SUMMARY
Many cells undergo symmetry-breaking polarization toward a randomly oriented “front” in the absence of spatial cues. In budding yeast, such polarization involves a positive feedback loop that enables amplification of stochastically arising clusters of polarity factors. Previous mathematical modeling suggested that, if more than one cluster were amplified, the clusters would compete for limiting resources and the largest would “win,” explaining why yeast cells always make one and only one bud. Here, using imaging with improved spatiotemporal resolution, we show the transient coexistence of multiple clusters during polarity establishment, as predicted by the model. Unexpectedly, we also find that initial polarity factor clustering is oscillatory, revealing the presence of a negative feedback loop that disperses the factors. Mathematical modeling predicts that negative feedback would confer robustness to the polarity circuit and make the kinetics of competition between polarity factor clusters relatively insensitive to polarity factor concentration. These predictions are confirmed experimentally.
INTRODUCTION
Polarity establishment employs an evolutionarily ancient machinery centered around the conserved Rho family GTPase Cdc42p (Park and Bi, 2007). During polarization, GTP-Cdc42p becomes concentrated at the cortical site destined to be the “front” of the cell. In response to cell-cycle cues, Saccharomyces cerevisiae cells concentrate polarity regulators at one of several predictable sites defined by landmark proteins (Park and Bi, 2007). In the absence of interpretable landmarks (e.g., in rsr1Δ mutants), however, yeast cells nevertheless polarize and bud at a single random site (Bender and Pringle, 1989; Chant and Herskowitz, 1991). Such “symmetry breaking” polarization requires the scaffold protein Bem1p, which associates with the Cdc42p-directed guanine nucleotide exchange factor (GEF), Cdc24p, and a p21-activated kinase (PAK) (Bose et al., 2001; Gulli et al., 2000; Irazoqui et al., 2003; Kozubowski et al., 2008). This complex is thought to mediate a positive feedback loop that enables small stochastic clusters of GTPCdc42p to become amplified (Kozubowski et al., 2008). Mathematical modeling suggested that, although more than one stochastic cluster could be amplified in this manner, Bem1p complexes would soon become depleted from the cytoplasm, after which the clusters would compete with each other and the largest one would “win” (Goryachev and Pokhilko, 2008; Howell et al., 2009). Thus, Bem1p-mediated positive feedback combined with competition for limiting Bem1p complexes could explain why rsr1Δ yeast cells polarize to one and only one site.
The competition hypothesis predicts that polarity establishment should frequently proceed via a transient intermediate stage with more than one polarity cluster, but there is limited experimental evidence for such intermediates, as only rare, fleeting two-cluster instances were identified in rsr1Δ cells (Howell et al., 2009). Thus, either competition occurs very rapidly, or some other mechanism ensures that only a single cluster develops. To distinguish between these possibilities, we developed higher-resolution filming conditions that circumvented the phototoxicity of previous protocols. We now document the frequent formation of more than one polarity cluster, and rapid competition between clusters, during symmetry-breaking polarization in rsr1Δ cells. Rapid filming of initial polarity establishment also revealed unexpected oscillatory clustering of polarity factors, indicative of negative feedback. Mathematical modeling suggested that negative feedback could confer advantageous features, including robustness and rapid competition between clusters even in the face of increasing polarity factor concentrations. Experimental tests confirmed these predictions, suggesting that negative feedback improves the robustness of the yeast polarity circuit.
RESULTS
Multicluster Intermediates En Route to a Single Polarization Site
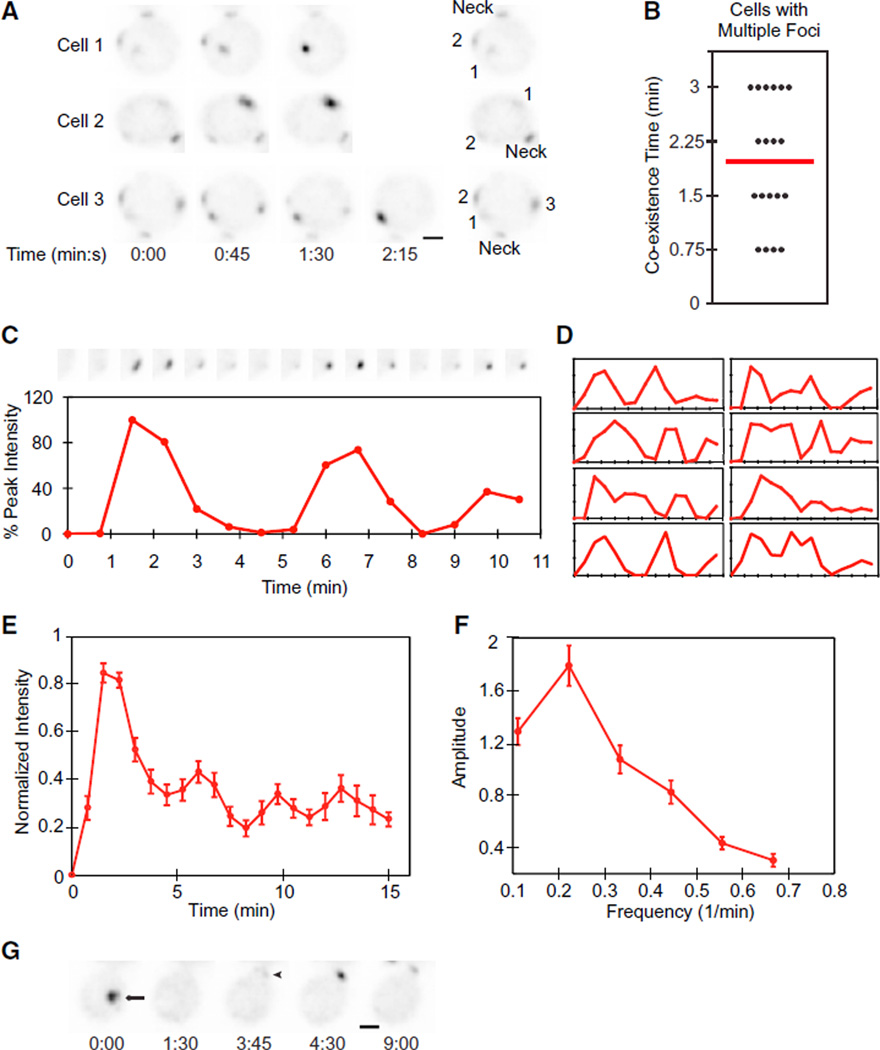
A major obstacle to filming cells at high resolution is phototoxicity (Carlton et al., 2010). We found that budding was delayed or blocked as light exposure was increased (Figure S1 available online). Reasoning that synchronizing cells would allow us to film for a shorter period prior to polarity establishment, thereby reducing total light exposure, we tested several synchrony protocols. We found that cells synchronized using hydroxyurea (HU) arrest/release were considerably less photosensitive than unsynchronized cells, allowing us to increase light exposure 4-fold without adverse effects (Figure S1). In diploid cells breaking symmetry, we detected two or three clusters of Bem1p-GFP in 28% of cells (n = 67) (Figure 1A and Movie S1). In all cases, either merging of nearby clusters (Johnson et al., 2011) or competition between clusters left only a single cluster at what became the incipient bud site (Figure 1A). Growth of the clusters and resolution to a single cluster occurred within 2 min on average (Figure 1B). Similar fast resolution of multiple clusters to one was detected in unsynchronized cells (Figure S1). Thus, formation of multiple polarization clusters is a common occurrence, and cells rapidly and efficiently resolve the clusters by merging and competition.
Figure 1. Dynamic Behaviors of Bem1p-GFP during Polarity Establishment.
Inverted images (so dark spots represent concentrations of Bem1p-GFP) from movies of cells breaking symmetry. Time in min:s. Scale bar, 2 µm. (Neck) The “old” neck signal in the attached daughter cell.
(A) Growth of multiple Bem1p clusters (numbered in the key at right) and resolution to a single cluster. t = 0 indicates the first detection of polarized signal.
(B) Coexistence time between the first detection of two to three faint clusters and the first frame showing a single cluster (n = 19).
(C) Oscillatory clustering of Bem1p. (Top) Cropped images of the polarization site at 45 s intervals. t = 0 is 45 s before the first detection of polarized signal, and trace ends at bud emergence. (Bottom) Amount of Bem1p-GFP in the cluster.
(D) Bem1p accumulation in eight other cells.
(E) Averaged plot from 36 cells aligned by the first peak.
(F) Power spectrum analysis of the 12 cells with the longest traces.
(G) Relocating cluster of Bem1p. An initial cluster (arrow) dispersed, and a new cluster appeared (arrowhead) at what became the bud site.
Error bars in (E) and (F) represent SEM. See also Figure S1.
Oscillations in Polarity Protein Concentration within Clusters
Faster filming also revealed an unexpected behavior: oscillations in Bem1p-GFP concentration at the polarization site (Figure 1C and Movie S1). Rapid initial clustering was followed by equally rapid dispersal of Bem1p-GFP. Subsequent behavior generally involved one or two further cycles of clustering and dispersal, often with lower amplitude, before stabilizing prior to bud emergence (Figure 1D). Similar though less dramatic oscillations occurred in unsynchronized cells (Figure S1). These data suggest that the initial polarization process is oscillatory but is then damped due to either intrinsic or extrinsic factors. Oscillations were also apparent in cells with multiple clusters, once the rapid resolution to a single cluster had occurred. Despite cell-to-cell variability, oscillation was still apparent in the averaged behavior of cells aligned using the first peak (Figure 1E). Power spectrum analysis suggested a dominant oscillation frequency of 0.22/min (Figure 1F).
In 15% of cells with a single cluster, the initial cluster disappeared entirely and a new cluster appeared at a different location (usually at what had been the mother-bud neck; Figure 1G). Similar “relocating” clusters were previously noted in cells overexpressing Bem1p-GFP (Howell et al., 2009). Both the oscillatory and relocation behaviors suggest that initial Bem1p clustering is antagonized by some form of negative feedback.
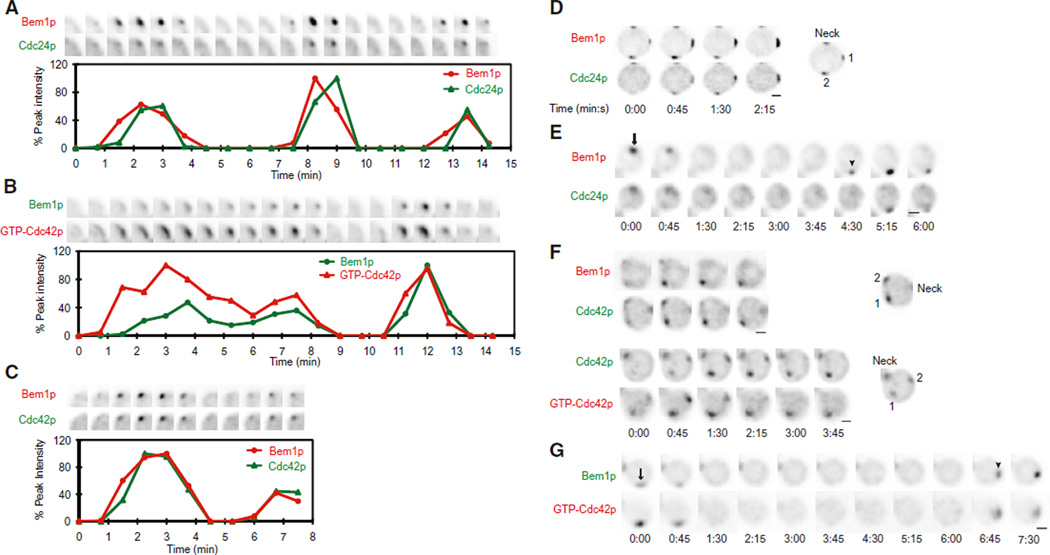
We next asked whether oscillatory and relocation behaviors were shared by other polarity regulators. The Cdc42p-directed GEF, Cdc24p, oscillated in parallel with Bem1p (Figure 2A and Movie S2). Fluorescent probes to detect GTP-Cdc42p (Tong et al., 2007) and total Cdc42p (Bi et al., 2000) also paralleled Bem1p behavior (Figures 2B and 2C and Movie S2). However, these probes were somewhat toxic (Figure S2), and the incidence of cells displaying high-amplitude oscillations was reduced when these probes were expressed. Cells displaying competing clusters, as well as relocation, were also observed with all probes (Figures 2D–2G), indicating that the core polarity regulators all concentrate, disperse, and reappear in concert.
Figure 2. Cdc42p and Cdc24p Cocluster and Disperse with Bem1p.
Two-color movies were processed as in Figure 1.
(A) Bem1p-tdTomato and Cdc24p-GFP oscillate in parallel. (Top) Cropped images of the polarization site. (Bottom) Quantification.
(B) Bem1p-GFP and GTP-Cdc42p (visualized using the PBD-tdTomato probe) oscillate in parallel.
(C) Bem1p-tdTomato and GFP-Cdc42p oscillate in parallel.
(D) Bem1p-tdTomato and Cdc24p-GFP cocluster and compete (clusters are numbered in the key at right).
(E) Bem1p-tdTomato and Cdc24p-GFP clusters relocate in parallel.
(F) Bem1p-tdTomato and GFP-Cdc42p (top), and GFP-Cdc42p and PBD-tdTomato (bottom) cocluster and compete.
(G) Bem1p-GFP and GTP-Cdc42p clusters relocate in parallel.
See also Figure S2.
A previous study on rsr1Δ mutants reported wave-like motion of broad crescents of polarity factors along the cell cortex (Ozbudak et al., 2005), unlike the focused, nonmotile, oscillatory clusters that we observed. We did detect cluster “movement” in G1-arrested rsr1Δ cells (data not shown) and in occasional cells expressing the GTP-Cdc42p-binding probe (Movie S3). In our filming conditions, all cells budded within 15 min of polarity establishment (Figure S1), but in the previous study cells took much longer (>50 min), raising the possibility that those cells were delayed in G1 (perhaps due to high light exposure and/or toxic fluorescent probes). We cannot rule out potential effects due to temperature or strain background differences, but we suspect that our filming conditions more accurately reflect physiological polarity establishment.
Oscillatory Polarization Is Not Due to Downstream F-Actin or Septin Action
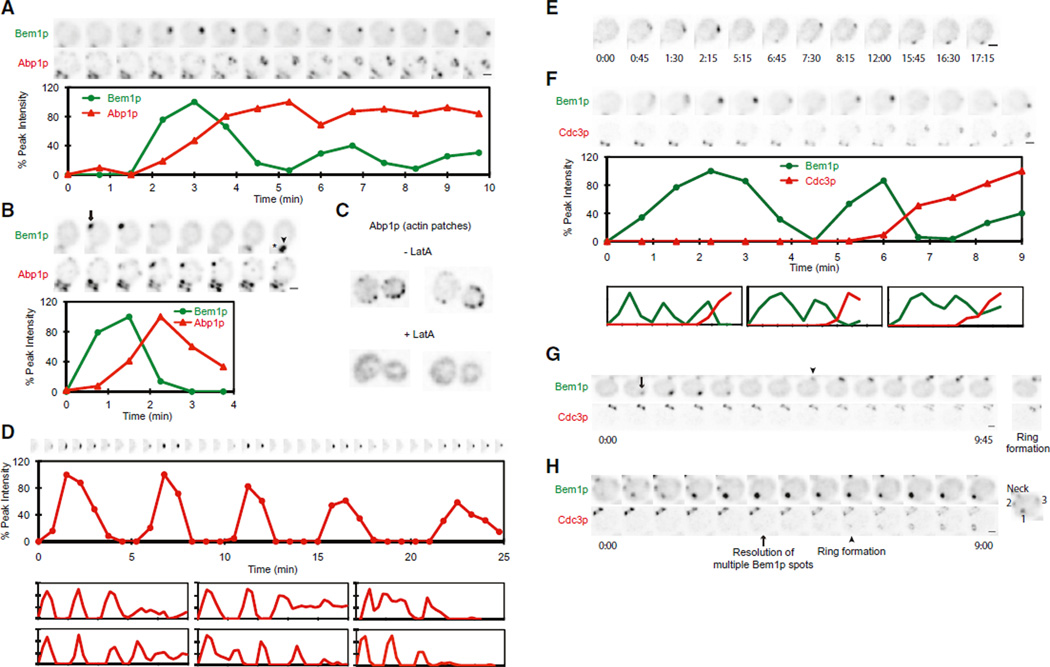
What could cause the oscillations in polarity factor localization? The initial clustering of Bem1p, Cdc24p, and Cdc42p is thought to occur via positive feedback (Goryachev and Pokhilko, 2008; Howell et al., 2009; Kozubowski et al., 2008), but positive feedback would not cause polarity proteins to disperse after they had clustered. Actin patches (labeled with Abp1p-mCherry) clustered at the polarization site ~90 s after Bem1p (Howell et al., 2009), correlating with Bem1p dispersal (Figures 3A and 3B and Movie S4). Actin-directed exocytosis could perturb polarity by inserting new membrane (Layton et al., 2011) or delivering Cdc42p-directed GAPs (Knaus et al., 2007; Ozbudak et al., 2005). Actin-mediated endocytosis could also disrupt polarity (Irazoqui et al., 2005; Yamamoto et al., 2010). However, treatment with Latrunculin A (Lat A) (Ayscough et al., 1997) to depolymerize actin did not abolish oscillatory Bem1p clustering or relocation (Figures 3C–3E and Movie S4). Similar behaviors may correspond to the “unstable” polarization of GFP-Cdc42p previously noted in Lat A-treated cells (Wedlich-Soldner et al., 2004). Indeed, Bem1p oscillations were prolonged in Lat A-treated compared to untreated cells. Thus, F-actin is not required for Bem1p oscillation but may contribute to damping such oscillation.
Figure 3. Actin and Septin Polarization Relative to Bem1p.
Movies were processed as in Figure 1.
(A and B) Abp1p-mCherry (marker for actin patches) clusters as Bem1p-GFP begins to disperse in oscillating (A) or relocating (B) clusters. (Asterisk) Old mother-bud neck.
(C) Actin patches are dispersed by Lat A.
(D) Bem1p-GFP oscillation is prolonged in Lat A. (Top) Cropped images of the polarization site. (Middle) Quantification of Bem1p-GFP. (Bottom) Six other examples.
(E) Relocation of Bem1p-GFP in Lat A-treated cell.
(F) Damping of Bem1p-GFP oscillation is correlated with septin (Cdc3p-mCherry) recruitment (plots as in D).
(G) Septin recruitment begins after relocation of Bem1p. No septin signal appears at the position of the first Bem1p-GFP cluster (arrow). Bem1p-GFP then relocates to the site of the old mother-bud neck (arrowhead) where remaining septins from cytokinesis obscure the new ring.
(H) Septin recruitment (arrowhead) begins after resolution of multicluster Bem1p intermediate (arrow).
The septins are filament-forming cytoskeletal proteins that are recruited by Cdc42p and form a ring surrounding the bud site (McMurray and Thorner, 2009; Oh and Bi, 2011). The septin Cdc3p accumulated ~4 min after initial Bem1p clustering (Chen et al., 2011), correlating with the damping of oscillation (Figure 3F and Movie S5). In cells that either relocated the Bem1p cluster or had multiple clusters, Cdc3p only began to accumulate after relocation or competition had occurred (Figures 3G and 3H and Movie S5). The timing of septin recruitment suggests that septins do not account for the initial Bem1p dynamics but may contribute to damping the oscillations. Because Lat A treatment can impair septin recruitment (Kadota et al., 2004; Kozubowski et al., 2005), the prolonged Bem1p oscillations in Lat A-treated cells may stem from delayed septin-mediated damping.
Interestingly, doubling the Cdc3p-mCherry gene dosage (in homozygous rather than heterozygous strains as used above) severely reduced the amplitude of Bem1p oscillations, consistent with a role of septins in damping such oscillation. However, exposure to ~20%–50% more light also reduced the oscillation amplitude. Thus, even a mildly increased stress from light and/or fluorescent probes partly obscures the oscillation. This low level of stress did not prevent polarization or timely budding, indicating that high-amplitude oscillation is not required for successful bud formation.
Including a Negative Feedback Loop in a Computational Model of Polarity Establishment Can Lead to Oscillatory Polarization
Our findings indicate that core polarity regulators initially cluster and disperse in an oscillatory manner. The oscillations do not require downstream F-actin and septin recruitment, although these cytoskeletal factors may well contribute to damping the oscillation. Thus, oscillation may be intrinsic to the core polarity machinery. Current models of polarity establishment invoke positive feedback loops to amplify small clusters of GTPCdc42p, but such models cannot explain oscillatory behavior. Instead, oscillatory phenomena in biology are generally due to the presence of negative feedback (Novák and Tyson, 2008). We therefore tested whether adding negative feedback to an existing mathematical model for polarity establishment in yeast would yield oscillatory clustering.
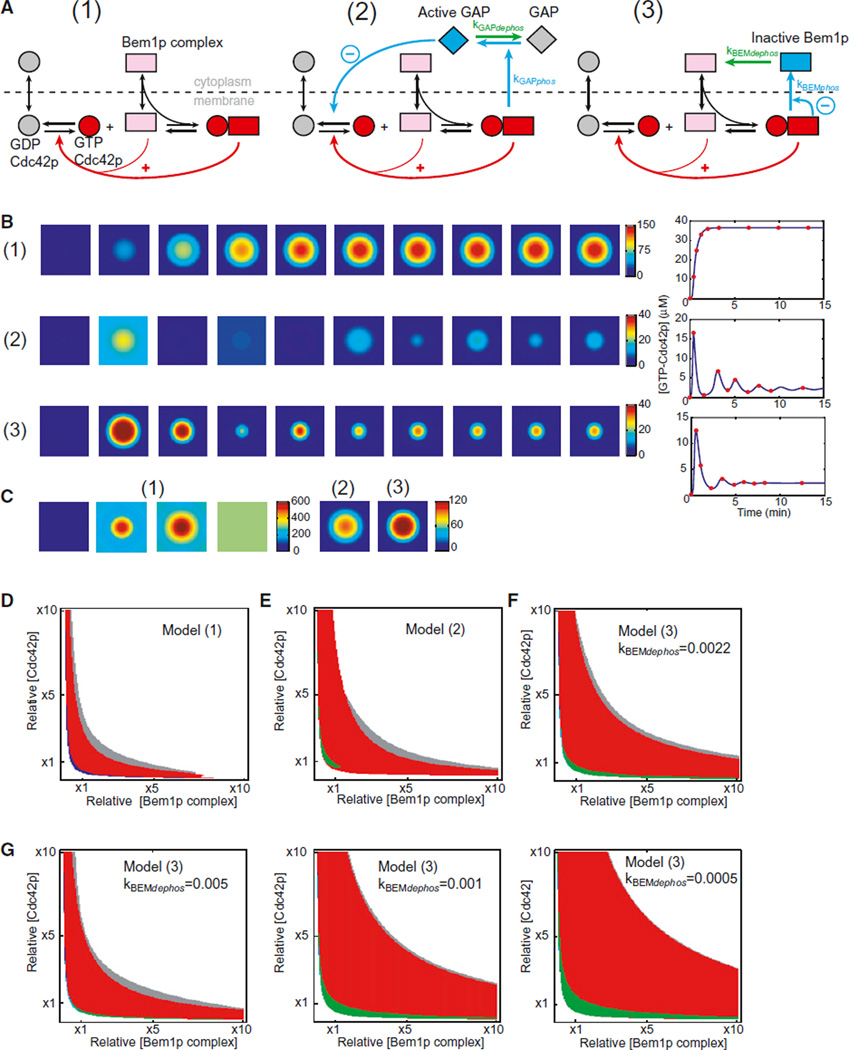
The starting model (1), developed based on genetic and biochemical data, incorporates Cdc42p, Rho-GDI, and a cytoplasmic Bem1p-scaffolded complex containing a PAK and the Cdc42p-directed GEF (Goryachev and Pokhilko, 2008; Howell et al., 2009; Kozubowski et al., 2008). We simplified the previous model by eliminating the Rho-GDI as a separate entity and subsuming its activity into the behavior of GDP-Cdc42p (Figure 4A). Positive feedback occurs because GTP-Cdc42p at the membrane can recruit the cytoplasmic Bem1p complex, which generates more GTP-Cdc42p from local GDP-Cdc42p by GEF-catalyzed exchange. We considered two simple negative feedback mechanisms, which operate by activating an inhibitor (2) or inhibiting an activator (3). In model 2, we assume that GTPCdc42p leads to the activation of a cytoplasmic Cdc42p-directed GAP, perhaps via PAK-mediated phosphorylation. Activated GAP antagonizes GTP-Cdc42p accumulation, dispersing the cluster. Dephosphorylation resets GAP activity, allowing another round of clustering. In model 3, we assume that GTP-Cdc42p leads to modification of the Bem1p complex, perhaps via PAK-mediated phosphorylation. Phosphorylated complex accumulates in the cytoplasm and cannot bind GTP-Cdc42p, reducing the amount of complex available for positive feedback and thereby allowing the cluster to disperse. Dephosphorylation resets the complex, allowing another round of clustering.
Figure 4. Negative Feedback Can Cause Oscillation and Improves Robustness of the Polarity Model.
(A) Diagram of the starting model (1) and two variants incorporating negative feedback via a Cdc42p-directed GAP (2) or the Bem1p complex (3). Positive feedback is indicated by red arrows and negative feedback by blue arrows. We assume that GTP-Cdc42p/PAK activates the GAP (blue GAP, model 2) or inactivates Bem1p complex components (blue complex, model 3). Phosphorylated proteins are then dephosphorylated in the cytoplasm (green arrows).
(B) Snapshots from simulations. The square represents a two-dimensional plasma membrane, and color indicates GTP-Cdc42p concentration. Snapshots are indicated by red dots in the tracings to the right, plotting GTP-Cdc42p concentration with time.
(C) Snapshots of simulations with 6.5-fold higher starting concentration of Cdc42p: model 1 spreads GTP-Cdc42p uniformly (left), whereas models 2 and 3 yield a polarized steady state (right).
(D) Behavior of model 1 at varying polarity protein concentration. (Red) Turing-unstable region: polarization occurs in response to small perturbation; (blue/gray) regions where both uniform and polarized states are stable: polarization occurs in response to large perturbation; (white) no polarized steady state.
(E) Behavior of model 2. (Green) Sustained oscillations.
(F) Behavior of model 3.
(G) Robustness, indicated by the area of red regions, varies with changing negative feedback parameters. Model 3 was analyzed at the indicated values of kBEMdephos.
See also Figures S3 and S4.
With appropriately tuned parameters, models 2 and 3 both exhibited damped oscillatory clustering (Figures 4B and S3). The oscillatory region of parameter space was significantly expanded in models with multistep negative feedback as compared to the simple one-step feedback loops modeled here (data not shown), presumably because additional steps introduce delay in negative feedback, facilitating oscillation. In the oscillatory region of parameter space, the dominant behavior predicted by the models involved periodic accumulation of GTP-Cdc42p over the entire cortex (Figure S3). However, addition of noise converted the spatially uniform oscillations into sustained oscillatory clustering. Moreover, simulations with noise exhibited both competing and relocating clusters (Figure S3). Thus, addition of negative feedback and noise can, in principle, promote all of the polarity dynamics observed in cells.
Negative Feedback Improves Robustness to Increased Cdc42p or GEF Concentrations
What advantage does a negative feedback loop impart to the polarity circuit? Intuitively, it would seem self-destructive to incorporate a mechanism that counteracts polarity establishment. Although the positive feedback-only mathematical model 1 is good at “growing” a GTP-Cdc42p cluster, we found that the ability of this model to polarize effectively was very sensitive to the concentrations of Cdc42p and the Bem1p-GEF-PAK complex. At higher concentrations, runaway growth of the cluster led to spreading of GTP-Cdc42p all over the plasma membrane, resulting in a uniform steady state (Figure 4C). This type of behavior is quite general for “substrate depletion” models in which limiting the amount of a polarity “substrate” is critical to prevent spreading of polarity factors over the entire cortex (Jilkine and Edelstein-Keshet, 2011). However, when negative feedback was incorporated, the models were able to produce a polarized Cdc42p profile despite the increase in polarity factor concentration (Figure 4C), suggesting that one benefit of negative feedback would be to make polarization more robust in the face of fluctuating concentrations of polarity factors.
To determine how the behavior of our models responded to a broader range of polarity protein concentrations, we performed linear stability analysis. The models displayed four main types of steady-state behaviors, color-coded in Figures 4D–4F. In the red region, the homogeneous state was unstable to small spatial perturbations (Turing instability), and with any noise, the models evolved to a polarized steady state. In the white region, the homogeneous state was stable, whereas polarized states were unstable and evolved to the unpolarized steady state. In the blue and gray regions, there was coexistence of uniform and polarized steady states. Polarization could not be triggered by small perturbations, but a sufficiently large polarizing perturbation would induce transition from the uniform to the polarized state. In the green region, in negative feedback-containing models, small perturbations could induce sustained oscillation (Figure S3). The simulations in Figure 4B are derived from the lower-left part of the red region. In other parts of the red region (i.e., with increased amounts of Cdc42p and/or the Bem1p complex), oscillation was muted (Figure S3).
For model 1, only a narrow slice of parameter space (including the previously employed Cdc42p and Bem1p complex concentrations, indicated as ×1) developed a polarized steady state (Figure 4D). Inclusion of either GAP-mediated (Figure 4E) or GEF-mediated (Figure 4F) negative feedback expanded the parameter space, yielding effective Cdc42p polarization (red region). Thus, inclusion of negative feedback makes the models more robust to changes in component concentrations.
The prediction that negative feedback would increase robustness was not sensitive to exact parameter values (Figure S4). However, the degree of increased robustness was dependent on the negative feedback parameters. For example, decreasing the Bem1p complex dephosphorylation rate in model 3 progressively broadened the polarization-competent red region (Figure 4G). Because similar predictions were obtained by modeling quite different mechanisms for negative feedback, it is likely that negative feedback would improve robustness regardless of the precise feedback mechanism.
Polarity Establishment Is Robust to Increased Cdc42p or GEF Concentration
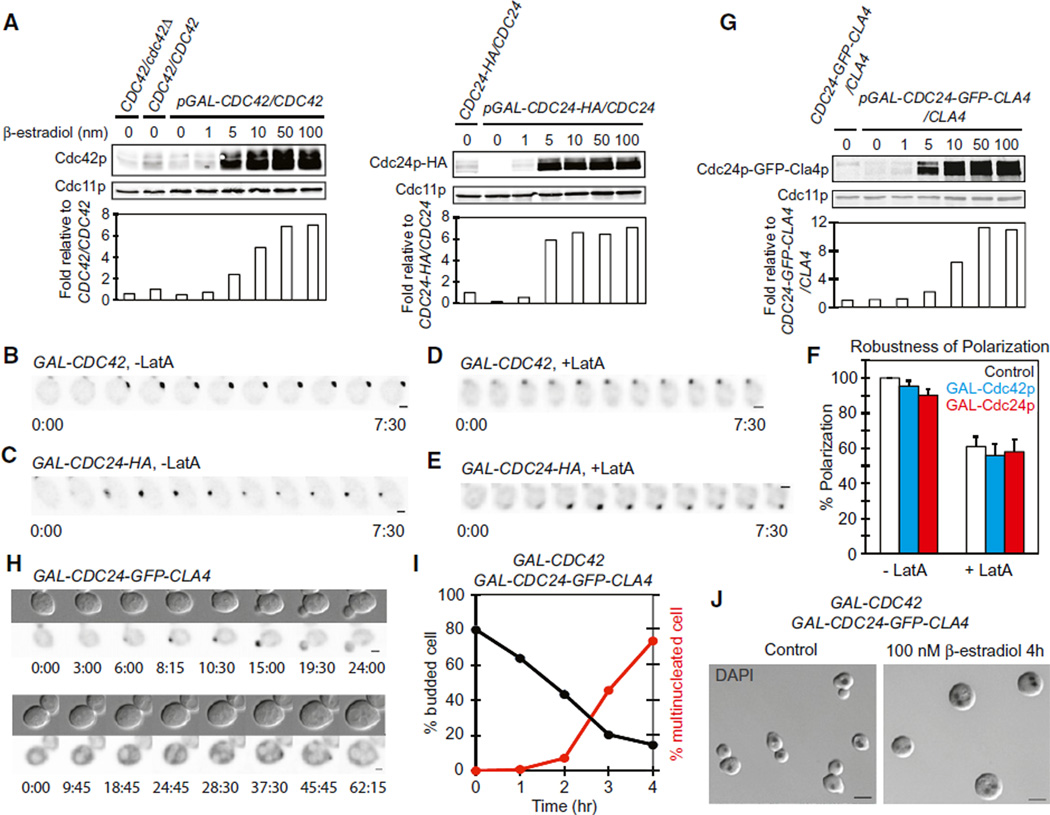
Is biological polarity establishment indeed robust to increases in Cdc42p or Bem1p complex concentration? To test this, we used a galactose-regulated promoter to overexpress either Cdc42p or its GEF, Cdc24p. Because cells are more photosensitive when grown in galactose (data not shown), we used an artificial transcription factor that allows induction by β-estradiol in glucose-containing media (Takahashi and Pryciak, 2008). Filming of Bem1p-GFP revealed robust polarization even following ~7-fold overexpression of Cdc42p or Cdc24p (Figures 5A–5C and Movie S6). A previous study reported that Cdc42p overexpression blocked polarity establishment in cells lacking F-actin (Altschuler et al., 2008), but we found that polarization occurred with comparable efficiency whether or not cells overexpressed Cdc42p or Cdc24p, even in cells treated with Lat A (Figures 5D–5F). Thus, polarity establishment in yeast is robust to increases in Cdc42p or Cdc24p concentration, to a degree that is at odds with the predictions of the positive feedback-only mathematical model.
Figure 5. Polarization Is Robust to Overexpression of Cdc42p or Cdc24p.
(A) Western blot and quantification of Cdc42p (left) and Cdc24p-HA (right) in response to b-estradiol (subsequent panels used 100 nM).
(B–E) Bem1p-GFP polarization in representative cells overexpressing Cdc42p (B and D) or Cdc24p-HA (C and E), in the absence (B and C) or presence (D and E) of Lat A.
(F) Quantification of the percentage of cells that polarized in control (white), Cdc42p-overexpressing (blue), or Cdc24p-HA-overexpressing (red) strains in the absence or presence of Lat A (mean ± SEM).
(G) Western blot and quantification of Cdc24p-GFP-Cla4p fusion. 1× indicates expression level from the CDC24 promoter.
(H) Cdc24p-GFP-Cla4p distribution in cells that do (top) or do not (bottom) polarize. Nuclei and vacuoles exclude the protein and appear light.
(I) Plot of budding index and frequency of multinucleate cells following induction of both Cdc24p-GFP-Cla4p and Cdc42p.
(J) Representative cells from (I) after 0 hr (left) or 4 hr (right) of induction. Overlay of inverted DAPI staining and DIC images. Scale bars, 5 µm.
Effect of Overexpressing a Cdc24p-Cla4p Fusion Protein
It is unclear to what degree overexpression of Cdc24p would raise the level of the full Bem1p complex, especially if (as posited in model 3) elements of the complex are subject to negative feedback. To circumvent potential controls on complex assembly, we expressed a Cdc24p-Cla4p fusion protein that mimics the full complex (Kozubowski et al., 2008) (Figure 5G). As previously reported, this fusion caused hyperpolarized growth in budded cells (Kozubowski et al., 2008), but here, we focus on its effects on initial polarity establishment. Time-lapse analysis indicated that a majority of cells expressing the fusion could polarize, but some cells were delayed in polarization and a few cells underwent a full cell cycle without establishing polarity (Figure 5H and Movie S7). Overexpression of Cdc42p together with the Cdc24p-Cla4p fusion blocked polarity establishment in a large majority of cells, leading to the accumulation of large, unbudded, multinucleate cells (Figures 5I and 5J). The simplest interpretation of these findings is that combined expression of Cdc42p and a fusion protein that mimics the full Bem1p complex drives the system into the “white” regime of parameter space, where GTP-Cdc42p spreads throughout the cortex.
Rapid Competition between Clusters and Buffering of GTP-Cdc42p
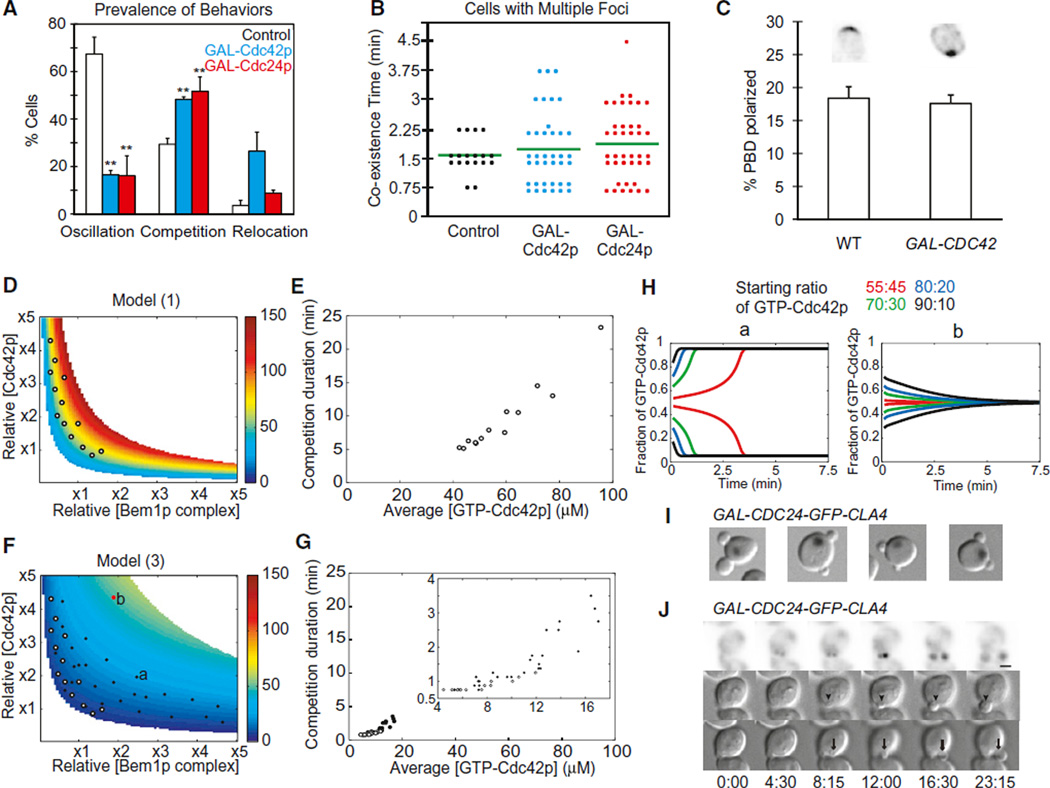
Overexpression of Cdc42p or Cdc24p altered the kinetics of polarization, damping oscillation and (in cells overexpressing Cdc42p) increasing the frequency of relocation (Figure 6A and Movie S6). Multiple clusters were more common in overexpressors (Figure 6A), but resolution to a single cluster still occurred rapidly (Figure 6B). This was surprising because we expected that cells overexpressing Cdc42p or Cdc24p would build larger clusters, which would then take longer to dismantle during competition (Howell et al., 2009). We assessed the amount of GTP-Cdc42p in the polarized clusters using the fluorescent GTP-Cdc42p-binding reporter (Tong et al., 2007). Despite considerable cell-to-cell variation in the total amount of reporter in the cell, a relatively consistent ~19% of the probe was polarized in late-G1 wild-type cells (Figure 6C). Strikingly, a similar fraction of the probe was polarized in cells overexpressing Cdc42p (Figure 6C), suggesting that cells are able to buffer the polarized cluster against Cdc42p overexpression, explaining why competition remained rapid.
Figure 6. Negative Feedback Buffers the Accumulation of GTP-Cdc42p and Can Accelerate or Abolish Competition between Clusters.
(A) Prevalence of high-amplitude oscillation (left), multicluster intermediates (middle), and relocating clusters (right) in control (white), Cdc42p-overexpressing (blue), or Cdc24p-HA-overexpressing (red) strains (mean ± SEM). **p < 0.01 by two-tailed t test; significant difference between overexpressors and controls.
(B) Quantification of the time taken to resolve multicluster intermediates.
(C) The fraction of the GTP-Cdc42p-binding probe (mean ± SEM) that is polarized in late G1 cells is similar with (right) or without (left) Cdc42p overexpression. Representative images are shown at top.
(D) Steady-state GTP-Cdc42p levels in model 1 change rapidly as component concentrations are increased. Color indicates steady-state GTP-Cdc42p concentration (calculated from the spatially uniform situation) in the parameter space displaying Turing instability. Circles indicate points used for simulations in (E).
(E) Correlation between GTP-Cdc42p concentration and the time taken to resolve competition. Each symbol represents a simulation, at parameter values from the circles in (D), of the competition between two unequal clusters (ratio 55:45), plotting the time taken to resolve competition (y axis) and the average GTP-Cdc42p concentration of the two-cluster starting state (x axis).
(F) Steady-state GTP-Cdc42p levels in model 3 are buffered against increases in component concentrations. Symbols indicate points used for simulations in (G). White circles are as in (D), whereas black symbols are in the expanded polarity region. Symbols labeled “a” and “b” indicate parameters used in (H).
(G) Negative feedback maintains rapid competition in a broad range of parameter space. Kinetics of competition between clusters (as in E), at parameter values indicated in (F). (Inset) Expanded view of lower-left quadrant.
(H) Negative feedback can lead to equalization of clusters instead of competition between clusters at high levels of polarity proteins. Simulations are as described in (E), with the indicated starting ratios between unequal clusters, using the parameter values from the symbols labeled “a” and “b” in (F).
(I) Examples of two-budded cells from a culture induced to express Cdc24p-Cla4p fusion for 4 hr. Overlay of inverted DAPI staining and DIC images.
(J) Simultaneous growth of two buds (arrow and arrowhead in different DIC z planes) and polarization of Cdc24p-Cla4p fusion to both buds. See also Figure S5.
To assess how models with and without negative feedback would impact competition times, we simulated competition between two clusters that started out with a 55:45 ratio of Cdc42p content. Model 1 predicted that elevating either Cdc42p or Bem1p complex concentration would elevate the steady-state level of GTP-Cdc42p (Figure 6D) and cause correspondingly slower resolution of competition (Figure 6E). However, model 3 predicted that negative feedback would buffer the GTP-Cdc42p level (Figure 6F) and that competition would remain rapid (Figure 6G), as observed experimentally (Figures 6B and 6C). Thus, an added advantage of negative feedback is that, when multiple clusters form, they compete rapidly even if component concentrations are increased.
The buffering effect of negative feedback significantly reduced competition times in the majority of the simulations, producing coexistence times that are consistent with the experimental observations (Figure 6G). However, this was not universally true in all parts of parameter space. Whereas in model 1, a larger cluster always (eventually) outcompeted and eliminated a smaller cluster, in model 3, competition failed at sufficiently high Cdc42p and Bem1p complex concentrations. With these parameters, the clusters equalized rather than competing (Figure 6H), and simulations evolved to a stable steady state containing two equal clusters (Figure S5). In cells, this would presumably lead to formation of two buds. Interestingly, occasional cells (~1%) expressing the Cdc24p-Cla4p fusion did make two buds (Figure 6I). Two-budded cells polarized stably to two sites (Figure 6J and Movie S7), though sometimes one site disappeared, leading to the development of unequal-sized buds.
DISCUSSION
Negative Feedback during Polarity Establishment
Filming of symmetry-breaking polarization at high resolution under low-light imaging conditions revealed that clusters of polarity factors congregated rapidly (often within 45 s) and then unexpectedly dispersed, subsequently reforming and dispersing up to three more times before stabilizing (Figures 1 and 2). The dispersal occurred even in cells that only displayed a single polarity cluster, indicating that dispersal is not due to competition between clusters. In some cases, the dispersal appeared to be complete, and cells went on to assemble a new polarity cluster elsewhere (“relocation”). Oscillatory clustering was not predicted by existing models of polarity establishment and suggests that positive feedback-mediated initial polarization is rapidly antagonized by a negative feedback loop. Mathematical modeling suggested that adding a negative feedback loop to a previous model for polarity establishment could lead to oscillatory clustering, and different negative feedback mechanisms (acting either through a Cdc42p-directed GAP or GEF) produced qualitatively similar results (Figure 4). The mechanism of negative feedback in cells remains to be determined.
The negative feedback models predicted that oscillation would be muted when the concentrations of polarity factors were increased (Figure S3), and this was confirmed experimentally (Figure 6A). The sustained oscillations predicted by the deterministic models consisted mainly of spatially uniform accumulation of GTP-Cdc42p all over the cortex followed by GTP hydrolysis and return of Cdc42p to the cytoplasm. However, addition of noise eliminated such uniform oscillations and instead produced oscillatory clustering (Figure S3). Noise-containing simulations exhibited rapid multicluster competition followed by oscillation, as well as relocation of clusters. Thus, in appropriate parameter regimes, models that incorporate negative feedback and noise in addition to the previously modeled positive feedback can reproduce all of the polarity dynamics that we observed in cells.
Negative feedback-containing models produced either sustained or intrinsically damped oscillations, depending on the concentrations of polarity factors (Figure S3). However, in cells, the oscillatory clustering was always damped. Damping was correlated with the arrival of septins at the polarization site and was delayed in the absence of F-actin (a condition that delays septin assembly) (Figure 3). Thus, it may be that the core polarity machinery has the capacity to produce sustained oscillatory clustering and that downstream cytoskeletal factors act to dampen the oscillation.
It is unclear what advantage could stem from high-amplitude oscillations in polarity factor concentration. When cells were exposed to more stressful imaging conditions, they exhibited lower-amplitude oscillation, as did cells that were filmed without the photoprotective hydroxyurea pretreatment (Howell et al., 2009). Given the sensitivity of the behavior to filming conditions and component concentrations, it seems unlikely that such oscillation is important in and of itself. Instead, oscillation may have arisen as a byproduct of homeostatic negative feedback. As discussed below, adding negative feedback to the polarity model improves its robustness. Interestingly, robustness could be further improved by lowering the rates at which a negative feedback-modified GEF or GAP returned to its baseline state. Lowering those rates introduces a delay (as the modified GEF/GAP accumulates rapidly but takes time to return to its basal state), which, in turn, favors oscillatory behavior. Thus, oscillations might arise as a byproduct of a negative feedback loop that is present to optimize robustness.
Oscillations in polarized growth (after polarity establishment) have been particularly well studied in plants (Hepler et al., 2001), in which the oscillatory growth of pollen tubes is thought to involve interlinked positive and negative feedback loops (Yan et al., 2009). It is unclear whether oscillation per se is advantageous, as pollen tubes switch from prolonged continuous growth to oscillatory growth without overt changes in overall elongation speed or morphology (Feijó et al., 2001). Thus, the use of negative feedback to promote homeostasis or robustness may lead in some cases to the appearance of unselected oscillations, which may or may not be beneficial in themselves (Cheong and Levchenko, 2010; Feijó et al., 2001).
Robustness of Polarity Establishment
Although capable of polarity establishment, a model that only contains positive feedback is fragile in that increasing concentrations of polarity factors quickly overwhelm the system, causing GTP-Cdc42p to spread all over the cortex. A benefit of negative feedback is improved robustness to such changes: the negative feedback prevents runaway accumulation of GTPCdc42p, so the model retains the ability to polarize over a much wider range of polarity factor concentrations. Similar robustness predictions were obtained regardless of the modeled feedback mechanism or specific parameters (Figures 4 and S4). Thus, consistent with the well-known homeostatic influence of negative feedback in well-mixed systems (Brandman and Meyer, 2008), negative feedback confers improved robustness regardless of the precise feedback mechanism.
The modeling results prompted us to test whether yeast polarization is indeed robust to increased levels of polarity factors, and we found that cells polarized just as efficiently when Cdc42p or Cdc24p were overexpressed. The robustness that we observed is consistent with older reports that Cdc42p overexpression is tolerated by yeast (Ziman and Johnson, 1994) but is contrary to the conclusion from a recent study suggesting that Cdc42p overexpression blocked polarity establishment in cells lacking F-actin (Altschuler et al., 2008). The apparent difference between those results and ours may stem from the fact that we overproduced wild-type Cdc42p whereas they used a myc-GFP-Cdc42p construct that is somewhat toxic (Figure S2). In addition, they used the same probe to score polarization, potentially making it difficult to detect a polarized signal above the high unpolarized background in overexpressing cells. We conclude that the yeast polarity establishment circuit is robust to increases in polarity factor concentration, even in cells lacking F-actin, and that robustness is likely to be conferred by negative feedback.
With the one-step negative feedback models that we considered, oscillations occur near the lower bound of the polarization-competent parameter regime, perhaps suggesting that cells sit near this boundary and would be very sensitive to any decrease in Cdc42p concentration. However, a 2-fold reduction in Cdc42p level (in hemizygous diploids) does not prevent polarization. Adding extra steps to lengthen the negative feedback loop can dramatically expand the region of parameter space capable of sustaining oscillations (data not shown) so that cells displaying oscillations would be robust to both increases and decreases in polarity factor concentrations.
Competition between Polarity Clusters
A long-standing question in the polarity field concerns why cells develop one and only one “front.” We recently suggested that, in yeast, positive feedback could give rise to more than one polarity cluster, but then the clusters would compete with each other so that a single winner would emerge (Howell et al., 2009). Alternatively, the small absolute numbers of a limiting polarity factor might make it unlikely that more than one cluster could develop (Altschuler et al., 2008). With previous filming protocols, it was difficult to detect the multicluster intermediates predicted by the competition hypothesis, but with improved imaging, we now document such intermediates in ~25% of cells breaking symmetry (rising to ~50% upon overexpression of Cdc24p or Cdc42p). These numbers represent a lower bound for the real incidence of such intermediates, as technical issues may prevent us from detecting small and/or short-lived clusters. Thus, multicluster intermediates are very frequent, and competition between polarity clusters is critical to prevent the development of more than one front.
Multicluster intermediates were short-lived, generally resolving to a single cluster within 2 min. Surprisingly, competition was similarly rapid even in cells overexpressing Cdc24p or Cdc42p, which were expected to build clusters containing more polarity proteins. As larger clusters take longer to dismantle during competition, it should take considerably longer to resolve the competition in favor of a single winner. Negative feedback can buffer the accumulation of polarity factors in clusters so that overexpression need not significantly increase the amount of Cdc42p or other factors in the cluster, explaining why competition did not take much longer in overexpressing cells than in controls. Thus, a second benefit of negative feedback in the polarity circuit is that, when more than one cluster forms, competition between clusters is more rapid.
The competition between clusters predicted by the modeling is biased such that larger clusters outcompete smaller ones (Howell et al., 2009). However, we detected rare instances in which a smaller cluster appeared to win (e.g., Figure 1A, cell 3). Negative feedback could, in principle, explain this observation if such feedback includes a partly localized component. That is, growth of a cluster may induce a negative feedback that is slightly stronger in the vicinity of that cluster than it is in the rest of the cell. If that were the case, then an initially stronger cluster might self-destruct, whereas a later-emerging distant cluster succeeds.
An unexpected prediction from mathematical modeling of polarity circuits with negative feedback was that, at high Cdc42p and Bem1p complex concentration, competition should fail to resolve polarity clusters. Instead, two clusters would tend to equalize so that each contains the same amount of polarity proteins. Presumably, this would lead to the formation of two buds in yeast, perhaps explaining the observation of occasional two-budded cells in strains overexpressing Bem1p (Howell et al., 2009) or a Cdc24p-Cla4p fusion (Figure 6). However, such cells might also arise if competition were drastically slowed (Howell et al., 2009).
We speculate that the cluster equalization predicted by the model at high polarity factor levels may be relevant to a currently unexplained behavior called tip-splitting or apical branching that occurs in a variety of fungi (Harris, 2008; Riquelme and Bartnicki-Garcia, 2004) and is particularly well studied in Ashbya gossypii (Knechtle et al., 2003). A. gossypii is an evolutionarily close relative of S. cerevisiae that uses related proteins to establish and maintain polarity but grows as a multinucleate filamentous fungus (Dietrich et al., 2004). As the hypha grows, accumulating more polarity factors, tip growth accelerates until, at some point, the polarization cluster expands (Schmitz et al., 2006) and splits into two equal clusters, generating a Y-shaped branch in the hypha. At the time of tip splitting, there are two neighboring polarity clusters that clearly do not compete with each other. A polarity circuit with built-in negative feedback may explain how these cells can sustain two equal clusters in close proximity and why they do not do so until a large size has been reached.
In conclusion, the oscillatory polarization observed under improved filming conditions reveals that the yeast polarity establishment circuit contains negative feedback. Modeling suggests that negative feedback confers robustness as well as the capacity for rapid competition between polarity clusters. The presence of negative feedback also raises the possibility that, in appropriate circumstances, the system could be tuned to produce several polarity axes, which may be required for generating the more complex morphologies observed in other eukaryotes.
EXPERIMENTAL PROCEDURES
Live Cell Microscopy
Prior to imaging, cells were grown in synthetic medium (MP Biomedicals) with dextrose. Cells were mounted on a slab composed of medium solidified with 2% agarose (Denville Scientific, Inc.). Images were acquired with an Axio Observer.Z1 (Zeiss) with outer environmental chamber (set to 30°C unless otherwise stated), a X-CITE 120XL metal halide fluorescence light source, and a 100×/1.46 Plan Apochromat oil-immersion objective controlled by MetaMorph software (Universal Imaging) (http://microscopy.duke.edu). Images were captured using a QuantEM backthinned EM-CCD camera (Photometrics). The fluorescence light source was used at ~50% maximal output, and a 2% ND filter was placed in the light path. An EM-Gain setting of 750 was used for the EM-CCD camera. Exposures were 250 ms (Bem1p-GFP, Bem1-tdTomato, or Cdc24p-GFP), 150 ms (Abp1p-mCherry), or 100 ms (Cdc3p-mCherry).
Deconvolution and Image Analysis
Images were deconvolved using Huygens Essential software (Scientific Volume Imaging). The classic maximum likelihood estimation and predicted point spread function method with signal-to-noise ratio 10 was used with a constant background across all images from the same day. The output format was 16-bit, unscaled images to enable comparison of pixel values. To detect polarity foci in different focal planes, maximum intensity projections were constructed and scored visually for the presence of more than one focus. The coexistence time is the interval between the first frame in which more than one spot was detected and the frame when only one spot was detected. Quantification of Bem1p-GFP intensities used Volocity (Improvision). A threshold was set that would only select the polarized signal, and the summed, polarized intensity was recorded. Changes in intensity are reported as percent of maximum for that cell. Images were processed for presentation using Metamorph and Photoshop (Adobe).
To quantitate polarization efficiency, we analyzed 1.5 hr movies of cells released from HU treatment as follows: for each mother-daughter pair that went through cytokinesis in the first 30 min (as indicated by neck localization of Bem1p-GFP), we scored the progeny cells as polarized if and only if Bem1p-GFP polarized within the duration of the movie. For Lat A-treated cells, we also scored single unbudded cells. As some Lat A-treated cells show transient Bem1p-GFP polarization, we only counted as polarized those cells in which polarized signal lasted > 15 min. Cytokinesis is often defective in Lat A-treated cells, largely accounting for the decrease in the scored efficiency of polarization.
To quantitate the frequency of high-amplitude oscillation, we set an arbitrary cutoff such that a cell was not scored unless the summed Bem1p-GFP intensity decreased to below 20% of the first peak before rising again.
Hydroxyurea Pretreatment
Cells growing in synthetic complete medium at 30°C were arrested with 200 mM HU (Sigma) for 3 hr, washed, released into fresh medium for 65 min, harvested, and mounted for live-cell microscopy. Due to the temperature sensitivity of the GFP-Cdc42p-containing strains, they were grown at 24°C, necessitating a 4 hr HU arrest and 2 hr release.
Latrunculin and β-Estradiol Treatment
Expression from the GAL1 promoter was induced by addition of β-estradiol. For HU-arrested cells, β-estradiol was added at the same time as the HU and was maintained in the subsequent media and filming slabs. Because Lat A treatment led to defective cytokinesis and frequent lysis of cells pretreated with HU, we did not synchronize the cells to be treated with Lat A. Instead, b-estradiol was added to exponentially growing cells 4 hr before cells were harvested and resuspended in medium with 200 µM Lat A (Invitrogen). Cells were then mounted on slabs containing both β-estradiol and Lat A for filming.
Supplementary Material
ACKNOWLEDGMENTS
We thank Nick Buchler, Beverle Errede, Steve Haase, Joe Heitman, Sally Kornbluth, and members of the Lew lab for stimulating discussions and comments on the manuscript. Thanks to Sam Johnson and the Light Microscopy Core Facility at Duke University for making experiments possible and for providing assistance. We thank Rong Li and Erfei Bi for providing Cdc42p probes and Peter Pryciak for the estrogen-responsive Gal4p. This work was supported by NIH grants GM79271 and GM84071 (T.C.E.) and GM62300 (D.J.L.), as well as a supplement for collaborative science (D.J.L. and T.C.E.).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures, five figures, three tables, and seven movies and can be found with this article online at doi:10.1016/j.cell.2012.03.012.
REFERENCES
- Altschuler SJ, Angenent SB, Wang Y, Wu LF. On the spontaneous emergence of cell polarity. Nature. 2008;454:886–889. doi: 10.1038/nature07119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayscough KR, Stryker J, Pokala N, Sanders M, Crews P, Drubin DG. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J. Cell Biol. 1997;137:399–416. doi: 10.1083/jcb.137.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender A, Pringle JR. Multicopy suppression of the cdc24 budding defect in yeast by CDC42 and three newly identified genes including the ras-related gene RSR1. Proc. Natl. Acad. Sci. USA. 1989;86:9976–9980. doi: 10.1073/pnas.86.24.9976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E, Chiavetta JB, Chen H, Chen GC, Chan CS, Pringle JR. Identification of novel, evolutionarily conserved Cdc42p-interacting proteins and of redundant pathways linking Cdc24p and Cdc42p to actin polarization in yeast. Mol. Biol. Cell. 2000;11:773–793. doi: 10.1091/mbc.11.2.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose I, Irazoqui JE, Moskow JJ, Bardes ES, Zyla TR, Lew DJ. Assembly of scaffold-mediated complexes containing Cdc42p, the exchange factor Cdc24p, and the effector Cla4p required for cell cycle-regulated phosphorylation of Cdc24p. J. Biol. Chem. 2001;276:7176–7186. doi: 10.1074/jbc.M010546200. [DOI] [PubMed] [Google Scholar]
- Brandman O, Meyer T. Feedback loops shape cellular signals in space and time. Science. 2008;322:390–395. doi: 10.1126/science.1160617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton PM, Boulanger J, Kervrann C, Sibarita JB, Salamero J, Gordon-Messer S, Bressan D, Haber JE, Haase S, Shao L, et al. Fast live simultaneous multiwavelength four-dimensional optical microscopy. Proc. Natl. Acad. Sci. USA. 2010;107:16016–16022. doi: 10.1073/pnas.1004037107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chant J, Herskowitz I. Genetic control of bud site selection in yeast by a set of gene products that constitute a morphogenetic pathway. Cell. 1991;65:1203–1212. doi: 10.1016/0092-8674(91)90015-q. [DOI] [PubMed] [Google Scholar]
- Chen H, Howell AS, Robeson A, Lew DJ. Dynamics of septin ring and collar formation in Saccharomyces cerevisiae. Biol. Chem. 2011;392:689–697. doi: 10.1515/BC.2011.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong R, Levchenko A. Oscillatory signaling processes: the how, the why and the where. Curr. Opin. Genet. Dev. 2010;20:665–669. doi: 10.1016/j.gde.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich FS, Voegeli S, Brachat S, Lerch A, Gates K, Steiner S, Mohr C, Pöhlmann R, Luedi P, Choi S, et al. The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science. 2004;304:304–307. doi: 10.1126/science.1095781. [DOI] [PubMed] [Google Scholar]
- Feijó JA, Sainhas J, Holdaway-Clarke T, Cordeiro MS, Kunkel JG, Hepler PK. Cellular oscillations and the regulation of growth: the pollen tube paradigm. Bioessays. 2001;23:86–94. doi: 10.1002/1521-1878(200101)23:1<86::AID-BIES1011>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Goryachev AB, Pokhilko AV. Dynamics of Cdc42 network embodies a Turing-type mechanism of yeast cell polarity. FEBS Lett. 2008;582:1437–1443. doi: 10.1016/j.febslet.2008.03.029. [DOI] [PubMed] [Google Scholar]
- Gulli MP, Jaquenoud M, Shimada Y, Niederhäuser G, Wiget P, Peter M. Phosphorylation of the Cdc42 exchange factor Cdc24 by the PAK-like kinase Cla4 may regulate polarized growth in yeast. Mol. Cell. 2000;6:1155–1167. doi: 10.1016/s1097-2765(00)00113-1. [DOI] [PubMed] [Google Scholar]
- Harris SD. Branching of fungal hyphae: regulation, mechanisms and comparison with other branching systems. Mycologia. 2008;100:823–832. doi: 10.3852/08-177. [DOI] [PubMed] [Google Scholar]
- Hepler PK, Vidali L, Cheung AY. Polarized cell growth in higher plants. Annu. Rev. Cell Dev. Biol. 2001;17:159–187. doi: 10.1146/annurev.cellbio.17.1.159. [DOI] [PubMed] [Google Scholar]
- Howell AS, Savage NS, Johnson SA, Bose I, Wagner AW, Zyla TR, Nijhout HF, Reed MC, Goryachev AB, Lew DJ. Singularity in polarization: rewiring yeast cells to make two buds. Cell. 2009;139:731–743. doi: 10.1016/j.cell.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irazoqui JE, Gladfelter AS, Lew DJ. Scaffold-mediated symmetry breaking by Cdc42p. Nat. Cell Biol. 2003;5:1062–1070. doi: 10.1038/ncb1068. [DOI] [PubMed] [Google Scholar]
- Irazoqui JE, Howell AS, Theesfeld CL, Lew DJ. Opposing roles for actin in Cdc42p polarization. Mol. Biol. Cell. 2005;16:1296–1304. doi: 10.1091/mbc.E04-05-0430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jilkine A, Edelstein-Keshet L. A comparison of mathematical models for polarization of single eukaryotic cells in response to guided cues. PLoS Comput. Biol. 2011;7:e1001121. doi: 10.1371/journal.pcbi.1001121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JM, Jin M, Lew DJ. Symmetry breaking and the establishment of cell polarity in budding yeast. Curr. Opin. Genet. Dev. 2011;21:740–746. doi: 10.1016/j.gde.2011.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadota J, Yamamoto T, Yoshiuchi S, Bi E, Tanaka K. Septin ring assembly requires concerted action of polarisome components, a PAK kinase Cla4p, and the actin cytoskeleton in Saccharomyces cerevisiae. Mol. Biol. Cell. 2004;15:5329–5345. doi: 10.1091/mbc.E04-03-0254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knaus M, Pelli-Gulli MP, van Drogen F, Springer S, Jaquenoud M, Peter M. Phosphorylation of Bem2p and Bem3p may contribute to local activation of Cdc42p at bud emergence. EMBO J. 2007;26:4501–4513. doi: 10.1038/sj.emboj.7601873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knechtle P, Dietrich F, Philippsen P. Maximal polar growth potential depends on the polarisome component AgSpa2 in the filamentous fungus Ashbya gossypii. Mol. Biol. Cell. 2003;14:4140–4154. doi: 10.1091/mbc.E03-03-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozubowski L, Larson JR, Tatchell K. Role of the septin ring in the asymmetric localization of proteins at the mother-bud neck in Saccharomyces cerevisiae. Mol. Biol. Cell. 2005;16:3455–3466. doi: 10.1091/mbc.E04-09-0764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozubowski L, Saito K, Johnson JM, Howell AS, Zyla TR, Lew DJ. Symmetry-breaking polarization driven by a Cdc42p GEF-PAK complex. Curr. Biol. 2008;18:1719–1726. doi: 10.1016/j.cub.2008.09.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layton AT, Savage NS, Howell AS, Carroll SY, Drubin DG, Lew DJ. Modeling vesicle traffic reveals unexpected consequences for Cdc42p-mediated polarity establishment. Curr. Biol. 2011;21:184–194. doi: 10.1016/j.cub.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurray MA, Thorner J. Septins: molecular partitioning and the generation of cellular asymmetry. Cell Div. 2009;4:18. doi: 10.1186/1747-1028-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novák B, Tyson JJ. Design principles of biochemical oscillators. Nat. Rev. Mol. Cell Biol. 2008;9:981–991. doi: 10.1038/nrm2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh Y, Bi E. Septin structure and function in yeast and beyond. Trends Cell Biol. 2011;21:141–148. doi: 10.1016/j.tcb.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozbudak EM, Becskei A, van Oudenaarden A. A system of counteracting feedback loops regulates Cdc42p activity during spontaneous cell polarization. Dev. Cell. 2005;9:565–571. doi: 10.1016/j.devcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- Park HO, Bi E. Central roles of small GTPases in the development of cell polarity in yeast and beyond. Microbiol. Mol. Biol. Rev. 2007;71:48–96. doi: 10.1128/MMBR.00028-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riquelme M, Bartnicki-Garcia S. Key differences between lateral and apical branching in hyphae of Neurospora crassa. Fungal Genet. Biol. 2004;41:842–851. doi: 10.1016/j.fgb.2004.04.006. [DOI] [PubMed] [Google Scholar]
- Schmitz HP, Kaufmann A, Köhli M, Laissue PP, Philippsen P. From function to shape: a novel role of a formin in morphogenesis of the fungus Ashbya gossypii. Mol. Biol. Cell. 2006;17:130–145. doi: 10.1091/mbc.E05-06-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Pryciak PM. Membrane localization of scaffold proteins promotes graded signaling in the yeast MAP kinase cascade. Curr. Biol. 2008;18:1184–1191. doi: 10.1016/j.cub.2008.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Z, Gao XD, Howell AS, Bose I, Lew DJ, Bi E. Adjacent positioning of cellular structures enabled by a Cdc42 GTPase-activating protein-mediated zone of inhibition. J. Cell Biol. 2007;179:1375–1384. doi: 10.1083/jcb.200705160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedlich-Soldner R, Wai SC, Schmidt T, Li R. Robust cell polarity is a dynamic state established by coupling transport and GTPase signaling. J. Cell Biol. 2004;166:889–900. doi: 10.1083/jcb.200405061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T, Mochida J, Kadota J, Takeda M, Bi E, Tanaka K. Initial polarized bud growth by endocytic recycling in the absence of actin cable-dependent vesicle transport in yeast. Mol. Biol. Cell. 2010;21:1237–1252. doi: 10.1091/mbc.E09-05-0412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan A, Xu G, Yang ZB. Calcium participates in feedback regulation of the oscillating ROP1 Rho GTPase in pollen tubes. Proc. Natl. Acad. Sci. USA. 2009;106:22002–22007. doi: 10.1073/pnas.0910811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziman M, Johnson DI. Genetic evidence for a functional interaction between Saccharomyces cerevisiae CDC24 and CDC42. Yeast. 1994;10:463–474. doi: 10.1002/yea.320100405. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.