Abstract
Background
Porcine circovirus type 2 (PCV2) is the causal agent of postweaning multisystemic wasting syndrome (PMWS), which has severely impacted the swine industry worldwide. PCV2 triggers a weak and atypical innate immune response, but the key genes and mechanisms by which the virus interferes with host innate immunity have not yet been elucidated. In this study, genes that control the response of primary porcine alveolar macrophages (PAMs), the main target of PCV2, were profiled in vitro.
Results
PAMs were successfully infected by PCV2-WH strain, as evidenced quantitative real-time polymerase chain reaction (qPCR) and immunofluorescence assay (IFA) results. Infection-related differential gene expression was investigated using pig microarrays from the US Pig Genome Coordination Program and validated by real-time PCR and enzyme-linked immunosorbent assay (ELISA). Microarray analysis at 24 and 48 hours post-infection (HPI) revealed 266 and 175 unique genes, respectively, that were differentially expressed (false discovery rate <0.05; fold-change >2). Only six genes were differentially expressed between 24 and 48 HPI. The up-regulated genes were principally related to immune response, cytokine activity, locomotion, regulation of cell proliferation, apoptosis, cell growth arrest, and antigen procession and presentation. The down-regulated genes were mainly involved in terpenoid biosynthesis, carbohydrate metabolism, translation, proteasome degradation, signal transducer activity, and ribosomal proteins, which were representative of the reduced vital activity of PCV2-infected cells.
Conclusions
PCV2 infection of PAMs causes up-regulation of genes related to inflammation, indicating that PCV2 may induce systematic inflammation. PCV2 persistently induced cytokines, mainly through the Toll-like receptor (TLR) 1 and TLR9 pathways, which may promote high levels of cytokine secretion. PCV2 may prevent apoptosis in PAMs by up-regulating SERPINB9 expression, possibly to lengthen the duration of PCV2 replication-permissive conditions. The observed gene expression profile may provide insights into the underlying immunological response and pathological changes that occur in pigs following PCV2 infection.
Background
Porcine circovirus 2 (PCV2) is a small, non-enveloped, single-stranded and closed-circular DNA virus that has been identified as the primary cause of postweaning multisystemic wasting syndrome (PMWS) in pigs. PMWS primarily affects pigs between the ages of five and 18 weeks. The clinical signs include progressive weight loss, jaundice, wasting, and respiratory disease, accompanied by increased mortality. The mortality rates of PMWS vary from 1-2%, but may reach up to 30% in complicated cases. PMWS has become endemic in many swine-producing countries, including China, Canada, the United States, Venezuela, Australia, and the United Kingdom [1-3].
DNA microarray technology, in combination with bioinformatics, has emerged as an efficient high-throughput tool that offers great advantages in the study of the genomic expression profiles of cells and host-microbe interactions [4]. This technology provides the ability to determine gene expression levels of thousands of different genes simultaneously. Indeed, high density gene arrays have already revealed the status of host gene expression following infection by several viruses and bacteria, including influenza A virus [5], monkeypox virus [6], Epstein-Barr virus [7], Streptococcus suis[8], and Haemophilus parasuis[9].
The major target cells of PCV2 are the porcine monocyte/macrophage lineage cells (MLCs) [10-13]. One type of MLC, porcine alveolar macrophages (PAMs), are the primary responders of the pulmonary innate immune system and defend against various pathogens [14]. While PAMs are known to be involved in PCV2 defense, the underlying molecular mechanisms of PCV2 pathogenicity remain to be completely elucidated. Furthermore, an immunosuppressive role has been characterized for PCV2 in porcine circovirus diseases (PCVD) [15,16], but little is known about the genes that are involved in this process. The presence of granulomatous inflammation and multinucleated giant cells in PMWS pigs [16] suggests a significant role for secreted cytokines and other immune-related signaling factors. Previous studies have investigated the cytokine and chemokine gene expression profile of PMWS by transcriptomic and genomic analyses. For example, the cytokine mRNA expression profile in lymph nodes of PMWS pigs was reported [17], as was the global transcriptome profile of colostrums-deprived piglets experimentally infected with PCV2 [18,19]. The genomic expression profile in lymph nodes of lymphoid-depleted PCV2-infected pigs was also reported [20]. However, these results can not fully illustrate the pathogenesis of PCV2.
Therefore, the objective of the present study was to analyze the PCV2-induced gene expression profiles in isolated PAMs in vitro in order to reveal specific mechanisms of the host innate immune response to PCV2. The temporal gene expression profiling using a two-channel microarray carrying thousands of genes was carried out with PAMs at 24 and 48 hours post-infection (HPI) to discriminate PCV2-induced early and late changes in gene expression.
Results
Confirmation of PCV2 infection in PAMs
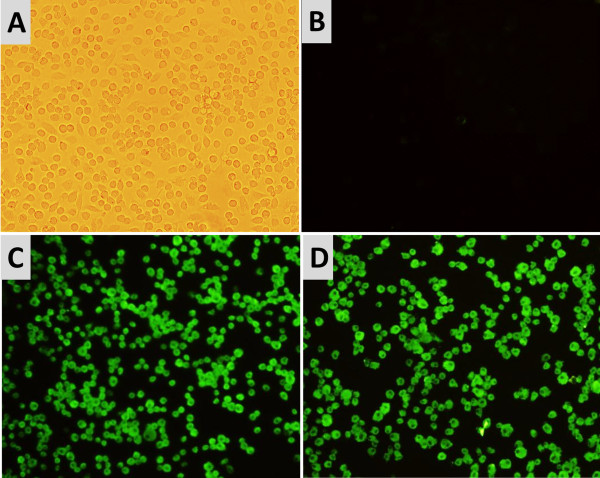
Immunofluorescence assay (IFA) revealed green fluorescent cells in the infected PAMs, indicating that the PAMs were successfully infected by PCV2 at 24 and 48 hours after inoculation (Figure 1). There were no significant differences in the levels of viral DNA or virus particles between the two time points (mean viral loads: 5.67±0.35 log10 and 5.72±0.49 log10). Neither PCV2 particles nor the PCV2 nucleotide signal were detected in the mock-inoculated PAMs at any of the experimental time points.
Figure 1.
Immunofluorescence antibody staining for PCV2-infected PAMs. Intracytoplasmic bright green fluorescence (fluorescein isothiocyanate) is seen in PCV2-infected PAMs. (A) Bright microscopy image of mock-inoculated PAMs. (B) Mock-inoculated PAMs. (C) PCV2-infected PAMs at 24 HPI. (D) PCV2-infected PAMs at 48 HPI. (B-D) were stained with PCV2 Cap protein-specific monoclonal antibody. Magnification: ×400.
Transcriptome analysis of the PAM response to PCV2 infection
The concentrations of RNA ranged from 0.67 to 1.97 μg/μL. All RNA samples were considered to be of high quality, based on RNA integrity number (RIN) values (ranging from 8.0 to 9.4). The isolated mRNA samples from 24 HPI and 48 HPI PAMs hybridized to 6345 and 5856 probe sets on the transcriptome microarray, respectively corresponding to 34.25% and 31.65% of all probe sets (Additional file 1: Table S1). After quantile normalization and statistical analysis, 1277 transcripts were identified (false discovery rate (FDR)-adjusted p-value <0.05). Furthermore, the criteria of a two-fold or greater change in differential expression and a FDR of 5% were chosen to determine up-regulated and down-regulated genes. According to these criteria, 266 and 175 genes were differentially expressed (DE) from baseline at 24 HPI and 48 HPI (Additional file 2: Table S2 and Additional file 3: Table S3). According to the corresponding biological process Gene Ontology (GO) terms, or potential biological processes reported in recent publications, the majority of these genes were related to Immune Response, Inflammatory Response, Antigen Processing and Presentation of Peptide or Polysaccharide Antigen via MHC Class II, Anti-viral Immune response, Cell Motility, and Negative Regulation of Apoptosis (Table 1). However, the differential expression between 24 HPI and 48 HPI reached the threshold of statistical significance for only six of those genes (Additional file 4: Table S4 and Additional file 5: Table S5). A total of 17 differentially expressed genes were selected to validate the microarray results by quantitative reverse transcription PCR (qRT-PCR) using the primers in Table 2. The qRT-PCR results confirmed, in general, the patterns of up-regulated and down-regulated expression of the 17 genes that was indicated by the microarray analysis (Table 3).
Table 1.
Functional grouping of genes differentially expressed (≥2-fold and P < 0.05, n = 3) between PCV2-infected and mock-infected control
| Functional classification | Gene name | Gene symbol | 24 HPI FC | Q-value, % (24hpi) | 48 HPI FC | Q-value, % (48hpi) | GenBank ID |
|---|---|---|---|---|---|---|---|
| Immune response | |||||||
| |
rho GDP dissociation inhibitor beta |
ARHGDIB |
2.16 |
0.41 |
1.21 |
13.11 |
NM_001009600 |
| |
CMRF35-like molecule 1 |
CLM1 |
2.35 |
0.95 |
1.38 |
13.11 |
AY457047 |
| |
cathepsin C |
CTSC |
2.12 |
3.06 |
1.74 |
11.46 |
NM_001814 |
| |
cathepsin S |
CTSS |
0.5 |
1.09 |
0.77 |
19.25 |
M90696.1 |
| |
Fc fragment of IgG low affinity II b receptor |
FCGR2B |
3.47 |
0.54 |
1.85 |
8.49 |
NM_001033013 |
| |
neutrophil cytosolic factor 4 |
NCF4 |
2.62 |
0 |
2.12 |
6.28 |
NM_000631 |
| Humoral immune response | |||||||
| |
chemokine ligand 16 |
CCL16 |
3.85 |
4.64 |
3.24 |
4.93 |
NM_004590; |
| |
CD74 molecule |
CD74 |
3.94 |
0 |
3.43 |
0.58 |
NM_213774 |
| |
complement factor D |
CFD |
0.36 |
0.41 |
0.59 |
2.88 |
NM_001928 |
| |
C-type lectin domain family 2, member B |
CLEC2B |
2.83 |
1.62 |
2.10 |
1.09 |
NM_005127 |
| Inflammatory response | |||||||
| |
chemokine ligand 3-like 3 |
CCL3L3 |
7.58 |
0 |
2.46 |
1.64 |
NM_001001437 |
| |
chemokine Ligand 5 |
CCL5 |
1.53 |
14.04 |
2.25 |
2.44 |
AK312212 |
| |
chemokine ligand 5 |
CXCL5 |
32.28 |
0 |
12.96 |
0 |
EU176356 |
| |
chemokine ligand 7 |
CXCL7 |
2.75 |
0.95 |
3.57 |
1.97 |
NM_213862.1 |
| |
interleukin-6 |
IL6 |
2.26 |
4.65 |
1.36 |
13.43 |
NM_214399.1 |
| |
S100 calcium binding protein A1 |
S100A1 |
0.31 |
0.41 |
0.40 |
0.71 |
NM_006271 |
| |
S100 calcium binding protein A12 |
S100A12 |
10.98 |
0 |
5.13 |
1.09 |
FJ263393.1 |
| |
S100 calcium binding protein A8 |
S100A8 |
12.96 |
0 |
6.83 |
0 |
FJ263391.1 |
| |
S100 calcium binding protein A9 |
S100A9 |
20.00 |
0 |
9.71 |
0 |
FJ263392.1 |
| |
serum amyloid A1 |
SAA1 |
14.59 |
0.54 |
1.82 |
24.35 |
NM_199161 |
| |
secreted phosphoprotein 1 |
SPP1 |
2.66 |
2.39 |
1.67 |
24.35 |
AK296035 |
| |
tumor necrosis factor, alpha-induced protein 6 |
TNFAIP6 |
6.54 |
0 |
3.26 |
0.58 |
NM_001159607 |
| Lipid biosynthesis | |||||||
| |
Lysophosphatidic acid acyltransferase, delta |
AGPAT4 |
2.34 |
0 |
1.48 |
0.58 |
NM_020133 |
| |
lysophosphatidic acid acyltransferase, zeta |
AGPAT6 |
1.56 |
0.54 |
2.03 |
0 |
NM_178819 |
| |
alkylglycerone phosphate synthase |
AGPS |
2.41 |
0.95 |
1.46 |
32.28 |
NM_003659 |
| |
farnesyl-diphosphate farnesyltransferase 1 |
FDFT1 |
0.77 |
14.04 |
0.45 |
1.82 |
AK297868 |
| |
isopentenyl-diphosphate delta isomerase 1 |
IDI1 |
0.84 |
26.53 |
0.46 |
2.88 |
NM_004508 |
| |
inositol-3-phosphate synthase 1 |
ISYNA1 |
2.12 |
0.54 |
1.65 |
1.44 |
NM_016368 |
| |
stearoyl-CoA desaturase |
SCD |
2.46 |
0.54 |
3.37 |
8.49 |
AB208982 |
| |
triosephosphate isomerase 1 |
TPI1 |
3.00 |
0 |
2.11 |
3.33 |
NM_000365 |
| Locomotion | |||||||
| |
CD97 molecule |
CD97 |
1.50 |
3.06 |
2.17 |
1.64 |
NM_001784 |
| |
coronin, actin binding protein, 1A |
CORO1A |
2.64 |
0.54 |
1.26 |
4.65 |
NM_007074 |
| |
chemokine ligand 10 |
CXCL10 |
7.69 |
4.93 |
2.58 |
1.07 |
NM_001565 |
| |
Kallmann syndrome 1 sequence |
KAL1 |
0.43 |
1.44 |
0.46 |
4.65 |
NM_000216 |
| |
mitogen-activated protein kinase 1 |
MAP2K1 |
2.03 |
0 |
1.17 |
1.7 |
NM_002755 |
| |
platelet factor 4 |
PF4 |
4.15 |
0 |
3.80 |
1.64 |
NM_002619 |
| |
phosphatidic acid phosphatase type 2B |
PPAP2B |
8.45 |
0.95 |
3.04 |
11.46 |
NM_003713 |
| |
prostaglandin-endoperoxide synthase 2 |
PTGS2 |
5.75 |
0 |
5.41 |
0 |
NM_001784 |
| Lyase activity | |||||||
| |
aldolase C, fructose-bisphosphate |
ALDOC |
3.08 |
0 |
2.18 |
3.33 |
AK294157 |
| |
carbonic anhydrase XIII |
CA13 |
0.41 |
0.82 |
0.40 |
0.71 |
NM_198584 |
| |
enolase 1, (alpha) |
ENO1 |
2.24 |
0 |
1.80 |
4.94 |
AK298600 |
| |
N-acetylneuraminate pyruvate lyase |
NPL |
1.71 |
1.62 |
2.28 |
0 |
AK297017 |
| |
serine dehydratase |
SDS |
7.58 |
0 |
9.14 |
0 |
NM_006843 |
| Antigen processing and presentation | |||||||
| |
Major histocompatibility complex, class II, DO alpha |
DOA |
2.29 |
0 |
1.42 |
13.11 |
NM_002119.3 |
| |
Major histocompatibility complex, class II, DO beta |
DOB |
17.85 |
2.39 |
3.78 |
1.64 |
NM_002120.3 |
| |
Major histocompatibility complex, class II, DQ alpha 1 |
DQA1 |
2.07 |
1.62 |
1.37 |
9.21 |
M33906.1 |
| |
Major histocompatibility complex, class II, DQ beta 1 |
DQB1 |
2.47 |
0 |
2.14 |
11.46 |
M32577 |
| |
Major histocompatibility complex, class II, DR alpha |
DRA |
7.87 |
0 |
3.69 |
1.08 |
NM_001134339 |
| |
Major histocompatibility complex, class II, DR beta 1 |
DRB1 |
5.83 |
0.95 |
2.17 |
1.17 |
AY770514.1 |
| Signal transducer activity | |||||||
| |
guanine nucleotide binding protein (G protein), gamma 2 |
GNG2 |
1.94 |
0.41 |
2.26 |
0.42 |
NM_0530 |
| |
regulator of G-protein signaling 1 |
RGS1 |
1.02 |
58.23 |
0.47 |
4.25 |
NM_002922 |
| |
transforming growth factor, alpha |
TGFA |
2.79 |
0 |
1.54 |
14.86 |
AK312899 |
| |
wingless-type MMTV integration site family, member 6 |
WNT6 |
0.49 |
1.09 |
0.64 |
21.46 |
NM_006522 |
| Antiviral immune response | |||||||
| |
chemokine ligand 8 |
CCL8 |
8.02 |
0 |
3.17 |
0 |
NM_214214.1 |
| |
extracellular matrix protein 1 |
ECM1 |
2.01 |
0.54 |
2.89 |
0 |
CB477333 |
| |
IFN-gamma |
IFNG |
0.49 |
0.41 |
3.18 |
0 |
NM_213948.1 |
| |
interleukin 18 |
IL18 |
2.58 |
0.95 |
5.84 |
19.25 |
NM_001562 |
| |
ISG15 ubiquitin-like modifier |
ISG15 |
1.56 |
10.92 |
2.51 |
0 |
EU584557.1 |
| |
interferon-stimulated gene 20 kDa protein |
ISG20 |
1.49 |
61.10 |
2.20 |
4.94 |
NM_002201 |
| |
vasoactive intestinal peptide |
VIP |
0.69 |
32.11 |
4.14 |
0 |
NM_194435 |
| Apoptosis | |||||||
| |
BCL2/adenovirus E1B 19kDa interacting protein 3 |
BNIP3 |
3.69 |
0 |
2.61 |
2.88 |
U15174.1 |
| |
CD14 molecule |
CD14 |
3.60 |
0 |
3.87 |
0 |
NM_001097445 |
| |
immediate early response 3 |
IER3 |
5.46 |
0.41 |
2.21 |
1.64 |
NM_003897 |
| |
lectin, galactoside-binding, soluble, 1 |
LGALS1 |
0.42 |
0.82 |
0.66 |
10.51 |
NM_002305 |
| |
interleukin 1 alpha |
IL1A |
21.31 |
0 |
8.47 |
0 |
NM_214029.1 |
| |
interleukin 1, beta |
IL1B |
18.69 |
0 |
10.33 |
2.88 |
M15330.1 |
| |
serine/threonine kinase 17a |
STK17A |
1.53 |
1.62 |
2 |
1.64 |
NM_00476 |
| |
tumor necrosis factor receptor superfamily, member 10d, decoy with truncated death domain |
TNFRSF10D |
2.26 |
0 |
2.21 |
61.11 |
NM_003840 |
| |
tumor necrosis factor alpha |
TNFα |
5.13 |
0.41 |
1.96 |
6.28 |
EU682384.1 |
| Cell proliferation | |||||||
| |
Betacellulin |
BTC |
2.26 |
0.95 |
1.78 |
1.97 |
NM_001729 |
| |
epithelial membrane protein 1 |
EMP1 |
0.49 |
0.82 |
0.62 |
2.88 |
NM_001423 |
| |
fatty acid binding protein 3, muscle and heart |
FABP3 |
1.88 |
0.95 |
2.57 |
3.09 |
NM_004102 |
| |
fms-related tyrosine kinase 1 |
FLT1 |
1.91 |
1.78 |
2.49 |
0 |
NM_002019 |
| |
interleukin 2 receptor, gamma |
IL2RG |
1.39 |
6.28 |
2.06 |
1.07 |
NM_000206 |
| |
interleukin 8 receptor, beta |
IL8RB |
1.33 |
45.16 |
3.41 |
0 |
NM_001557 |
| |
v-myc myelocytomatosis viral oncogene homolog |
MYC |
0.75 |
14.04 |
0.29 |
0.71 |
NM_002467 |
| pro-platelet basic protein | PPBP | 2.75 | 0.95 | 3.57 | 1.97 | NM_002704 | |
Note: The DE genes are presented with putative functions assigned by the GO term and manual annotation. Many genes with multiple functions were only listed in one category according to specific biology processes. FC: fold change; FC≥2: up-regulation (infection/control); FC≤0.5: down-regulation.
Table 2.
Priers used for qRT-PCR validation and additional expression profiling
| Gene | Primer sequence, 5′-3′ | Amplicon length, bp | GenBank accession number | |
|---|---|---|---|---|
| HPRT-1 a |
Forward: |
TCATTATGCCGAGGATTTGGA |
90 |
DQ136030 |
| |
Reverse: |
CTCTTTCATCACATCTCGAGCAA |
|
|
| GAPDH b |
Forward: |
TGCCAACGTGTCGGTTGT |
120 |
NG_007073.2 |
| |
Reverse: |
TGTCATCATATTTGGCAGGTTT |
|
|
| CD14 |
Forward: |
AGAGTTCAAAGAGTAGGGAA |
86 |
NM_001097445.2 |
| |
Reverse: |
AACGCTATCTGTCCTCAC |
|
|
| GPNMB |
Forward: |
CCCTCACGAGCACCCTTGT |
91 |
NM_001098584 |
| |
Reverse: |
AAGAAGCCAACGAAGACCAGGAT |
|
|
| ICAM1 |
Forward: |
GCAAGAAGATAGCCAACCA |
106 |
NM_000201.2 |
| |
Reverse: |
TGCCAGTTCCACCCGTTC |
|
|
| IFNγ |
Forward: |
AAAGATAACCAGCCCATTC |
93 |
NM_213948.1 |
| |
Reverse: |
GTCATTCAGTTTCCCAGA |
|
|
| IL6 |
Forward: |
CCTTCAGTCCAGTCGCCTTCTCC |
97 |
NM_214399.1 |
| |
Reverse: |
GCATCACCTTTGGCATCTTCTTCC |
|
|
| IL8 |
Forward: |
CACTGTGAAAATTCAGAAATCATTGTTA |
105 |
NM_213867.1 |
| |
Reverse: |
CTTCACAAATACCTGCACAACCTTC |
|
|
| ISG15 |
Forward: |
GGAGGGTAGGGAGGAGGGA |
108 |
EU584557.1 |
| |
Reverse: |
ATGGGCGTCACACAGGCT |
|
|
| S100A1 |
Forward: |
GGGAGAACAGTTGAGCAGATGGT |
104 |
NM_006271.1 |
| |
Reverse: |
GGAGGGAACAGCGGGATGAA |
|
|
| S100A12 c |
Forward: |
GGCATTATGACACCCTTATC |
168 |
NM_001160272.1 |
| |
Reverse: |
GTCACCAGGACCACGAAT |
|
|
| S100A4 |
Forward: |
GGGGAAAAGGACGGATGAAGC |
96 |
XM_001929560 |
| |
Reverse: |
GCAGGACAGGAAGACGCAGTA |
|
|
| S100A8 c |
Forward: |
GCGTAGATGGCGTGGTAA |
155 |
FJ263391.1 |
| |
Reverse: |
GCCCTGCATGTGCTTTGT |
|
|
| S100A9 c |
Forward: |
CCAGGATGTGGTTTATGGCTTTC |
186 |
NM_001177906.1 |
| |
Reverse: |
CGGACCAAATGTCGCAGA |
|
|
| TLR1 b |
Forward: |
TGCTGGATGCTAACGGATGTC |
102 |
NM_001031775.1 |
| |
Reverse: |
AAGTGGTTTCAATGTTGTTCAAAGTC |
|
|
| TLR2 b |
Forward: |
TCACTTGTCTAACTTATCATCCTCTTG |
162 |
AB085935.1 |
| |
Reverse: |
TCAGCGAAGGTGTCATTATTGC |
|
|
| TLR3 b |
Forward: |
AGTAAATGAATCACCCTGCCTAGCA |
112 |
DQ647698.1 |
| |
Reverse: |
GCCGTTGACAAAACACATAAGGACT |
|
|
| TLR4 b |
Forward: |
GCCATCGCTGCTAACATCATC |
108 |
AY535422.1 |
| |
Reverse: |
CTCATACTCAAAGATACACCATCGG |
|
|
| TLR5 |
Forward: |
CTCGCCCACCACATTA |
156 |
FJ668383 |
| |
Reverse: |
TGAGGGTCCCAAAGAGT |
|
|
| TLR6 b |
Forward: |
AACCTACTGTCATAAGCCTTCATTC |
95 |
NM_213760.1 |
| |
Reverse: |
GTCTACCACAAATTCACTTTCTTCAG |
|
|
| TLR8 b |
Forward: |
AAGACCACCACCAACTTAGCC |
105 |
NM_214187.1 |
| |
Reverse: |
GACCCTCAGATTCTCATCCATCC |
|
|
| TLR9 b |
Forward: |
CACGACAGCCGAATAGCAC |
122 |
AY859728.1 |
| |
Reverse: |
GGGAACAGGGAGCAGAGC |
|
|
| TNFα |
Forward: |
TGGTGGTGCCGACAGATGG |
102 |
EU682384.1 |
| |
Reverse: |
GGCTGATGGTGTGAGTGAGGAA |
|
|
| VCAM1 |
Forward: |
TCAGGGAGGACACAAAGAAGGG |
135 |
NM_213891.1 |
| Reverse: | AAACGGCAAACACCATCCAAAGT | |||
Table 3.
qRT-PCR validation of representative genes that are differentially expressed between control and PCV2-infected PAMs
|
Gene |
GenBank ID |
24 HPI |
48 HPI |
||
|---|---|---|---|---|---|
| Microarray FC | qRT-PCR FC | Microarray FC | qRT-PCR FC | ||
| CD14 |
NM_001097445.2 |
3.60 |
2.35(0.041) |
3.87 |
2.71(0.003) |
| GPNMB |
NM_001098584 |
−2.13 |
−4.04(<0.0001) |
−2.58 |
−7.89(0.002) |
| ICAM1 |
NM_000201.2 |
4.02 |
3.59(<0.0001) |
3.20 |
2.21(0.003) |
| IFNγ |
NM_213948.1 |
−2.04 |
−2.76(0.005) |
3.18 |
5.09(0.006) |
| IL6 |
NM_214399.1 |
2.26 |
4.60(0.001) |
1.36 |
2.47(0.001) |
| IL8 |
NM_213867.1 |
4.86 |
8.65(0.001) |
5.03 |
5.36(0.008) |
| ISG15 |
EU584557.1 |
1.56 |
1.37(0.006) |
2.51 |
2.83(0.031) |
| S100A1 |
NM_006271 |
−3.20 |
−4.42 (0.001) |
−2.49 |
−1.87 (0.01) |
| S100A12 |
NM_001160272.1 |
10.98 |
8.49(0.006) |
5.13 |
3.98(0.005) |
| S100A4 |
XM_001929560 |
−3.14 |
−4.26(0.002) |
−1.98 |
−3.27(0.001) |
| S100A8 |
FJ263391.1 |
12.96 |
15.47(<0.0001) |
6.83 |
4.34(0.003) |
| S100A9 |
NM_001177906.1 |
20.00 |
16.56(0.007) |
9.71 |
6.21(0.025) |
| TLR1 |
NM_001031775.1 |
2.76 |
2.29 (0.032) |
5.83 |
4.89(0.016) |
| TLR8 |
NM_214187.1 |
−4.35 |
−3.01(0.002) |
−1.14 |
−1.72(0.004) |
| TLR9 |
AY859728.1 |
2.47 |
4.66(0.012) |
1.43 |
2.24(0.0001) |
| TNFα |
EU682384.1 |
5.13 |
5.36(0.016) |
1.96 |
2.53(0.022) |
| VCAM1 | NM_213891 | −2.33 | −7.54(0.001) | −2.90 | −4.83 (0.005) |
All reactions were performed in triplicate. FC: fold-change.
Functional GO analysis of DE genes in PCV2-infected PAMs
The Ingenuity pathway analysis (IPA) program was used to determine the relevance of PAM gene functionalities and gene networks associated with PCV2 infection. While 266 (24 HPI) and 175 (48 HPI) DE genes were imputed into IPA, only 173 and 124 were characterized with specific cellular functions.
The 173 DE genes from 24 HPI PAMs were placed into 19 functional groups (Additional file 6: Table S6 and Additional file 7: Figure S8). The most frequently represented category of gene functionality was Inflammatory Response (Cellular Movement, and Immune Cell Trafficking) with 22 genes, followed by Cell-To-Cell Signaling and Interaction, Cellular Growth and Proliferation, and Cellular Compromise.
The 124 DE genes from 48 HPI PAMs were placed into 16 functional groups (Additional file 8: Table S7 and Additional file 9: Figure S9). The most frequently represented category of gene functionality was also Inflammatory Response with 25 genes, followed by Antigen Presentation, Cellular Movement, Lipid Metabolism, Cell Cycle, and Cellular Growth and Proliferation.
qRT-PCR validated DE genes in PCV2-infected PAMs
In order to validate the differential expression of some of the DE genes identified by microarray, eleven up-regulated genes and six down-regulated genes were selected for qRT-PCR analysis. All the selected genes were successfully amplified and the expression patterns corresponded to those observed by microarray. Although the extent of fold-change (FC) varied between the qRT-PCR and microarray results (Table 3 and Figure 2), the differential expression patterns were coincident, indicating the reliability of the microarray analysis.
Figure 2.
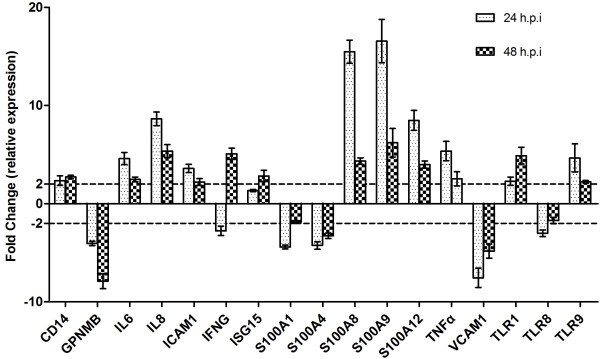
Validation of microarray data by qRT-PCR analysis. Fold-changes of gene expression in the PAMs following PCV2 infection compared with the control group are shown. The fold-changes were calculated by the formula of 2-ΔΔ Ct. Data is presented as mean ± SD of triplicate reactions for each gene transcript.
Toll-like receptors pathway analysis
Activation of the innate immune response is controlled in large part by the TLR family of pattern-recognition receptors (PRRs). A previous study showed that the DNA of PCV2 could impair the plasmacytoid- and monocyte-derived dendritic cells ability to induce interferon (IFN)-α and TNF-α by interacting with TLR7 and TLR9 [23]. In the current study, we verified the expression of TLR1, TLR2, TLR3, TLR4, CD14, TLR5, TLR6, TLR8, and TLR9 (the primers are listed in Table 2. Surprisingly, the two time points examined showed significant differences in up-regulation of TLR1 (2.29-fold vs. 4.89 fold), TLR4 cofactor CD14 (2.35-fold vs. 2.71-fold) and TLR9 (4.66-fold vs. 2.24-fold), and down-regulation of TLR8 (3.01-fold vs. 1.72-fold).
Profile of PAM secreted cytokines in response to PCV2-infection
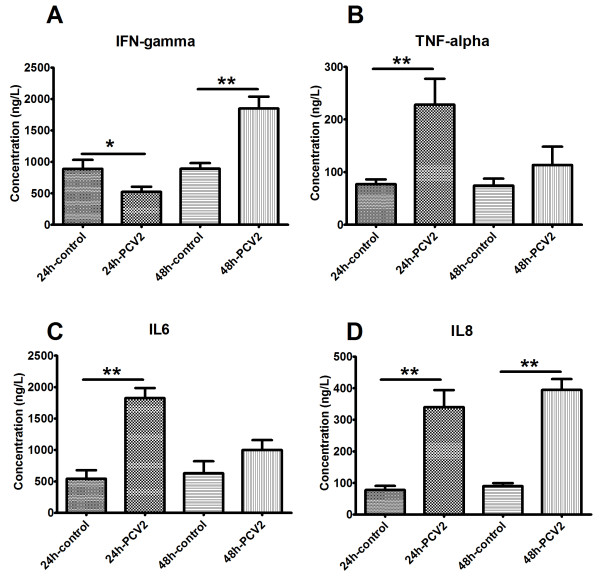
When compared with the supernatant of mock-inoculated PAMs, PCV2-infected PAMs secreted significantly higher levels of interleukin (IL)-8 (340.09 ± 53.64 vs 77.43 ± 13.15, by 4~6-fold; Figure 3D). Tumor necrosis factor-alpha (TNFα) (228.13 ± 49.36 vs 77.12 ± 9.10) and IL-6 (1827.02 ± 157.32 vs 542.70 ± 137.05) levels were significantly higher in supernatants of PCV2-inoculated PAMs at 24 HPI (2~3-fold vs. mock-inoculated PAMs; Figure 3B-C). Interferon-gamma (IFNγ), however, appeared to be significantly down-regulated (552.87 ± 83.25 vs 889.99 ± 141.03, by 1.70-fold) at 24 HPI but significantly up-regulated (1848.10 ± 190.19 vs 892.37 ± 89.66, by 2-fold) at 48 HPI (vs. mock-inoculated; Figure 3A). The relative mRNA levels of porcine IL-8, TNFα, IL-6, and IFNγ showed the same DE trends as the secreted protein levels (Table 3).
Figure 3.
Validation of microarray data by ELISA. PCV2-induced changes in secreted levels of IFNγ (A), TNFα (B), IL-6 (C) and IL-8 (D). The protein levels in culture supernatants of mock-inoculated and PCV2-infected PAMs were measured by ELISA. Data are expressed as ng/L of three independent experiments. * 0.01 <p <0.05 and ** p<0.01 for PCV2-infected vs. mock-inoculated groups for the corresponding HPI.
Discussion
PCV2 is an important pathogen of swine that has effects on both the pork industry and animal welfare. Despite the high mortality rates and widespread incidence of this pathogen, a definitive cure has not yet been found. An effective vaccine was created by Merial in 2005 (Circovac®; Merial, France) [24], and entered into widespread use in 2009. Despite the fact that the current vaccine strategy can efficiently control PCV2 infection, immunization failures exist in the field, and the molecular mechanisms underlying PMWS and porcine dermatitis and nephropathy syndrome (PDNS) caused by PCV2 remain largely unknown. The present study aimed to identify genes involved in the immune response against PCV2 in primary alveolar macrophages. Furthermore, by generating a comprehensive transcriptomic profile of the temporal PCV2 pathogenic process in the target host cells, we hoped to gain insights into the underlying molecular interactions and signaling pathways that may represent novel targets of improved preventative and therapeutic strategies.
Toll-like receptors
Toll-like receptors are essential sensors of microbial infection that are involved in the recognition of a variety of microbial products [25-27]. To date, 13 TLRs have been identified and characterized for their particular cognate ligands and signaling cascades to trigger pathogen-targeted immune responses [28]. For example, TLR7 recognizes guanosine- and uridine-rich sequences derived from ssRNA viruses [29], while TLR9 recognizes single-stranded DNA (ssDNA) in endosomal compartments and signals to induce the production of type 1 interferons (IFNs) and pro-inflammatory cytokines and chemokines; PCV2 DNA has been shown to modulate cytokine secretion thorough interaction with TLR9 [30]. In our study of the circular, single-stranded DNA virus, PCV2, differential gene expression of TLRs was found. Specifically, PCV2 infection of PAMs led to increased expression of TLR1, TLR4 cofactor CD14, and TLR9. Monocytic CD14 signaling is usually activated upon exposure to bacterial LPS, and this molecule’s up-regulation may induce ineffective antibacterial immunity. In addition, a similar mechanism has been demonstrated in human monocytic cells upon exposure to the severe acute respiratory syndrome (SARS) coronavirus [31]. These receptors’ up-regulation may contribute to the increased sensitivity of macrophages to other pathogens, thereby augmenting the inflammation and host immune response. Such an enhanced immune response may be beneficial, but an over-stimulation of the innate response could result in tissue damage. For example, the same phenomenon was observed upon macrophage exposure to tobacco smoke, wherein the expression of TLR3 was increased and severe inflammation was observed after treated with TLR3 ligand [32].
MHC class II molecules
MHC class II molecules play a central role in the initiation of the immune response by binding and presenting immunogenic peptides to CD4+ Th lymphocytes. However, only a few cell types express MHC class II molecules, such as the antigen presenting cell types that include macrophages, B lymphocytes, Langerhans cells, and dendritic cells. The very complex and tight regulation of MHC class II gene expression has direct implications for T lymphocyte activation. These molecules include DOA, DOB, DQA1, DQB1, DRA, and DRB1, and their molecule chaperone CD74; all of which were up-regulated in PCV2-infected PAMs. This expression pattern contradicts the host response observed for other viruses, such as African swine fever virus (CSFV) [33], classical swine fever virus [34] and influenza A virus [35], and that was observed in follicles and PAMs of PMWS piglets [36,37]. Increased expression of MHC class II molecular have been shown to be correlated to increased interferon production in pigs infection infected with hog cholera virus [38]. IFN-γ up-regulation have been shown after 48 HPI, its up-regulation could benefit PCV2 replication [39]. The aberrant expression of class II antigens could also compromise production of immunoregulatory cytokines, compromising immune function and perhaps favor viral persistence through escape from immune recognition.
Apoptosis and growth arrest
Apoptosis is considered to be an important host mechanism that interrupts viral replication and eliminates virus-infected cells. Viruses often kill infected cells by inducing apoptosis rather than necrosis, but repress apoptosis to prolong the life of the cell and increase the yield of progeny virions. SERPINB9, also called PI-9, belongs to the large superfamily of serine proteinase inhibitors, which bind to and inactivate serine proteinases, and acts to protect against granzyme B-mediated apoptosis [40]. PCV2-infected PAMs were found to have enhanced SERPINB9 expression, which may serve to prevent apoptosis and allow for longer-term replication and more virion production. BNIP3, an apoptosis-inducing dimeric mitochondrial protein [41], was also found to be increased in PCV2-infected PAMs. It is possible that this factor may represent the host’s counter response to PCV2, by which host-induced apoptosis would act to limit the amount of infected cells. It is also possible that these BNIP3 overexpressing cells represent a subset of PAMs that have reached full capacity of virus load, in which the virus itself may have manipulated the host gene expression to facilitate cell death and release of the virus progeny.
Actin alpha 2 (ACTA2) is a smooth muscle α-actin, whose expression is transformation-sensitive to growth signals in normal cells. Actively proliferating fibroblasts and smooth muscle cells have low levels of ACTA2, but the inhibition of cell proliferation by density arrest or treatment with antimitotic agents induces the smooth muscle α-actin promoter [42]. Thus, the ACTA2 up-regulation that was detected in PCV2-infected PAMs may be the result of growth arrest induced by the virus The same phenomenon was found in Rhabdomyosarcoma cells after EV71 infection [43].
ENO1 is involved in the induction of cell death in fibroblasts and has been implicated in tumorigenicity of human breast carcinoma cells [44]. Its regulation of cell growth is further suggested by the presence of functional transcriptional repression domains [45]. PCV2-infected PAMs showed enhanced ENO1 expression. It is possible that up-regulation of this gene may enhance growth arrest in PCV2-infected cells, thereby culminating in cell death.
Inflammatory response
Serum amyloid A 1 (SAA1) is a member of the SAA family of apolipoproteins synthesized in response to cytokines released by activated macrophages, which often serve as clinical markers of acute and chronic inflammatory diseases [46]. The pattern of increased SAA1 expression observed in PCV2-infected PAMs suggested that PCV2 induced an acute inflammatory response at the early time point (24 HPI) of infection. TNF-α and IL-1β are acute-phase proinflammatory mediators that promote inflammation and induce fever, tissue destruction, and, in some cases, shock and death [47]. Our study showed that PCV2 could induce TNF-α and IL-1β expression (Table 1) in PAMs at 24 HPI and 48 HPI. These results are in good agreement with those from a previous study of lymphoid tissues and peripheral blood mononuclear cells (PBMCs) of PMWS pigs [17,48]. SAA1, TNF-α, and IL-1β up-regulation after PCV2 infection suggests the occurrence of systemic inflammation following PCV2 infection.
Lymphohistiocytic to granulomatous pneumonia is a common histopathological finding in naturally-acquired PMWS and experimentally-infected PCV2 pigs [49,50]. Chemokines, such as MCP-1/CCL-2, were found to be up-regulated in the PCV2-infected PAMs. In a previous study, MCP-1 was suggested to play an important role in the pathogenesis of granulomatous inflammation in PMWS-affected pigs [51-53]. The progressive granulomatous inflammation has been speculated to be compromise organ function and result in PMWS-associated fatalities [54]. Furthermore, the MCP-1 cytokine is known to be an important chemokine for the recruitment of monocytes from the blood [55]. Thus, macrophage infiltration has been suggested as an important feature of the pathogenesis and progression of PMWS [56].
Recruitment of inflammatory cells in injured tissue is regulated by various cytokines and chemokines. PMWS-affected pigs present with increased expression of MIP-1, CCL-2, IL-1β, IL-8, and TNF-α mRNA in the lymphoid tissues and PBMCs, suggesting that such cytokines and chemokines may contribute to the development of granulomatous lymphadenitis and recruitment of macrophages or other inflammatory cells to the PCV2-infected tissues [17,52,57]. The PCV2-infected PAMs in our study showed up-regulation of MIP-1, CCL-2, IL-1β, IL-8 and TNF-α mRNA expression. TNF-α is known to activate microvascular endothelium and cause a pyrexic response [47]. Increased production of TNF-α may induce fever and respiratory distress in PCV2-infected pigs [58,59]. IL-8 is a chemoattractant for neutrophils and other polymorphonuclear leukocytes (PMNs) that are produced following acute infection [60]. Interaction of IL-8 and MCP-1/CCL-2 is also known to trigger tight adhesion of monocytes to vascular endothelium under flow conditions [61]. The ability of PCV2 to induce IL-8 production in PAMs or PBMCs has been previously reported [57,62,63]. Similar to IL-8, the PAM-derived chemokine AMCF-II/CXCL5 [64] is able to recruit PMNs to a lesion area [65]. Unlike the PMN-derived chemoattractants discussed above, MCP-1 is a powerful chemoattractant for monocytes and its activities can be further enhanced by IL-8 [61]. It is reasonable to speculate that the development of interstitial pneumonia in pigs suffering from naturally-acquired PMWS or experimental infection of PCV2 may result from the recruitment of acute phase inflammatory cells and subsequent mononuclear phagocytic cells from blood vessels to interalveolar septa and alveolar spaces by up-regulated cytokines and chemokines released from PCV2-containing PAMs.
Antiviral immune response
IFNγ is a Th1-specific cytokine produced by macrophages, NK cells, and other cell types during the onset of viral infection. It has a potent antiviral property that contributes to the control of acute viral infections, and is an important mediator of cellular responses. IFNγ mRNA was found to be slightly down-regulated at the early stage of in vitro PCV2 infection (24 HPI) of PAMs. This repressed expression may be indicative of suppressed Th1 responses, which may facilitate viral persistence and delayed viral clearance. At the late stage of in vitro infection (48 HPI), however, the IFNγ expression level increased. Such an up-regulation of expression may indicate a virus-modulated mechanism to enhance infection and replication at the late stage of PCV2 infection in vivo[39].
Conclusions
This is the first study to evaluate the gene expression profile of PCV2-infected PAMs in vitro. Microarray analysis showed that expression of 266 and 175 PAM genes was altered after 24 and 48 hours of PCV2 infection, respectively. Among these, several genes related to the inflammatory response were up-regulated, suggesting that PCV2 may induce systemic inflammation. Collectively, this work established a comprehensive differential transcription profile of early and late PCV2 infection in PAMs, which revealed that PCV2 can induce persistent cytokine production mainly through the TLR1 and TLR9 pathways.
Methods
Virus and cells
The PCV2-WH strain (GenBank accession number: FJ870967) was isolated from pooled samples of spleen and lymph nodes of a PMWS-affected pig. The virus was amplified by passaging in PCV-free porcine kidney (PK-15) cells and confirmed as PCV2 by nucleotide sequence analysis and reactivity with PCV2 Cap protein-specific monoclonal antibody [66]. The titer of the PCV2 inoculum was 1.0×107 TCID50/mL, as determined by titration in PCV-free PK-15 cells.
All animals’ tissue collection procedures were performed according to protocols approved by the Hubei Province PR China for Biological Studies Animal Care and Use Committee. Three 3-week-old Large White piglets were obtained from a herd in Hubei province, which tested negative for both PCV2 and porcine reproductive and respiratory syndrome virus (PRRSV), and were raised in controlled lab conditions. For the first three days of containment, the pigs were administered daily doses of enrofloxacin (1 mL of 5% solution) and lincospectin/spectinomycin (1 mL of 5 or 10% solution) to eliminate any residual bacterial pathogens [67]. Seven days later, the piglets were sacrificed. PAMs were collected by bronchoalveolar lavage and frozen in liquid nitrogen, as previously described [68]. Prior to use, the cells were confirmed as negative for PCV1, PCV2, parvovirus, pseudorabies virus (PRV), classical swine fever virus (CSFV), and PRRSV variously by PCR and RT-PCR [69,70]. The PAM phenotype was confirmed by flow cytometric detection of the macrophage markers SWC3, CD169, and SLAII [68].
RNA isolation and IFA
PAMs were thawed and cultured for 48 h before treatment, as previously described [71,72]. One primary culture from each animal was split into two for experimental analysis. The first was infected with PCV2-WH from the 11th passage at a multiplicity of infection (MOI) of 1. The second was mock-inoculated with DMEM to serve as a control. Virus uptake by the cells was determined by IFA using a PCV2 Cap protein-specific monoclonal antibody [66] and by quantitative real-time PCR (qPCR) [73]. Briefly, DNA was extracted from the cells using the E.Z.N.A.™ Viral DNA Kit (OMEGA, USA) according to the manufacturer’s instructions. The amplification was performed in a 25 μL reaction mixture containing 12.5 μL of 2× THUNDERBIRD Probe qPCR Mix (TOYOBO, Japan), 9.45 μL sterile, nuclease-free water, 10 pmol of forward primer (5′-CCAGGAGGGCGTTCTGACT-3'), 10 pmol of reverse primer (5′-CGTTACCGCTGGAGAAGGAA-3′), 4 pmol of TaqMan probe (5′-AATGGCATCTTCAACACCCGCCTCT-3′), 0.05 μL of 50× ROX reference dye, and 2 μL of the extracted DNA. The reaction was run in a real-time thermocycler (7500 PCR System; Applied BioSystems Inc.) with the following conditions: 1 cycle at 50°C for 2 min, 1 cycle at 95°C for 10 min, and 45 cycles at 95°C for 15 s and 62°C for 60 s (real time).
At 24 HPI and 48 HPI, the cells were collected for RNA extraction with the TRIzol reagent (Invitrogen Life Technologies, USA). The extracted RNA was further purified with an RNA clean-up kit (Macherey-Nagel, Germany) and applied to a Bioanalyzer 2100 spectrophotometer (Agilent Technologies, USA) to assess the integrity, quality, and quantity of the sample. The corresponding supernatants were collected for subsequent protein analysis and stored at −80°C until use.
RNA labeling, microarray hybridization, and data analysis
RNA was converted to aminoallyl-RNA with the Amino Allyl II MessageAmp™ aRNA Amplification Kit (Ambion, USA) and labeled with either Cy3 or Cy5 (Reactuve Dye Pack; Amersham, Sweden). Each Cy3- (or Cy5-) coupled PAM sample was then hybridized to its corresponding Cy5- (or Cy3-) coupled reference sample anchored on a pig microarray slide containing 20,400 oligonucleotides (US Pig Genome Coordination Program; http://www.animalgenome.org/pig/resources/array_request.html). Hybridizations and analyses were carried out according to a previously published protocol from CapitalBio Corporation (Beijing, China) [74]. Raw data was extracted from the TIFF images using LuxScan 3.0 software (CapitalBio). A spot-exclusion method was adopted to filter out faint spots, with signal intensities in the lowest 50%, and exclude the corresponding genes from further analysis. An intensity-dependent LOWESS program in the R language package was used to normalize the two-channel ratio values. As a measure of technical replication, each experiment was conducted with the corresponding dye swap.
Gene Cluster 3.0 and Eisen’s Treeview software (Stanford University, USA) were used to compare similarities among individual samples. Cy3/Cy5 ratios were log-transformed (base 2), median centered by arrays and genes, and hierarchically clustered (average linkage correlation metric). To determine gene products with significantly up-regulated expression, the PAM 24 HPI data sets were assessed for increases in average intensity of at least 2-fold. Statistical comparisons were performed by the one-class method in the significance analysis of microarray (SAM software version 3.0, Stanford University) [1]. All the microarray results from this study were deposited in the NCBI Gene Expression Omnibus database under the following accession numbers: platform, GPL13968; Series, GSE30918; Samples, GSM766444, GSM766445, GSM766446, GSM766447, GSM766448, GSM766449.
Functional interpretation of microarray data as well as pathway and network analysis
Differentially expressed genes were selected for pathway exploration using the IPA web-based query system (Ingenuity Systems, http://www.ingenuity.com), this system can generate the functional analysis and networks that are most significant to the data set, IPA compares proteins in the input group and displays a rank-ordered list of pathways and networks, whose activities are most likely affected. Ficher’s exact test was used to calculate a p-value determining the probability that each biofunction assigned to that data set is due to chance alone. Genes from the dataset that met the fold change cut-off of 2.0 were considered for the analysis. The score is the probability that a collection of proteins equal to or greater than the number in a network could be achieved by chance alone. A score of 3 indicates that there is a 1/1000 chance that the focus proteins are in a network due to random chance, therefore, score of 3 or higher have a 99.9% confidence of not being generated by random chance alone.
Quantitative RT-PCR
Equal amounts of RNA (1 μg) from PAMs was used to generate cDNA with oligo (dT) primers using the SuperScript II RNase H Reverse Transcriptase Kit (Invitrogen, USA). Quantitative RT-PCR reactions were set up in triplicate using reagents from the SYBR Green I PCR Kit (Toyobo) and 10 μM primers (designed by the Beacon Designer 7.0 program, http://www.premierbiosoft.com; Table 2). The amplification reactions were performed on a LightCycler 480 (Roche) with the following conditions: 30 s at 95°C, 40 cycles of 95°C for 10 s, 60°C for 5 s and 72°C for 10 s. Subsequently, melting curve analysis and quantitative analysis of the data were performed using the Roche LightCycler 480 software version 1.5.0. Each sample was run in triplicate. Samples were normalized at each respective time point, using mock-infected PAMs as calibrators and the hypoxantine phosphoribosyltransferase (HPRT-1) and glyceraldehyde-3-phosphatedehydrogenase (GAPDH) as the reference genes.. The average threshold cycles of the replicates (each gene was detected three times) were used to calculate the fold-changes between PCV2-infected or mock-inoculated PAMs by using the formula of 2-delta-delta Ct described by Livak and Schmittgen [75].
Cytokine detection in culture supernatant
The secreted protein levels of IFNγ, IL-6, IL-8 and TNFα were determined by testing culture supernatants with the respective commercial ELISA kits (R&D Systems, USA). All samples were tested in triplicate and read at 450 nm using an ELISA plate reader (Bio-Tek, USA).
Statistical analysis of quantitative PCR data
Statistical analyses were carried out using Microsoft Excel 2007 (Microsoft Co., USA). Differences between groups were assessed by one-way repeated measures analysis of variance followed by Tukey’s multiple comparison tests. P-values less than 0.05 were considered to indicate statistical significance.
Abbreviations
CSFV: Classical swine fever virus; DE: Differentially expressed; FC: Fold-change; FDR: False discovery rate; GO: Gene ontology; HPI: Hours post infection; IFNγ: Interferon-gamma; IL: Interleukin; IPA: Ingenuity pathway analysis; ORF: Open reading frame; PAMs: Porcine alveolar macrophages; PBMC: Peripheral blood mononuclear cell; PCR: Polymerase chain reaction; PCV: Porcine circovirus; PMN: Polymorphonuclear leukocytes; PRRSV: Porcine reproductive and respiratory syndrome virus; qPCR: Quantitative PCR; qRT-PCR: Quantitative reverse transcription PCR; TLR: Toll-like receptor; TNFα: Tumor necrosis factor-alpha.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
LWT carried out most of the experiments and wrote the manuscript. HQG, CHC, CC, and JCA critically revised the manuscript and the experimental design. LSQ, WY, DF, YWD, and YK helped with the experiments. All of the authors read and approved the final version of the manuscript.
Supplementary Material
PAM transcriptome analysis following PCV2 infection at 24 and 48 hours post-infection in comparation to the mock-infected control.
DE transcripts (n=267) after PCV2 infection at 24 hours post-infection (q-value<5%, FC≥2.0 or ≤0.5). Fold changes represent the difference in expression levels in infected and mock-infected control.
DE transcripts (n=175) after PCV2 infection at 48 hours post-infection, (q-value<5%, FC≥2.0 or ≤0.5). Fold changes represent the difference in expression levels in infected and mock-infected control.
Six DE transcripts at 48 HPI, as compared to 24 hours post-infection (q-value<5%, FC≥2.0 or ≤0.5).
Complete list of the DE transcripts at 24 and 48 hours post-infection (q-value<5%, FC≥2.0 or ≤0.5).
Gene list of DE genes grouped by IPA based on gene function after 24 hours post-infection. The table contains columns with names of genes involved in the network, the score value and the top functions. Focus genes are shown in red.
Top networks of interacting genes from the DE genes at 24 hours post-infection analyzed by IPA.
Gene list of DE genes grouped by IPA based on gene function after 48 hours post-infection. The table contains columns with names of genes involved in the network, the score value and the top functions. Focus genes are shown in red.
Top networks of interacting genes from the DE genes at 48 hours post-infection analyzed by IPA.
Contributor Information
Wentao Li, Email: wentaovet@yahoo.com.
Shuqing Liu, Email: liushuqing112@yahoo.com.
Yang Wang, Email: andywangseven@yahoo.com.cn.
Feng Deng, Email: dengfeng112@gmail.com.
Weidong Yan, Email: yanwd2007@163.com.
Kun Yang, Email: yangkun369@126.com.
Huanchun Chen, Email: chenhch@mail.hzau.edu.cn.
Qigai He, Email: heqigai@yahoo.com.
Catherine Charreyre, Email: Catherine.Charreyre@merial.com.
Jean-Christophe Audoneet, Email: Jean-Christophe.AUDONNET@merial.com.
Acknowledgements
This work was supported by grants from China Agricultural Research System (No. CARS-36) and Merial Limited.
References
- Cano J, Rodr¨ªguez-Cari o C, Sogbe E, Utrera V, D¨ªaz C, Segal¨¦s J, Olvera A, Calsamiglia M. Postweaning multisystemic wasting syndrome in pigs in Venezuela. Vet Rec. 2005;156(19):620. doi: 10.1136/vr.156.19.620-a. [DOI] [PubMed] [Google Scholar]
- Muhling J, Raye WS, Buddle JR, Wilcox GE. Genetic characterisation of Australian strains of porcine circovirus types 1 and 2. Aust Vet J. 2006;84(12):421–425. doi: 10.1111/j.1751-0813.2006.00081.x. [DOI] [PubMed] [Google Scholar]
- Done S, Gresham A, Potter R, Chennells D. PMWS and PDNS-two recently recognised diseases of pigs in the UK. In Pract. 2001;23(1):14. doi: 10.1136/inpract.23.1.14. [DOI] [Google Scholar]
- Solis M, Wilkinson P, Romieu R, Hernandez E, Wainberg MA, Hiscott J. Gene expression profiling of the host response to HIV-1 B, C, or A/E infection in monocyte-derived dendritic cells. Virology. 2006;352(1):86–99. doi: 10.1016/j.virol.2006.04.010. [DOI] [PubMed] [Google Scholar]
- Chakrabarti AK, Vipat VC, Mukherjee S, Singh R, Pawar SD, Mishra AC. Host gene expression profiling in influenza A virus-infected lung epithelial (A549) cells: a comparative analysis between highly pathogenic and modified H5N1 viruses. Virol J. 2010;7:219. doi: 10.1186/1743-422X-7-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkhalil A, Hammamieh R, Hardick J, Ichou MA, Jett M, Ibrahim S. Gene expression profiling of monkeypox virus-infected cells reveals novel interfaces for host-virus interactions. Virol J. 2010;7:173. doi: 10.1186/1743-422X-7-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernasconi M, Berger C, Sigrist JA, Bonanomi A, Sobek J, Niggli FK, Nadal D. Quantitative profiling of housekeeping and Epstein-Barr virus gene transcription in Burkitt lymphoma cell lines using an oligonucleotide microarray. Virol J. 2006;3:43. doi: 10.1186/1743-422X-3-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Zhang A, Chen B, Teng L, Wang Y, Chen H, Jin M. Response of swine spleen to Streptococcus suis infection revealed by transcription analysis. BMC Genomics. 2010;11:556. doi: 10.1186/1471-2164-11-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Li C, Fang M, Zhu M, Li X, Zhou R, Li K, Zhao S. Understanding Haemophilus parasuis infection in porcine spleen through a transcriptomics approach. BMC Genomics. 2009;10:64. doi: 10.1186/1471-2164-10-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilpin DF, McCullough K, Meehan BM, McNeilly F, McNair I, Stevenson LS, Foster JC, Ellis JA, Krakowka S, Adair BM. et al. In vitro studies on the infection and replication of porcine circovirus type 2 in cells of the porcine immune system. Vet Immunol Immunop. 2003;94(3–4):149–161. doi: 10.1016/s0165-2427(03)00087-4. [DOI] [PubMed] [Google Scholar]
- Chang HW, Pang VF, Chen LJ, Chia MY, Tsai YC, Jeng CR. Bacterial lipopolysaccharide induces porcine circovirus type 2 replication in swine alveolar macrophages. Vet Microbiol. 2006;115(4):311–319. doi: 10.1016/j.vetmic.2006.03.010. [DOI] [PubMed] [Google Scholar]
- Allan GM, Ellis JA. Porcine circoviruses: a review. J Vet Diagn Invest. 2000;12(1):3–14. doi: 10.1177/104063870001200102. [DOI] [PubMed] [Google Scholar]
- Choi C, Chae C. Distribution of porcine parvovirus in porcine circovirus 2-infected pigs with postweaning multisystemic wasting syndrome as shown by in-situ hybridization. J Comp Pathol. 2000;123(4):302–305. doi: 10.1053/jcpa.2000.0421. [DOI] [PubMed] [Google Scholar]
- McGuire K, Glass EJ. The expanding role of microarrays in the investigation of macrophage responses to pathogens. Vet Immunol Immunop. 2005;105(3):259–275. doi: 10.1016/j.vetimm.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Chae C. Postweaning multisystemic wasting syndrome: a review of aetiology, diagnosis and pathology. Vet J. 2004;168(1):41–49. doi: 10.1016/j.tvjl.2003.09.018. [DOI] [PubMed] [Google Scholar]
- Chae C. A review of porcine circovirus 2-associated syndromes and diseases. Vet J. 2005;169(3):326–336. doi: 10.1016/j.tvjl.2004.01.012. [DOI] [PubMed] [Google Scholar]
- Darwich L, Pie S, Rovira A, Segales J, Domingo M, Oswald IP, Mateu E. Cytokine mRNA expression profiles in lymphoid tissues of pigs naturally affected by postweaning multisystemic wasting syndrome. J Gen Virol. 2003;84:2117–2125. doi: 10.1099/vir.0.19124-0. [DOI] [PubMed] [Google Scholar]
- Tomas A, Fernandes LT, Sanchez A, Segales J. Time course differential gene expression in response to porcine circovirus type 2 subclinical infection. Vet Res. 2010;41(1):12. doi: 10.1051/vetres/2009060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes LT, Tomas A, Bensaid A, Perez-Enciso M, Sibila M, Sanchez A, Segales J. Exploratory Study on the Transcriptional Profile of Pigs Subclinically Infected with Porcine Circovirus Type 2. Anim Biotechnol. 2009;20(3):96–109. doi: 10.1080/10495390902885785. [DOI] [PubMed] [Google Scholar]
- Lee G, Han D, Song JY, Lee YS, Kang KS, Yoon S. Genomic expression profiling in lymph nodes with lymphoid depletion from porcine circovirus 2-infected pigs. J Gen Virol. 2010;91(Pt 10):2585–2591. doi: 10.1099/vir.0.022608-0. [DOI] [PubMed] [Google Scholar]
- Tomás A, Fernandes LT, Sánchez A, Segalés J. Time course differential gene expression in response to porcine circovirus type 2 subclinical infection. Vet Res. 2009;41(1):12–12. doi: 10.1051/vetres/2009060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S, Zhang M, Li W, Wang Y, Liu Y, He Q. Proteomic analysis of porcine alveolar macrophages infected with porcine circovirus type 2. J Proteomics. 2012;75(11):3258–3269. doi: 10.1016/j.jprot.2012.03.039. [DOI] [PubMed] [Google Scholar]
- Vincent IE, Balmelli C, Meehan B, Allan G, Summerfield A, McCullough KC. Silencing of natural interferon producing cell activation by porcine circovirus type 2 DNA. Immunology. 2007;120(1):47–56. doi: 10.1111/j.1365-2567.2006.02476.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charreyre C, Bésème S, Brun A, Bublot M, Joisel F, Lapostolle B, Sierra P, Vaganay A. Protection of piglets against a PCV2 experimental challenge by vaccinating gilts with inactivated adjuvanted PCV2 vaccine. Proceedings of the 20th IPVS. 2005. pp. 26–30.
- Janeway CA, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S. TLR signaling. Cell Death Differ. 2006;13(5):816–825. doi: 10.1038/sj.cdd.4401850. [DOI] [PubMed] [Google Scholar]
- Takeda K, Kaisho T, Akira S. Toll-like receptors. Annu Rev Immunol. 2003. pp. 335–376. [DOI] [PubMed]
- Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34(5):637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- Lund J, Alexopoulou L, Sato A, Karow M, Adams N, Gale N, Iwasaki A, Flavell R. Recognition of single-stranded RNA viruses by Toll-like receptor 7. P Natl Acad Sci USA. 2004;101(15):5598. doi: 10.1073/pnas.0400937101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmelli C, Steiner E, Moulin H, Peduto N, Herrmann B, Summerfield A, McCullough K. Porcine circovirus type 2 DNA influences cytoskeleton rearrangements in plasmacytoid and monocyte‒derived dendritic cells. Immunology. 2011;132(1):57–65. doi: 10.1111/j.1365-2567.2010.03339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Yen Y-T, Singh S, Kao C-L, Wu-Hsieh BA. SARS-CoV Regulates Immune Function-Related Gene Expression in Human Monocytic Cells. Viral Immunol. 2012;25(4):277–288. doi: 10.1089/vim.2011.0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koarai A, Yanagisawa S, Sugiura H, Ichikawa T, Akamatsu K, Hirano T, Nakanishi M, Matsunaga K, Minakata Y, Ichinose M. Cigarette smoke augments the expression and responses of toll‒like receptor 3 in human macrophages. Respirology. 2012;17(6):1018–1025. doi: 10.1111/j.1440-1843.2012.02198.x. [DOI] [PubMed] [Google Scholar]
- Juarrero MG, Mebus CA, Pan R, Revilla Y, Alonso JM, Lunney JK. Swine leukocyte antigen and macrophage marker expression on both African swine fever virus-infected and non-infected primary porcine macrophage cultures. Vet Immunol Immunop. 1992;32(3–4):243–259. doi: 10.1016/0165-2427(92)90049-V. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Yu YJ, Feng L, Cai XB, Tang HB, Sun SK, Zhang HY, Liang JJ, Luo TR. Global transcriptional profiles in peripheral blood mononuclear cell during classical swine fever virus infection. Virus Res. 2010;148(1–2):60–70. doi: 10.1016/j.virusres.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Liang Q, Luo J, Zhou K, Dong J, He H. Immune-related gene expression in response to H5N1 avian influenza virus infection in chicken and duck embryonic fibroblasts. Mol Immunol. 2011;48(6-7):924–930. doi: 10.1016/j.molimm.2010.12.011. [DOI] [PubMed] [Google Scholar]
- Mandrioli L, Sarli G, Zengarini M, Panarese S, Marcato P. Immunohistochemical MHC-II and interleukin 2-R (CD25) expression in lymph nodes of pigs with spontaneous postweaning multisystemic wasting syndrome (PMWS) Vet Pathol. 2006;43(6):993–997. doi: 10.1354/vp.43-6-993. [DOI] [PubMed] [Google Scholar]
- McNeilly F, Allan G, Foster J, Adair B, McNulty M. Effect of porcine circovirus infection on porcine alveolar macrophage function. Vet Immunol Immunop. 1996;49(4):295–306. doi: 10.1016/0165-2427(95)05476-6. [DOI] [PubMed] [Google Scholar]
- Torlone V, Titoli F, Gialletti L. Circulating interferon production in pigs infected with hog cholera virus. Life Sci. 1965;4(17):1707. doi: 10.1016/0024-3205(65)90218-3. [DOI] [PubMed] [Google Scholar]
- Meerts P, Misinzo G, Nauwynck HJ. Enhancement of porcine circovirus 2 replication in porcine cell lines by IFN-gamma before and after treatment and by IFN-alpha after treatment. J Interf Cytok Res. 2005;25(11):684–693. doi: 10.1089/jir.2005.25.684. [DOI] [PubMed] [Google Scholar]
- Ekert PG, Silke J, Vaux DL. Caspase inhibitors. Cell Death Differ. 1999;6(11):1081–1086. doi: 10.1038/sj.cdd.4400594. [DOI] [PubMed] [Google Scholar]
- Chen G, Ray R, Dubik D, Shi L, Cizeau J, Bleackley RC, Saxena S, Gietz RD, Greenberg AH. The E1B 19K/Bcl-2–binding Protein Nip3 is a Dimeric Mitochondrial Protein that Activates Apoptosis. J Exp Med. 1997;186(12):1975–1983. doi: 10.1084/jem.186.12.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar CC, Kim JH, Bushel P, Armstrong L, Catino JJ. Activation of smooth muscle alpha-actin promoter in ras-transformed cells by treatments with antimitotic agents: correlation with stimulation of SRF:SRE mediated gene transcription. J Biochem. 1995;118(6):1285–1292. doi: 10.1093/oxfordjournals.jbchem.a125020. [DOI] [PubMed] [Google Scholar]
- Leong WF, Chow VTK. Transcriptomic and proteomic analyses of rhabdomyosarcoma cells reveal differential cellular gene expression in response to enterovirus 71 infection. Cell Microbiol. 2006;8(4):565–580. doi: 10.1111/j.1462-5822.2005.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray RB, Steele R, Seftor E, Hendrix M. Human breast carcinoma cells transfected with the gene encoding a c-myc promoter-binding protein (MBP-1) inhibits tumors in nude mice. Cancer Res. 1995;55(17):3747–3751. [PubMed] [Google Scholar]
- Ghosh AK, Steele R, Ray RB. Functional domains of c-myc promoter binding protein 1 involved in transcriptional repression and cell growth regulation. Mol Cell Biol. 1999;19(4):2880–2886. doi: 10.1128/mcb.19.4.2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malle E, De Beer FC. Human serum amyloid A (SAA) protein: a prominent acute-phase reactant for clinical practice. Eur J Clin Invest. 1996;26(6):427–435. doi: 10.1046/j.1365-2362.1996.159291.x. [DOI] [PubMed] [Google Scholar]
- Dinarello CA. Proinflammatory cytokines. Chest. 2000;118(2):503–508. doi: 10.1378/chest.118.2.503. [DOI] [PubMed] [Google Scholar]
- Sipos W, Duvigneau JC, Willheim M, Schilcher F, Hartl RT, Hofbauer G, Exel B, Pietschmann P, Schmoll F. Systemic cytokine profile in feeder pigs suffering from natural postweaning multisystemic wasting syndrome (PMWS) as determined by semiquantitative RT-PCR and flow cytometric intracellular cytokine detection. Vet Immunol Immunop. 2004;99(1–2):63–71. doi: 10.1016/j.vetimm.2004.01.001. [DOI] [PubMed] [Google Scholar]
- Opriessnig T, Halbur PG. Concurrent infections are important for expression of porcine circovirus associated disease. Virus Res. 2012;164(1-2):20–32. doi: 10.1016/j.virusres.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magar R, Larochelle R, Thibault S, Lamontagne L. Experimental transmission of porcine circovirus type 2 (PCV2) in weaned pigs: a sequential study. J Comp Pathol. 2000;123(4):258–269. doi: 10.1053/jcpa.2000.0413. [DOI] [PubMed] [Google Scholar]
- Kim J, Chae C. Expression of monocyte chemoattractant protein-1 but not interleukin-8 in granulomatous lesions in lymph nodes from pigs with naturally occurring postweaning multisystemic wasting syndrome. Vet Pathol. 2003;40(2):181–186. doi: 10.1354/vp.40-2-181. [DOI] [PubMed] [Google Scholar]
- Kim J, Chae C. Expression of monocyte chemoattractant protein-1 and macrophage inflammatory protein-1 in porcine circovirus 2-induced granulomatous inflammation. J Comp Pathol. 2004;131(2–3):121–126. doi: 10.1016/j.jcpa.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Tsai Y, Jeng C, Hsiao S, Chang H, Liu J, Chang C, Lin C, Chia M, Pang V. Porcine circovirus type 2 (PCV2) induces cell proliferation, fusion, and chemokine expression in swine monocytic cells in vitro. Vet Res. 2010;41(5):60. doi: 10.1051/vetres/2010032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krakowka S, Ellis J, McNeilly F, Gilpin D, Meehan B, McCullough K, Allan G. Immunologic features of porcine circovirus type 2 infection. Viral Immunol. 2002;15(4):567–582. doi: 10.1089/088282402320914511. [DOI] [PubMed] [Google Scholar]
- Mantovani A, Bonecchi R, Locati M. Tuning inflammation and immunity by chemokine sequestration: decoys and more. Nat Rev Immunol. 2006;6(12):907–918. doi: 10.1038/nri1964. [DOI] [PubMed] [Google Scholar]
- Segalés J, Domingo M, Chianini F, Majó N, Domínguez J, Darwich L, Mateu E. Immunosuppression in postweaning multisystemic wasting syndrome affected pigs. Vet Microbiol. 2004;98(2):151–158. doi: 10.1016/j.vetmic.2003.10.007. [DOI] [PubMed] [Google Scholar]
- Darwich L, Balasch M, Plana-Duran J, Segales J, Domingo M, Mateu E. Cytokine profiles of peripheral blood mononuclear cells from pigs with postweaning multisystemic wasting syndrome in response to mitogen, superantigen or recall viral antigens. J Gen Virol. 2003;84:3453–3457. doi: 10.1099/vir.0.19364-0. [DOI] [PubMed] [Google Scholar]
- Albina E, Truong C, Hutet E, Blanchard P, Cariolet R, L’Hospitalier R, Mahe D, Allee C, Morvan H, Amenna N. et al. An experimental model for post-weaning multisystemic wasting syndrome (PMWS) in growing piglets. J Comp Pathol. 2001;125(4):292–303. doi: 10.1053/jcpa.2001.0508. [DOI] [PubMed] [Google Scholar]
- Rovira A, Balasch M, Segales J, Garcia L, Plana-Duran J, Rosell C, Ellerbrok H, Mankertz A, Domingo M. Experimental inoculation of conventional pigs with porcine reproductive and respiratory syndrome virus and porcine circovirus 2. J Virol. 2002;76(7):3232–3239. doi: 10.1128/JVI.76.7.3232-3239.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggiolini M, Dewald B, Moser B. Interleukin-8 and related chemotactic cytokines-CXC and CC chemokines. Adv Immunol. 1994;55:97–179. [PubMed] [Google Scholar]
- Gerszten RE, Garcia-Zepeda EA, Lim YC, Yoshida M, Ding HA, Gimbrone MA Jr, Luster AD, Luscinskas FW, Rosenzweig A. MCP-1 and IL-8 trigger firm adhesion of monocytes to vascular endothelium under flow conditions. Nature. 1999;398(6729):718–723. doi: 10.1038/19546. [DOI] [PubMed] [Google Scholar]
- Chang HW, Jeng CR, Lin TL, Liu JJ, Chiou MT, Tsai YC, Chia MY, Jan TR, Pang VF. Immunopathological effects of porcine circovirus type 2 (PCV2) on swine alveolar macrophages by in vitro inoculation. Vet Immunol Immunop. 2006;110(3–4):207–219. doi: 10.1016/j.vetimm.2005.09.016. [DOI] [PubMed] [Google Scholar]
- Forta M, Fernandes LT, Nofrarias M, Diaz I, Sibila M, Pujols J, Mateu E, Segales J. Development of cell-mediated immunity to porcine circovirus type 2 (PCV2) in caesarean-derived, colostrum-deprived piglets. Vet Immunol Immunop. 2009;129(1–2):101–107. doi: 10.1016/j.vetimm.2008.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JB, Rovai LE, Herschman HR. Sequence similarities of a subgroup of CXC chemokines related to murine LIX: implications for the interpretation of evolutionary relationships among chemokines. J Leukoc Biol. 1997;62(5):598–603. doi: 10.1002/jlb.62.5.598. [DOI] [PubMed] [Google Scholar]
- Goodman RB, Forstrom JW, Osborn SG, Chi EY, Martin TR. Identification of two neutrophil chemotactic peptides produced by porcine alveolar macrophages. J Biol Chem. 1991;266(13):8455–8463. [PubMed] [Google Scholar]
- Cao S, Chen H, Zhao J, Lu J, Xiao S, Jin M, Guo A, Wu B, He Q. Detection of porcine circovirus type 2, porcine parvovirus and porcine pseudorabies virus from pigs with postweaning multisystemic wasting syndrome by multiplex PCR. Vet Res Commun. 2005;29(3):263–269. doi: 10.1023/b:verc.0000047501.78615.0b. [DOI] [PubMed] [Google Scholar]
- Genini S, Delputte PL, Malinverni R, Cecere M, Stella A, Nauwynck HJ, Giuffra E. Genome-wide transcriptional response of primary alveolar macrophages following infection with porcine reproductive and respiratory syndrome virus. J Gen Virol. 2008;89(Pt 10):2550–2564. doi: 10.1099/vir.0.2008/003244-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wensvoort G, Terpstra C, Pol JM, Ter Laak EA, Bloemraad M, De Kluyver EP, Kragten C, Van Buiten L, Den Besten A, Wagenaar F. et al. Mystery swine disease in The Netherlands: the isolation of Lelystad virus. Vet Q. 1991;13(3):121–130. doi: 10.1080/01652176.1991.9694296. [DOI] [PubMed] [Google Scholar]
- Ogawa H, Taira O, Hirai T, Takeuchi H, Nagao A, Ishikawa Y, Tuchiya K, Nunoya T, Ueda S. Multiplex PCR and multiplex RT-PCR for inclusive detection of major swine DNA and RNA viruses in pigs with multiple infections. J Virol Methods. 2009;160(1–2):210–214. doi: 10.1016/j.jviromet.2009.05.010. [DOI] [PubMed] [Google Scholar]
- Choi KS, Chae JS. Genetic characterization of porcine circovirus type 2 in Republic of Korea. Res Vet Sci. 2008;84(3):497–501. doi: 10.1016/j.rvsc.2007.05.017. [DOI] [PubMed] [Google Scholar]
- Delputte PL, Nauwynck HJ. Porcine arterivirus infection of alveolar macrophages is mediated by sialic acid on the virus. J Virol. 2004;78(15):8094–8101. doi: 10.1128/JVI.78.15.8094-8101.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chueh LL, Lee KH, Wang FI, Pang VF, Weng CN. Sequence analysis of the nucleocapsid protein gene of the porcine reproductive and respiratory syndrome virus Taiwan MD-001 strain. Adv Exp Med Biol. 1998;440:795–799. doi: 10.1007/978-1-4615-5331-1_103. [DOI] [PubMed] [Google Scholar]
- Yang K, Li W, Niu H, Yan W, Liu X, Wang Y, Cheng S, Ku X, He Q. Efficacy of single dose of an inactivated porcine circovirus type 2 (PCV2) whole-virus vaccine with oil adjuvant in piglets. Acta Vet Scand. 2012;54(1):67. doi: 10.1186/1751-0147-54-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson TA, Lobenhofer EK, Fulmer-Smentek SB, Collins PJ, Chu TM, Bao WJ, Fang H, Kawasaki ES, Hager J, Tikhonova IR. et al. Performance comparison of one-color and two-color platforms within the MicroArray Quality Control (MAQC) project. Nat Biotechnol. 2006;24(9):1140–1150. doi: 10.1038/nbt1242. [DOI] [PubMed] [Google Scholar]
- Livak K, Schmittgen T. Analysis of relative gene expression data using real-time quantitative PCR and the 2-[Delta][Delta] CT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PAM transcriptome analysis following PCV2 infection at 24 and 48 hours post-infection in comparation to the mock-infected control.
DE transcripts (n=267) after PCV2 infection at 24 hours post-infection (q-value<5%, FC≥2.0 or ≤0.5). Fold changes represent the difference in expression levels in infected and mock-infected control.
DE transcripts (n=175) after PCV2 infection at 48 hours post-infection, (q-value<5%, FC≥2.0 or ≤0.5). Fold changes represent the difference in expression levels in infected and mock-infected control.
Six DE transcripts at 48 HPI, as compared to 24 hours post-infection (q-value<5%, FC≥2.0 or ≤0.5).
Complete list of the DE transcripts at 24 and 48 hours post-infection (q-value<5%, FC≥2.0 or ≤0.5).
Gene list of DE genes grouped by IPA based on gene function after 24 hours post-infection. The table contains columns with names of genes involved in the network, the score value and the top functions. Focus genes are shown in red.
Top networks of interacting genes from the DE genes at 24 hours post-infection analyzed by IPA.
Gene list of DE genes grouped by IPA based on gene function after 48 hours post-infection. The table contains columns with names of genes involved in the network, the score value and the top functions. Focus genes are shown in red.
Top networks of interacting genes from the DE genes at 48 hours post-infection analyzed by IPA.