Abstract
Background
Particular cytokine gene polymorphisms are involved in the regulation of the antibody production. The consequences of already described IL-4, IL-10 and IL-13 gene polymorphisms on biological parameters and antibody levels were investigated among 576 mothers at delivery and their newborns in the context of P. falciparum placental malaria infection.
Methods
The study took place in the semi-rural area of Tori-Bossito, in south-west Benin, where malaria is meso-endemic. Six biallelic polymorphisms were determined by quantitative PCR using TaqMan® Pre-Designed SNP Genotyping Assays, in IL-4 (rs2243250, rs2070874), IL-10 (rs1800896, rs1800871, rs1800872) and IL-13 (rs1800925) genes. Antibody responses directed to P. falciparum MSP-1, MSP-2, MSP-3, GLURP-R0, GLURP-R2 and AMA-1 recombinant proteins were determined by ELISA.
Results
The maternal IL-4−590*T/IL-4+33*T haplotype (one or two copies) was associated with favorable maternal condition at delivery (high haemoglobin levels, absence of placental parasites) and one of its component, the IL-4−590TT genotype, was related to low IgG levels to MSP-1, MSP-2/3D7 and MSP-2/FC27. Inversely, the maternal IL-10−1082AA was positively associated with P. falciparum placenta infection at delivery. As a consequence, the IL-10−819*T allele (in CT and TT genotypes) as well as the IL-10−1082*A/IL-10−819*T/IL-10−592*A haplotype (one or two copies) in which it is included, were related to an increased risk for anaemia in newborns. The maternal IL-10−1082AA genotype was related to high IgG levels to MSP-2/3D7 and AMA-1 in mothers and newborns, respectively. The IL-13 gene polymorphism was only involved in the newborn’s antibody response to AMA-1.
Conclusion
These data revealed that IL-4 and IL-10 maternal gene polymorphisms are likely to play a role in the regulation of biological parameters in pregnant women at delivery (anaemia, P. falciparum placenta infection) and in newborns (anaemia). Moreover, IL-4, IL-10 and IL-13 maternal gene polymorphisms were related to IgG responses to MSP-1, MSP-2/3D7 and MSP-2/FC27 in mothers as well as to AMA-1 in newborns.
Keywords: Malaria, P. falciparum, Cytokine gene polymorphisms, IL-4, IL-10, IL-13, Pregnancy, Cord blood, Recombinant proteins, Specific antibodies
Background
Host susceptibility to malaria is attributable to a number of factors, which include the genetic background of both host and pathogen. Indeed host genetic factors are involved in the regulation of the individual’s immunological competence. Several chromosomal regions containing genes coding for cytokines or cytokine receptors have been implicated in the control of P. falciparum infection levels [1-3]. The 5q31-q33 region located in chromosome 5 notably contains a number of genes initially found associated with the immune response directed to Schistosoma haematobium[4]. In the case of malaria, studies have provided evidence for linkage between the 5q31-q33 region and blood infection levels in populations originating from Burkina Faso [5], Cameroon [6] and Senegal [7]. Interleukin (IL) genes such as IL-3, IL-4, IL-5, IL-9 and IL-13 are notably clustered in this chromosome region [5,8].
The products of the pleiotropic IL-4 gene intervene in multiple immune modulating functions depending on a variety of cell types [9]. IL-4 is defined as a cytokine produced by Th2 cells, and is involved in the regulation of the humoral immune response. It is a key factor for the differentiation of precursor T helper cells into Th2 cells that induce IgE production by plasmocytes. This cytokine is an important regulator in the isotype switching from IgM/IgG to IgE [10]. Finally, IL-4 plays a critical role in the regulation of the antibody response induced by Plasmodium parasites [8,11]. A gene located in the 1q32.1 region of chromosome 1 encodes IL-10 which synergizes the production of antibody isotypes (IgG, IgA and IgM) induced by IL-4 [12]. IL-4 and IL-10 have been shown to be important for parasite clearance in later antibody-mediated phases of infection [13]. Among the genes located in the 5q31.1 region, IL-13 encodes a cytokine which is a central mediator of the physiological changes induced by allergic inflammation. The functions of IL-13 considerably overlap those of IL-4, especially with regard to their role on erythropoiesis. IL-13 has anti-inflammatory properties and induces IgE secretion from activated human B cells [14].
Several polymorphisms affecting IL-4, IL-10, IL-13 genes, lead to changes in cytokine production levels that may impact isotype switching as well as cell interaction and thus be associated with immune-related diseases such as malaria [15,16]. A variant at position IL-4−590C/T has been shown to enhance IL-4 and IgE production [17]. Studies in Burkina Faso [18] and Mali [19,20] revealed differences in the distribution of IL-4−590C/T allele and genotype frequencies between Fulani and non Fulani ethnic groups. In both studies, the IL-4−590*T allele was associated with high levels of anti-malaria IgG levels among Fulani. A study conducted among Ghanaian children showed that carriers of both IL-4−590*T and IL-4+33*T alleles presenting cerebral malaria had elevated IgE compared to non carriers [21].
There is evidence for associations between IL-10 gene polymorphisms grouped into haplotypes and IL-10 levels as well as antibody levels. The IL-10−1082G/A polymorphism has notably been associated with variable IL-10 production [22]. The IL-10−592*A allele has been linked to low levels of total IgE and the IL-10−1082*A/IL-10−592*A haplotype has been negatively associated with specific IgE and IgG4 [23]. IL-10−819*C was associated with skin lesions induced by leishmaniasis, in relation with high IL-10 levels produced by carriers of the IL-10−819CC genotype [24]. Besides, low IL-10 producing IL-10−1082*A/IL-10−819*T/IL-10−592*A haplotype carriers may be at risk of severe malarial anaemia, among pediatric populations [25]. Finally, the IL-13−1055*T allele is found in individuals who present enhanced IL-13 production and seem protected against severe malaria [16].
In endemic areas, children aged less than 5 years are particularly affected by malaria because of their lack of specific acquired immunity. Several authors have reported that the susceptibility of the child aged less than one year is linked to malaria infections during pregnancy. Thus children born from mothers with a P. falciparum infected placenta at delivery have an increased risk of developing malaria during their first years of life [26-28]. Despite this information, knowledge on the genetic and immunological mechanisms related to increased susceptibility of pregnant women and their children to malaria is still lacking. We hypothesized that if specific maternal cytokine gene polymorphisms lead to an increased production of specific antimalarial antibodies at the individual level, they may also help to lower the in utero sensitization of the fetus to plasmodial antigens, and therefore contribute to delay the appearance of the first malaria attacks during the first months of life.
Studies conducted in different malaria endemic areas have reported that IgG to various malaria antigens were associated with clinical protection [29-31]. Passive antibody transfer to non-immune individuals showed that protective immunity among adults in endemic areas is at least partly humoral [32]. Cytophilic antibodies to merozoite surface protein-3 (MSP-3) and glutamate-rich protein (GLURP) were shown to be predominant in protected individuals, contrary to non-cytophilic antibodies [33]. Studies have also shown that antibodies directed to MSP-1−19 and apical membrane antigen 1 (AMA-1) were associated with a reduced risk of clinical malaria [34]. Antibodies to AMA-1 have been reported to exhibit parasite growth inhibitory activity in a growth inhibition assay [35]. Antibodies to merozoite surface protein-2 (MSP-2) of P. falciparum have been associated with protection from clinical malaria in independent studies [36]. Each of these antigens (AMA-1, MSP-119, MSP-2, MSP-3, and GLURP) is considered as a candidate for inclusion into a multivalent malaria vaccine.
The aim of the present study was to verify the consequences of known cytokine gene polymorphisms on maternal biological parameters and specific antibody levels at delivery and to investigate their role on fetal health and immunity, taking into account the presence or absence of placental infection by P. falciparum. The simultaneous measurement of the antibody response directed against a panel of plasmodial antigens will allow drawing a picture of the specific immune status at the individual level.
Methods
Study area
The study was conducted in the rural area of Tori-Bossito, (45,000 inhabitants, 40 km west of Cotonou, Benin). Malaria transmission is perennial, with two seasonal peaks corresponding to rainy seasons, from April to July and September to November. P. falciparum is the main cause of infection and an average entomological inoculation rate (EIR) of 15.5 infective bites per person per year has been measured [37].
Study population and sampling
A cohort of 660 mothers and their children was constituted from June 2007 to June 2008 in three maternities from the Tori-Bossito area, welcoming parturient women from nine surrounding villages [37]. In the present study, 576 mothers and their newborns were investigated, excluding twins and children who died before D7. At delivery, peripheral blood was drawn from all mothers and cords in EDTA Vacutainer® tubes (Becton Dickinson, Meylan, France). Plasma cytokine levels were measured from blood collected for 80 mothers and cords in supplementary heparinized Vacutainer® tubes. After collection, plasma samples were stored at −80°C and buffy-coats at −20°C.
Thick blood smears were prepared from circulating blood of mothers, cord blood and placenta samples. Staining with Giemsa allowed determining P. falciparum parasitaemia by microscopy. Leukocytes and parasites were counted simultaneously until leukocyte or parasite numbers reached 500. A thick blood smear was declared negative if no parasite was found after 500 leucocytes had been counted. The following variables were measured for each mother: (1) village of residence; (2) age; (3) parity and (4) estimated exposure to malaria transmission during pregnancy assessed by means of mosquito catches precisely described in Cottrell et al. [38]; (5) circulating parasite density and (6) presence of placental parasites at delivery.
Biological assays were assessed in part in the laboratory established at Cotonou by the UMR 216 in partnership with the Faculté des Sciences de la Santé (FSS) and the Institut des Sciences Biomédicales Appliquées (ISBA) in Benin, and in the laboratory of the UMR 216 in Paris, France. For each included parturient woman, a written informed consent was obtained. Both the Ethical Committee of the FSS in Benin and the Consultative Committee on Professional Conduct and Ethics (CCDE) of the Institut de Recherche pour le Développement (IRD) approved the protocol.
Human genotyping
Genomic DNA was extracted from maternal buffy-coats containing mononuclear cells, using Qiamp® DNA blood midi kits according to the manufacturer’s instructions. Six cytokine gene polymorphisms were determined by quantitative PCR using TaqMan® Pre-Designed SNP (Single Nucleotide Polymorphism) Genotyping Assays, including the C/T mutation located at the −590 (rs2243250,) and +33 (rs2070874) positions of the IL-4 gene, the G/A mutation at the −1082 (rs1800896), the C/T mutation at the −819 (rs1800871) and the C/A mutation at the −592 (rs1800872) positions of the IL-10 gene, and the C/T mutation at the −1055 position (rs1800925) of the IL-13 gene. Each TaqMan genotyping assay contained two primers for amplifying the sequence of interest and two TaqMan® MGB probes for detecting alleles. The presence of two probe pairs in each reaction allowed genotyping of the two possible variant alleles at the SNP site in the DNA target sequence. The genotyping assay determined the presence or absence of a SNP based on the change in fluorescence of the dyes associated with the probes. Amplification of each locus was performed in a total volume of 10 μl, containing 0.25 μM of each primer, 2 μl genomic DNA and 2× PCR Master Mix (Applied Biosystems master mix contains AmpliTaq Gold® DNA Polymerase, Ultra Pure water, deoxyribonucleotide triphosphates (dNTPs), ROX™ Passive Reference and Buffer components) in a programmable thermocycler (Applied Biosystems PRISM 7900HT®).
Malaria antigens
Seven recombinant proteins corresponding to antigens of P. falciparum expressed on the surface of merozoites and candidates for inclusion in a multivalent asexual stage vaccine [39] were chosen. The recombinant proteins were obtained free of charge through an international network of collaborators. MSP-119 (Uganda-Palo-Alto strain) (gift from Pasteur Institute, Paris, France) was expressed in a Baculovirus/insect cell system [40]; AMA-1 25–545 (FVO strain) (gift from the Biomedical Primate Research Centre, Rijswijk, The Netherlands) was expressed in Pichia pastoris[41]; MSP-2/3D7 and MSP-2/FC27 (gift from La Trobe University, Melbourne, Australia) [42] as well as MSP-3 212–380 (F32 strain) [43], GLURP 25–514 (F32 strain) and GLURP 706–1178 (F32 strain) [44] (all from the Statens Serum Institute, Copenhagen, Denmark) were expressed in E. coli. GLURP-R0 (amino acids 25–514) and -R2 (amino acids 706–1178) corresponded to the N-terminal nonrepeat region and to the C-terminal repeat region of the protein, respectively [30].
Antibody measurements
An Enzyme-Linked Immuno-Sorbent Assay (ELISA) following a standardized methodology described in the Afro-immunoassay network standard operating procedure (procedures AIA-001-01 and −02) was used to assess the antibody response directed to the panel of P. falciparum recombinant proteins.
ELISA plates were coated overnight with 100 μl of recombinant protein solutions at a final concentration of 1 μg/ml in 1X PBS. Blocking buffer (3% milk powder in PBS - 0.1% Tween 20) was added (150 μl per well) and plates were kept at room temperature for 1 hour. Plasma samples were diluted in 1X PBS - 1% milk powder - 0.1% Tween 20-0.02% sodium azide. For IgG measurements, maternal and cord plasma samples were diluted 1:200 for all recombinant proteins except for AMA-1 where a dilution 1:2000 was used. For IgM measurements, plasma samples from maternal blood were diluted 1:200 and those of cord blood 1:50.
Two polyclonal antibodies conjugated to HRP were used: a goat anti-human IgG (Caltag H10007) and a goat anti-human IgM (Caltag H15007) both diluted 1: 3000. Bound enzyme was detected with TMB and the reaction was stopped with 0.2 M H2SO4 (100 μl/well). Plates were extensively washed between each incubation period with 1X PBS - 0.1% Tween 20-0.5 M NaCl. The optical density (OD) was read at 450 nm (reference filter 620 nm). Positive-control plasma samples from Gabonese individuals and negative-control plasma samples from Dutch individuals were included in each plate. A Microsoft Excel based curve fitting program (ADAMSEL FPL b039) containing a collection of macros and worksheets was used for calculating antibody concentrations (μg/ml) from the OD values, using the standard curves obtained from each ELISA test plate (http://www.emvda.bio.ed.ac.uk/software.php).
Cytokine assays
IL-4, IL-10 and IL-13 plasma levels were measured in a sub-sample of 80 mothers presenting no difference in any criterion compared to the whole group of 576 mothers. Four of them presented P. falciparum parasites both in circulating blood and placenta. The DuoSet® ELISA tests (R&D systems) were used and results were expressed in pg/ml by reference to standard curves prepared in each plate with recombinant cytokines. Thresholds of sensitivity were 31 pg/ml for IL-4 and IL-10, and 94 pg/ml for IL-13.
Statistical analysis
The significance of deviation from Hardy-Weinberg equilibrium was tested using the χ2 goodness-of-fit test. Strength of linkage disequilibrium (LD) between pairs of markers located along a same chromosome was measured as r2[45], using the Haploview software v4.1 [46]. Regions of strongly associated markers (LD blocks) were inferred using the confidence-interval method proposed by Gabriel et al. [47]. For SNPs located within a same LD block, haplotypes were inferred from the unphased genotype data using the Bayesian method implemented in PHASE v.2.1.1 [48], using default parameters. To avoid the convergence of the algorithm to a local maximum, we ran it 10 times with different random seeds and kept the output from the run with the best average value. The most probable haplotype constitution of each sample was also inferred with a maximum likelihood method using the Expectation-Maximization (EM) algorithm implemented in Arlequin v3.1 [49]. All samples had the same pair of haplotypes estimated by the EM and PHASE methods, therefore ensuring the reliability of the inference procedure.
For univariate analyses, differences in proportions were analyzed using the Chi square test. Differences in means were tested by the nonparametric Mann–Whitney U-test or Kruskal-Wallis test (for more than 2 groups to be compared). Statview 5.0 (SAS Institute Inc., Cary, NC) was used for these calculations. Results of the univariate analyses involving maternal cytokine gene polymorphisms are reported in the Additional files 1 and 2 (for biological parameters of either the mothers or the newborns) and in the Additional files 3 and 4 (for P. falciparum specific antibody levels of either the mothers or the newborns). Variables with P values < 0.20, were further considered in the multivariate analysis.
Multivariate analysis of the association between anaemia, placental infection and covariates was performed using multiple logistic regressions. For haemoglobin level, multivariate analyses were conducted using multiple linear regressions. Concerning antibody levels, due to non Gaussian distribution (even when data were log-transformed), median regression models were used. These models estimate standard errors via bootstrapping. We specified 1,000 replications for each analysis, and when significant (P < 0.05) a new analysis under the same model was carried out using 5,000 replications. Associations between biological and immunological phenotypes and each cytokine gene polymorphism were tested under additive, dominant and recessive genetic models.
The four tables reporting results of association, listed only cytokine gene polymorphisms for which multivariate analysis concluded to significant differences in relation to mothers and/or newborns biological and/or immunological parameters. For all tests, P values of less than 0.05 were considered significant. All analyses were performed with Stata software, version 11.0 (StatCorp LP, College Station, TX, USA).
Results
Study population, clinical and parasitological characteristics
Haematological and parasitological data of the 576 included mothers and their newborns are presented in Table 1. Mothers distributed into 88 primigravidae (15%, mean age = 25.0 years) and 488 multigravidae (85%, mean age = 27.8 years). Primigravidae were more numerous than multigravidae to present circulating parasites at delivery (15 (17.0%) vs. 34 (7.0%), P = 0.002) but parasite densities did not differ (median parasite density/μl (IQR): 27 (5–514) vs. 5 (2–77), P = 0.08). Among the 563 mothers for whom placenta blood smears were available, 63 (10.9%) mothers presented a P. falciparum infected placenta at delivery. More positive smears were identified in primigravidae compared to multigravidae (17 (19.8%) vs. 46 (9.6%), P = 0.006). Anaemia was frequent, with 39.1% and 61.5% of mothers and newborns presenting haemoglobin values < 11 g/dl and < 15 g/dl, respectively. The mean birth weight (± SD) was 2985 g (± 388 g) with 53 newborns (9.2%) presenting a low birth weight (< 2500 g). The 5 newborns presenting parasites in cord blood were born of mothers with both circulating P. falciparum at delivery and presence of placental malaria infection.
Table 1.
Haematological and parasitological data among 576 mothers and their newborns
| Parameters | Mothers (peripheral blood) | Newborns (cord blood) |
|---|---|---|
| Haemoglobin (g/dl) |
11.3 (± 1.5) |
14.5 (± 2.0) |
| mean (± SD) a | ||
| Anaemia |
224 (39.1) |
351 (61.5) |
| n (%) b | ||
|
P. falciparum prevalence |
49 (8.5) |
5 (0.9) |
| n (%) c | ||
| Parasite density d | 556 (159–9990) | 367 (235–1492) |
a 3 (5) missing values for mothers (newborns).
b anaemia defined as Hb < 11 g/dl (mothers) and Hb < 15 g/dl (newborns).
c 2 missing values.
d median parasite density (/μl) (25th-75th percentiles), zero values excluded.
Maternal IL-4, IL-10 and IL-13 gene polymorphisms
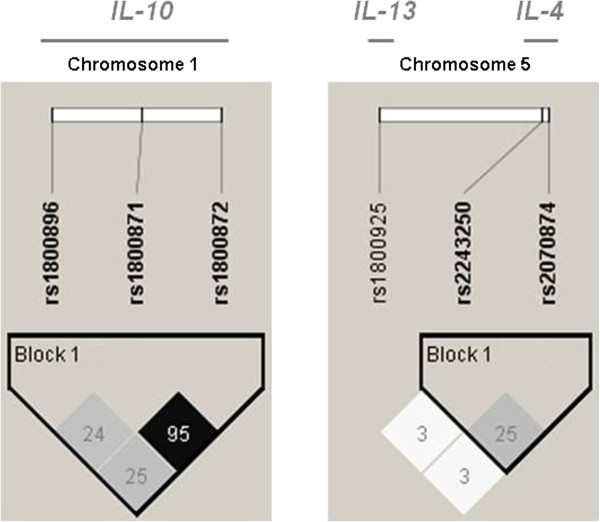
Cytokine gene polymorphisms were determined for the whole group of 576 mothers. No statistically significant deviation from Hardy-Weinberg equilibrium was observed for any polymorphism (all P-values > 0.44, Table 2). The heterozygous carriage was predominant (at least 50%), except for IL-4−590 and IL-10−1082. Since the IL-13−1055 SNP presented virtually no LD with the two IL-4 SNPs in position −590 and +33 along the chromosome 5 (r2 = 0.03 with both SNPs, Figure 1), only haplotypes including the two IL-4 SNPs were reconstructed and the IL-13 SNP was treated independently in statistical analyses. By contrast, the three IL-10 SNPs located on chromosome 1 belong to a single block of LD (Figure 1) and were thus combined into haplotypes. On the basis of literature data, the particular IL-4−590*T/IL-4+33*T and IL-10−1082*A/IL-10−819*T/IL-10−592*A haplotypes were selected, in order to examine their relationships with biological and immunological parameters. This allowed to consider three groups of mothers bearing either two copies (n = 140), one copy (n = 291) or no copy (n = 145) of IL-4−590*T/IL-4+33*T haplotype (referred to as IL4-TT haplotype). Similarly, three groups of mothers were defined, who carried either two copies (n = 87), one copy (n = 293) or no copy (n = 196) of IL-10−1082*A/IL-10−819*T/IL-10−592*A haplotype (referred to as IL10-ATA haplotype).
Table 2.
Distribution of cytokine gene polymorphisms in the study population of 576 mothers
| Gene | SNPs | Location in the gene | Genotype | Frequency in the study population | Hardy-Weinberg equilibrium test χ2(P-value, df = 2) |
|---|---|---|---|---|---|
|
IL-4 |
rs2243250 |
−590 C/T |
CC |
25 (4%) |
χ 2 = 0.03 (P = 0.98) |
| |
|
|
CT |
193 (34%) |
|
| |
|
|
TT |
358 (62%) |
|
| |
rs2070874 |
+33 C/T |
CC |
145 (25%) |
χ 2 = 0.08 (P = 0.96) |
| |
|
|
CT |
290 (50%) |
|
| |
|
|
TT |
141 (25%) |
|
|
IL-10 |
rs1800896 |
−1082 G/A |
GG |
47 (8%) |
χ 2 = 0.63 (P = 0.73) |
| |
|
|
GA |
223 (39%) |
|
| |
|
|
AA |
306 (53%) |
|
| |
rs1800871 a |
−819 C/T |
CC |
184 (33%) |
χ 2 = 1.09 (P = 0.58) |
| |
|
|
CT |
281 (51%) |
|
| |
|
|
TT |
90 (16%) |
|
| |
rs1800872 |
−592 C/A |
CC |
191 (33%) |
χ 2 = 1.64 (P = 0.44) |
| |
|
|
CA |
294 (51%) |
|
| |
|
|
AA |
91 (16%) |
|
|
IL-13 |
rs1800925 |
−1055 C/T |
CC |
186 (32%) |
χ 2 = 1.48 (P = 0.48) |
| |
|
|
CT |
295 (51%) |
|
| TT | 95 (17%) |
a 21 missing values.
Figure 1.
Patterns of linkage disequilibrium between IL-10 SNPs and between IL-4 and IL-13 SNPs. High pairwise LD (r2) between markers is illustrated with dark shading. The r2 values (× 100) for the marker pairs are listed in the corresponding boxes.
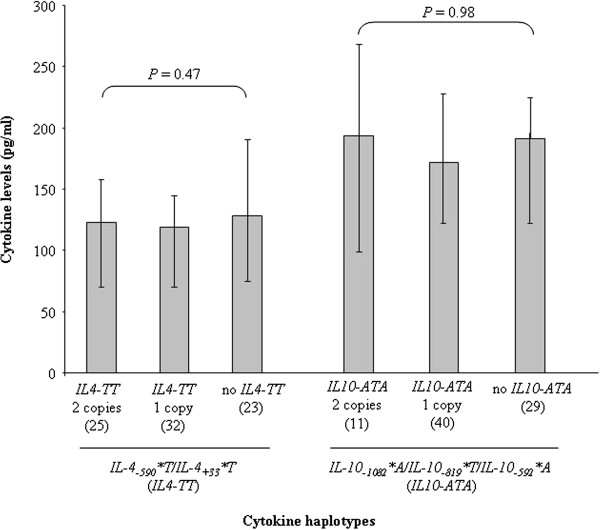
Genotype-dependent plasma levels of cytokines are presented in Figure 2 in a sub-sample of 80 mothers, where it appears that similar IL-4, IL-10 or IL-13 levels were secreted in the plasma of mothers harboring each of the three genotypes of the corresponding cytokine gene polymorphisms, except for IL-10−1082, for which mothers with the GA genotype presented lower IL-10 plasma levels than their counterparts with the GG genotype (P = 0.006). But this difference did not impact the cytokine levels among the different IL-4 and IL-10 haplotype groups, as illustrated in Figure 3.
Figure 2.
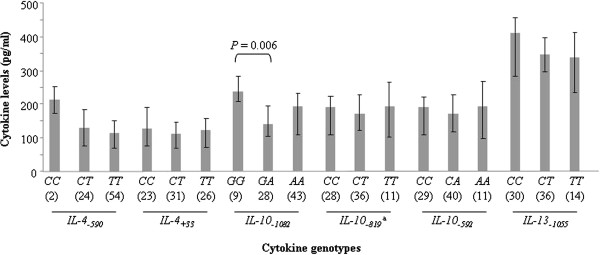
Plasma levels of cytokines according to cytokine genotypes, in 80 mothers at delivery.a 5 missing values. Boxes delimit median values and bars denote the 25th and 75th percentiles. Differences in cytokine levels between genotypes were examined for each cytokine variant with the Mann–Whitney U test.
Figure 3.
Plasma levels of cytokines according to cytokine haplotypes, in 80 mothers at delivery. Boxes delimit median values and bars denote the 25th and 75th percentiles. Differences in cytokine levels between haplotypes were examined with the Kruskal-Wallis test.
Association between maternal cytokine gene polymorphisms and biological data of mothers
Maternal haemoglobin levels at delivery were impacted by IL-4 gene polymorphisms (Table 3, on the basis of the results of the univariate analysis presented in the Additional file 1). Indeed, presence of the IL-4+33*T allele in genotypes CT and TT was associated with higher haemoglobin levels than those found in mothers carrying the CC genotype (P = 0.03). This observation was reinforced when examining the association of the IL4-TT haplotype under a dominant effect (P = 0.03). The same associations remain valid when considering maternal anaemia, defined by an Hb level < 11 g/dl, where it appeared that IL-4+33CT and TT carriers as well as carriers of one or two copies of IL4-TT haplotype were less at risk of anaemia than IL-4+33CC and non IL4-TT carriers, respectively (both analyses, OR = 0.66, P = 0.04).
Table 3.
Association between maternal cytokine gene polymorphisms and mothers’ biological parameters: multivariate analysis
| Maternal cytokine gene (n) | Haemoglobin (g/dl) (SD)a | Pb | Anaemia OR [CI95]c | Pb | P. falciparumplacenta infection OR [CI95]d | Pe |
|---|---|---|---|---|---|---|
|
IL-4−590 genotypes: |
|
|
|
|
|
|
| CC (25) |
11.2 (0.3) |
|
1 |
|
|
|
| CT (193) |
11.1 (0.3) |
0.88 |
1.13 [0.48-2.65] |
0.77 |
|
|
| TT (358) |
11.3 (0.3) |
0.65 |
0.87 [0.38-1.98] |
0.73 |
OR CT and TT vs. CC = 0.36 [0.14-0.93] |
0.03 |
|
IL-4+33 genotypes: |
|
|
|
|
|
|
| CC (145) |
11.3 (0.2) |
|
|
|
|
|
| CT and TT (431) |
11.6 (0.1) |
0.03 |
OR CT and TT vs. CC = 0.66 [0.44-0.97] |
0.04 |
OR CT and TT vs. CC = 0.54 [0.31-0.94] |
0.03 |
|
IL-4−590/IL-4+33 haplotypes: |
|
|
|
|
|
|
| No IL4-TT (145) |
11.3 (0.2) |
|
|
|
|
|
| 1 or 2 copies IL4-TT (431) |
11.6 (0.1) |
0.03 |
OR 1 or 2 copies IL4-TT vs. no IL4-TT = 0.66 [0.44-0.97] |
0.04 |
OR 1 or 2 copies IL4-TT vs. no IL4-TT = 0.54 [0.31-0.94] |
0.03 |
|
IL-10−1082 genotypes: |
|
|
|
|
|
|
| GG (47) |
11.4 (0.2) |
|
1 |
|
|
|
| GA (223) |
11.3 (0.2) |
0.53 |
1.01 [0.52-1.94] |
0.98 |
|
|
| AA (306) | 11.3 (0.2) | 0.59 | 1.23 [0.65-2.34] | 0.52 | OR AA vs GG and GA = 1.95 [1.11-3.42] | 0.02 |
a 3 missing values.
b analysis adjusted on village.
c anaemia defined as Hb < 11 g/dl.
d 13 missing values.
e analysis adjusted on parity and village.
P values < 0.05 are in bold.
Village localization, young mother’s age, primigravidity and IL-4 gene polymorphisms were associated with placental malaria infection under a dominant model (Table 3). Indeed, mothers harboring either the IL-4−590*T allele in genotypes CT and TT or the IL-4+33*T allele in genotypes CT and TT were less numerous to present placental malaria infection at delivery than mothers with CC genotype (OR = 0.36, P = 0.03 and OR = 0.54, P = 0.03, respectively). The same pattern was observed for the IL4-TT haplotype: a lesser prevalence of placental malaria infection was confirmed in mothers carrying one or two copies of the IL4-TT haplotype when compared to non IL4-TT haplotype carriers (OR = 0.54, P = 0.03).
Village localization, primigravidity and IL-10−1082 polymorphism were associated with placental malaria infection under a recessive model, with more IL-10−1082AA mothers presenting placental malaria infection at delivery in comparison to IL-10−1082GG and GA mothers (OR = 1.95 P = 0.02) (Table 3). Due to the strong correlation between circulating parasite densities and prevalence of placental infection at delivery, maternal age, village, primigravidity and the same IL-10−1082AA genotype were positively associated under a recessive model with the level of circulating parasite densities (OR [CI95] = 2.34 [1.23-4.48], P = 0.01).
Association between maternal cytokine gene polymorphisms and biological data of newborns
Newborn’s anaemia, defined by an Hb level < 15 g/dl, but not cord haemoglobin levels considered as a continuous variable, was impacted by IL-10 gene polymorphisms (Table 4, on the basis of the results of the univariate analysis presented in the Additional file 2). Indeed, maternal anaemia, young maternal age and IL-10−819 polymorphism were associated with newborn’s anaemia under a dominant model: the presence of the IL-10−819*T allele in the CT and TT genotypes was related to a higher risk of newborn’s anaemia compared to its absence in CC carriers (OR = 1.46, P = 0.04). Combined to a borderline effect towards a higher risk of anaemia in infants born to IL-10−1082GA and AA mothers compared to GG ones (OR = 1.81, P = 0.06), this effect persisted in infants issued from mothers carrying one or two copies of IL10-ATA haplotype compared to mothers without the IL10-ATA haplotype (OR = 1.45, P = 0.04).
Table 4.
Association between maternal cytokine gene polymorphisms and newborns’ biological parameters: multivariate analysis
| Maternal cytokine gene (n) | Haemoglobin (g/dl) (SD)a | Pb | Anaemia OR [CI95]c | Pd |
|---|---|---|---|---|
|
IL-10−1082 genotypes: |
|
|
|
|
| GG (47) |
15.1 (0.3) |
|
|
|
| GA and AA (529) |
14.6 (0.3) |
0.07 |
OR GA and AA vs GG = 1.81 [0.98-3.32] |
0.06 |
|
IL-10−819 genotypes: |
|
|
|
|
| CC (191) |
14.8 (0.2) |
|
|
|
| CT and TT (385) |
14.6 (0.2) |
0.14 |
OR CT and TT vs CC = 1.46 [1.02-2.10] |
0.04 |
|
IL-10−592 genotypes: |
|
|
|
|
| CC (191) |
14.7 (0.1) |
|
|
|
| CA and AA (385) |
14.4 (0.2) |
0.13 |
OR CA and AA vs CC = 1.36 [0.95-1.95] |
0.09 |
|
IL-10−1082/IL-10−819/IL-10−592 haplotypes: |
|
|
|
|
| No IL10-ATA (196) |
14.8 (0.2) |
|
|
|
| 1 or 2 copies IL10-ATA (380) | 14.6 (0.2) | 0.13 | OR 1 or 2 copies IL10-ATA vs. no IL10-ATA = 1.45 [1.02-2.08] | 0.04 |
a 5 missing values.
b analysis adjusted on maternal anaemia (Hb < 11 g/dl) and low birthweight (< 2500 g).
c newborn’s anaemia defined as Hb < 15 g/dl.
d analysis adjusted on maternal anaemia and age.
P values < 0.05 are in bold.
Birthweight, either considered as a continuous variable or dichotomized into less or more than 2500 g, was not influenced by the maternal cytokine gene polymorphisms under study.
Association between maternal cytokine gene polymorphisms and antibody responses of mothers
IgG levels to the seven P. falciparum recombinant proteins under investigation are illustrated in Figure 4, as well as IgM to AMA-1. In all cases, specific antibody levels were higher in the circulating blood of mothers at delivery than in the cord blood of their newborns (all P < 0.02). The AMA-1 recombinant protein elicited the highest antibody levels in both mother and child groups.
Figure 4.
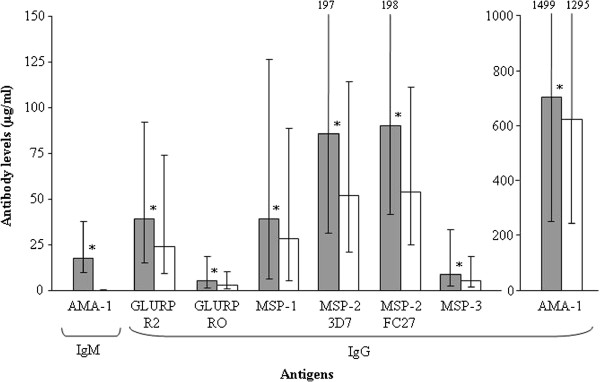
Antibody levels to P. falciparum recombinant proteins in 576 mothers at delivery and their newborns. Boxes delimit median values and bars denote the 25th and 75th percentiles. Asterisks (*) indicate that mothers have higher antibody levels than their newborns (Mann Whitney U-test, all P < 0.02) Symbols used for bars: mothers (grey) and infants (white).
MSP-1, MSP-2/3D7 and MSP-2/FC27 were the only antigens for which specific antibody responses of mothers were clearly associated with cytokine gene polymorphisms (Table 5 on the basis of the results of the univariate analysis presented in the Additional file 3). More precisely, IL-4−590TT carriers presented lower IgG levels to MSP-1 (P = 0.02) and MSP-2/3D7 (P = 0.03) than mothers with CC and CT genotypes. The same IL-4−590 polymorphism was related to the antibody response to MSP-2/FC27 under an additive model, with decreasing IgG levels from mothers bearing CC to CT and TT genotypes (P = 0.02). Finally, IgG levels to MSP-2/3D7 were higher for mothers with the IL-10−1082AA genotype than for those with GG and GA genotypes (P = 0.04).
Table 5.
Association between maternal cytokine gene polymorphisms and antibody levels to P. falciparum antigens in mothers: multivariate analysis
| Antibody response | Maternal cytokine gene (n) | Antibody levels (μg/ml)a(SD) | P |
|---|---|---|---|
| IgM to AMA-1 |
|
|
|
| |
IL-10−1082 genotypes: |
|
|
| |
GG (47) |
13.9 (3.3) |
|
| |
GA (223) |
17.0 (3.5) |
0.08 b |
| |
AA (306) |
20.0 (3.7) |
|
| IgG to MSP-1 |
|
|
|
| |
IL-4−590 genotypes: |
|
|
| |
CC and CT (218) |
49.0 (5.5) |
|
| |
TT (358) |
32.8 (7.0) |
0.02c |
| IgG to MSP-2/3D7 |
|
|
|
| |
IL-4−590 genotypes: |
|
|
| |
CC and CT (218) |
95.5 (10.3) |
|
| |
TT (358) |
70.3 (11.9) |
0.03d |
| |
IL-10−1082 genotypes: |
|
|
| |
GG and GA (270) |
68.2 (6.2) |
|
| |
AA (306) |
92.9 (12.2) |
0.04d |
| IgG to MSP-2/FC27 |
IL-4−590 genotypes: |
|
|
| |
CC (25) |
147.7 (61.7) |
|
| |
CT (193) |
98.0 (62.7) |
|
| |
TT (358) |
85.4 (62.8) |
0.02b |
| IgG to GLURP-R2 e |
|
|
|
| |
IL-4−590 genotypes: |
|
|
| |
CC (25) |
23.2 (6.9) |
|
| |
CT and TT (551) |
38.3 (7.6) |
0.05 d |
| |
IL-10−1082/IL-10−819/IL-10−592 haplotypes: |
|
|
| |
No or 1 copy IL10-ATA (489) |
38.6 (3.0) |
|
| 2 copies IL10-ATA (87) | 24.6 (7.6) | 0.07 d |
a median value.
b analysis adjusted on village.
c analysis adjusted on presence of P. falciparum circulating parasites in mothers and presence of placental parasites.
d analysis adjusted on presence of placental parasites.
e 5 missing values.
P values < 0.05 are in bold.
Association between maternal cytokine gene polymorphisms and antibody responses of newborns
Only cord blood IgG levels directed to AMA-1 were impacted by maternal cytokine gene polymorphisms (Table 6, on the basis of the results of the univariate analysis presented in the Additional file 4). Namely, an association involved IgG levels to AMA-1 and IL-10−1082 under a recessive effect, with higher IgG levels in newborns from IL-10−1082AA vs. GG and GA mothers (P = 0.04). In contrast, maternal IL-13−1055 was associated with lower IgG levels to AMA-1 under a recessive effect, concerning newborns from IL-13−1055TT vs. CC and CT mothers (P = 0.03).
Table 6.
Association between maternal cytokine gene polymorphisms and antibody levels to P. falciparum antigens in newborns: multivariate analysis
| Antibody response | Maternal cytokine gene (n) | Antibody levels (μg/ml)a(SD) | P |
|---|---|---|---|
| IgG to AMA-1 |
|
|
|
| |
IL-10−1082 genotypes: |
|
|
| |
GG and GA (270) |
1497.3 (214.0) |
|
| |
AA (306) |
1638.9 (68.3) |
0.04b |
| |
IL-13−1055 genotypes: |
|
|
| |
CC and CT (481) |
1596.0 (230.8) |
|
| |
TT (95) |
1444.0 (69.9) |
0.03b |
| IgG to MSP-2/3D7 |
|
|
|
| |
IL-10−1082 genotypes: |
|
|
| |
GG and GA (270) |
46.0 (4.1) |
|
| |
AA (306) |
57.6 (6.5) |
0.08 c |
| IgG to GLURP-R0 |
|
|
|
| |
IL-10−592 genotypes: |
|
|
| |
CC and CA (485) |
3.2 (0.3) |
|
| |
AA (91) |
2.2 (0.5) |
0.06 c |
| |
IL-10−1082/IL-10−819/IL-10−592 haplotypes: |
|
|
| |
No or 1 copy IL10-ATA (489) |
3.2 (0.3) |
|
| 2 copies IL10-ATA (87) | 2.2 (0.5) | 0.05 c |
a median value.
b analysis adjusted on maternal age, parity and estimated exposure to malaria transmission during pregnancy.
c analysis adjusted on presence of P. falciparum circulating parasites in mothers.
P values < 0.05 are in bold.
Discussion
The present study aimed at exploring the impact of maternal cytokine gene polymorphisms on biological parameters and antimalarial antibody responses of mothers at delivery and their newborns. Cytokines involved in antibody production such as IL-4, IL-10 and IL-13 were investigated. A clear impact of IL-4 gene polymorphisms on biological parameters of mothers at delivery was revealed. Indeed, the IL4-TT haplotype (one or two copies) was found negatively associated with P. falciparum placenta infection and maternal anaemia. The IL-4−590TT genotype was related to lower maternal IgG levels to MSP-1, MSP-2/3D7 and MSP-2/FC27 than the CC and CT genotypes. The maternal IL-4 gene polymorphism did impact neither biological parameters nor antibody responses of the newborns. IL-10−1082AA genotypes were positively associated with both P. falciparum placenta infection and circulating parasites in mothers. Similarly, IL-10−819CT and TT genotypes as well as the IL10-ATA haplotype (one or two copies) were positively related to anaemia in newborns. Concerning antibody responses, IL-10−1082AA genotypes were associated with higher IgG levels to MSP-2/3D7 and AMA-1 in mothers and in newborns, respectively. The only association involving IL-13−1055 concerned the IgG response to AMA-1 in cord blood, with lower levels in newborns from mothers with the TT vs. CC and CT genotypes. IL-4 and IL-10 haplotypes were not related to plasma cytokine levels, probably due to the small sample size of the sub-population in which cytokine levels were measured.
Around 40% of the mothers and more than 60% of the newborns presented anaemia. The relationships between anaemia during pregnancy and birth outcomes were studied in depth in this cohort by Koura et al., who noticed that these prevalence rates were in agreement with those already recorded in Benin, and observed that newborn’s anaemia was related to maternal anaemia [50].
Our finding of a lower prevalence of placental malaria infection in mothers with either the IL-4−590 or IL-4+33CT and TT genotypes, or still the IL4-TT haplotype, is in accordance with recent results indicating reduced malaria risk in IL4-TT carriers from Indian tribal populations [51]. On the contrary, the IL-4−590*T allele was related to high prevalence of P. falciparum infection in asymptomatic Fulani of Mali, but not in a sympatric group of Dogon [19]. The lesser prevalence of placental parasites in mothers carrying one or two copies of the IL4-TT haplotype was consistent with the higher haemoglobin values (and decreased risk of anaemia) observed in these mothers at delivery, probably due to a lesser impact of malaria-induced anaemia. A complementary explanation would be that IL-4 is known for its role in the promotion of erythropoiesis [52]. It would be coherent that these favorable maternal condition should be accompanied by strong antimalarial antibody responses, as it has been already demonstrated for specific IgG in Fulani individuals with the IL-4−590*T allele [18]. This would be in agreement with a previous observation made in malaria-infected individuals, of an association between the activation of IL-4 producing T-cell subsets and the production of antimalarial specific antibodies [11]. Those antibodies would reflect an effective response to infecting parasites [53], allowing to combat infection and therefore to prevent its evolution toward high parasite densities and sequestration into the placenta. Nonetheless, in the present study, the IL-4−590TT genotype was uniformly associated negatively with maternal IgG levels to MSP-1, MSP-2/3D7 and MSP-2/FC27. The most obvious hypothesis for this observation rests on a decreased induction of antimalarial antibodies in IL-4−590TT mothers as a result of a lighter weight of P. falciparum placenta infection in these mothers. It should be reminded that the Beninese women under study cumulated previous malaria experience, and/or that co-infection with other parasitic or bacterial pathogens not investigated in the present study may have occurred, thereby preventing a clear relationship. Some regulatory factors placed under genetic control and in LD with IL-4 gene polymorphisms may also impact the data [54]. In fact, studies on the relationships between IL-4 gene polymorphisms and IgG levels are limited and conflicting [18,55]. The Th2 cytokine induces Ig class switching from IgM/IgG to IgE [9,10]. IL-4 has been incriminated in the aggravation of cerebral malaria due to its role in an increase in parasite mass [56], in an infiltration by monocytes, basophils, and eosinophils and an increase in parasite sequestration [57]. In the context of our study, independently of any morbidity linked to malaria, no biological or immunological advantage could be deduced for the newborns from the IL-4 genetic characteristics of their respective mothers. Comparatively to IgG which are mainly of maternal origin in the cord blood due to the existence of a passive transfer of antibodies during the pregnancy, IgM do not cross the placenta and are therefore synthesized by the fetus, as a result of a sensitization to malaria antigens in utero[58,59]. Nevertheless, the low levels and prevalence rates of newborns’ IgM to AMA-1 hampered any observation in link with maternal cytokine gene polymorphisms.
As for IL-4, data from the literature indicate that IL-10 gene polymorphisms need to be analyzed combined into haplotypes and no longer at the limiting scale of single nucleotide polymorphisms. We thus studied the effects of the candidate IL-10−1082G/A, IL-10−819C/T and IL-10−592C/A polymorphisms simultaneously, through a haplotype analysis, in relation to the biological and immunological data recorded in both the mothers and their newborns. As for IL-4, were only considered the final results issued from a multivariate analysis taking into account specific maternal parameters such as age, parity, residence place, the estimated entomological inoculation rate during the whole pregnancy period and/or the presence of circulating parasites as well as placental parasites at delivery. Special attention was paid to the IL-10−1082*A/IL-10−819*T/IL-10−592*A (IL10-ATA) haplotype carriers, assimilated to low IL-10 producers [60,61]. It emerges from this study that infants born from IL10-ATA (and IL-10−819CT and TT) mothers inherit poorer physiological conditions than others, with a greater risk of anaemia, possibly in relation to the greater risk of placenta malarial infection as well as presence of circulating parasites observed in IL-10−1082AA mothers.
IL-10 is an important immune modulating factor with anti-inflammatory activities that lead to inhibition of the activation of antigen-presenting cells, resulting in their reduced ability to induce T cell responses. But IL-10 has also some pro-inflammatory activity since it promotes activation and differentiation of B cells and induces immunoglobulin synthesis [62]. It is therefore expected that low IL-10 producers such as the IL10-ATA carriers present lower antibody levels compared to no IL10-ATA carriers. For example, a previous study showed that the IL-10−1082*A/IL-10−592*A haplotype was associated with decreased specific IgE and IgG4 levels [23]. But conversely to this expectation, IL-10−1082AA mothers of the present study had higher IgG to MSP-2/3D7 than GG and GA ones. This pattern of response found an immunological echo in the newborns, as those born from mothers with the IL-10−1082AA genotype presented higher IgG levels to AMA-1 compared to newborns from GG and GA mothers. Two hypotheses can be put forward to explain this observation. Firstly, the lower degree of control of P. falciparum infection in IL-10−1082AA mothers may result in a strongest antibody response in both mothers and their newborns (via the in utero transfer of maternal IgG and/or the production of intrinsic IgG in response to parasitic soluble antigens encountered in utero). Secondly, there is no dogma regarding a decreased IL-10 production in individuals with IL10-ATA haplotype, as the combination of a particular IL-1 genotype with the IL10-ATA haplotype was shown to result in increased IL-10 plasma levels in healthy Finnish individuals, demonstrating the importance of the individual genetic background [63].
Turner et al. demonstrated a difference in IL-10 secretion in association with the presence or absence of of the IL-10−1082*A allele in the human IL-10 promoter, after stimulation of peripheral blood mononuclear cells [22]. This is in accordance with our observation of lower IL-10 plasma levels in mothers with the IL-10−1082GA genotype compared to those with the GG genotype. Nevertheless, no difference in the plasma IL-10 levels according to IL-10 haplotypes was recorded in the sub-sample of 80 mothers for whom cytokine levels were measured. It should be noted that individual differences in the levels of the IL-10 measured at a specific moment may not only result from host genetic factors predisposing to high or low production, but also for a great part from the physiological condition at that time, as well as from global immunity.
In Thailand, the functional IL-13−1055*T allele has been shown to enhance resistance to severe malaria through the alteration of IL-13 production [16]. The same allele has been associated with resistance to S. mansoni[64,65] and inversely with susceptibility to S. haematobium infection [66]. Isnard et al. established that the IL-13−1055C/T polymorphism is in LD with other polymorphisms [66], suggesting that only weak effects may be revealed when investigating the single polymorphism in relation to clinical or immunological phenotypes. This may explain why IL-13−1055C/T alleles were not found associated with any parameter in the present study, except lower IgG levels to AMA-1 in newborns from IL-13−1055TT vs. CC and TT mothers. It would be unwise to comment this observation, as it did not present any biological corollary (either in mothers or newborns) in the present study.
Besides the main results, it is interesting to note that the distribution of the cytokine genotypes in our study population from south Benin (most participants belonging to the Tori ethnic group) is similar to that observed in the Yoruba (YRI) sample from the 1000 Genomes project [67], located in the neighboring country (Nigeria), and the opposite of the one observed in the European (CEU) sample of the same database (data not shown). This distribution profile may suggest a selective genetic advantage conferred by the secular pressure exerted by infectious diseases such as malaria [68].
Conclusion
This study shows that IL-4 and IL-10 gene polymorphisms are likely to play a role in the regulation of mothers’ and newborns’ control of malaria infection. This is the first demonstration of an impact of maternal IL-10 gene polymorphisms on the newborn specific IgG production, opening hypotheses about the consequences that this may have later in the life of the child for the elaboration of his own IgG responses. These results may provide a contribution to a better understanding of the immunopathogenic mechanisms underlying maternal-fetal interactions. Further studies are required to strengthen the knowledge of the impact of maternal gene polymorphisms on the immunoglobulin isotypes synthesized by the fetus and thereafter the infant.
Abbreviations
AMA: Apical membrane antigen; ELISA: Enzyme-linked immunosorbent assay; GLURP: Glutamate rich protein; Ig: Immunoglobulin; IL: Interleukin; LD: Linkage disequilibrium; MSP: Merozoite surface protein; OR: Odds ratio; PCR: Polymerase chain reaction; SNP: Single nucleotide polymorphism.
Competing interests
The authors declare no conflict of interest.
Authors’ contributions
LAG, GA and MNF designed the study. LAG, GA, MNF, DC, CD, LPA, AC, FB and MA recruited the children and the adults and conducted the field work. LR, NJ, BD and AC performed microscopic reading while LAG, DC, BA, CD, LR and MNF realized the laboratory assays. LAG, GA, SA and MNF analyzed the data and drafted the manuscript. The final manuscript was read and approved by all authors.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
Association between maternal cytokine gene polymorphisms and mothers’ biological parameters (n = 576): univariate analysis. This table summarizes the results of the univariate analysis performed for examining differences between maternal cytokine genotypes or haplotypes and biological parameters (haemoglobin, prevalence of circulating P. falciparum parasites, prevalence of P. falciparum placenta infection) of mothers.
Association between maternal cytokine gene polymorphisms and newborns’ biological parameters (n = 576): univariate analysis. This table summarizes the results of the univariate analysis performed for examining differences between maternal cytokine genotypes or haplotypes and biological parameters (birthweight, haemoglobin) of newborns.
Association between maternal cytokine gene polymorphisms and mothers’ antibody levels to P. falciparum antigens (n = 576): univariate analysis. This table summarizes the results of the univariate analysis performed for examining differences between maternal cytokine genotypes or haplotypes and mothers’ IgG levels to 7 recombinant proteins of P. falciparum asexual stage antigens as well as IgM levels to 1 of these antigens.
Association between maternal cytokine gene polymorphisms and newborns’ antibody levels to P. falciparum antigens (n = 576): univariate analysis. This table summarizes the results of the univariate analysis performed for examining differences between maternal cytokine genotypes or haplotypes and newborns’ IgG levels to 7 recombinant proteins of P. falciparum asexual stage antigens as well as IgM levels to 1 of these antigens.
Contributor Information
Adjimon Gatien Lokossou, Email: gbenalog@yahoo.fr.
Célia Dechavanne, Email: celia.dechavanne@gmail.com.
Aziz Bouraïma, Email: azlesage@yahoo.fr.
David Courtin, Email: david.courtin@ird.fr.
Agnès Le Port, Email: agnesleport@yahoo.fr.
Rodolphe Ladékpo, Email: ladekpo@yahoo.fr.
Julien Noukpo, Email: noukpovi@yahoo.fr.
Désiré Bonou, Email: bondesire@yahoo.fr.
Claude Ahouangninou, Email: cahoun83@yahoo.fr.
Audrey Sabbagh, Email: audrey.sabbagh@ird.fr.
Benjamin Fayomi, Email: bfayomi2@yahoo.fr.
Achille Massougbodji, Email: massougbodjiachille@yahoo.fr.
André Garcia, Email: Andre.Garcia@ird.fr.
Florence Migot-Nabias, Email: florence.migot-nabias@ird.fr.
Acknowledgements
We thank all women and infants of Tori Bossito who agreed to participate to the study as well as the UMR 216 team of Cotonou and the staff of the medical health centre of Tori Bossito (F Loumedjinon and M Daoudou) for performing field activities. We are grateful to R Anders (La Trobe University, Melbourne, Australia), S Longacre (Pasteur Institute, Paris, France), E Remarque (Biomedical Primate Research Centre, Rijswijk, The Netherlands) and M Theisen (Statens Serum Institute, Copenhagen, Denmark) for their gift of P. falciparum recombinant proteins. We thank the Faculté des Sciences de la Santé (FSS), the Institut des Sciences Biomédicales Appliquées de Cotonou (ISBA) and the Programme National de Lutte contre le Paludisme (PNLP) for their institutional support.
Financial support
The study was funded by the Institut de Recherche pour le Développement (IRD) and the Agence Nationale de la Recherche (ANR). A PhD scholarship was awarded by AIRD-DPF to A Lokossou.
References
- Dessein AJ, Chevillard C, Marquet S, Henri S, Hillaire D, Dessein H. Genetics of parasitic infections. Drug Metab Dispos. 2001;29(4 Pt 2):484–488. [PubMed] [Google Scholar]
- Flori L, Kumulungui B, Aucan C, Esnault C, Traore AS, Fumoux F, Rihet P. Linkage and association between Plasmodium falciparum blood infection levels and chromosome 5q31-q33. Genes Immun. 2003;4(4):265–268. doi: 10.1038/sj.gene.6363960. [DOI] [PubMed] [Google Scholar]
- Garcia A, Cot M, Chippaux J-P, Ranque S, Feingold J, Demenais F, Abel L. Genetic control of blood infection levels in human malaria: evidence for a complex genetic model. AmJTrop Med Hyg. 1998;58(4):480–488. doi: 10.4269/ajtmh.1998.58.480. [DOI] [PubMed] [Google Scholar]
- Marquet S, Abel L, Hillaire D, Dessein H, Kalil J, Feingold J, Weissenbach J, Dessein AJ. Genetic localization of a locus controlling the intensity of infection by Schistosoma mansoni on chromosome 5q31-q33. Nat Genet. 1996;14(2):181–184. doi: 10.1038/ng1096-181. [DOI] [PubMed] [Google Scholar]
- Rihet P, Abel L, Traoré Y, Traoré-Leroux T, Aucan C, Fumoux F. Human malaria: segregation analysis of blood infection levels in a suburban area and a rural area in Burkina Faso. Genet Epidemiol. 1998;15(5):435–450. doi: 10.1002/(SICI)1098-2272(1998)15:5<435::AID-GEPI1>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Garcia A, Marquet S, Bucheton B, Hillaire D, Cot M, Fievet N, Dessein AJ, Abel L. Linkage analysis of blood Plasmodium falciparum levels: interest of the 5q31-q33 chromosome region. AmJTrop Med Hyg. 1998;58(6):705–709. doi: 10.4269/ajtmh.1998.58.705. [DOI] [PubMed] [Google Scholar]
- Sakuntabhai A, Ndiaye R, Casadémont I, Peerapittayamongkol C, Rogier C, Tortevoye P, Tall A, Paul R, Turbpaiboon C, Phimpraphi W, Trape JF, Spiegel A, Heath S, Mercereau-Puijalon O, Dieye A, Julier C. Genetic determination and linkage mapping of Plasmodium falciparum malaria related traits in Senegal. PLoS One. 2008;3(4):e2000. doi: 10.1371/journal.pone.0002000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malaguarnera L, Musumeci S. The immune response to Plasmodium falciparum malaria. Lancet Infect Dis. 2002;2(8):472–478. doi: 10.1016/S1473-3099(02)00344-4. [DOI] [PubMed] [Google Scholar]
- Marsh DG, Neely JD, Breazeale DR, Ghosh B, Freidhoff LR, Ehrlich-Kautzky E, Schou C, Krishnaswamy G, Beaty TH. Linkage analysis of IL4 and other chromosome 5q31.1 markers and total serum immunoglobulin E concentrations. Science. 1994;264(5162):1152–1156. doi: 10.1126/science.8178175. [DOI] [PubMed] [Google Scholar]
- Vercelli D. Regulation of IgE synthesis. Allergy Proc. 1993;14(6):413–416. doi: 10.2500/108854193778792795. [DOI] [PubMed] [Google Scholar]
- Troye-Blomberg M, Riley EM, Kabilan L, Holmberg M, Perlmann H, Andersson U, Heusser CH, Perlmann P. Production by activated human T cells of interleukin 4 but not interferon-gamma is associated with elevated levels of serum antibodies to activating malaria antigens. Proc Natl Acad Sci USA. 1990;87(14):5484–5488. doi: 10.1073/pnas.87.14.5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousset F, Garcia E, Defrance T, Péronne C, Vezzio N, Hsu DH, Kastelein R, Moore KW, Banchereau J. Interleukin 10 is a potent growth and differentiation factor for activated human B lymphocytes. Proc Natl Acad Sci USA. 1992;89(5):1890–1893. doi: 10.1073/pnas.89.5.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troye-Blomberg M, Berzins K, Perlmann P. T-cell control of immunity to the asexual blood stages of the malaria parasite. Crit Rev Immunol. 1994;14(2):131–155. doi: 10.1615/CritRevImmunol.v14.i2.20. [DOI] [PubMed] [Google Scholar]
- Wynn TA. IL-13 effector functions. Annu Rev Immunol. 2003;21:425–456. doi: 10.1146/annurev.immunol.21.120601.141142. [DOI] [PubMed] [Google Scholar]
- de Mendonca VR, Goncalves MS, Barral-Netto M. The host genetic diversity in malaria infection. J Trop Med. 2012;2012:940616. doi: 10.1155/2012/940616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi J, Naka I, Patarapotikul J, Hananantachai H, Looareesuwan S, Tokunaga K. A single-nucleotide substitution from C to T at position −1055 in the IL-13 promoter is associated with protection from severe malaria in Thailand. Genes Immun. 2003;4(7):528–531. doi: 10.1038/sj.gene.6364010. [DOI] [PubMed] [Google Scholar]
- Rosenwasser LJ, Klemm DJ, Dresback JK, Inamura H, Mascali JJ, Klinnert M, Borish L. Promoter polymorphisms in the chromosome 5 gene cluster in asthma and atopy. Clin Exp Allergy. 1995;25(Suppl 2):74–78. doi: 10.1111/j.1365-2222.1995.tb00428.x. discussion 95–6. [DOI] [PubMed] [Google Scholar]
- Luoni G, Verra F, Arcà B, Sirima BS, Troye-Blomberg M, Coluzzi M, Kwiatkowski D, Modiano D. Antimalarial antibody levels and IL4 polymorphism in the Fulani of West Africa. Genes Immun. 2001;2(7):411–414. doi: 10.1038/sj.gene.6363797. [DOI] [PubMed] [Google Scholar]
- Vafa M, Maiga B, Berzins K, Hayano M, Bereczky S, Dolo A, Daou M, Arama C, Kouriba B, Färnert A, Doumbo OK, Troye-Blomberg M. Associations between the IL-4–590 T allele and Plasmodium falciparum infection prevalence in asymptomatic Fulani of Mali. Microbes Infect. 2007;9(9):1043–1048. doi: 10.1016/j.micinf.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Vafa M, Maiga B, Israelsson E, Dolo A, Doumbo OK, Troye-Blomberg M. Impact of the IL-4–590 C/T transition on the levels of Plasmodium falciparum specific IgE, IgG, IgG subclasses and total IgE in two sympatric ethnic groups living in Mali. Microbes Infect. 2009;11(8–9):779–784. doi: 10.1016/j.micinf.2009.04.017. [DOI] [PubMed] [Google Scholar]
- Gyan BA, Goka B, Cvetkovic JT, Kurtzhals JL, Adabayeri V, Perlmann H, Lefvert AK, Akanmori BD, Troye-Blomberg M. Allelic polymorphisms in the repeat and promoter regions of the interleukin-4 gene and malaria severity in Ghanaian children. Clin Exp Immunol. 2004;138(1):145–150. doi: 10.1111/j.1365-2249.2004.02590.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner DM, Williams DM, Sankaran D, Lazarus M, Sinnott PJ, Hutchinson IV. An investigation of polymorphism in the interleukin-10 gene promoter. Eur J Immunogenet. 1997;24(1):1–8. doi: 10.1111/j.1365-2370.1997.tb00001.x. [DOI] [PubMed] [Google Scholar]
- Carpenter D, Abushama H, Bereczky S, Färnert A, Rooth I, Troye-Blomberg M, Quinnell RJ, Shaw MA. Immunogenetic control of antibody responsiveness in a malaria endemic area. Hum Immunol. 2007;68(3):165–169. doi: 10.1016/j.humimm.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Salhi A, Rodrigues V Jr, Santoro F, Dessein H, Romano A, Castellano LR, Sertorio M, Rafati S, Chevillard C, Prata A, Alcaïs A, Argiro L, Dessein A. Immunological and genetic evidence for a crucial role of IL-10 in cutaneous lesions in humans infected with Leishmania braziliensis. J Immunol. 2008;180(9):6139–6148. doi: 10.4049/jimmunol.180.9.6139. [DOI] [PubMed] [Google Scholar]
- Ouma C, Davenport GC, Were T, Otieno MF, Hittner JB, Vulule JM, Martinson J, Ong’echa JM, Ferrell RE, Perkins DJ. Haplotypes of IL-10 promoter variants are associated with susceptibility to severe malarial anemia and functional changes in IL-10 production. Hum Genet. 2008;124(5):515–524. doi: 10.1007/s00439-008-0578-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hesran JY, Cot M, Personne P, Fievet N, Dubois B, Beyemé M, Boudin C, Deloron P. Maternal placental infection with Plasmodium falciparum and malaria morbidity during the first 2 years of life. Am J Epidemiol. 1997;146(10):826–831. doi: 10.1093/oxfordjournals.aje.a009200. [DOI] [PubMed] [Google Scholar]
- Mutabingwa TK, Bolla MC, Li JL, Domingo GJ, Li X, Fried M, Duffy PE. Maternal malaria and gravidity interact to modify infant susceptibility to malaria. PLoS Med. 2005;2(12):e407. doi: 10.1371/journal.pmed.0020407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz NG, Adegnika AA, Breitling LP, Gabor J, Agnandji ST, Newman RD, Lell B, Issifou S, Yazdanbakhsh M, Luty AJ, Kremsner PG, Grobusch MP. Placental malaria increases malaria risk in the first 30 months of life. Clin Infect Dis. 2008;47(8):1017–1025. doi: 10.1086/591968. [DOI] [PubMed] [Google Scholar]
- Courtin D, Oesterholt M, Huismans H, Kusi K, Milet J, Badaut C, Gaye O, Roeffen W, Remarque EJ, Sauerwein R, Garcia A, Luty AJ. The quantity and quality of African children’s IgG responses to merozoite surface antigens reflect protection against Plasmodium falciparum malaria. PLoS One. 2009;4(10):e7590. doi: 10.1371/journal.pone.0007590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodoo D, Theisen M, Kurtzhals JA, Akanmori BD, Koram KA, Jepsen S, Nkrumah FK, Theander TG, Hviid L. Naturally acquired antibodies to the glutamate-rich protein are associated with protection against Plasmodium falciparum malaria. J Infect Dis. 2000;181(3):1202–1205. doi: 10.1086/315341. [DOI] [PubMed] [Google Scholar]
- Oeuvray C, Bouharoun-Tayoun H, Gras-Masse H, Bottius E, Kaidoh T, Aikawa M, Filgueira MC, Tartar A, Druilhe P. Merozoite surface protein-3: a malaria protein inducing antibodies that promote Plasmodium falciparum killing by cooperation with blood monocytes. Blood. 1994;84(5):1594–1602. [PubMed] [Google Scholar]
- McGregor IA, Wilson ME, Billewicz WZ. Malaria infection of the placenta in The Gambia, West Africa; its incidence and relationship to stillbirth, birthweight and placental weight. Trans R Soc Trop Med Hyg. 1983;77(2):232–244. doi: 10.1016/0035-9203(83)90081-0. [DOI] [PubMed] [Google Scholar]
- Theisen M, Soe S, Brunstedt K, Follmann F, Bredmose L, Israelsen H, Madsen SM, Druilhe P. A Plasmodium falciparum GLURP-MSP3 chimeric protein; expression in Lactococcus lactis, immunogenicity and induction of biologically active antibodies. Vaccine. 2004;22(9–10):1188–1198. doi: 10.1016/j.vaccine.2003.09.017. [DOI] [PubMed] [Google Scholar]
- Branch OH, Udhayakumar V, Hightower AW, Oloo AJ, Hawley WA, Nahlen BL, Bloland PB, Kaslow DC, Lal AA. A longitudinal investigation of IgG and IgM antibody responses to the merozoite surface protein-1 19-kiloDalton domain of Plasmodium falciparum in pregnant women and infants: associations with febrile illness, parasitemia, and anemia. AmJTrop Med Hyg. 1998;58(2):211–219. doi: 10.4269/ajtmh.1998.58.211. [DOI] [PubMed] [Google Scholar]
- Pan W, Huang D, Zhang Q, Qu L, Zhang D, Zhang X, Xue X, Qian F. Fusion of two malaria vaccine candidate antigens enhances product yield, immunogenicity, and antibody-mediated inhibition of parasite growth in vitro. J Immunol. 2004;172(10):6167–6174. doi: 10.4049/jimmunol.172.10.6167. [DOI] [PubMed] [Google Scholar]
- Osier FH, Fegan G, Polley SD, Murungi L, Verra F, Tetteh KK, Lowe B, Mwangi T, Bull PC, Thomas AW, Cavanagh DR, McBride JS, Lanar DE, Mackinnon MJ, Conway DJ, Marsh K. Breadth and magnitude of antibody responses to multiple Plasmodium falciparum merozoite antigens are associated with protection from clinical malaria. Infect Immun. 2008;76(5):2240–2248. doi: 10.1128/IAI.01585-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Port A, Cottrell G, Martin-Prevel Y, Migot-Nabias F, Cot M, Garcia A. First malaria infections in a cohort of infants in Benin: biological, environmental and genetic determinants. Description of the study site, population methods and preliminary results. BMJ Open. 2012;2(2):e000342. doi: 10.1136/bmjopen-2011-000342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottrell G, Kouwaye B, Pierrat C, Le Port A, Bouraïma A, Fonton N, Hounkonnou MN, Massougbodji A, Corbel V, Garcia A. Modeling the influence of local environmental factors on malaria transmission in Benin and its implications for cohort study. PLoS One. 2012;7(1):e28812. doi: 10.1371/journal.pone.0028812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebie I, Diarra A, Ouedraogo A, Soulama I, Bougouma EC, Tiono AB, Konate AT, Chilengi R, Theisen M, Dodoo D, Remarque E, Bosomprah S, Milligan P, Sirima SB. Humoral responses to Plasmodium falciparum blood-stage antigens and association with incidence of clinical malaria in children living in an area of seasonal malaria transmission in Burkina Faso. West Africa. Infect Immun. 2008;76(2):759–766. doi: 10.1128/IAI.01147-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet S, Pêtres S, Holm I, Fontaine T, Rosario S, Roth C, Longacre S. Soluble and glyco-lipid modified baculovirus Plasmodium falciparum C-terminal merozoite surface protein 1, two forms of a leading malaria vaccine candidate. Vaccine. 2006;24(33–34):5997–6008. doi: 10.1016/j.vaccine.2006.04.069. [DOI] [PubMed] [Google Scholar]
- Kocken CH, Withers-Martinez C, Dubbeld MA, van der Wel A, Hackett F, Valderrama A, Blackman MJ, Thomas AW. High-level expression of the malaria blood-stage vaccine candidate Plasmodium falciparum apical membrane antigen 1 and induction of antibodies that inhibit erythrocyte invasion. Infect Immun. 2002;70(8):4471–4476. doi: 10.1128/IAI.70.8.4471-4476.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy JS, Marjason J, Elliott S, Fahey P, Bang G, Malkin E, Tierney E, Aked-Hurditch H, Adda C, Cross N, Richards JS, Fowkes FJ, Boyle MJ, Long C, Druilhe P, Beeson JG, Anders RF. A phase 1 trial of MSP2-C1, a blood-stage malaria vaccine containing 2 isoforms of MSP2 formulated with Montanide® ISA 720. PLoS One. 2011;6(9):e24413. doi: 10.1371/journal.pone.0024413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho LJ, Oliveira SG, Theisen M, Alves FA, Andrade MC, Zanini GM, Brígido MC, Oeuvray C, Póvoa MM, Muniz JA, Druilhe P, Daniel-Ribeiro CT. Immunization of Saimiri sciureus monkeys with Plasmodium falciparum merozoite surface protein-3 and glutamate-rich protein suggests that protection is related to antibody levels. Scand J Immunol. 2004;59(4):363–372. doi: 10.1111/j.0300-9475.2004.01409.x. [DOI] [PubMed] [Google Scholar]
- Theisen M, Vuust J, Gottschau A, Jepsen S, Høgh B. Antigenicity and immunogenicity of recombinant glutamate-rich protein of Plasmodium falciparum expressed in Escherichia coli. Clin Diagn Lab Immunol. 1995;2(1):30–34. doi: 10.1128/cdli.2.1.30-34.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill WG, Robertson A. Linkage disequilibrium in finite populations. Theor Appl Genet. 1968;38:226–231. doi: 10.1007/BF01245622. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D. The structure of haplotype blocks in the human genome. Science. 2002;296(5576):2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- Stephens M, Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73(5):1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform Online. 2007;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Koura GK, Ouedraogo S, Le Port A, Watier L, Cottrell G, Guerra J, Choudat I, Rachas A, Bouscaillou J, Massougbodji A, Garcia A. Anaemia during pregnancy: impact on birth outcome and infant haemoglobin level during the first 18 months of life. Trop Med Int Health. 2012;17(3):283–291. doi: 10.1111/j.1365-3156.2011.02932.x. [DOI] [PubMed] [Google Scholar]
- Jha AN, Singh VK, Kumari N, Singh A, Antony J, van Tong H, Singh S, Pati SS, Patra PK, Singh R, Toan NL, le Song H, Assaf A, Messias-Reason IJ, Velavan TP, Singh L, Thangaraj K. IL-4 haplotype -590T, -34T and intron-3 VNTR R2 is associated with reduced malaria risk among ancestral indian tribal populations. PLoS One. 2012;7(10):e48136. doi: 10.1371/journal.pone.0048136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dybedal I, Larsen S, Jacobsen SE. IL-12 directly enhances in vitro murine erythropoiesis in combination with IL-4 and stem cell factor. J Immunol. 1995;154(10):4950–4955. [PubMed] [Google Scholar]
- Metzger WG, Okenu DM, Cavanagh DR, Robinson JV, Bojang KA, Weiss HA, McBride JS, Greenwood BM, Conway DJ. Serum IgG3 to the Plasmodium falciparum merozoite surface protein 2 is strongly associated with a reduced prospective risk of malaria. Parasite Immunol. 2003;25(6):307–312. doi: 10.1046/j.1365-3024.2003.00636.x. [DOI] [PubMed] [Google Scholar]
- Tangteerawatana P, Perlmann H, Hayano M, Kalambaheti T, Troye-Blomberg M, Khusmith S. IL4 gene polymorphism and previous malaria experiences manipulate anti-Plasmodium falciparum antibody isotype profiles in complicated and uncomplicated malaria. Malar J. 2009;8:286. doi: 10.1186/1475-2875-8-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidyarani M, Selvaraj P, Prabhu Anand S, Jawahar MS, Adhilakshmi AR, Narayanan PR. Interferon gamma (IFNgamma) &interleukin-4 (IL-4) gene variants & cytokine levels in pulmonary tuberculosis. Indian J Med Res. 2006;124(4):403–410. [PubMed] [Google Scholar]
- Kumaratilake LM, Ferrante A, Jaeger T, Rzepczyk C. GM-CSF-induced priming of human neutrophils for enhanced phagocytosis and killing of asexual blood stages of Plasmodium falciparum: synergistic effects of GM-CSF and TNF. Parasite Immunol. 1996;18(3):115–123. doi: 10.1046/j.1365-3024.1996.d01-64.x. [DOI] [PubMed] [Google Scholar]
- Cabantous S, Poudiougou B, Oumar AA, Traore A, Barry A, Vitte J, Bongrand P, Marquet S, Doumbo O, Dessein AJ. Genetic evidence for the aggravation of Plasmodium falciparum malaria by interleukin 4. J Infect Dis. 2009;200(10):1530–1539. doi: 10.1086/644600. [DOI] [PubMed] [Google Scholar]
- King CL, Malhotra I, Wamachi A, Kioko J, Mungai P, Wahab SA, Koech D, Zimmerman P, Ouma J, Kazura JW. Acquired immune responses to Plasmodium falciparum merozoite surface protein-1 in the human fetus. J Immunol. 2002;168(1):356–364. doi: 10.4049/jimmunol.168.1.356. [DOI] [PubMed] [Google Scholar]
- Xi G, Leke RG, Thuita LW, Zhou A, Leke RJ, Mbu R, Taylor DW. Congenital exposure to Plasmodium falciparum antigens: prevalence and antigenic specificity of in utero-produced antimalarial immunoglobulin M antibodies. Infect Immun. 2003;71(3):1242–1246. doi: 10.1128/IAI.71.3.1242-1246.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koss K, Satsangi J, Fanning GC, Welsh KI, Jewell DP. Cytokine (TNF alpha, LT alpha and IL-10) polymorphisms in inflammatory bowel diseases and normal controls: differential effects on production and allele frequencies. Genes Immun. 2000;1(3):185–190. doi: 10.1038/sj.gene.6363657. [DOI] [PubMed] [Google Scholar]
- Mangia A, Santoro R, Piattelli M, Pazienza V, Grifa G, Iacobellis A, Andriulli A. IL-10 haplotypes as possible predictors of spontaneous clearance of HCV infection. Cytokine. 2004;25(3):103–109. doi: 10.1016/j.cyto.2003.10.005. [DOI] [PubMed] [Google Scholar]
- Moore KW, de Waal MR, Coffman RL, O’Garra A. Interleukin-10 and the interleukin-10 receptor. Annu Rev Immunol. 2001;19:683–765. doi: 10.1146/annurev.immunol.19.1.683. [DOI] [PubMed] [Google Scholar]
- Kilpinen S, Huhtala H, Hurme M. The combination of the interleukin-1alpha (IL-1alpha-889) genotype and the interleukin-10 (IL-10 ATA) haplotype is associated with increased interleukin-10 (IL-10) plasma levels in healthy individuals. Eur Cytokine Netw. 2002;13(1):66–71. [PubMed] [Google Scholar]
- Gatlin MR, Black CL, Mwinzi PN, Secor WE, Karanja DM, Colley DG. Association of the gene polymorphisms IFN-gamma +874, IL-13–1055 and IL-4–590 with patterns of reinfection with Schistosoma mansoni. PLoS Negl Trop Dis. 2009;3(2):e375. doi: 10.1371/journal.pntd.0000375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouriba B, Traore HA, Dabo A, Sangare L, Guindo H, Keita AS, Reimert CM, van Dam GJ, Deelder AM, Doumbo O, Dessein AJ. Urinary disease in 2 Dogon populations with different exposure to Schistosoma haematobium infection: progression of bladder and kidney diseases in children and adults. J Infect Dis. 2005;192(12):2152–2159. doi: 10.1086/498214. [DOI] [PubMed] [Google Scholar]
- Isnard A, Kouriba B, Doumbo O, Chevillard C. Association of rs7719175, located in the IL13 gene promoter, with Schistosoma haematobium infection levels and identification of a susceptibility haplotype. Genes Immun. 2011;12(1):31–39. doi: 10.1038/gene.2010.43. [DOI] [PubMed] [Google Scholar]
- Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, McVean GA. 1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh K. Evolution and selection of human leukocyte antigen alleles by Plasmodium falciparum infection. Hum Immunol. 2008;69(12):856–860. doi: 10.1016/j.humimm.2008.08.294. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Association between maternal cytokine gene polymorphisms and mothers’ biological parameters (n = 576): univariate analysis. This table summarizes the results of the univariate analysis performed for examining differences between maternal cytokine genotypes or haplotypes and biological parameters (haemoglobin, prevalence of circulating P. falciparum parasites, prevalence of P. falciparum placenta infection) of mothers.
Association between maternal cytokine gene polymorphisms and newborns’ biological parameters (n = 576): univariate analysis. This table summarizes the results of the univariate analysis performed for examining differences between maternal cytokine genotypes or haplotypes and biological parameters (birthweight, haemoglobin) of newborns.
Association between maternal cytokine gene polymorphisms and mothers’ antibody levels to P. falciparum antigens (n = 576): univariate analysis. This table summarizes the results of the univariate analysis performed for examining differences between maternal cytokine genotypes or haplotypes and mothers’ IgG levels to 7 recombinant proteins of P. falciparum asexual stage antigens as well as IgM levels to 1 of these antigens.
Association between maternal cytokine gene polymorphisms and newborns’ antibody levels to P. falciparum antigens (n = 576): univariate analysis. This table summarizes the results of the univariate analysis performed for examining differences between maternal cytokine genotypes or haplotypes and newborns’ IgG levels to 7 recombinant proteins of P. falciparum asexual stage antigens as well as IgM levels to 1 of these antigens.