Abstract
The ATR kinase is a master regulator of the DNA damage response. Yet, how ATR is activated towards different substrates is still poorly understood. Here, we show that ATR phosphorylates Chk1 and RPA32 through distinct mechanisms at replication-associated DNA double-stranded breaks (DSBs). In contrast to the rapid phosphorylation of Chk1, RPA32 is progressively phosphorylated by ATR at Ser33 during DSB resection prior to the phosphorylation of Ser4/Ser8 by DNA-PKcs. Surprisingly, despite its reliance on ATR and TopBP1, substantial RPA32 Ser33 phosphorylation occurs in a Rad17-independent but Nbs1-dependent manner in vivo and in vitro. Importantly, the role of Nbs1 in RPA32 phosphorylation can be separated from ATM activation and DSB resection, and is dependent upon its interaction with RPA. An Nbs1 mutant unable to bind RPA fails to support proper recovery of collapsed replication forks, suggesting that the Nbs1-mediated mode of ATR activation is important for the repair of replication-associated DSBs.
Introduction
The ability of cells to sense and signal DNA damage in their genomes is crucial for genomic stability. In human cells, the ataxia telangiectasia mutated (ATM) and the ATM- and Rad3-related (ATR) kinases are two master regulators of DNA damage signaling (Ciccia and Elledge, 2010). ATM, ATR, and their related DNA-dependent protein kinase (DNA-PKcs) belong to the PI3K-like kinase (PIKK) family. While ATM and DNA-PKcs are primarily activated by DNA double-stranded breaks (DSBs), ATR responds to a broad spectrum of DNA damage (Cimprich and Cortez, 2008; Flynn and Zou, 2011). Unlike ATM and DNA-PKcs, ATR is essential for cell survival even in the absence of extrinsic DNA damage, underscoring the critical function of ATR in coping with intrinsic genomic stress (Barlow et al., 2013; Brown and Baltimore, 2000; Murga et al., 2009; Toledo et al., 2011). Although the DNA damage specificities and functions of ATM, ATR, and DNA-PKcs are clearly distinct, how they distinguish different types of DNA damage and execute their unique functions are still poorly understood. In particular, how ATR is activated by different types of DNA damage and replication stress is still largely unknown.
Studies in different organisms have revealed some of the key principles of ATR activation. In response to DNA damage and replication stress, the complex of ATR and its functional partner ATRIP is recruited to sites of DNA damage and stalled replication forks by RPA-coated single-stranded DNA (RPA-ssDNA) (Byun et al., 2005; Costanzo et al., 2003; Zou and Elledge, 2003). The activation of ATR-ATRIP requires additional regulators, such as the Rad17-RFC complex, the Rad9-Rad1-Hus1 (9-1-1) complex, and TopBP1 (Kumagai et al., 2006; Lin et al., 2012; Navadgi-Patil and Burgers, 2009; Zou et al., 2002). Independently of the recruitment of ATR-ATRIP to RPA-ssDNA, the Rad17-RFC complex recognizes the junctions of ssDNA and dsDNA (double-stranded DNA) and loads 9-1-1 complexes onto dsDNA (Ellison and Stillman, 2003; Zou et al., 2003). Through a process that is still not fully understood, TopBP1 is recruited to damaged DNA and interacts with Rad17, 9-1-1, and autophosphorylated ATR (Cotta-Ramusino et al., 2011; Delacroix et al., 2007; Lee and Dunphy, 2010; Lee et al., 2007; Liu et al., 2011; Wang et al., 2011; Yan and Michael, 2009). The engagement of TopBP1 with ATR-ATRIP allows TopBP1 to stimulate the kinase activity of ATR and facilitate ATR to recognize its substrates (Kumagai et al., 2006; Liu et al., 2011; Mordes et al., 2008). In this model of ATR activation, ATR is activated by Rad17 and TopBP1 around ssDNA/dsDNA junctions. Indeed, Chk1, an effector kinase of ATR critical for the replication stress response and cell cycle arrest, is phosphorylated by ATR in a Rad17-, TopBP1-, and ssDNA/dsDNA junction-dependent manner (Liu et al., 2006; MacDougall et al., 2007; Van et al., 2010; Yamane et al., 2003; Zou et al., 2002). However, it is important to note that although Chk1 phosphorylation has been widely used as a surrogate for ATR activation, it remains unclear whether Chk1 phosphorylation accurately evinces the active mode of ATR in all situations.
In this study, we asked if ATR is always activated by the Rad17-TopBP1 circuitry after DNA damage. In particular, we wondered if ATR is activated by Rad17 and TopBP1 at extensively resected DSBs, such as those generated in S phase at collapsed replication forks. When long ssDNA is generated at DSBs by resection, a fraction of ATR could be recruited to the RPA-ssDNA distal to ssDNA/dsDNA junctions, raising a question as to whether and how this fraction of ATR is activated on RPA-ssDNA. To address this question, we analyzed the activation of ATR by camptothecin (CPT), which induces replication-associated DSBs that undergo rapid and efficient resection (Avemann et al., 1988; Sartori et al., 2007). We found that ATR is activated in two distinct modes towards Chk1 and RPA32. In one mode, ATR phosphorylates Chk1 rapidly, whereas in the other mode, ATR phosphorylates RPA32 Ser33 progressively during resection. The activation of ATR towards RPA32 is driven by resection and requires TopBP1. Surprisingly, Nbs1, a component of the Mre1-Rad50-Nbs1 (MRN) complex (Carney et al., 1998; Costanzo et al., 2001; Difilippantonio et al., 2005; Stracker and Petrini, 2011), plays a more important role than Rad17 in the phosphorylation of RPA32. The function of Nbs1 in RPA32 phosphorylation can be separated from ATM activation and DSB resection, and is dependent upon a direct interaction between Nbs1 and RPA. An Nbs1 mutant unable to bind RPA is compromised in its ability to support recovery of collapsed replication forks. Together, these results suggest that Nbs1 mediates a TopBP1-dependent, but Rad17-independent mode of ATR activation on RPA-ssDNA, allowing ATR to phosphorylate substrates such as RPA32 independently of ssDNA/dsDNA junctions and promote repair of replication-associated DSBs.
Results
ATR phosphorylates Chk1 and RPA32 Ser33 via distinct mechanisms
ATR is known to be activated by the replication-associated DSBs induced by CPT (Avemann et al., 1988; Sartori et al., 2007). Using VE-821, a specific ATR inhibitor (Reaper et al., 2011), we confirmed that a number of ATR substrates, including Chk1, Rad17, RPA32, and ATR itself, were phosphorylated in an ATR-dependent manner after CPT treatment (Fig. S1A). In contrast to these ATR substrates, ATM and DNA-PKcs underwent efficient autophosphorylation in the presence of VE-821 (Fig. S1B). Furthermore, the phosphorylation of several ATM and/or DNA-PKcs substrates, including Chk2, p53, and H2AX, was not affected by VE-821 (Fig. S1B). These results confirm that VE-821 specifically inhibits ATR, but not ATM and DNA-PKcs, in CPT treated cells.
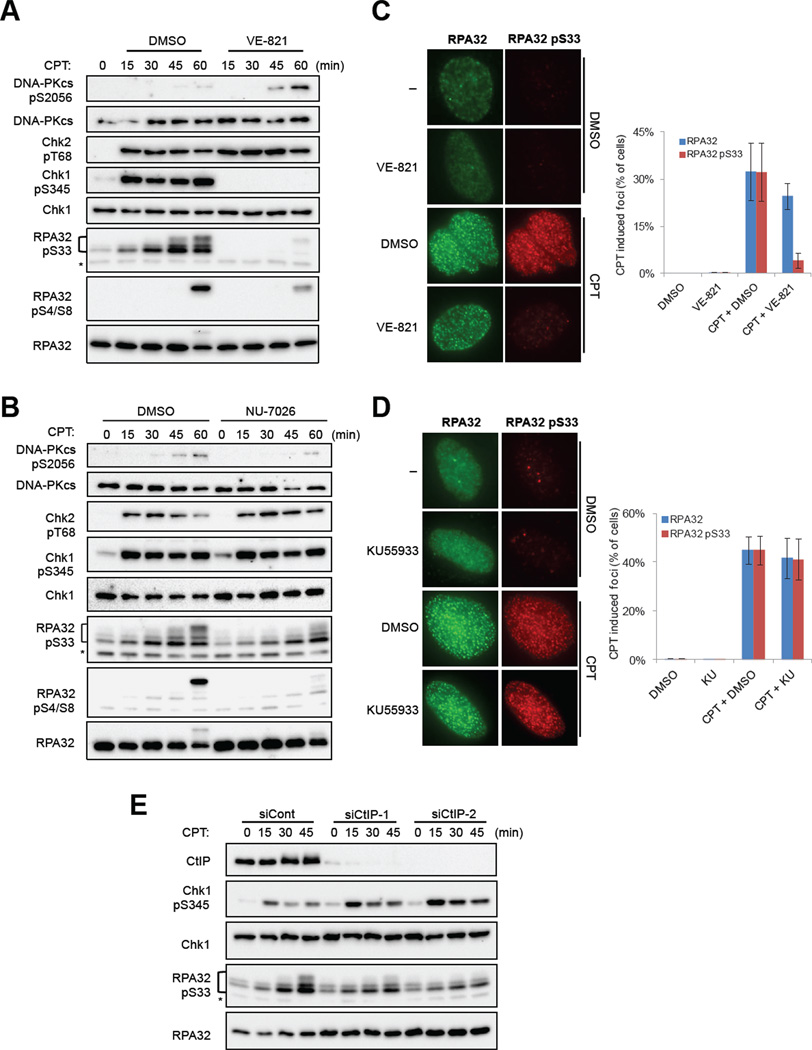
In response to CPT, RPA32 is phosphorylated at multiple sites, including Ser4/Ser8, Thr21, and Ser33 (Anantha et al., 2007; Block et al., 2004; Sartori et al., 2007). Using phospho-specific antibodies, we found that RPA32 Ser33 was phosphorylated within 15 min after CPT treatment, whereas RPA32 Ser4/Ser8 was phosphorylated at approximately 60 min (Fig. 1A). Furthermore, the phosphorylation of RPA32 Ser33 was abolished by VE-821, whereas RPA32 Ser4/Ser8 phosphorylation was only partially reduced (Fig. 1A). The phosphorylation of RPA32 Ser4/Ser8 was shown to be DNA-PKcs-dependent (Anantha et al., 2007; Liaw et al., 2011; Liu et al., 2012). Indeed, the CPT-induced phosphorylation of RPA32 Ser4/Ser8 was abolished by the DNA-PKcs inhibitor NU7026 (Fig. 1B). In contrast, RPA32 Ser33 phosphorylation was not affected by NU7026 (Fig. 1B). Thus, compared with the phosphorylation of RPA32 Ser4/Ser8, the phosphorylation of RPA32 Ser33 is a more direct and reliable marker for ATR activation.
Fig. 1. ATR-mediated RPA32 Ser33 phosphorylation in response to CPT-associated DSBs.
(A) U2OS cells were treated with 1 µM CPT in the presence or absence of 10 µM VE-821 and collected at the indicated times. The levels of the indicated proteins and the CPT-induced phosphorylation events were analyzed by Western blot. *: a protein cross-reacting to the phospho-Ser33 antibody. (B) Cells pre-treated with 10µM NU-7026 or DMSO for 1 hr were exposed to 1 µM CPT in the presence or absence of 10 µM NU-7026. Extracts were analyzed as in (A). (C) Cells were treated with 1 µM CPT for 1 hr, or mock treated in the absence or presence of VE-821. Immunofluorescence analysis of RPA32 and phospho-Ser33 was performed with specific antibodies. Representative images of cells are shown in the left panel. The fractions of cells that displayed RPA32 or phospho-Ser33 foci are shown in the right panel. Error bars: standard deviations from three independent experiments (n=3). (D) Cells were treated with 1 µM CPT for 2 hr, or mock treated, in the absence or presence of KU55933. Immunofluorescence analysis was performed and quantified as in (C). Error bars: standard deviations from three independent experiments (n=3). (E) U2OS cells transfected with control or CtIP siRNAs were treated with 1 µM CPT and collected at indicated time points. Extracts were analyzed as in (A).
The phosphorylation of RPA32 has been linked to DSB resection (Sartori et al., 2007). A recent study suggested that ATR promotes resection in Xenopus extracts (Peterson et al., 2013). Consistent with this finding, at 1 hr after CPT treatment, the formation of RPA32 foci was reduced by VE-821 (Fig. 1C). Nevertheless, a significant fraction of VE-821-treated cells displayed RPA32 foci, suggesting that ATR is not essential for resection (Fig. 1C). Importantly, the majority of VE-821-treated cells that displayed RPA32 foci were negative for phospho-Ser33 foci, suggesting that ATR is required for RPA32 Ser33 phosphorylation after resection (Fig. 1C). In contrast to VE-821, the ATM inhibitor KU55933 drastically diminished the foci of both RPA32 and phospho-Ser33 1 hr after CPT treatment (Fig. S1C). At 2 hr, KU55933-treated cells no longer displayed a significant reduction in RPA32 foci and phospho-Ser33 foci (Fig. 1D), showing that inhibition of ATM delays resection transiently. In KU55933-treated cells, RPA32 foci were always colocalized with phospho-Ser33 foci (Fig. 1D), suggesting that ATM does not have a direct role in RPA32 Ser33 phosphorylation post resection. Consistent with RPA32 Ser33 phosphorylation being a specific marker of ATR activation, this phosphorylation event was clearly detected in the ATM-deficient AT cells and the DNA-PKcs-deficient M059J cells, even when they were treated with NU7026 and KU55933, respectively (Figs. S1D–E).
Having established RPA32 Ser33 phosphorylation as a marker for CPT-induced ATR activation, we asked whether this phosphorylation event is regulated in the same way as Chk1 phosphorylation. In response to CPT, Chk1 was rapidly phosphorylated at Ser345 in an ATR-dependent manner (Fig. 1A). In contrast to the phosphorylation of RPA32 Ser33, which gradually accumulated during the first 60 min after CPT treatment, Chk1 phosphorylation plateaued within 15 min (Fig. 1A). Thus, Chk1 and RPA32 Ser33 are phosphorylated by ATR with distinct kinetics. Consistent with the sequential phosphorylation of RPA32 by ATR and DNA-PKcs (Anantha et al., 2007), the phosphorylation of RPA32 Ser4/Ser8 by DNA-PKcs occurred significantly later than Ser33 phosphorylation and Chk1 phosphorylation (Fig. 1A) (Kousholt et al., 2012).
The phosphorylation of RPA32 Ser4/Ser8 by DNA-PKcs requires CtIP, which is critical for resection (Sartori et al., 2007). Prior to the abrupt phosphorylation of Ser4/Ser8, Ser33 was gradually phosphorylated by ATR (Fig. 1A), suggesting that Ser33 phosphorylation may be the initial phosphorylation event on RPA32 that is driven by resection. Indeed, knockdown of CtIP with two independent siRNAs drastically reduced the phosphorylation of RPA32 Ser33 during the first 60 min after CPT treatment (Fig. 1E). On the other hand, consistent with a recent report (Kousholt et al., 2012), depletion of CtIP did not significantly affect the phosphorylation of Chk1 during this early phase of the CPT response. Thus, in contrast to the rapid phosphorylation of Chk1, the phosphorylation of RPA32 Ser33 by ATR occurs progressively during resection.
Distinct roles of Rad17 and Nbs1 in RPA phosphorylation
The distinct kinetics and resection dependency of Chk1 and RPA32 phosphorylation prompted us to investigate whether ATR is activated towards these two substrates in different ways. To dissect the genetic requirements for RPA32 Ser33 phosphorylation, we used siRNAs to knock down the upstream regulators of the ATR pathway. Consistent with the effects of VE-821, knockdown of ATR abolished the phosphorylation of both Chk1 and RPA32 Ser33 (Fig. S2A). Two independent siRNAs targeting TopBP1, the key activator of ATR, also drastically reduced the phosphorylation of Chk1 and RPA32 Ser33 (Fig. 2A). Therefore, similar to Chk1 phosphorylation, RPA32 Ser33 phosphorylation requires both ATR and TopBP1.
Fig. 2. RPA32 and Chk1 are phosphorylated by ATR through distinct mechanisms.
(A–C) U2OS cells transfected with control, TopBP1 (A), Rad17 (B), or Nbs1 (C) siRNAs were treated with 1 µM CPT and collected at the indicated time points. The levels of the indicated proteins and the CPT-induced phosphorylation events were analyzed by Western blot.
TopBP1 and Rad17 function together in Chk1 phosphorylation (Delacroix et al., 2007; Lee and Dunphy, 2010). As expected, knockdown of Rad17 with two independent siRNAs significantly reduced Chk1 phosphorylation (Fig. 2B). Surprisingly, however, substantial RPA32 Ser33 phosphorylation was detected in Rad17 knockdown cells (Fig. 2B), suggesting the existence of a Rad17-independent mechanism of RPA32 Ser33 phosphorylation. The MRN complex has been implicated in ATR regulation and RPA phosphorylation (Jazayeri et al., 2006; Manthey et al., 2007; Myers and Cortez, 2006; Olson et al., 2007a; Stiff et al., 2005; Yoo et al., 2009; Zhong et al., 2005). Knockdown of Nbs1 with two independent siRNAs clearly reduced RPA32 Ser33 phosphorylation without altering cell-cycle distribution (Figs. 2C, S2B). Consistent with Nbs1 knockdown, the Nbs1-deficient NBN-ILB1 cells displayed much lower RPA32 Ser33 phosphorylation than the Nbs1-complemented cells (Fig. S2C). Compared with Rad17 knockdown, Nbs1 knockdown reduced RPA32 Ser33 phosphorylation to a greater extent. When both Rad17 and Nbs1 were knocked down, RPA32 Ser33 phosphorylation was further reduced (Fig. S2D), suggesting that Rad17 and Nbs1 function in parallel to activate ATR towards RPA32 Ser33. Interestingly, while Nbs1 knockdown clearly reduced RPA32 Ser33 phosphorylation, its effects on Chk1 phosphorylation were modest (Fig. 2C). Thus, in contrast to Rad17, Nbs1 plays a more important role in RPA phosphorylation than in Chk1 phosphorylation.
Nbs1 mediates RPA32 phosphorylation independently of Rad17 and ssDNA/dsDNA junctions
The MRN complex regulates ATM activation and DSB resection (Symington and Gautier, 2011; Uziel et al., 2003). Furthermore, the MRN complex is implicated in the removal of TopI from CPT-induced DSBs (Sacho and Maizels, 2011). To determine whether Nbs1 regulates RPA32 phosphorylation directly or indirectly, we analyzed the function of Nbs1 in RPA32 phosphorylation using an in vitro assay that we recently developed (Shiotani and Zou, 2009). In this assay, linear dsDNA is resected by T7 exonuclease or exonuclease III to generate ssDNA overhangs, and subsequently added to HeLa cell nuclear extracts to activate ATR (Shiotani and Zou, 2011). Using this assay, we have shown that RPA32 Ser33 is phosphorylated in an ATR-dependent manner. Since both ATM and DNA-PKcs are inhibited in extracts and dsDNA is already resected before it is added to extracts, this assay provides a unique opportunity to test if Nbs1 has an ATM-, resection-, and end processing-independent function in RPA32 phosphorylation.
Consistent with its dependency on TopBP1 in cells, RPA32 Ser33 phosphorylation was not induced by resected dsDNA in extracts derived from TopBP1 knockdown cells (Fig. 3A). In contrast, RPA32 Ser33 phosphorylation occurred efficiently in extracts from CtIP knockdown cells, confirming that the role of MRN-CtIP in DSB resection is bypassed by the use of pre-resected dsDNA in this assay (Fig. 3B). Also consistent with the observation in cells, the phosphorylation of RPA32 Ser33 in extracts was only modestly reduced by Rad17 depletion (Fig. 3C). Strikingly, even in this resection-independent in vitro assay, Nbs1 was clearly required for RPA32 Ser33 phosphorylation (Fig. 3D). These results reveal a post-resection function of Nbs1 in RPA32 phosphorylation.
Fig. 3. Nbs1 regulates RPA32 phosphorylation independently of ATM activation and DSB resection.
(A) Nuclear extracts were prepared from HeLa cells transfected with TopBP1 siRNA or mock transfected. A 6-kb dsDNA fragment was resected with T7 exonuclease or exonuclease III as described in the Experimental Procedures. Resected or unresected dsDNA was added to cell extracts, and the resected dsDNA-induced phosphorylation of RPA32 Ser33 was analyzed with phospho-specific antibody. (B) The resected dsDNA-induced phosphorylation of RPA32 Ser33 was analyzed in extracts of CtIP knockdown cells and control cells as in (A). (C) The resected dsDNA-induced phosphorylation of RPA32 Ser33 was analyzed in extracts of Rad17 knockdown cells and control cells as in (A). (D) The resected dsDNA-induced phosphorylation of RPA32 Ser33 was analyzed in extracts of Nbs1 knockdown cells and control cells as in (A).
Since substantial RPA32 Ser33 phosphorylation occurred independently of Rad17 in vivo and in vitro, we asked if this phosphorylation event is absolutely dependent upon ssDNA/dsDNA junctions. In nuclear extracts, ssDNA alone was sufficient to induce RPA32 Ser33 phosphorylation in a length-dependent manner (Fig. 4A). We noted that unlike resected dsDNA, ssDNA did not induce mobility shift of RPA32 in protein gels. The reason for this difference is currently unknown. Importantly, the ssDNA-induced RPA32 Ser33 phosphorylation was dependent upon ATR, TopBP1, and Nbs1 (Figs. 4B–D), suggesting that a process driven by ssDNA lengthening and mediated by Nbs1 and TopBP1 is able to activate ATR towards RPA independently of ssDNA/dsDNA junctions.
Fig. 4. ssDNA is sufficient to induce RPA32 phosphorylation.
(A) ssDNA of the indicated length was added to nuclear extracts, and the ssDNA-induced phosphorylation of RPA32 Ser33 was analyzed with phospho-specific antibody. (B) The ssDNA-induced phosphorylation of RPA32 Ser33 was analyzed in extracts of ATR knockdown cells and control cells as in (A). (C) The ssDNA-induced phosphorylation of RPA32 Ser33 was analyzed in extracts of TopBP1 knockdown cells and control cells as in (A). (D) The ssDNA-induced phosphorylation of RPA32 Ser33 was analyzed in extracts of Nbs1 knockdown cells and control cells as in (A).
Nbs1 targets the MRN complex to RPA-ssDNA
We next asked how Nbs1 executes its post-resection function in ATR activation. The MRN complex interacts with RPA (Oakley et al., 2009; Olson et al., 2007b). Consistent with previous studies, we found that in extracts Mre11, Rad50, and Nbs1 all bound to ssDNA in an RPA-dependent manner (Fig. 5A). The RPA complex containing the RPA70 t-11 mutant, whose N-terminal OB fold was disrupted by point mutations, was unable to recruit MRN to ssDNA (Fig. 5B). Furthermore, purified MRN complex bound to ssDNA in an RPA-dependent manner, showing that MRN associates with RPA-ssDNA directly (Fig. 5C). Even in the absence of Mre11 and Rad50, purified Nbs1 bound to ssDNA in an RPA-dependent manner, suggesting a direct interaction between Nbs1 and RPA-ssDNA (Fig. 5D).
Fig. 5. Nbs1 targets the MRN complex to RPA-ssDNA.
(A) Biotinylated 70-nucleotide ssDNA (5 pmole) was coated with increasing amounts of purified RPA (0, 5, 10, and 20 pmole) and then incubated in cell extracts. The proteins bound to RPA-ssDNA were retrieved by streptavidin-coated beads and analyzed by Western blot using the indicated antibodies. All components of the Mre11-Rad50-Nbs1 complex bound to ssDNA in an RPA-dependent manner. (B) Biotinylated ssDNA was coated with either wild-type RPA or the RPA t-11 mutant complex purified from E. coli. The binding of Rad50 and Mre11 to RPA-ssDNA was tested as in (A). (C) Biotinylated ssDNA coated with RPA was incubated with purified MRN complex. The MRN components bound to RPA-ssDNA were analyzed by Western blot. (D) The binding of purified Nbs1 to RPA-ssDNA was tested as in (C). (E) Extracts were prepared from cells transfected with control or Nbs1-2 siRNA. Biotinylated ssDNA coated with RPA was incubated in the extracts, and the proteins bound to RPA-ssDNA were analyzed by Western blot using the indicated antibodies.
To determine whether the recruitment of MRN to RPA-ssDNA is dependent on Nbs1, we generated extracts from Nbs1 knockdown cells. Consistent with previous studies, loss of Nbs1 did not affect the levels of Mre11 and Rad50 (Fig. 5E) (Stewart et al., 1999). However, in extracts of Nbs1 knockdown cells, neither Mre11 nor Rad50 bound to RPA-ssDNA efficiently (Fig. 5E). In contrast to Mre11 and Rad50, ATRIP still associated with RPA-ssDNA efficiently in the absence of Nbs1 (Fig. 5E). Together, these results suggest that Nbs1 targets the MRN complex to RPA-ssDNA.
Nbs1 binds RPA-ssDNA via a previously uncharacterized domain
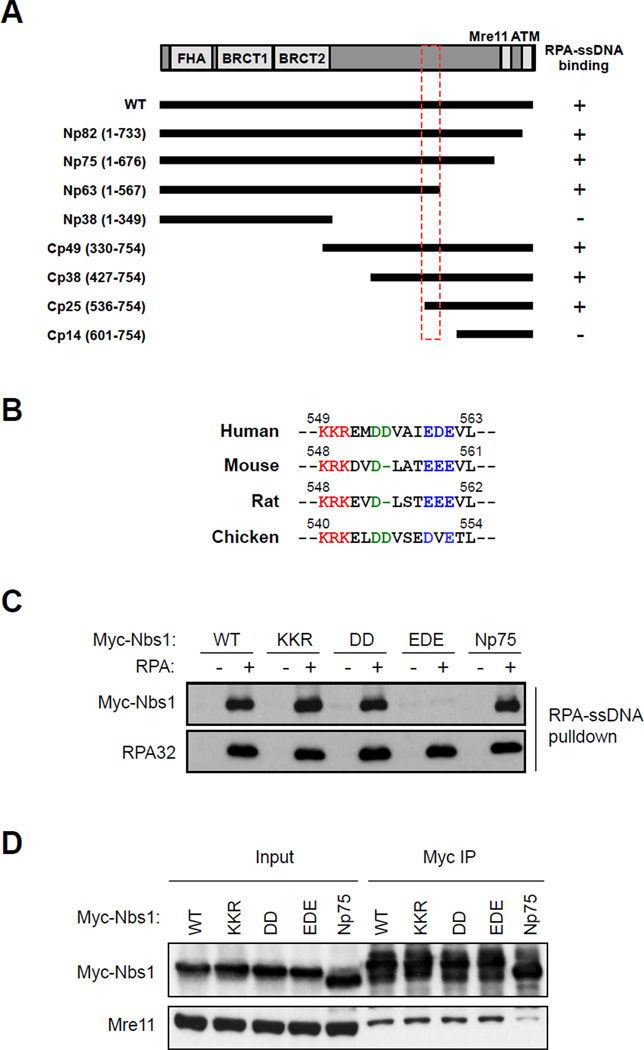
To determine if the interaction between Nbs1 and RPA-ssDNA is important for the phosphorylation of RPA32 Ser33 by ATR, we sought to map the RPA-ssDNA-interacting domain of Nbs1. A set of myc-tagged Nbs1 fragments were expressed in cells and tested for binding to biotinylated ssDNA coated with RPA (Figs. 6A, S3A–B). An Nbs1 fragment containing the N terminal FHA and BRCT domains (Np38) was unable to interact with RPA-ssDNA (Fig. S3A). On the other hand, two Nbs1 fragments lacking the FHA and BRCT domains (Cp49 and Cp38) retained the ability to interact with RPA-ssDNA (Fig. S3B). Moreover, an Nbs1 fragment lacking the C terminal ATM and Mre11 binding domains (Np75) still bound to RPA-ssDNA (Fig. S3A). Thus, the RPA-ssDNA-interacting domain of Nbs1 is distinct from its known functional domains.
Fig. 6. Mapping the RPA-ssDNA-interacting domain of Nbs1.
(A) Schematic of the Nbs1 fragments used in this study and their ability to bind RPA-ssDNA. (B) The RPA-ssDNA-binding domain of human Nbs1 is aligned to the corresponding regions in the Nbs1 of other higher vertebrates. The conserved charged amino acids are highlighted in colors. (C) The conserved charged amino acids in the RPA-ssDNA-binding domain of Nbs1 were mutated in myc-tagged, full-length Nbs1 (KKR to EEG, DD to AA, and EDE to AAA). These Nbs1 mutants were transiently expressed in 293T cells, and their ability to bind RPA-ssDNA was tested in cell extracts using biotinylated ssDNA coated with RPA. Wild-type Nbs1 (WT) and an Nbs1 mutant lacking the Mre11- and ATM-binding domains (Np75) were also tested as controls. (D) Myc-tagged, full-length Nbs1 (WT) and its mutant derivatives (KKR, DD, EDE, and Np75) were transiently expressed in 293T cells. The myc-tagged Nbs1 proteins were immunoprecipitated with anti-myc antibody and the Mre11 co-precipitated was analyzed by Western blot using anti-Mre11 antibody.
Through testing the binding of additional Nbs1 fragments to RPA-ssDNA (Figs. S3A–B), we found that all the Nbs1 fragments that were able to bind RPA-ssDNA encompassed the amino acids 537–567 (Fig. 6A). Alignment of the sequences of Nbs1 from different species revealed that a stretch of charged amino acids in this region is conserved in higher vertebrates (Fig. 6B). To pinpoint the residues that are critical for RPA-ssDNA binding, we mutated the conserved residues KKR, DD, and EDE to EEG, AA, and AAA in full-length Nbs1, respectively (Fig. 6B; Nbs1KKR, Nbs1DD, and Nbs1EDE). Only the Nbs1EDE mutant, but not the Nbs1KKR and Nbs1DD mutants, lost the ability to bind RPA-ssDNA (Fig. 6C). The Nbs1EDE mutant still bound to Mre11 efficiently (Fig. 6D), suggesting that loss of the EDE motif of Nbs1 specifically disrupts its binding to RPA-ssDNA.
The recognition of RPA-ssDNA by Nbs1 is important for RPA32 Ser33 phosphorylation and recovery of collapsed forks
To determine whether the binding of Nbs1 to RPA-ssDNA is important for RPA32 Ser33 phosphorylation, we first tested the Nbs1EDE mutant in vitro. We expressed siRNA-resistant myc-Nbs1WT and myc-Nbs1EDE in cells where endogenous Nbs1 was knocked down and prepared extracts from these cells. In myc-Nbs1WT-containing extracts, resected dsDNA induced RPA32 Ser33 phosphorylation efficiently (Fig. 7A). In contrast, RPA32 Ser33 phosphorylation was poorly induced by resected dsDNA in extracts containing myc-Nbs1EDE (Fig. 7A), suggesting that the ability of Nbs1 to recognize RPA-ssDNA is important for its post-resection function in RPA32 Ser33 phosphorylation in vitro.
Fig. 7. The interaction of Nbs1 and RPA is required for Nbs1-mediated RPA32 phosphorylation and recovery from collapsed replication forks.
(A) U2OS-derivative cell lines were established to express myc-tagged Nbs1WT and Nbs1EDE conditionally. As indicated, cells were treated with 1 µg/ml doxycycline (Dox) to induce myc-tagged Nbs1 proteins, and transfected with siNbs1-2 to knock down endogenous Nbs1. Nuclear extracts were prepared from cells, and the ability of resected dsDNA to induce RPA32 Ser33 phosphorylation was analyzed with phospho-specific antibody. (B) Myc-tagged Nbs1WT and Nbs1EDE were used to replace endogenous Nbs1 as in (A). Cells were synchronized in S phase as described in Experimental Procedure and subsequently treated with CPT (1 µM) for 45 min or mock treated. The levels of the indicated proteins and the phosphorylation of RPA32 Ser33 were analyzed by Western blot. (C) Myc-tagged Nbs1WT and Nbs1EDE were used to replace endogenous Nbs1 as in (A). Cells were synchronized in S phase, treated with CPT (1 µM) for 1 h or mock treated, and then released into CPT-free medium. After 24 h, cells were immunostained with antibody to γH2AX, and the fractions of cells with γH2AX foci were quantified. Error bars: standard deviations from three independent experiments (n=3). (D) A model that depicts the two distinct modes of ATR activation at resected DSBs.
Next, we tested whether the Nbs1EDE mutant is able to support RPA32 Ser33 phosphorylation in cells. We generated stable cell lines in which siRNA-resistant myc-Nbs1WT or myc-Nbs1EDE can be inducibly expressed. Upon induction of myc-Nbs1WT in Nbs1 knockdown cells, RPA32 Ser33 phosphorylation was clearly induced by CPT (Fig. 7B, lanes 3–4). In contrast, the levels of RPA32 Ser33 phosphorylation were not increased by CPT in cells expressing the myc-Nbs1EDE mutant (Fig. 7B, lanes 7–8). Thus, consistent with our in vitro experiments, the Nbs1EDE mutant also fails to support RPA32 Ser33 phosphorylation in cells.
Finally, we sought to determine whether the recognition of RPA-ssDNA by Nbs1 is important for the DNA damage response. Since CPT induces replication-associated DSBs, we focused on the recovery of collapsed replication forks. We synchronized cells in S phase with thymidine and released them into CPT-containing media. As expected, γH2AX foci were induced in cells regardless of whether endogenous Nbs1 was replaced by myc-Nbs1WT or myc-Nbs1EDE (Fig. 7C). After the myc-Nbs1WT expressing cells were released from CPT, γH2AX foci were largely gone in 24 hr (Fig. 7C), indicating the recovery of collapsed forks. In contrast, a significant fraction of the myc-Nbs1EDE expressing cells retained γH2AX foci 24 hr after the release from CPT (Fig. 7C), suggesting that the recovery of collapsed forks was compromised. These results establish a functional link from the Nbs1-mediated ATR activation to the repair of replication-associated DSBs.
Discussion
The activation of ATR has been extensively studied using Chk1 phosphorylation as a readout. Both in vivo and in vitro experiments have suggested that Rad17 and TopBP1 are two key regulators of the activation of ATR towards Chk1 (Cimprich and Cortez, 2008; Flynn and Zou, 2011). In the current model of ATR activation, both RPA-ssDNA and ssDNA/dsDNA junctions are critical determinants for Chk1 phosphorylation (MacDougall et al, 2007). While RPA-ssDNA presents a platform for ATR-ATRIP recruitment, the Rad17-RFC complex and 9-1-1 complexes are localized to ssDNA/dsDNA junctions. The co-localization of ATR-ATRIP, Rad17, 9-1-1, and TopBP1 around ssDNA/dsDNA junctions may create a protein-DNA assembly that allows TopBP1 to activate the ATR-ATRIP kinase complex and enable it to recognize its downstream substrates. This mechanism of ATR activation towards Chk1 is consistent with the fact that ssDNA is not sufficient to trigger checkpoint signaling through Chk1 (Guo and Dunphy, 2000; MacDougall et al., 2007).
Although ssDNA and ssDNA/dsDNA junctions are commonly induced by DNA damage and replication stress, the relative abundance of these two DNA structures may vary in different contexts. For example, at replication forks stalled by aphidicolin, an increased number of primers and ssDNA/dsDNA junctions may be generated, which enhance the activation of ATR towards Chk1 (Van et al., 2010). On the other hand, when DSBs undergo extensive resection in S phase, long stretches of ssDNA may be generated without increasing the number of ssDNA/dsDNA junctions. We found that RPA32 Ser33 is progressively phosphorylated by ATR during the resection of CPT-induced DSBs. Although an ATR-mediated event, RPA32 Ser33 phosphorylation is regulated differently from Chk1 activation. In addition to its delayed kinetics and greater dependency on resection, RPA32 Ser33 phosphorylation is more dependent on Nbs1 but less dependent on Rad17 compared with Chk1 phosphorylation. Furthermore, unlike Chk1 phosphorylation, RPA32 Ser33 phosphorylation can occur on ssDNA in a length-dependent manner independently of ssDNA/dsDNA junctions. These findings suggest that, in a resection-driven manner, Nbs1 mediates a distinct mode of ATR activation towards RPA32.
How do Rad17 and Nbs1 regulate the activation of ATR during resection? As resection is initiated at DSBs and ssDNA/dsDNA junctions are generated, the Rad17-RFC complex recognizes these junctions and loads 9-1-1 complexes onto dsDNA (Fig. 7D). This Rad17- and ssDNA/dsDNA junction-mediated process promotes ATR activation and Chk1 phosphorylation around the ssDNA/dsDNA junctions. In addition, a fraction of RPA adjacent to ssDNA/dsDNA junctions is phosphorylated by ATR in a Rad17-dependent manner during this early phase of resection. As resection continues, ssDNA is gradually lengthened, placing increasing amounts of RPA and ATR-ATRIP on the ssDNA distal to ssDNA/dsDNA junctions (Fig. 7D). In the late phase of resection, a fraction of RPA and ATR-ATRIP on ssDNA becomes “out of reach” for the Rad17 and 9-1-1 complexes at ssDNA/dsDNA junctions. In this situation, the MRN complex may directly recognize RPA-ssDNA via Nbs1, and activate ATR-ATRIP by recruiting TopBP1 (Yoo et al., 2009). Consistent with this model, substantial RPA32 Ser33 phosphorylation is driven by resection and dependent upon both Nbs1 and TopBP1 in vivo and in vitro. Furthermore, the direct interaction between Nbs1 and RPA-ssDNA is needed for efficient RPA32 Ser33 phosphorylation both in vivo and in vitro. Since the Nbs1-mediated mode of ATR activation is independent of Rad17, which interacts with Claspin to promote Chk1 activation (Wang et al., 2006), this mode of ATR activation is specifically directed to RPA but not Chk1.
When and where does Nbs1 activate ATR in cells? In laser microirradiated cells, a fraction of Nbs1 precisely colocalizes with RPA in the ssDNA sub-compartments at DSBs (Bekker-Jensen et al., 2006). In the context of DNA replication, Mre11 partially colocalizes with PCNA during S phase (Maser et al., 2001). Although RPA32 Ser33 phosphorylation is rapidly induced by CPT, it does not occur robustly in hydroxyurea (HU)-treated cells until DSBs become detectable in these cells (Figs. 1A, S1F) (Sakasai et al., 2006). This observation, together with the dependency of RPA32 Ser33 phosphorylation on resection, suggests that RPA32 Ser33 phosphorylation occurs primarily at replication-associated DSBs. Indeed, the phosphorylation of RPA32 has been implicated in the progression and repair of stressed replication forks by previous studies (Liu et al., 2012; Shi et al., 2010; Vassin et al., 2009). We found that when the interaction between Nbs1 and RPA-ssDNA was disrupted, the recovery of collapsed replication forks was compromised (Fig. 7C). These findings suggest that the Nbs1-mediated mode of ATR activation may be particularly important for the repair of replication-associated DSBs. Consistent with this idea, the MRN complex was shown to co-localize with RPA at collapsed forks (Manthey et al., 2007). Furthermore, the MRN complex is required for preventing accumulation of DSBs during replication (Costanzo et al., 2001). In addition to the mechanism that we propose, recent studies suggested that Mre11 degrades stalled replication forks in the absence of BRCA1/2 and Fanconi anemia proteins, and promotes ATR activation via a nuclease-dependent mechanism at stalled forks (Lee and Dunphy, 2013; Schlacher et al., 2011; Schlacher et al., 2012). The MRN complex may regulate the ATR response and protect replication forks through multiple mechanisms.
It is important to note that although the phosphorylation of RPA32 Ser33 is a marker for Nbs1-mediated ATR activation, ATR may have additional substrates when it is activated in this mode. Many of the known or putative substrates of ATR, including a number of DNA repair proteins, interact with RPA and may accumulate on RPA-ssDNA during resection (Matsuoka et al., 2007). The Nbs1-mediated mode of ATR activation may contribute to the phosphorylation of these ATR substrates and to DNA repair, particularly during the late phase of the DNA damage response. Together, Rad17 and Nbs1 may orchestrate the phosphorylation of two distinct sets of ATR substrates in two sequential phases of the DNA damage response. Whereas Rad17 may promote DNA damage signaling, cell cycle arrest, and the early events of DNA repair through Chk1 phosphorylation, Nbs1 may promote the late events of DNA repair through progressive phosphorylation of ATR substrates on RPA-ssDNA. The coordination of these two distinct mechanisms of ATR activation may be important for the full function of ATR in the DNA damage response.
Experimental Procedures
Cell culture and drug treatment
HeLa, U2OS and 293T cells were cultured in DMEM supplemented with 10% FBS. NBS-ILB1 cells were cultured in DMEM supplemented with 15% FBS. NBS1-ILB1 cells stably expressing the full-length Nbs1 transgene were maintained in the same medium containing 500 µg/ml G418. For RNA interference, cells were transfected with 40 nM siRNA using RNAi MAX transfection reagent (Invitrogen). The sequences of the siRNAs used in this study are listed in the Supplemental Information. Plasmid transfection of 293T cells was performed with Lipofectamine 2000 (Invitrogen) and the transfected cells were analyzed after 48 hr. U2OS derivative cell lines stably expressing myc-Nbs1 or mutants were cultured in DMEM with 10% FBS and 100 µg/mL Zeocin (Invitrogen). Expression of myc-Nbs1 was induced with 1µg/mL doxycycline (Sigma). To synchronize cells in S phase, cells were treated with 2.5 mM thymidine (Sigma) for 20 h, washed thoroughly with PBS, and released into thymidine-free medium for 3h. To assess the recovery of collapsed replication forks, synchronized S-phase cells were treated with CPT for 1 h and then released into CPT-free medium. Kinase inhibitors VE-821, KU55933, and NU7024 were used at 10 µM. Camptothecin (CPT) was used at 1 µM.
Antibodies
The phospho-specific antibodies to ATR pT1989 were previously described (Liu et al, 2011). ATR, Nbs1, Mre11, Rad50, TopBP1, phospho-Rad17 (Ser645), and phospho-RPA32 (Ser33) and (Ser4/Ser8) antibodies are from Bethyl. Rad17 and Chk1 antibodies are from Santa Cruz. Phospho-Chk1 (Ser345), phospho-Chk2 (Thr68), and phospho-p53 (Ser15) antibodies are from Cell Signaling. Phospho-H2AX (Ser139) antibody is from Millipore. Phospho-ATM (Ser1981) antibody is from Epitomics. RPA32 antibody is from Thermo Scientific. Myc antibody is from Medical & Biological Laboratories. Phospho-DNA-PKcs (Ser2056) and CtIP antibodies are kindly provided by Drs. B. Chen and R. Baer, respectively.
Extract-based ATR activation assay
Extract-based ATR activation assay were performed as described previously (Shiotani and Zou, 2011). To analyze single-stranded DNA-induced ATR activation, nuclear extracts were pre-treated with 10 µM of KU-55933 and NU7026 for 15 min on ice to inhibit ATM and DNA-PKcs, and supplemented with the reaction buffer (buffer R), which brings the final buffer compositions to 10 mM HEPES (pH 7.6), 50 mM KCl, 0.1 mM MgCl2, 1 mM PMSF, 0.5 mM DTT, 1 mM ATP, 10 µg/ml creatine kinase, and 5 mM phosphocreatine. ssDNA of various lengths was incubated in the extracts for 30 min at 37 °C. The sequences of the DNA oligonucleotides used in this study are listed in the Supplemental Data.
RPA-ssDNA binding assay
Biotinylated ssDNA (5’TGCAGCTGGCACGACAGGTTTTAATGAATCGGCCA-ACGCGCGGGGAGAGGCGGTTTGCGTATTGGGCGCT-biotin-3’) were attached to streptavidin-coated magnetic beads in the binding buffer [10 mM Tris-HCl (pH 7.5), 100 mM NaCl, 10% glycerol, 0.01% NP-40, and 10 µg/ml bovine serum albumin] followed by incubation with or without purified RPA at room temperature for 30 min. To analyze the binding of various proteins to RPA-ssDNA, RPA-ssDNA was incubated with purified MRN, Nbs1, or cell lysates in the binding buffer. To map the RPA-ssDNA-interacting domain of Nbs1, lysates prepared from cells expressing various myc-Nbs1 mutants were used. After a 30-min incubation at room temperature, beads were retrieved and washed twice with the binding buffer. The proteins bound to beads were denatured in the SDS sample buffer, separated on SDS-PAGE, and analyzed by Western blot. Recombinant RPA complex was purified from E. coli as previously described (Henricksen et al, 1994). The MRN complex and Nbs1 were purified from insect cells infected with baculoviruses expressing Mre11, Rad50, and Nbs1 (Paull and Gellert, 1999).
Supplementary Material
Highlights.
ATR phosphorylates RPA32 Ser33 progressively during DSB resection
RPA32 Ser33 phosphorylation is largely independent of Rad17 but dependent on Nbs1
Nbs1 promotes RPA32 Ser33 phosphorylation independently of ATM and resection
The interaction of Nbs1 and RPA is important for RPA32 Ser33 phosphorylation
Acknowledgement
We thank Dr. K. Cimprich for communicating unpublished results, Drs. S. Elledge, A. Nussenzweig, T. Paull, X. Wu, B. Chen, R. Baer, P. Concannon, and K. Komatsu for reagents, and members of the Zou lab for helpful discussion. P. H. was supported by a postdoctoral fellowship from the Swedish Research Council. A. M. is supported by a FRSQ fellowship. L. Z. is a Jim & Ann Orr Massachusetts General Hospital Research Scholar. This work was supported by grants from the Japanese Ministry of Education, Culture, Sports, Science and Technology to B. S. (KAKENHI, 24800044) and H. T. (KAKENHI, 23300363), and grants from NIH (GM076388), ACS (RSG-08-297), and the Federal Share of MGH Proton Program to L. Z.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anantha RW, Vassin VM, Borowiec JA. Sequential and synergistic modification of human RPA stimulates chromosomal DNA repair. J Biol Chem. 2007;282:35910–35923. doi: 10.1074/jbc.M704645200. [DOI] [PubMed] [Google Scholar]
- Avemann K, Knippers R, Koller T, Sogo JM. Camptothecin, a specific inhibitor of type I DNA topoisomerase, induces DNA breakage at replication forks. Mol Cell Biol. 1988;8:3026–3034. doi: 10.1128/mcb.8.8.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow JH, Faryabi RB, Callen E, Wong N, Malhowski A, Chen HT, Gutierrez-Cruz G, Sun HW, McKinnon P, Wright G, et al. Identification of early replicating fragile sites that contribute to genome instability. Cell. 2013;152:620–632. doi: 10.1016/j.cell.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekker-Jensen S, Lukas C, Kitagawa R, Melander F, Kastan MB, Bartek J, Lukas J. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J Cell Biol. 2006;173:195–206. doi: 10.1083/jcb.200510130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block WD, Yu Y, Lees-Miller SP. Phosphatidyl inositol 3-kinase-like serine/threonine protein kinases (PIKKs) are required for DNA damage-induced phosphorylation of the 32 kDa subunit of replication protein A at threonine 21. Nucleic Acids Res. 2004;32:997–1005. doi: 10.1093/nar/gkh265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown EJ, Baltimore D. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev. 2000;14:397–402. [PMC free article] [PubMed] [Google Scholar]
- Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005;19:1040–1052. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney JP, Maser RS, Olivares H, Davis EM, Le Beau M, Yates JR, 3rd, Hays L, Morgan WF, Petrini JH. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell. 1998;93:477–486. doi: 10.1016/s0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo V, Robertson K, Bibikova M, Kim E, Grieco D, Gottesman M, Carroll D, Gautier J. Mre11 protein complex prevents double-strand break accumulation during chromosomal DNA replication. Mol Cell. 2001;8:137–147. doi: 10.1016/s1097-2765(01)00294-5. [DOI] [PubMed] [Google Scholar]
- Costanzo V, Shechter D, Lupardus PJ, Cimprich KA, Gottesman M, Gautier J. An ATR- and Cdc7-dependent DNA damage checkpoint that inhibits initiation of DNA replication. Mol Cell. 2003;11:203–213. doi: 10.1016/s1097-2765(02)00799-2. [DOI] [PubMed] [Google Scholar]
- Cotta-Ramusino C, McDonald ER, 3rd, Hurov K, Sowa ME, Harper JW, Elledge SJ. A DNA damage response screen identifies RHINO, a 9-1-1 and TopBP1 interacting protein required for ATR signaling. Science. 2011;332:1313–1317. doi: 10.1126/science.1203430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delacroix S, Wagner JM, Kobayashi M, Yamamoto K, Karnitz LM. The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev. 2007;21:1472–1477. doi: 10.1101/gad.1547007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Difilippantonio S, Celeste A, Fernandez-Capetillo O, Chen HT, Reina San Martin B, Van Laethem F, Yang YP, Petukhova GV, Eckhaus M, Feigenbaum L, et al. Role of Nbs1 in the activation of the Atm kinase revealed in humanized mouse models. Nat Cell Biol. 2005;7:675–685. doi: 10.1038/ncb1270. [DOI] [PubMed] [Google Scholar]
- Ellison V, Stillman B. Biochemical characterization of DNA damage checkpoint complexes: clamp loader and clamp complexes with specificity for 5' recessed DNA. PLoS Biol. 2003;1:E33. doi: 10.1371/journal.pbio.0000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn RL, Zou L. ATR: a master conductor of cellular responses to DNA replication stress. Trends Biochem Sci. 2011;36:133–140. doi: 10.1016/j.tibs.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Dunphy WG. Response of Xenopus Cds1 in cell-free extracts to DNA templates with double-stranded ends. Mol Biol Cell. 2000;11:1535–1546. doi: 10.1091/mbc.11.5.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henricksen LA, Umbricht CB, Wold MS. Recombinant replication protein A: expression, complex formation, and functional characterization. J Biol Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]
- Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukas J, Jackson SP. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol. 2006;8:37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- Kousholt AN, Fugger K, Hoffmann S, Larsen BD, Menzel T, Sartori AA, Sorensen CS. CtIP-dependent DNA resection is required for DNA damage checkpoint maintenance but not initiation. J Cell Biol. 2012;197:869–876. doi: 10.1083/jcb.201111065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A, Lee J, Yoo HY, Dunphy WG. TopBP1 activates the ATR-ATRIP complex. Cell. 2006;124:943–955. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- Lee J, Dunphy WG. Rad17 plays a central role in establishment of the interaction between TopBP1 and the Rad9-Hus1-Rad1 complex at stalled replication forks. Mol Biol Cell. 2010;21:926–935. doi: 10.1091/mbc.E09-11-0958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Dunphy WG. The Mre11-Rad50-Nbs1 (MRN) complex has a specific role in the activation of Chk1 in response to stalled replication forks. Mol Biol Cell. 2013 doi: 10.1091/mbc.E13-01-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Kumagai A, Dunphy WG. The Rad9-Hus1-Rad1 checkpoint clamp regulates interaction of TopBP1 with ATR. J Biol Chem. 2007;282:28036–28044. doi: 10.1074/jbc.M704635200. [DOI] [PubMed] [Google Scholar]
- Liaw H, Lee D, Myung K. DNA-PK-dependent RPA2 hyperphosphorylation facilitates DNA repair and suppresses sister chromatid exchange. PLoS One. 2011;6:e21424. doi: 10.1371/journal.pone.0021424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SJ, Wardlaw CP, Morishita T, Miyabe I, Chahwan C, Caspari T, Schmidt U, Carr AM, Garcia V. The Rad4(TopBP1) ATR-activation domain functions in G1/S phase in a chromatin-dependent manner. PLoS Genet. 2012;8:e1002801. doi: 10.1371/journal.pgen.1002801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Bekker-Jensen S, Mailand N, Lukas C, Bartek J, Lukas J. Claspin operates downstream of TopBP1 to direct ATR signaling towards Chk1 activation. Mol Cell Biol. 2006;26:6056–6064. doi: 10.1128/MCB.00492-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Opiyo SO, Manthey K, Glanzer JG, Ashley AK, Amerin C, Troksa K, Shrivastav M, Nickoloff JA, Oakley GG. Distinct roles for DNA-PK, ATM and ATR in RPA phosphorylation and checkpoint activation in response to replication stress. Nucleic Acids Res. 2012;40:10780–10794. doi: 10.1093/nar/gks849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Shiotani B, Lahiri M, Marechal A, Tse A, Leung CC, Glover JN, Yang XH, Zou L. ATR autophosphorylation as a molecular switch for checkpoint activation. Mol Cell. 2011;43:192–202. doi: 10.1016/j.molcel.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDougall CA, Byun TS, Van C, Yee MC, Cimprich KA. The structural determinants of checkpoint activation. Genes Dev. 2007;21:898–903. doi: 10.1101/gad.1522607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manthey KC, Opiyo S, Glanzer JG, Dimitrova D, Elliott J, Oakley GG. NBS1 mediates ATR-dependent RPA hyperphosphorylation following replication-fork stall and collapse. J Cell Sci. 2007;120:4221–4229. doi: 10.1242/jcs.004580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maser RS, Mirzoeva OK, Wells J, Olivares H, Williams BR, Zinkel RA, Farnham PJ, Petrini JH. Mre11 complex and DNA replication: linkage to E2F and sites of DNA synthesis. Mol Cell Biol. 2001;21:6006–6016. doi: 10.1128/MCB.21.17.6006-6016.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- Mordes DA, Glick GG, Zhao R, Cortez D. TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes Dev. 2008;22:1478–1489. doi: 10.1101/gad.1666208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murga M, Bunting S, Montana MF, Soria R, Mulero F, Canamero M, Lee Y, McKinnon PJ, Nussenzweig A, Fernandez-Capetillo O. A mouse model of ATR-Seckel shows embryonic replicative stress and accelerated aging. Nat Genet. 2009;41:891–898. doi: 10.1038/ng.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers JS, Cortez D. Rapid activation of ATR by ionizing radiation requires ATM and Mre11. J Biol Chem. 2006;281:9346–9350. doi: 10.1074/jbc.M513265200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navadgi-Patil VM, Burgers PM. The unstructured C-terminal tail of the 9-1-1 clamp subunit Ddc1 activates Mec1/ATR via two distinct mechanisms. Mol Cell. 2009;36:743–753. doi: 10.1016/j.molcel.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley GG, Tillison K, Opiyo SA, Glanzer JG, Horn JM, Patrick SM. Physical interaction between replication protein A (RPA) and MRN: involvement of RPA2 phosphorylation and the N-terminus of RPA1. Biochemistry. 2009;48:7473–7481. doi: 10.1021/bi900694p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson E, Nievera CJ, Lee AY, Chen L, Wu X. The Mre11-Rad50-Nbs1 complex acts both upstream and downstream of ataxia telangiectasia mutated and Rad3-related protein (ATR) to regulate the S-phase checkpoint following UV treatment. J Biol Chem. 2007a;282:22939–22952. doi: 10.1074/jbc.M702162200. [DOI] [PubMed] [Google Scholar]
- Olson E, Nievera CJ, Liu E, Lee AY, Chen L, Wu X. The Mre11 complex mediates the S-phase checkpoint through an interaction with replication protein A. Mol Cell Biol. 2007b;27:6053–6067. doi: 10.1128/MCB.00532-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull TT, Gellert M. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 1999;13:1276–1288. doi: 10.1101/gad.13.10.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson SE, Li Y, Wu-Baer F, Chait BT, Baer R, Yan H, Gottesman ME, Gautier J. Activation of DSB Processing Requires Phosphorylation of CtIP by ATR. Mol Cell. 2013;49:657–667. doi: 10.1016/j.molcel.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reaper PM, Griffiths MR, Long JM, Charrier JD, Maccormick S, Charlton PA, Golec JM, Pollard JR. Selective killing of ATM- or p53-deficient cancer cells through inhibition of ATR. Nat Chem Biol. 2011;7:428–430. doi: 10.1038/nchembio.573. [DOI] [PubMed] [Google Scholar]
- Sacho EJ, Maizels N. DNA repair factor MRE11/RAD50 cleaves 3'-phosphotyrosyl bonds and resects DNA to repair damage caused by topoisomerase 1 poisons. J Biol Chem. 2011;286:44945–44951. doi: 10.1074/jbc.M111.299347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakasai R, Shinohe K, Ichijima Y, Okita N, Shibata A, Asahina K, Teraoka H. Differential involvement of phosphatidylinositol 3-kinase-related protein kinases in hyperphosphorylation of replication protein A2 in response to replication-mediated DNA double-strand breaks. Genes Cells. 2006;11:237–246. doi: 10.1111/j.1365-2443.2006.00942.x. [DOI] [PubMed] [Google Scholar]
- Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011;145:529–542. doi: 10.1016/j.cell.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K, Wu H, Jasin M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 2012;22:106–116. doi: 10.1016/j.ccr.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W, Feng Z, Zhang J, Gonzalez-Suarez I, Vanderwaal RP, Wu X, Powell SN, Roti Roti JL, Gonzalo S. The role of RPA2 phosphorylation in homologous recombination in response to replication arrest. Carcinogenesis. 2010;31:994–1002. doi: 10.1093/carcin/bgq035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiotani B, Zou L. Single-stranded DNA orchestrates an ATM-to-ATR switch at DNA breaks. Mol Cell. 2009;33:547–558. doi: 10.1016/j.molcel.2009.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiotani B, Zou L. A human cell extract-based assay for the activation of ATM and ATR checkpoint kinases. Methods Mol Biol. 2011;782:181–191. doi: 10.1007/978-1-61779-273-1_13. [DOI] [PubMed] [Google Scholar]
- Stewart GS, Maser RS, Stankovic T, Bressan DA, Kaplan MI, Jaspers NG, Raams A, Byrd PJ, Petrini JH, Taylor AM. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell. 1999;99:577–587. doi: 10.1016/s0092-8674(00)81547-0. [DOI] [PubMed] [Google Scholar]
- Stiff T, Reis C, Alderton GK, Woodbine L, O'Driscoll M, Jeggo PA. Nbs1 is required for ATR-dependent phosphorylation events. Embo J. 2005;24:199–208. doi: 10.1038/sj.emboj.7600504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracker TH, Petrini JH. The MRE11 complex: starting from the ends. Nat Rev Mol Cell Biol. 2011;12:90–103. doi: 10.1038/nrm3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annu Rev Genet. 2011;45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- Toledo LI, Murga M, Zur R, Soria R, Rodriguez A, Martinez S, Oyarzabal J, Pastor J, Bischoff JR, Fernandez-Capetillo O. A cell-based screen identifies ATR inhibitors with synthetic lethal properties for cancer-associated mutations. Nat Struct Mol Biol. 2011;18:721–727. doi: 10.1038/nsmb.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uziel T, Lerenthal Y, Moyal L, Andegeko Y, Mittelman L, Shiloh Y. Requirement of the MRN complex for ATM activation by DNA damage. Embo J. 2003;22:5612–5621. doi: 10.1093/emboj/cdg541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van C, Yan S, Michael WM, Waga S, Cimprich KA. Continued primer synthesis at stalled replication forks contributes to checkpoint activation. J Cell Biol. 2010;189:233–246. doi: 10.1083/jcb.200909105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassin VM, Anantha RW, Sokolova E, Kanner S, Borowiec JA. Human RPA phosphorylation by ATR stimulates DNA synthesis and prevents ssDNA accumulation during DNA-replication stress. J Cell Sci. 2009;122:4070–4080. doi: 10.1242/jcs.053702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Gong Z, Chen J. MDC1 collaborates with TopBP1 in DNA replication checkpoint control. J Cell Biol. 2011;193:267–273. doi: 10.1083/jcb.201010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zou L, Lu T, Bao S, Hurov KE, Hittelman WN, Elledge SJ, Li L. Rad17 phosphorylation is required for claspin recruitment and Chk1 activation in response to replication stress. Mol Cell. 2006;23:331–341. doi: 10.1016/j.molcel.2006.06.022. [DOI] [PubMed] [Google Scholar]
- Yamane K, Chen J, Kinsella TJ. Both DNA topoisomerase II-binding protein 1 and BRCA1 regulate the G2-M cell cycle checkpoint. Cancer Res. 2003;63:3049–3053. [PubMed] [Google Scholar]
- Yan S, Michael WM. TopBP1 and DNA polymerase-alpha directly recruit the 9-1-1 complex to stalled DNA replication forks. J Cell Biol. 2009;184:793–804. doi: 10.1083/jcb.200810185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo HY, Kumagai A, Shevchenko A, Dunphy WG. The Mre11-Rad50-Nbs1 complex mediates activation of TopBP1 by ATM. Mol Biol Cell. 2009;20:2351–2360. doi: 10.1091/mbc.E08-12-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong H, Bryson A, Eckersdorff M, Ferguson DO. Rad50 depletion impacts upon ATR-dependent DNA damage responses. Hum Mol Genet. 2005;14:2685–2693. doi: 10.1093/hmg/ddi302. [DOI] [PubMed] [Google Scholar]
- Zou L, Cortez D, Elledge SJ. Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes Dev. 2002;16:198–208. doi: 10.1101/gad.950302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- Zou L, Liu D, Elledge SJ. Replication protein A-mediated recruitment and activation of Rad17 complexes. Proc Natl Acad Sci U S A. 2003;100:13827–13832. doi: 10.1073/pnas.2336100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.