Abstract
Background
Small breast epithelial mucin (SBEM) has been implicated in tumor genesis and micrometastasis in breast cancer. Triple-negative breast cancer (TNBC) was characterized by high incidence in young women,early relapse and a very poor prognosis. The aim of this study was to evaluate the association of SBEM expression in tissues of TNBC with disease-free survival (DFS) and overall survival (OS).
Methods
SBEM protein expression was detected in 87 available formalin-fixed paraffin-embedded (FFPE) tissue specimens from TNBC patients by means of immunohistochemistry (IHC). We analyzed the correlation between the SBEM protein expression and DFS and OS during a 5 year follow-up period, respectively. And a SBEM cut-off value of prognosis was established associated with DFS and OS. SBEM was analyzed against other risk factors in multivariate analysis.
Results
SBEM 3+ score was cut-off value of prognosis and significantly correlated with DFS (p = 0.000) and OS (p = 0.001) in TNBC patients. There was a marked associations (p <0.05) between SBEM 3+ score and tumor size, grade, node status, TNM stage and Ki67. Multivariate analysis showed that patients with SBEM 3+ represented a higher risk of recurrence and mortality than those with a lower SBEM expression (HR = 3.370 with p = 0.008 for DFS and HR = 4.185 with p = 0.004 for OS).
Conclusions
SBEM is an independent risk predictor and may offer utility as a prognostic marker in TNBC patients.
Virtual Slides
http://www.diagnosticpathology.diagnomx.eu/vs/1624613061936917
Keywords: SBEM, DFS, OS, IHC, TNBC
Introduction
Breast cancer remains to be an important public health problem. Triple-negative breast cancer (TNBC), one of five molecular subtypes recognized in 2000 [1,2], lacks estrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor 2 (HER2) expression. TNBC is characterized by high incidence in young women, early recurrence and shows a relative sensitivity of chemotherapy. Most studies [3-5] showed that the prognosis of TNBC was less favorable than that of non- TNBC. Therefore, more attention is being paid to unraveling the oncogenes that expressing at inappropriately high levels or being altered to have novel properties, which leads to invasion and metastasis of carcinoma cells [6]. Small breast epithelial mucin (SBEM) is a newly cloned gene and expresses in breast cancer cell lines rather than in cell lines of non-breast origin [7]. SBEM was only expressed in mammary and salivary glands [7]. High SBEM expression was found to be strongly associated with the histopathological detection of lymph node metastases [7]. Several laboratories showed that SBEM expression correlated with higher tumor grade [8], TNM staging and lymph node metastasis at both mRNA and protein levels [9]. SBEM protein was more frequently observed in ER- than in ER + breast cancers [8,10] and SBEM expression showed a trend towards an association with decreased OS and DFS in SBEM + patients [8]. Valladares Ayerbes et al. [10] studied the expression profiles of SBEM gene in silico and in vitro, and demonstrated that SBEM-mRNA could serve as a marker for bone marrow micro metastasis in breast cancer patients [10]. Researchers also reported that SBEM had the potential to be a specific marker for predicting hematogenous micro metastasis and responses to neo-adjuvant chemotherapy in breast cancer [9].
Based on the above information, SBEM might play an important role in progression and metastasis of breast cancer, especially in TNBC. The aim of this study was to analyze association of SBEM expression in tissue of TNBC with clinical- pathological features, DFS and OS, and to identify new prognostic and/or predictive biomarkers for TNBC patients.
Materials and methods
The study examined cases from 126 patients diagnosed between 2006 and 2008 in our hospital. 39 cases without evaluable tumor tissue were excluded from the analysis. The final database for analysis included 87 cases with histological confirmation. Clinical data of all the cases were reviewed retrospectively from medical records in our hospital. All patients were females and had a minimum 5 years’ follow-up records. All the patients underwent operational treatment according to clinical practice guidelines of National Comprehensive Cancer Network (NCCN) of the United States. None of the patients received neo-adjuvant therapy. Statistic and analysis of clinicopathological parameters, including age at diagnosis, disease stage, tumor size, tumor grade, lymph node status and Ki67, were listed in Table 1.
Table 1.
Clinicopathological characteristics of patients
| Parameter | Number (n) | Subgroup cut-offs |
SBEM < 3 score |
SBEM = 3 score |
P-value | ||
|---|---|---|---|---|---|---|---|
| Number (n) | % | Number (n) | % | ||||
| Age |
87 |
|
76 |
72.4 |
11 |
63.6 |
0.809 |
| X > 35 |
55 |
27.6 |
7 |
36.4 |
|||
| X ≤ 35 |
21 |
|
4 |
|
|||
| TNM staging |
87 |
|
76 |
|
11 |
|
0.047 |
| I (1) |
36 |
47.4 |
2 |
18.2 |
|||
| II (2) |
29 |
38.2 |
4 |
36.4 |
|||
| III (3) |
11 |
14.5 |
5 |
45.5 |
|||
| P53 |
49 |
|
40 |
|
9 |
|
0.435 |
| Mutated |
21 |
52.5 |
7 |
75 |
|||
| No-mutated |
19 |
47.5 |
2 |
25 |
|||
| Node |
87 |
|
76 |
|
11 |
|
0.034 |
| + |
26 |
34.2 |
8 |
72.7 |
|||
| _ |
50 |
65.8 |
3 |
27.3 |
|||
| Grade |
67 |
|
57 |
|
9 |
|
0.471 |
| Low (1) |
20 |
35.1 |
2 |
22.2 |
|||
| Mod (2) |
23 |
40.4 |
3 |
33.3 |
|||
| High (3) |
14 |
24.6 |
4 |
44.4 |
|||
| Size |
87 |
|
76 |
|
11 |
|
0.494 |
| X > 20 mm |
40 |
52.6 |
7 |
63.6 |
|||
| X ≤ 20 mm |
36 |
47.4 |
4 |
36.4 |
|||
| Ki67 | 71 | |
62 |
|
9 |
|
0.027 |
| X > 35 |
19 |
32.8 |
7 |
77.8 |
|||
| X ≤ 35 | 39 | 67.2 | 2 | 22.2 | |||
All cases examined were ER and PR negative by IHC. HER2 status was considered negative if the immunohistochemical score was 0 or 1+, or if the score was 2+ but non-amplification by fluorescence in situ hybridization (FISH), and positive if the score was 3 +.
SBEM expression and evaluation of IHC
All tissues were collected surgically under the supervision of an experienced pathologist. SBEM expression was measured by IHC on FFPE samples.
Streptavidinperosidase (S-P) IHC staining was performed using SBEM antibody of mouse monoclonal (diluted 1/800, Abcam plc. Cambridge, UK). The detailed procedures were done as described by Jennbacken [11].
PBS was used to replace the primary antibody in negative controls. SBEM was a secreted protein and it was mainly located in cell membrane, subordinately in cytoplasm. Normal breast tissues were in general weakly or negative. So it was evaluated and scored if cell membrane and/or cytoplasm reactivity were observed [8]. But there was no relevant clinical cut-off point and the standard evaluation methods reported for SBEM in the literature. According to our data and TMA IHC grading method by Serrero G [12] and Pan [13], our scoring was semiquantitatively categorized as: ≤5% of tumor cells staining with/without weakly stained was negative (0), followed by a score of 1 (>5% of tumor cells and with weak/focal positive staining or ≤5% of tumor cells with strongly stained), 2 (>5% of tumor cells and with moderate/focal positive staining), 3 (>5% of tumor cells and with strong/diffuse positive staining).
Statistical analysis
The correlation between SBEM, Clinicopathological characteristics and survival outcomes was compared by Pearson’s Χ2 test. Survival analyses, including DFS and OS, were performed with the log rank test and all results were displayed in Kaplan–Meier. DFS was defined as the time interval from date of diagnosis to the time of last disease-free follow-up or at death for those patients who died without a previous recurrence. OS was defined as the time interval from date of diagnosis to time of last follow-up or death [14]. Time to recurrence (local, regional and distant) was censored at time of last disease-free follow-up, and at death for those patients who died without a previous recurrence [14]. Statistical significance was defined as P value < 0.05. SPSS17.0 software package was used for all statistical analyses.
In order to observe whether the SBEM expression had an independent prognostic value with conventional risk factors, the risk factor alone, or along with SBEM was analyzed with Cox’s proportional hazards models. Multivariate analysis was performed to determine the association of SBEM with all combined clinical risk factors on DFS and OS.
Results
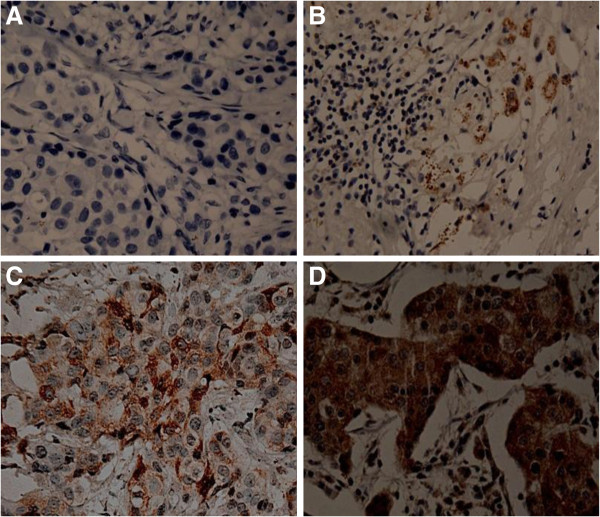
The clinicopathological characteristics of patients were described in Table 1. In the whole group, the median age was 42 (from 24 to 67) years old, the median DFS was 23 months (from 2 to 54 months) and median OS was 34 months (from 6 to 60 months). Figure 1 showed photomicrograph examples of SBEM expression with different scores in TNBC tissues determined by IHC.
Figure 1.
Immunohistochemical staining for SBEM. A: 0+: ≤5% of tumor cells staining with/without weakly stained was negative (original magnification × 200); B: 1+: >5% of tumor cells and with weak/focal positive staining or ≤5% of tumor cells with strongly stained (original magnification × 100); C: 2+: >5% of tumor cells and with moderate/focal positive staining (original magnification × 200); D: 3+: >5% of tumor cells and with strong/diffuse positive staining (original magnification × 200).
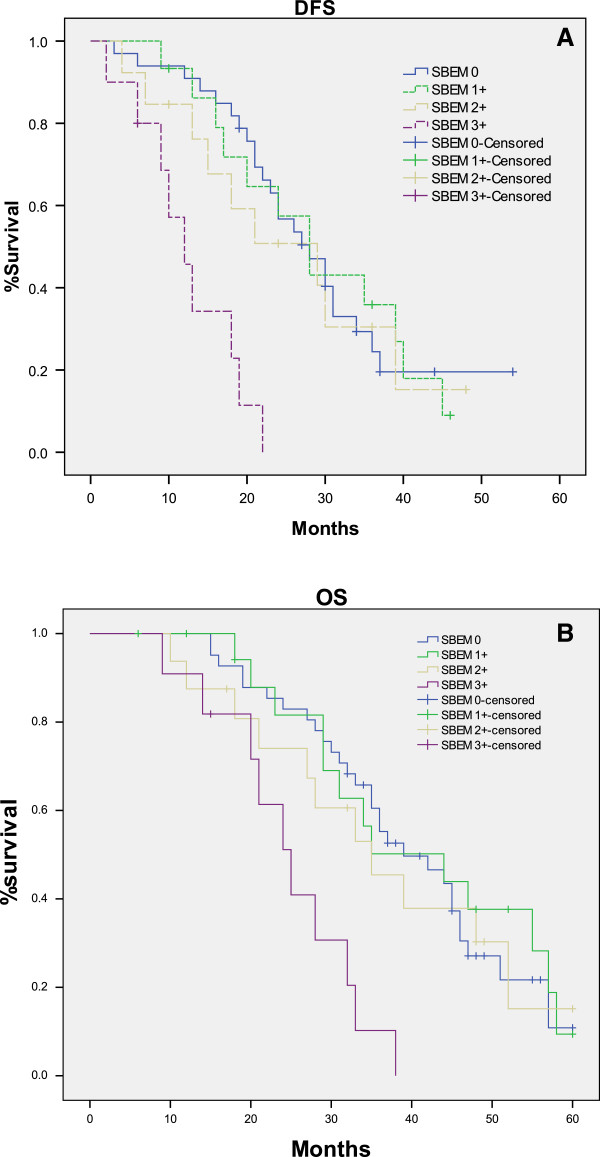
To evaluate SBEM prognostic significance, we analyzed SBEM score (0, 1+, 2+, and 3+) in relation with DFS and OS in TNBC patients. No significant difference was found between DFS or OS and each group (SBEM score of 0, 1+ and 2+) by pairwise comparison methods (p >0.05). But, there was a marked associations between SBEM 3+ score and SBEM score of 0, 1+ and 2+ (p <0.05) (Figure 2). The results of log-rank testing for SBEM different scores were showed in Table 2.
Figure 2.
Kaplan-Meier estimates for DFS and OS by SBEM scores. No significant difference was found between different SBEM score (0, 1+ and 2+) and DFS (A) or OS (B) (p >0.05). But, there was a marked difference between SBEM 3+ score and SBEM score of 0, 1+ and 2+ (p <0.05) on DFS (A) and OS (B).
Table 2.
Log-rank testing for SBEM different scores
| SBEM scores |
DFS |
OS |
||
|---|---|---|---|---|
| Chi-squared | P Value | Chi-squared | P Value | |
|
0 and 1 |
0.020 |
0.887 |
0.124 |
0.724 |
|
0 and 2 |
0.147 |
0.701 |
0.160 |
0.689 |
|
0 and 3 |
22.641 |
0.000 |
15.472 |
0.000 |
|
1 and 2 |
0.087 |
0.768 |
0.623 |
0.430 |
|
1 and 3 |
12.501 |
0.000 |
9.431 |
0.002 |
| 2 and 3 | 6.578 | 0.010 | 5.512 | 0.019 |
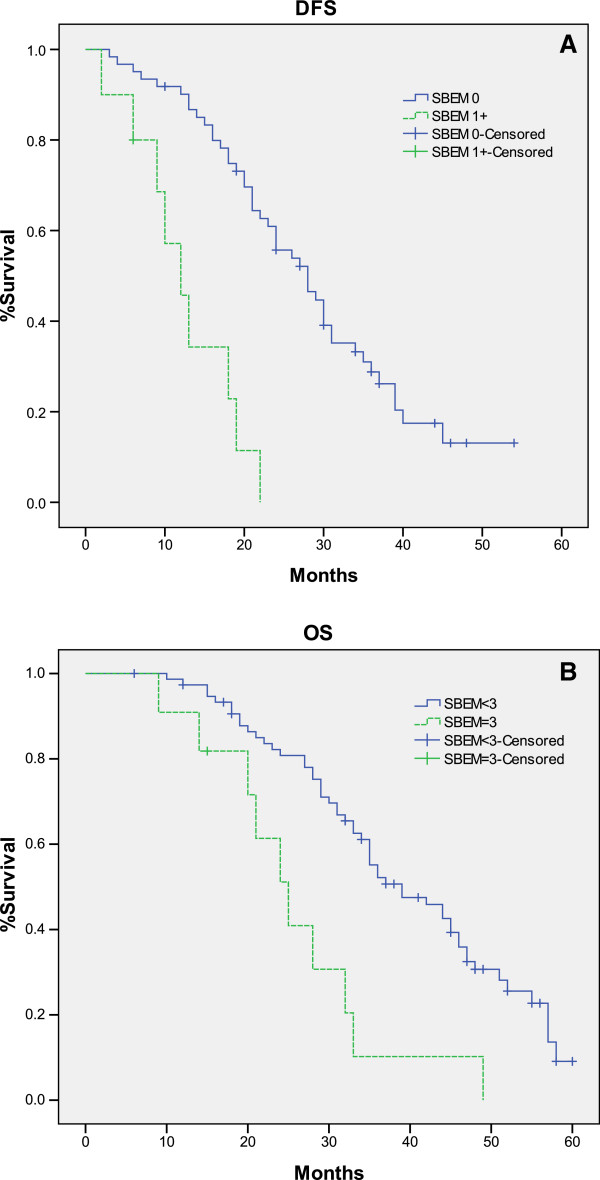
We observed that high SBEM expression with SBEM 3+ score was consistent with high recurrence and death rates, while lower SBEM expression (0, 1+ and 2+) was reversed. Based on the statistics above, we believed that SBEM expression with SBEM 3+ score might be the SBEM cut-off value of prognosis. We divided the cases into two groups, one is the SBEM < 3+ group, the other is SBEM = 3+ group. From Figure 3, we found that DFS and OS function curves showed the large separation between SBEM < 3+ group and SBEM = 3+ group. The log-rank tests confirmed that SBEM score of 3+ was significant associated with DFS and OS (p = 0.000, p = 0.001, respectively). The results of log-rank testing for SBEM score cut-off were showed in Table 3. According to these results, the threshold effect of SBEM score 3+ was verified.
Figure 3.
Kaplan-Meier estimates for DFS and OS by SBEM < 3+ group and SBEM = 3+ group. There was a significant difference between SBEM =3 + group and SBEM score <3+ group on DFS (A) and OS (B) (p = 0.000, p = 0.001, respectively).
Table 3.
Log-rank testing for SBEM score cut-off establishment
| SBEM scores |
DFS |
OS |
||
|---|---|---|---|---|
| Chi-squared | P Value | Chi-squared | P Value | |
|
0 and 1 |
0.020 |
0.887 |
0.124 |
0.724 |
|
0,1 and 2 |
0.120 |
0.729 |
0.399 |
0.528 |
| 0,1,2 and 3 | 23.524 | 0.000 | 11.595 | 0.001 |
On the basis of the cut-off established, 87 patients were divided into 2 groups. Table 1 showed that there were significant associations between SBEM 3+ score and nodal involvement, TNM stage and Ki67 (p < 0.05). Neither SBEM < 3+ group nor SBEM = 3+ group, SBEM expression indicated no significant correlations with age, tumor size and grade. Based on the Kaplan Meier curves for DFS and OS function, the median DFS and OS of SBEM < 3+ group were 28 and 39 months, respectively, while those of SBEM = 3+ group were only 12 and 25 months, respectively. Patients with high SBEM expression had poor clinical outcomes. In SBEM = 3+ group, no patients could survive over 5 years. The longest time of OS was 38 months. In comparison, the patients in SBEM < 3+ group had the higher survival probabilities and longer OS than those in SBEM = 3+ group. We inferred that patients in SBEM < 3+ group had a higher risk of recurrence or death than those in SBEM < 3+ group.
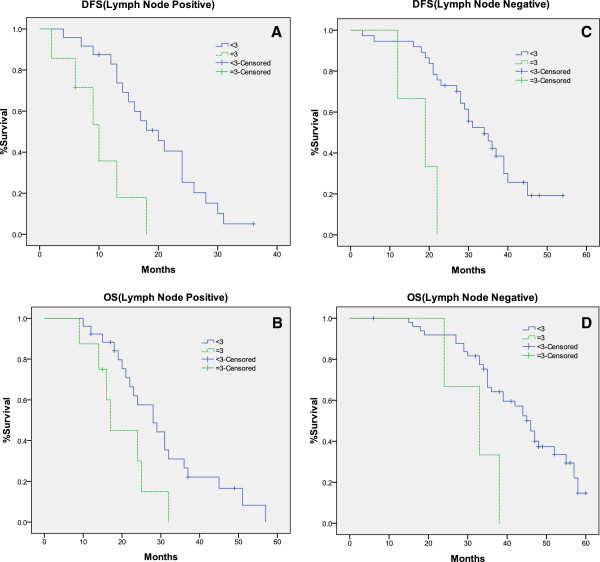
Similar observations of Kaplan Meier curves for DFS and OS function were obtained when the cases were segregated into lymph node negative and positive group (Figure 4). The results of log-rank testing for lymph node positive group and negative group were showed in Table 4.
Figure 4.
Kaplan-Meier estimates for DFS and OS in lymph node positive and negative group. SBEM score of 3+ was significant associated with DFS and OS in both lymph node positive (A, B) and negative group (C, D) (p <0.05).
Table 4.
Log-rank testing for lymph node positive group and lymph node negative group
| Lymph node | DFS | OS | ||
|---|---|---|---|---|
| |
Chi-squared |
P Value |
Chi-squared |
P Value |
|
positive |
9.392 |
0.002 |
4.525 |
0.033 |
| negative | 9.960 | 0.002 | 5.524 | 0.019 |
Univariate analysis showed that SBEM 3+ scores were associated with an HR of 5.768 for DFS and 4.113 for OS in Table 5 and Table 6. It was demonstrated that grade, age, disease stage, lymph node status and Ki67 for DFS and OS were other predictors of survival. From Table 5, we found that there was no marginally change when SBEM was added to each covariate in DFS. Similar results were showed for OS in Table 6.
Table 5.
The impact of SBEM and other risk factors on DFS by Cox’s proportional hazards analysis
| Covariate |
P value |
Hazard radio |
95% Confidence interval |
|||
|---|---|---|---|---|---|---|
| Univariate | Added SBEM | Univariate | Added SBEM | Univariate | Added SBEM | |
| SBEM (3+ vs. <3+) |
0.000 |
0.000 |
5.768 |
5.768 |
2.584-12.876 |
2.584-12.876 |
| Lymph Node (Pos. vs. Neg.) |
0.000 |
0.000 |
3.589 |
3.433 |
1.996-6.451 |
1.893-6.228 |
| Age (>35Y vs. ≤35Y) |
0.749 |
0.785 |
1.103 |
0.918 |
0.606-2.006 |
0.495-1.702 |
| Tumor size (>20 mm vs. ≤20 mm) |
0.457 |
0.583 |
0.814 |
0.858 |
0.473-1.400 |
0.498-1.480 |
| Grade (>1 vs. =1) |
0.035 |
0.053 |
2.052 |
1.911 |
1.053-3.999 |
0.991-3.799 |
| Grade (=3 vs. ≤2) |
0.000 |
0.001 |
3.930 |
3.549 |
1.905-8.110 |
1.699-7.413 |
| TNM (>1 vs. =1) |
0.000 |
0.000 |
3.439 |
3.148 |
1.925-6.143 |
1.816-5.459 |
| TNM (=3 vs. ≤2) |
0.000 |
0.000 |
5.647 |
5.446 |
2.601-12.264 |
2.401-12.352 |
| Ki67 (≤35 vs. >35) | 0.001 | 0.004 | 3.060 | 2.714 | 1.576-5.942 | 1.366-5.393 |
Table 6.
The impact of SBEM and other risk factors on OS by Cox’s proportional hazards analysis
| Covariate |
P value |
Hazard radio |
95% Confidence interval |
|||
|---|---|---|---|---|---|---|
| Univariate | Added SBEM | Univariate | Added SBEM | Univariate | Added SBEM | |
| SBEM (3+ vs. <3+) |
0.000 |
0.000 |
4.113 |
4.113 |
2.004-8.440 |
2.004-8.440 |
| Lymph Node (Pos. vs. Neg.) |
0.000 |
0.000 |
2.972 |
2.734 |
1.777-4.970 |
1.617-4.623 |
| Age (>35Y vs. ≤35Y) |
0.646 |
0.942 |
1.140 |
0.979 |
0.652-1.992 |
0.551-1.738 |
| Tumor size (>20 mm vs. ≤20 mm) |
0.308 |
0.367 |
0.771 |
0.794 |
0.468-1.271 |
0.481-1.311 |
| Grade (>1 vs. =1) |
0.066 |
0.085 |
0.563 |
1.720 |
0.305-1.040 |
0.928-3.188 |
| Grade (=3 vs. ≤2) |
0.001 |
0.004 |
3.036 |
2.688 |
1.572-5.864 |
1.371-5.273 |
| TNM (>1 vs. =1) |
0.000 |
0.000 |
3.164 |
2.769 |
1.918-5.218 |
1.705-4.498 |
| TNM (=3 vs. ≤2) |
0.000 |
0.001 |
4.000 |
3.565 |
1.987-8.055 |
1.734-7.332 |
| Ki67 (≤35 vs. > 35) | 0.001 | 0.004 | 2.956 | 2.597 | 1.585-5.512 | 1.362-4.950 |
Finally, SBEM and other clinical risk factors, including grade, disease stage, lymph node status and Ki67, were combined to determine the associations by multivariate analysis. Table 7 showed that HR for SBEM adjusted for the other risk factors remained unaffected and significant (HR = 3.370 with p = 0.008 for DFS and HR = 4.185 with p = 0.004 for OS).
Table 7.
Multivariate analysis results of SBEM with other risk factors on DFS and OS
| Covariate | DFS | OS | ||||
|---|---|---|---|---|---|---|
| |
P value |
Hazard radio |
95% Confidence interval |
P value |
Hazard radio |
95% Confidence interval |
| SBEM (3+ vs. <3+) |
0.008 |
3.370 |
1.382-8.218 |
0.004 |
4.185 |
1.587-11.039 |
| Lymph Node (Pos. vs. Neg.) |
0.751 |
1.195 |
0.398-3.585 |
0.198 |
2.097 |
0.680-6.469 |
| Grade (=3 vs ≤2) |
0.934 |
1.054 |
0.306-3.630 |
0.473 |
1.519 |
0.486-4.750 |
| TNM (>1 vs. =1) |
0.489 |
1.441 |
0.513-4.050 |
0.006 |
3.309 |
1.421-7.708 |
| TNM (=3 vs. ≤2) |
0.001 |
3.851 |
1.744-8.500 |
0.015 |
3.529 |
1.274-9.772 |
| Ki67 (≤35 vs. >35) | 0.152 | 2.022 | 0.771-5.301 | 0.917 | 1.065 | 0.327-3.463 |
Discussion
Breast cancer is pushed into first place in the United States and many other parts of world. Breast cancer alone is expected to account for 29% (226,870) of all new cancer cases among women [15]. Although incidence rate of breast cancer remains relatively stable in recent 5 years, its death rate declines by 34% because of the development of diagnosis and targeting medication. TNBC accounts for about 15% of all breast cancers. Patients with TNBC are more likely to experience death and distant recurrence compared to those with other cancers, and the median time to death/distant recurrence is significantly shortened. TNBC is one of solid tumors which are sensitive to chemotherapy, but other modalities, such as endocrine and targeted therapy, are not applicable. So, it is crucial to find specific markers to detect micro metastases and provide useful information to guide early therapeutic methods of TNBC patients.
Although various biological markers had been proposed for the detection of breast cancer cells, they were often affected by tumor differentiation, lower specificity and detection rate. Cyclin D1 was an effective marker for the differential diagnosis of other papillary lesions. Because Cyclin D1expressed in both lesions, it could not be used to distinguish between papilloma and papillary carcinoma lesions [16]. Petra Barros et al. found that the expression of β1 integrin had an impact in disease-specific survival (number of months from diagnosis to the time of death due to breast cancer) and could be a marker of poor prognosis in breast cancer [17]. But, β1 integrin played a role in predicting the clinical course and prognosis of several types of cancers [18], especially abundant expressing in Non-small-cell lung carcinoma [19]. By the same token, carcinoembryonic antigen (CEA) was not a specific marker of breast cancer because it was expressed at high levels in a variety of human tissues including lung, breast, and colorectal cancer. Although BRCA1 was associated with the genesis, progression, and prognosis of young breast cancer patients [20], they only accounted for about 5% of breast cancer occurrences [21]. So, researchers are searching for more promising genes to improve the screen, diagnosis and prognosis predicting of breast cancer.
SBEM was tissue-specific protein and only expresses in mammary and salivary glands [7]. SBEM could serve as a useful marker for breast nodal metastasis, and for detection of micro metastatic cells within lymph nodes. Also, it was used for the differential diagnosis of the primary origin of an unknown metastasis, especially in high grade and ER/PR-negative tumors [7]. Hinde et al. confirmed that the predictive power of IHC criteria appeared to be similar to that of gene expression analysis. The IHC information could be used to improve therapeutic decisions, mainly for luminal B, Her2- over-expressing and basal-like subtypes [22]. So, we examined SBEM levels in FFPE tissue sections by IHC test and then analyzed the correlation of SBEM expression with DFS and OS. Liu et al. reported that SBEM protein expression correlated with tumor size, TNM staging and lymph node metastasis [9]. Ki67 is used to assess the prognosis of cancer patients [23]. It would be indicated that SBEM were related to prognostic value with Ki67. Overall these risk factors, including age, grade, size, disease stage, lymph node status, and Ki67, were analyzed in our study. Our data showed that the detection rate of SBEM in FFPE tissue of TNBC was 58%, which was higher than previous report [8]. SBEM expression levels positively correlated with DFS and OS in TNBC patients. The Cox’s proportional hazards regression model showed that SBEM was independent for grade, age, disease stage, lymph node status, and Ki67. When we adjusted SBEM to combine with each clinical risk factor, SBEM expression still remained significant. Multivariate analysis showed that patients with a high SBEM expression of 3+ represented a higher risk of recurrence and mortality than those with a low SBEM expression (HR = 3.370 with p = 0.008 for DFS and HR = 4.185 with p = 0.004 for OS). SBEM could be regarded as an independent prognostic factor in TNBC.
We believed that SBEM would show much more advantages than other protein–based biomarkers and would be used as prognostic indicator. Meanwhile, SBEM expression in PB (Peripheral blood) of breast cancer patients was markedly higher than that of healthy donors and other cancer patients [9]. Determination of SBEM protein in tissue and mRNA expression in PB of TNBC patients maybe helpful for early diagnosis, choice of treatment, decision of the degree of malignancy and risk prediction of recurrence. However, it is necessary to determine the function of a certain gene by carrying out large sample studies or a large meta-analysis in different institutions and hospitals. In the past decade, the findings about the relationship between Catechol-O-methyltransferase Val158Met (COMT Val108/158Met) polymorphism and breast cancer risk were inconsistent. A large meta-analysis conducted by Xue Qin confirmed that COMT Val108/158Met polymorphism may not be associated with breast cancer risk [24]. Similarly, the prognostic significance of SBEM needs further evidence in TNBC patients.
Due to different breast cancer subtype are associated with different gene expression patterns, it is significant to identify the particular gene to suit the proper biological characteristic of a certain type of primary tumor. SBEM over-expression maybe the special characteristic of tumor cells in TNBC. In conclusion, we have done some really nice research in which SBEM shows its prognostic value in TNBC. Our findings may eventually lead to wide application of SBEM as a tumor marker or a target gene for therapy and rapid development in the diagnostic and therapeutic products for TNBC patients in future.
Conclusion
We have demonstrated that SBEM is differentially expressed in TNBC patients by immunostaining and SBEM may be best viewed as an independent prognostic factor of DFS and OS. The expression of SBEM does significantly correlate with a DFS and an OS of TNBC patients. We suggest that SBEM could be a promising prognostic biomarker in TNBC patients for cancer diagnostics, as well as be a possible target for the treatment of TNBC patients.
Consent
Written informed consent was obtained from the patient for publication of this report and any accompanying images.
Abbreviations
SBEM: Small breast epithelial mucin; TNBC: Triple-negative breast cancer; ER: Estrogen receptor; PR: Progesterone receptor; HER2: Human epidermal growth factor receptor-2; OS: Overall survival; DFS: Disease-free survival; IHC: IHC; PB: Peripheral blood; RT–PCR: Reverse transcription polymerase chain reaction; TNM: Tumor node metastasis; FISH: Fluorescence in situ hybridization; NCCN: National comprehensive cancer network; CEA: Carcinoembryonic antigen
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
LL contributed to the study design, data acquisition and analysis and drafted the manuscript; ZL was involved in data acquisition and revision of the manuscript; SQ, ZZ and YL worked on aspects of data acquisition and analysis; XX contributed to the study design and data-analysis, developed the algorithm and coordinated the study; FS conceived and coordinated the study. All authors read and approved the final manuscript.
Contributor Information
Liang Liu, Email: jinshatanwo@163.com.
Zhaozhe Liu, Email: liuzhaozhe@163.com.
Shuxian Qu, Email: qushuxian@163.com.
Zhendong Zheng, Email: zhengzhendong@163.com.
Yongye Liu, Email: liuyongye@163.com.
Xiaodong Xie, Email: wantouliuliang@163.com.
Fulin Song, Email: songfulinliuliang@163.com.
Acknowledgments
We thank Dr. Yang Ming and DR. Jing Qing for expert technical assistance with IHC. We thank Dr. Piao Ying, Dr. Ding Zhenyu and Dr. Guo Fang for secretarial and organizational support in our experiments. We also thank Dr. Liu Yunen and Dr. Wang Xu for critical revision of the manuscript.
References
- Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lønning PE, Børresen-Dale AL, Brown PO, Botstein D. Molecular portraits of human breast tumors. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Lønning PE, Børresen-Dale AL. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, Karaca G, Troester MA, Tse CK, Edmiston S, Deming SL, Geradts J, Cheang MC, Nielsen TO, Moorman PG, Earp HS, Millikan RC. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006;295:2492–2502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- Liedtke C, Mazouni C, Hess KR, André F, Tordai A, Mejia JA, Symmans WF, Gonzalez-Angulo AM, Hennessy B, Green M, Cristofanilli M, Hortobagyi GN, Pusztai L. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol. 2008;26:1275–1281. doi: 10.1200/JCO.2007.14.4147. [DOI] [PubMed] [Google Scholar]
- Millar EK, Graham PH, O'Toole SA, McNeil CM, Browne L, Morey AL, Eggleton S, Beretov J, Theocharous C, Capp A, Nasser E, Kearsley JH, Delaney G, Papadatos G, Fox C, Sutherland RL. Prediction of local recurrence, distant metastases, and death after breast conserving therapy in early-stage invasive breast cancer using a five-biomarker panel. J Clin Oncol. 2009;27:4701–4708. doi: 10.1200/JCO.2008.21.7075. [DOI] [PubMed] [Google Scholar]
- Knudson AG. Two genetic hits (more or less) to cancer. Nat Rev Cancer. 2001;1(2):157–162. doi: 10.1038/35101031. [DOI] [PubMed] [Google Scholar]
- Miksicek RJ, Myal Y, Watson PH, Walker C, Murphy LC, Leygue E. Identification of a novel breast-and salivary gland specific, mucin-like gene strongly expressed in normal and tumor human mammary epithelium. Cancer Res. 2002;62(10):2736–2740. [PubMed] [Google Scholar]
- Skliris GP, Hubé F, Gheorghiu I, Mutawe MM, Penner C, Watson PH, Murphy LC, Leygue E, Myal Y. Expression of small breast epithelial mucin (SBEM) protein in tissue microarrays (TMAs) of primary invasive breast cancers. Histopathology. 2008;52(3):355–369. doi: 10.1111/j.1365-2559.2007.02955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ZZ, Xie XD, Qu SX, Zheng ZD, Wang YK. Small breast epithelial mucin (SBEM) has the potential to be a marker for predicting hematogenous micrometastasis and response to neoadjuvant chemotherapy in breast cancer. Clin Exp Metastasis. 2010;27:251–259. doi: 10.1007/s10585-010-9323-2. [DOI] [PubMed] [Google Scholar]
- Valladares-Ayerbes M, Iglesias-Díaz P, Díaz-Prado S, Ayude D, Medina V, Haz M, Reboredo M, Antolín S, Calvo L, Antón-Aparicio LM. Diagnostic accuracy of small breast epithelial mucin mRNA as a marker for bone marrow micrometastasis in breast cancer: a pilot study. J Cancer Res Clin Oncol. 2009;135(9):1185–1195. doi: 10.1007/s00432-009-0559-7. [DOI] [PubMed] [Google Scholar]
- Jennbacken K, Vallbo C, Wang W, Damber JE. Expression of Vascular Endothelial Growth Factor C (VEGF-C) and VEGF Receptor-3 in human prostate cancer are associated with regional lymph node metastasis. Prostate. 2005;65(2):110–116. doi: 10.1002/pros.20276. [DOI] [PubMed] [Google Scholar]
- Serrero G, Ioffe O. Expression of the novel autocrine growth factor PC-Cell Derived Growth Factor in human breast cancer tissue. Hum Pathol. 2003;34:1148–1154. doi: 10.1016/S0046-8177(03)00425-8. [DOI] [PubMed] [Google Scholar]
- Pan AP, Huang GY, Chen J. Relationship between hepatitis B virus covalently closed circular DNA and HBx protein expression in hepatocellular carcinoma and its significance. World Chin J Digestol. 2009;17:712–715. [Google Scholar]
- Serrero G, Hawkins DM, Yue B, Ioffe O, Bejarano P, Phillips JT, Head JF, Elliott RL, Tkaczuk KR, Godwin AK, Weaver J, Kim WE. Progranulin (GP88) tumor tissue expression is associated with increased risk of recurrence in breast cancer patients diagnosed with estrogen receptor positive invasive ductal carcinoma. Breast Cancer Res. 2012;14:R26. doi: 10.1186/bcr3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA: A Cancer Journal for Clinicians. 2012;62(1):10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhu J-f, Liu Y-y, Han G-p. An analysis of Cyclin D1, Cytokeratin 5/6 and Cytokeratin 8/18 expression in breast papillomas and papillary carcinomas. Diagn Pathol. 2013;8:8. doi: 10.1186/1746-1596-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petra Barros Dos S, Zanetti JS, Alfredo R-S, Eduardo IC B. Beta integrin predicts survival in breast cancer: a clinicopathological and immunohistochemical study. Diagn Pathol. 2012;7:104. doi: 10.1186/1746-1596-7-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao ES, Zhang H, Chen YY, Lee B, Chew K, Moore D, Park C. Increased beta1 integrin is associated with decreased survival in invasive breast cancer. Cancer Res. 2007;67(2):659–664. doi: 10.1158/0008-5472.CAN-06-2768. [DOI] [PubMed] [Google Scholar]
- Dingemans AM, van den Boogaart V, Vosse BA, van Suylen RJ, Griffioen AW, Thijssen VL. Integrin expression profiling identifies integrin alpha 5 and beta 1 as prognostic factors in early stage non-small cell lung cancer. Mol Cancer. 2010;9(152):1–9. doi: 10.1186/1476-4598-9-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qingli Z, Qinghui Z, Hua C, Xiaoli Z. The ectopic expression of BRCA1 is associated with genesis, progression, and prognosis of breast cancer in young patients. Diagn Pathol. 2012;7:181. doi: 10.1186/1746-1596-7-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lux MP, Fasching PA, Beckmann MW. Hereditary breast and ovarian cancer: review and future perspectives. J Mol Med (Berl) 2006;84(1):16–28. doi: 10.1007/s00109-005-0696-7. [DOI] [PubMed] [Google Scholar]
- Hinde EI F, Sanae C, Sofia J, Kaoutar M, My Abdelilah M, Abdelaziz B, Omar M, Afaf A. Luminal B tumors are the most frequent molecular subtype in breast cancer of North African women: an immunohistochemical profilestudy from Morocco. Diagn Pathol. 2012;7:170. doi: 10.1186/1746-1596-7-170. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Weigel MT, Dowsett M. Current and emerging biomarkers in breast cancer: prognosis and prediction. Endocr Relat Cancer. 2010;17:R245–R262. doi: 10.1677/ERC-10-0136. [DOI] [PubMed] [Google Scholar]
- Xue Q, Qiliu P, Aiping Q, Zhiping C, Liwen L, Yan D, Li X, Juanjuan X, Haiwei L, Taijie L, Shan L, Jinmin Z. Association of COMT Val158Met polymorphism and breast cancer risk: an updated meta-analysis. Diagn Pathol. 2012;7:136. doi: 10.1186/1746-1596-7-136. [DOI] [PMC free article] [PubMed] [Google Scholar]