Abstract
Background
H19 is a paternally imprinted gene that has been shown to be highly expressed in the trophoblast tissue. Results from previous studies have initiated a debate as to whether noncoding RNA H19 acts as a tumor suppressor or as a tumor promotor in trophoblast tissue. In the present study, we developed lentiviral vectors expressing H19-specific small interfering RNA (siRNA) to specifically block the expression of H19 in the human choriocarcinoma cell line JAR. Using this approach, we investigated the impact of the H19 gene on the proliferation, invasion and apoptosis of JAR cells. Moreover, we examined the effect of H19 knockdown on the expression of insulin-like growth factor 2 (IGF2), hairy and enhancer of split homologue-1 (HES-1) and dual-specific phosphatase 5 (DUSP5) genes.
Results
H19 knockdown inhibited apoptosis and proliferation of JAR cells, but had no significant impact on cell invasion. In addition, H19 knockdown resulted in significant upregulation of HES-1 and DUSP5 expression, but not IGF2 expression in JAR cells.
Conclusions
The finding that H19 downregulation could simultaneously inhibit proliferation and apoptosis of JAR cells highlights a putative dual function for H19 in choriocarcinoma and may explain the debate on whether H19 acts as a tumor suppressor or a tumor promotor in trophoblast tissue. Furthermore, upregulation of HES-1 and DUSP5 may mediate H19 downregulation-induced suppression of proliferation and apoptosis of JAR cells.
Keywords: H19, JAR cells, Choriocarcinoma, HES-1, DUSP5, IGF2
Background
H19 is a paternally imprinted gene encoding a noncoding RNA [1]. As one of the first imprinted genes discovered, H19 was initially identified as a tumor suppressor because embryo-derived tumor cells overexpressing H19 exhibited growth retardation, morphological changes and abrogated clonogenicity in soft agar, as well as suppressed tumorigenicity in nude mice [2]. Downregulation of H19 gene expression was identified as an early event in the formation of several tumor types [3-8]. Recently it was demonstrated that mice lacking H19 showed an overgrowth phenotype, while transgenic mice overexpressing H19 showed postnatal growth reduction [9].
In contrast, overexpression or loss of imprinting (LOI) of H19 has been observed in a wide variety of tumors, including bladder carcinoma [10,11], epithelial ovarian cancer [12], esophageal cancer [13], lung cancer [14], breast adenocarcinoma [15,16], endometrial cancer [17], and invasive cervical carcinoma [18]. H19 knockdown significantly decreased the clonogenicity and anchorage-independent growth property of several breast and lung cancer cell lines [19]. A link between H19 and several tumorigenesis-related genes, such as c-Myc[19], thioredoxin[20] and E2F1[21], has been well established. Thus, it remains controversial whether H19 functions as a tumor promotor or a tumor suppressor, and it is possible that H19 plays differential roles depending on tissue type and/or developmental stage [13].
H19 is highly expressed in trophoblast tissue, predominantly but not exclusively from the maternal allele [22,23]. Complete hydatidiform moles (CHM) showed high-level expression of H19 from the paternal allele, while choriocarcinomas developing from CHMs had reduced numbers of H19-positive cells [23]. H19 downregulation was also observed in gestational trophoblastic tumors [24]. These observations appear to support the notion that H19 functions as a tumor suppressor in trophoblast tissue. However, Ariel I et al. found that H19 expression was found to be abundant, in a decreasing order, in the intermediate trophoblast (villous and interstitial), the cytotrophoblast, and the syncytiotrophoblast. And prominent expression of H19 was found in placental site trophoblastic tumor and gestational choriocarcinoma [25]. After choriocarcinoma-derived JEG-3 cells were subcutaneously injected into nude mice, a five-fold increase in H19 RNA level was detected in the resulting tumors, and cells highly expressing H19 were more tumorigenic [26]. Therefore, the exact role of H19 in the trophoblast is yet undetermined.
In previous studies, we demonstrated that DNA methyltransferase inhibitor 5-aza-2′-deoxycytidine could demethylate the promoter region of the H19 gene, upregulate the expression of H19 transcript, and reduce the proliferation, migration and invasion properties of choriocarcinoma-derived JEG-3 cells [27,28]. As the demethylating effects of 5-aza-2′-deoxycytidine are unspecific, other genes may be demethylated in tandem and complicate the biological effects. To further clarify our understanding of the function of the H19 gene in choriocarcinoma, we developed lentiviral vectors expressing H19-specific small interfering RNA (siRNA) for use in human choriocarcinoma cells. Considering that the human choriocarcinoma cell line JAR is more easily infected by lentiviruses than the cell line JEG-3 (our unpublished observation), JAR cells were used in the present study. Using the siRNA technology, we investigated the impact of H19 knockdown on the proliferation, invasion, and apoptosis of JAR cells. Furthermore, we examined the effects of H19 knockdown on the expression of the insulin-like growth factor 2 (IGF2), hairy and enhancer of split homologue-1 (HES-1), and dual-specific phosphatase 5 (DUSP5) genes, which were chosen based on the previous evidence that H19 is a negative regulator of IGF2[29] and our unpublished observation that the expression of HES-1 and DUSP5 genes was altered in JEG-3 cells transfected with a eukaryotic expression vector carrying the full-length H19 cDNA.
Methods
Cell culture
Human choriocarcinoma cell line JAR was purchased from American Type Culture Collection (USA). After thawing, cells were allowed to grow in Dulbecco’s modified Eagle’s medium/Ham’s nutrient mixture F12 (DMEM/F12; Gibco-BRL, USA) medium supplemented with 10% fetal bovine serum (Gibco-BRL, USA) at 37°C in a humidified atmosphere containing 5% CO2. After three passages, cells were used for viral infection.
Construction and screening of lentiviral vectors harboring H19-specific siRNA
The following siRNA target sequences in the human H19 gene (GenBank accession No. NR_002196) were selected: TARGET1, GCCTTCAAGCATTCCATTA; TARGET2, GGAGAGTTAGCAAAGGTGA; TARGET3, CGTGACAAGCAGGACATGA; and TARGET4, GAGGAACCAGACCTCATCA. Four pairs of complementary oligonucleotides were then designed (Additional file 1: Table S1). The stem-loop oligonucleotides were synthesized and cloned into a lentivirus-based vector carrying the green fluorescent protein (GFP) gene (pGCSIL-GFP, Genechem, Shanghai, China). A universal sequence (PSC-NC: TTCTCCGAACGTGTCACGT) was used as a negative control for RNA interference. Lentiviral particles were prepared as previously described [30].
The four siRNA-carrying lentiviral vector constructs were used to infect JAR cells at a multiplicity of infection (MOI) of 20 (low MOI) and 40 (high MOI). Three days after infection, GFP expression was detected to calculate the infection efficiency. Five days after infection, cells were harvested. Real-time reverse transcription-polymerase chain reaction (RT-PCR) was performed to determine H19 knockdown efficiency and screen for the siRNA with the highest knockdown efficiency which was then used for subsequent experiments.
RNA isolation, reverse transcription and quantitative PCR
Total RNA isolation using Trizol Reagent (Invitrogen, USA) and reverse transcription using Moloney murine leukemia virus (M-MLV) reverse transcriptase (Promega, USA) were carried out according to the manufacturer’s instructions. Quantitative PCR was performed in a final volume of 20 μl containing 1 μl cDNA, 0.5 μl primers (2.5 μM) and 10 μl SYBR premix exTaq (TaKaRa, Dalian, China). The cycling conditions were: 95°C for 15 min; 40 cycles of 95°C for 5 s and 60°C for 30 s; and one cycle of 95°C for 60 s, 55°C for 60 s and 95°C for 60 s. The sequences of primers used in real-time RT-PCR were as follows: forward 5′-TCCCAGAACCCACAACATGAA-3′ and reverse 5′-TTCACCTTCCAGAGCCGATTC-3′ for H19 (150 bp); forward 5′-TCCTCACCTCGCTACTCG-3′ and reverse 5′-ACATCCACGCAACACTCAG-3′ for DUSP5 (106 bp); forward 5′-GAGAGGCGGCTAAGGTGTTTG-3′ and reverse 5′-CTGGTGTAGACGGGGATGAC-3′ for HES-1 (121 bp); forward 5′-CCTCCAGTTCGTCTGTGGG-3′ and reverse 5′-CACGTCCCTCTCGGACTTG-3′ for IGF2 (163 bp); and forward 5′-GGCGGCACCACCATGTACCCT-3′ and reverse 5′-AGGGGCCGGACTCGTCATACT -3′ for beta-actin (202 bp). Data analysis was performed using the 2-ΔΔCt method. The experiment was independently repeated three times.
Cell cycle analysis
Cells were seeded onto six-well plates and cultured. Upon reaching 80% confluence, cells were released by digestion with trypsin and harvested. After centrifugation, the cell pellet was washed twice with pre-cooled phosphate buffered saline (PBS) and fixed with pre-cooled 70% ethanol. Then, cells were resuspended in PBS, filtered through a 400-mesh sieve, and stained with propidium iodide (PI, 50 μg/mL, 100 μg/mL RNase in PBS) at 37°C for 30 min. The cell cycle was then determined by flow cytometry as previously described [31].
Apoptosis assay
Apoptosis was detected using the ApoScreen Annexin V Apoptosis Kit (SouthernBiotech, USA) according to the manufacturer’s instructions. In addition, one more group of cells was incubated with 30 μg/ml diaminedichloroplatinum (DDP) for 16 hours and subjected to apoptosis assay. The experiment was independently repeated three times.
Methyl thiazolyl tetrazolium (MTT) assay
Cells were seeded in sextuplicate for each condition in 96-well plates at a density of 2×104/mL in 100 μl per well. After 24 hours of culture, medium was replaced. Cells were then further cultured in this manner for 1–6 days. Four hours before the termination of culture, MTT (5 mg/mL; Sigma, USA) was added at a volume of 10 μL per well. Afterwards, the entire supernatant was discarded and dimethyl sulfoxide (DMSO) was added at a volume of 100 μL per well and incubated in an air bath shaker at 37°C for 10 min. The absorbance at 570 nm of each well was determined using the 1420 Multilabel Counter (PerkinElmer, USA). The experiment was independently repeated three times.
Cell invasion assay
The cell invasion assay was performed using a commercial kit (Chemicon, Canada), according to the manufacturer’s instructions. The CHEMICON Cell Invasion Assay is performed in an Invasion Chamber, a 24-well tissue culture plate with 12 cell culture inserts. The inserts contain an 8 μm pore size polycarbonate membrane, over which a thin layer of ECMatrixTM is dried. The ECMatrixTM layer serves as an in vitro reconstituted basement membrane and occludes the membrane pores, blocking non-invasive cells from migrating through. Invasive cells, on the other hand, migrate through the ECM layer and cling to the bottom of the polycarbonate membrane. Before JAR cells were plated at a density of 1 × 104 cells in each insert, 300 μL of warm serum free media were added to the interior of the inserts to rehydrate the ECM layer for 1–2 hours at room temperature. Cells were incubated at 37°C in 5% CO2 for 72 h. A cotton-tipped swab was used to gently remove non-invading cells as well as the ECMatrix gel from the interior of the inserts. Five hundred microliters of staining solution were added to the unoccupied wells of the plate. Invasive cells on lower surface of the membrane were stained by dipping inserts in the staining solution for 20 minutes. Inserts were dipped in a beaker of water several times to rinse and air dried. Stained cells were dissolved in 10% acetic acid and OD at 560 nm was detected by colorimetric reading. Each assay was performed in triplicate and repeated three times.
Statistical analysis
Statistical analysis was performed using SPSS 13.0 software package (SPSS Inc, Chicago, IL, USA). Numerical data are expressed as mean ± standard deviation. Multiple means were compared using analysis of variance and the Tukey post hoc test, while two means were compared using Student’s t-test. A P value of less than 0.05 was considered statistically significant.
Results
Selection of the lentiviral vector harboring siRNA with the highest knockdown efficiency
The four lentiviral constructs harboring different siRNAs (KD1, KD2, KD3 and KD4) were used to infect JAR cells. In parallel, a negative control (NC) was run. The infection efficiencies of these lentiviral vectors were all above 80% as revealed by fluorescence microscopy (Figure 1A). Real-time RT-PCR assay showed that all four constructs, whether they were used at a high or low MOI, could significantly downregulate H19 gene expression in JAR cells (Figure 1B, C). The highest knockdown efficiency was achieved using KD3 (low MOI: 93% relative to the NC group; high MOI: 95%), which was subsequently designated as H19-RNAi-LV-3.
Figure 1.
Selection of the lentiviral vector harboring siRNA with the highest knockdown efficiency. A, Fluorescence microscopy examination of the infection efficiencies of different lentiviral vectors in JAR cells (magnification 200 ×). a. JAR cells infected with lentiviral particles harboring a non-targeting control siRNA (NC group) in the light microscope; b. JAR cells of NC group in the fluorescence microscope; c. Fusion image of the top two images; d. JAR cells infected with KD3-harboring lentiviral particles (KD group)at a low MOI in the light microscope; e. JAR cells of KD group at a low MOI in the fluorescence microscope; f. Fusion image of the top two images; g. JAR cells infected with KD3-harboring lentiviral particles at a high MOI in the light microscope; h. JAR cells of KD group at a high MOI in the fluorescence microscope. i. Fusion image of the top two images. The fluorescence expression in cells infected with KD1, KD2 and KD4 was similar to that in cells infected with KD3. The infection efficiencies of these lentiviral vectors were about to be above 80%. B, Relative levels of H19 in JAR cells infected with different groups of lentiviral particles at a low MOI. C, Relative levels of H19 in JAR cells infected with different groups of lentiviral particles at a high MOI. Compared to the NC group, H19 expression was significantly downregulated in JAR cells infected with different groups of lentiviral particles, at either a low or high MOI. Beta-actin was used to normalize the PCR data. The highest knockdown efficiency was achieved using KD3-harboring lentiviral particles.
H19 Knockdown inhibited the proliferation of JAR cells
The percentage of cells in G1 phase was significantly higher in the H19 knockdown group than in the NC group (P < 0.01), while the percentages of cells in G2/M phase and S phase respectively showed no significant changes between the two groups (Figure 2A). The MTT assay showed that cell proliferation was significantly inhibited in the H19 knockdown group as compared to that in the NC group (P < 0.01) (Figure 2B).
Figure 2.

Effect of H19 knockdown on the growth of JAR cells. A, Comparison of the percentages of cells in G1, G2/M and S phase between NC and KD groups. The percentage of cells in G1 phase was significantly higher in the KD group than in the NC group, while the percentages of cells in G2/M phase and S phase respectively showed no significant changes between the two groups. B, Growth curves of cells in each group. MTT analysis showed that H19 knockdown significantly inhibited cell proliferation (n = 18). ** denotes P < 0.01.
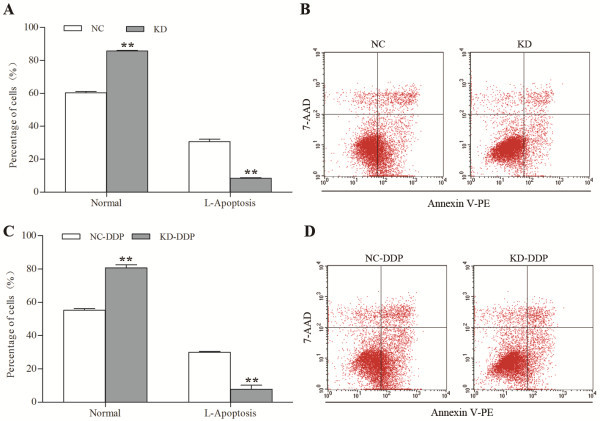
H19 Knockdown inhibited apoptosis of JAR cells
As shown in Figure 3, H19 knockdown significantly increased the percentage of normal cells but decreased the percentage of early apoptotic cells when compared to the negative control group (85.70% ± 0.41% vs. 60.4% ± 0.70% and 8.46% ± 0.37% vs. 30.74% ± 1.45%, respectively; Ps < 0.01 for both). After incubation with diaminedichloroplatinum (an apoptosis inducer) for 16 hours, the percentage of normal cells was still significantly higher in the H19 knockdown group than in the NC group (80.70% ± 1.88% vs. 55.20% ± 0.98%, P < 0.01), while the percentage of early apoptotic cells was still significantly lower in the H19 knockdown group than in the NC group (7.80% ± 2.39% vs. 30.00% ± 0.46%, P < 0.01).
Figure 3.
Effect of H19 knockdown on the apoptosis of JAR cells. A. Comparison of the percentages of normal cells and early apoptotic cells between the NC group and the KD group. H19 knockdown significantly increased the percentage of normal cells but decreased the percentage of early apoptotic cells when compared with the NC group; B. Flow cytometric analysis of JAR cells in the NC group and the KD group; C. Comparison of the percentages of normal cells and early apoptotic cells between the two groups with DDP treatment. H19 knockdown also significantly increased the percentage of normal cells but decreased the percentage of early apoptotic cells when compared with the NC-DDP group; D. Flow cytometric analysis of JAR cells in the NC-DDP group and the KD-DDP group. NC-DDP is the negative control vector-harboring cells incubated with DDP; KD-DDP, H19-RNAi-LV-3-infected cells incubated with DDP. ** denotes P < 0.01.
H19 Knockdown does not alter the invasion of JAR cells
To determine if H19 knockdown was able to affect the invasion properties of JAR cells, the cell invasion assay was performed. The invasion rate of cells in the H19 knockdown group and the NC group were 68.85% ± 3.04% and 79.50% ± 5.52%, respectively. H19 knockdown had no significant impact on the invasion rate of JAR cells. The result was shown in Additional file 1: Figure S1.
H19 Knockdown upregulated HES-1 and DUSP5 expression but not IGF2 expression
To gain insight into how H19 knockdown alters the phenotype of human choriocarcinoma cells, we examined the expression of IGF2, HES-1 and DUSP5 mRNAs by real-time RT-PCR. The results showed that, compared with the negative control group, H19 knockdown significantly upregulated the expression of HES-1 and DUSP5 mRNAs (1.24 ± 0.06 vs. 1.01 ± 0.14 and 2.81 ± 0.30 vs. 1.00 ± 0.05; P < 0.05 and 0.01, respectively), but had no significant impact on the expression of IGF2 mRNA (1.22 ± 0.09 vs. 1.02 ± 0.21; P > 0.05, Figure 4). The Ct values were shown in Additional file 1: Table S2.
Figure 4.

Effect of H19 knockdown on the expression of IGF2, HES-1 and DUSP5 mRNAs in JAR cells.H19 knockdown significantly upregulated the expression of HES-1 and DUSP5 mRNAs, but had no significant impact on the expression of IGF2 mRNA. KD, H19 knockdown group. * denotes P < 0.05 and ** denotes P < 0.01.
Discussion
Using a lentiviral vector harboring H19-specific siRNA, the expression of H19 in JAR cells was specifically downregulated. We showed that the invasion of JAR cells was not affected in response to H19 downregulation, which is inconsistent with our previous studies indicating that 5-aza-2′-deoxycytidine-mediated H19 upregulation could reduce the invasion capacity of JEG-3 cells [27,28]. This may be due to unspecific demethylating effects of 5-aza-2′-deoxycytidine or differences among the cell types studied. Furthermore, we also demonstrated that H19 knockdown inhibited the apoptosis and proliferation properties of JAR cells. The results of the present study indicate that H19 exerts both tumor-promoting and tumor-suppressing effects in choriocarcinoma cells, at least in part, by regulating cell proliferation and apoptosis.
It has been hypothesized for many years that a potential tumor suppressor gene is present at chromosome 11p15.5 where H19 is located, because a loss of heterozygosity at this locus is observed in certain embryonal tumors [2]. Subsequent experiments implicated H19 may function as a tumor suppressor [2,8]. However, other studies provided conflicting evidence that supported the idea that H19 RNA harbors oncogenic properties [10-20]. Similar observations have been made in trophoblast tissue; some studies revealed that H19 has tumor-suppressing activity [23,24], while others suggested that it has tumor-promoting activity [26]. Therefore, H19 may play differential roles under different physiologic conditions. In the present study, we demonstrated that H19 knockdown could simultaneously inhibit the proliferation and apoptosis of JAR cells, suggesting that H19 expression may have both proliferation-inducing and apoptosis-inducing activity in choriocarcinoma-derived cells. Interestingly, a key event in tumorigenesis is the loss of equilibrium between cell proliferation and cell death [32]. Thus, our observation that H19 can simultaneously induce proliferation and apoptosis highlights a putative dual role of H19 in choriocarcinoma cell lines and may explain the debate over whether H19 is a tumor suppressor or a tumor promotor in trophoblast tissue. Shoshani O et al. discovered that direct knockdown of H19 expression in diploid tumorigenic mouse mesenchymal stromal cells(MSCs) resulted in acquisition of polyploid cell traits. And artificial tetraploidization of diploid MSCs reduces tumorigenic potential in conjunction with suppression of H19 expression. This establishs a link between H19 expression and cell ploidy. Poluploidization might either have a cancer promoting or a cancer-preventing effect as is the case in aneuploidy [33]. Then, further experiments should determine whether the inhibition of proliferation and apoptosis by H19 knockdown is associated with the polyploidy of trophoblast cells. However, since the human choriocarcinoma cell line JAR may already have a different H19 gene regulatory pattern from normal trophoblasts, the extrapolation of results from JAR cells to the normal trophoblast must be cautious. Further studies in normal trophoblast cells should be done to address this issue.
Like H19, IGF2 is an imprinted gene and the two are closely linked. However, while H19 is expressed from the maternal allele, IGF2 is transcribed from the paternal allele in most tissues [34]. A growing body of evidence has indicated that H19 RNA is a negative regulator of IGF2. Gabory et al.[9] demonstrated that H19 transgenic or knockout mice developed abnormal phenotypes due to alterations in IGF2 mRNA levels. Esquiliano et al.[35] found that targeted disruption of the H19 gene in mice induced the placental phenotype via upregulation of IGF2 expression. In the present study, we have demonstrated that H19 knockdown had no significant impact on the expression of IGF2 mRNA. Nevertheless, this result is not too surprising because numerous studies have not been able to provide a consistent relationship between the expression levels and imprinting status of H19 and IGF2 in all tissues. Adam et al.[36] discovered that H19 showed biallelic expression in extravillous cytotrophoblasts, while IGF2 showed monoallelic expression. Lustig-Yariv et al.[37] showed that H19 and IGF-2 RNA levels differed greatly among different JAR or JEG-3 clones. Moreover, we also found that both H19 and IGF-2 RNAs were highly expressed in normal term placenta and JEG-3 cells, and H19 overexpression did not alter IGF2 expression in JEG-3 cells (unpublished data). Therefore, these data suggest that IGF2 may not be involved in H19 downregulation-mediated inhibition of proliferation and apoptosis of JAR cells.
Our results did show that H19 knockdown significantly upregulated expression of the HES-1 gene in JAR cells. HES-1 gene is an important effector of the Notch pathway that has been implicated in regulating the proliferation, differentiation and apoptosis in multiple tumor cell types. Kunnimalaiyaan et al.[38] found that increased HES-1 expression suppressed the proliferation of carcinoid tumor cells. Nefedova et al.[39] demonstrated that overexpression of HES-1 abrogated gamma-secretase inhibitor-induced apoptosis in multiple myeloma cells. Importantly, we also found that H19 overexpression could downregulate the expression of HES-1 in JEG-3 cells (about 2.27 fold) (unpublished data). Thus, we speculated that HES-1 may mediate H19 downregulation-induced suppression of proliferation and apoptosis of JAR cells. Although it remains unclear how H19 knockdown upregulates HES-1 expression in JAR cells, the recent finding that H19 can negatively regulate an imprinted gene network may provide some clues to the mechanisms of regulation of HES-1 expression by H19[9].
Furthermore, we demonstrated that H19 knockdown could also significantly upregulate the expression of DUSP5 gene in JAR cells. The DUSP5 gene encodes a mitogen-activated protein kinase phosphatase (MKP). It has been shown that extracellular signal regulated kinase 2 (ERK2) is a specific substrate of DUSP5[40]. As ERK2 plays an important role in regulating cell proliferation and apoptosis, it is reasonable to surmise that DUSP5 may also be involved in such processes. Importantly, we also found that H19 overexpression could downregulate the expression of DUSP5 in JEG-3 cells (about 2.63 fold) (unpublished data). Therefore, DUSP5 may also be involved in H19 downregulation-induced suppression of proliferation and apoptosis of JAR cells.
Conclusions
The present study provides evidence that H19 knockdown does not alter invasion but inhibits proliferation and apoptosis of JAR cells. The observation that H19 downregulation simultaneously suppresses proliferation and apoptosis of JAR cells suggests a putative dual functionality of H19 in choriocarcinoma cell lines and may explain the debate over whether H19 is a tumor suppressor or a tumor promotor in trophoblast tissue. Moreover, we also found that H19 knockdown upregulates the expression of the HES-1 and DUSP5 genes but not of the IGF-2 gene, suggesting that HES-1 and DUSP5 may mediate H19 downregulation-induced suppression of proliferation and apoptosis of JAR cells.
Abbreviations
CHM: Complete hydatidiform moles; DDP: Diamminedichloroplatinum; DMSO: Dimethyl sulfoxide; DUSP5: Dual-specific phosphatase 5; ERK2: Extracellular signal regulated kinase 2; GFP: Green fluorescent protein; HES-1: Hairy and enhancer of split homologue-1; IGF2: Insulin-like growth factor 2; LOI: Loss of imprinting; MKP: Mitogen-activated protein kinase phosphatase; M-MLV: Moloney murine leukemia virus; MOI: Multiplicity of infection; MTT: Methyl thiazolyl tetrazolium; NC: Negative control; PBS: Phosphate buffered saline; PI: Propidium iodide; RT-PCR: Reverse transcription-polymerase chain reaction; siRNA: Small interfering RNA
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
LL and MC contributed to the overall conceptual design and also analyzed the data, contributed to the interpretation of findings and wrote the first draft of the manuscript. LY and KC was the principal investigator and contributed with the idea, as well as to the interpretation of the results and the writing of the manuscript.CN, JS, FM, LM, AN, JK, GM, JO, LG, CR and BB were involved in the experiment of the study and revision of the manuscript. All authors have read and approved the final manuscript.
Supplementary Material
The structure of siRNAs in lentiviral vectors. Table S2 The Ct values from quantitative PCR reactions. Figure S1: Effect of H19 knockdown on the invasion of JAR cells. The invasion rate of cells in the H19 knockdown group and the NC group were 68.85% ± 3.04% and 79.50% ± 5.52%, respectively. H19 knockdown had no significant impact on the invasion rate of JAR cells.
Contributor Information
Li-Li Yu, Email: yulili0413@126.com.
Kai Chang, Email: changkai0203@163.com.
Lin-Shan Lu, Email: lulinshan0413@126.com.
Dan Zhao, Email: zhandan0413@126.com.
Jian Han, Email: hanjian0413@126.com.
Ying-Ru Zheng, Email: zhengyingru0413@126.com.
Yao-Hua Yan, Email: yanyaohua0413@126.com.
Ping Yi, Email: yiping0413@126.com.
Jian-Xin Guo, Email: guojianxin11@126.com.
Yuan-Guo Zhou, Email: zhouyuanguo11@126.com.
Ming Chen, Email: chenming1971@yahoo.com.
Li Li, Email: lililiyuyu@sina.com.
Acknowledgements
We are grateful to Wen Zhao and Xingyun Chen for their technical assistance. This work was supported by a grant (No. 81070505) from the National Natural Science Foundation of China.
References
- Zhang Y, Tycko B. Monoallelic expression of the human H19 gene. Nat Genet. 1992;1:40–44. doi: 10.1038/ng0492-40. [DOI] [PubMed] [Google Scholar]
- Hao Y, Crenshaw T, Moulton T, Newcomb E, Tycko B. Tumour-suppressor activity of H19 RNA. Nature. 1993;365:764–767. doi: 10.1038/365764a0. [DOI] [PubMed] [Google Scholar]
- Steenman MJ, Rainier S, Dobry CJ, Grundy P, Horon IL, Feinberg AP. Loss of imprinting of IGF2 is linked to reduced expression and abnormal methylation of H19 in Wilms’ tumour. Nat Genet. 1994;7:433–439. doi: 10.1038/ng0794-433. [DOI] [PubMed] [Google Scholar]
- Cui H, Hedborg F, He L, Nordenskjöld A, Sandstedt B, Pfeifer-Ohlsson S, Ohlsson R. Inactivation of H19, an imprinted and putative tumor repressor gene, is a preneoplastic event during Wilms’ tumorigenesis. Cancer Res. 1997;57:4469–4473. [PubMed] [Google Scholar]
- Riccio A, Sparago A, Verde G, De Crescenzo A, Citro V, Cubellis MV, Ferrero GB, Silengo MC, Russo S, Larizza L, Cerrato F. Inherited and sporadic epimutations at the IGF2-H19 locus in Beckwith-Wiedemann syndrome and Wilms’ tumor. Endocr Dev. 2009;14:1–9. doi: 10.1159/000207461. [DOI] [PubMed] [Google Scholar]
- Moulton T, Crenshaw T, Hao Y, Moosikasuwan J, Lin N, Dembitzer F, Hensle T, Weiss L, McMorrow L, Loew T. Epigenetic lesions at the H19 locus in Wilms’ tumour patients. Nat Genet. 1994;7:440–447. doi: 10.1038/ng0794-440. [DOI] [PubMed] [Google Scholar]
- Isfort RJ, Cody DB, Kerckaert GA, Tycko B, LeBoeuf RA. Role of the H19 gene in Syrian hamster embryo cell tumorigenicity. Mol Carcinog. 1997;20:189–193. doi: 10.1002/(SICI)1098-2744(199710)20:2<189::AID-MC5>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Yoshimizu T, Miroglio A, Ripoche MA, Gabory A, Vernucci M, Riccio A, Colnot S, Godard C, Terris B, Jammes H, Dandolo L. The H19 locus acts in vivo as a tumor suppressor. Proc Natl Acad Sci USA. 2008;105:12417–12422. doi: 10.1073/pnas.0801540105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabory A, Ripoche MA, Le Digarcher A, Watrin F, Ziyyat A, Forné T, Jammes H, Ainscough JF, Surani MA, Journot L, Dandolo L. H19 acts as a trans regulator of the imprinted gene network controlling growth in mice. Development. 2009;136:3413–3421. doi: 10.1242/dev.036061. [DOI] [PubMed] [Google Scholar]
- Ariel I, Lustig O, Schneider T, Pizov G, Sappir M, De-Groot N, Hochberg A. The imprinted H19 gene as a tumor marker in bladder carcinoma. Urology. 1995;45:335–338. doi: 10.1016/0090-4295(95)80030-1. [DOI] [PubMed] [Google Scholar]
- Ariel I, Sughayer M, Fellig Y, Pizov G, Ayesh S, Podeh D, Libdeh BA, Levy C, Birman T, Tykocinski ML, de Groot N, Hochberg A. The imprinted H19 gene is a marker of early recurrence in human bladder carcinoma. Mol Pathol. 2000;53:320–323. doi: 10.1136/mp.53.6.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanos V, Prus D, Ayesh S, Weinstein D, Tykocinski ML, De-Groot N, Hochberg A, Ariel I. Expression of the imprinted H19 oncofetal RNA in epithelial ovarian cancer. Eur J Obstet Gynecol Reprod Biol. 1999;85:7–11. doi: 10.1016/S0301-2115(98)00275-9. [DOI] [PubMed] [Google Scholar]
- Hibi K, Nakamura H, Hirai A, Fujikake Y, Kasai Y, Akiyama S, Ito K, Takagi H. Loss of H19 imprinting in esophageal cancer. Cancer Res. 1996;56:480–482. [PubMed] [Google Scholar]
- Kondo M, Suzuki H, Ueda R, Osada H, Takagi K, Takahashi T. Frequent loss of imprinting of the H19 gene is often associated with its overexpression in human lung cancers. Oncogene. 1995;10:1193–1198. [PubMed] [Google Scholar]
- Adriaenssens E, Dumont L, Lottin S, Bolle D, Leprêtre A, Delobelle A, Coll J, Curgy JJ. H19 overexpression in breast adenocarcinoma stromal cells is associated with tumor values and steroid receptor status but independent of p53 and Ki-67 expression. Am J Pathol. 1998;153:1597–1607. doi: 10.1016/S0002-9440(10)65748-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lottin S, Adriaenssens E, Dupressoir T, Berteaux N, Montpellier C, Coll J, Dugimont T, Curgy JJ. Overexpression of an ectopic H19 gene enhances the tumorigenic properties of breast cancer cells. Carcinogenesis. 2002;23:1885–1895. doi: 10.1093/carcin/23.11.1885. [DOI] [PubMed] [Google Scholar]
- Tanos V, Ariel I, Prus D, De-Groot N, Hochberg A. H19 and IGF2 gene expression in human normal, hyperplastic, and malignant endometrium. Int J Gynecol Cancer. 2004;14:521–525. doi: 10.1111/j.1048-891x.2004.014314.x. [DOI] [PubMed] [Google Scholar]
- Douc-Rasy S, Barrois M, Fogel S, Ahomadegbe JC, Stéhelin D, Coll J, Riou G. High incidence of loss of heterozygosity and abnormal imprinting of H19 and IGF2 genes in invasive cervical carcinomas. Uncoupling of H19 and IGF2 expression and biallelic hypomethylation of H19. Oncogene. 1996;12:423–430. [PubMed] [Google Scholar]
- Barsyte-Lovejoy D, Lau SK, Boutros PC, Khosravi F, Jurisica I, Andrulis IL, Tsao MS, Penn LZ. The c-Myc oncogene directly induces the H19 noncoding RNA by allele-specific binding to potentiate tumorigenesis. Cancer Res. 2006;66:5330–5337. doi: 10.1158/0008-5472.CAN-06-0037. [DOI] [PubMed] [Google Scholar]
- Lottin S, Vercoutter-Edouart AS, Adriaenssens E, Czeszak X, Lemoine J, Roudbaraki M, Coll J, Hondermarck H, Dugimont T, Curgy JJ. Thioredoxin post-transcriptional regulation by H19 provides a new function to mRNA-like non-coding RNA. Oncogene. 2002;21:1625–1631. doi: 10.1038/sj.onc.1205233. [DOI] [PubMed] [Google Scholar]
- Berteaux N, Lottin S, Monté D, Pinte S, Quatannens B, Coll J, Hondermarck H, Curgy JJ, Dugimont T, Adriaenssens E. H19 mRNA-like noncoding RNA promotes breast cancer cell proliferation through positive control by E2F1. J Biol Chem. 2005;280:29625–29636. doi: 10.1074/jbc.M504033200. [DOI] [PubMed] [Google Scholar]
- Goshen R, Rachmilewitz J, Schneider T, de Groot N, Ariel I, Palti Z. The expression of the H19 and IGF-2 genes during human embryogenesis and placental development. Mol Reprod Dev. 1993;34((4):374–379. doi: 10.1002/mrd.1080340405. [DOI] [PubMed] [Google Scholar]
- Walsh C, Miller SJ, Flam F, Fisher RA, Ohlsson R. Paternally derived H19 is differentially expressed in malignant and nonmalignant trophoblast. Cancer Res. 1995;55:1111–1119. [PubMed] [Google Scholar]
- Kim SJ, Park SE, Lee C, Lee SY, Kim IH, An HJ, Oh YK. Altered imprinting, promoter usage, and expression of insulin-like growth factor-II gene in gestational trophoblastic diseases. Gynecol Oncol. 2003;88:411–418. doi: 10.1016/S0090-8258(02)00143-9. [DOI] [PubMed] [Google Scholar]
- Ariel I, Lustig O, Oyer CE, Elkin M, Gonik B, Rachmilewitz J, Biran H, Goshen R, de Groot N, Hochberg A. Relaxation of imprinting in trophoblastic disease. Gynecol Oncol. 1994;53(2):212–219. doi: 10.1006/gyno.1994.1118. [DOI] [PubMed] [Google Scholar]
- Rachmilewitz J, Elkin M, Rosensaft J, Gelman-Kohan Z, Ariel I, Lustig O, Schneider T, Goshen R, Biran H, de Groot N. H19 expression and tumorigenicity of choriocarcinoma derived cell lines. Oncogene. 1995;11:863–870. [PubMed] [Google Scholar]
- Lu L, Li L, Yu L, Yi P, Li P, Chen X. The expression of imprinted gene H19 and the demethylation effect of 5-aza-2′-deoxycytidine in a choriocarcinoma cell line. Prog Obstet Gynecol. 2008;17:342–345. [Google Scholar]
- Lu L, Li L, Yu L, Zhao D, Yi P, Li P. Effect of 5-aza-2′-deoxycytidine on the invasion of human JEG-3 choriocarcinoma cells and preliminary study on related mechanism. Chongqing Medicine. 2008;37:933–934. [Google Scholar]
- Wilkin F, Paquette J, Ledru E, Hamelin C, Pollak M, Deal CL. H19 sense and antisense transgenes modify insulin-like growth factor-II mRNA levels. Eur J Biochem. 2000;267(13):4020–4027. doi: 10.1046/j.1432-1327.2000.01438.x. [DOI] [PubMed] [Google Scholar]
- Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- Bae SN, Kim J, Lee YS, Kim JD, Kim MY, Park LO. Cytotoxic effect of zinc-citrate compound on choriocarcinoma cell lines. Placenta. 2007;28(1):22–30. doi: 10.1016/j.placenta.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Fausto N, Vail ME, Pierce RH, Franklin CC, Campbell J. In: Malignant Liver Tumours: Basic Concepts and Clinical Management. proceedings of, Falk Workshop; 2002:256–275. Berr F, editor. Dordrecht: Kluwer Academic Publishers; 2003. Interactions between cell proliferation and apoptosis in the development and progression of liver tumours; pp. 20–26. [Google Scholar]
- Shoshani O, Massalha H, Shani N, Kagan S, Ravid O, Madar S, Trakhtenbrot L, Leshkowitz D, Rechavi G, Zipori D. Polyploidization of murine mesenchymal cells is associated with suppression of the long noncoding RNA H19 and reduced tumorigenicity. Cancer Res. 2012;72(24):6403–6413. doi: 10.1158/0008-5472.CAN-12-1155. [DOI] [PubMed] [Google Scholar]
- Viville S, Surani MA. Towards unraveling the Igf2/H19 imprinted domain. Bioessays. 1995;17:835–838. doi: 10.1002/bies.950171004. [DOI] [PubMed] [Google Scholar]
- Esquiliano DR, Guo W, Liang L, Dikkes P, Lopez MF. Placental glycogen stores are increased in mice with H19 null mutations but not in those with insulin or IGF type 1 receptor mutations. Placenta. 2009;30:693–699. doi: 10.1016/j.placenta.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam GR, Cui H, Miller SJ, Flam F, Ohlsson R. Allele-specific in situ hybridization (ASISH) analysis: a novel technique which resolves differential allelic usage of H19 within the same cell lineage during human placental development. Development. 1995;122:839–847. doi: 10.1242/dev.122.3.839. [DOI] [PubMed] [Google Scholar]
- Lustig-Yariv O, Schulze E, Komitowski D, Erdmann V, Schneider T, de Groot N, Hochberg A. The expression of the imprinted genes H19 and IGF-2 in choriocarcinoma cell lines. Is H19 a tumor suppressor gene? Oncogene. 1997;15:169–177. doi: 10.1038/sj.onc.1201175. [DOI] [PubMed] [Google Scholar]
- Kunnimalaiyaan M, Yan S, Wong F, Zhang Y, Chen H. Hairy Enhancer of Split-1 (HES-1), a Notchl effector, inhibits the growth of carcinoid tumor cells. Surgery. 2005;138:1137–1142. doi: 10.1016/j.surg.2005.05.027. [DOI] [PubMed] [Google Scholar]
- Nefedova Y, Sullivan DM, Bolick SC, Dalton WS, Gabrilovich DI. Inhibition of Notch signaling induces apoptosis of myeloma cells and enhances sensitivity to chemotherapy. Blood. 2008;111:2220–2229. doi: 10.1182/blood-2007-07-102632. [DOI] [PubMed] [Google Scholar]
- Mandl M, Slack DN, Keyse SM. Specific inactivation and nuclear anchoring of extracellular signal-regulated kinase 2 by the inducible dual-specificity protein phosphatase DUSP5. Mol Cell Biol. 2005;25:1830–1845. doi: 10.1128/MCB.25.5.1830-1845.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The structure of siRNAs in lentiviral vectors. Table S2 The Ct values from quantitative PCR reactions. Figure S1: Effect of H19 knockdown on the invasion of JAR cells. The invasion rate of cells in the H19 knockdown group and the NC group were 68.85% ± 3.04% and 79.50% ± 5.52%, respectively. H19 knockdown had no significant impact on the invasion rate of JAR cells.