Abstract
Background
16S rRNA methylase-producing Gram-negative bacteria are highly resistant to all clinically important aminoglycosides. We analyzed clinical strains of 16S rRNA methylase-producing Acinetobactor baumannii and Pseudomonas aeruginosa obtained from clinical isolates in medical settings in Vietnam.
Methods
From 2008 to 2011, 101 clinical strains of A. baumannii and 15 of P. aeruginosa were isolated from patients in intensive care units (ICUs) in two medical settings in Vietnam. Antimicrobial susceptibilities were determined using the microdilution method and epidemiological analysis was performed by pulsed-field gel electrophoresis and MLST. Genes encoding the 16S rRNA methylases, OXAs and CTX-Ms were analyzed by PCR and sequence analysis.
Results
16S rRNA methylase-producing Gram-negative pathogens were detected in two hospitals in Vietnam. Of the 101 clinical isolates of A. baumannii and the 15 of P. aeruginosa isolated from two ICUs in these hospitals, 72 (71.3%) were highly resistant to amikacin, arbekacin and gentamicin, with MICs greater than 1,024 mg/L. The 16S rRNA methylases ArmA and RmtB were produced by 61 and 9 isolates of A. baumannii, respectively, and RmtB was produced by 2 isolates of P. aeruginosa. Moreover, 52 of the A. baumannii isolates producing 16S rRNA methylases harbored both blaOXA-23-like and blaOXA-51-like genes. Most A. baumannii isolates producing 16S rRNA methylase obtained in hospital A in Hanoi were ST91 and ST231, whereas most from hospital B in Ho Chi Minh City were ST136, ST195, and ST254.
The two P. aeruginosa isolates harboring rmtB showed different patterns on PFGE, one each corresponding to ST217 and ST313.
Conclusions
Gram-negative bacteria producing the 16S rRNA methylases ArmA and RmtB are emerging in medical settings in Vietnam. A. baumannii isolates in northern and southern regions of Vietnam may be of different lineages.
Keywords: Acinetobacter baumannii, Pseudomonas aeruginosa, Aminoglycoside resistance, Intensive care unit, 16S rRNA methylase
Background
Aminoglycosides widely used to treat infectious diseases caused by Gram-negative bacteria have a high affinity for the 16S rRNA of the bacterial 30S ribosome and block protein synthesis [1]. Enzymatic modification [1] and the methylation of 16S rRNA makes these bacteria highly resistant to all clinically important aminoglycosides [2]. In 2003, clinical isolates of highly aminoglycoside-resistant Gram-negative bacteria producing16S rRNA methylase were identified in France [3] and Japan [4]. Since then, 16S rRNA methylase-producing Gram-negative bacteria have been isolated in other parts of the world, including Asian countries such as Afghanistan, Bangladesh, China, Hong Kong, India, Japan, Korea, Oman and Pakistan [5]. To date, however, there have been no reports of these isolates from South-East Asian countries, including Vietnam.
Since 2003, eight plasmid-associated 16S rRNA methylase genes, armA, rmtA, rmtB, rmtC, rmtD, rmtE, rmtF and npmA, have been identified in clinical and veterinary isolates from various geographic areas, including East Asia, Europe and the Americas, since 2003 [5,6].
Methods
Bacterial strains
From 2008 to 2011, 50 clinical strains of A. baumannii and 15 of P. aeruginosa were isolated from patients in an ICU in hospital A in Hanoi, Vietnam; and 51 strains of A. baumannii were isolated from patients in an ICU in hospital B in Ho Chi Minh City, Vietnam. Of the 101 A. baumannii strains isolated, 98 were from patients’ respiratory tracts and 3 from blood. Of the 15 P. aeruginosa strains, 14 were from respiratory tracts and 1 from pus. Most patients were on ventilators, and the samples were mostly aspirates from ventilation tubes. All clinical isolates used in this study were obtained during standard patient care.
Antimicrobial susceptibility and pulsed-field gel electrophoresis (PFGE)
MICs of all bacteria to amikacin (Sigma-Aldrich, St. Louis, MO), arbekacin (Meiji Seika Pharma Co., Tokyo, Japan), aztreonam (Eizai, Tokyo, Japan), ceftadizime (Sigma-Aldrich), ciprofloxacin (Daiichi Pharmaceutical Co, Tokyo, Japan), colistin (Sigma-Aldrich), gentamicin (Nacalai Tesque, Kyoto, Japan), imipenem (Banyu Pharmaceutical Co, Tokyo, Japan), meropenem (Sumitomo Pharmaceutical Co., Osaka, Japan), piperacillin (Sigma-Aldrich) and pipiracillin/tazobactam (Toyama Chemical Co., Tokyo, Japan) were determined using the microdilution method, according to the guidelines of the Clinical and Laboratory Standards Institute (M07-A9). A. baumannii DNA was digested with the restriction enzyme ApaI and P. aeruginosa DNA was digested with SpeI, followed by pulsed-field gel electrophoresis (PFGE). PFGE analysis was performed as described previously [7]. Fingerprinting patterns were analyzed by the unweighted-pair-group method using Molecular Analyst Fingerprinting Plus software (Bio-Rad Laboratories, Hercules, CA, USA) to create an average linkage-based dendrogram.
Multilocus sequence typing (MLST)
MLST of 16S rRNA methylase-producing pathogens was performed according to the protocols described on the A. baumannii (http://pubmlst.org/abaumannii/) and P. aeruginosa (http://pubmlst.org/paeruginosa/) MLST Database websites. Seven chromosomal genes were PCR amplified and sequenced, with their nucleotide sequences compared with the sequences submitted to the MLST database to determine allele numbers and STs.
Detection of aminoglycoside-resistant genes
PCR with 16S rRNA methylase gene specific primers [2,8,9] was performed to detect the armA, rmtA, rmtB, rmtC, rmtD, rmt E and npmA genes. All PCR amplicons were sequenced using an ABI PRISM 3130 sequencer (Applied Biosystems, Foster City, CA, USA).
Whole genomes of methylase-negative A. baumannii and P. aeruginosa, which had MICs 128 mg/L to amikacin, 32 mg/L to arbekacin and 128 mg/L to gentamicin, were extracted by DNeasy Blood & Tissue kit (QIAGEN, Tokyo, Japan) and sequenced by MiSeq (Illumina, San Diego, CA). The sequence data were used to confirm aminoglycoside-resistant genes.
Detection of OXAs and CTX-Ms encoding genes
The presence of blaOXA-23-like, blaOXA-24-like, blaOXA-51-like, blaOXA-58-like and blaCTX-Ms in 16S rRNA methylase-producing isolates were investigated by PCR [10,11].
Determination of the genetic environment surrounding rmtB
A draft genome sequence of an isolate of A. baumannii, NCGM36, harboring rmtB was determined using the GS Junior System (Roche Diagnostics K.K, Tokyo).
Ethical approval
This study was approved in 2007 by Ministry of Health, Bach Mai Hospital (Memorandum of agreement for the collaborative research project on epidemiology of nosocomial infections at the Bach Mai Hospital) and in 2011 by Cho Ray Hospital (approval number: 1644/QD-BVCR), and by the Biosafety Committee, National Center for Global Health and Medicine (approval number: 23-M-49).
Results
Antimicrobial susceptibility and aminoglycoside-resistant genes
The MICs at which 50% and 90% of the 101 A. baumannii and 15 P. aeruginosa isolates were inhibited (MIC50 and MIC90, respectively) were determined (Table 1). Seventy of the 101 A. baumannii isolates (71.3%) had MICs >1,024 mg/L to all aminoglycosides tested, including amikacin, arbekacin and gentamicin. All 70 isolates had 16S rRNA methylases, with 61 having armA and the remaining 9 having rmtB (Figure 1). The remaining 31 isolates had MICs ≤128 mg/L to amikacin, ≤32 mg/L to arbekacin and ≤128 mg/L to gentamicin and no methylase genes. Whole genome sequencing of 2 methylase-negative isolates showing relative resistance to aminoglycosides revealed that one had aac(6’)-IIb and aadB and that the other had aac(6’)-IIb and aadA2.
Table 1.
MIC50 and MIC90 values and percent antimicrobial resistance for A. baumannii and P. aeruginosa clinical isolates
| Antimicrobial agent | |
A. baumannii (n = 101) |
P. aeruginosa (n = 15) |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Breakpoint forresistancea (mg/L) | %Resistance | Range(mg/L) | MIC50(mg/L) | MIC90(mg/L) | Breakpoint forresistancea (mg/L) | %Resistance | Range(mg/L) | MIC50(mg/L) | MIC90(mg/L) | |
| Amikacin |
≥64 |
85 |
<2- > 1024 |
>1024 |
>1024 |
≥64 |
60 |
<2- > 1024 |
64 |
>1024 |
| Arbekacin |
- |
- |
<2- > 1024 |
>1024 |
>1024 |
- |
- |
2- > 1024 |
2 |
>1024 |
| Aztreonam |
- |
- |
<2- > 256 |
128 |
>256 |
≥32 |
46 |
4- > 256 |
16 |
>256 |
| Ceftazidime |
≥32 |
55 |
<4- > 512 |
>512 |
>512 |
≥32 |
46 |
<4- > 512 |
16 |
>512 |
| Ciprofloxacin |
≥4 |
98 |
<1- > 128 |
128 |
>128 |
≥4 |
66 |
<1-64 |
8 |
32 |
| Colistin |
≥4 |
6 |
<0.25-4 |
2 |
2 |
≥8 |
0 |
<0.25-0.5 |
<0.25 |
0.5 |
| Gentamicin |
≥16 |
91 |
<2- > 1024 |
>1024 |
>1024 |
≥16 |
66 |
<2- > 1024 |
256 |
>1024 |
| Imipenem |
≥16 |
48 |
<4-128 |
16 |
32 |
≥8 |
53 |
<4-64 |
16 |
32 |
| Meropenem |
≥16 |
51 |
<4- > 256 |
32 |
64 |
≥8 |
53 |
<4-32 |
16 |
32 |
| Piperacillin |
≥128 |
100 |
<4- > 512 |
>512 |
>512 |
≥128 |
46 |
<4- > 512 |
32 |
256 |
| Piperacillin/Tazobactam | ≥128/4 | 52 | <4- > 512 | 256 | 512 | ≥128/4 | 13 | <4-512 | 32 | 128 |
aBreakpoints for antimicrobial resistance were determined according to the guidelines of the Clinical and Laboratory Standards Institute (M07-A9).
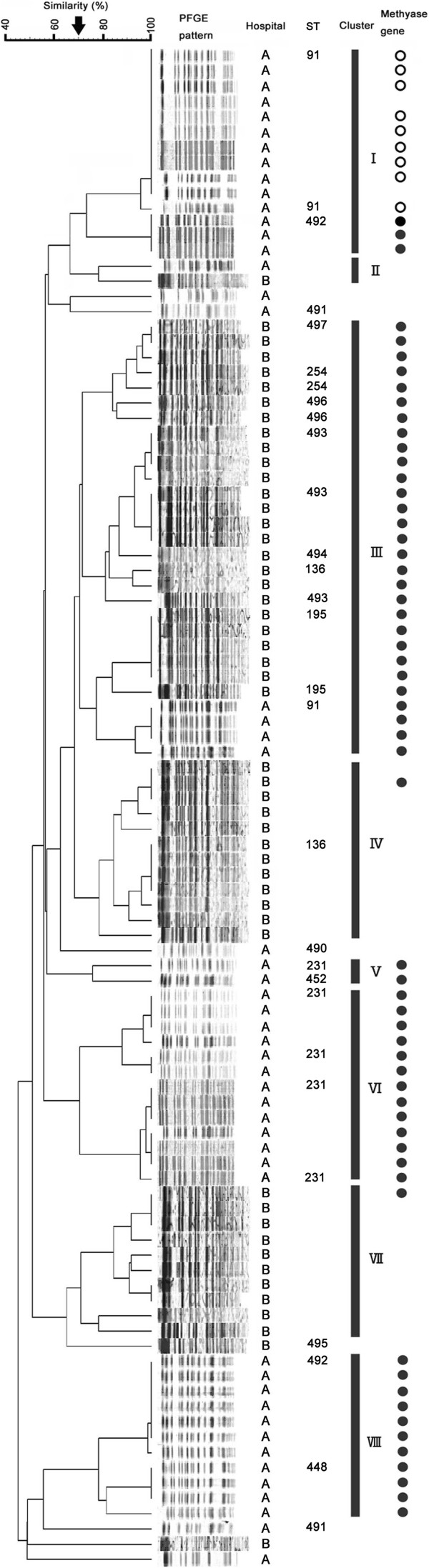
Figure 1.
PFGE pattern and MLST analysis of 101 Acinetobacter baumannii isolates. Eight clusters (I-VIII) with more than 70% similarity were identified. Isolates harboring armA (●) and rmtB (○) are shown in the column on the right.
Of the 15 P. aeruginosa isolates, 2 had MICs >1,024 mg/L to amikacin, arbekacin and gentamicin, and harbored the 16S rRNA methylase rmtB (Figure 2). The 13 methylase-negative isolates had MICs <2 - 256 mg/L to amikacin (MIC50 64 mg/L and MIC90 128 mg/L), 1–32 mg/L to arbekacin (MIC50 2 mg/L and MIC90 4 mg/L), and 1–32 mg/L to <0.5 - 512 mg/L to gentamicin (MIC50 256 mg/L and MIC90 512 mg/L). The remaining 13 did not have any methylase genes (Figure 2).
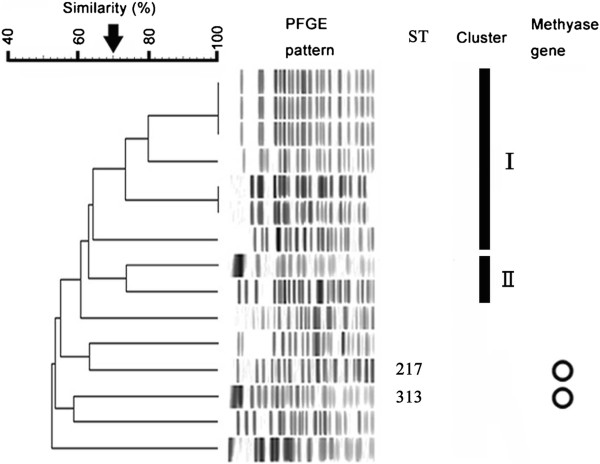
Figure 2.
PFGE pattern and MLST analysis analysis of 15 Pseudomonas aeruginosa isolates. Eight clusters with more than 70% similarity were detected. Two clusters (I and II) with more than 70% similarity were identified. Isolates harboring rmtB (○) are shown in the column on the right.
OXAs and CTX-Ms encoding genes in 16S rRNA methylase-producing isolates
Of the 61 A. baumannii isolates harboring armA, 1 had blaOXA-23-like, blaOXA-51-like and blaCTX-Ms genes, 51 had blaOXA-23-like and blaOXA-51-like genes, and 8 had blaOXA-51-like genes. All 9 A. baumannii isolates harboring rmtB had blaOXA-23-like and blaOXA-51-like genes. In contrast, the 2 P. aeruginosa isolates harboring16S rRNA methylase genes had neither the blaOXAs nor the blaCTX-Ms gene.
PFGE analysis and MLST
PFGE analysis of the 101 A. baumannii isolates revealed 8 clusters (Figure 1). Isolates from Clusters I, III, IV, V, VI, VII, and VIII were obtained from either one or the other hospital, whereas isolates from Clusters II and III were obtained from both. These results indicate that A. baumannii isolates had expanded in a clonal manner in both hospitals and that some isolates may spread among hospitals in Vietnam.
The16S rRNA methylase-encoding the rmtB gene was detected in Cluster I A. baumannii isolates, whereas armA was present in isolates from Clusters I, III, IV, V, VI, VII, and VIII. Isolates harboring rmtB were obtained from one hospital and isolates harboring armA were from both hospitals.
The A. baumannii isolates producing 16S rRNA methylase belonged to ST254, ST231, ST195, ST136, ST91 and 8 new STs, ST490, ST491, ST492, ST493, ST494, ST495, ST496 and ST497 (Figure 1). Most of the A. baumannii isolates producing 16S rRNA methylase from hospital A in Hanoi were ST91 and ST231, whereas most from hospital B in Ho Chi Minh City were ST136, ST195 and ST254.
The two isolates harboring rmtB showed different patterns on PFGE, belonging to ST217 and ST313.
Genetic environment surrounding rmtB
The rmtB gene was associated with an ISCR3 mobile element upstream and a Tn3 transposon structure blaTEM-1-tnpR-tnpA downstream (data not shown). The genetic environment of rmtB had more than 99.9% nucleotide sequence identity, from nucleotide 1 to 8,337, to plasmid pXD2 (Gen bank accession no. JN315966) in E. coli, which causes bovine milk mastitis in China [12]. NCGM36, which harbored rmtB, had the blaOXA-23 and blaOXA-68 genes, but had neither the aac(6’)-Ib-cr nor the blaCTX-Ms gene.
Discussion
The high prevalence of 16S rRNA methylase producing Gram-negative bacteria in hospitals in Vietnam may have resulted from the high rate of use of aminoglycosides. It has been estimated that 67.4% of hospitalized patients in Vietnam received antibiotics, including 18.9% who received aminoglycosides, with many 30.8% of these prescriptions considered inappropriate [13]. This rate of antibiotic use was much higher than in European countries (17.8%-32.0%) [14,15]. Moreover, the rate of inappropriate indications for antibiotic prescriptions in hospitals in Vietnam was much higher than rates reported in Malaysia (4.0%) [16], Turkey (14.0%) [17], and Hong Kong (20.0%) [18].
A. baumannii isolates from the northern and southern regions of Vietnam may be of different lineages. To date, 2 strains of A. baumannii showing ST91 and 3 showing ST136 have been isolated in China; 6 strains showing ST195 have been isolated, 1 in Norway, 2 in Thailand, 2 in Malaysia and 1 in China;, 5 strains showing ST231 have been isolated in Brazil and 1 strain showing ST254 has been isolated in China (http://pubmlst.org/abaumannii/). ST136 and ST195 belong to clonal complex 92, the most widely disseminated complex worldwide [19]. Two strains of P. aeruginosa producing RmtB, showing ST217 and ST313, may have originally derived from Europe or Australia, because, to date, P. aeruginosa ST217 isolates were obtained only in the United Kingdom and ST313 isolates only in Australia, France and Hungary [20] (http://pubmlst.org/paeruginosa/).
To our knowledge, this is the first report showing that A. baumannii strains harboring a 16S rRNA methylase (ArmA or RmtB) and with blaOXA-23-like and blaOXA-51-like genes are emerging in medical settings in Vietnam. ArmA and OXA-23-like producing Gram-negative pathogens have been reported in Bulgaria [21], France [22], India [23], Korea [24], Norway [25] and the United States of America [26], and ArmA and OXA-51-like producing strains have been reported in Japan [27]. Moreover, armA and rmtB have been linked to blaCTX-Ms [28,29], but almost all ArmA producing isolates in Vietnam did not harbor blaCTX-Ms.
We found that some A. baumannii clinical isolates harbored rmtB. The genetic environment of the rmtB regions was very similar to the nucleotide sequence, from nt 1 to nt 8,337, of the plasmid pXD2. However, the plasmid of NCGM36 likely differs from pXD2 (Gen bank accession no. JN315966), in that the former NCGM36 did not have aac(6’)-Ib or blaCTX-Ms.
Since 16S rRNA methylase genes in A. baumannii and P. aeruginosa are located in transferable plasmids [5], the absence of methylase genes was found in the same PFGE clusters. The details of these plasmids will be reported elsewhere.
We plan to survey Gram-negative pathogens producing 16S rRNA methylases in 2013 in Vietnam, since more than 9 Gram-negative bacteria producing 16S rRNA methylases have been reported, including A. baumannii, Citrobacter freundii, Enterobacter spp (including E. cloacae), Escherichia coli, Klebsiella pneumonia, Morganella morganii, Proteus mirabilis, Providencia spp (including P. stuartii), and P. aeruginosa[5].
Conclusions
This is the first report describing the presence of methylase producing Gram-negative bacteria in medical settings in Southeast Asia, specifically in Vietnam. A. baumannii isolates from northern and southern regions of Vietnam may be of different lineages.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
TT: Performed PCR and sequencing, analyzed data and drafted the manuscript. TMA: Performed MLST analyses. YK, NO, NT and TAT: Performed epidemiological analysis at BMH. NVH, NGB and NQA: Designed protocols and supervised this study at BMH. DMP, TTTN and PHT: Performed clinical bacterial analyses. PTX: Performed epidemiological analysis at CRH. LTAT and NTS: Designed protocols and supervised this study at CRH. TK: Designed protocols and supervised this study. All authors read and approved the final version manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Contributor Information
Tatsuya Tada, Email: ttada@ri.ncgm.go.jp.
Tohru Miyoshi-Akiyama, Email: takiyam@ri.ncgm.go.jp.
Yasuyuki Kato, Email: ykato@hosp.ncgm.go.jp.
Norio Ohmagari, Email: nohmagari@hosp.ncgm.go.jp.
Nozomi Takeshita, Email: nozomitake@gmail.com.
Nguyen Viet Hung, Email: hungcnk@yahoo.com.
Doan Mai Phuong, Email: doanmaiphuong@yahoo.com.
Truong Anh Thu, Email: truonganhthu2704@yahoo.com.
Nguyen Gia Binh, Email: drbinhicu@gmail.com.
Nguyen Quoc Anh, Email: huong74bm@gmail.com.
Tran Thi Thanh Nga, Email: ngatrancrh@gmail.com.
Pham Hong Truong, Email: truongpham_dh54@yahoo.com.
Phan Thi Xuan, Email: phanthixuan@gmail.com.
Le Thi Anh Thu, Email: letathu@yahoo.com.
Nguyen Truong Son, Email: truongson_cr@yahoo.com.vn.
Teruo Kirikae, Email: tkirikae@ri.ncgm.go.jp.
Acknowledgments
This study was supported by a grant (H24-Shinko-ippan-010) from the Ministry of Health, Labor and Welfare of Japan, and JSPS KAKENHI Grant Number 24790432. We thank Mrs. Kayo Shimada for PFGE analyses.
References
- Jana S, Deb JK. Molecular understanding of aminoglycoside action and resistance. Appl Microbiol Biotechnol. 2006;70(2):140–150. doi: 10.1007/s00253-005-0279-0. [DOI] [PubMed] [Google Scholar]
- Doi Y, Arakawa Y. 16S ribosomal RNA methylation: emerging resistance mechanism against aminoglycosides. Clin Infect Dis. 2007;45(1):88–94. doi: 10.1086/518605. [DOI] [PubMed] [Google Scholar]
- Galimand M, Courvalin P, Lambert T. Plasmid-mediated high-level resistance to aminoglycosides in Enterobacteriaceae due to 16S rRNA methylation. Antimicrob Agents Chemother. 2003;47(8):2565–2571. doi: 10.1128/AAC.47.8.2565-2571.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane K, Wachino J, Suzuki S, Shibata N, Kato H, Shibayama K, Kimura K, Kai K, Ishikawa S, Ozawa Y, Konda T, Arakawa Y. 16S rRNA methylase-producing, gram-negative pathogens, Japan. Emerg Infect Dis. 2007;13(4):642–646. doi: 10.3201/eid1304.060501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachino J, Arakawa Y. Exogenously acquired 16S rRNA methyltransferases found in aminoglycoside-resistant pathogenic Gram-negative bacteria: An update. Drug Resist Updat. 2012;15(3):133–148. doi: 10.1016/j.drup.2012.05.001. [DOI] [PubMed] [Google Scholar]
- Yokoyama K, Doi Y, Yamane K, Kurokawa H, Shibata N, Shibayama K, Yagi T, Kato H, Arakawa Y. Acquisition of 16S rRNA methylase gene in Pseudomonas aeruginosa. Lancet. 2003;362(9399):1888–1893. doi: 10.1016/S0140-6736(03)14959-8. [DOI] [PubMed] [Google Scholar]
- Sekiguchi J, Asagi T, Miyoshi-Akiyama T, Kasai A, Mizuguchi Y, Araake M, Fujino T, Kikuchi H, Sasaki S, Watari H, Kojima T, Miki H, Kanemitsu K, Kunishima H, Kikuchi Y, Kaku M, Yoshikura H, Kuratsuji T, Kirikae T. Outbreaks of multidrug-resistant Pseudomonas aeruginosa in community hospitals in Japan. J Clin Microbiol. 2007;45(3):979–989. doi: 10.1128/JCM.01772-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MA, Baker KN, Orfe LH, Shah DH, Besser TE, Call DR. Discovery of a gene conferring multiple-aminoglycoside resistance in Escherichia coli. Antimicrob Agents Chemother. 2010;54(6):2666–2669. doi: 10.1128/AAC.01743-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Yu H, Guo Q, Xu X, Ye X, Wu S, Guo Y, Wang M. Distribution of 16S rRNA methylases among different species of Gram-negative bacilli with high-level resistance to aminoglycosides. Eur J Clin Microbiol Infect Dis. 2010;29(11):1349–1353. doi: 10.1007/s10096-010-1004-1. [DOI] [PubMed] [Google Scholar]
- Woodford N, Ellington MJ, Coelho JM, Turton JF, Ward ME, Brown S, Amyes SG, Livermore DM. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int J Antimicrob Agents. 2006;27(4):351–353. doi: 10.1016/j.ijantimicag.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Monstein HJ, Ostholm-Balkhed A, Nilsson MV, Nilsson M, Dornbusch K, Nilsson LE. Multiplex PCR amplification assay for the detection of blaSHV, blaTEM and blaCTX-M genes in Enterobacteriaceae. APMIS. 2007;115(12):1400–1408. doi: 10.1111/j.1600-0463.2007.00722.x. [DOI] [PubMed] [Google Scholar]
- Li DX, Zhang SM, Hu GZ, Wang Y, Liu HB, Wu CM, Shang YH, Chen YX, Du XD. Tn3-associated rmtB together with qnrS1, aac(6′)-Ib-cr and bla CTX-M-15 are co-located on an F49:A-:B- plasmid in an Escherichia coli ST10 strain in China. J Antimicrob Chemother. 2012;67(1):236–238. doi: 10.1093/jac/dkr428. [DOI] [PubMed] [Google Scholar]
- Thu TA, Rahman M, Coffin S, Harun-Or-Rashid M, Sakamoto J, Hung NV. Antibiotic use in Vietnamese hospitals: A multicenter point-prevalence study. Am J Infect Control. 2012;40(9):840–844. doi: 10.1016/j.ajic.2011.10.020. [DOI] [PubMed] [Google Scholar]
- Hulscher ME, Grol RP, van der Meer JW. Antibiotic prescribing in hospitals: a social and behavioural scientific approach. Lancet Infect Dis. 2010;10(3):167–175. doi: 10.1016/S1473-3099(10)70027-X. [DOI] [PubMed] [Google Scholar]
- Kritsotakis EI, Dimitriadis I, Roumbelaki M, Vounou E, Kontou M, Papakyriakou P, Koliou-Mazeri M, Varthalitis I, Vrouchos G, Troulakis G, Gikas A. Case-mix adjustment approach to benchmarking prevalence rates of nosocomial infection in hospitals in Cyprus and Greece. Infect Control Hosp Epidemiol. 2008;29(8):685–692. doi: 10.1086/588704. [DOI] [PubMed] [Google Scholar]
- Hughes AJ, Ariffin N, Huat TL, Abdul Molok H, Hashim S, Sarijo J, Abd Latif NH, Abu Hanifah Y, Kamarulzaman A. Prevalence of nosocomial infection and antibiotic use at a university medical center in Malaysia. Infect Control Hosp Epidemiol. 2005;26(1):100–104. doi: 10.1086/502494. [DOI] [PubMed] [Google Scholar]
- Ceyhan M, Yildirim I, Ecevit C, Aydogan A, Ornek A, Salman N, Somer A, Hatipoglu N, Camcioglu Y, Alhan E, Celik U, Hacimustafaoglu M, Celebi S, Inan D, Kurt N, Oner AF, Gulumser O, Gunes A, Coskun Y. Inappropriate antimicrobial use in Turkish pediatric hospitals: a multicenter point prevalence survey. Int J Infect Dis. 2010;14(1):e55–e61. doi: 10.1016/j.ijid.2009.03.013. [DOI] [PubMed] [Google Scholar]
- Lee MK, Chiu CS, Chow VC, Lam RK, Lai RW. Prevalence of hospital infection and antibiotic use at a university medical center in Hong Kong. J Hosp Infect. 2007;65(4):341–347. doi: 10.1016/j.jhin.2006.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamouda A, Evans BA, Towner KJ, Amyes SG. Characterization of epidemiologically unrelated Acinetobacter baumannii isolates from four continents by use of multilocus sequence typing, pulsed-field gel electrophoresis, and sequence-based typing of bla(OXA-51-like) genes. J Clin Microbiol. 2010;48(7):2476–2483. doi: 10.1128/JCM.02431-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libisch B, Watine J, Balogh B, Gacs M, Muzslay M, Szabo G, Fuzi M. Molecular typing indicates an important role for two international clonal complexes in dissemination of VIM-producing Pseudomonas aeruginosa clinical isolates in Hungary. Res Microbiol. 2008;159(3):162–168. doi: 10.1016/j.resmic.2007.12.008. [DOI] [PubMed] [Google Scholar]
- Strateva T, Markova B, Marteva-Proevska Y, Ivanova D, Mitov I. Widespread dissemination of multidrug-resistant Acinetobacter baumannii producing OXA-23 carbapenemase and ArmA 16S ribosomal RNA methylase in a Bulgarian university hospital. Braz J Infect Dis. 2012;16(3):307–310. [PubMed] [Google Scholar]
- Bonnin RA, Potron A, Poirel L, Lecuyer H, Neri R, Nordmann P. PER-7, an extended-spectrum beta-lactamase with increased activity toward broad-spectrum cephalosporins in Acinetobacter baumannii. Antimicrob Agents Chemother. 2011;55(5):2424–2427. doi: 10.1128/AAC.01795-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthikeyan K, Thirunarayan MA, Krishnan P. Coexistence of blaOXA-23 with blaNDM-1 and armA in clinical isolates of Acinetobacter baumannii from India. J Antimicrob Chemother. 2010;65(10):2253–2254. doi: 10.1093/jac/dkq273. [DOI] [PubMed] [Google Scholar]
- Kim JW, Heo ST, Jin JS, Choi CH, Lee YC, Jeong YG, Kim SJ, Lee JC. Characterization of Acinetobacter baumannii carrying bla(OXA-23), bla(PER-1) and armA in a Korean hospital. Clin Microbiol Infect. 2008;14(7):716–718. doi: 10.1111/j.1469-0691.2008.02022.x. [DOI] [PubMed] [Google Scholar]
- Karah N, Haldorsen B, Hermansen NO, Tveten Y, Ragnhildstveit E, Skutlaberg DH, Tofteland S, Sundsfjord A, Samuelsen O. Emergence of OXA-carbapenemase- and 16S rRNA methylase-producing international clones of Acinetobacter baumannii in Norway. J Med Microbiol. 2011;60(Pt 4):515–521. doi: 10.1099/jmm.0.028340-0. [DOI] [PubMed] [Google Scholar]
- Doi Y, Adams JM, Yamane K, Paterson DL. Identification of 16S rRNA methylase-producing Acinetobacter baumannii clinical strains in North America. Antimicrob Agents Chemother. 2007;51(11):4209–4210. doi: 10.1128/AAC.00560-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y, Suwabe A. Diverse carbapenem-resistant mechanisms in 16S rRNA methylase-producing Acinetobacter baumannii. J Med Microbiol. 2013;62(4):618–622. doi: 10.1099/jmm.0.048991-0. [DOI] [PubMed] [Google Scholar]
- Galimand M, Sabtcheva S, Courvalin P, Lambert T. Worldwide disseminated armA aminoglycoside resistance methylase gene is borne by composite transposon Tn1548. Antimicrob Agents Chemother. 2005;49(7):2949–2953. doi: 10.1128/AAC.49.7.2949-2953.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Lin CJ, Chen JH, Fung CP, Chang FY, Lai YK, Lin JC, Siu LK. Taiwan Surveillance of Antimicrobial Resistance Project: Widespread dissemination of aminoglycoside resistance genes armA and rmtB in Klebsiella pneumoniae isolates in Taiwan producing CTX-M-type extended-spectrum beta-lactamases. Antimicrob Agents Chemother. 2009;53(1):104–111. doi: 10.1128/AAC.00852-08. [DOI] [PMC free article] [PubMed] [Google Scholar]