Abstract
Purpose.
MicroRNA-155 (miR-155) and STAT3 are implicated in uveitis and pathogenic mechanisms of CNS autoimmune diseases. In our study, we used miR-155−/− mice and mice with targeted STAT3 deletion in T cells (CD4-STAT3KO) to investigate roles of miR-155 and STAT3 in the development of experimental autoimmune uveitis (EAU), a mouse model of human uveitis.
Methods.
We induced EAU in WT, miR-155−/−, or CD4-STAT3KO mice by immunization with interphotoreceptor retinoid-binding protein/complete Freund's adjuvant (IRBP/CFA) or adoptive transfer of T cells. EAU was assessed by funduscopy and histology. RNA expression was analyzed by quantitative PCR (qPCR), while cytokine production was assessed by fluorescence-activated cell sorting (FACS).
Results.
We used a combination of genomic and genetic tools to provide the first evidence that STAT3 binds directly to the miR-155 locus and that STAT3 is required for miR-155 expression. Furthermore, STAT3-dependent increase in miR-155 expression in vivo correlated temporally with onset of EAU, and miR-155−/− or CD4-STAT3KO mice did not suffer EAU. CD4+ lymph node cells from IRBP-immunized WT mice transferred EAU to naïve wild-type (WT) and miR-155−/− mice, while miR-155−/− IRBP-specific T cells did not.
Conclusions.
Although miR-155 and STAT3 have been implicated in the etiology of multiple sclerosis (MS), uveitis, or rheumatoid arthritis, their exact roles in these diseases are unclear. We show here for the first time to our knowledge that STAT3 regulates miR-155 expression in Th17 cells. We show further that STAT3 and miR-155 form an axis that promotes the expansion of pathogenic Th17 cells that mediate uveitis. Thus, STAT3 and miR-155 may be therapeutic targets for treating uveitis and other Th17-mediated inflammatory disorders.
Keywords: EAU, STAT3, miR-155, uveitis, Th17 cells
We show for the first time, to our knowledge, that STAT3 regulates miR-155 expression by Th17 cells and a STAT3/miR-155 axis mediates uveitis by promoting Th17 expansion. Data suggest that therapeutic strategies that combine miR-155 inhibition with blockade of STAT3 signaling may ameliorate Th17-mediated autoimmune disease.
Introduction
Mammalian microRNAs (miRNAs) regulate diverse cellular processes, including inflammation by targeting cognate mRNAs for deadenylation or translational repression.1,2 miR-155-5p (miR-155) now has emerged as an important regulator of the stability and fitness of regulatory T cells,3 and recent studies indicate a genetic association and altered gene expression of miR-155 in multiple sclerosis patients.4 Active lesions in the brain of patients with MS are characterized by a strong upregulation of miR-1555 and miR-155 also has been implicated in arthritis.6 Support for the involvement of miR-155 in etiology of organ-specific autoimmune diseases has come from studies of experimental autoimmune encephalomyelitis (EAE) and experimental arthritis models, and in a recent report it was shown that miR-155−/− mice are highly resistant to EAE.2 However, in contrast to the findings in rheumatoid arthritis, multiple sclerosis (MS), EAE, and experimental arthritis, miR-155 knockout mice suffer from an exaggerated autoimmune response in the lungs, with marked leukocyte invasion and increased lung airway remodeling,7 suggesting that miR-155 may confer protection in asthma. In the eye, expression of miR-155 has been detected in the retinal pigment epithelium, a tissue that has a critical role in the pathogenesis of retinal degenerative diseases.8 Microarray analysis revealed substantial upregulation of miR-155 expression in retinal pigment epithelial cells in response to the inflammatory cytokines IFN-γ, TNF-α, and IL-1β, indicating a potential role of miR-155 in ocular inflammatory diseases.8 Moreover, in a recent study a 2-fold reduction of miR-155 was observed in peripheral blood mononuclear cells (PBMCs) and dendritic cells (DCs) of patients with active ocular Behcet's disease,9 suggesting the potential involvement of miR-155 in human uveitis. Collectively, these observations suggest that miR-155 may be regulated differentially in distinct autoimmune disorders and cell types, but its specific roles have yet to be delineated clearly.
Given that the direct targets of miR-155 may include a variety of genes, such as TAB2, c-Maf, PU.1, SHIP1, AGTR1, MMP3, AID, SOCS1, BACH1, CEBPB, CSFR, and JARID2 (available in the public domain at http://www.targetscan.org/ and http://pictar.mdc-berlin.de/),2 with distinct functions in lymphocytes and antigen presenting cells, it still is not clear what signaling pathways or transcription factors regulate miR-155 expression and functions, or how miR-155 promotes autoimmune inflammation. However, the involvement of Th17 cells and miR-155 in the pathogenesis of clinical and experimental uveitis, rheumatoid arthritis, and multiple sclerosis suggests that a common set of factors may regulate Th17 development and miR-155 expression.
In our study, we sought to clarify the role of miR-155 and STAT3 in ocular inflammation using experimental autoimmune uveitis (EAU), the mouse model of human uveitis. Since mice with targeted deletion of STAT3 in CD4+ T cells cannot generate Th17 and do not suffer EAE or EAU, and miR-155 knockout mice are resistant to EAE, we examined possible convergence of miR-155- and STAT3-induced pathways in regulating Th17 cells and autoimmune pathologies. We show here that the expression of miR-155 and IL-17 by Th17 cells is dependent on STAT3, and that enhancement of autoimmune pathology in EAU requires STAT3 and miR-155.
Materials and Methods
Mice
miR-155−/− mice were described previously10 and purchased from The Jackson Laboratory (Bar Harbor, ME). For this study, miR-155−/− mouse strain (B6.Cg-Mir155tm1.1Rsky/J, stock #007745; The Jackson Laboratory) were backcrossed further to C57BL/6NTac (Taconic model #B6-F and B6-M; Taconic, Hudson, NY) for a total of 12 generations. WT C57BL/6NTac mice were used as controls. Mice with conditional deletion of Stat3 in CD4+ T cells (CD4-STAT3KO) were derived by breeding Stat3fl/fl (MGI: 2384272; The Jackson Laboratory) with CD4-Cre (Taconic model #4196-F/B6-Cg-Tg[CD4-cre]1Cwi N9; Taconic) mouse strains, and both strains have been backcrossed extensively to C57BL/6J background.11–13 Littermate Stat3fl/fl mice, in C57BL/6J background, were used as wild type (WT) controls. Mice were maintained and treated in accordance with National Eye Institute (NEI), National Institute for Allergies and Infectious Diseases (NIAID), and Animal Care and Use Committee guidelines. All animal studies conformed to the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research.
Induction of EAU by Active Immunization
Mice were immunized with 150 μg interphotoreceptor retinoid-binding protein (IRBP) and 300 μg of human IRBP peptide (1–20) in 0.2 mL emulsion 1:1 vol/vol with Complete Freund's adjuvant (CFA) containing Mycobacterium tuberculosis strain H37RA (2.5 mg/mL). The mice also received Bordetella pertussis toxin (0.2 μg/mouse) concurrent with immunization and clinical disease was established by funduscopy as described previously.14 Eyes for histologic or funduscopic evaluation were harvested 0, 7, 10, 14, or 21 days after immunization. For histology, eyes were harvested, fixed in 4% glutaraldehyde for 30 minutes, and transferred to 10% buffered formalin. After adequate fixation, specimens were dehydrated through graded alcohols and embedded in methacrylate. Serial vertical sections through the papillary–optic nerve plane were cut and stained with hematoxylin and eosin as described previously.14 Photographs of representative sections were taken on a photomicroscope (Carl Zeiss AG, Oberkochen, Germany). Clinical disease was established and scored by funduscopy as described.14,15 Briefly, following intraperitoneal (IP) injection of ketamine (1.4 mg/mouse) and xylazine (0.12 mg/mouse), pupils were dilated by topical administration of 1% tropicamide ophthalmic solution (Alcon, Inc., Fort Worth, TX). To avoid a subjective bias, evaluation of the fundus photographs was conducted without knowledge of the mouse identity by a masked observer. At least six images (2 posterior central retinal views, 4 peripheral retinal views) were taken from each eye by positioning the endoscope, and viewing from superior, inferior, lateral, and medial fields, and each individual lesion was identified, mapped, and recorded. The clinical grading system for retinal inflammation was as established previously.16
Induction of EAU in miR-155−/− Mice by Adoptive Transfer of WT Uveitogenic T Cells
EAU was induced in WT C57BL/6 mice by immunization with IRBP in CFA, and disease was confirmed by funduscopy. Donor mice were sacrificed, and CD4+ T cells were isolated from draining lymph nodes and spleen of mice with EAU, and stimulated with IRBP (20 μg/mL) for 3 days. The IRBP-specific T cells then were transferred intravenously (IV) to naive miR-155−/− recipient mice at 10 × 106 cells/mouse. Ten days after the T-cell transfer, disease was assessed by funduscopy.
Isolation, Propagation and Characterization of CD4+ T cells
CD4+ T cells were activated with plate-bound anti-CD3 Ab (3 μg/mL; BD BioSciences, San Jose, CA) and anti-CD28 Ab (3 μg/mL) in complete medium. For propagation under Th1 condition, medium was supplemented with anti–IL-4 Ab (10 μg/mL) and IL-12 (10 ng/mL; Pepro Tech, Inc., Rocky Hill, NJ). For Th2 condition the medium contained IL-4 (10 ng/mL), anti–IFN-γ Ab (10 μg/mL), and anti–IL-12 Ab (10 μg/mL). For Th17 condition the medium contained TGF-β (2 ng/mL), IL-6 (10 ng/mL), IL-1β (10 ng/mL), anti–IFN-γ Ab (10 μg/mL), and anti–IL-4 Ab (10 μg/mL), while Treg polarization medium contained TGF-β (5 ng/mL) and IL-2 (10 ng/mL). Where indicated CD4+ T cells were analyzed without stimulation, while most cultures were stimulated for 4 days. For intracellular cytokine detection, freshly isolated or cultured CD4+ T cells were restimulated for 5 hours with PMA (20 ng/mL)/ionomycin (1 μM) in the presence of Golgi-stop at the recommended concentrations (BD Pharmingen, San Diego, CA). The cells were surface stained with labeled antibodies (Abs), fixed, permeabilized, and stained with the requisite Abs using the BD Biosciences Cytofix/Cytoperm kit according to the manufacturer's instructions. Fluorescence-activated cell sorting (FACS) analysis using anti-CD3, CD4, IFN-γ, IL-17, Foxp3, mAbs, and corresponding isotype control Abs (BD Pharmingen) was performed on Becton Dickinson FACSCalibur or LSRII (BD Biosciences) as described previously.15
Quantitative (qPCR) and Semi-Quantitative RT-PCR Analysis
Complementary DNA (cDNA) was generated as described previously17 and each gene-specific primer pair used for RT-PCR analysis spans at least an intron. RT-qPCR analysis was performed using primers and probes from Applied Biosystems, Inc. (Foster City, CA). The relative mRNA expression levels were normalized to the levels of the housekeeping gene, GAPDH.
miRNA Analysis
Expression profiling of miRNAs by deep sequencing was described previously.18 For microRNA expression validation, total RNA was isolated from cell samples using mirVana (Applied Biosystems, Inc.) isolation kit, and cDNA was prepared using TaqMan microRNA reverse transcription kit (Applied Biosystems, Inc.) with primer and probe sets for miR-155, and U6 as a housekeeping control. qPCR was performed on a 7900HT fast real time PCR system (Applied Biosystems, Inc.).
Chromatin Immunoprecipitation (ChIP) Analyses
We downloaded GSE26552 from National Center for Biotechnology Information Gene Expression Omnibus (NCBI GEO, available in the public domain at http://www.ncbi.nlm.nih.gov/geo/) to view STAT3 ChIP-seq data that were reported previously.19 ChIP-seq reads were realigned to the mouse reference genome (NCBI 37, mm9) using BowTie version 0.12.7. Duplicate reads were removed using SAMtools 0.1.16. Only reads with unique matches were retained and converted into BED files for visualization on a local mirror of the University of California, Santa Cruz (UCSC) Genome Browser (available in the public domain at http://genome.ucsc.edu/). ChIP validation experiments were performed as described previously.20 qPCR was performed using these primers: 5′-TGAACCGTGGCTGTGTTA and 5′-GAAGGCAAGGTAGACTTCAGC.
Results
miR-155−/− Mice are Resistant to EAU
Th17 cells are implicated in pathogenesis of EAU and EAE that are animal models of uveitis and multiple sclerosis, respectively.13,21 In a recent report, miR-155 expression was linked to the development of Th17 cells and miR-155−/− mice were found to be highly resistant to EAE, suggesting a role for miR-155 in CNS autoimmune diseases.2 In our study, we have used the mouse EAU model to investigate the potential involvement of miR-155 in uveitis. Despite global deletion of miR-155 and the potential functional defects in multiple cell types, the miR-155 KO mice do not suffer from spontaneous autoimmunity or inflammatory disease, and, therefore, were suitable for our study. EAU was induced in WT C57BL/6NTac or miR-155−/− mice (on C57BL/6NTac background) and disease severity was assessed by histologic analysis of the retina. Histology of eye sections 21 days after immunization with IRBP revealed that all WT mice had severe EAU, characterized by substantial infiltration of inflammatory cells into the retina (Fig. 1A). Development of numerous massive retinal folds (indicated by a blue asterisk), a hallmark of severe uveitis, is consistent with the higher disease scores noted for the WT mice (Fig. 1B). In contrast, none of the miR-155−/− mice had EAU as evidenced by the apparent absence of inflammatory cells in vitreous or retina (Fig. 1A). The very low EAU scores of miR-155−/− retinas coincided with absence of retinal folds, a hallmark of severe EAU (Fig. 1B). However, it is important to note that our studies were performed on mice that were on the C57Bl/6N, as well as, the C57BL/6J backgrounds, and we cannot rule out an influence of background differences between these 2 strains.22 To investigate whether the resistance of miR-155−/− mice to EAU was due to the loss of miR-155 in T helper cells, we isolated T cells from the lymph node (LN) and spleen of WT mice with EAU, restimulated them in vitro with IRBP, and transferred the uveitogenic cells into unimmunized WT or miR-155−/− mice. Ten days after adoptive transfer of the uveitogenic WT-cells, the WT and miR-155−/− mouse strains exhibited signs of EAU (Fig. 1C), and had comparable EAU scores (Fig. 1D) as determined by funduscopy. Since uveitis develops upon transfer of WT uveitogenic T cells into miR-155−/− mice, one major defect in the absence of miR-155 is in generating sufficient numbers of pathogenic T cells.
Figure 1. .
miR-155−/− mice are resistant to EAU. (A) WT or miR-155−/− mice were immunized with IRBP in CFA and their eyes were enucleated 21 days after immunization, and histologic sections through the retina were stained with H&E. White arrows indicate presence of inflammatory cells in the vitreous (V) and blue star depicts pathologic foci characterized by the presence of retinal folds and hemorrhage. OpN, optic nerve; GCL, ganglion cell layer; INL, inner nuclear layer; ONL, outer nuclear layer. (B) Clinical scores were determined from histologic analysis of pathology slides of day-21 post-immunized mice. (C) Uveitogenic CD4+ T cells from the LN and spleen of WT mice with EAU were transferred into unimmunized WT or miR-155−/− mice (10 × 106 per mouse, n = 5). Ten days after adoptive cell transfer, funduscopy was performed. Fundus images were taken using an otoendoscopic imaging system, and they revealed the development of papillitis (black arrows), retinal vasculitis (blue arrows), and inflammatory infiltrates (white arrows). (D) Clinical scores and assessment of disease severity was based on changes at the optic nerve disc and retinal tissues as described previously.16 Data represent at least 3 independent experiments. Statistical significance determined using unpaired Student's t-test (**P < 0.01). NS, not significant.
Resistance of miR-155−/− to EAU Derives From Defects in Inducing Expansion of Th1 and Th17 Cells
We next examined whether the resistance of miR-155−/− mice to EAU derived from inability of the miR-155−/− mice to mount immunologic responses to the autoantigen, IRBP. LN and spleen cells were harvested from naïve unimmunized mice or from WT or miR-155−/− mice at day 21 after immunization with IRBP. The cells were cultured for 2 or 4 days in the presence or absence of IRBP, and lymphocyte proliferative responses were assessed by the thymidine incorporation assay. As indicated, WT and miR-155−/− lymphocytes proliferated significantly in response to stimulation by IRBP, although a greater response was mounted by WT compared to the miR-155−/− cells (Fig. 2A). This result is consistent with the decreased size of the miR-155−/− LN and spleen (Fig. 2B). However, the miR-155−/− mouse LN contained more lymphocytes than the unimmunized C57BL/6 WT LN (Fig. 2C), indicating that the miR-155−/− mouse is immunocompetent. Despite the significant increase in inflammatory cells in the LN of miR-155−/− mice following immunization with IRBP/CFA, the total numbers of LN mononuclear cells were comparable to numbers of these cells detected in LN of unimmunized mice (Fig. 2C), suggesting that one main impact of the loss of miR-155 is on lymphocyte numbers.
Figure 2.
Autoantigen-induced expansion of CD4+ T cells is defective in miR-155–deficient mice. (A) LN and spleen cells from unimmunized mice, or from WT or miR-155−/− mice 21 days after immunization with IRBP were cultured with or without IRBP, and lymphocyte proliferative responses were assessed by the thymidine incorporation assay. (B) Eight-week-old WT or miR-155−/− mice were euthanized and LN or spleen was harvested. Picture and graph illustrate differences in size and weight of these tissues following immunization of the two mouse strains with IRBP/CFA. (C) Left: CD4+ T cells isolated from the spleen and LN of mice described in (A). Right: total lymphocytes isolated for LN of mice were quantified on a Vi-CELL Cell Viability Analyzer (Beckman Coulter, Inc., Sykesville, MD). Data represent at least 3 independent experiments. Statistical significance was determined using Student's t-test (*P < 0.05, **P < 0.01).
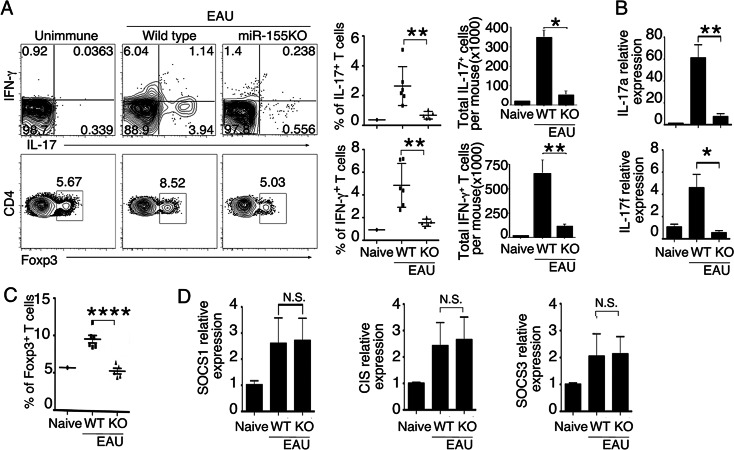
During EAU in mice, Th1 and Th17 cells are expanded and recruited into the retina, and their levels vary during the course of the disease.21,23 Therefore, we examined whether the resistance of miR-155−/− mice to EAU derived in part from defects in generation of Th1 and Th17 cells. Freshly isolated LN (without stimulation) from WT or miR-155−/− mice 21 days after immunization with IRBP/CFA were analyzed by the intracellular cytokine assay. In line with previous reports,13,21 onset of EAU pathology was correlated temporally with increase of IL-17A+ Th17 or IFN-γ+ Th1 cells (Fig. 3A). However, there was a marked defect in Th1 and Th17 development in miR-155−/− mice. RNA analysis showed further that the Th17 cells were defective in IL-17A and IL-17F expression in the absence of miR-155 (Fig. 3B). Consistent with reports showing that increase in Treg cells may contribute to mechanisms that mediate recovery from EAU,24 the frequency of Foxp3-expressing T cells was elevated significantly in LN of the WT EAU mice (Fig. 3C). Our results also are consistent with a defect in proliferative fitness of miR-155−/− Treg cells as described previously.3 Despite this reduction in Treg cells in the absence of miR-155, uveitis did not develop. In contrast, LN of unimmunized and IRBP-immunized miR-155-deficient mice contained similar frequencies of Treg cells (Figs. 3A, 3C), indicating that miR-155 may be required for expansion of Treg cells during EAU, but not required for development of Tregs. Taken together, these results indicated that, while Tregs may have a role in the recovery from uveitis, disease etiology appear to derive ultimately from the role of miR-155 or other factors that mediate the generation and expansion of uveitogenic effector T cells. Recent studies suggest that miR-155 also may regulate host immunity by targeting genes that regulate T-helper cell lineage decisions.25 For example, miR-155 was found to prolong IL-2–induced STAT5 signaling and confer competitive fitness to Treg cells by targeting the negative feedback regulator of cytokine signaling, Suppressor of Cytokine Signaling 1 (SOCS1).3 To verify this observation, we analyzed CD4+ T cells from WT or miR-155−/− mice for the expression of three SOCS family members induced in T-helper cells during CD4+ T-cell differentiation. However, we found comparable levels of SOCS1, SOCS3, or cytokine-induced SH2-containing protein 1 (CIS1) expression by WT or miR-155–deficient T cells (Fig. 3D). Taken together, these results suggested that miR-155 has wide-ranging effects on all CD4+ lymphocytes, as well as, on many aspects of their immunoregulatory functions.
Figure 3. .
miR-155−/− mice immunized with IRBP/CFA exhibit defective production of inflammatory cytokines. (A) CD4+ T cells from LN of WT or miR-155 KO mice (day 21 after immunization) were analyzed by intracellular cytokine staining assay. CD4+ T cells were gated, and numbers in quadrants indicate percentage of CD4+IL-17A+, CD4+IFN-γ+, or CD4+Foxp3+ T cells. (B) RT-qPCR analysis of IL-17A and IL-17F mRNA in cells described in (A). (C) Percentages of CD4+Foxp3+ T cells. (D) RT-qPCR analysis of SOCS1, CIS, and SOCS3 mRNA in cells described in (A) enriched for CD4+ T cells by MACS. Data represent at least 3 independent experiments. Statistical significance was determined using Student's t-test (*P < 0.05, **P < 0.01, ****P < 0.0001).
Resistance of miR-155−/− Mice to EAU Correlates With Defects in Inducing Expansion of CD8+ Cells
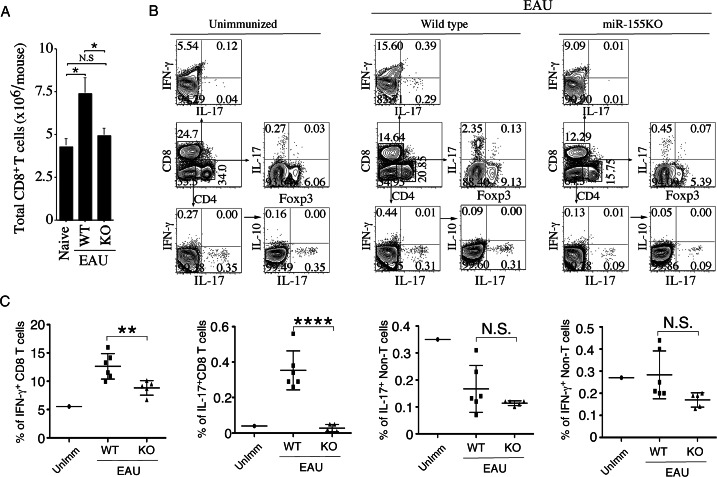
We also investigated whether the loss of miR-155 affected the expansion of IFN-γ–expressing CD8+ cells, as well as other inflammatory cells. LN and spleen cells were isolated from WT or miR-155−/− mice 21 days after immunization with IRBP/CFA, and the freshly isolated cells (without stimulation) were analyzed by the intracellular cytokine assay. Analysis of draining lymph node cells of WT or miR-155−/− mice showed that LN of IRBP-immunized miR-155−/− mouse contained more lymphocytes than the unimmunized C57BL/6 WT LN (Fig. 2C). However, the total numbers of CD8+ T cells were comparable to numbers of these cells detected in LN of unimmunized mice (Fig. 4A). Compared to the WT, the percentage of IFN-γ– and IL-17–expressing CD8 T cells was significantly low (Figs. 4B, 4C), indicating that miR-155 contributes to Ag-induced expansion and functions of CD8+ T cells. On the other hand, the levels of non–T-lymphocytes expressing IL-10, IL-17, or IFN-γ were unaffected (Fig. 4C).
Figure 4. .
Autoantigen-induced expansion of CD8+ T cells is defective in miR-155−/− mice. (A) CD8+ T cells isolated from the spleens and LNs of unimmunized WT mice, and WT or miR-155–deficient mice immunized with IRBP/CFA (day 21 after immunization) were quantified on a Vi-CELL Cell Viability Analyzer (Beckman Coulter, Inc.). (B) LNs of WT or miR-155 KO mice described in (A) were analyzed by intracellular cytokine staining assay. CD8+ T cells were gated and numbers in quadrants indicate percentage of CD8+ cells expressing IFN-γ or IL-17A. CD4+ T cells were gated and numbers in quadrants indicate percentage of CD4+ cells expressing IL-17A or Foxp3. Non–T cells (CD4−CD8− cells) were gated and numbers in quadrants indicate percentage of non–T cells expressing IFN-γ, IL-17A, or IL-10. (C) Statistical analysis of the percentages of CD8+ and non–T cells expressing IFN-γ or IL-17A in the LN and spleen of mice described in (A). Data represent at least 3 independent experiments. Statistical significance was determined using Student's t-test (*P < 0.05, **P < 0.01, ****P < 0.0001).
miR-155 Expression in CD4+ T Cells Is Regulated by STAT3
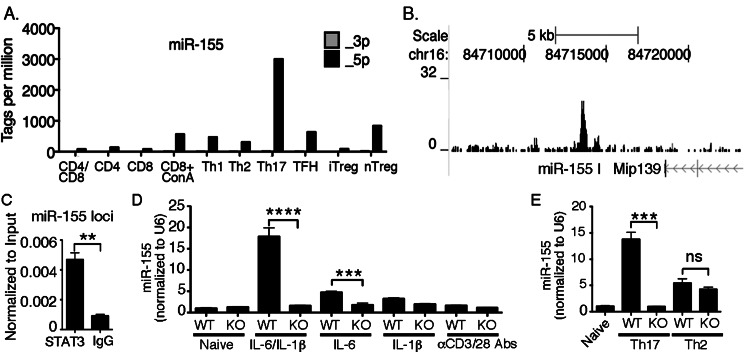
To determine the expression profile of miR-155 among T-cell subsets, we took advantage of a previously reported microRNA expression profiling dataset.18 As a result, we were able to determine that miR-155 expression was highest in Th17 cells (Fig. 5A, Kuchen et al.,18 and Escobar T, Muljo S, unpublished data, 2012). Since a transcriptional program induced by cytokines that activate STAT3 characterizes Th17 differentiation, we reanalyzed a previously reported ChIP-seq dataset19 to determine whether STAT3 can bind directly to the Mir155 locus. This revealed enrichment of STAT3 around the promoter region of Mir155 (Fig. 5B).19 We validated this finding by STAT3 ChIP-qPCR (Fig. 5C). To establish further a functional link between STAT3 and miR-155 in the regulation of Th17 differentiation, we examined whether cytokines that polarize naïve CD4+ T cells toward a Th17 developmental program could induce miR-155 expression through STAT3-dependent mechanisms. We focused on IL-1β and IL-6, which signal via the NFκB or STAT3 pathway, respectively. RT-qPCR analysis revealed marked induction of miR-155 expression by IL-6 and IL-1β (Fig. 5D). Notably, IL-6 and IL-1β synergized to induce dramatic elevation of mature miR-155 expression in a STAT3 dependent manner (Fig. 5D). Consistent with the requirement of STAT3 for Th17 differentiation and miR-155 expression, we found that STAT3-deficient T cells cultured under Th17 polarization condition failed to induce miR-155 expression, whereas there was no such STAT3 dependency under Th2 condition (Fig. 5E).
Figure 5.
Preferential miR-155 expression in Th17 cells is activated by STAT3. (A) Relative abundance of mature miR-155-5p and -3p, guide, and passenger strand respectively, among T-cell subsets were obtained from http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE21630. (B) STAT3 ChIP-seq analysis in Th17 cells using data from http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSM652877. (C) STAT3 ChIP-qPCR in Th17 cells using anti-STAT3 or nonspecific IgG control and primers that span the peak in (B). (D) Naïve CD4+ T cells from WT or CD4-STAT3KO mice were stimulated with anti-CD3/anti-CD28 antibodies for 4 days in medium containing IL-1β (10 ng/mL), IL-6 (10 ng/mL), or both cytokines, and miR-155 expression was examined using RT-qPCR. (E) Naïve CD4+ from WT and CD4-STAT3KO mice were stimulated under Th17 or Th2 polarization condition for 4 days, and miR-155 expression was assayed using RT-qPCR. Data represent at least 3 independent experiments. Statistical significance was determined using Student's t-test (**P < 0.01, ***P < 0.001, ****P < 0.0001).
Resistance of CD4-STAT3−/− Mice to EAU Correlated With Defective Expression of miR-155
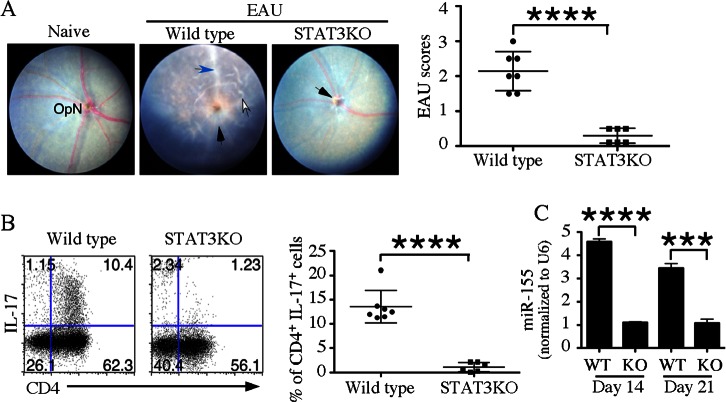
To examine whether resistance of CD4-STAT3KO mice to EAU derives from defects in miR-155 expression, we induced EAU in WT or CD4-STAT3KO mice by active immunization with IRBP. WT mice had full-blown EAU, while CD4-STAT3KO mice were resistant to EAU (Fig. 6A). In the absence of STAT3, we also observed a marked reduction in the percentage of IL-17A+ T cells (Fig. 6B) and in the expression of miR-155 in CD4+ T cells isolated from the LN at two time points during disease (Fig. 6C). Thus, our findings revealed a STAT3/miR-155 axis that is required for EAU development.
Figure 6.
Resistance of CD4-STAT3KO mice to EAU correlates with defective expression of miR-155. (A) WT or CD4-STAT3KO mice were immunized with IRBP in CFA and 21 days after immunization eyes were examined by funduscopy. (B) Freshly isolated LN cells from WT or CD4-STAT3KO mice (day 21 after immunization) were analyzed by the intracellular cytokine staining. CD4+ T cells were gated and numbers in quadrants indicate percentage of CD4+ or IL-17A+ T cells. (C) LN cells were isolated at 2 time points after immunization from mice described and their RNA was analyzed for the expression of miR-155 by RT-qPCR. Data represent at least 2 independent batches of experiments (n = 5 per experiment). Statistical significance was determined using Student's t-test (***P < 0.001 and ****P < 0.0001).
Discussion
Consistent with the involvement of miR-155 in the development of several T-cell–mediated autoimmune diseases, we have shown here that miR-155−/− mice are resistant to EAU. Th17 cells are implicated in uveitis and MS, and loss of STAT3 in CD4+ T cells in mice prevents the generation of Th17 cells and development of EAU or EAE.13 Similarly, miR-155−/− mice are resistant to EAE, but it was not clear whether this is related to STAT3. STAT3 has a key role in Th17 differentiation by binding to thousands of sites throughout the genome and regulating a highly orchestrated gene expression program; however, it was not appreciated previously that one key target of STAT3 could be a miRNA. Here, we demonstrated a convergence of miR-155– and STAT3-dependent pathways in the development of uveitis. We showed for the first time to our knowledge that resistance of CD4-STAT3KO mice to EAU derived in part from defective expression of miR-155 by T-helper cells. We also showed that the resistance of miR-155−/− mice to EAU derives in part from defects in the development of Th1 and Th17 cells during EAU. In addition, the contribution of miR-155 in the susceptibility to EAU appears to be CD4+ T-cell intrinsic. However, miR-155 may have wide ranging effects in the immune system and we do not yet have a complete understanding of its functions. On the one hand, it is essential for Treg function, but it also is crucial for effector T-cell function. The ability to delete miR-155 conditionally in specific cell types would demonstrate its contribution in various immune responses. In addition, systematic identification of miR-155 targets in individual cell types is key toward understanding its mechanisms of action, particularly since its targets may vary depending on the cell type.
Interestingly, we found that among T cells, the Th17 subset expressed the highest level of miR-155, and the expression of IL-17A and IL-17F was significantly down-regulated in miR-155−/− T cells, consistent with involvement of miR-155 in the development and/or function of Th17 subset (Fig. 3B). By ChIP analyses, we have demonstrated that STAT3 binds directly to the miR-155 locus and that mature miR-155 expression is defective in the absence of STAT3. Toward elucidating the roles of STAT3 and miR-155 in the pathophysiology of autoinflammatory diseases, we showed here that the increase of STAT3-dependent miR-155 expression correlated temporally with the onset and progression of EAU. Taken together, these observations support the notion that the resistance of CD4-STAT3 KO mice to EAU stems in part from the inability of STAT3-deficient T cells to induce expression of miR-155. Thus, during Th17 differentiation, activation of STAT3 may drive high miR-155 expression while sustained expression of miR-155 might promote autoimmune inflammation by enhancing the stability of the Th17 phenotype. In humans, autosomal dominant hyper IgE or Job's syndrome frequently is due to a dominant negative mutation in STAT3 and results in defective Th17 cell differentiation.26 Our findings suggested that part of the Th17 defect in this human primary immunodeficiency may stem from a failure to induce miR-155 expression.
In humans, uveitis often is characterized by repeated cycles of remission and recurrent intraocular inflammation,27,28 and it accounts for more than 10% of severe visual handicaps in the United States.29 Although steroids and other anti-inflammatory drugs are effective therapy, renal toxicity or other adverse effects preclude prolonged use.27 This has led to significant interest in identifying new therapeutic targets that can form the basis for developing biologics or small molecule inhibitors for treating uveitis. In our study, we have shown that IRBP-specific T cells that express miR-155 can induce autoimmune uveitis, while IRBP-specific T cells that lack miR-155 cannot. We have established a strong correlation between increase in miR-155 expression in T cells in vivo and temporal onset of EAU, and shown that CD4-STAT3KO mice that do not develop EAU also are defective in miR-155 expression. These results thus, provide mechanistic link between miR-155 levels in T cells and susceptibility to EAU. In EAU, STAT3 is required for the development of Th17 cells, but also for miR-155 expression in Th17 cells, suggesting that STAT3 and miR-155 are potential therapeutic targets for treating uveitis. In context of therapeutic application of our finding, we believe that we have shown that STAT3 and miR-155 form an axis that promotes the expansion of pathogenic Th17 cells that mediate uveitis. Thus, targeting STAT3 and miR-155 may be beneficial for treating uveitis and other Th17-mediated inflammatory disorders. We already have demonstrated partial efficacy of STAT3 inhibitors.30 If pursued as a therapy for uveitis, we ideally would propose topical administration onto the eye a formulation consisting of miR-155 and/or STAT3 inhibitors to avoid undesirable systemic effects. However, this will depend on results of proof-of-concept studies in mice.
Acknowledgments
This work benefited greatly from data deposited in NCBI GEO. We thank Cuong Nguyen for reanalysis of ChIP-seq data and Yves-Olivier Guettard for preliminary STAT3 ChIP analyses.
Supported by the Intramural Research Program of the NEI and NIAID, National Institutes of Health (NIH). The authors alone are responsible for the content and writing of the paper.
Disclosure: T. Escobar, None; C.-R. Yu, None; S.A. Muljo, None; C.E. Egwuagu, None
References
- 1. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004; 116: 281–297 [DOI] [PubMed] [Google Scholar]
- 2. O'Connell RM, Kahn D, Gibson WS, et al. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity. 2010; 33: 607–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lu LF, Thai TH, Calado DP, et al. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity. 2009; 30: 80–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Paraboschi EM, Solda G, Gemmati D, et al. Genetic association and altered gene expression of mir-155 in multiple sclerosis patients. Int J Mol Sci. 2011; 12: 8695–8712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Thamilarasan M, Koczan D, Hecker M, Paap B, Zettl UK. MicroRNAs in multiple sclerosis and experimental autoimmune encephalomyelitis. Autoimmun Rev. 2012; 11: 174–179 [DOI] [PubMed] [Google Scholar]
- 6. Kurowska-Stolarska M, Alivernini S, Ballantine LE, et al. MicroRNA-155 as a proinflammatory regulator in clinical and experimental arthritis. Proc Natl Acad Sci U S A. 2011; 108: 11193–11198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rodriguez A, Vigorito E, Clare S, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007; 316: 608–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kutty RK, Nagineni CN, Samuel W, Vijayasarathy C, Hooks JJ, Redmond TM. Inflammatory cytokines regulate microRNA-155 expression in human retinal pigment epithelial cells by activating JAK/STAT pathway. Biochem Biophys Res Commun. 2010; 402: 390–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhou Q, Xiao X, Wang C, et al. Decreased microRNA-155 expression in ocular Behcet's disease but not in Vogt Koyanagi Harada syndrome. Invest Ophthalmol Vis Sci. 2012; 53: 5665–5674 [DOI] [PubMed] [Google Scholar]
- 10. Thai TH, Calado DP, Casola S, et al. Regulation of the germinal center response by microRNA-155. Science. 2007; 316: 604–608 [DOI] [PubMed] [Google Scholar]
- 11. Lee CK, Raz R, Gimeno R, et al. STAT3 is a negative regulator of granulopoiesis but is not required for G-CSF-dependent differentiation. Immunity. 2002; 17: 63–72 [DOI] [PubMed] [Google Scholar]
- 12. Raz R, Lee CK, Cannizzaro LA, d'Eustachio P, Levy DE. Essential role of STAT3 for embryonic stem cell pluripotency. Proc Natl Acad Sci U S A. 1999; 96: 2846–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu X, Lee YS, Yu CR, Egwuagu CE. Loss of STAT3 in CD4+ T cells prevents development of experimental autoimmune diseases. J Immunol. 2008; 180: 6070–6076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Oh HM, Yu CR, Lee Y, Chan CC, Maminishkis A, Egwuagu CE. Autoreactive memory CD4+ T lymphocytes that mediate chronic uveitis reside in the bone marrow through STAT3-dependent mechanisms. J Immunol. 2011; 187: 3338–3346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yu CR, Mahdi RR, Oh HM, et al. Suppressor of cytokine signaling-1 inhibits lymphocyte recruitment into the retina and protects SOCS1 transgenic rats and mice from ocular inflammation. Invest Ophthalmol Vis Sci. 2011; 52: 6978–6986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xu H, Koch P, Chen M, Lau A, Reid DM, Forrester JV. A clinical grading system for retinal inflammation in the chronic model of experimental autoimmune uveoretinitis using digital fundus images. Exp Eye Res. 2008; 87: 319–326 [DOI] [PubMed] [Google Scholar]
- 17. Egwuagu CE, Yu CR, Zhang M, Mahdi RM, Kim SJ, Gery I. Suppressors of cytokine signaling proteins are differentially expressed in Th1 and Th2 cells: implications for Th cell lineage commitment and maintenance. J Immunol. 2002; 168: 3181–3187 [DOI] [PubMed] [Google Scholar]
- 18. Kuchen S, Resch W, Yamane A, et al. Regulation of microRNA expression and abundance during lymphopoiesis. Immunity. 2010; 32: 828–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang XP, Ghoreschi K, Steward-Tharp SM, et al. Opposing regulation of the locus encoding IL-17 through direct, reciprocal actions of STAT3 and STAT5. Nat Immunol. 2011; 12: 247–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wei G, Wei L, Zhu J, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009; 30: 155–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Amadi-Obi A, Yu CR, Liu X, et al. T(H)17 cells contribute to uveitis and scleritis and are expanded by IL-2 and inhibited by IL-27/STAT1. Nat Med. 2007; 13: 711–718 [DOI] [PubMed] [Google Scholar]
- 22. Mattapallil MJ, Wawrousek EF, Chan CC, et al. The Rd8 mutation of the Crb1 gene is present in vendor lines of C57BL/6N mice and embryonic stem cells, and confounds ocular induced mutant phenotypes. Invest Ophthalmol Vis Sci. 2012; 53: 2921–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Luger D, Silver PB, Tang J, et al. Either a Th17 or a Th1 effector response can drive autoimmunity: conditions of disease induction affect dominant effector category. J Exp Med. 2008; 205: 799–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sun M, Yang P, Du L, Zhou H, Ren X, Kijlstra A. Contribution of CD4+CD25+ T cells to the regression phase of experimental autoimmune uveoretinitis. Invest Ophthalmol Vis Sci. 2010; 51: 383–389 [DOI] [PubMed] [Google Scholar]
- 25. Lu LF, Boldin MP, Chaudhry A, et al. Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell. 2010; 142: 914–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Milner JD, Brenchley JM, Laurence A, et al. Impaired T(H)17 cell differentiation in subjects with autosomal dominant hyper-IgE syndrome. Nature. 2008; 452: 773–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nussenblatt RB. Bench to bedside: new approaches to the immunotherapy of uveitic disease. Int Rev Immunol. 2002; 21: 273–289 [DOI] [PubMed] [Google Scholar]
- 28. Nussenblatt RB, Fortin E, Schiffman R, et al. Treatment of noninfectious intermediate and posterior uveitis with the humanized anti-Tac mAb: a phase I/II clinical trial. Proc Natl Acad Sci U S A. 1999; 96: 7462–7466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nussenblatt RB. The natural history of uveitis. Int Ophthalmol. 1990; 14: 303–308 [DOI] [PubMed] [Google Scholar]
- 30. Yu CR, Lee YS, Mahdi RM, Surendran N, Egwuagu CE. Therapeutic targeting of STAT3 (Signal Transducers and Activators of Transcription 3) pathway inhibits experimental autoimmune uveitis. PLoS One. 2012; 7: e29742 [DOI] [PMC free article] [PubMed] [Google Scholar]