferreira A, Ashburner M. The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PLoS Biol. 2008;6:e2. doi: 10.1371/journal.pbio.1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherry EJ. T cell exhaustion. Nat. Immunol. 2011;12:492–499. doi: 10.1038/ni.2035. [DOI] [PubMed] [Google Scholar]
- Wherry EJ, Ha SJ, Kaech SM, Haining WN, Sarkar S, Kalia V, Subramaniam S, Blattman JN, Barber DL, Ahmed R. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity. 2007;27:670–684. doi: 10.1016/j.immuni.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, Littman DR, Benoist C, Mathis D. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.