Abstract
The design, manufacture and application of safer products and manufacturing processes have been important goals over the last decade and will advance in the future under the umbrella of "Green Chemistry". In this review, we focus on the burgeoning diversity of new engineered nanomaterials (ENMs) and the prescient need for a nanotoxicology paradigm that quickly identifies potentially hazardous nanochemistries. Advances in predictive toxicological modeling in the developing zebrafish offer the most immediate translation to human hazard that is practically achievable with high throughput approaches. Translation in a vertebrate model that is also a low cost alternative to rodents for hazard prediction has been a desirable but elusive testing paradigm. The utility of zebrafish, if applied early in the ENM discovery pipeline, could greatly enhance efforts toward greener and more efficient nanoscience. Early pipeline detection of human and environmental health impacts will quickly inform decisions in the design and production of safer commercial ENMs.
1. Introduction
Recent market data indicate that the global chemical industry produced more than 100,000 chemical products in 2008, sustaining a $2.7 trillion market.1 Sources of chemicals are ever evolving and increasing to meet consumer demands. Along with the deluge of new chemicals has come increased uncertainty about the potential for new chemical hazard. Toxicology simply has not matched the pace of chemical innovation. While new chemistries are generally derivations and expansions of old, toxicological knowledge of the core moieties of new chemistries is hardly a sufficient basis for ranking hazard potential. Seemingly small structural changes can cause what look like disproportionately large toxicological responses, far beyond what one might have predicted from the change. This is simply because intimate knowledge of the chemical's interaction with target biomolecules is inadequate. The more diverse our chemical production becomes, the more biotargets we are likely to inadvertently hit. And for those chemistries in mass production, we can inevitably expect them to make their way into the environment where the complexities of matrix, oxidation and metabolism, just for starters, make determination of their hazard potential difficult indeed. For these reasons, concerns over the safety of engineered nanomaterials (ENMs) have emerged as calls for proactive and predictive toxicological study to define their potential environmental health and safety of ENMs (nanoEHS), also known as nanotoxicology.
ENMs exhibit physicochemical properties that are markedly differ from conventional macromolecules. Over the last decade, the unique chemical, mechanical and electrical properties of ENMs have been widely applied to various technologies, from consumer products and environmental remediation, to military science and medicine.2 It is not surprising that nanotechnology (NT) is the rapid transition from the discovery phase to the commercialization phase. The number of 1015 NT-based consumer products released into the market in 2009 was a 379% rise from 2005 (http://www.nanotechproject.org/consumerproducts). This trend is expected to generate $1 trillion worldwide by 2015.2 ENMs are rapidly being incorporated into an ever longer list of consumer products, but supporting toxicology for this chemistry has not even begun to keep pace with the innovation. Yet, it must if we are to avoid repeating past blunders. DDT and PCBs are the classic examples of miracle chemicals: cheap to manufacture, extremely effective, used heavily and widely, but with tremendous unforeseen hazards. As their risk to human health and the environment became evident, the U.S. Congress banned their commercial utility in 1976 under the Toxic Substances Control Act (TSCA).3,4
Adequately financed and scientifically sound efforts at systematically identifying the hazard potential of ENMs, prior to their commercialization, are the regulatory and ENM industry goals. Though such efforts have not yet been widely implemented, nanotoxicology is rapidly building a larger repertoire of industry and regulatory stakeholders. On the environmental health and safety side, there are model precedents for a large scale nanotoxicology initiative. For example, Tox21 is a collaboration among multiple federal Agencies - EPA's National Center for Computational Toxicology, the National Institutes of Environmental Health Sciences (NIEHS)/National Toxicology Program (NTP), the National Institutes of Health/National Human Genome Research Institute/NIH Chemical Genomics Center (NCGC), and the U.S. Food and Drug Administration (FDA) - to develop, validate and translate innovative, high-throughput, mostly in vitro chemical screening methods to prioritize substances for further in-depth toxicological evaluation, identify mechanisms of action for further investigation, and develop predictive models for in vivo biological response (http://www.epa.gov/ncct/dsstox/sdf_tox21s.html). The European Registration, Evaluation, Authorization, and Restriction of Chemical substances (REACH), is the European counterpart to Tox21.5 Right now, these toxicology initiatives center mostly on rapid cell-based assays, but in vitro assays are still unlikely to translate well into hazard prediction. The pressing need remains for efficient vertebrate toxicology, a model with which to address the backlog of tens of thousands of conventional chemicals and ENMs, and for which assay results translate effectively and reliably to human hazard.6 The backlog is tremendous, but use of an appropriate model could enable regulatory concerns to bring much of it under their knowledge umbrella. Doing so would have the obvious secondary benefit of establishing a large enough quantitative structure-activity relationship (QSAR) database that toxicity prediction in silico would become an increasingly effective tool.
2. The merits of in vitro and in vivo approaches for chemical hazard prediction
Over the last decade, great technological leaps in molecular and cellular biology have firmly established 21st century toxicology as an essential melding of both in vitro and in vivo approaches. Hazard potential is based largely on a compound’s ability to bioaccumulate, and adsorption, distribution, metabolism, and excretion (ADME) characteristics. These are not very predictive parameters when generated in vitro, as the biological space queried is quite limited and thus, outcomes are not easily translatable to a human response. But in vitro models are a far less expensive means of rapidly querying at least some key parts of biological space with many chemicals, at multiple doses. The strength of in vitro approaches to toxicology is that they are generally adaptable to very high throughput screening efforts and, in many cases, they provide direct read out of perturbation in known gene targets or pathways. In theory, a large and diverse enough battery of in vitro assays on many different human cell types would probe enough biological space to yield hits with very low false positive and false negative rates. The biological space would be truly comprehensive and the pace of discovery truly faster. When conducted for a structurally diverse array of several thousand compounds, this approach will allow for the rapid and mechanism-based prediction of in vivo biological responses. This is a key goal of Tox21: to refine traditional toxicology assays, develop rapid mechanism-based predictive screens, and improve the overall utility of data for making public health decisions.7–9 The use of diverse human-derived cell lines has the potential to add benefit of novel biomarker discovery. Biomarkers of human exposure are highly sought because, once established, they are easily screened in human populations.
The power of the in vivo approach is that, by definition, screening in the whole animal covers all of biological space. Yet, extrapolating mouse and rat toxicology data - the gold standard - across dose, species and life stages has traditionally been fraught with uncertainty because these models did not provide mechanistic information on how and why chemicals exert toxicity. But this was a function of the model and not inherent to the in vivo approach. Huge technological advances in and concomitant declining costs of whole-genome omics screens have made essentially all of gene space available for query in the whole animal. The remaining challenges are to anchor whole genome responses to relevant in vivo endpoints, and to validate pathway-based predictions with targeted studies in the whole animals.10 The latter need is also true of in vitro screening approaches, and, overall, a rather daunting challenge in rodent models. However, this is far less of a challenge, indeed underway and building momentum for at least a decade, in an alternative vertebrate model, the zebrafish.
3. Green chemistry, Toxicology and Green nanotechnology - Common Goals
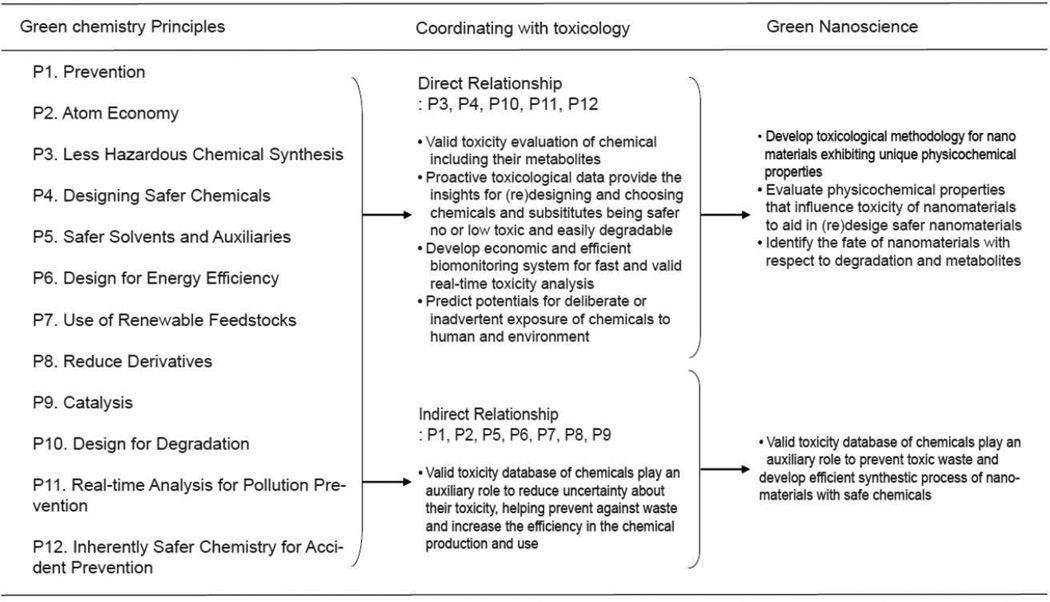
Increased awareness that green chemistry will be ultimately more sustainable, as well as a necessity, has focused attention on reducing or eliminating the use and generation of hazardous substances in the design, manufacture and application of chemical products.11 Figure 1 shows the relationships between toxicology and the 12 principles of Green Chemistry. Half of the 12 principles (P1, P3, P4, P7, P10 and P12) are related directly to the field of toxicology. Greener designs and less hazardous syntheses (P3 and P4), while laudable principles; have to be guided by solid science and practical economics. A strong commitment to toxicity testing in the right model at every step of development can achieve both. This also applies to designing for degradation (P7 and P10). The objectives of toxicology regarding the remaining principles are to reduce waste and to increase materials efficiency. Collectively, these principles are aimed at reducing the uncertainty of toxicity caused by any chemicals generated during and after the synthesis process. We note that nanotoxicology, to be relevant, must also aim to identify which, and understand how, ENM physicochemical properties determine biological hazards (P3, P4 and P12), thereby guiding the (re)design of less toxic and easily degradable, yet optimally functional ENMs (Figure 2) (P4, P7 and P10). Predictive vertebrate toxicology, nanotoxicology and green chemistry principles thus substantially overlap in their goals (Figure 1).
Figure 1.
Relationship between the 12 green chemistry principles and toxicology. The role of toxicology to attain 12 green chemistry principles are described. PX, where X= principles 1–12
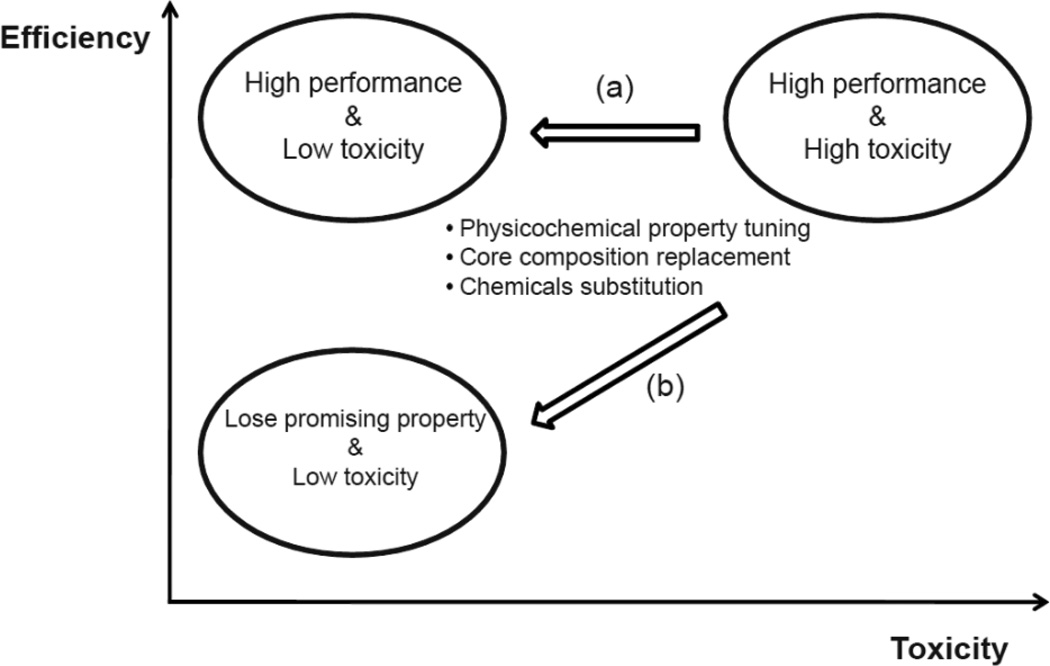
Figure 2.
Efficiency versus Toxicity. When evolving materials having highly efficient functions but high toxicity, the direction (a) that derive the development of high performance and no or low toxicity of materials is more desirable than the direction (b), a collateral loss of initially promising properties.
We have established that an alarming lack of toxicological data for most ENMs has led to numerous concerns about nanoEHS. Rather than skirting peril while unilaterally forging ahead with commercialization, this represents a golden opportunity: Both the ENMs industry and regulatory concerns can partner in leveraging tools like zebrafish and the peaking interests of federal funding sources to addresses the challenges of nanoEHS, and dramatically reduce long range liability. The opportunity has been recognized by some NT stakeholders, but not enough. For example, the Federal Government developed a National Nanotechnology Initiative (NNI) - Environmental, Health, and Safety (EHS) Research Strategy (http://www.nano.gov/sites/default/files/pub_resource/nni_2011_ehs_research_strategy.pdf). It outlines a comprehensive approach to ensuring the safe, effective and responsible development and use of NT with six core research areas: (1) Nanomaterial Measurement Infrastructure, (2) Human Exposure Assessment, (3) Human Health, (4) Environment, (5) Risk Assessment and Risk Management Methods, with consideration of ethical, legal, and societal implications of NT and (6) Informatics and Modeling. However, NT industry stakeholders have not yet extensively partnered with the NNI-EHS research strategy. Much remains to be implemented before smart regulatory decisions can happen.
The characteristics of ENMs are fundamentally different than the characteristics of conventional small molecules. Indeed, it is difficult to overstate the degree to which ENM unique physicochemical behaviors influence their hazard potential, and to understate how naive the current understanding of nano structure-activity links remains.2,12–15 A number of studies with various toxicological approaches are building a knowledge base of nanoEHS. These studies highlighted that little is known about how ENMs interact with biological components such as proteins, membranes, phospholipids, endocytic vesicles, organelles, DNA and biological fluids.16 Furthermore, the environmental fate and transport of ENMs remain largely unknown so there is much uncertainty regarding the scope of potential ENM hazard to humans and other organisms in the environment.17 It is noted that Ferry et al. recently reported the transfer of gold nanoparticles (AuNPs) from the water column to the marine food web.18 The uses for and production of ENMs have exploded, far outpacing an effective nanoEHS oversight. Wide reaching and absurdly generalized regulation could be adopted, but it should never be the objective of nanotoxicology to limit ENM innovation or use while regulatory concerns, employing obsolete approaches to toxicology, engage in a futile attempt at catch-up safety assessments. Rather, the goal of nanotoxicology should be to provide ENM developers with a means to practically and efficiently parallel their R&D and commercialization efforts with rapid and predictive toxicological assessments. One of the best available means of doing this is with the zebrafish developmental toxicology model.
4. Zebrafish: A powerful in vivo toxicity model
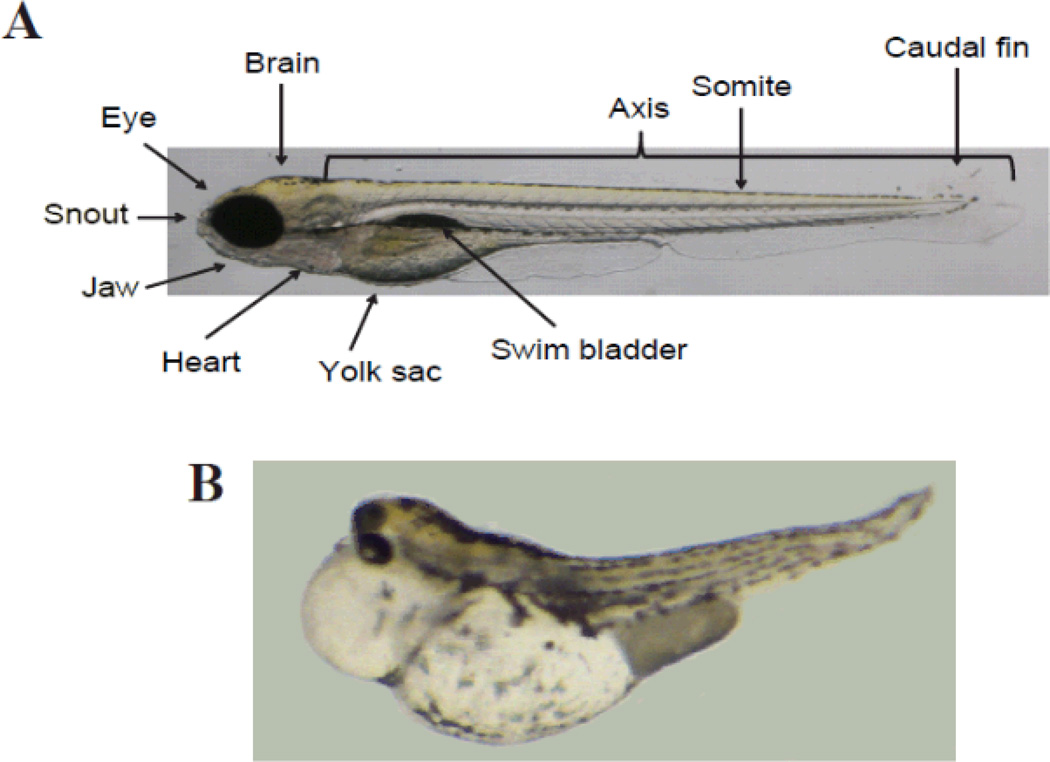
We have emphasized the importance of a rapid and efficient vertebrate model with sufficient relevance to humans to be a predictive toxicology tool. The embryonic zebrafish (Danio rerio) has emerged as the best choice, offering marked advantages over developmental rodent models in both sensitivity to toxins and quicker identification of the toxic mechanism.19–22 A fast track to identifying developmental toxicity mechanisms is possible using zebrafish because an induced phenotype, whether morphological, physiological or behavioral, can quickly be dissected using molecular approaches. Zebrafish are also a viable alternative model with respect to Replacement, Reduction, and Refinement of animal toxicity testing.23 Zebrafish are resource-efficient due to their small size, high fecundity and relatively inexpensive husbandry. A pair of adult zebrafish can produce 200–300 fertile eggs every week. Gastrulation to the segmentation stage of somitogenesis only takes 24 hours and development is complete by 72 hours after fertilization. During the first 48 to 60 hours of development zebrafish embryos are optically clear, allowing observation of chemical-induced effects in the organs, an enormous advantage over assays of rodent development. Their small size also translates into a much smaller requirement of test chemicals for toxicity evaluation. Figure 3 displays an example of 120 hours post fertilization zebrafish embryos, with and without developmental defects in the brain, eye, snout, jaw, somite and caudal fin. Examples of yolk sac and pericardial edemas and a bent spine are also apparent.
Figure 3.
Zebrafish embryo images showing no effect (a) and positive effect (b) on organs at 120 hours post-fertilization (hpf).
The versatility of the zebrafish model is further bolstered by the availability of many mutant lines, diverse and robust transgenic lines and the simplicity of performing transient antisense RNA knockdowns. The near complete annotation of the genome has enabled the integration of zebrafish toxicology, genomics and toxicogenomics. With increased through put, it is now possible to rapidly screen the exposure related phenotypes and identify the gene expression changes that underlie the effect. Thus, toxicogenomics is a powerful approach for uncovering the mechanisms of toxicity by identifying the developmental pathways perturbed by toxicants. The availability of many zebrafish mutant lines and transgenic reporter zebrafish has been enormously powerful for understanding the function of zebrafish genes and their targets. Transgenic reporter lines expressing a fluorescent protein under the control of a tissue-specific promoter allow easy visualization (even naked-eye during development) of gene expression patterns and tissue structural changes at any life stage using fluorescence microscopy. Toxicological evaluation of cadmium24 and arsenite25 was performed using the heat shock 70 protein promoter to drive expression of green fluorescent protein (GFP). A zebrafish line with luciferase-GFP driven by an electrophile-responsive element enabled investigators to describe the mechanism of oxidative stress response to mercury.26
Zebrafish mutations are proven to be useful for uncovering the genetic targets of some toxicants. Screening mutants against a variety of chemicals that elicit a known phenotype will uncover non-responders to some chemicals which, by definition, are operant via a target or pathway that includes the mutant gene product. A recent example of this approach is the identification of the zebrafish persephone mutant that is resistant to mechanosensory hair cell death caused by different aminoglycoside compounds that also cause hair cell death in humans.27 For example, a zebrafish aryl hydrocarbon receptor (AHR2) null line was developed for toxicity assessments of polycyclic aromatic hydrocarbons (PAH) as a means of distinguishing AHR-dependent from AHR-independent PAH structures.28 Details on materials and protocols for mutant and transgenic zebrafish are provided on the websites that include the Zebrafish International Resource Center (ZIRC), Zebrafish Information Network (ZFIN), Sanger zebrafish sequencing project, ZF-Models, Sanger Zebrafish Mutation Resource and EuFishBiomed.29
Gene knockdown using morpholino oligonucleotides (MOs) is a powerful and rapid means of transiently suppressing a gene's expression to reveal its function. The developing zebrafish are particularly suited to this technology.30 MOs are synthetic antisense oligonuclotides injected into the one cell embryo and evenly dispersed by cell division throughout the developing embryos. MOs are effective until 96 to 120 hours post fertilization by binding their target mRNA and either preventing translation of the message or blocking the correct splicing of the message to its mature form. When a zebrafish morphant recapitulates the phenotype of a chemical exposure, by definition a target gene or pathway component has been identified. The role of AHR2 and cytochrome P451A (CYP1A) in PAH toxicity was elucidated in part with transient MO-knockdown zebrafish.31 The use of a nuclear factor erythroid 2-related factor 2 (Nrf2) MO in zebrafish identified the role of Nrf2 in the expression of cytoprotective enzymes of heme oxygenase-1 (HO-1) against oxidative stress induced by perfluorooctane sulfonate (PFOS) exposure.32
To overcome the transient nature of MO knockdown, stable knockouts are also readily achievable in zebrafish. Zinc Finger Nuclease (ZFN) targeting33,34 and Transcription Activator-Like Effector Nucleases (TALENs)35,36 are the two most widely used methods of generating stable knockout zebrafish. These designer nucleases bind to and cleave DNA at particular target sites, inducing error-prone repair that results in insertion or deletion mutations. Use of TALENs is the more efficient and precise approach of the two36,37 and has already been used to create dozens of zebrafish knockout lines, each available to the research community.
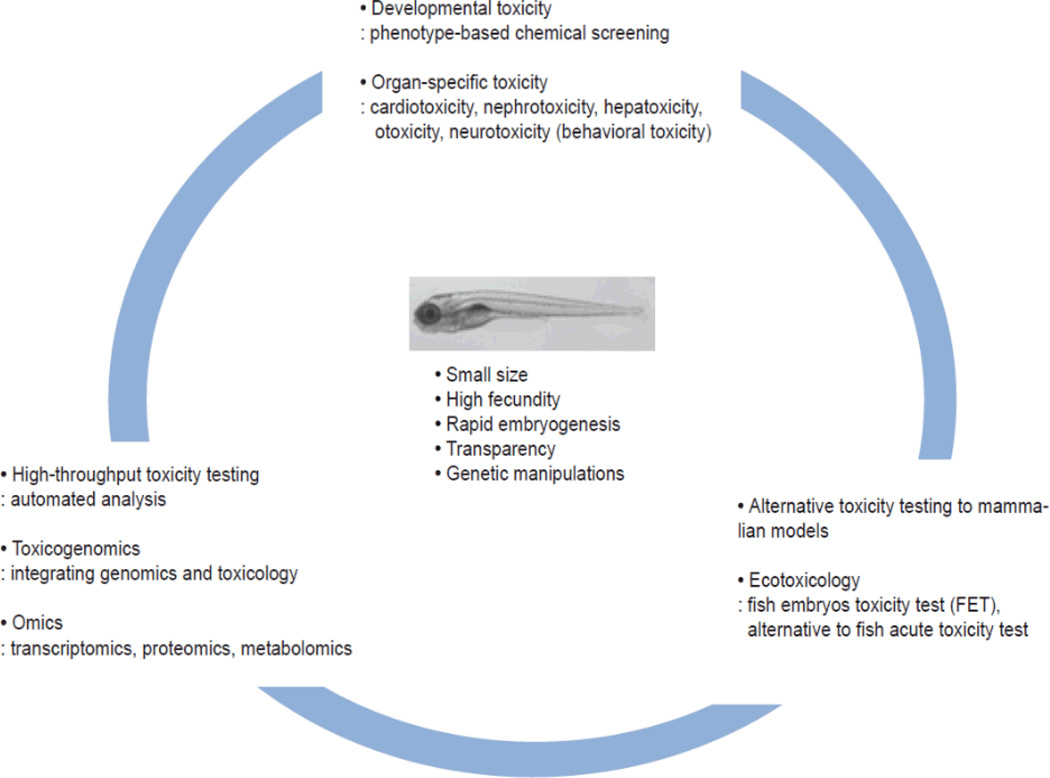
The molecular signaling underlying the effects of toxicant perturbations are all highly tractable with the tools available for zebrafish. These ‘fly and worm-like’ approaches in a vertebrate model have made high throughput toxicology, with human relevance, a reality (Figure 4). It is expected that zebrafish will be the go-to model for addressing the > 40,000 compound backlog of chemicals already in production while increasingly building a more predictive knowledgebase.
Figure 4.
The advantages of zebrafish over a wide range of toxicology testing including developmental, organ-specific, high-throughput and integrated mechanistic toxicology approaches as an alternative animal toxicity model.
5. How zebrafish developmental toxicology can drive green nanotechnology
The Tox21 strategy for screening thousands of toxicants at multiple doses via a comprehensive battery of high throughput in vitro assays in many different cell types is unprecedented.7 Even more unprecedented is that this approach is already underway in vivo using zebrafish to screen the developmental toxicity of the EPA ToxCast portfolio of compounds.38,39 The utility of zebrafish allows whole animal biological space to be probed and the physical characteristics described render zebrafish assays highly amenable to automation.40 Lam et al. demonstrated that the zebrafish is a desirable model for a high-throughput chemogenomics.41 The zebrafish embryo toxicity test (ZFET) can already substitute for the acute fish test in ecotoxicology.
Toxicological studies indicate that the zebrafish is well suited for use in developing nanoEHS.42–45 George et al. utilized zebrafish as a robust high-throughput platform for nanoEHS and transformed zebrafish toxicology data into in silico analyses for hazard ranking of ENMs through comparison with in vitro results.44 Recently, we developed a rapid and cost-effective automated toxicity screening system from embryo preparation, exposure to toxicants, digital imaging to developmental toxicity phenotype analysis.40 Using this system, we quickly anchored a defective eye phenotypic response of developing embryos exposed to AuNPs with relevant transcriptional (mRNA) responses and concluded that AuNPs perturbed specific developmental pathways required for eye embryogenesis and pigmentation (manuscript under review).46 Such zebrafish in vivo toxicity studies for are rapid and highly effective. More generally, in vivo toxicity data for ENMs are extremely rare. It is estimated that the cost for testing existing ENMs will be $249 million, but could exceed a $ billion if long-term in vivo testing becomes a requirement of commercially producing ENMs.47 The utilization of zebrafish will make it possible to rapidly and efficiently characterize in vivo toxicity and predict likely hazards to humans.
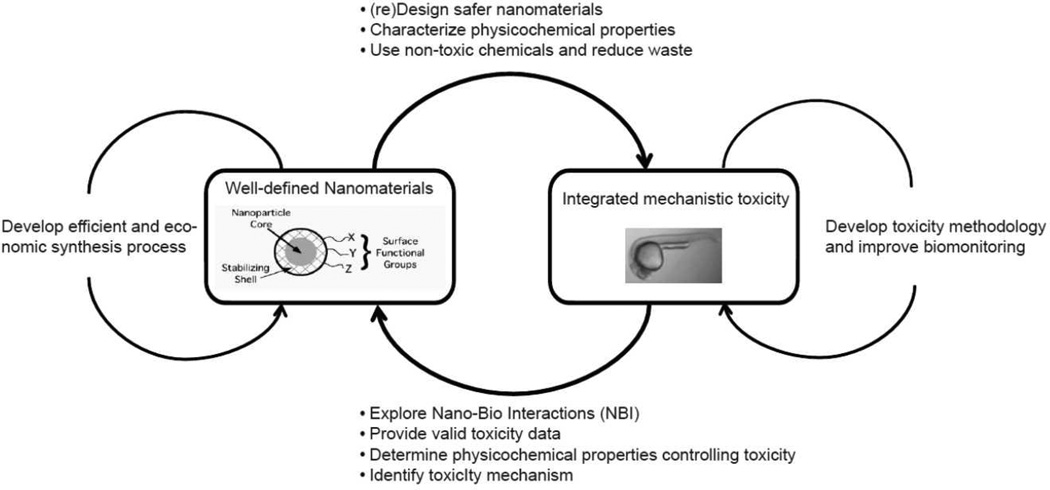
Toxicological studies for nanoEHS will be directed for green nanoscience, safer ENMs production and efficient synthetic process development. However, there is the gap between toxicology and material sciences for nanoEHS. To attain green nanoscience, close interdisciplinary cooperation will be necessary as illustrated in Figure 5. The consistent testing of well-defined ENM preparations is a critical first step. Informative QSARs are impossible if tight control is not exercised over the formulations under scrutiny. Moreover, consumers must expect that manufacturer requirements will force adherence to ENM quality and purity standards for which safety data is already in place.48 Upon successful synthesis of well-defined ENMs with the desired core composition, size, shape, surface coating and surface chemistry, toxicity assessment should immediately follow. Zebrafish developmental toxicology done in parallel with ENM discovery and development will build insights for the design of safer novel ENMs, addressing the current uncertainty about safety of ENMs, and dramatically reducing commercialization liability by avoiding formulations that are later found to be human and environmental hazards. Two approaches to investigate nanotoxicology are to examine biomarkers of molecular activity or pathway disruption by ENMs in vivo and determine the specific structural properties of ENMs driving any observed toxicity. This integration of structure and hazardous activity assessments can then be used to guide the selection of ENM core composition, size, shape, surface coating and surface chemistry with lower hazard potential. In the part of toxicology, the toxicity methodologies optimized for nanoEHS are developed to be gradually contiguous to true exposure scenarios and toxicity. On the other hand, in the part of materials science, the efficient and economic synthesis process is devised to produce reproducible high purified ENMs. In our lab we have tested the biological responses of zebrafish embryos exposed to well-defined, structurally-diverse AuNPs,45 finding that their toxicity, uptake and elimination were dependent on surface chemistry and core size. These findings were reported to the Safer Nanomaterials and Nanomanufacturing Initiative (SNNI) (http://www.greennano.org/) as a preliminary step toward cooperative toxicology and materials science to develop high performance, non-toxic AuNPs as shown in Figure 2. AuNPs are already used for a growing range of biomedical applications, such as cancer chemotherapy, imaging, and gene delivery.49–51 Despite an intensive body of research on AuNPs,18,51–53 there is still much that we do not understand about the bioactivity AuNPs. Because of their utility and the precision with which AuNPs are easily engineered, they were an effective ENM with which begin comprehensive toxicology in the zebrafish model. Table 1 summarizes examples of toxicity results from several types of AuNPs in the zebrafish embryonic toxicity assay carried out by our laboratory.42,45,46,54 The embryonic toxicities of AuNPs were heavily dependent on surface charge modification. Non-functionalized AuNPs is not toxic. Exposure to positively and negatively charged AuNPs cause embryonic mortality and malformations; while neutrally charged AuNPs are non-toxic, indicating that this assay can effectively discriminate between toxic and non-toxic ENMs. Schaeublin et al. conducted in vitro assays to evaluate cytoxicity of the same AuNPs as our laboratory using a human keratinocyte cell line (HaCaT).55 They also found that the surface charged AuNPs were more toxic than neutral AuNPs, inducing significant damage in the mitochondrial membrane not observed with neutral AuNPs.
Figure 5.
Interdisciplinary activity between toxicology and materials synthesis. The close reciprocal feedback is necessary to design and develop chemicals satisfying requirements for the commercialization (i.e., high performance and no or low toxicity). The exchange of information can devise and enhance their own technologies such as toxicity methodology and synthesis process.
Table 1.
Actual toxicity of several types of gold nanoparticles (AuNPs) in the zebrafish embryonic assay
| Gold nanoparticles (AuNPs) | General Characteristics | Toxicity (mass basis) | References |
|---|---|---|---|
| 2-mercaptoethanesulfonic acid (MES-AuNPs) |
Ligand stabilized with anionic MES Negative surface charge Core size: 1.5 nm |
50% mortality, 30% malformation at 250 mg/L |
45 |
| Mercaptoethoxyethoxyethanol (MEEE-AuNPs) |
Ligand stabilized with neutral MEEE Neutral surface charge Core size: 1.5 nm |
Non-toxic | 45 |
| N,N,Ntrimethylammoniumethanethiol (TMAT-AuNPs) |
Ligand stabilized with cationic TMAT Positive surface charge Core size: 1.5 nm |
50% mortality at 30 mgL | 45,46 |
| 3-mercaptopropionic acid (3-MPA-AuNPs) |
Ligand stabilized with 3-MPA Core size: 1.2 nm |
75% mortality at 50 mg/L | 54 |
| Bare AuNPs | Not functionalized | Non-toxic | 42,46 |
Comprehensive ENMs purity evaluations and characterization must be completed in order to define the physicochemical properties that influence biocompatibility. Impurities, for example, can often greatly influence toxicity results. Contaminating organic solvents, catalysts and coating materials used in the synthesis process could cause toxicity falsely ascribed to the ENMs. For example, a tetrahydrofuran (THF) degradation product was attributed to toxicity of aqueous C60 in zebrafish,58 and cetyltrimethyl-ammoniumbromide (CTAB), widely used for surface coating of AuNPs, was also proven to be toxic.51 Finally, Jakubek et al. demonstrated that small amounts of the catalyst yttrium released form carbon nanotubes efficiently inhibited calcium ion channels to produce in vitro toxicity.59
Careful characterization of the ENMs in the dry state and also in relevant aqueous biological fluids is essential as it is now widely appreciated that the environment greatly influences the physical properties of ENMs 56,57 In particular, ENMs tend to rapidly agglomerate in solution and test media consisting of higher ionic strength, which can unpredictably alter toxicity. For example, when ENM agglomerates affect bioavailability it is challenging to interpret toxicological results. We have found that precision engineered functionalized AuNPs can be generated to maintain their desired sizes in both nanopure water and zebrafish standard medium, thus we can associate the nanomaterial size and charge to its toxicity with great certainty.45 Using small-angle X-ray scattering (SAXS) it is now possible to accurately measure the core sizes of AuNPs in the one nanometer range in solution. These small particles are difficult to measure using dynamic light scattering (DLS), as DLS is not efficient for sizes less than 10 nm.46 Another approach that we employed to control agglomerate size during toxicity testing in the zebrafish assay was to dilute the standard medium. We demonstrated that zebrafish can develop in reduced ionic strength solutions. We found surprising results using 1.2 nm 3-mercaptopropionic acid-functionalized AuNPs (1.2 nm 3-MPA-AuNPs) which rapidly agglomerate in standard testing solutions.54 We found that the toxicity of 1.2 nm 3-MPA-AuNPs was significantly higher in low ionic medium where the 1.2 nm 3-MPA-AuNPs remained highly dispersed. This suggests that there is a high probability to detect false negative response when testing ENMs that tend to agglomerate. This is because the test solution itself can profoundly alter the size, surface area and charge of the primary test materials, leading to reduced availability and uptake. In summary, the use of appropriate ENM characterization methods must be applied before and during the exposure to avoid some of the critical errors made in early days of nanotoxicology.
Conclusions and future perspectives
The toxicology tools and approaches necessary to evaluate the hazard potentials of ENMs and solidly support green nanoscience are already available. With intensive application of the developing zebrafish and advanced in vitro assays, nanotoxicology will migrate away from “testing everything” and instead understand the ENM features that influence biocompatibility and be in a position to predictive biocompatibility. When we achieve predictive authority, green design, production, use and disposal of ENMs, with close regard for their environmental health and safety, will be a practical reality.
Acknowledgments
This review was supported by National Institute of Environmental Health Sciences (NIEHS) grants P3000210 and ES016896.
Biographies

Kitae Kim
Dr. Kitae Kim received his BS in Environmental Engineering in 2003 from Kwangwoon University - Seoul, Korea and PhD in Environmental Science and Engineering from Gwangju Institute of Science and Technology (GIST) - Gwangju, Korea in 2009. He joined in Prof. Robert Tanguay group as a post-doctoral researcher at Oregon State University in 2010. His research currently involves the identification of toxicity mechanism of gold nanoparticles and the determination of physicochemical properties that influence biological responses to Zebrafish.

Robert Tanguay
Dr. Robert Tanguay is a Distinguished Professor in the Department of Environmental and Molecular Toxicology and the Director of the Sinnhuber Aquatic Research Laboratory. He received his BA in Biology from California State University - San Bernardino (1988) and his PhD in Biochemistry from the University of California-Riverside (1995) and postdoctoral training in developmental toxicology from the University of Wisconsin-Madison (1996–1999). He has been at Oregon State University since 2003. His laboratory has broad research interest in the areas of system toxicology, nanotechnology and tissue regeneration.
References
- 1.Opportunities in chemical distribution: Optimizing marketing and sales channels, managing complexity, and redefining the role of distributors. Boston: The Boston Consulting Group; 2010. [Google Scholar]
- 2.Nel A, Xia T, Madler L, Li N. Science. 2006;311:622–627. doi: 10.1126/science.1114397. [DOI] [PubMed] [Google Scholar]
- 3.Robertson LW, Hansen LG. Kentucky: The University Press of Kentucky; 2001. [Google Scholar]
- 4.U. S.D. o. H. a. H. Services, ed. U. S. D. o. H. a. H. Services. 2002. [Google Scholar]
- 5.Matus KJ, Zimmerman JB, Beach E. Environ. Sci. Technol. 2010;44:6022. doi: 10.1021/es102149j. [DOI] [PubMed] [Google Scholar]
- 6.Meng H, Xia T, George S, Nel AE. ACS Nano. 2009;3:1620–1627. doi: 10.1021/nn9005973. [DOI] [PubMed] [Google Scholar]
- 7.Toxicity Testing in the 21st Century: A vision and a Strategy, National Research Council. Washington, DC: Natl. Acad. Press; 2007. [Google Scholar]
- 8.Krewski D, Andersen ME, Mantus E, Zeise L. Risk Anal. 2009;29:474–479. doi: 10.1111/j.1539-6924.2008.01150.x. [DOI] [PubMed] [Google Scholar]
- 9.Kavlock RJ, Austin CP, Tice RR. Risk Anal. 2009;29:485–487. doi: 10.1111/j.1539-6924.2008.01168.x. discussion 492-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knudsen TB, Kavlock RJ, Daston GP, Stedman D, Hixon M, Kim JH. Birth Defects Res., Part B. 2011;92:413–420. doi: 10.1002/bdrb.20315. [DOI] [PubMed] [Google Scholar]
- 11.Tucker JL. Org. Process Res. Dev. 2010;14:328–331. [Google Scholar]
- 12.Colvin VL. Nat. Biotechnol. 2003;21:1166–1170. doi: 10.1038/nbt875. [DOI] [PubMed] [Google Scholar]
- 13.Dreher KL. Toxicol. Sci. 2004;77:3–5. doi: 10.1093/toxsci/kfh041. [DOI] [PubMed] [Google Scholar]
- 14.Oberdorster E, Zhu S, Blickley TM, McClellan-Green P, Haasch ML. Carbon. 2005;44:1112–1120. [Google Scholar]
- 15.Maynard AD, Aitken RJ, Butz T, Colvin V, Donaldson K, Oberdorster G, Philbert MA, Ryan J, Seaton A, Stone V, Tinkle SS, Tran L, Walker NJ, Warheit DB. Nature. 2006;444:267–269. doi: 10.1038/444267a. [DOI] [PubMed] [Google Scholar]
- 16.Nel AE, Madler L, Velegol D, Xia T, Hoek EMV, Somasundaran P, Klaessig F, Castranova V, Thompson M. Nat. Mater. 2009;8:543–557. doi: 10.1038/nmat2442. [DOI] [PubMed] [Google Scholar]
- 17.Klaine SJ, Alvarez PJ, Batley GE, Fernandes TF, Handy RD, Lyon DY, Mahendra S, McLaughlin MJ, Lead JR. Environ. Toxicol. Chem. 2008;27:1825–1851. doi: 10.1897/08-090.1. [DOI] [PubMed] [Google Scholar]
- 18.Ferry JL, Craig P, Hexel C, Sisco P, Frey R, Pennington PL, Fulton MH, Scott IG, Decho AW, Kashiwada S, Murphy CJ, Shaw TJ. Nat. Nanotechnol. 2009;4:441–444. doi: 10.1038/nnano.2009.157. [DOI] [PubMed] [Google Scholar]
- 19.Spitsbergen J, Kent M. Toxicol. Pathol. 2003;31:62–87. doi: 10.1080/01926230390174959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abikoff H, McGough J, Vitiello B, McCracken J, Davies M, Walkup J, Riddle M, Oatis M, Greenhill L, Skrobala A, March J, Gammon P, Robinson J, Lazell R, McMahon DJ, Ritz L. J. Am. Acad. Child Adolesc. Psychiatr. 2005;44:418–427. doi: 10.1097/01.chi.0000155320.52322.37. [DOI] [PubMed] [Google Scholar]
- 21.Nagel R. ALTEX. 2002;19:38–48. [PubMed] [Google Scholar]
- 22.Spence R, Gerlach G, Lawrence C, Smith C. Biol. Rev. Camb. Philos. Soc. 2008;83:13–34. doi: 10.1111/j.1469-185X.2007.00030.x. [DOI] [PubMed] [Google Scholar]
- 23.Embry MR, Belanger SE, Braunbeck TA, Galay-Burgos M, Halder M, Hinton DE, Leonard MA, Lillicrap A, Norberg-King T, Whale G. Aquatic toxicology (Amsterdam, Netherlands) 2010;97:79–87. doi: 10.1016/j.aquatox.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 24.Blechinger SR, Warren JT, Jr, Kuwada JY, Krone PH. Environ. Health Perspect. 2002;110:1041–1046. doi: 10.1289/ehp.021101041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seok SH, Baek MW, Lee HY, Kim DJ, Na YR, Noh KJ, Park SH, Lee HK, Lee BH, Ryu DY, Park JH. Toxicol. in Vitro. 2007;21:870–877. doi: 10.1016/j.tiv.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 26.Kusik BW, Carvan MJ, 3rd, Udvadia AJ. Mar. Biotechnol. 2008;10:750–757. doi: 10.1007/s10126-008-9113-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hailey DW, Roberts B, Owens KN, Stewart AK, Linbo T, Pujol R, Alper SL, Rubel EW, Raible DW. PLoS Genet. 2012;8:e1002971. doi: 10.1371/journal.pgen.1002971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goodale BC, La Du JK, Bisson WH, Janszen DB, Waters KM, Tanguay RL. PLoS ONE. 2012;7:e29346. doi: 10.1371/journal.pone.0029346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang L, Ho NY, Alshut R, Legradi J, Weiss C, Reischl M, Mikut R, Liebel U, Muller F, Strahle U. Reproductive toxicology (Elmsford, N.Y.) 2009;28:245–253. doi: 10.1016/j.reprotox.2009.04.013. [DOI] [PubMed] [Google Scholar]
- 30.Nasevicius A, Ekker SC. Nat. Genet. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- 31.Billiard S, Timme-Laragy AR, Wassenberg DM, Cockman C, Di Giulio RT. Toxicol. Sci. 2006 doi: 10.1093/toxsci/kfl011. [DOI] [PubMed] [Google Scholar]
- 32.Shi X, Zhou B. Toxicol. Sci. 2010;115:391–400. doi: 10.1093/toxsci/kfq066. [DOI] [PubMed] [Google Scholar]
- 33.Amacher SL. Brief. Funct. Genomic Proteomic. 2008;7:460–464. doi: 10.1093/bfgp/eln043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ekker SC. Zebrafish. 2008;5:121–123. doi: 10.1089/zeb.2008.9988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sander JD, Cade L, Khayter C, Reyon D, Peterson RT, Joung JK, Yeh JR. Nat. Biotechnol. 2011;29:697–698. doi: 10.1038/nbt.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moore FE, Reyon D, Sander JD, Martinez SA, Blackburn JS, Khayter C, Ramirez CL, Joung JK, Langenau DM. PLoS ONE. 2012;7:e37877. doi: 10.1371/journal.pone.0037877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bedell VM, Wang Y, Campbell JM, Poshusta TL, Starker CG, Krug Ii RG, Tan W, Penheiter SG, Ma AC, Leung AY, Fahrenkrug SC, Carlson DF, Voytas DF, Clark KJ, Essner JJ, Ekker SC. Nature. 2012 doi: 10.1038/nature11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sipes NS, Padilla S, Knudsen TB. Birth Defects Res., Part C. 2011;93:256–267. doi: 10.1002/bdrc.20214. [DOI] [PubMed] [Google Scholar]
- 39.Padilla S, Corum D, Padnos B, Hunter DL, Beam A, Houck KA, Sipes N, Kleinstreuer N, Knudsen T, Dix DJ, Reif DM. Reproductive toxicology (Elmsford, N.Y.) 2012;33:174–187. doi: 10.1016/j.reprotox.2011.10.018. [DOI] [PubMed] [Google Scholar]
- 40.Mandrell D, Truong L, Jephson C, Sarker MR, Moore A, Lang C, Simonich MT, Tanguay RL. J. Lab. Autom. 2012;17:66–74. doi: 10.1177/2211068211432197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lam SH, Mathavan S, Tong Y, Li H, Karuturi RK, Wu Y, Vega VB, Liu ET, Gong Z. PLoS Genet. 2008;4:e1000121. doi: 10.1371/journal.pgen.1000121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bar-Ilan O, Albrecht RM, Fako VE, Furgeson DY. Small. 2009;5:1897–1910. doi: 10.1002/smll.200801716. [DOI] [PubMed] [Google Scholar]
- 43.Fako VE, Furgeson DY. Adv. Drug Deliv. Rev. 2009;61:478–486. doi: 10.1016/j.addr.2009.03.008. [DOI] [PubMed] [Google Scholar]
- 44.George S, Xia T, Rallo R, Zhao Y, Ji Z, Lin S, Wang X, Zhang H, France B, Schoenfeld D, Damoiseaux R, Liu R, Bradley KA, Cohen Y, Nel AE. ACS Nano. 2011;5:1805–1817. doi: 10.1021/nn102734s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harper SL, Carriere JL, Miller JM, Hutchison JE, Maddux BL, Tanguay RL. ACS Nano. 2011;5:4688–4697. doi: 10.1021/nn200546k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim K, Zaikova T, Hutchison JE, Tanguay RL. Toxicol. Sci. doi: 10.1093/toxsci/kft081. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Choi JY, Ramachandran G, Kandlikar M. Environ. Sci. Technol. 2009;43:3030–3034. doi: 10.1021/es802388s. [DOI] [PubMed] [Google Scholar]
- 48.Hussain SM, Braydich-Stolle LK, Schrand AM, Murdock RC, Yu KO, Mattie DM, Schlager JJ, Terrones M. Adv. Mater. 2009;21:1549–1559. [Google Scholar]
- 49.Basu S, Harfouche R, Soni S, Chimote G, Mashelkar RA, Sengupta S. Proc. Natl. Acad. Sci. U S A. 2009;106:7957–7961. doi: 10.1073/pnas.0902857106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ghosh P, Han G, De M, Kim CK, Rotello VM. Adv. Drug Deliv. Rev. 2008;60:1307–1315. doi: 10.1016/j.addr.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 51.Alkilany AM, Nagaria PK, Hexel CR, Shaw TJ, Murphy CJ, Wyatt MD. Small. 2009;5:701–708. doi: 10.1002/smll.200801546. [DOI] [PubMed] [Google Scholar]
- 52.Dykman L, Khlebtsov N. Chem. Soc. Rev. 2012;41:2256–2282. doi: 10.1039/c1cs15166e. [DOI] [PubMed] [Google Scholar]
- 53.Giljohann DA, Seferos DS, Daniel WL, Massich MD, Patel PC, Mirkin CA. Angew. Chem. Int. Ed. 2010;49:3280–3294. doi: 10.1002/anie.200904359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Truong L, Zaikova T, Richman EK, Hutchison JE, Tanguay RL. Nanotoxicology. 2011;6:691–699. doi: 10.3109/17435390.2011.604440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schaeublin NM, Braydich-Stolle LK, Schrand AM, Miller JM, Hutchison J, Schlager JJ, Hussain SM. Nanoscale. 2011;3:410–420. doi: 10.1039/c0nr00478b. [DOI] [PubMed] [Google Scholar]
- 56.Murdock RC, Braydich-Stolle L, Schrand AM, Schlager JJ, Hussain SM. Toxicol. Sci. 2008;101:239–253. doi: 10.1093/toxsci/kfm240. [DOI] [PubMed] [Google Scholar]
- 57.Soenen SJ, Rivera-Gil P, Montenegro J, Parak WJ, De Smedt SC, Braeckmans K. Nanotoday. 2011;6:446–465. [Google Scholar]
- 58.Henry TB, Menn FM, Fleming JT, Wilgus J, Compton RN, Sayler GS. Environ. Health Perspect. 2007;115:1059–1065. doi: 10.1289/ehp.9757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jakubek LM, Marangoudakis S, Raingo J, Liu X, Lipscombe D, Hurt RH. Biomaterials. 2009;30:6351–6357. doi: 10.1016/j.biomaterials.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]