Abstract
To gain an understanding of the genetic structure and dispersal dynamics of T. infestans populations, we analyzed the multilocus genotype of 10 microsatellite loci for 352 T. infestans collected in 21 houses of 11 rural communities in October 2002. Genetic structure was analyzed at the community and house compound levels. Analysis revealed that vector control actions affected the genetic structure of T. infestans populations. Bug populations from communities under sustained vector control (core area) were highly structured and genetic differentiation between neighboring house compounds was significant. In contrast, bug populations from communities with sporadic vector control actions were more homogeneous and lacked defined genetic clusters. Genetic differentiation between population pairs did not fit a model of isolation by distance at the microgeographical level. Evidence consistent with flight or walking bug dispersal was detected within and among communities, dispersal was more female-biased in the core area and results suggested that houses received immigrants from more than one source. Putative sources and mechanisms of re-infestation are described. These data may be use to design improved vector control strategies
Keywords: Chagas disease, Triatoma infestans, vector control, population structure, microsatellite loci
INTRODUCTION
The current Chagas disease control program in southern South America, the Southern Cone Initiative, calls for the elimination of Triatoma infestans by residual spraying with insecticides. To date, T. infestans-mediated transmission of Trypanosoma cruzi has been interrupted in Uruguay, Chile and Brazil (Dias et al., 2002; Schofield et al., 2006), but the program has been less successful in the Gran Chaco region (northern Argentina, Bolivia and Paraguay), mainly due to high levels of bug reinfestation after spraying (Gürtler, 2007; Gürtler et al., 2007). A key question to be answered is whether recurrent infestations are due to residual domestic populations that survive insecticide spraying, or to re-invasion of bugs from external sources (either from unsprayed communities or from sylvatic habitats). A better understanding of the spatio-temporal structure of vector populations is needed to help answer such questions and to design successful control campaigns.
Peridomestic sites were found to be the most important sources of T. infestans bugs, re-invading houses after residual insecticide spraying within a community in northern Argentina (Cecere et al., 2002; Gürtler et al., 2004). Depending on the distance among sites, history of infestation and relative timing of control actions, the reinfestation process in a rural community might be initiated from internal sites within the community or from external sites up to 1,500 m away (Cecere et al., 2006). Wing geometric morphometry studies of T. infestans and spatio-temporal analysis of the reinfestation process detected spatial population substructure within the rural community of Amamá (Santiago del Estero, Argentina), but the methods applied in those studies lacked the resolution needed to explain the underlying mechanisms that generated that pattern (Cecere et al., 2004; Schachter-Broide et al., 2004). Definition of triatomine population boundaries, geographic distribution, dispersal dynamics and gene flow among populations require molecular markers with enough resolution power to detect differences among recently diverged populations within a species.
Microsatellites are short-tandem sequence repeats that evolve at rates 105–108 higher than single mutations depending on the taxonomic group (Lia, 2004). At such rates, significant amounts of polymorphism in the number of tandem repetitions occur among individuals within a population and therefore microsatellites are widely used in population genetic studies. Microsatellite loci have been described for several species of triatomine bugs: Rhodnius pallescens (Harry et al., 1998), R. prolixus (Harry et al., 2008b), Triatoma dimidiata (Anderson et al., 2002), T. infestans (García et al., 2004; Marcet et al., 2006), and T. pseudomaculata (Harry et al., 2008a) and were recently applied in regional and microgeographical studies of T. infestans populations from Argentina and Bolivia (Perez de Rosas et al., 2007a; 2007b; Richer et al., 2007; Pizarro et al., 2008).
Although different definitions of T. infestans population have been used in different studies (i.e., locality, habitat, household), the correct panmitic unit for this species is yet to be established, (Pizarro et al., 2008). High levels of substructure within a locality, including differentiation among neighboring households and between domestic and peridomestic habitats, were detected (Perez de Rosas et al., 2007a; 2007b; Pizarro et al., 2008). Moreover, results on wing morphometric of T. infestans indicated that individual capture sites represented the discrete unit where metric differentiation took place (Schachter-Broide et al., 2004). These results emphasize the need to study T. infestans population genetic structure at a microgeographical level, within a community.
Populations under demographic stability present a genetic structure pattern compatible with a process of differentiation under mutation-drift equilibrium (Slatkin & Hudson, 1991; Slatkin, 1993), in which a model of isolation by distance (IBD) should explain the degree of genetic divergence between populations. In contrast, growing empirical evidence showed that several species have not yet reached a migration–drift equilibrium and their genetic structure pattern reflects historical populations proceses rather than the current pattern of geneflow (Slatkin, 1993; Garnier et al., 2004). T. infestans populations subjected to vector control actions are largely disturbed, subjected to repeated bottle-neck events and are susceptible to local differentiation by genetic drift (Perez de Rosas et al., 2007b). We expect, therefore, that T. infestans populations under high control pressure present higher gene flow restrictions and higher differentiation among them that undisturbed populations.
The present work is part of a longitudinal study on the ecoepidemiology of Chagas disease in a well-defined rural area in northwestern Argentina under sustained vector surveillance and selective insecticide sprays. In this study, we applied 10 microsatellite loci to examine closely related T. infestans populations from two neighboring rural areas in Santiago del Estero, northwest Argentina, which had been under different frequencies of residual insecticide spraying over the previous decade. We describe and compare the patterns of population structure observed in both intervention areas and discuss the mechanisms that may have produced such patterns. Micro-geographical population genetic analysis applied here aims to detect the presence of first generation immigrants, to determine whether immigrants come from internal or external sources from a community (Cecere et al., 2006) and to assess putative sources and mechanisms of reinfestation.
MATERIALS AND METHODS
Insect origin
Triatomine collections were conducted in 300 houses in Amamá (27° 12’ 33’’S, 63° 02’ 10’’W) and 35 neighboring communities, Moreno department, province of Santiago del Estero, northern Argentina, in October 2002. The area and the history of previous vector control actions have been described in detail elsewhere (Cecere et al., 2004; Ceballos et al., 2005; Gürtler et al., 2007). Our study included samples from two areas that differed in their history of vector control: the core area (Amamá, Trinidad, Mercedes, Villa Matilde and Pampa Pozo), subjected to recurrent vector control actions in the framework of a community-based program supervised jointly by the research team and the National Vector Control Program (NVCP), and the peripheral area (i.e, communities surrounding the core area) where unsupervised community-based insecticide applications promoted by the NCVP were sporadically conducted (Cardinal et al., 2007). Long-term interventions followed by sustained, supervised community-based actions in the core area caused significant reductions in bug infestation and re-colonization rates; suppression of the domestic reestablishment of bug populations and reduction of T. cruzi infection prevalence in bugs and dogs, all of which led to interruption of local human transmission (Cardinal et al., 2007; Gürtler et al., 2007).
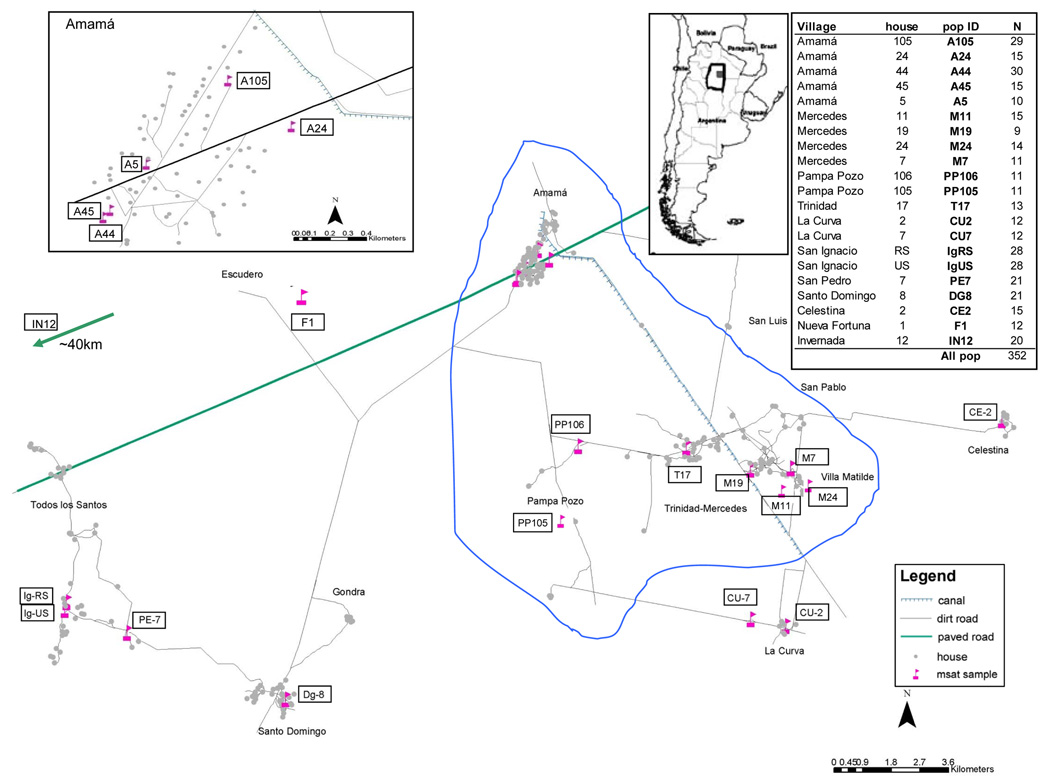
Experienced collectors from NVCP searched for triatomine bugs in all the houses and peridomestic sites, using a dislodging spray and following a standardized procedure (Gürtler et al., 1999). The 352 T. infestans used in this study were from 21 houses (in 11 communities) where at least 10 bugs were captured at the same house compound (Fig. 1). For this microgeographic study, we considered bugs captured in the same capture site (i.e. pig corral, storeroom, etc) or within the same house compound (i.e., different peridomestic structures within a house) as one population. A sample from Invernada (IN12, Figueroa department, Santiago del Estero) captured in February 2004 was also included as an external group for comparisons. Each individual capture site was geo-referenced using a GPS (GeoExplorer II; Trimble Sunnyvale, CA), and non-linear geographic pairwise distances between sites were calculated with ArcGIS 8.1 (Environmental Systems Research Institute, Redlands, CA).
Figure 1.
Map of the study area including location of collection sites. Inserts: a) Detail of Amamá; b) Location of Moreno department, Santiago del Estero, in Argentina; and c) House compounds and number of individuals analyzed per site. Enclosed in blue is the CORE area.
Multilocus genotyping
DNA extraction and PCR amplification conditions are described in Marcet et al. (2006). Nine loci described therein were used for this analysis (Tims3, Tims5, Tims22, Tims23, Tims27, Tims42, Tims56, Tims64, Tims65) plus a tenth locus (Tims19) isolated following the same procedures described in Marcet et al.(2006). Multiplex PCR was carried out in pairs of loci with the same annealing temperature and different dye colors (Tinf_ms5 with Tinf_ms42 and Tinf_ms22 with Tinf_ms27). DNA fragment detection with 1 bp resolution was performed with an automated DNA sequencer ABI 3100 and size determination was obtained with GeneScan 3.7 and Genotyper 3.7 (Applied Biosystems).
Analysis
Linkage Disequilibrium (LD) exact tests between microsatellite loci pairs were computed with GENEPOP3.4 (Raymond & Rousset, 1995). The fit to Hardy-Weinberg (HW) equilibrium expectations was evaluated using the U score test available in GENEPOP3.4, under the null hypothesis of random union of gametes. Allele number per locus and population were obtained directly after the binning, and mean allele number among all loci were compared among populations. Fis-W&C (Weir & Cockerham, 1984); Allele Richness (aRch), which is a sample-size independent measure of allele number (El Mousadik & Petit, 1996); Gene Diversity or expected heterozygosity (He) and observed heterozygosity (Ho) (Nei, 1987) were obtained with FSTAT2.9.3.2 (Goudet, 1995) or GENEPOP3.4. Bonferroni correction for multiple comparisons was applied when needed (Rice, 1989).
Hierarchical Analysis of Molecular Variance (AMOVA) (Excoffier et al., 1992) was performed at three levels of population structure: among communities, among populations (house compounds) and among individuals in a population. AMOVA calculations (based on the average values for the 10 loci) were performed with ARLEQUIN 3.01(Excoffier et al., 2005).
Pairwise Fst values and significance levels were calculated with ARLEQUIN 3.01 as in Weir & Cockerham (1984). To determine whether the genetic differentiation among population pairs fit a model of isolation by distance (IBD), the FST / (1- FST) values were regressed against the natural logarithm of pairwise geographic distances between populations (Rousset, 1997). A Mantel test with 100,000 permutations was performed with GENEPOP 3.4.
To further study the genetic structure of our dataset, we used a Bayesian assignment approach to determine genetic clusters within the entire dataset. The test implemented in STRUCTURE 2.2 (Pritchard et al., 2000; Falush et al., 2003; 2007) uses the individual multilocus genotypes to determine the number of K distinguishable populations (or genetic clusters) within the sample analyzed, and simultaneously assigns individuals to each cluster without using prior information of the sampling locations. Considering the small geographical scale we studied, we assumed a model of admixed populations (which allows for the possibility that individuals may have mixed ancestry in more than one of the K populations) and correlated allele frequencies, as suggested for closely related populations (Falush et al., 2003). In each run, a burn-in period of 50,000 and 100,000 length was applied. The number of genetic clusters (K) was determined according to Pritchard and collaborators (2000). Individuals were assigned to a specific cluster if the probability of assignment was ≥ 70%. For individuals that were not assigned to any cluster by this method, we used the assignment-exclusion a posteriori test implemented in GENECLASS 2 (Piry et al., 2004). At this stage, the genetic clusters obtained with STRUCTURE were used as reference populations. Populations were not excluded as possible sources of individual origin if the marginal probability was ≥ 0.05.
Each bug population (i.e., from each house compound) was examined to identify potential first-generation immigrants using the assignment test implemented in GENECLASS 2 (Piry et al., 2004). We used a Bayesian assignment criteria (Rannala & Mountain, 1997) and the Monte-Carlo resampling method (Paetkau et al., 2004) to obtain, for each individual, the marginal probability of being a first-generation migrant (with 10,000 replicates and significance level at p ≤ 0.01). We further evaluated the probability that the putative population source of the detected first generation immigrants was among the sampled house compounds, using the assignment-exclusion test implemented in GENECLASS2 and the same model used to detect the immigrants.
To examine whether there was a pattern of sex-biased dispersal, we used the test implemented in FSTAT 2.9.3.2 (Goudet, 1995) based on the algorithm described in Goudet et al. (2002). This test uses a randomization approach to compare the genetic structure of males and females in order to identify potential immigrants of either sex across all populations. Estimated values of Fis and Fst (Weir & Cockerham, 1984), Relatedness [2Fst/(1+Fst)], and ad-hoc assignment index (Alc) and its variance (vAlc) (Favre et al., 1997) were compared between sexes by the randomization approach applied in FSTAT 2.9.3.2. If there exists a significant sex bias in dispersal, the dispersing sex should be less genetically structured (i.e., smaller values of Fst), present a larger heterozygote deficit (i.e., positive and higher Fis) and a low AIc value with a higher variance of this index (vAIc).
RESULTS
Overall loci amplification success of the 10 microsatellite loci was greater than 94%, and results were highly repeatable. Fisher’s exact tests for linkage disequilibrium were not significant for any pair of loci when tested across all populations, supporting the assumption of loci independence. Significant deviations from Hardy-Weinberg (HW) equilibrium were observed for loci Tims5, Tims19, Tims27, Tims64 and Tims65 when analyzed across all populations (Table 1). All deviations were due to heterozygote deficit. The mean number of alleles per locus among all capture sites was 17 (range, 6–31), and mean allele richness (aRch) was 4.6 (range 3–6.2). Tims5, Tims42, Tims65 and Tims19 presented the highest number of alleles and allele richness (Table 1).
Table 1.
Genetic variability summary per loci and per population.
| House compounds | |||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A5 | A24 | A44 | A45 | A105 | M7 | M11 | M19 | M24 | PP105 | PP106 | T17 | CE2 | CU2 | CU7 | F1 | DG8 | PE7 | IgRS | IgUS | IN12 | overall houses | ||
| Fis W&C | 0.273 | - | 0.041 | −0.172 | −0.084 | −0.071 | −0.04 | −0.194 | 0.2 | −0.328* | −0.01 | −0.031 | −0.232 | −0.386 | −0.294 | 0.114 | −0.254 | −0.172 | −0.049 | −0.121 | −0.232 | −0.11 | |
| He | 0.45 | 0.45 | 0.51 | 0.2 | 0.26 | 0.14 | 0.47 | 0.53 | 0.69 | 0.45 | 0.73 | 0.58 | 0.49 | 0.39 | 0.56 | 0.64 | 0.51 | 0.6 | 0.57 | 0.6 | 0.49 | ||
| [Ho] | [0.33] | - | [0.43] | [0.6] | [0.21] | [0.27] | [0.14] | [0.56] | [0.43] | [0.9] | [0.45] | [0.75] | [0.71] | [0.67] | [0.5] | [0.5] | [0.8] | [0.6] | [0.63] | [0.64] | [0.74] | [0.54] | |
| aRich [Na] | 2.5[3] | 1[1] | 2.5[3] | 2.8[3] | 1.8[3] | 2.2[3] | 1.6[2] | 3[4] | 2.7[3] | 3.5[4] | 2.6[3] | 3.7[4] | 3.2[4] | 2.4[3] | 2[2] | 3.1[4] | 3.2[4] | 3[4] | 2.8[4] | 3[4] | 3.6[5] | 3[6] | |
| Fis W&C | 0.104 | −0.057 | 0.166* | 0.087 | 0.094 | −0.011 | 0.195 | −0.244 | 0.333 | 0 | −0.11 | −0.026 | 0.055 | 0.265 | −0.143 | 0.243 | 0.065 | 0.216* | 0.372** | 0.214 | −0.011 | 0.086** | |
| He | 0.79 | 0.89 | 0.85 | 0.82 | 0.83 | 0.81 | 0.67 | 0.82 | 0.74 | 0.83 | 0.82 | 0.71 | 0.81 | 0.81 | 0.8 | 0.83 | 0.82 | 0.77 | 0.79 | 0.81 | 0.78 | 0.8 | |
| [Ho] | [0.71] | [0.93] | [0.71] | [0.75] | [0.75] | [0.82] | [0.55] | [1] | [0.5] | [0.83] | [0.9] | [0.73] | [0.77] | [0.6] | [0.91] | [0.64] | [0.76] | [0.61] | [0.5] | [0.64] | [0.79] | [0.73] | |
| aRich [Na] | 5.1[6] | 6.2[9] | 5.7[9] | 5.1[6] | 5.4[9] | 4.7[6] | 4[5] | 4.9[6] | 4.4[6] | 6.2[7] | 4.6[5] | 3.4[4] | 4.9[7] | 4.9[7] | 5.1[8] | 5[6] | 5.1[8] | 4.4[6] | 4.5[6] | 5.1[10] | 4.6[7] | 6.2[18] | |
| Fis W&C | 0.205 | 0.411* | 0.702** | 0.591* | 0.444** | 0.535** | 0.885** | 0.467* | 0.538** | −0.066 | −0.042 | 0.059 | 0.659* | 0.257* | 0.125 | 0.328 | 0.051 | 0.518* | 0.23* | 0.5** | 0.26 | 0.364** | |
| He | 0.78 | 0.9 | 0.86 | 0.85 | 0.77 | 0.84 | 0.7 | 0.81 | 0.88 | 0.85 | 0.87 | 0.71 | 0.61 | 0.55 | 0.72 | 0.49 | 0.72 | 0.43 | 0.72 | 0.71 | 0.81 | 0.74 | |
| [Ho] | [0.63] | [0.54] | [0.26] | [0.36] | [0.43] | [0.4] | [0.08] | [0.44] | [0.42] | [0.9] | [0.91] | [0.67] | [0.21] | [0.42] | [0.64] | [0.33] | [0.68] | [0.21] | [0.56] | [0.36] | [0.6] | [0.48] | |
| aRich [Na] | 5.1[7] | 6.5[10] | 5.7[9] | 5.9[10] | 4.6[9] | 5.4[8] | 3.6[4] | 5.1[6] | 6.1[9] | 5.6[8] | 6[8] | 3.4[4] | 3.9[6] | 3.8[6] | 4.5[6] | 2.4[3] | 4.3[8] | 2.6[4] | 4.3[8] | 4.5[8] | 4.9[8] | 5.8[30] | |
| Fis W&C | 0.45* | 0.034 | 0.048 | 0.191 | 0.053 | 0.042 | 0.031 | 0.02 | 0.341** | 0.169 | 0.111 | −0.054 | 0.373 | 0.823* | 0.038 | 0.529 | 0.185 | −0.118 | 0.031 | −0.267 | −0.202 | 0.133 | |
| He | 0.71 | 0.69 | 0.8 | 0.82 | 0.73 | 0.76 | 0.69 | 0.68 | 0.75 | 0.53 | 0.81 | 0.63 | 0.63 | 0.45 | 0.47 | 0.52 | 0.67 | 0.6 | 0.65 | 0.58 | 0.39 | 0.65 | |
| [Ho] | [0.4] | [0.67] | [0.77] | [0.67] | [0.69] | [0.73] | [0.67] | [0.67] | [0.5] | [0.44] | [0.73] | [0.67] | [0.4] | [0.08] | [0.45] | [0.25] | [0.55] | [0.67] | [0.63] | [0.73] | [0.47] | [0.56] | |
| aRich [Na] | 4.1[5] | 3.3[4] | 4.9[7] | 5[8] | 4.1[7] | 4.7[6] | 3.8[5] | 4.4[6] | 4.1[5] | 2.9[3] | 4.7[6] | 3.8[5] | 3.2[5] | 2.4[3] | 3.3[5] | 2[2] | 3.6[5] | 2.8[4] | 2.9[3] | 2.7[5] | 2.5[4] | 4.2[14] | |
| Fis W&C | - | 0.649 | −0.087 | - | −0.042 | −0.17 | −0.182 | 0.326 | 0.304 | −0.143 | 0.167 | 0.214 | −0.04 | −0.048 | −0.111 | −0.023 | 0.07 | −0.041 | 0.138 | −0.08 | 0.017 | 0.08 | |
| He | 0.2 | 0.2 | 0.08 | 0.38 | 0.09 | 0.3 | 0.65 | 0.3 | 0.35 | 0.76 | 0.63 | 0.14 | 0.24 | 0.38 | 0.16 | 0.38 | 0.38 | 0.26 | 0.25 | 0.46 | 0.33 | ||
| [Ho] | - | [0.07] | [0.22] | [0.08] | [0.39] | [0.09] | [0.36] | [0.44] | [0.21] | [0.4] | [0.64] | [0.5] | [0.14] | [0.25] | [0.42] | [0.17] | [0.35] | [0.4] | [0.22] | [0.27] | [0.45] | [0.3] | |
| aRich [Na] | 1[1] | 1.8[2] | 1.8[3] | 1.4[2] | 2.2[3] | 1.5[2] | 1.9[2] | 3.5[4] | 1.9[2] | 2.4[3] | 4.1[5] | 3.2[4] | 1.6[2] | 2.3[4] | 2.9[5] | 1.8[3] | 2.3[3] | 2[2] | 1.8[2] | 2.2[4] | 2.5[4] | 2.5[9] | |
| Fis W&C | −0.007 | 0.067 | −0.038 | 0.202 | 0.309** | −0.029 | 0.24* | −0.032 | 0.044 | 0.213 | −0.034 | 0.27* | 0.12 | 0.327 | 0.281* | −0.163 | −0.041 | 0.17 | 0.071 | 0.248* | 0.426* | 0.122** | |
| He | 0.79 | 0.71 | 0.86 | 0.89 | 0.82 | 0.8 | 0.61 | 0.86 | 0.8 | 0.78 | 0.53 | 0.73 | 0.73 | 0.73 | 0.75 | 0.87 | 0.77 | 0.34 | 0.73 | 0.81 | 0.71 | 0.74 | |
| [Ho] | [0.8] | [0.67] | [0.9] | [0.71] | [0.57] | [0.82] | [0.47] | [0.89] | [0.77] | [0.63] | [0.55] | [0.54] | [0.64] | [0.5] | [0.55] | [1] | [0.8] | [0.29] | [0.68] | [0.62] | [0.41] | [0.66] | |
| aRich [Na] | 5.2[8] | 4.1[6] | 5.7[8] | 6.3[9] | 5.2[9] | 4.7[6] | 3.6[5] | 5.6[7] | 4.7[6] | 5[6] | 3.5[5] | 4.1[5] | 3.6[4] | 3.8[5] | 4.1[5] | 5.6[7] | 4.8[7] | 2.7[5] | 4.4[8] | 5.2[8] | 3.9[6] | 5.5[15] | |
| Fis W&C | 0.318 | 0.212 | 0.221 | 0.133 | 0.079 | −0.17 | −0.157 | 0.127 | 0.04 | −0.103 | −0.1 | 0.127 | 0.295 | 0.12 | 0.206 | 0.066 | 0.357 | 0.098 | 0.057 | 0.033 | 0.23 | 0.106 | |
| He | 0.72 | 0.81 | 0.84 | 0.9 | 0.72 | 0.86 | 0.54 | 0.65 | 0.82 | 0.73 | 0.91 | 0.47 | 0.7 | 0.85 | 0.73 | 0.8 | 0.81 | 0.77 | 0.83 | 0.79 | 0.71 | 0.76 | |
| [Ho] | [0.5] | [0.64] | [0.66] | [0.79] | [0.67] | [1] | [0.62] | [0.57] | [0.79] | [0.8] | [1] | [0.42] | [0.5] | [0.75] | [0.58] | [0.75] | [0.53] | [0.7] | [0.79] | [0.77] | [0.55] | [0.68] | |
| aRich [Na] | 4.5[6] | 5[7] | 5.4[9] | 6.7[11] | 4.4[8] | 5.8[8] | 3.1[5] | 3.6[4] | 5.3[9] | 4[4] | 6.8[10] | 2.7[3] | 3.9[6] | 5.4[7] | 3.8[5] | 5.1[8] | 5.1[9] | 4.3[6] | 5.5[11] | 5[11] | 3.9[6] | 5.6[23] | |
| Fis W&C | 0.193 | 0.388 | 0.205 | −0.078 | −0.142* | −0.139 | 0.125 | −0.358 | −0.109 | 0.077 | 0.111 | −0.016 | −0.017 | 0.181 | −0.125 | −0.076 | 0.155 | −0.091 | −0.016 | 0.109 | −0.257 | 0.006 | |
| He | 0.82 | 0.69 | 0.72 | 0.78 | 0.62 | 0.72 | 0.68 | 0.75 | 0.71 | 0.65 | 0.61 | 0.61 | 0.63 | 0.71 | 0.67 | 0.7 | 0.59 | 0.52 | 0.7 | 0.62 | 0.68 | 0.68 | |
| [Ho] | [0.67] | [0.43] | [0.57] | [0.83] | [0.7] | [0.82] | [0.6] | [1] | [0.79] | [0.6] | [0.55] | [0.62] | [0.64] | [0.58] | [0.75] | [0.75] | [0.5] | [0.57] | [0.71] | [0.56] | [0.85] | [0.67] | |
| aRich [Na] | 4.9[6] | 3.8[6] | 3.6[5] | 4.2[5] | 3.5[5] | 3.8[4] | 3.6[4] | 3.8[4] | 3.3[4] | 3.3[4] | 2.9[3] | 3[4] | 3.1[4] | 3.4[4] | 3.3[4] | 3.9[5] | 2.8[4] | 2.8[4] | 3.8[5] | 3.5[5] | 3.4[4] | 4.1[8] | |
| Fis W&C | 0.323 | 0.736* | 0.33* | 0.076* | 0.655** | −0.101 | −0.037 | 0.816* | 0.471* | 0.304 | 1* | 0.625 | −0.017 | −0.023 | 0.306 | 0.163 | 0.471* | 0.082 | 0.334* | 0.338 | 0.146 | 0.358** | |
| He | 0.73 | 0.57 | 0.49 | 0.62 | 0.75 | 0.5 | 0.13 | 0.64 | 0.77 | 0.52 | 0.7 | 0.36 | 0.72 | 0.49 | 0.65 | 0.69 | 0.56 | 0.72 | 0.66 | 0.65 | 0.47 | 0.59 | |
| [Ho] | [0.5] | [0.15] | [0.33] | [0.57] | [0.26] | [0.55] | [0.13] | [0.13] | [0.42] | [0.36] | [0] | [0.14] | [0.73] | [0.5] | [0.45] | [0.58] | [0.3] | [0.67] | [0.44] | [0.43] | [0.4] | [0.38] | |
| aRich [Na] | 3.5[4] | 2.8[3] | 2.8[4] | 3.3[4] | 4.2[6] | 2.7[3] | 1.6[2] | 3.5[4] | 4.6[6] | 3.1[4] | 3[3] | 2[2] | 4[6] | 3.2[4] | 3.2[4] | 3.5[4] | 3[4] | 3.6[4] | 4.1[8] | 3.4[5] | 2[2] | 3.7[11] | |
| Fis W&C | 0.616* | 0.449* | 0.707** | 0.476* | 0.286* | 0.143 | 0.432* | −0.032 | 0.553* | 0.556 | −0.264 | −0.149 | 0.221 | 0.574* | 0.833* | 0.135* | 0.224* | 0.579** | 0.284* | 0.429* | 0.067 | 0.33** | |
| He | 0.84 | 0.76 | 0.76 | 0.78 | 0.86 | 0.77 | 0.53 | 0.86 | 0.73 | 0.62 | 0.8 | 0.8 | 0.42 | 0.62 | 0.57 | 0.86 | 0.6 | 0.7 | 0.75 | 0.63 | 0.59 | 0.71 | |
| [Ho] | [0.33] | [0.43] | [0.23] | [0.42] | [0.62] | [0.67] | [0.31] | [0.89] | [0.33] | [0.29] | [1] | [0.91] | [0.33] | [0.27] | [0.1] | [0.75] | [0.47] | [0.3] | [0.54] | [0.36] | [0.55] | [0.48] | |
| aRich [Na] | 5.4[7] | 5.2[8] | 4.4[7] | 4.2[5] | 6[11] | 4.8[6] | 3[4] | 5.5[7] | 4.2[6] | 2.9[3] | 4.8[6] | 4.7[6] | 2.7[4] | 3.7[5] | 3.8[5] | 5.9[9] | 4[8] | 4.2[6] | 4.6[9] | 3.8[7] | 3.8[8] | 5.6[31] | |
|
|
Fis W&C | 0.276** | 0.279 | 0.263 | 0.187** | 0.209** | 0.04 | 0.222** | 0.09 | 0.277** | 0.064 | 0.082 | 0.074 | 0.154 | 0.23** | 0.133 | 0.123 | 0.127 | 0.135 | 0.151** | 0.167** | 0.064 | 0.159** |
| Av(aRich) | 4.1 | 3.96 | 4.25 | 4.48 | 4.15 | 3.41 | 3.53 | 3.61 | 3.81 | 3.83 | 4.01 | 2.97 | 4.29 | 4.14 | 3.88 | 4.30 | 3.24 | 3.87 | 3.83 | 3.41 | 3.50 | 4.6 | |
| Av(He) | 0.7 | 0.63 | 0.69 | 0.71 | 0.67 | 0.60 | 0.60 | 0.62 | 0.66 | 0.65 | 0.64 | 0.50 | 0.72 | 0.71 | 0.66 | 0.73 | 0.58 | 0.67 | 0.65 | 0.64 | 0.62 | 0.72 | |
| Av(aRich [Na]) | 4.1[5] | 4[6] | 4.3[6] | 4.5[6] | 4.1[7] | 3.4[5] | 3.5[5] | 3.6[5] | 3.8[6] | 3.8[5] | 4[5] | 3[4] | 4.3[5] | 4.1[6] | 3.9[5] | 4.3[5] | 3.2[5] | 3.9[6] | 3.8[7] | 3.4[4] | 3.5[5] | 4.6[17] | |
Fis ( W&C) = Fis value as Weir & Cockerman (1984); He = Expected heterozygosis or Gene diversity (Weir & Cockerham, 1984); Ho=Observed heterozygosis; aRch= allele richness independent of sample size (El Mousadik & Petit, 1996); Na = Allele number per loci per population; Av = average across 10 loci.
Significant deviation from Hardy-Weinberg equilibrium (U-score test p≤ 0.05)
bold: significant deviation after sequential Bonferroni correction ( p≤0.0002) (Holm, 1979).
Analysis of the 21 house compounds revealed that in eight (A5, A45, A105, M11, M24, CU2, IgRS and IgUS), the null hypothesis of HW equilibrium was rejected after Bonferroni correction (P ≤ 0.0002) when analyzed across the 10 loci (Table 1). All deviations from HW equilibrium were due to heterozygote deficit, and evidenced high to moderate population sub-structure as reflected on their Fis values (0.15–0.28) (Table 1). All house compounds showed similar levels of within-population variability. Differences in allele richness (aRich) or gene diversity (He) among population pairs were not significant (ANOVA and Kruskal-Wallis tests, P > 0.05). Fixed alleles were found at loci Tims23 and Tims3 in the populations A5 and A24, respectively.
Pairwise comparisons of Fst values at the community level showed highly significant genetic differentiation for most communities except for some communities in the periphery (Table 2). At the house compound level, significant genetic differentiation was detected among most houses, except for a few house compounds within a community (A44-A45-A24 in Amamá and CU2-CU7 from La Curva) and among some house compounds from communities in the periphery (Table 3). Fst values from the core area were on average higher (0.11) than among populations pairs from the periphery (0.06). There was no significant fit to an IBD model at the household level in the total sample (Mantel r = 0.0035, p = 0.74, Figure 2a), nor by discriminating between core and peripheral areas (Figure 2b).
Table 2.
Genetic differentiation among community pairs measured by pairwise Fst (Weird & Cockerman, 1984). Underlined numbers are non significant Fst values after Bonferroni correction (p> 0.0009)
| A | CE | CU | Dg | F | M | PP | PE | Ig | T | |
|---|---|---|---|---|---|---|---|---|---|---|
| Amama | * | |||||||||
| Celestina | 0.080 | * | ||||||||
| LaCurva | 0.075 | 0.043 | * | |||||||
| StoDomingo | 0.065 | 0.078 | 0.049 | * | ||||||
| LaFavorina | 0.043 | 0.067 | 0.077 | 0.077 | * | |||||
| Mercedes | 0.031 | 0.074 | 0.055 | 0.053 | 0.063 | * | ||||
| PampaPozo | 0.051 | 0.117 | 0.085 | 0.083 | 0.097 | 0.049 | * | |||
| San Pedro | 0.119 | 0.075 | 0.084 | 0.058 | 0.114 | 0.104 | 0.150 | * | ||
| San Ignacio | 0.043 | 0.027 | 0.039 | 0.032 | 0.033 | 0.036 | 0.076 | 0.062 | * | |
| Trinidad | 0.139 | 0.192 | 0.137 | 0.135 | 0.163 | 0.128 | 0.094 | 0.208 | 0.137 | * |
| Invernada | 0.078 | 0.040 | 0.018 | 0.073 | 0.071 | 0.050 | 0.090 | 0.098 | 0.028 | 0.128 |
Table 3.
Genetic differentiation matrix (Fst, below diagonal) and pairwise distance in km (above diagonal). Bold underlined values are not differentiated population pairs (Fisher exact tests p≥0.05) and underlined values are not differentiated pairs after applying sequential Bonferroni correction (p≥ 0.00023). The house of Invernada, situated over 60km appart from the communities analyzed here, was not included in the geographical-genetic regression and therefore distances are not included in the matrix.
| House compound | A5 | A24 | A44 | A45 | A105 | CE2 | CU2 | CU7 | DG8 | F1 | M7 | M11 | M19 | M24 | PP105 | PP106 | PE7 | Ig-RS | Ig-US | T17 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A5 | * | 0.82 | 0.45 | 0.34 | 0.66 | 15.77 | 14 | 13.21 | 15.94 | 10.34 | 10.58 | 10.45 | 9.54 | 10.96 | 8.10 | 5.91 | 16.96 | 18.02 | 17.80 | 7.67 |
| A24 | 0.071 | * | 1.15 | 1.09 | 0.45 | 15.08 | 13.72 | 12.98 | 16.52 | 10.92 | 10.09 | 9.99 | 9.11 | 10.47 | 8.20 | 5.93 | 17.69 | 18.78 | 18.57 | 7.33 |
| A44 | 0.060 | 0.065 | * | 0.11 | 1.09 | 15.81 | 13.77 | 12.96 | 15.50 | 10.42 | 10.48 | 10.34 | 9.41 | 10.85 | 7.73 | 5.58 | 16.55 | 17.64 | 17.42 | 7.50 |
| A45 | 0.056 | 0.026 | 0.019 | * | 0.99 | 15.84 | 13.86 | 13.05 | 15.60 | 10.36 | 10.54 | 10.40 | 9.48 | 10.91 | 7.84 | 5.69 | 16.63 | 17.70 | 17.49 | 7.57 |
| A105 | 0.081 | 0.081 | 0.083 | 0.063 | * | 15.51 | 14.15 | 13.39 | 16.58 | 10.47 | 10.54 | 10.44 | 9.56 | 10.92 | 8.51 | 6.27 | 17.61 | 18.66 | 18.45 | 7.76 |
| CE2 | 0.137 | 0.106 | 0.109 | 0.075 | 0.109 | * | 9.35 | 10.03 | 24.39 | 25.91 | 6.73 | 7.08 | 8.01 | 6.56 | 13.85 | 13.29 | 28.24 | 30.04 | 29.92 | 9.88 |
| CU2 | 0.137 | 0.086 | 0.101 | 0.070 | 0.146 | 0.065 | * | 1.10 | 16.07 | 23.91 | 4.95 | 4.75 | 5.10 | 4.68 | 7.53 | 8.62 | 20.68 | 22.70 | 22.64 | 6.42 |
| CU7 | 0.131 | 0.071 | 0.110 | 0.063 | 0.107 | 0.036 | 0.027 | * | 15.05 | 23.00 | 4.91 | 4.64 | 4.76 | 4.70 | 6.47 | 7.68 | 19.60 | 21.61 | 21.55 | 5.78 |
| DG8 | 0.103 | 0.091 | 0.081 | 0.056 | 0.112 | 0.078 | 0.043 | 0.070 | * | 19.55 | 17.78 | 17.41 | 16.61 | 17.88 | 10.93 | 12.54 | 5.43 | 7.57 | 7.63 | 15.24 |
| F1 | 0.091 | 0.093 | 0.067 | 0.028 | 0.053 | 0.067 | 0.093 | 0.067 | 0.077 | * | 20.89 | 20.76 | 19.86 | 21.27 | 13.22 | 15.31 | 17.43 | 17.24 | 16.96 | 17.85 |
| M7 | 0.117 | 0.092 | 0.085 | 0.046 | 0.107 | 0.160 | 0.132 | 0.129 | 0.141 | 0.099 | * | 0.37 | 1.32 | 0.39 | 7.13 | 6.75 | 21.51 | 23.32 | 23.20 | 3.36 |
| M11 | 0.235 | 0.107 | 0.141 | 0.105 | 0.197 | 0.138 | 0.087 | 0.105 | 0.127 | 0.139 | 0.137 | * | 1.03 | 0.57 | 6.78 | 6.46 | 21.16 | 22.97 | 22.85 | 3.10 |
| M19 | 0.087 | 0.073 | 0.055 | 0.040 | 0.091 | 0.123 | 0.139 | 0.117 | 0.077 | 0.062 | 0.094 | 0.160 | * | 1.60 | 5.86 | 5.43 | 20.24 | 22.03 | 21.91 | 2.08 |
| M24 | 0.085 | 0.066 | 0.079 | 0.055 | 0.097 | 0.092 | 0.094 | 0.089 | 0.068 | 0.107 | 0.114 | 0.142 | 0.079 | * | 7.31 | 7.03 | 21.69 | 23.51 | 23.39 | 3.67 |
| PP105 | 0.096 | 0.043 | 0.049 | 0.030 | 0.128 | 0.079 | 0.011 | 0.031 | 0.065 | 0.061 | 0.089 | 0.120 | 0.100 | 0.055 | * | 2.32 | 14.39 | 16.19 | 16.08 | 4.31 |
| PP106 | 0.120 | 0.127 | 0.133 | 0.107 | 0.148 | 0.197 | 0.190 | 0.182 | 0.150 | 0.165 | 0.131 | 0.226 | 0.102 | 0.136 | 0.121 | * | 15.35 | 16.98 | 16.83 | 3.42 |
| PE7 | 0.166 | 0.163 | 0.156 | 0.122 | 0.125 | 0.075 | 0.103 | 0.079 | 0.058 | 0.114 | 0.187 | 0.194 | 0.126 | 0.106 | 0.147 | 0.203 | * | 2.15 | 2.20 | 18.54 |
| IgRS | 0.107 | 0.087 | 0.063 | 0.037 | 0.079 | 0.056 | 0.073 | 0.059 | 0.043 | 0.039 | 0.102 | 0.108 | 0.066 | 0.074 | 0.038 | 0.168 | 0.075 | * | 0.28 | 20.25 |
| IgUS | 0.110 | 0.082 | 0.078 | 0.038 | 0.078 | 0.017 | 0.041 | 0.037 | 0.041 | 0.044 | 0.116 | 0.080 | 0.100 | 0.077 | 0.039 | 0.167 | 0.069 | 0.035 | * | 20.12 |
| T17 | 0.175 | 0.162 | 0.164 | 0.122 | 0.184 | 0.192 | 0.156 | 0.135 | 0.135 | 0.163 | 0.183 | 0.216 | 0.145 | 0.166 | 0.090 | 0.154 | 0.208 | 0.140 | 0.155 | * |
| IN12 | 0.147 | 0.085 | 0.094 | 0.053 | 0.120 | 0.040 | 0.039 | 0.008 | 0.073 | 0.071 | 0.120 | 0.067 | 0.113 | 0.095 | 0.015 | 0.189 | 0.098 | 0.044 | 0.029 | 0.128 |
Figure 2.
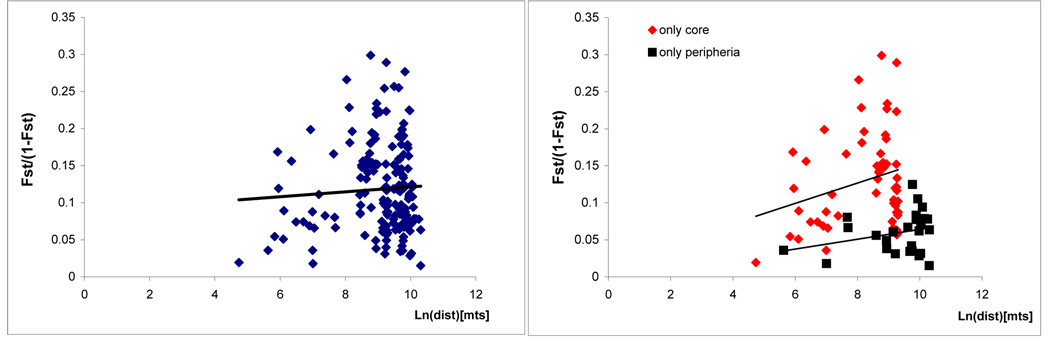
Isolation by distance test results. Pairwise genetic distance plotted against the natural logarithm of the geographic distance among house compound pairs (Rousset, 1997). a) All house compound pairs considered (Mantel r = 0.0035, p = 0.74); b) Discriminating house compounds from the core area and the periphery (core: Mantel r = 0.0679, p= 0.73 ; periphery: Mantel r = 0.0773, p= 0.89).
Remarkably, bug populations A105 from Amamá, T17 from Trinidad, PP106 from Pampa Pozo and the houses from Mercedes were highly differentiated from their neighboring house compounds despite their proximity. The population from PP105 was not differentiated from almost any other household. This result may be attributed to biases caused by small sample size or to missing data for some loci (probably due to poor DNA quality of some samples) and therefore, this population was not considered for further analysis. Within the A105 house compound, the bug populations from two peridomestic structures were significantly differentiated according to the Fst (0.075, P=0.04), and comparisons of genic and genotype distributions (Fisher’s exact tests, P < 0.0001). HW equilibrium hypothesis was not rejected for either of the two sites.
Hierarchical AMOVA showed the highest variance component at the intra-house compound level (>90% of the total variance) whereas the inter-house variability accounted for 9.8% of the total variance (Table 4). Analyzing the sites grouped by community, the inter-house variability accounted for 7.2% of the total variance whereas the community level only accounted for 2.9% of the total variance. Variance component distribution was different in the core area than in the periphery. In the core, the variance explained by the house compound level (10.6%) was almost twice as much as that of the periphery (5.8%), or 3-fold higher (8.9 vs 3.3) when considering the community level. The community component of the variance was similar between the two areas (2.5 and 2.7) but was not significant in the periphery. The percentage of variance among house compounds was maximum (13.6%) among houses from Mercedes, Trinidad and Pampa Pozo.
Table 4.
Summarized results of hierarchical analysis of molecular variance (AMOVA) at ten microsatellite loci.
| Source of variation | df | percentage of variation | F index | p* |
|---|---|---|---|---|
| Among all house compounds | ||||
| Among house compounds | 19 | 9.8 | 0.098 | < 0.0000 |
| Within house compound | 644 | 90.2 | ||
| Among communities | ||||
| Among communities | 9 | 2.9 | 0.029 | 0.005 |
| Among house compound within communities | 10 | 7.2 | 0.074 | < 0.0000 |
| Within house compound | 644 | 89.9 | 0.101 | < 0.0000 |
| Core vs Periphery | ||||
| Between Core and Periphery | 1 | 2.7 | 0.026 | < 0.0000 |
| Among house compound within CORE and PERI | 18 | 8.3 | 0.085 | < 0.0000 |
| Within house compound | 644 | 89.1 | 0.109 | < 0.0000 |
| Among communities within the CORE | ||||
| Among communities (A-M-TPP) | 2 | 2.5 | 0.025 | < 0.0000 |
| Among house compound within communities | 9 | 8.9 | 0.091 | < 0.0000 |
| Within house compound | 354 | 88.6 | 0.114 | < 0.0000 |
| Among communities within the Periphery | ||||
| Among communities | 5 | 2.7 | 0.027 | 0.06 |
| Among house compound within communities | 2 | 3.3 | 0.034 | < 0.0000 |
| Within house compound | 290 | 94.0 | 0.06 | < 0.0000 |
| Households within CORE | ||||
| Among house compounds | 11 | 10.6 | 0.106 | < 0.0000 |
| Within house compound | 354 | 89.4 | ||
| Within Amamá | ||||
| Among house compound | 4 | 6.4 | 0.064 | < 0.0000 |
| Within house compound | 193 | 93.6 | ||
| Within Mercedes-Trinidad-Pampa Pozo | ||||
| Among house compound | 6 | 13.6 | 0.136 | < 0.0000 |
| Within house compound | 161 | 86.4 | ||
| Within the periphery | ||||
| Among house compound | 8 | 5.6 | 0.056 | < 0.0000 |
| Within house compound | 329 | 94.4 |
df = degrees of freedom
significance level based on 10000 permutations. F index = fixation index (Weird & Cockerman, 1984).
Bayesian assignment of individual genotypes determined an optimal number of eight genetic clusters for the entire sample (excluding Invernada), and a total of 177 individuals (54%) was assigned to the eight clusters. Posterior exclusions tests assigned a total of 260 T. infestans (78.3%) to the eight genetic clusters in Table 5. Four of these clusters contained mostly bugs from Trinidad (T), Mercedes (M) and Pampa Pozo (PP), and two clusters contained bugs exclusively from houses T17 and M7 respectively. Most samples from Amamá were assigned to two clusters. Samples from the periphery were assigned to the remaining two clusters, although one also contained most of the samples from house M11 and a few other specimens from Mercedes and Amamá.
Table 5.
Assignment of individual genotypes of T. infestans from 20 house compounds from the core (above the line) and the periphery areas. Bayesian algorithm implemented in STRUCTURE (Pritchard, et al. 2000) determined 8 clusters and the numbers in the cells are the number of individual per house compound assigned in the specific cluster with probability ≥0.70.
| House compound | clu1 | clu2 | clu3 | clu4 | clu5 | clu6 | clu7 | clu8 | N | Total assigned | NO assigned (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| A5 | 8 | 1 | 1 | 10 | 10 | - | |||||
| A24 | 5 | 6 | 1 | 15 | 12 | 3 (20%) | |||||
| A44 | 2 | 21 | 30 | 23 | 7 (23.3%) | ||||||
| A45 | 4 | 6 | 1 | 15 | 11 | 4 (26.7%) | |||||
| A105 | 25 | 1 | 29 | 26 | 3 (10.3%) | ||||||
| M7 | 9 | 1 | 1 | 11 | 11 | - | |||||
| M11 | 1 | 14 | 15 | 15 | - | ||||||
| M19 | 4 | 2 | 9 | 6 | 3 (33.3%) | ||||||
| M24 | 6 | 1 | 1 | 1 | 14 | 9 | 5 (35.7%) | ||||
| PP105 | 1 | 1 | 1 | 3 | 1 | 11 | 7 | 4 (36.4%) | |||
| PP106 | 11 | 11 | 11 | - | |||||||
| T17 | 12 | 13 | 12 | 1 (7.7%) | |||||||
| CE2 | 1 | 3 | 1 | 7 | 15 | 12 | 3 (20%) | ||||
| CU2 | 3 | 3 | 12 | 6 | 6 (50%) | ||||||
| CU7 | 1 | 1 | 3 | 4 | 12 | 9 | 3 (25%) | ||||
| F1 | 4 | 2 | 1 | 12 | 7 | 5 (41.7%) | |||||
| DG8 | 1 | 12 | 1 | 21 | 14 | 7 (33.3%) | |||||
| PE7 | 1 | 17 | 21 | 18 | 3 (14.3%) | ||||||
| Ig-RS | 8 | 12 | 28 | 20 | 8 (28.6%) | ||||||
| Ig-US | 2 | 10 | 1 | 8 | 28 | 21 | 7 (25%) | ||||
| Total | 12 | 9 | 17 | 10 | 50 | 64 | 41 | 57 | 332 | 260 (78.3%) | 72 (21.7%) |
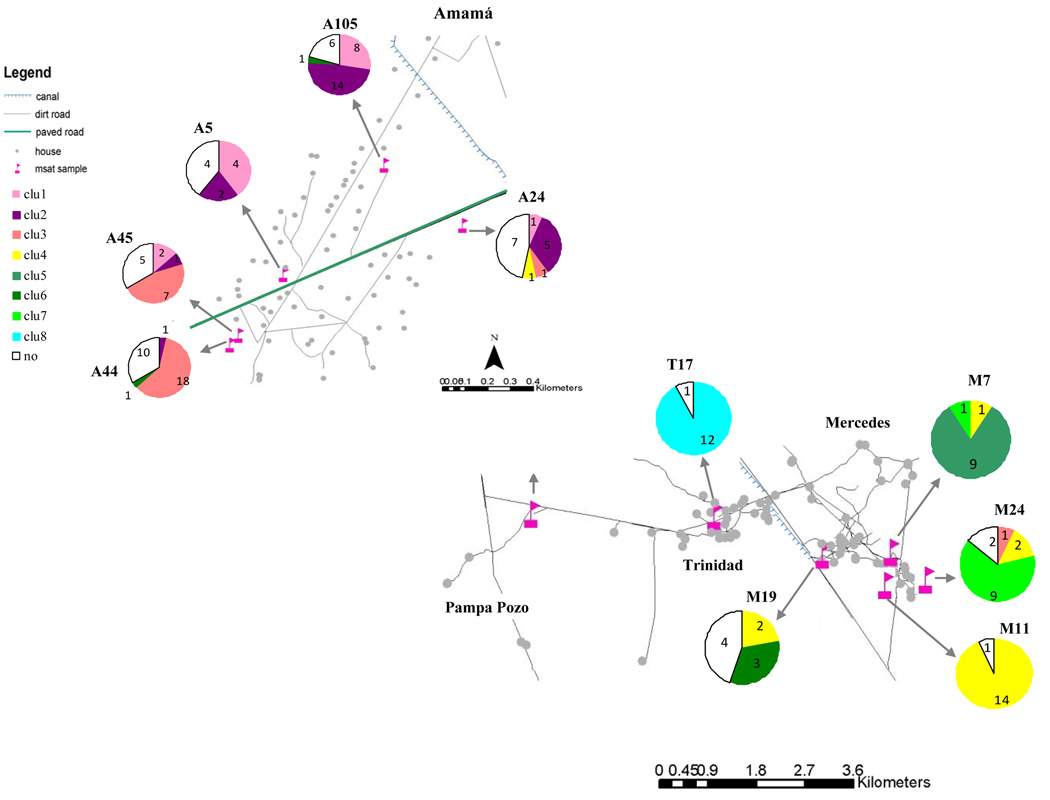
The Bayesian assignment procedure was repeated separately for samples from the core and the periphery. The test failed to detect population structure among house compounds in the periphery (i.e., bugs from all the sites were assigned to a unique cluster). However, the efficiency of assignment and the Ln (probability of data) improved when only the sites of the core area were considered. The number of significant clusters in the core area was eight, and the assignment efficiency was 76%. Five clusters contained samples from T, M and PP (Figure 3b) and three of them were exclusive for individual capture sites (M7, M24 and T17). The samples from Amamá were grouped into three clusters, two of which contained mostly bugs from A105 (a goat corral and a chicken nest within the same house compound); the third cluster included bugs from A44 and A45 (Fig. 3a). In Amamá, 32% of the bugs were not assigned to any cluster whereas only 12% of the M, T, and PP bugs were not assigned.
Figure 3.
a, b: Map of the core area (a) Amamá; (b)Trinidad, Mercedes and Pampa Pozo. Charts show bayesian assigment results per house compound. Each color represents a genetic cluster and the numbers in the charts are the number of individuals per household assigned to a specific cluster with probability >75%.
A total of 22 putative first generation migrants was identified in the core area (1–3 per household) whereas only 4 putative migrants were detected in four houses from the periphery. The putative migrants were 14 males, 5 females, 5 nymphs V and 2 nymphs IV. Exclusion tests determined that all putative migrants but 4 could have originated from one or more house compounds sampled in the current study. The 4 unassigned putative first-generation migrants were collected at houses A44 (male), A105 (female), M19 (V) and T17 (male) are likely to be migrants from populations not included in this study.
A statistically significant tendency toward female-biased dispersal was observed in the core area and in the whole sample if including the periphery houses pooled together (Table 6). Fst and relatedness values for males were significantly higher than those for females (reflecting higher structure in males) and, as expected for the sex dispersing the most, the coefficient of relatedness for females was negative and showed a higher variance (Table 6). No significant sex-biased dispersal was detected in the periphery tested separately.
Table 6.
Sex biased dispersion test results.
| N | Fis | Fst | Related ness | Alc | vAlc | ||
|---|---|---|---|---|---|---|---|
|
|
Females | 40 | 0.23 | 0.038 | 0.061 | –1.47 | 34.76 |
| Males | 71 | 0.27 | 0.086 | 0.128 | 0.83 | 21.47 | |
| overall | 111 | 0.26 | 0.061* | 0.093* | * | - | |
 |
Females | 63 | 0.23 | 0.038 | 0.061 | –0.66 | 31.58 |
| Males | 133 | 0.23 | 0.056 | 0.089 | 0.31 | 18.13 | |
| overall | 196 | 0.23 | 0.0484* | 0.077* | - | - | |
| Females | 23 | 0.21 | 0.059 | 0.094 | 0.65 | 16.58 | |
| Males | 62 | 0.17 | 0.027 | 0.035 | –0.24 | 12.86 | |
| overall | 85 | 0.18 | 0.027 | 0.044 | - | - | |
Fis - Fst: Fixation index (Weir & Cockerham, 1984); Relatedness [2Fst/(1+Fst)], Alc: ad-hoc assignment index (Favre et al., 1997); vAlc: variance of Alc
Randomization test significant (p<0.05).
DISCUSSION
The results of this study suggest that long-term vector control interventions affected the population structure of T. infestans. In the periphery, little differentiation among distant populations and failure to detect substructure among the more established bug populations was observed. In the core area, however, strong differentiation among neighboring households and higher population substructure was observed, despite the evidence of frequent active flight dispersal among and within house compounds empirically corroborated in this area (Vazquez-Prokopec et al., 2004; 2006). Substructure within houses (probably due to the presence of migrants from other populations) accounts for the strong deviations from HW equilibrium. Most migration toward the studied populations was originated from other sites within the community (internal sites) but in at least 4 cases, the immigrants genotype could not be assigned to any of the populations studied here (external sources).
Effects of vector control actions on the genetic structure of T. infestans populations
Populations of T. infestans from the periphery appeared to be less structured and less differentiated than those from the core area. The cumulative effect of migration among neighboring populations for several generations (because vector control interventions were sporadic, asynchronous and with partial coverage) might account for the apparent pattern of high gene-flow among the sampled populations in the periphery. Active flight dispersal of T. infestans may account for undifferentiated neighboring sites (Cu2-Cu7-Ce2) but would not explain the similarity among populations from houses located farther than 1.5 km, the observed dispersal distance reported for T. infestans (Schweigmann et al., 1988). Passive bug dispersal in household goods (bags, suitcases, firewood) may explain gene-flow among distant communities. However, passive dispersal in our study area may explain individual cases of house infestation, but is not likely to account for the genetic similarity observed between communities that were >40 km apart and between which there would be very little or no contact through visits or exchange of goods (e.g., between La Curva or Celestina and Invernada).
The bug populations from the study houses in the core area became established up to 4 years prior to the current study samples (e.g., house A45), according to entomological evaluations performed regularly since 1992 (Gürtler et al., 2007). Considering that a few insects (migrants or local survivors) that carry a few allele variants can establish a population (founder effect) and that flight dispersal of T. infestans varies seasonally (Ceballos et al., 2005; Vazquez- Prokopec et al., 2006), the time elapsed since establishment of each bug colony might not have been sufficient for genic frequencies to homogenize among houses (Boileau et al., 1992). Moreover, the effects of genetic drift in small, recently established populations could randomly fix certain allele variants in different populations if migration among them is not significant; this process would increase differentiation among populations.
Genetic differentiation and geographic distance among populations
The amount of divergence between populations (measured by Fst values) did not correlate with geographical distance among houses at the micro-geographical level studied here (pairwise distance < 30 km). Lack of fit to a model of IBD was also observed in T. infestans populations studied with microsatellites (Richer et al., 2007; Pizarro et al., 2008), and among some localities in a macrogeographical study in which data fit to a IBD (Perez de Rosas et al., 2007b). Genetic structure of T. infestans populations recovering after massive insecticide spraying reflect populations that are established discretely at an individual structure and exchange more migrants with closer sites than with sites located further away. In this context, T. infestans populations in our study area may not reach migration-drift equilibrium (which would be reflected in an IBD pattern) because of frequent residual spraying with insecticides, seasonal variations in bug population size, and variations in rates and sources of dispersal. Moreover, when considering distances among suitable ecotopes for T. infestans (i.e., different houses or peridomestic structures) located within its dispersal range, bug dispersal among sites might be determined by factors such as host availability or microclimatic conditions within a structure (Lehane et al., 1992; Ceballos et al., 2005; Vazquez-Prokopec et al., 2006) rather than distance among sites.
Sex-biased dispersal
Female-biased dispersal was observed in bug populations of this study which agrees with evidence showing higher flight initiation probabilities in females than males (Lehane & Schofield, 1982; Williams & Schofield, 1985; Canale & Carcavallo, 1988; Lehane et al., 1992; Galvao et al., 2001; Gurevitz et al., 2006). Moreover, Gurevitz et al. (2007) reported larger flight muscle mass in females, which would imply that females have higher capacity than males to carry heavier loads as well as have longer and broader flight ranges. Nevertheless, flight dispersal of T. infestans is not sex-restricted; more males than females were collected by light-trapping in small-scale field studies (Vazquez-Prokopec et al., 2004). In our study, among the putative first-generation migrants detected, the number of males (14) was almost three times higher than females (5). However, it must be noted that most of the T. infestans in this study were male, as was also the case in light-trap and timed bug collections (Vazquez-Prokopec et al., 2004).
Substructure within a house compound
Substructure within houses accounts for the strong deviations from HW equilibrium observed in some houses. The effect of pooling together individuals from different sub-populations (Wahlund effect) might be the cause of significant heterozygote deficit. The presence of migrants (probably from more than one source), whose genic frequencies have not yet reached the values expected under random mating, might be reflected in the observed deviation from HW equilibrium. Significant differentiation between two peridomestic structures of the same house (A105) was observed, which leads to the hypothesis of independent sources of reinfestation acting simultaneously.
Inbreeding and the effect of null alleles may also produce heterozygote deficit. However, the effects of inbreeding would have caused heterozygote deficit in all or most loci, which did not occur in any population studied here. The presence of null alleles cannot be completely ruled out. However, we did not detect individual samples that repeatedly failed to amplify a particular locus while amplifying the remaining loci. Therefore it is highly unlikely that the presence of homozygotes due to the presence of null alleles could cause biases in our results.
T. infestans population genetic structure and reinfestation process after residual spraying
The pattern of genetic structure and results of assignment tests in Amamá indicate the occurrence of active dispersal of bugs among particular house compounds within the community, whereas the presence of sub-structuring within the community reflects different degrees of gene flow among houses. T. infestans populations from two houses separated by 115 m in southern Amamá (A44 and A45, Fig. 5a) were not differentiated from each other by microsatellite markers or by wing geometric morphometry (Schachter-Broide et al., 2004). Light-trap collections performed around the infested sites provided evidence of active dispersal of T. infestans (Vazquez-Prokopec et al., 2004). Moreover, in the pig corral from A46 (house not used in this study), 2 bugs (a female and one fifth-instar nymph) of T. infestans were captured in October 2002 and the multilocus genotype for the female was assigned to the genetic cluster conformed mostly by bugs from A44 and A45 (data not shown).
Five months after a massive residual insecticide spraying in Amamá in December 1997, a few nymphs were found in the kitchen of house A45 and a colony was detected there one year later. Seventeen months after the spraying T. infestans bugs were found at two corrals of the neighboring house A44 (found newly infested since before the 1997 insecticide spraying). The genetic cluster from samples of the southern Amamá area supports the hypothesis of a unique source of reinfestation (most likely local survivors after insecticide spraying) and subsequent active bug dispersal among sites. These results are consistent with the spatial analysis of reinfestations from 1993 to 1997 that showed that in southern Amamá, a single pig corral was the most likely source of bugs for sites within a 400 m radius (Cecere et al., 2004).
In contrast to the latter pattern, the bug population from house A105 (Fig. 5a) was significantly differentiated from other study populations in Amamá. Our results indicate that despite the exchange of dispersants between A105 and populations from southern Amamá, most A105 bugs did not belong to the same genetic cluster. The A105 population presented 13 private alleles in 8 loci, 10 of them not found in other houses in Amamá and 3 not found in any other community evaluated herein. This strongly suggests an independent origin of A105 populations. Both the spatio-temporal analysis of reinfestation in Amamá during 1993–1997 and the wing geometric morphometry of T. infestans in year 2000 showed that the northern infestation source appeared to be independent from the southern sources (Cecere et al., 2004; Schachter-Broide et al., 2004). Unfortunately, bug samples from northern houses were not available for analysis to corroborate if A105 belongs to a northern cluster.
The genic and genotype profile of bug populations from A5 and A24 houses reflect an admixed origin, possibly due to immigration from independent sources, and a posterior mixture of migrants with local individuals. Both sites presented many bugs (40 and 47%, respectively) that were not assigned to a particular genetic cluster. These insects are interpreted as migrants from sites external to the samples analyzed here or insects that could be assigned with the same probability to more than one cluster. The latter might be a second-generation product of a cross between migrant and local insects relative to the site where they were captured. The absence of private alleles and results of exclusion tests for the putative origin of first-generation migrants support the second option.
The stronger differentiation among populations from Trinidad, Mercedes and Pampa Pozo (situated at 8 km from Amamá) may be explained by the relative isolation of houses, and therefore, the lower probability of active dispersal with consequent decrease in gene flow occurring among them. Significant differences in the number of flight-dispersing adult T. infestans between Amamá and Trinidad villages were observed in light-trap studies (Vazquez- Prokopec et al., 2004). Morphometric analyses of wings were in agreement with these results, showing that individual bugs from Mercedes, Trinidad and San Pablo were clearly grouped by populations whereas bugs from Amamá overlapped with bugs from all sample sites from all communities (Schachter-Broide et al., unpublished results).
Bug populations from La Curva and Celestina (belonging to the peripheral area but geographically close to Trinidad, Mercedes and Pampa Pozo) were significantly differentiated from all populations of the latter three villages. This finding lends supports to the hypothesis of reinfestation due to local survivors within the village rather than to the invasion from external sources in the case of Trinidad and Pampa Pozo (Cecere et al., 2006). Nevertheless, infrequent events of bug introduction from external sites, with subsequent local differentiation as a result of both isolation and genetic drift, may also account for this pattern. Further analysis of a larger number of bug populations before and after residual insecticide spraying with adequate follow up over time combined with morphometric and spatial analysis studies, will lead to better insight of the observed population structure patterns and disentangle underlying mechanisms.
The results presented in this work provide detailed and reliable information on natural populations of T. infestans at the capture site level, which on the one side corroborated the genetic heterogeneity of populations, indicating that the micro-geographical scale is the best approach for population analysis, and on the other, provides specific insights on each population that together with the detailed eco-epidemiological information gathered for this area, allowed to establish the potential sources of reinfestation and the interactions among populations. This knowledge would be applied in upcoming vector control interventions evaluations in this area as well as a baseline information for populations genetic studies in different endemic regions.
ACKNOWLEDGMENTS
The authors would like to thank: G. Lawrence, L.A. Ceballos, M.V. Cardinal, M.C. Cecere, G.M. Vazquez-Prokopec, D.M. Canale and R. Stariolo who participated in field or laboratory work; to A. Peixoto, A.G. Schijman, V. Confalonieri and two anonymous referees for valuable comments and suggestions on earlier versions of the manuscript. To the CDC core facilities for providing the oligonucleotides. This study was supported by awards from the National Institutes of Health/National Science Foundation Ecology of Infectious Disease program award R01 TW05836 funded by the Fogarty International Center and the National Institute of Environmental Health Sciences to U. Kitron and R.E. Gürtler, the Agencia Nacional de Promoción Científica y Técnica (Argentina), and University of Buenos Aires to R.E. Gürtler. R.E. Gürtler is member of CONICET Researcher’s Career.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Anderson JM, Lai JE, Dotson EM, Cordon-Rosales C, Ponce C, Norris DE, Beard CB. Identification and characterization of microsatellite markers in the Chagas disease vector Triatoma dimidiata. Infect. Genet. Evol. 2002;1(3):243–248. doi: 10.1016/s1567-1348(02)00033-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boileau MG, Hebert PDN, Schwartz SS. Non-equilibrium gene frequency divergence: persistent founder effects in natural populations. Journal of Evolutionary Biology. 1992;5(1):25–39. [Google Scholar]
- Canale DM, Carcavallo RU. Triatoma infestans (Klug) In: Carcavallo RR, JE, Tonn RJ, editors. Factores biológicos y ecológicos en la Enfermedad de Chagas. Buenos Aires, Argentina: Ministerio de Salud y Acción Social de Argentina; 1988. [Google Scholar]
- Cardinal MV, Lauricella MA, Marcet PL, Orozco MM, Kitron U, Gürtler RE. Impact of community-based vector control on house infestation and Trypanosoma cruzi infection in Triatoma infestans, dogs and cats in the Argentine Chaco. Acta Trop. 2007;103(3):201–211. doi: 10.1016/j.actatropica.2007.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceballos LA, Vazquez-Prokopec GM, Cecere MC, Marcet PL, Gürtler RE. Feeding rates, nutritional status and flight dispersal potential of peridomestic populations of Triatoma infestans in rural northwestern Argentina. Acta Trop. 2005;95(2):149–159. doi: 10.1016/j.actatropica.2005.05.010. [DOI] [PubMed] [Google Scholar]
- Cecere MC, Gürtler RE, Canale DM, Chuit R, Cohen JE. Effects of partial housing improvement and insecticide spraying on the reinfestation dynamics of Triatoma infestans in rural northwestern Argentina. Acta Trop. 2002;84(2):101–116. doi: 10.1016/s0001-706x(02)00183-3. [DOI] [PubMed] [Google Scholar]
- Cecere MC, Vazquez-Prokopec GM, Gürtler RE, Kitron U. Spatio-temporal analysis of reinfestation by Triatoma infestans (Hemiptera: Reduviidae) following insecticide spraying in a rural community in northwestern Argentina. Am. J. Trop. Med. Hyg. 2004;71(6):803–810. [PMC free article] [PubMed] [Google Scholar]
- Cecere MC, Vazquez-Prokopec GM, Gürtler RE, Kitron U. Reinfestation sources for Chagas disease vector, Triatoma infestans, Argentina. Emerging Infect. Dis. 2006;12(7):1096–1102. doi: 10.3201/eid1207.051445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias JC, Silveira AC, Schofield CJ. The impact of Chagas disease control in Latin America: a review. Mem. Inst. Oswaldo Cruz. 2002;97(5):603–612. doi: 10.1590/s0074-02762002000500002. [DOI] [PubMed] [Google Scholar]
- El Mousadik A, Petit R. High level of genetic differentiation for allelic richness among populations of the argan tree (Argania spinosa (L.) Skeels) endemic to Morocco. Theor. Appl. Genet. 1996;92:832–839. doi: 10.1007/BF00221895. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131(2):479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier LG, Laval G, Schneider S. Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evol. Bioinform. Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164(4):1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Molecular Ecology Notes. 2007 doi: 10.1111/j.1471-8286.2007.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favre L, Balloux F, Goudet J, Perrin N. Female-biased dispersal in the monogamous mammal Crocidura russula: evidence from field data and microsatellite patterns. Proc Biol Sci. 1997;264(1378):127–132. doi: 10.1098/rspb.1997.0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvao C, Rocha DS, Jurberg J, Carcavallo R. Flight initiation in Triatoma infestans and T. melanosoma (Hemiptera, Reduviidae) Mem. Inst. Oswaldo Cruz. 2001;96(1):137–140. doi: 10.1590/s0074-02762001000100017. [DOI] [PubMed] [Google Scholar]
- García BA, Zheng LO, Pérez de Rosas AR, Segura EL. Primer note. Isolation and characterization of polymorphic microsatellite loci in the Chagas’ disease vector Triatoma infestans (Hemiptera: Reduviidae) Mol. Ecol. Notes. 2004;4:568–571. [Google Scholar]
- Garnier S, Alibert P, Audiot P, Prieur B, Rasplus J-Y. Isolation by distance and sharp discontinuities in gene frequencies: implications for the phylogeography of an alpine insect species, Carabus solieri. Mol. Ecol. 2004;13:1883–1897. doi: 10.1111/j.1365-294X.2004.02212.x. [DOI] [PubMed] [Google Scholar]
- Goudet J. Fstat version 1.2: a computer program to calculate F-statistics. J. Hered. 1995;86:485–486. [Google Scholar]
- Goudet J, Perrin N, Waser P. Tests for sex-biased dispersal using bi-parentally inherited genetic markers. Mol. Ecol. 2002;11(6):1103–1114. doi: 10.1046/j.1365-294x.2002.01496.x. [DOI] [PubMed] [Google Scholar]
- Gurevitz JM, Ceballos LA, Kitron U, Gürtler RE. Flight initiation of Triatoma infestans (Hemiptera: Reduviidae) under natural climatic conditions. J. Med. Entomol. 2006;43(2):143–150. doi: 10.1603/0022-2585(2006)043[0143:fiotih]2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevitz JM, Kitron U, Gürtler RE. Flight muscle dimorphism and heterogeneity in flight initiation of field-collected Triatoma infestans (Hemiptera: Reduviidae) J. Med. Entomol. 2007;44(2):186–191. doi: 10.1603/0022-2585(2007)44[186:fmdahi]2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gürtler RE, Cecere MC, Canale DM, Castanera MB, Chuit R, Cohen JE. Monitoring house reinfestation by vectors of Chagas disease: a comparative trial of detection methods during a four-year follow-up. Acta Trop. 1999;72(2):213–234. doi: 10.1016/s0001-706x(98)00096-5. [DOI] [PubMed] [Google Scholar]
- Gürtler RE, Canale DM, Spillmann C, Stariolo R, Salomon OD, Blanco S, Segura EL. Effectiveness of residual spraying of peridomestic ecotopes with deltamethrin and permethrin on Triatoma infestans in rural western Argentina: a district-wide randomized trial. Bull. World Health Organ. 2004;82(3):196–205. [PMC free article] [PubMed] [Google Scholar]
- Gürtler RE. Eco-epidemiología regional de la transmisión vectorial: Enfermedad de Chagas en el Gran Chaco. In: Silveira AC, editor. La enfermedad de Chagas. A la puerta de los 100 años del conocimiento de una endemia americana ancestral. Balance y futuro, 1909–2006. Chagas, hacia el Siglo XXI. Organización Panamericana de la Salud; 2007. [Google Scholar]
- Gürtler RE, Kitron U, Cecere MC, Segura EL, Cohen JE. Sustainable vector control and management of Chagas disease in the Gran Chaco, Argentina. Proc. Natl. Acad. Sci. U. S. A. 2007;104(41):16194–16199. doi: 10.1073/pnas.0700863104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harry M, Poyet G, Romana CA, Solignac M. Isolation and characterization of microsatellite markers in the bloodsucking bug Rhodnius pallescens (Heteroptera, Reduviidae) Mol. Ecol. 1998;7(12):1784–1786. doi: 10.1046/j.1365-294x.1998.00519.x. [DOI] [PubMed] [Google Scholar]
- Harry M, Dupont L, Romana C, Demanche C, Mercier A, Livet A, Diotaiuti L, Noireau F, Emperaire L. Microsatellite markers in Triatoma pseudomaculata (Hemiptera, Reduviidae, Triatominae), Chagas' disease vector in Brazil. Infect Genet Evol. 2008a doi: 10.1016/j.meegid.2008.03.005. [DOI] [PubMed] [Google Scholar]
- Harry M, Roose CL, Vautrin D, Noireau F, Romaña CA, Solignac M. Microsatellite markers from the Chagas disease vector, Rhodnius prolixus (Hemiptera, Reduviidae), and their applicability to Rhodnius species. Infection, Genetics and Evolution. 2008b;8(3):381–385. doi: 10.1016/j.meegid.2008.01.006. [DOI] [PubMed] [Google Scholar]
- Lehane M, Schofield CJ. Flight initiation in Triatoma infestans (Klug) (Hemiptera: Reduviidae) Bull. Entomol. Res. 1982;72:497–510. [Google Scholar]
- Lehane MJ, McEwen PK, Whitaker CJ, Schofield CJ. The role of temperature and nutritional status in flight initiation by Triatoma infestans. Acta Trop. 1992;52:27–38. doi: 10.1016/0001-706x(92)90004-h. [DOI] [PubMed] [Google Scholar]
- Lia V. Doctoral Thesis. Dept. Ecología, Genética y Evolución. Buenos Aires, Argentina: Univ. de Buenos Aires; 2004. Diversidad genética y estructura poblacional en razas nativas de maíz (Zea mays ssp. mays) del Noroeste Argentino: presente y pasado del germoplasma autóctono; p. 171. [Google Scholar]
- Marcet PL, Lehmann T, Groner G, Gürtler RE, Kitron U, Dotson EM. Identification and characterization of microsatellite markers in the Chagas disease vector Triatoma infestans (Heteroptera: Reduviidae) Infect. Genet. Evol. 2006;6(1):32–37. doi: 10.1016/j.meegid.2005.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. Molecular Evolutionary Genetics. New York: Columbia University Press; 1987. [Google Scholar]
- Paetkau D, Slade R, Burden M, Estoup A. Genetic assignment methods for the direct, real-time estimation of migration rate: a simulation-based exploration of accuracy and power. Mol. Ecol. 2004;13(1):55–65. doi: 10.1046/j.1365-294x.2004.02008.x. [DOI] [PubMed] [Google Scholar]
- Perez de Rosas AR, Segura EL, Fichera L, Garcia BA. Macrogeographic and microgeographic genetic structure of the Chagas' disease vector Triatoma infestans (Hemiptera: Reduviidae) from Catamarca, Argentina. Genetica. 2007a doi: 10.1007/s10709-007-9208-8. [DOI] [PubMed] [Google Scholar]
- Perez de Rosas AR, Segura EL, Garcia BA. Microsatellite analysis of genetic structure in natural Triatoma infestans (Hemiptera: Reduviidae) populations from Argentina: its implication in assessing the effectiveness of Chagas' disease vector control programmes. Mol. Ecol. 2007b;16(7):1401–1412. doi: 10.1111/j.1365-294X.2007.03251.x. [DOI] [PubMed] [Google Scholar]
- Piry S, Alapetite A, Cornuet JM, Paetkau D, Baudouin L, Estoup A. GENECLASS2: a software for genetic assignment and first-generation migrant detection. J. Hered. 2004;95(6):536–539. doi: 10.1093/jhered/esh074. [DOI] [PubMed] [Google Scholar]
- Pizarro JC, Gilligan LM, Stevens L. Microsatellites reveal a high population structure in Triatoma infestans from Chuquisaca, Bolivia. PLoS Neglected Tropical Diseases. 2008;2(3):e202. doi: 10.1371/journal.pntd.0000202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155(2):945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rannala B, Mountain JL. Detecting immigration by using multilocus genotypes. Proc. Natl. Acad. Sci. U. S. A. 1997;94(17):9197–9201. doi: 10.1073/pnas.94.17.9197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond M, Rousset F. GENEPOP. (version 1.2): population genetics software for exact tests and ecumenicism. J. Heredity. 1995;86:248–249. [Google Scholar]
- Rice W. Analyzing tables of statistical tests. Evolution. 1989;43:223–225. doi: 10.1111/j.1558-5646.1989.tb04220.x. [DOI] [PubMed] [Google Scholar]
- Richer W, Kengne P, Cortez MR, Perrineau MM, Cohuet A, Fontenille D, Noireau F. Active dispersal by wild Triatoma infestans in the Bolivian Andes. Trop. Med. Int. Health. 2007;12(6):759–764. doi: 10.1111/j.1365-3156.2007.01846.x. [DOI] [PubMed] [Google Scholar]
- Rousset F. Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics. 1997;145(4):1219–1228. doi: 10.1093/genetics/145.4.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachter-Broide J, Dujardin JP, Kitron U, Gürtler RE. Spatial structuring of Triatoma infestans (Hemiptera, Reduviidae) populations from northwestern Argentina using wing geometric morphometry. J. Med. Entomol. 2004;41(4):643–649. doi: 10.1603/0022-2585-41.4.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield CJ. Control of Chagas'Disease vectors. Brit Med Bull. 1985;41(2):187–194. doi: 10.1093/oxfordjournals.bmb.a072048. [DOI] [PubMed] [Google Scholar]
- Schofield CJ, Jannin J, Salvatella R. The future of Chagas disease control. Trends Parasitol. 2006;22(12):583–588. doi: 10.1016/j.pt.2006.09.011. [DOI] [PubMed] [Google Scholar]
- Schweigmann N, Vallvé S, Muscio O, Ghillini M, Alberti A, Wisnivesky - Colli C. Dispersal flight by Triatoma infestans in an arid area of Argentina. Med. Vet. Entomol. 1988;2 doi: 10.1111/j.1365-2915.1988.tb00215.x. [DOI] [PubMed] [Google Scholar]
- Slatkin M, Hudson RR. Pairwise comparisons of mitochondrial DNA sequences in stable and exponentially growing populations. Genetics. 1991;129(2):555–562. doi: 10.1093/genetics/129.2.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M. Isolation by distance in equilibrium and non-equilibrium populations. Evolution. 1993;47(1):264–279. doi: 10.1111/j.1558-5646.1993.tb01215.x. [DOI] [PubMed] [Google Scholar]
- Vazquez-Prokopec GM, Ceballos LA, Kitron U, Gürtler RE. Active dispersal of natural populations of Triatoma infestans (Hemiptera: Reduviidae) in rural north-western Argentina. J. Med. Entomol. 2004;41(4):614–621. doi: 10.1603/0022-2585-41.4.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Prokopec GM, Ceballos LA, Marcet PL, Cecere MC, Cardinal MV, Kitron U, Gürtler RE. Seasonal variations in active dispersal of natural populations of Triatoma infestans in rural northwestern Argentina. Med. Vet. Entomol. 2006;20(3):273–279. doi: 10.1111/j.1365-2915.2006.00637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Williams NG, Schofield CJ. The role of temperature in flight initiation of triatomine bugs. Trans. R. Soc. Trop. Med. Hyg. 1985;79:282. [Google Scholar]