Abstract
The microenvironment of human follicular lymphoma (FL), an incurable B-cell non-Hodgkin lymphoma, is thought to play a major role in its pathogenesis and course. Microenvironmental cells of likely importance include follicular helper T cells (TFH) and regulatory T cells (Tregs), and understanding their interactions with FL tumor cells is necessary to develop novel therapeutic strategies. We found that IL-4 and CD40L are expressed by intratumoral TFH and induce production of CCL17 and CCL22 by FL tumor cells. IL-4 alone induces only CCL17, but enhances stimulation by CD40L of both CCL17 and CCL22. Consistent with our in vitro results, mRNA transcripts of IL-4 correlated with CCL17 but not CCL22 in gene expression profiling studies of FL biopsies, whereas CD40L correlated with both CCL17 and CCL22. Tumor supernatants induced preferential migration of Tregs and IL-4–producing T cells rather than IFN-γ–producing T cells, and antibodies to CCR4 significantly abrogated the migration of Tregs. Our results suggest that through two distinct mechanisms, intratumoral TFH induce production of CCL17 and CCL22 by FL tumor cells and facilitate active recruitment of Tregs and IL-4–producing T cells, which in turn may stimulate more chemokine production in a feed-forward cycle. Thus, TFH appear to play a major role in generating an immunosuppressive tumor microenvironment that promotes immune escape and tumor survival and growth. Our results provide novel insights into the cross talk between TFH, tumor cells, and Tregs in FL and offer potential targets for development of therapeutic strategies to overcome immune evasion.
INTRODUCTION
Follicular lymphoma (FL) is the most common indolent B-cell lymphoma and comprises 22% of all non-Hodgkin’s lymphomas worldwide.1 FL is derived from germinal center B cells and is characterized by hyperexpression of the anti-apoptotic Bcl-2 oncoprotein as a consequence of the t(14;18) BCL2/JH translocation.2 However, the t(14;18) translocation does not appear to be sufficient for lymphomagenesis, as B cells with the t(14;18) translocation can be found in a substantial proportion of healthy individuals.3,4 Moreover, lymphomas develop in only 10%–15% of transgenic mice in which BCL2 expression was driven by an IgH enhancer (Eμ).5 Therefore, growth factors such as cytokines and other protumor factors present in the tumor microenvironment may be necessary for the pathogenesis and progression of FL.6 Recently, using proteomic profiling of tumor lysates, Calvo and colleagues found that IL-4 levels were significantly higher in FL tissues than in tissues from follicular hyperplasia.7 Furthermore, they demonstrated increased basal phosphorylation of downstream targets of IL-4, STAT6 and the mitogen-activated protein (MAP) kinase extracellular signal-related kinase (Erk), in FL tissues as compared with benign follicular hyperplasia in tonsils. Additional reports showed that follicular helper T cells (TFH) express high levels of IL-4 and CD40 ligand (CD40L) mRNA in FL and may be involved in promoting the survival of tumor B cells via IL-4 and CD40L8,9 consistent with other in vitro studies.10,11 Together, these reports suggest that IL-4 and CD40L expressed by TFH may act as protumor factors and may play a role in the pathogenesis of FL.
Evidence in the literature suggests that the FL tumor microenvironment also contains antitumor factors.6 The indolent nature of FL,12 induction of spontaneous remissions in patients who are observed without therapy,12 isolation of antitumor T cells from the tumor microenvironment,13,14 and correlation of survival with the gene expression signature of tumor-infiltrating immune cells in FL patients15 all support the assertion that antitumor factors are present in the tumor microenvironment in FL and suggest that FL is naturally immunogenic. Furthermore, the induction of antitumor immune responses in most FL patients after idiotypic vaccination,16,17 the high clinical response rates observed with the anti-CD20 monoclonal antibody rituximab,18,19 and prolonged progression-free survival (PFS) after nonmyeloablative allogeneic stem-cell transplantation20 suggest that FL is highly immune-responsive. However, immunosuppressive cells such as forkhead box P3 (Foxp3)+ regulatory T cells (Tregs) and macrophages present in the FL tumor microenvironment may limit the efficacy of antitumor immune responses that are both naturally and therapeutically induced, and thus may exert a protumor effect.21 Consequently, the natural history of FL in patients who are observed without therapy as well as clinical outcome of patients undergoing therapeutic intervention are likely to depend on the relative dominance of the protumor and antitumor factors within the tumor microenvironment. Characterization of such factors and studying the dynamic interactions between the tumor and microenvironmental cells is necessary to provide a better understanding of the pathogenesis and course of FL.
Regulatory T cells are among the most potent suppressors of effector T cells and other immune cells.22 Tregs have been shown to inhibit T-cell responses against both foreign antigens and self-antigens such as tumor antigens. Several reports have suggested that Tregs are increased in number in the tumor microenvironment of a variety of human cancers including follicular lymphoma22–26, and intratumoral Tregs from B-cell lymphoma patients have been shown to inhibit the function of antitumor CD4+ and CD8+ T cells.23,27,28 Tregs may be increased in number in FL via induction from conventional CD4+ T cells by the tumor cells.26,29,30 Alternatively, Tregs may also be actively recruited to the tumor site by chemokine expression in the tumor microenvironment. In FL, CCL17 (thymus and activation-regulated chemokine) and CCL22 (macrophage-derived chemokine) produced by the tumor cells have been proposed to play a role in recruiting Tregs into the tumor microenvironment.23,30 However, the mechanism leading to the increased production of these chemokines by FL tumor cells is unknown. CCL17 is constitutively expressed by thymocytes, monocyte-derived dendritic cells, and keratinocytes31,32, and CCL22 is constitutively expressed by macrophages, dendritic cells, keratinocytes, and epithelial cells.33 However, B cells are not known to constitutively express either of these chemokines.
Here, we demonstrate that CCL17 and CCL22 are produced by tumor cells in FL as a result of crosstalk with TFH. Furthermore, we show that these chemokines induce preferential migration of Tregs and IL-4–producing CD4+ T cells that in turn might stimulate more chemokine production, thus creating a vicious cycle and an immunosuppressive tumor microenvironment that promotes immune evasion and tumor survival and growth. These results provide novel insights into the role of TFH in immune evasion in FL and offer potential targets for the development of therapeutic strategies.
MATERIALS AND METHODS
Patient samples
All blood and tissue samples were obtained after informed consent from patients through an institutional review board-approved protocol. Lymph node biopsies were obtained from patients with FL at the time of their initial diagnosis prior to therapy. Tonsils were obtained from children who had undergone elective tonsillectomy. The use of tonsils with benign follicular hyperplasia as controls for FL has generally been accepted in the scientific literature.7,15,34 Surgical samples were stored as formalin-fixed tissue sections in paraffin and/or processed into a single-cell suspension and cryopreserved. Due to limited number of cells obtained from each tumor or tonsil, the same samples could not be used in all experiments. Altogether, 21 tumors and 14 tonsil samples were used for the various in vitro studies. Serum samples were obtained from healthy donors and patients with FL at the time of their initial diagnosis (Supplementary Table 1). All FL patients were treated uniformly with prednisone, doxorubicin, cyclophosphamide, and etoposide chemotherapy followed by autologous tumor-derived idiotype vaccination.16,35
Cell lines
Mantle cell lymphoma cell lines (Mino and Granta519), transformed FL cell lines that carry t(14;18) (RL, DHL-16, DOHH-2), diffuse large B-cell lymphoma cell lines (EJ-1 and SKI), germinal center B-cell-derived diffuse large B-cell lymphoma cell lines (SU-DHL-4, PFEIFFER, and TOLEDO), and Burkitt’s lymphoma cell lines (Raji and Daudi) were cultured in RPMI Medium 1640 supplemented with 10% fetal bovine serum, 10 mM Hepes, 1× GlutaMAX, 50μM β-mercaptoethanol, 1 mM sodium pyruvate, 100 U/ml penicillin + 100 μg/ml streptomycin, and 10 μg/ml gentamicin (all from Invitrogen) at 37°C and 5% CO2 in air.
Reagents
CD3 Allophycocyanin (APC), CD4 Peridinin-chlorophyll proteins (PerCP)-Cy5.5, CD8 PerCP-Cy5.5, CD10 APC, CD20 fluorescein isothiocyanate (FITC), CD25 FITC, CD127 PerCP-Cy5.5, IL-4Rα Phycoerythrin (PE), IL-4 PE, IL-13 PE, IFN-γ APC, phospho-STAT6 APC (Tyr641), and CCR4 PE were all obtained from BD Biosciences. Kappa FITC and lambda FITC were obtained from Invitrogen. Foxp3 PE (clone PCH101) was obtained from eBioscience. ELISA kits for CCL17 and CCL22 were obtained from R&D Systems. Neutralizing antibodies against CCL17 and CCL22 were obtained from R&D Systems. PMA and ionomycin were obtained from Sigma-Aldrich. Recombinant human IL-4, recombinant human IL-13, recombinant human CCL17, recombinant human CCL22, and CCL17 and CCL22 antibodies for immunohistochemical staining were obtained from Peprotech. IL-4, phospho-STAT6 (Tyr641), and Foxp3 antibodies for immunohistochemical staining were obtained from Abcam. Anti-mouse IgG antibody (BA-2000) and anti-rabbit antibody (BA-1000) were obtained from Vector Laboratories. STAT6, phospho-STAT6 (Tyr641), and β-actin antibodies for Western blot analysis were obtained from Cell Signaling Technology. For TFH staining, CD4 Amcyan, CD3 APC-Cy7, CXCR5 Alexa488, BCL6 PE, ICOS APC, CD40L PerCP-Cy5.5 were obtained from BD Biosciences and PD1 PE from eBioscience.
Magnetic cell separation (MACS)
Tumor cells were isolated from single-cell suspensions of lymph node biopsies or from surgical samples from patients with lymphoma by depleting T cells using CD3 magnetic beads (Miltenyi Biotec) as described previously.17 The procedure yielded tumor B cells of >99% purity as determined by surface staining for CD10, CD20, and monoclonal light-chain restriction. Regulatory T cells were isolated by MACS using Regulatory T Cell Isolation Kit (Miltenyi Biotec) according to the manufacturer’s instructions. To isolate non-Tregs, we isolated CD4+ T cells using CD4+ T Cell Isolation Kit and then depleted that cell fraction of Tregs using CD25 magnetic beads (Miltenyi Biotec). These procedures yielded >90% purity of Tregs (CD4+CD25+CD127loFoxp3+) and non-Tregs (CD4+CD25−Foxp3−) as determined by flow cytometric analysis.
Flow cytometric analysis
Surface staining and Foxp3 (eBioscience) and phospho-STAT6 (BD Biosciences) staining for flow cytometric analysis were performed according to the manufacturer’s instructions. Samples were acquired using a BD FACS Calibur system (BD Biosciences) and analyzed using Cell Quest Pro (BD Biosciences) or FlowJo (Tree Star, Inc.) software. TFH identified as CD3+CD4+CD25−PD1hiCXCR5hi were sorted using FACSAria (Becton Dickinson, San Diego, CA, USA). Purity of sorted TFH was >98%.
Real-time PCR
Total RNA was extracted from purified tumor and tonsil B-cells using Trizol reagent (Invitrogen), followed by treatment with DNase (Ambion). Real-time PCR was performed on the ABI Prism 7900 using Power SYBR Green RNA-to-CT 1-Step Kit according to the manufacturer’s instructions (Applied Biosystems). Primers for CCL17 (Hs00171074) and CCL22 (Hs00171080) were obtained from Applied Biosystems. GAPDH was used as an endogenous control (Integrated DNA Technologies). The δδCt method was used to calculate the proportional difference in CCL17 and CCL22 expression in tumor B cells and tonsil B cells.
Immunohistochemical analysis
Formalin-fixed paraffin-embedded tissue sections were deparaffinized and rehydrated, and antigen retrieval was performed according to the manufacturer’s protocol (Vector Laboratories). Tissue sections were treated for 5 min with 0.3% hydrogen peroxide solution to block endogenous peroxidase and then incubated with a blocking buffer (1% BSA in PBS; Sigma) for 5 min at room temperature. Next, slides were incubated with primary antibodies overnight at 4°C at a dilution of 1:100 in blocking buffer; then the slides were washed and incubated for 30 min with appropriate secondary antibodies in blocking buffer. The sections were stained using DAB solution (Vector Laboratories). Digital photomicrographs were acquired using DP Controller software (Olympus) mounted on a BX41 Clinical Microscope (Olympus).
Analysis of cDNA microarray data
Expression of chemokine genes, IL-4, IL-13, IL-4 receptor, and IL-13 receptor in tumor cells isolated from FL biopsies and B cells isolated from reactive tonsils were obtained from a publicly available microarray dataset (http://icg.cpmc.columbia.edu/Web_Data/JCI/2003_JCI.htm).34 Log2-transformed data were analyzed for relative levels of gene expression. Data from the Oncomine database (https://www.oncomine.org/resource/login.html) were also similarly displayed for expression of CCL17 and CCL22 mRNA in multiple lymphoproliferative disorders. Whole-genome gene expression data from the study of Dave et al15 were obtained from a publicly available source (http://llmpp.nih.gov/FL/). After log2 normalization and reannotation of the microarray version used (06/09/2011; Release 32, http://www.affymetrix.com), data for selected genes were examined for pairwise correlation by Pearson’s correlation and Spearman’s rank correlation coefficients, with significance for the Pearson’s correlation coefficient (PCC) determined assuming no relationship in the population. The PCC for selected genes of relevance to immune cell types were calculated and displayed as a correlation matrix heat map, as pioneered by Tosolini et al.36
Cytokine and chemokine induction
Tumor cells or intratumoral T cells were cultured in the presence or absence of the indicated stimuli, and cytokines or chemokine levels were assessed in the supernatants after 48 h by ELISA or multiplex assays according to the manufacturer’s instructions. For intracellular cytokine assay, cells were cultured in the absence or presence of PMA (50 ng/ml) and ionomycin (500 ng/ml) for 14 h. Brefeldin A (5 μg/ml, Sigma-Aldrich) was added for the last 12 h of the incubation, and the cells were harvested, fixed, permeabilized, and stained for surface markers and intracellular cytokines by fluorochrome-labeled monoclonal antibodies as previously described.37
Chemotaxis assay
Tumor cells were cultured in the absence or presence of IL-4 (20 ng/ml) and supernatants were harvested after 48 h for chemotaxis assay. Tumor supernatants or recombinant CCL17 and CCL22 (200 ng/ml each) were plated in triplicate in the lower chambers of a transwell plate, and Tregs or non-Tregs (2 × 105 cells/well) were plated in triplicate in the upper chambers of a transwell plate separated by a 5-μm pore polycarbonate filter (Corning) and cultured for 2 h. Cells that migrated across the filter were recovered and counted using the trypan blue exclusion method. The percentage of cells that had migrated was calculated using the following formula: (number of cells migrated into the lower chamber/number of cells seeded in the corresponding upper chamber) × 100. The inhibition of chemotaxis was assessed using anti-CCR4–blocking antibody (20 μg/ml). Chemotaxis inhibition was calculated as a percentage using the following formula: [(X − Y)/X] × 100, where X is the fraction of cells that migrated in the absence of antibodies and Y is the fraction of cells that migrated in the presence of antibodies.
Western blot analysis
Approximately 2–4 × 106 tumor cells were lysed in a buffer composed of 50 mM Tris-Cl (pH, 7.4), 5 mM EDTA, 150 mM NaCl, 0.5% Triton-X 100, 1 mg/ml leupeptin and aprotinin, and 1 mM PMSF. Protein content of the cell lysates was quantified using the Bradford assay (Bio-Rad), and 25 μg of total protein was dissolved in Laemmli SDS-PAGE sample buffer prior to separation in 10% SDS-PAGE gels. After electrophoresis, proteins were transferred to a nitrocellulose membrane (Bio-Rad) for Western blot analysis with anti-STAT6 or anti-phospho-STAT6 antibodies (1:1000 dilution each) for 1 h at room temperature or at 4°C overnight. Bound antibodies were detected with horseradish peroxidase-conjugated secondary antibodies diluted at 1:2000 and with SuperSignal West Pico Substrate (Thermo Scientific).
Knockdown of STAT6 with siRNA
Granta519 cells were transfected with STAT6-specific siRNA (5′GGGAGAAGAUGUGUGAAACUCUGAA-3′ and 5′-GAAUCCGGGAUCUUGCU CAGC UCAA-3′)38 or a Universal Stealth siRNA negative control (40 nmol each, Invitrogen) or transfection control pmaxGFP (Amaxa) using Nucleofector Kit V (Amaxa) according to manufacturer’s instructions. After 24 h, transfection efficiency was determined by flow cytometric analysis, and 55%–60% of the transfected cells were found to be positive for GFP. Knockdown of STAT6 and phospho-STAT6 was determined by Western blot analysis. Transfected cells were incubated with IL-4 (20 ng/ml) for an additional 24 h, and chemokine production was assessed in the supernatants by ELISA.
Statistical analysis
Differences between chemokine levels between unpaired experimental groups were determined using the Wilcoxon rank-sum test using Prism software (Graphpad Software, Inc). Differences between paired test and control samples were assessed by two-sided paired Student’s t-test using Microsoft Excel 2007. P values < 0.05 were considered statistically significant. Progression-free survival was defined as time from initiation of treatment to the time of progression or death, whichever occurred first, or to the time of last contact. Progression-free survival was estimated using the Kaplan-Meier method. Log-rank test was performed to test the difference in survival times between groups. Multivariate regression models were developed to examine the effect of chemokine level adjusted for Follicular Lymphoma International Prognostic Index (FLIPI) score.39 Regression analyses of PFS data was conducted using the Cox proportional hazards model. Due to skewed nature of distribution of the two chemokines CCL17 and CCL22, a logarithm (base 2) transformation was used in the analysis when included in the regression models as a continuous variable. SAS version 9.2 and S-Plus version 8.04 were used to carry out the statistical analyses.
RESULTS
IL-4–producing T cells are increased in the tumor microenvironment in FL
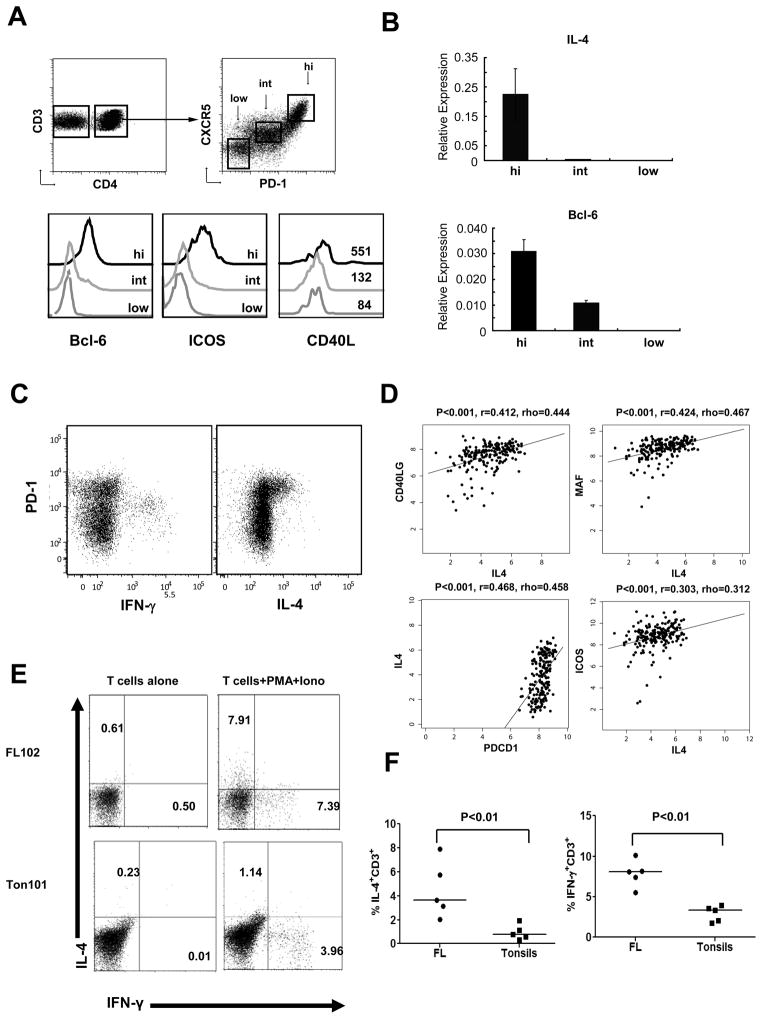
Calvo et al described increase in IL-4 in FL tumor lysates but did not identify the source of IL-4.7 Pangault et al showed that TFH cells in FL have high levels of IL-4 mRNA but did not confirm IL-4 expression at protein level. Moreover, expression of Bcl6, the master transcription factor for TFH differentiation, was not assessed.8 Here, we conclusively demonstrated that the CD4+PD-1hiCXCR5hi T cells are the major source of IL-4 in FL and expressed high levels of Bcl6 as well as ICOS and CD40L, suggesting that they represented TFH (Figures 1A–C). Consistent with our in vitro results, we found a moderate degree of correlation between IL-4 mRNA and genes associated with TFH such as CD40L, MAF, PDCD1, and ICOS in gene expression arrays of whole tumor tissues from FL patients reported previously by Dave et al15 (Figure 1D). From the same dataset, we calculated a self-correlation matrix of genes associated with a number of different immune cell subsets, which showed that IL-4 is most closely associated with a cluster of TFH-associated genes that is distinct from a TH1/TC1 cluster as well as genes associated with TH2, TH17, NK cells, macrophages, myeloid DC and plasmacytoid DC (Supplementary Figure 1A). These results suggest that the observed correlations between IL-4 and TFH genes are indeed specific. Consistent with this, IL-4 production was observed in T-cell cultures but was negligible in non-T cell cultures (Supplementary Figures 1B and 1C). More importantly, we found that IL-4 expressing T cells are increased in FL compared with tonsil controls (Figure 1E and 1F). To determine whether FL tumor cells respond to IL-4 produced in the tumor microenvironment, we evaluated the expression of IL-4Rα by flow cytometric analysis. We found that FL tumor cells and a subset of intratumoral T cells expressed IL-4Rα (Supplementary Figure 1D). Moreover, IL-4Rα mRNA was hyperexpressed in FL tumor cells compared with germinal center B cells (both centrocytes and centroblasts) from reactive tonsils (Supplementary Figures 1E and 1F). Taken together, these results indicate that IL-4 is produced predominantly by TFH and IL-4 expressing T cells are increased in FL.
Figure 1. IL-4–producing T cells are increased in the FL tumor microenvironment.
(A) Multiparametric flow cytometry was performed on intratumoral T cells. Intratumoral CD4+ T cells were gated into three subsets based on PD-1 and CXCR5 (PD-1hiCXCR5hi, PD-1intCXCR5int, and PD-1loCXCR5lo), and expression of Bcl6, ICOS, and CD40L was determined. Mean fluorescence intensity of CD40L is indicated in the histogram for the three subsets. (B) Intratumoral CD4+ T cells were sorted into PD-1hiCXCR5hi, PD-1intCXCR5int, and PD-1loCXCR5lo subsets and expression of IL-4 and Bcl6 was determined by real-time PCR. (C) Intratumoral CD4+ T cells were incubated for 12 h in the presence or absence of PMA and ionomycin. Brefeldin A was added for the last 10 h, and expression of IFN-γ and IL-4 vs PD-1 expression is shown in CD4+ T cells by flow cytometry. (D) The correlation between mRNA transcripts of IL-4 and that of CD40L, MAF, PDCD1 and ICOS was determined in the microarray dataset of 191 FL tumors published previously by Dave et al.15 Pearson’s correlation co-efficient (r) and Spearman’s rank correlation co-efficient (rho) are shown. (E and F) T cells isolated from FL biopsy samples or reactive tonsils (n=5 each) were stimulated as in panel C. IL-4 and IFN-γ production was assessed in CD3+ T cells by intracellular cytokine staining and the percentage of T cells producing cytokine was determined. Representative data (E) and data from 5 pairs of samples (F) are shown. Statistical significance was determined by Wilcoxon rank-sum test.
Phosphorylation of STAT6 is increased in FL tumors
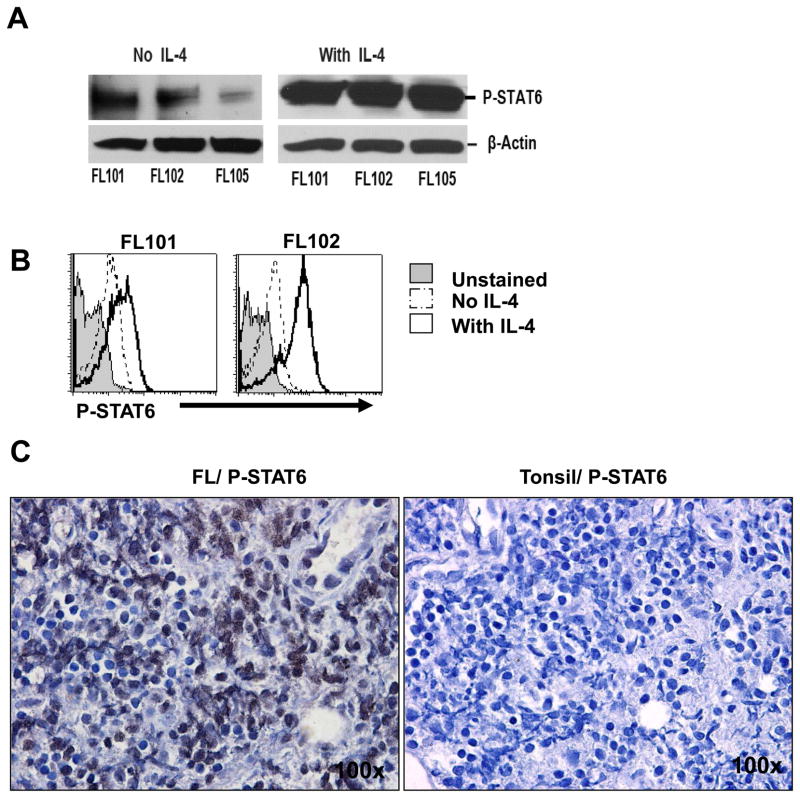
Binding of IL-4 to its receptor in hematopoietic cells activates the tyrosine kinases Jak1 and Jak2; this process results in recruitment and phosphorylation of STAT6, leading to STAT6’s dimerization and translocation to the nucleus where it activates STAT6-responsive genes.40,41 Consistent with the fact that IL-4 was produced in the tumor microenvironment in FL, we observed that phospho-STAT6 was present at low levels in tumor cells from primary FL samples but was substantially enhanced by IL-4 (Figures 2A and 2B). Furthermore, using immunohistochemical staining we found that phospho-STAT6 was increased in primary FL biopsy samples compared with tonsil controls (Figure 2C). Collectively, these results suggested that the increase in IL-4 induced enhanced phosphorylation of STAT6 in FL tumors compared with tonsils.
Figure 2. IL-4 induces phosphorylation of STAT6 in FL tumor B cells.
(A and B) Tumor cells isolated from FL biopsy samples were incubated in the absence or presence of IL-4 (20 ng/ml) for 15 min, and expression of phospho-STAT6 (P-STAT6) was determined by Western blot analysis (A) or phosflow (B). Data shown are representative of three independent experiments (A) and eight FL patient samples tested (B). (C) Immunohistochemical staining for phospho-STAT6 (dark brown) was performed on formalin-fixed, paraffin-embedded FL (left) and tonsil (right) tissues. Images shown are the original magnification × 100.
CCL17 and CCL22 are increased in B-cell lymphomas
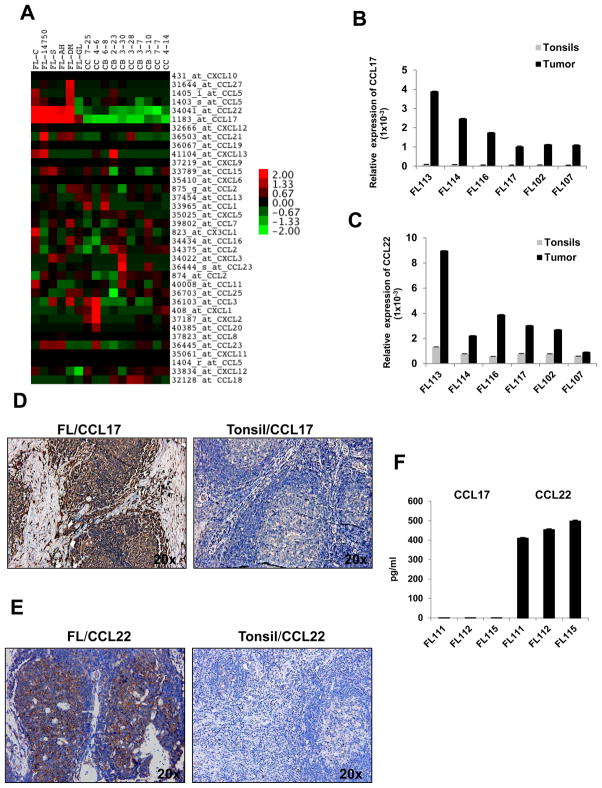
Since CCL17 and CCL22 are STAT6-responsive genes,41 we used publicly available cDNA microarray data (available from http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE2350 and from https://www.oncomine.org/resource/login.html),34 to determine whether these genes are hyperexpressed in B-cell lymphomas relative to other chemokines. We compared the expression patterns of 32 chemokine mRNAs in FL tumors and germinal center B cells (both centrocytes and centroblasts) from tonsil controls and observed that CCL17 and CCL22 were hyperexpressed in 5 of 6 FL tumor B cell specimens (Figure 3A and Supplementary Figure 2A). CCL17 and/or CCL22 were also hyperexpressed in Hodgkin lymphoma, Burkitt’s lymphoma, chronic lymphocytic leukemia, diffuse large B-cell lymphoma, and mantle cell lymphoma (Supplementary Figures 2B and 2C). The hyperexpression of CCL17 and CCL22 in FL tumor cells was confirmed at the mRNA level by real-time PCR (Figures 3B and 3C) and at the protein level by immunohistochemical analysis (Figures 3D and 3E). However, in ex vivo cultures, CCL22 but not CCL17 was spontaneously produced, which indicates that the secretion of CCL17 by FL tumor cells may be dependent on paracrine signals from the tumor microenvironment (Figure 3F).
Figure 3. CCL17 and CCL22 are increased in FL.
(A) Expression of relative chemokine mRNA levels in FL tumors (n = 6) and germinal center B cells (centrocytes, CC and centroblasts, CB) from reactive tonsils (n = 5 each) that were purified by depleting T cells, NK cells, and monocytes/macrophages using immunomagnetic beads were determined using publicly available cDNA microarray data (available from http://icg.cpmc.columbia.edu/Web_Data/JCI/2003_JCI.htm) and shown as a heat map. Log2-transformed data are median-centered for each gene, and variation from the median is indicated by the color bar. (B and C) Real-time PCR was performed to assess the expression of CCL17 (B) and CCL22 (C) mRNA in purified FL tumors (n = 6; 95.7–97.9% pure) and reactive tonsil B cells (n = 6; 98.7–99.6% pure). Expression of the chemokine genes relative to GAPDH in each sample is shown. (D and E) Immunohistochemical staining for CCL17 (D) and CCL22 (E) was performed on formalin-fixed paraffin-embedded FL (left) and tonsil (right) tissues. Images shown are the original magnification × 20 and are representative of data from three different FL tumors. (F) Tumor cells isolated from three FL biopsy samples were incubated for 48 h, and spontaneous production of CCL17 and CCL22 was assessed in the culture supernatants by ELISA.
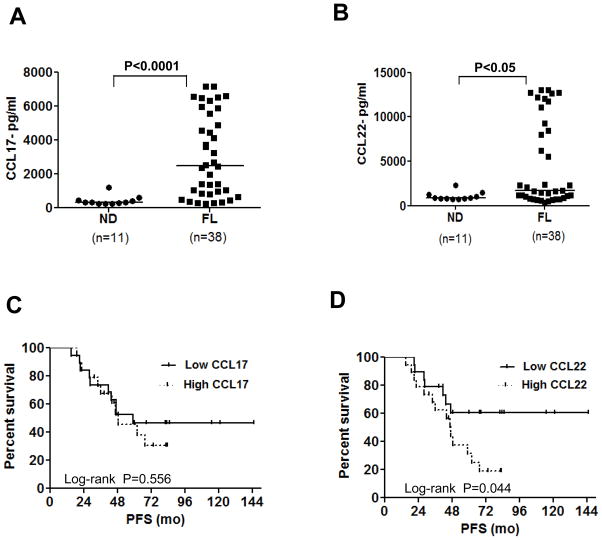
CCL17 and CCL22 were also significantly increased in the serum of FL patients at diagnosis compared with normal donors (Figures 4A and 4B; Supplementary Table 1) and interestingly, high levels of serum CCL22 but not CCL17 at diagnosis correlated with poor PFS compared to patients with low levels of CCL22 (median PFS of 47 mo vs not reached, log-rank p = 0.044) (Figures 4C and 4D). With the adjustment of FLIPI risk score (low-intermediate vs high) in multivariate Cox proportional hazard model, CCL22 remained significant. Patients with higher CCL22 had larger hazard of progression than those with lower CCL22 (Hazard Ratio = 1.38 (95% CI: 1.013 to 1.887) per unit increase of CCL22 on a log base 2 scale; p = 0.041). Correlation with overall survival could not be performed, as there were only two deaths in the entire cohort of 38 patients. Neither the CCL17 nor CCL22 levels were significantly different (p > 0.05) between the FLIPI risk groups (low-intermediate vs high) suggesting that the serum chemokine levels were not just markers of tumor burden.
Figure 4. CCL17 and CCL22 are increased in the serum of FL patients and CCL22 is predictive of inferior progression-free survival.
The levels of CCL17 (A) and CCL22 (B) were determined by ELISA in the serum of FL patients at diagnosis (n = 38) and in normal donors (ND, n = 11). The horizontal bars represent the mean values for each group. Statistical significance was determined by Wilcoxon rank-sum test. (C and D) Kaplan-Meier survival curves for progression free survival (PFS) in FL patients with high (> median, n = 19) and low (≤ median, n = 19) serum levels of CCL17 and CCL22 at diagnosis and treated uniformly with prednisone, doxorubicin, cyclophosphamide, and etoposide chemotherapy followed by autologous tumor-derived idiotype vaccination are shown. P value was calculated by log-rank test.
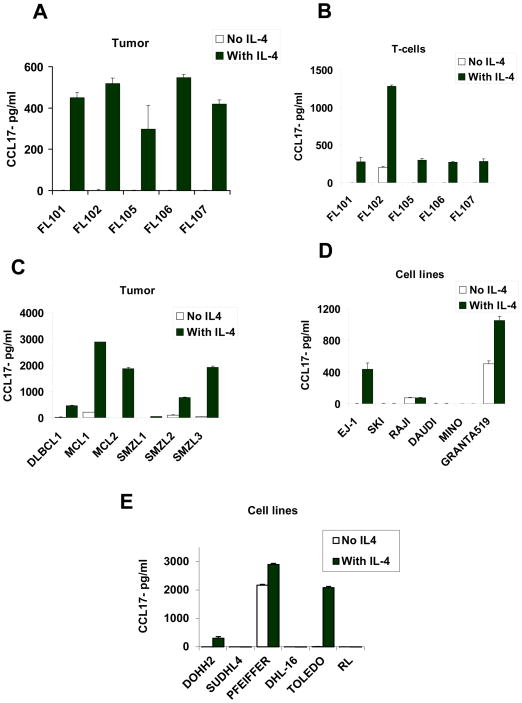
IL-4 induces production of CCL17 by tumor cells from FL and other B-cell lymphomas
Since IL-4 was previously shown to induce CCL17 in myeloid cells via STAT6,42 we determined whether IL-4 induced production of CCL17 by FL tumor cells. We cultured purified tumor B cells in the presence or absence of IL-4 and assessed the production of CCL17 in the culture supernatants. Since IL-4Rα was also expressed on a subset of intratumoral T cells (Supplementary Figure 1D), we also tested the effect of IL-4 on T cells. We observed that IL-4 induced significant production of CCL17 by both tumor cells (Figure 5A) and intratumoral T cells (Figure 5B) in FL. Of interest, IL-4 also induced significant production of CCL17 in primary tumor cells derived from diffuse large B-cell lymphoma, mantle cell lymphoma, and splenic marginal-zone lymphoma (Figure 5C) as well as a transformed FL cell line (DOHH-2), diffuse large B-cell lymphoma cell lines (EJ-1, PFEIFFER, and TOLEDO) and a mantle cell lymphoma cell line (Granta519) (Figure 5D and 5E). However, IL-4 had minimal effect on the production of CCL22 in FL (Supplementary Figures 3A and 3B) and in other B-cell lymphomas and cell lines (Supplementary Figures 3C–E). Together, these results indicated that CCL17 was induced by IL-4 in FL and other B-cell lymphomas.
Figure 5. IL-4 induced production of CCL17 by lymphoma tumor cells.
(A–E) Tumor cells (A and C) and intratumoral T cells (B) isolated from lymphoma biopsy samples (A–C) or lymphoma cell lines (D and E) were incubated in triplicate in the presence or absence of IL-4 (20 ng/ml). After 48 h, CCL17 production was assessed in the culture supernatants by ELISA. FL, follicular lymphoma; DLBCL, diffuse large B-cell lymphoma; MCL, mantle cell lymphoma; SMZL, splenic marginal-zone lymphoma; EJ-1, SKI, SUDHL4, PFEIFFER, and TOLEDO, DLBCL cell lines; RL, DHL-16, and DOHH2, transformed FL cell lines; Raji and Daudi, Burkitt’s lymphoma cell lines; and Mino and Granta519, MCL cell lines.
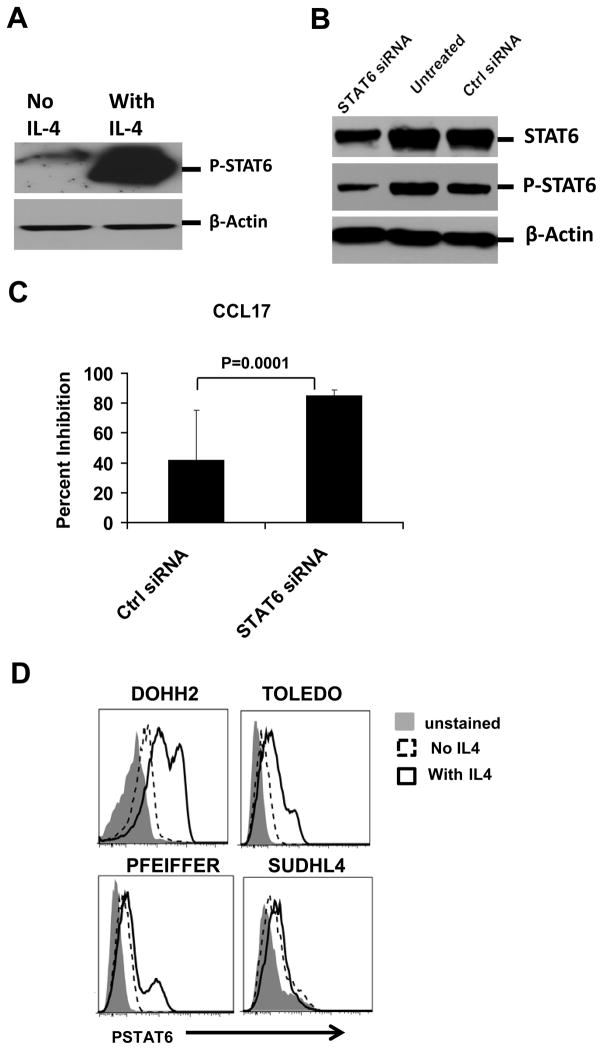
Induction of CCL17 by IL-4 in tumor B cells is STAT6 dependent
To determine whether the induction of CCL17 by IL-4 is STAT6 dependent in B-cell lymphomas, we assessed the effect of STAT6 knockdown on CCL17 production in Granta519 cells. We found that IL-4 enhanced the phosphorylation of STAT6 in the mantle cell lymphoma cell line Granta519 (Figure 6A). Using STAT6-specific small interfering RNA (siRNA) but not control siRNA, we were able to substantially knock down STAT6 and phospho-STAT6 in Granta519 (Figure 6B). After we cultured these transfected cells in the presence of IL-4, we observed that CCL17 production was markedly inhibited in cells transfected with STAT6-specific siRNA compared to control siRNA (Figure 6C). Induction of CCL17 by IL-4 in diffuse large B-cell lines (DOHH2, TOLEDO, and PFEIFFER) was also associated with phosphorylation of STAT6 (Figure 6D). In contrast, the SUDHL4 cell line did not produce CCL17 in response to IL-4 (Figure 5E) and had minimal induction of phospho-STAT6 (Figure 6D). Together, these results indicated that IL-4 induced CCL17 in B-cell lymphomas by activating phospho-STAT6.
Figure 6. Induction of CCL17 by IL-4 in tumor B cells was STAT6 dependent.
(A and D) Granta519 cells (A) and other lymphoma cell lines (D) were incubated in the absence or presence of IL-4 (20 ng/ml) for 15 min, and expression of phospho-STAT6 (P-STAT6) was determined by Western blot analysis (A) or phosflow (D). (B) Granta519 cells were transfected with STAT6-specific siRNA or Universal Stealth siRNA negative control; after 24 h, levels of STAT6 and phospho-STAT6 were determined by Western blot analysis in untreated and transfected cells. (C) Granta519 cells were transfected as above in triplicate and incubated with IL-4 for an additional 24 h, and CCL17 production was assessed in the supernatants by ELISA. Percent inhibition of CCL17 production as compared to untransfected cells is shown. Statistical significance was determined by two-sided paired Student’s t-test.
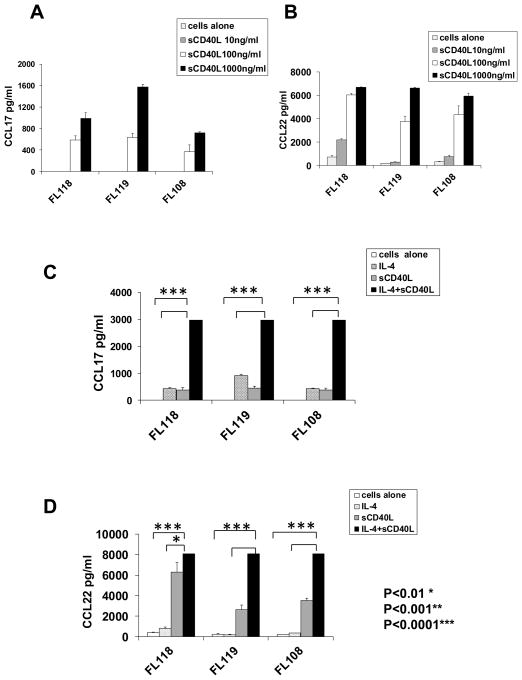
CD40L induces production of both CCL17 and CCL22 from FL tumor cells and has synergistic effects with IL-4
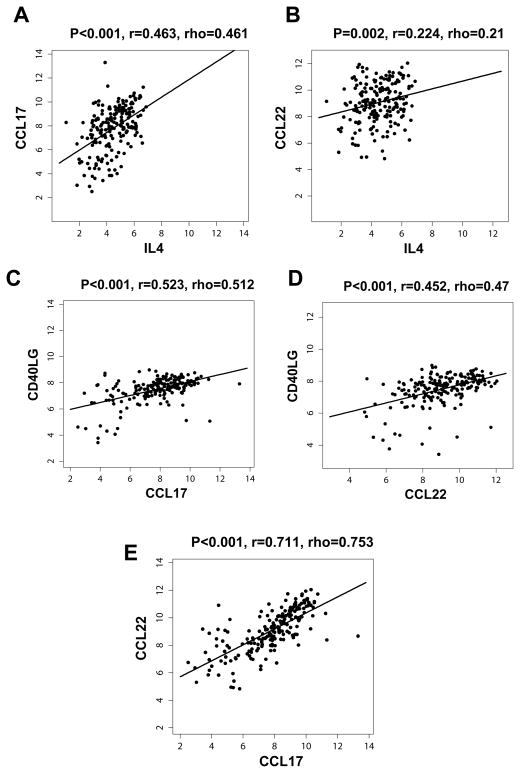
Since CD40L can stimulate production of CCL17 and CCL22 from B cells43,44 and is hyperexpressed on the IL-4-producing TFH (Figure 1C), we determined whether it might play a role in the induction of these chemokines in FL. We found that CD40L induced significant production of CCL17 and CCL22 by tumor cells (Figure 7A and 7B) in FL. Interestingly, there was marked synergism between CD40L and IL-4 in the production of both chemokines (Figure 7C and 7D). Consistent with our in vitro results (Figures 5A, 7, and Supplementary Figure 3A), we found a moderate degree of correlation between mRNA transcripts of IL-4 and CCL17 but not CCL22 in gene expression microarray studies of FL tumors (Figures 8A and 8B). In contrast, CD40L mRNA transcripts correlated with both CCL17 and CCL22 (Figures 8C and 8D). This suggests that the expression of IL-4 and CD40L may be differentially regulated in TFH and/or expression of CD40L by non-TFH, although at lower levels (Figure 1A) may contribute to the expression of these chemokines. We also found a strong correlation between CCL17 and CCL22 suggesting that these genes may be co-regulated (Figure 8E).
Figure 7. CD40L induced production of CCL17 and CCL22 and has synergistic effects with IL-4.
Tumor cells isolated from three FL biopsy samples were incubated in triplicate (1×106 tumor cells/well) in the presence or absence of varying concentrations of recombinant human soluble CD40L (sCD40L) alone (A and B) or at 100 ng/ml of sCD40L (C and D), IL-4 alone (20 ng/ml), or both (C and D). After 48 h, CCL17 and CCL22 production was assessed in the culture supernatants by ELISA. All P values were calculated using paired Student’s t test.
Figure 8. IL4 mRNA transcripts correlated with CCL17 and CD40L correlated with both CCL17 and CCL22.
The correlation between mRNA transcripts of IL-4 (A and B) and CD40L (C and D) vs. CCL17 (A and C) and CCL22 (B and D) was determined in the microarray dataset of 191 FL tumors published previously by Dave et al.15 (E) Correlation between CCL17 and CCL22 mRNA transcripts for the same dataset. Pearson’s correlation coefficient (r) and Spearman’s rank correlation coefficient (rho) are shown.
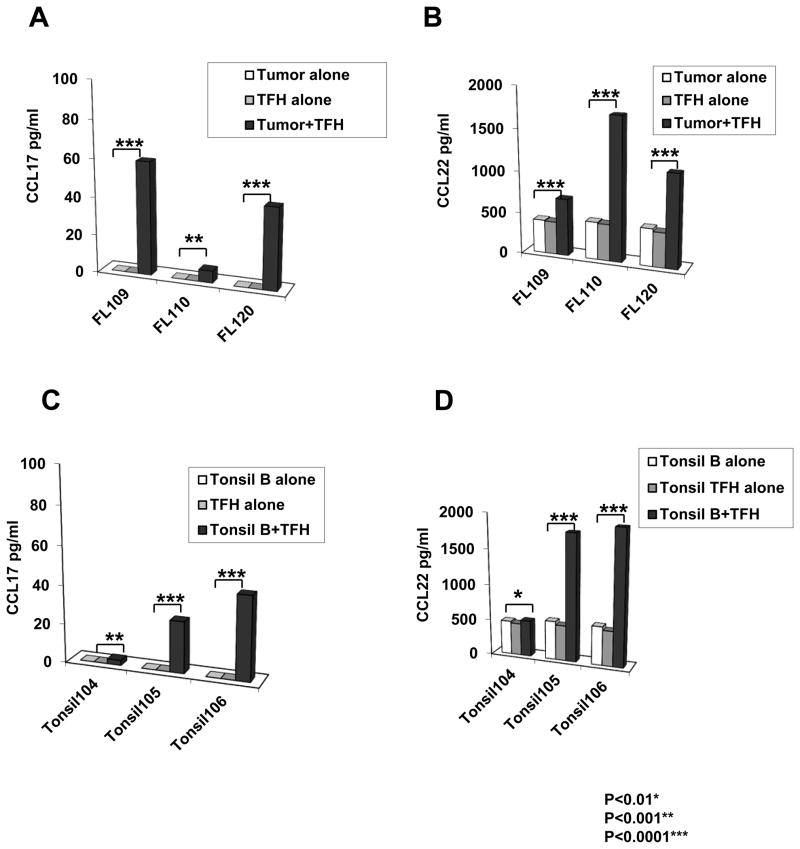
In agreement with the above results, co-culture of FL tumor cells with autologous intratumoral TFH showed production of both CCL17 and CCL22 into the culture supernatants (Figures 9A and 9B). We also observed similar production of CCL17 and CCL22 when B cells from tonsils were co-cultured with autologous TFH (Figures 9C and 9D). The lower amount of chemokine production seen in the tumor-TFH co-cultures (Figures 9A and 9B) compared with recombinant proteins (Figure 7) is probably due to the lower number of tumor cells used per well (Figures 9A and 9B) relative to the experiment with recombinant IL-4 and CD40L (Figure 7), and due to the likely suboptimal function of cryopreserved TFH compared to freshly isolated TFH or TFH in vivo.45 Nevertheless, these results suggested that TFH express IL-4 and CD40L, presumably after cognate recognition of an MHC-associated peptide presented on the surface of tumor B cells, and induce production of CCL17 and CCL22 by FL tumor cells.
Figure 9. CCL17 and CCL22 are induced in autologous tumor-TFH co-cultures.
TFH (CD3+CD4+CD25−PD1hiCXCR5hi) were sorted by FACS from three cryopreserved FL biopsy samples and three tonsil samples. They were cultured in the absence or presence of autologous tumor cells or tonsil B cells (0.35×106 tumor cells/well). After 96 h, CCL17 (A and C) and CCL22 (B and D) production was assessed in the culture supernatants by ELISA. All P values were calculated using paired Student’s t test.
CCL17 and CCL22 induce preferential migration of Tregs and IL-4-producing T cells
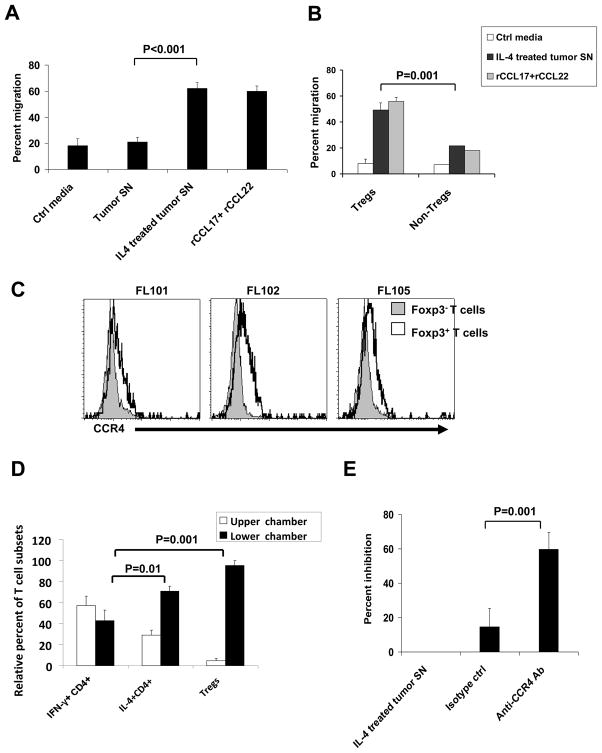
Since CCR4 is the only known chemokine receptor for CCL17 and CCL22 and is overexpressed on Tregs46 and TH2 cells,47 we determined whether IL-4–treated tumor cells induced preferential migration of these cells. First, we cultured FL tumor cells in the presence or absence of IL-4 for 48 h and tested the ability of the culture supernatants to induce chemotaxis of T cell subsets. We found that culture supernatants from IL-4–treated tumor cells and recombinant CCL17 and CCL22 chemokines induced significant migration of Tregs in a chemotaxis assay. In contrast, the migration of Tregs in response to culture supernatants obtained from tumor cells alone was not significantly different from that observed with control media (Figure 10A). Treg migration was also observed when recombinant CCL17 and CCL22 chemokines were used alone and the migration was not significantly different between chemokine concentrations of 2, 20, and 200 ng/ml for CCL17 and CCL22 (data not shown). More importantly, culture supernatants from IL-4–treated tumor cells induced preferential migration of Tregs compared with non-Tregs (Figure 10B), and this preferential migration was correlated with a higher expression of CCR4 in Tregs than in non-Tregs (Figure 10C). This result may offer one mechanism for increased number of Foxp3+ Tregs in FL tumors compared with tonsils.23–26
Figure 10. Preferential migration of Tregs and IL-4-producing CD4+ T cells.
(A, B, and E) Tumor cells isolated from FL biopsy samples were incubated in the presence or absence of IL-4 (20 ng/ml) for 48 h, and culture supernatants (SN) were harvested for use in chemotaxis assays. (A and B) Tregs and non-Tregs were isolated to >90% purity from PBMC of normal donors by MACS and used in the chemotaxis assay in the presence or absence of control (Ctrl) media, tumor supernatants, and recombinant CCL17 and CCL22 (200 ng/ml each) as described in the Materials and Methods. Representative data on the percentage of migration of Tregs and/or non-Tregs from four independent experiments are shown. (C) Expression of CCR4 was determined by flow cytometric analysis on CD4+Foxp3− (gray closed histograms) and CD4+Foxp3+ (black open histograms) T cells from three FL biopsy samples. Representative data from three of eight FL samples tested are shown. (D) Intratumoral T cells were isolated to >90% purity from FL102 tumor biopsy sample by MACS using Pan T Cell Isolation Kit and used in a chemotaxis assay in triplicate in the presence of control media (not shown) or recombinant CCL17 and CCL22 (200 ng/ml each). After 2 hours, cells were harvested separately from upper and lower chambers and intracellular cytokine assay and Foxp3 staining were performed as described in Materials and Methods. The relative percentage of CD4+IFN-γ+IL-4−, CD4+IFN-γ−IL-4+, and Treg (CD4+CD25+CD127loFoxp3+) T-cell subsets in the upper and lower chambers was determined using the following formula: (Percent T-cell subset in upper or lower chamber × 100)/(Total percent of T-cell subset in upper and lower chambers). (E) Chemotaxis assay was performed with isolated Tregs as in panel A. Percentage of inhibition of migration of Tregs by isotype control antibody and CCR4 blocking antibody was determined and shown as compared with IL-4–treated tumor SN. Data shown are representative of three independent experiments. All P values were calculated using paired Student’s t test.
To determine the relative migration of Tregs, IFN-γ-producing (TH1), and IL-4-producing (TH2 or TFH) T-cell subsets, we tested intratumoral T cells of FL in a chemotaxis assay. We observed that Tregs and IL-4–producing T cells migrated significantly more in response to CCL17 and CCL22 chemokines than IFN-γ–producing T cells (Figure 10D). The migration of Tregs in response to tumor supernatants was significantly inhibited by anti-CCR4 blocking antibody but not by isotype control antibody (Figure 10E). Taken together, these results suggested that Tregs and IL-4–producing T cells might be recruited preferentially into the FL tumor microenvironment.
DISCUSSION
Our results provide novel insights into the cross talk between TFH, tumor cells, and Tregs in FL. Prior reports suggested that TFH might play a role in promoting the survival and growth of FL tumor cells through IL-4 and CD40L.10,11,42,45 Our data suggest that IL-4 and CD40L expressed by intratumoral TFH may have additional effects. We show that they induce expression of CCL17 and CCL22 through paracrine effects on tumor cells in FL. Although we observed similar induction of chemokines by tonsils samples in vitro (Figures 9A–D), CCL17 and CCL22 were increased in FL tissues. This may be explained by the increased numbers of IL-4-producing T cells in FL (Figure 1F). More importantly, we observed that these chemokines induce preferential recruitment of Tregs and IL-4–producing T cells rather than IFN-γ–producing T cells into the tumor microenvironment, thus creating a positive feedback cycle that may lead to further production of these chemokines and an immunosuppressive tumor microenvironment that facilitates immune escape and promotes the survival and growth of the tumor. This vicious cycle appears to be primarily driven by the increased number of IL-4–producing T cells in the tumor microenvironment. Although additional studies with larger number of patients are needed, the potential role of CCL22 in promoting tumor growth and escape is supported by our observation that FL patients with high serum level of this chemokine at diagnosis have inferior PFS (Figure 4D). The lack of correlation between CCL17 and PFS suggests that CCL22 might play a more dominant role in the tumor microenvironment. Although we found increased levels of CCL17 and CCL22 in the serum of FL patients, their concentrations are likely to be higher in the tumor where they are produced compared with peripheral blood. Therefore, it is very probable that the chemokine gradient is in favor of active recruitment of Tregs and IL-4–producing T cells from the peripheral blood into the tumor microenvironment. The recruited Tregs may in turn suppress the function of tumor-specific effector T cells and other immune cells such as natural killer cells and dendritic cells, and promote immune evasion by the tumor.
The increase in IL-4-producing T cells (Figure 1F), and the basal phosphorylation of STAT6 in tumor cells observed in our studies (Figures 2A–C) are consistent with recent reports.7,42 IL-4+IFN-γ− CD4+ T cells have been classically described as TH2 cells, but recent reports suggest that IL-4 in secondary lymphoid organs is almost entirely produced by TFH cells; classical IL-4–producing TH2 cells migrate from the lymph node after activation and mediate effector function in the peripheral tissues.48 In agreement with this and other reports,42,45 we observed that IL-4-producing T cells were TFH in FL (Figures 1A–C). Recent reports also suggest that CXCR5+CD4+ T cells in the peripheral blood could be precursors of germinal center TFH cells seen in secondary lymphoid organs.49 More interestingly, circulating CXCR5+CD4+ T cells appear to be comprised of TH1-, TH2-, and TH17-like subsets that produce IFN-γ, IL-4, and IL-17, respectively.50 In FL patients, IL-4–producing CD4+ T cells recruited from the peripheral blood into the FL tumor microenvironment may be predominantly TH2-like CXCR5+CD4+ TFH precursor cells that may transform into IL-4–producing TFH cells after migration into the lymph node. IL-4 may also be produced by Vδ1 T lymphocytes in the tumor microenvironment; IL-4–producing Vδ1 T lymphocytes were shown to be increased in number in the peripheral blood in patients with FL, mantle cell lymphoma, and marginal-zone lymphoma when compared with healthy donors, and Vδ1 T lymphocytes were detectable in these tumors.51 Our results and these reports indicate that increased IL-4 in the FL tumor microenvironment is due to TFH cells, with a possible additional contribution from Vδ1 T cells. In Hodgkin lymphoma, in contrast, IL-13 is the cytokine predominantly responsible for STAT6 activation and CCL17 production.38,52 In our studies, IL-13 was not hyperexpressed in FL tissues compared with reactive tonsils and had minimal or no effect on the production of CCL17 and CCL22 (data not shown).
Our results suggest that chemokine-driven active recruitment of Tregs is one reason for their increased number in the FL tumor microenvironment. However, Tregs can be induced from conventional T cells by tumor cells in B-cell lymphomas.26,29 Therefore, development of therapeutic approaches that inhibit both the recruitment and induction of Tregs may be necessary to decrease the number of Tregs in the tumor microenvironment. In light of the differential expression of CCR4 on Tregs46 and non-Tregs (Figure 10C) and the knowledge that CCR4 is the only known receptor for CCL17 and CCL22, our data support the development of blocking antibodies or small-molecule antagonists against CCR453,54 that can be used in combination with other immunotherapeutic strategies in therapies for FL and other B-cell malignancies. Such agents are likely to inhibit the recruitment of immunosuppressive Tregs as well as tumor-promoting, IL-4–producing T cells and shift the balance in favor of antitumor immunity mediated by TH1 and TC1 cells in the tumor microenvironment, thereby enhancing the efficacy of therapeutic vaccines16,17 and other immunomodulatory drugs.
Although intratumoral Tregs have been shown to be immunosuppressive by in vitro studies, there is uncertainty regarding the benefit of Treg reduction therapy because increased numbers of Foxp3+ cells in FL tumors have been associated with improved survival in FL.24,55 However, Farinha et al. observed that only the architectural pattern of Foxp3+ cells within FL tumors, rather than their number, influenced outcome; specifically, an intrafollicular localization pattern of Tregs correlated with decreased survival.25 This suggests the presence of distinct Treg subsets within the FL tumor microenvironment, and recent studies in basic immunology have shown that Tregs may differentiate into distinct subsets, based on various cues from their microenvironment, to regulate distinct effector cells such as TH1, TH2, TH17, and TFH.56–60 In FL, Tregs are present in both follicular and interfollicular regions, potentially representing distinct subsets.25 Tregs that localize to intrafollicular regions (follicular regulatory T cells or TFR) in normal lymphoid tissues express high levels of CXCR5 and Bcl6 similar to TFH and were shown to be responsible for regulating germinal center reactions.59,60 Indeed, TFR were recently demonstrated to be present in FL tissues.45 Whether other Treg subsets exist in FL tumors and whether CCL17 and CCL22 expressed by tumor cells have differential effects on recruitment of different Treg subsets needs to be studied in the future.
In summary, our results provide novel insights into the interactions between TFH, tumor cells, and Tregs in FL. We showed that intratumoral TFH induce production of CCL17 and CCL22 by tumor cells via two distinct mechanisms involving IL-4 and CD40L. Overexpression of CCL17 and CCL22 in the tumor microenvironment induces preferential recruitment of Tregs and IL-4–producing T cells that in turn may stimulate more chemokine production, thus creating a vicious cycle and an immunosuppressive tumor microenvironment that promotes immune evasion and tumor survival and growth. Therapeutic strategies that interrupt this cycle may lead to inhibition of FL tumor growth by depleting survival and growth factors provided by the IL-4 and CD40L expressed by TFH. Furthermore, they may alter the tumor microenvironment by skewing the balance from immunosuppression to antitumor immunity; thereby enhancing the efficacy of both naturally induced and therapeutically induced antitumor immune responses and leading to improved clinical outcomes in patients with FL and possibly other B-cell malignancies.
Supplementary Material
Acknowledgments
This work was supported by the Doris Duke Charitable Foundation (S.S.N.), R01 CA155143 (S.S.N. and R.E.D.), Lymphoma SPORE Developmental Research Program (S.S.N. and R.E.D.), The University of Texas MD Anderson Cancer Center Institutional Research Grant (S.S.N.), and Leukemia and Lymphoma Society Specialized Center of Research grant 7262–08 (C.D.). The University of Texas MD Anderson Cancer Center Lymphoma Tissue Bank is supported by the National Institutes of Health Lymphoma SPORE grant P50CA136411 and Fredrick B. Hagemeister Research Fund. The Flow Cytometry Core Facility is supported by The University of Texas MD Anderson Cancer Center Support Grant from National Institutes of Health CA16672.
We thank Kristi M. Speights for editorial assistance.
ABBREVIATIONS
- BCL2
B-cell lymphoma 2
- FL
follicular lymphoma
- PFS
progression-free survival
- TFH
follicular helper T cells
- Tregs
regulatory T cells
- TFR
follicular regulatory cells
Footnotes
AUTHOR CONTRIBITIONS
SR and SSN designed the study, analyzed the data, and wrote the manuscript. SR, FC, HJP, DN, SK, RS, DD, TC, and CD designed and/or performed the experiments. NF and AL provided patient samples. FV provided cell lines. HYL and VB performed statistical analyses. MZ and RED analyzed the gene expression profiling data. All authors reviewed and approved the manuscript.
DISCLOSURE OF CONFLICTS OF INTERESTS
The authors declare that there are no competing financial interests.
References
- 1.A clinical evaluation of the International Lymphoma Study Group classification of non-Hodgkin’s lymphoma. The Non-Hodgkin’s Lymphoma Classification Project. Blood. 1997;89:3909–18. [PubMed] [Google Scholar]
- 2.Tsujimoto Y, Cossman J, Jaffe E, et al. Involvement of the bcl-2 gene in human follicular lymphoma. Science. 1985;228:1440–3. doi: 10.1126/science.3874430. [DOI] [PubMed] [Google Scholar]
- 3.Dolken G, Dolken L, Hirt C, et al. Age-dependent prevalence and frequency of circulating t(14;18)-positive cells in the peripheral blood of healthy individuals. J Natl Cancer Inst Monogr. 2008:44–7. doi: 10.1093/jncimonographs/lgn005. [DOI] [PubMed] [Google Scholar]
- 4.Dolken G, Illerhaus G, Hirt C, et al. BCL-2/JH rearrangements in circulating B cells of healthy blood donors and patients with nonmalignant diseases. J Clin Oncol. 1996;14:1333–44. doi: 10.1200/JCO.1996.14.4.1333. [DOI] [PubMed] [Google Scholar]
- 5.McDonnell TJ, Korsmeyer SJ. Progression from lymphoid hyperplasia to high-grade malignant lymphoma in mice transgenic for the t(14; 18) Nature. 1991;349:254–6. doi: 10.1038/349254a0. [DOI] [PubMed] [Google Scholar]
- 6.Kuppers R. Mechanisms of B-cell lymphoma pathogenesis. Nat Rev Cancer. 2005;5:251–62. doi: 10.1038/nrc1589. [DOI] [PubMed] [Google Scholar]
- 7.Calvo KR, Dabir B, Kovach A, et al. IL-4 protein expression and basal activation of Erk in vivo in follicular lymphoma. Blood. 2008;112:3818–26. doi: 10.1182/blood-2008-02-138933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pangault C, Ame-Thomas P, Ruminy P, et al. Follicular lymphoma cell niche: identification of a preeminent IL-4-dependent T(FH)-B cell axis. Leukemia. 2010;24:2080–9. doi: 10.1038/leu.2010.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ame-Thomas P, Le Priol J, Yssel H, et al. Characterization of intratumoral follicular helper T cells in follicular lymphoma: role in the survival of malignant B cells. Leukemia. 2012;26:1053–63. doi: 10.1038/leu.2011.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buske C, Twiling A, Gogowski G, et al. In vitro activation of low-grade non-Hodgkin’s lymphoma by murine fibroblasts, IL-4, anti-CD40 antibodies and the soluble CD40 ligand. Leukemia. 1997;11:1862–7. doi: 10.1038/sj.leu.2400822. [DOI] [PubMed] [Google Scholar]
- 11.Schmitter D, Koss M, Niederer E, et al. T-cell derived cytokines co-stimulate proliferation of CD40-activated germinal centre as well as follicular lymphoma cells. Hematol Oncol. 1997;15:197–207. doi: 10.1002/(sici)1099-1069(199711)15:4<197::aid-hon614>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 12.Horning SJ, Rosenberg SA. The natural history of initially untreated low-grade non-Hodgkin’s lymphomas. N Engl J Med. 1984;311:1471–5. doi: 10.1056/NEJM198412063112303. [DOI] [PubMed] [Google Scholar]
- 13.Schultze JL, Seamon MJ, Michalak S, et al. Autologous tumor infiltrating T cells cytotoxic for follicular lymphoma cells can be expanded in vitro. Blood. 1997;89:3806–16. [PubMed] [Google Scholar]
- 14.Lee ST, Liu S, Radvanyi L, et al. A novel strategy for rapid and efficient isolation of human tumor-specific CD4(+) and CD8(+) T-cell clones. J Immunol Methods. 2008;331:13–26. doi: 10.1016/j.jim.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dave SS, Wright G, Tan B, et al. Prediction of survival in follicular lymphoma based on molecular features of tumor-infiltrating immune cells. N Engl J Med. 2004;351:2159–69. doi: 10.1056/NEJMoa041869. [DOI] [PubMed] [Google Scholar]
- 16.Bendandi M, Gocke CD, Kobrin CB, et al. Complete molecular remissions induced by patient-specific vaccination plus granulocyte-monocyte colony-stimulating factor against lymphoma. Nat Med. 1999;5:1171–7. doi: 10.1038/13928. [DOI] [PubMed] [Google Scholar]
- 17.Neelapu SS, Baskar S, Gause BL, et al. Human autologous tumor-specific T-cell responses induced by liposomal delivery of a lymphoma antigen. Clin Cancer Res. 2004;10:8309–17. doi: 10.1158/1078-0432.CCR-04-1071. [DOI] [PubMed] [Google Scholar]
- 18.McLaughlin P, Grillo-Lopez AJ, Link BK, et al. Rituximab chimeric anti-CD20 monoclonal antibody therapy for relapsed indolent lymphoma: half of patients respond to a four-dose treatment program. J Clin Oncol. 1998;16:2825–33. doi: 10.1200/JCO.1998.16.8.2825. [DOI] [PubMed] [Google Scholar]
- 19.Witzig TE, Vukov AM, Habermann TM, et al. Rituximab therapy for patients with newly diagnosed, advanced-stage, follicular grade I non-Hodgkin’s lymphoma: a phase II trial in the North Central Cancer Treatment Group. J Clin Oncol. 2005;23:1103–8. doi: 10.1200/JCO.2005.12.052. [DOI] [PubMed] [Google Scholar]
- 20.Khouri IF, McLaughlin P, Saliba RM, et al. Eight-year experience with allogeneic stem cell transplantation for relapsed follicular lymphoma after nonmyeloablative conditioning with fludarabine, cyclophosphamide, and rituximab. Blood. 2008;111:5530–6. doi: 10.1182/blood-2008-01-136242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zou W. Immunosuppressive networks in the tumour environment and their therapeutic relevance. Nat Rev Cancer. 2005;5:263–74. doi: 10.1038/nrc1586. [DOI] [PubMed] [Google Scholar]
- 22.Zou W. Regulatory T cells, tumour immunity and immunotherapy. Nat Rev Immunol. 2006;6:295–307. doi: 10.1038/nri1806. [DOI] [PubMed] [Google Scholar]
- 23.Yang ZZ, Novak AJ, Stenson MJ, et al. Intratumoral CD4+CD25+ regulatory T-cell-mediated suppression of infiltrating CD4+ T cells in B-cell non-Hodgkin lymphoma. Blood. 2006;107:3639–46. doi: 10.1182/blood-2005-08-3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carreras J, Lopez-Guillermo A, Fox BC, et al. High numbers of tumor-infiltrating FOXP3-positive regulatory T cells are associated with improved overall survival in follicular lymphoma. Blood. 2006;108:2957–64. doi: 10.1182/blood-2006-04-018218. [DOI] [PubMed] [Google Scholar]
- 25.Farinha P, Al-Tourah A, Gill K, et al. The architectural pattern of FOXP3-positive T cells in follicular lymphoma is an independent predictor of survival and histologic transformation. Blood. 115:289–95. doi: 10.1182/blood-2009-07-235598. [DOI] [PubMed] [Google Scholar]
- 26.Mittal S, Marshall NA, Duncan L, et al. Local and systemic induction of CD4+CD25+ regulatory T-cell population by non-Hodgkin lymphoma. Blood. 2008;111:5359–70. doi: 10.1182/blood-2007-08-105395. [DOI] [PubMed] [Google Scholar]
- 27.Yang ZZ, Novak AJ, Ziesmer SC, et al. Attenuation of CD8(+) T-cell function by CD4(+)CD25(+) regulatory T cells in B-cell non-Hodgkin’s lymphoma. Cancer Res. 2006;66:10145–52. doi: 10.1158/0008-5472.CAN-06-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hilchey SP, De A, Rimsza LM, et al. Follicular lymphoma intratumoral CD4+CD25+GITR+ regulatory T cells potently suppress CD3/CD28-costimulated autologous and allogeneic CD8+CD25- and CD4+CD25- T cells. J Immunol. 2007;178:4051–61. doi: 10.4049/jimmunol.178.7.4051. [DOI] [PubMed] [Google Scholar]
- 29.Yang ZZ, Novak AJ, Ziesmer SC, et al. CD70+ non-Hodgkin lymphoma B cells induce Foxp3 expression and regulatory function in intratumoral CD4+CD25 T cells. Blood. 2007;110:2537–44. doi: 10.1182/blood-2007-03-082578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ai WZ, Hou JZ, Zeiser R, et al. Follicular lymphoma B cells induce the conversion of conventional CD4+ T cells to T-regulatory cells. Int J Cancer. 2009;124:239–44. doi: 10.1002/ijc.23881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sallusto F, Schaerli P, Loetscher P, et al. Rapid and coordinated switch in chemokine receptor expression during dendritic cell maturation. Eur J Immunol. 1998;28:2760–9. doi: 10.1002/(SICI)1521-4141(199809)28:09<2760::AID-IMMU2760>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 32.Saeki H, Tamaki K. Thymus and activation regulated chemokine (TARC)/CCL17 and skin diseases. J Dermatol Sci. 2006;43:75–84. doi: 10.1016/j.jdermsci.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 33.Mantovani A, Gray PA, Van Damme J, et al. Macrophage-derived chemokine (MDC) J Leukoc Biol. 2000;68:400–4. [PubMed] [Google Scholar]
- 34.Kuppers R, Klein U, Schwering I, et al. Identification of Hodgkin and Reed-Sternberg cell-specific genes by gene expression profiling. J Clin Invest. 2003;111:529–37. doi: 10.1172/JCI16624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schuster SJ, Neelapu SS, Gause BL, et al. Vaccination with patient-specific tumor-derived antigen in first remission improves disease-free survival in follicular lymphoma. J Clin Oncol. 2011;29:2787–94. doi: 10.1200/JCO.2010.33.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tosolini M, Kirilovsky A, Mlecnik B, et al. Clinical impact of different classes of infiltrating T cytotoxic and helper cells (Th1, th2, treg, th17) in patients with colorectal cancer. Cancer Res. 2011;71:1263–71. doi: 10.1158/0008-5472.CAN-10-2907. [DOI] [PubMed] [Google Scholar]
- 37.Neelapu SS, Baskar S, Kwak LW. Detection of keyhole limpet hemocyanin (KLH)-specific immune responses by intracellular cytokine assay in patients vaccinated with idiotype-KLH vaccine. J Cancer Res Clin Oncol. 2001;127 (Suppl 2):R14–9. doi: 10.1007/BF01470994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buglio D, Georgakis GV, Hanabuchi S, et al. Vorinostat inhibits STAT6-mediated TH2 cytokine and TARC production and induces cell death in Hodgkin lymphoma cell lines. Blood. 2008;112:1424–33. doi: 10.1182/blood-2008-01-133769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Solal-Celigny P, Roy P, Colombat P, et al. Follicular lymphoma international prognostic index. Blood. 2004;104:1258–65. doi: 10.1182/blood-2003-12-4434. [DOI] [PubMed] [Google Scholar]
- 40.Colgan J, Rothman P. Manipulation of signaling to control allergic inflammation. Curr Opin Allergy Clin Immunol. 2007;7:51–6. doi: 10.1097/ACI.0b013e32801297e6. [DOI] [PubMed] [Google Scholar]
- 41.Hebenstreit D, Wirnsberger G, Horejs-Hoeck J, et al. Signaling mechanisms, interaction partners, and target genes of STAT6. Cytokine Growth Factor Rev. 2006;17:173–88. doi: 10.1016/j.cytogfr.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 42.Liddiard K, Welch JS, Lozach J, et al. Interleukin-4 induction of the CC chemokine TARC (CCL17) in murine macrophages is mediated by multiple STAT6 sites in the TARC gene promoter. BMC Mol Biol. 2006;7:45. doi: 10.1186/1471-2199-7-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ghia P, Transidico P, Veiga JP, et al. Chemoattractants MDC and TARC are secreted by malignant B-cell precursors following CD40 ligation and support the migration of leukemia-specific T cells. Blood. 2001;98:533–40. doi: 10.1182/blood.v98.3.533. [DOI] [PubMed] [Google Scholar]
- 44.Lin L, Nonoyama S, Oshiba A, et al. TARC and MDC are produced by CD40 activated human B cells and are elevated in the sera of infantile atopic dermatitis patients. J Med Dent Sci. 2003;50:27–33. [PubMed] [Google Scholar]
- 45.Owen RE, Sinclair E, Emu B, et al. Loss of T cell responses following long-term cryopreservation. J Immunol Methods. 2007;326:93–115. doi: 10.1016/j.jim.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iellem A, Mariani M, Lang R, et al. Unique chemotactic response profile and specific expression of chemokine receptors CCR4 and CCR8 by CD4(+)CD25(+) regulatory T cells. J Exp Med. 2001;194:847–53. doi: 10.1084/jem.194.6.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bonecchi R, Bianchi G, Bordignon PP, et al. Differential expression of chemokine receptors and chemotactic responsiveness of type 1 T helper cells (Th1s) and Th2s. J Exp Med. 1998;187:129–34. doi: 10.1084/jem.187.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reinhardt RL, Liang HE, Locksley RM. Cytokine-secreting follicular T cells shape the antibody repertoire. Nat Immunol. 2009;10:385–93. doi: 10.1038/ni.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu D, Vinuesa CG. The elusive identity of T follicular helper cells. Trends Immunol. 31:377–83. doi: 10.1016/j.it.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 50.Morita R, Schmitt N, Bentebibel SE, et al. Human blood CXCR5(+)CD4(+) T cells are counterparts of T follicular cells and contain specific subsets that differentially support antibody secretion. Immunity. 34:108–21. doi: 10.1016/j.immuni.2010.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Catellani S, Poggi A, Bruzzone A, et al. Expansion of Vdelta1 T lymphocytes producing IL-4 in low-grade non-Hodgkin lymphomas expressing UL-16-binding proteins. Blood. 2007;109:2078–85. doi: 10.1182/blood-2006-06-028985. [DOI] [PubMed] [Google Scholar]
- 52.Skinnider BF, Kapp U, Mak TW. The role of interleukin 13 in classical Hodgkin lymphoma. Leuk Lymphoma. 2002;43:1203–10. doi: 10.1080/10428190290026259. [DOI] [PubMed] [Google Scholar]
- 53.Ishida T, Ueda R. CCR4 as a novel molecular target for immunotherapy of cancer. Cancer Sci. 2006;97:1139–46. doi: 10.1111/j.1349-7006.2006.00307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bayry J, Tchilian EZ, Davies MN, et al. In silico identified CCR4 antagonists target regulatory T cells and exert adjuvant activity in vaccination. Proc Natl Acad Sci U S A. 2008;105:10221–6. doi: 10.1073/pnas.0803453105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alvaro T, Lejeune M, Salvado MT, et al. Immunohistochemical patterns of reactive microenvironment are associated with clinicobiologic behavior in follicular lymphoma patients. J Clin Oncol. 2006;24:5350–7. doi: 10.1200/JCO.2006.06.4766. [DOI] [PubMed] [Google Scholar]
- 56.Koch MA, Tucker-Heard G, Perdue NR, et al. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nature immunology. 2009;10:595–602. doi: 10.1038/ni.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chaudhry A, Rudra D, Treuting P, et al. CD4+ regulatory T cells control TH17 responses in a Stat3-dependent manner. Science. 2009;326:986–91. doi: 10.1126/science.1172702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zheng Y, Chaudhry A, Kas A, et al. Regulatory T-cell suppressor program coopts transcription factor IRF4 to control T(H)2 responses. Nature. 2009;458:351–6. doi: 10.1038/nature07674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chung Y, Tanaka S, Chu F, et al. Follicular regulatory T cells expressing Foxp3 and Bcl-6 suppress germinal center reactions. Nature medicine. 2011;17:983–8. doi: 10.1038/nm.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Linterman MA, Pierson W, Lee SK, et al. Foxp3+ follicular regulatory T cells control the germinal center response. Nature medicine. 2011;17:975–82. doi: 10.1038/nm.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.