Abstract
LBP [LPS (lipopolysaccharide)-binding protein] and BPI (bactericidal/permeability-increasing protein) are components of the immune system that have been principally studied in mammals for their involvement in defense against bacterial pathogens. These proteins share a basic architecture and residues involved in LPS binding. Putative orthologues, i.e., proteins encoded by similar genes that diverged from a common ancestor, have been found in a number of non-mammalian vertebrate species and several non-vertebrates. Similar to other aspects of immunity, such as the activity of Toll-like receptors and NOD (nucleotide-binding oligomerization domain) proteins, analysis of the conservation of LBPs and BPIs in the invertebrates promises to provide insight into features essential to the form and function of these molecules. This review considers state-of-the-art knowledge in the diversity of the LBP/BPI proteins across the eukaryotes and also considers their role in mutualistic symbioses. Recent studies of the LBPs and BPIs in an invertebrate model of beneficial associations, the Hawaiian bobtail squid Euprymna scolopes’ alliance with the marine luminous bacterium Vibrio fischeri, are discussed as an example of the use of non-vertebrate models for the study of LBPs and BPIs.
Keywords: LBP, BPI, mutualism, invertebrate, Euprymna scolopes, Vibrio fischeri
Introduction
LBP [LPS (lipopolysaccharide)-binding protein] and BPI (bactericidal/permeability-increasing protein) are closely related proteins involved in innate immunity. LBP, which is produced largely by hepatocytes, is secreted into the bloodstream, where it binds LPS and catalyzes the extraction and transfer of individual LPS molecules to CD14, forming a monomeric LPS-CD14 complex that is a key intermediate in delivery of LPS to MD-2/TLR4 (Toll-like receptor 4) and TLR4-dependent cell activation. BPI, which is produced by neutrophils, has higher affinity for LPS and bacteria, is bactericidal and represses inflammation by preventing LBP from delivering LPS to CD14 [1]. Much of our knowledge of the function of LBP and BPI relates to their roles in host response to acute pathogenesis involving Gram-negative bacteria or LPS in the bloodstream. LBP is also believed to play a role in the handling of LPS that has been absorbed across the intestinal barrier (a more common process with high-fat diets), helping to shuttle LPS to lipoproteins and chylomicrons, modulating monocyte/macrophage activation and pro-inflammatory cytokine secretion [2].
Members of this protein family and their relatives also function in other sites that interact with bacteria, most notably along the mucosal surfaces. For example, BPI is not only found in neutrophils and their secretions; it is also produced in human and mouse epithelia, including intestinal epithelia. As in the blood, it has bactericidal effects and blocks endotoxin signaling [3] [4]. Pro-inflammatory mediators have little effect on its expression, but an anti-inflammatory eicosanoid of the lipoxin pathway has been shown to increase epithelial BPI expression [4]. Because diverse assemblages of bacteria promote health of the mucosa, it is likely that the activities of the LBPs, BPIs and their relatives at these sites function not only in defense but also to modulate responses to the essential microbial partners. Although these immune proteins have not been studied in this capacity, their principal ligand, LPS, has. Specifically, host recognition of the normal microbiota in the gut is important for resistance to epithelial injury and critical for gut homeostasis. Oral administration of LPS can mimic the effects of intact bacteria [5]. The circumstantial evidence taken together suggests that members of LPB/BPI protein family participate in control of the normal microbiota.
LBP and BPI have been most extensively studied in mammals, but myriad examples of related proteins occur throughout the animal kingdom and even in other eukaryotes. Several invertebrate species that have LBP and BPI orthologues provide simpler, more tractable models of bacterial interaction with animal epithelia. Unlike the mammalian mucosal surfaces, which typically harbour hundreds to thousands of bacterial phylotypes, these sites in the invertebrates often support partnerships of low diversity, with single to a few microbial phylotypes [6].
Several experimentally tractable invertebrate systems are being exploited as models of animal-bacterial interactions [7] and promise to shed light on the role of LBP and BPI in both pathogenic and non-pathogenic associations. In addition to offering relative simplicity, the study of invertebrate systems reveals features that are evolutionarily conserved across the animal kingdom and, as such, can provide insight into the essential functional features of proteins, such as LBP and BPI. Analogous contributions to the study of toll-like receptors (TLRs) in humans were the result of the discovery and characterization of these proteins in the fruit fly Drosophila melanogaster. Further, similarities between regulation of the transcription pathway in Drosophila development and NF-κB activation in the mammalian immune system prompted investigations that revealed the immune role of Toll in Drosophila and spurred advances in the study of mammalian immune systems [8].
This review will first discuss the relevant features of mammalian LBP and BPI to set the stage for comparisons across the animal kingdom. Then, we will introduce examples of similar proteins in the non-mammalian vertebrates, the invertebrates, and in other eukaryotes. Finally, we will discuss applications of several of these as models, with special emphasis on the squid-vibrio system, for research on the basic nature of the biochemistry and physiology of members of the LBP and BPI protein family.
Characteristics of LBP and BPI in mammals
LBP and BPI have a characteristic, conserved two-domain “boomerang” structure, with an N-terminal domain and a C-terminal domain that share little sequence identity, but are very similar in overall architecture. The N-terminal domain carries out binding of LPS, but the precise LPS-binding site is still a matter of some conjecture. LBP and BPI are part of a wider family of lipid-binding proteins that includes members whose functions are not directly related to bacterial pathogenesis, such as CETP (cholesteryl ester transfer protein) and PLTP (phospholipid transfer protein). While these proteins are not the focus of this discussion, it is worth mentioning that they share this basic architecture [9]. Certain PLUNCs (palate, lung, and nasal epithelium clones) have only a single BPI domain [10].
More basic residues occur in the N-terminus of human BPI than in LBP, a pattern common to the other mammalian LBP/BPI proteins; this feature is believed to promote improved binding to bacterial membranes [1] [11]. However, several basic residues are conserved between LBP and BPI of humans, cattle, mice, rats, and rabbits. These occur at the positions corresponding to human LBP residues R42, R48, K92, K95, and K99 [11]. R94 has also been implicated as important for LPS binding [12], though it is not universally conserved. Based on the crystal structure of BPI [13] [14], these residues are near each other, close to the tip of the protein’s N-terminal domain [11]. Four of these residues (all but K95) form a structural motif for binding LPS that is also present in proteins such as Escherichia coli FhuA, lactoferrin, lysozyme, and LALF (Limulus anti-bacterial and anti-LPS factor) [15], which are proteins that also interact with the surfaces of Gram-negative bacteria [16] [17] [18].
Data from mutagenesis experiments generally support the importance of the conserved residues. The replacement of three positively charged human LBP residues, K92, R94 and K95, with alanine dramatically reduces the protein’s LPS binding capability [12] [19]. The positive charge corresponding to LBP R94 is common, but not universally conserved among mammalian LBP/BPI proteins: in the aligned sequence of human BPI, a glutamine residue fills this position instead [11]. These residues are not necessarily present in more distant members of this protein family, such as human CETP and PLTP, even where the basic architecture of BPI is believed to be retained [9].
Human BPI contains two apolar lipid-binding pockets. In the crystal structure, these contain phosphatidylcholine, but their role in interactions with LPS is unclear [14]. Unlike CD14 and MD-2, complexes of BPI (or LBP) with individual LPS monomers have not been described. Existing evidence strongly suggests that the primary interaction of BPI and LBP with LPS is with interfaces containing large numbers of LPS molecules packed closely together, such as aggregates of purified LPS, outer membranes of Gram-negative bacteria or shed outer membrane vesicles [1]. These interactions appear to be driven by electrostatic interactions between multiple anionic groups clustered within the inner core/lipid A region of LPS and clusters of cationic residues concentrated at the tip of the N-terminal domain. The higher concentration of net basicity of BPI against LBP in this region correlates with the higher affinity of BPI (compared with LBP) for these LPS-rich interfaces [1] [11–14].
Analysis of amino acid sequences of other proteins in the LBP/BPI family suggests that they contain these pockets as well [11]. The LBP/BPI family has a conserved disulfide bond, which is present between residues C132 and C175 of the N-terminal domain in human BPI [11]. This disulfide bond and associated residues are also present in related proteins, such as CETP and PLTP [13] [20].
Beyond mammalian LBP/BPI
In recent years, the study of LBP and BPI has expanded beyond mammals. This family of proteins appears to be ancient. Proteins with BPI-like domains occur even outside the metazoans, such as in Monosiga brevicollis, which belongs to a group of marine choanoflagellate protists considered ancestral to the metazoans [21][22]. Phylostratigraphic analysis, a method in which all available sequence information for the biological world is considered in the construction of phylogenetic relationships, suggests that the CETP family emerged before the last common ancestor of today’s eukaryotes, although this analysis does not make mention of the point at which LBP and BPI emerged [23].
Apart from mammals, members of the LBP/BPI family have been reported in other vertebrates, including fish such as the rainbow trout Onchorhynchus mykiss [24] and Atlantic cod Gadus morhua [20], and birds, such as the chicken, Gallus gallus, which has a BPI but not LBP [25] [26]. They also exist in various invertebrates. Caenorhabditis elegans has multiple proteins with high sequence similarity to LBP [25], and the freshwater snail Biomphalaria glabrata has several variants as well [27] [28]. These few examples serve to demonstrate that LBPs and BPIs are widespread among the non-mammalian animal groups.
A few themes emerge upon analysis of the structure of these non-mammalian proteins. The BPI disulfide bond is ubiquitous, being present in BPI family members as distant as the tunicate Ciona intestinalis [20] and, a simple ClustalW alignment suggests, as far away as the protist M. brevicollis. Many non-mammalian LBP/BPI proteins are predicted to have the basic “boomerang” two-domain fold of human LBP and BPI [25]. It is generally believed that the ancestor of LBP and BPI was a single-domain protein whose gene was duplicated [25], though the two-domain structure is common. Cases of LBP/BPI proteins with one domain have been reported, such as one of three BPI-family proteins in the sponge Amphimedon queenslandica [29]. However, in this case it is believed to have arisen from a neighboring, two-domain LBP/BPI gene.
Some animals have abandoned LBP/BPI altogether. For example, D. melanogaster does not have an LBP/BPI and uses PGRPs (peptidoglycan recognition proteins) to detect Gram-negative bacteria [22]. D. melanogaster also has a GNBP (Gram-negative bacterial-binding protein) capable of binding lipopolysaccharide and β-1,3-glucan [30]. GNBP and the related LGBP (lipopolysaccharide- and β-1,3-glucan-binding protein) are present in numerous arthropods and molluscs, e.g., the mosquito Anopheles gambiae [31] and freshwater crayfish Pacifastacus leniusculus [32]. They are similar to CD14 and resemble defective β-1,3 or β-1,3–1,4 glucanases, possessing a functioning β-1,3 glucan binding site, but missing, in at least some cases, two glutamate residues believed to be important for catalytic activity [31] [33].
In animals that do have an LBP/BPI-like protein, there may be divergence from characteristics conserved in these proteins in mammals. Some features of mammalian LBP and BPI are better conserved than others. The Atlantic cod G. morhua has positively charged residues corresponding to positions 42, 48, 92, and 99 of human BPI or LBP, but not position 95, i.e. it retains most sites believed to be important to LBP/BPI function [20]. More distantly, a protein of this family in the oyster Crassotrea gigas lacks basic residues corresponding to R42 and R48 in human LBP, although it does share basic residues by alignment with LBP and BPI in other positions, including the positions corresponding to K92, K95 and K99 [34]. In contrast, a BPI-like protein obtained from haemocytes of the snail B. glabrata [27] [28], when subjected to a ClustalW alignment by the authors of this review, did not obviously retain any of the positive charges corresponding to human LBP residues R42, R48, K92, K95, and K99, although in several cases positively charged residues were close and may be functionally relevant in LPS binding. These invertebrate proteins also have small insertions or deletions relative to human LBP and BPI, and their structures are not available, and so it is difficult to know exactly how these differences affect binding of LPS and other factors. Both of the invertebrate proteins mentioned in this paragraph appear to retain the conserved disulfide bond. Nevertheless, this variation across the animals does raise questions about how these proteins function, and what elements are required for LBP or BPI activity.
Because of differences from canonical residues in the non-mammalian proteins, and because mammalian LBP and BPI are believed to have arisen from a gene duplication after the radiation of the mammals [24], one might plausibly question whether LBP and BPI are meaningful as distinct categories outside of mammals. Some insight may be obtained from considering other aspects of the proteins’ chemistry and activity. Human BPI has a higher pI (roughly 9.4) than human LBP (roughly 6.3) [35] [36], and it is generally assumed that invertebrate proteins will have a similar pattern. The most effective approach to determine whether isoelectric point correlates with antibacterial activity typical of BPI-like proteins, and thereby to distinguish candidate BPI from an LBP-like protein, is experimental analysis. The protein of the cod G. morhua mentioned above, for example, is up-regulated in the blood and peritoneum of the fish after intraperitoneal injection of bacteria. Its expression pattern is more similar to mammalian BPI than LBP, though to the best of our knowledge it has not been determined whether the protein has bactericidal activity like BPI [37]; the predicted pI of this protein based on information provided by Solstad et al. and computed using Expasy ProtParam [36], is roughly 10, supporting its identity as a BPI. The C. gigas protein binds LPS, is bactericidal and has a predicted isoelectic point of 9.3, making it more functionally similar to human BPI than LBP. This protein is constitutively expressed in epithelial tissues and is also up-regulated in haemocytes in response to bacterial challenge from non-pathogenic marine organisms [34].
We are not aware of a comparably well-characterized non-mammalian protein that functions more similarly to human LBP than BPI, although there are related invertebrate proteins with a charged residue profile more similar to human LBP than human BPI, such as those in the sponge A. queenslandica [29]. It is possible that comparing the LPS-binding functionality of LBP/BPI variants, between and within species, will contribute to improved knowledge of the molecular activities of these proteins.
Toward models of LBP/BPI function in mutualism and development
Several of the animals mentioned above offer the opportunity to study LBP and BPI function in experimentally tractable systems. For example, researchers have begun using molluscs to study LBP and BPI in development. In the snail B. glabrata, egg mass fluid contains significant quantities of LBP/BPI, suggesting its utilization in parental immune protection of offspring [28]. Notably, this protein’s sequence differed from a previously characterized LBP/BPI from haemocytes of the same species, suggesting that different isoforms serve alternative functions in B. glabrata [27] [28]. In the oyster C. gigas, larvae develop as free-swimming forms in the plankton, exposed to ~106 bacteria/ml of seawater [38]. BPI transcript was detected throughout larval development of this species, increasing markedly around the time of the differentiation of epithelia. Experiments in which these larvae were challenged with both Gram-positive and Gram-negative non-pathogenic bacteria, including two Vibrio species and Micrococcus luteus, demonstrated that transcription of this BPI increases in the larval stage in a bacterial-dose dependent manner [39]. These observations suggest a role for the BPI in immune defense during the developmental process, a time when the larvae are highly vulnerable to bacterial settlement.
Another system ripe for study of LBP and BPI is the Hawaiian bobtail squid Euprymna scolopes and its partner, the bioluminescent bacterium Vibrio fischeri. These organisms form a binary (one animal, one bacterial species), easily evaluated and easily manipulated symbiosis, in which bacteria colonize the squid’s light organ and produce light. As such, these organisms provide an excellent model system for a number of research questions, among them the role of LPS in chronic, beneficial colonization of an epithelial surface by Gram-negative bacteria [40].
During the process of bacterial colonization of the light organ, the MAMPs (microbe-associated molecular patterns) of V. fischeri play a central role in symbiont-host communication. Most notably, these molecules induce the transformation from a host-organ morphology that promotes symbiont colonization to one that facilitates the organ’s mature function in bioluminescence production. The most conspicuous feature of this process is the loss of a superficial ciliated field that facilitates harvesting of the symbionts from the seawater. This developmental program involves a series of cellular and biochemical events, including haemocyte trafficking, apoptosis and attenuation of the levels of nitric oxide synthase and its product, nitric oxide. V. fischeri MAMPs work in synergy to trigger all of these events [41] [42]. Specifically, lipid A induces early-stage apoptosis of E. scolopes epithelial cells, as the light organ adapts to the presence of the symbiont [43], and LPS, working in concert with the PGN (peptidoglycan) monomer, triggers the completion of the apoptotic process. This pattern of LPS-induced apoptosis is not unique to the Euprymna/Vibrio system, as the ability of LPS to induce host cell apoptosis has been investigated in other species [44] [45], generally with a focus on pathogenesis rather than mutualistic association.
Many of the details of this morphogenetic process in the squid-Vibrio symbiosis, however, are still incompletely understood. Among these, given the apparent importance of LPS in the developmental process, is how E. scolopes detects the presence of bacterial LPS in the light organ and how LPS functions in synergy with the PGN monomer. As it turns out, E. scolopes possesses at least three light-organ proteins in the LBP/BPI family, which have recently been sequenced (Fig. 1). The expression of one of these proteins, EsLBP1, has been evaluated in the context of establishment of mutualism. Eighteen hours into colonization, at which point apoptosis and epithelial regression are well underway, eslbp1 mRNA is up-regulated roughly 9-fold over comparable, symbiont-free control animals. EsLBP1 protein itself has been detected in the animal’s bacteria-containing crypt spaces at the same time point (Fig. 2) [46].
Fig. 1.
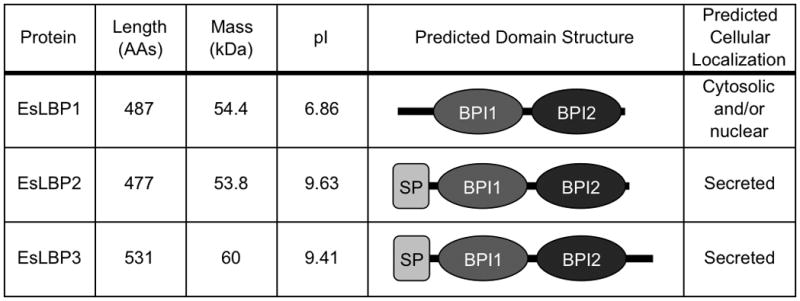
Characteristics of the predicted E. scolopes LBP proteins. The derived amino acid sequence of each of the EsLBP cDNAs was analyzed for biochemical parameters and protein family domains using ExPASy ProtParam [36] and the SMART algorithm [50]. AAs, amino acids. NCBI accession numbers for sequences: JF514880, JF514881, JF514882.
Fig. 2.
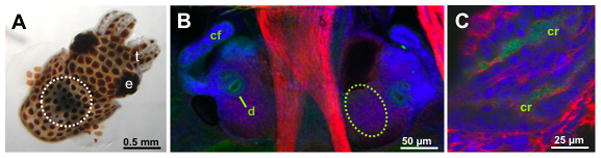
Immunocytochemical localization of EsLBP1 in the light organ of juvenile E. scolopes. A. The position of the organ in the whole, live juvenile squid. The organ can be seen through the translucent dorsal surface of the animal as a dark region in the center of the body (white dashed circle). B. Confocal microscopy image of the ventral surface of the juvenile light organ. Each lateral lobe of the organ bears a complex ciliated field (cf), which promotes harvesting of V. fischeri from the environment, and ducts (d), passageways through which the symbiont cells enter host tissues. Anti-EsLBP1 antibodies (green) label regions along the apical surfaces of the epithelia of the cells of the ciliated fields and the pores. The dashed oval region is where deeper images were taken in (C). C. Confocal image of the deep crypt (cr) region of the light organ. Anti-EsLBP1 antibodies (green) label the deep crypt spaces where the symbiont cells reside following colonization of the organ. In (B, C), nuclei, blue (TOTO-3); actin, red (rhodamine phalloidin). e, eye; t, tentacles. For confocal microscopy methods, see [46].
The patterns of occurrence of three distinct EsLBPs in the light organ may enable the tissues to use LPS to signal the variety of processes in which it is implicated. This type of strategy has already been described in this system. Specifically, previous studies of the symbiosis have characterized some of the EsPGRPs in the light organ; genes encoding four members of this family are expressed in the hatchling organ. In-depth analyses of two of the proteins, EsPGRP1 and 2, have demonstrated that a mechanism by which the animal can respond to PGN over the trajectory of early development is to deploy the isoforms at different times and in different locations in the organ [47] [48]. The biochemical properties of both the EsLBPs and the EsPGRPs, along with their presence and relative importance in haemocytes, the light organ epithelium and extracellular crypts, are at present being evaluated. The results of such studies promise to shed light on how animal epithelia interact with the LPS and PGN as individual MAMPs, as well as how these MAMPs synergize to trigger host responses.
Conclusion
In light of the fact that all animal body plans arose in the Cambrian, some 520–540 million years ago [49], in the context of the bacteria-rich environment of the oceans, it is not surprising that the animals have developed mechanisms to respond to the surface molecules of members of the bacterioplankton. These responses may involve exploiting bacterial molecules for defensive purposes or as signals for normal development and homeostasis. Analyses of the current genomic databases are revealing that members of the LBP/BPI family of proteins represent an ancient means of recognizing MAMPs. Proteins similar to mammalian LBP and BPI exist in diverse organisms, with some residues strictly conserved and others forming common motifs. Opportunities exist to examine these proteins and determine core features of LBP and BPI. Additionally, these discoveries are occurring as we become increasingly aware of the full spectrum of symbiosis, from pathogenesis to mutualism.
Acknowledgments
We wish to thank Natacha Kremer, Mallory Agard and Ned Ruby for assistance and advice. This work was supported by grants from the National Institutes of Health (R01 AI50661) and National Science Foundation (IOS 0817232).
Abbreviations
- BPI
Bactericidal/permeability-increasing protein
- CETP
Cholesteryl ester transfer protein
- LBP
Lipopolysaccharide-binding protein
- MAMP
Microbe-associated molecular pattern
- PGN
Peptidoglycan
- PLTP
Phospholipid transfer protein
- TLR
Toll-like receptor
References
- 1.Weiss J. Bactericidal/permeability-increasing protein (BPI) and lipopolysaccharide-binding protein (LBP): Structure, function and regulation in host defence against gram-negative bacteria. Biochem Soc Trans. 2003;31:785–790. doi: 10.1042/bst0310785. [DOI] [PubMed] [Google Scholar]
- 2.Manco M, Putignani L, Bottazzo GF. Gut microbiota, lipopolysaccharides, and innate immunity in the pathogenesis of obesity and cardiovascular risk. Endocr Rev. 2010;31:817–844. doi: 10.1210/er.2009-0030. [DOI] [PubMed] [Google Scholar]
- 3.Canny G, Levy O, Furuta GT, Narravula-Alipati S, Sisson RB, Serhan CN, Colgan SP. Lipid mediator-induced expression of bactericidal/permeability-increasing protein (BPI) in human mucosal epithelia. Proc Natl Acad Sci U S A. 2002;99:3902–3907. doi: 10.1073/pnas.052533799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Canny G, Cario E, Lennartsson A, Gullberg U, Brennan C, Levy O, Colgan SP. Functional and biochemical characterization of epithelial bactericidal/permeability-increasing protein. Am J Physiol Gastrointest Liver Physiol. 2006;290:G557–67. doi: 10.1152/ajpgi.00347.2005. [DOI] [PubMed] [Google Scholar]
- 5.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 6.Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449:811–818. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosenstiel P, Philipp EE, Schreiber S, Bosch TC. Evolution and function of innate immune receptors--insights from marine invertebrates. J Innate Immun. 2009;1:291–300. doi: 10.1159/000211193. [DOI] [PubMed] [Google Scholar]
- 8.Imler J, Hoffmann JA. Toll receptors in innate immunity. Trends Cell Biol. 2001;11:304–311. doi: 10.1016/s0962-8924(01)02004-9. [DOI] [PubMed] [Google Scholar]
- 9.Qiu X, Mistry A, Ammirati MJ, Chrunyk BA, Clark RW, Cong Y, Culp JS, Danley DE, Freeman TB, Geoghegan KF, Griffor MC, Hawrylik SJ, Hayward CM, Hensley P, Hoth LR, Karam GA, Lira ME, Lloyd DB, McGrath KM, Stutzman-Engwall KJ, Subashi AK, Subashi TA, Thompson JF, Wang IK, Zhao H, Seddon AP. Crystal structure of cholesteryl ester transfer protein reveals a long tunnel and four bound lipid molecules. Nat Struct Mol Biol. 2007;14:106–113. doi: 10.1038/nsmb1197. [DOI] [PubMed] [Google Scholar]
- 10.Bingle CD, LeClair EE, Havard S, Bingle L, Gillingham P, Craven CJ. Phylogenetic and evolutionary analysis of the PLUNC gene family. Protein Sci. 2004;13:422–430. doi: 10.1110/ps.03332704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beamer LJ, Carroll SF, Eisenberg D. The BPI/LBP family of proteins: A structural analysis of conserved regions. Protein Sci. 1998;7:906–914. doi: 10.1002/pro.5560070408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lamping N, Hoess A, Yu B, Park TC, Kirschning CJ, Pfeil D, Reuter D, Wright SD, Herrmann F, Schumann RR. Effects of site-directed mutagenesis of basic residues (arg 94, lys 95, lys 99) of lipopolysaccharide (LPS)-binding protein on binding and transfer of LPS and subsequent immune cell activation. J Immunol. 1996;157:4648–4656. [PubMed] [Google Scholar]
- 13.Beamer LJ, Carroll SF, Eisenberg D. Crystal structure of human BPI and two bound phospholipids at 2.4 angstrom resolution. Science. 1997;276:1861–1864. doi: 10.1126/science.276.5320.1861. [DOI] [PubMed] [Google Scholar]
- 14.Beamer LJ, Carroll SF, Eisenberg D. The three-dimensional structure of human bactericidal/permeability-increasing protein: Implications for understanding protein-lipopolysaccharide interactions. Biochem Pharmacol. 1999;57:225–229. doi: 10.1016/s0006-2952(98)00279-2. [DOI] [PubMed] [Google Scholar]
- 15.Ferguson AD, Welte W, Hofmann E, Lindner B, Holst O, Coulton JW, Diederichs K. A conserved structural motif for lipopolysaccharide recognition by procaryotic and eucaryotic proteins. Structure. 2000;8:585–592. doi: 10.1016/s0969-2126(00)00143-x. [DOI] [PubMed] [Google Scholar]
- 16.Ellison RT, Giehl TJ. Killing of gram-negative bacteria by lactoferrin and lysozyme. J Clin Invest. 1991;88:1080–1091. doi: 10.1172/JCI115407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Killmann H, Benz R, Braun V. Properties of the FhuA channel in the escherichia coli outer membrane after deletion of FhuA portions within and outside the predicted gating loop. J Bacteriol. 1996;178:6913–6920. doi: 10.1128/jb.178.23.6913-6920.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weiss CA, Wasiluk KR, Kellogg TA, Dunn DL. Bactericidal and endotoxin neutralizing activity of a peptide derived from limulus antilipopolysaccharide factor. Surgery. 2000;128:339–344. doi: 10.1067/msy.2000.108061. [DOI] [PubMed] [Google Scholar]
- 19.Reyes O, Vallespi MG, Garay HE, Cruz LJ, Gonzalez LJ, Chinea G, Buurman W, Arana MJ. Identification of single amino acid residues essential for the binding of lipopolysaccharide (LPS) to LPS binding protein (LBP) residues 86–99 by using an ala-scanning library. J Pept Sci. 2002;8:144–150. doi: 10.1002/psc.375. [DOI] [PubMed] [Google Scholar]
- 20.Stenvik J, Solstad T, Strand C, Leiros I, Jorgensen TTO. Cloning and analyses of a BPI/LBP cDNA of the atlantic cod (gadus morhua L.) Dev Comp Immunol. 2004;28:307–323. doi: 10.1016/j.dci.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 21.King N, Westbrook MJ, Young SL, Kuo A, Abedin M, Chapman J, Fairclough S, Hellsten U, Isogai Y, Letunic I, Marr M, Pincus D, Putnam N, Rokas A, Wright KJ, Zuzow R, Dirks W, Good M, Goodstein D, Lemons D, Li W, Lyons JB, Morris A, Nichols S, Richter DJ, Salamov A, Sequencing JG, Bork P, Lim WA, Manning G, Miller WT, McGinnis W, Shapiro H, Tjian R, Grigoriev IV, Rokhsar D. The genome of the choanoflagellate monosiga brevicollis and the origin of metazoans. Nature. 2008;451:783–788. doi: 10.1038/nature06617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Irazoqui JE, Urbach JM, Ausubel FM. Evolution of host innate defence: Insights from caenorhabditis elegans and primitive invertebrates. Nat Rev Immunol. 2010;10:47–58. doi: 10.1038/nri2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Domazet-Loso T, Brajkovic J, Tautz D. A phylostratigraphy approach to uncover the genomic history of major adaptations in metazoan lineages. Trends Genet. 2007;23:533–539. doi: 10.1016/j.tig.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 24.Inagawa H, Honda T, Kohchi C, Nishizawa T, Yoshiura Y, Nakanishi T, Yokomizo Y, Soma G. Cloning and characterization of the homolog of mammalian lipopolysaccharide-binding protein and bactericidal permeability-increasing protein in rainbow trout oncorhynchus mykiss. J Immunol. 2002;168:5638–5644. doi: 10.4049/jimmunol.168.11.5638. [DOI] [PubMed] [Google Scholar]
- 25.Beamer LJ, Fischer D, Eisenberg D. Detecting distant relatives of mammalian LPS-binding and lipid transport proteins. Protein Sci. 1998;7:1643–1646. doi: 10.1002/pro.5560070721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chiang SC, Veldhuizen EJ, Barnes FA, Craven CJ, Haagsman HP, Bingle CD. Identification and characterisation of the BPI/LBP/PLUNC-like gene repertoire in chickens reveals the absence of a LBP gene. Dev Comp Immunol. 2010 doi: 10.1016/j.dci.2010.09.013. [In press] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitta G, Galinier R, Tisseyre P, Allienne JF, Girerd-Chambaz Y, Guillou F, Bouchut A, Coustau C. Gene discovery and expression analysis of immune-relevant genes from biomphalaria glabrata hemocytes. Dev Comp Immunol. 2005;29:393–407. doi: 10.1016/j.dci.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 28.Hathaway JJ, Adema CM, Stout BA, Mobarak CD, Loker ES. Identification of protein components of egg masses indicates parental investment in immunoprotection of offspring by biomphalaria glabrata (gastropoda, mollusca) Dev Comp Immunol. 2010;34:425–435. doi: 10.1016/j.dci.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gauthier ME, Du Pasquier L, Degnan BM. The genome of the sponge amphimedon queenslandica provides new perspectives into the origin of toll-like and interleukin 1 receptor pathways. Evol Dev. 2010;12:519–533. doi: 10.1111/j.1525-142X.2010.00436.x. [DOI] [PubMed] [Google Scholar]
- 30.Kim YS, Ryu JH, Han SJ, Choi KH, Nam KB, Jang IH, Lemaitre B, Brey PT, Lee WJ. Gram-negative bacteria-binding protein, a pattern recognition receptor for lipopolysaccharide and beta-1,3-glucan that mediates the signaling for the induction of innate immune genes in drosophila melanogaster cells. J Biol Chem. 2000;275:32721–32727. doi: 10.1074/jbc.M003934200. [DOI] [PubMed] [Google Scholar]
- 31.Warr E, Das S, Dong Y, Dimopoulos G. The gram-negative bacteria-binding protein gene family: Its role in the innate immune system of anopheles gambiae and in anti-plasmodium defence. Insect Mol Biol. 2008;17:39–51. doi: 10.1111/j.1365-2583.2008.00778.x. [DOI] [PubMed] [Google Scholar]
- 32.Lee SY, Wang R, Soderhall K. A lipopolysaccharide- and beta-1,3-glucan-binding protein from hemocytes of the freshwater crayfish pacifastacus leniusculus, purification, characterization, and cDNA cloning. J Biol Chem. 2000;275:1337–1343. doi: 10.1074/jbc.275.2.1337. [DOI] [PubMed] [Google Scholar]
- 33.Lee WJ, Lee JD, Kravchenko VV, Ulevitch RJ, Brey PT. Purification and molecular cloning of an inducible gram-negative bacteria-binding protein from the silkworm, bombyx mori. Proc Natl Acad Sci U S A. 1996;93:7888–7893. doi: 10.1073/pnas.93.15.7888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gonzalez M, Gueguen Y, Destoumieux-Garzon D, Romestand B, Fievet J, Pugniere M, Roquet F, Escoubas JM, Vandenbulcke F, Levy O, Saune L, Bulet P, Bachere E. Evidence of a bactericidal permeability increasing protein in an invertebrate, the crassostrea gigas cg-BPI. Proc Natl Acad Sci U S A. 2007;104:17759–17764. doi: 10.1073/pnas.0702281104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eilers B, Mayer-Scholl A, Walker T, Tang C, Weinrauch Y, Zychlinsky A. Neutrophil antimicrobial proteins enhance shigella flexneri adhesion and invasion. Cell Microbiol. 2010;12:1134–1143. doi: 10.1111/j.1462-5822.2010.01459.x. [DOI] [PubMed] [Google Scholar]
- 36.Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Solstad T, Stenvik J, Jorgensen TO. mRNA expression patterns of the BPI/LBP molecule in the atlantic cod (gadus morhua L.) Fish Shellfish Immunol. 2007;23:260–271. doi: 10.1016/j.fsi.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 38.Glavin DP, Cleaves HJ, Schubert M, Aubrey A, Bada JL. New method for estimating bacterial cell abundances in natural samples by use of sublimation. Appl Environ Microbiol. 2004;70:5923–5928. doi: 10.1128/AEM.70.10.5923-5928.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tirape A, Bacque C, Brizard R, Vandenbulcke F, Boulo V. Expression of immune-related genes in the oyster crassostrea gigas during ontogenesis. Dev Comp Immunol. 2007;31:859–873. doi: 10.1016/j.dci.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 40.McFall-Ngai M. Host-microbe symbiosis: The squid-vibrio association--a naturally occurring, experimental model of animal/bacterial partnerships. Adv Exp Med Biol. 2008;635:102–112. doi: 10.1007/978-0-387-09550-9_9. [DOI] [PubMed] [Google Scholar]
- 41.Koropatnick TA, Engle JT, Apicella MA, Stabb EV, Goldman WE, McFall-Ngai MJ. Microbial factor-mediated development in a host-bacterial mutualism. Science. 2004;306:1186–1188. doi: 10.1126/science.1102218. [DOI] [PubMed] [Google Scholar]
- 42.Altura MA, Stabb EV, Goldman W, Apicella MA, McFall-Ngai MJ. Attenuation of host NO production by MAMPs potentiates development of the host in the squid-vibrio symbiosis. 2011. [In press] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Foster JS, Apicella MA, McFall-Ngai MJ. Vibrio fischeri lipopolysaccharide induces developmental apoptosis, but not complete morphogenesis, of the euprymna scolopes symbiotic light organ. Dev Biol. 2000;226:242–254. doi: 10.1006/dbio.2000.9868. [DOI] [PubMed] [Google Scholar]
- 44.Norimatsu M, Ono T, Aoki A, Ohishi K, Tamura Y. In-vivo induction of apoptosis in murine lymphocytes by bacterial lipopolysaccharides. J Med Microbiol. 1995;43:251–257. doi: 10.1099/00222615-43-4-251. [DOI] [PubMed] [Google Scholar]
- 45.Norimatsu M, Ono T, Aoki A, Ohishi K, Takahashi T, Watanabe G, Taya K, Sasamoto S, Tamura Y. Lipopolysaccharide-induced apoptosis in swine lymphocytes in vivo. Infect Immun. 1995;63:1122–1126. doi: 10.1128/iai.63.3.1122-1126.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chun CK, Troll JV, Koroleva I, Brown B, Manzella L, Snir E, Almabrazi H, Scheetz TE, Bonaldo Mde F, Casavant TL, Soares MB, Ruby EG, McFall-Ngai MJ. Effects of colonization, luminescence, and autoinducer on host transcription during development of the squid-vibrio association. Proc Natl Acad Sci U S A. 2008;105:11323–11328. doi: 10.1073/pnas.0802369105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Troll JV, Bent EH, Pacquette N, Wier AM, Goldman WE, Silverman N, McFall-Ngai MJ. Taming the symbiont for coexistence: A host PGRP neutralizes a bacterial symbiont toxin. Environ Microbiol. 2010 doi: 10.1111/j.1462-2920.2009.02121.x. [In press] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Troll JV, Adin DM, Wier AM, Paquette N, Silverman N, Goldman WE, Stadermann FJ, Stabb EV, McFall-Ngai MJ. Peptidoglycan induces loss of a nuclear peptidoglycan recognition protein during host tissue development in a beneficial animal-bacterial symbiosis. Cell Microbiol. 2009;11:1114–1127. doi: 10.1111/j.1462-5822.2009.01315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Budd GE. The earliest fossil record of the animals and its significance. Philos Trans R Soc Lond B Biol Sci. 2008;363:1425–1434. doi: 10.1098/rstb.2007.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Letunic I, Doerks T, Bork P. SMART 6: recent updates and new developments. Nucleic Acids Res. 2009;37(Database issue):D229–32. doi: 10.1093/nar/gkn808. [DOI] [PMC free article] [PubMed] [Google Scholar]


