Abstract
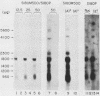
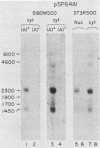
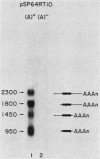
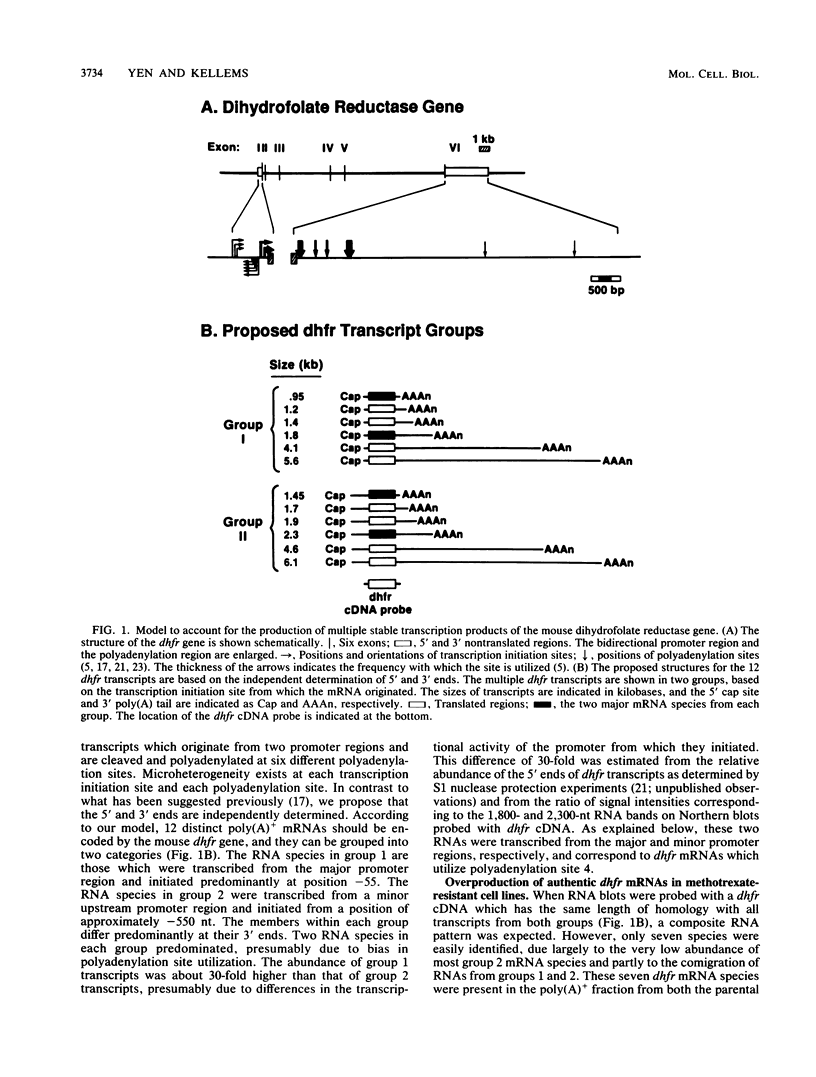
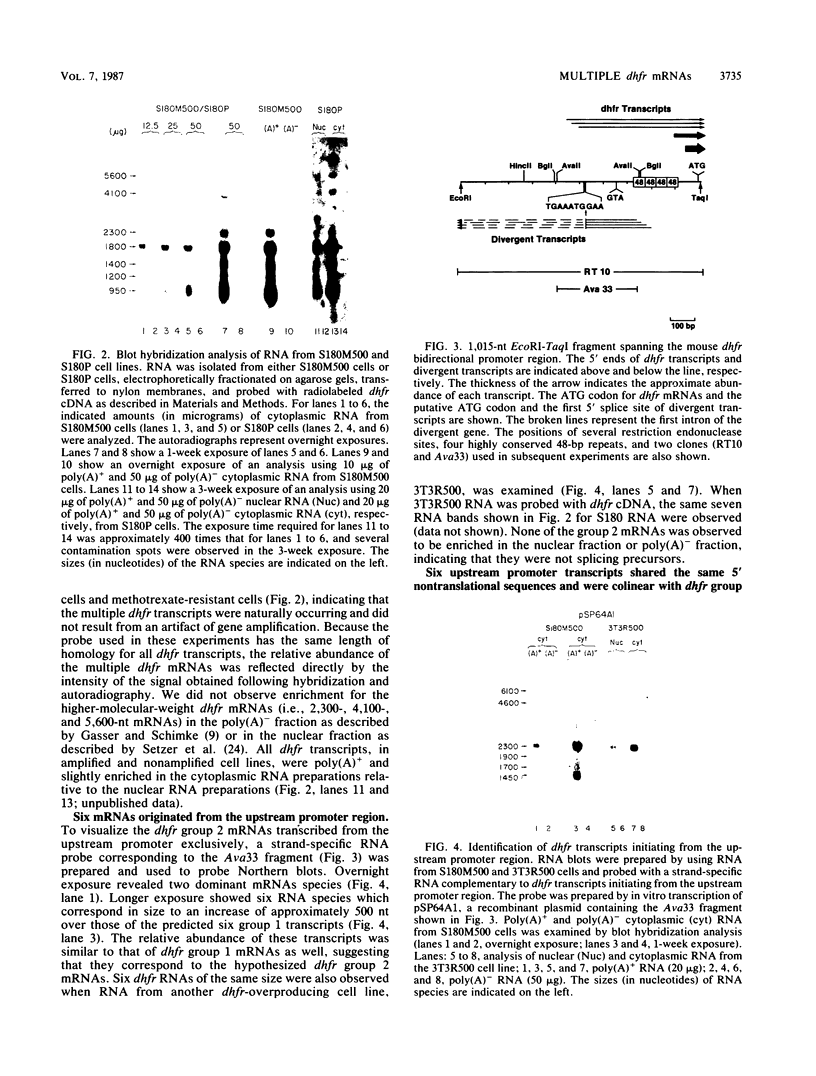
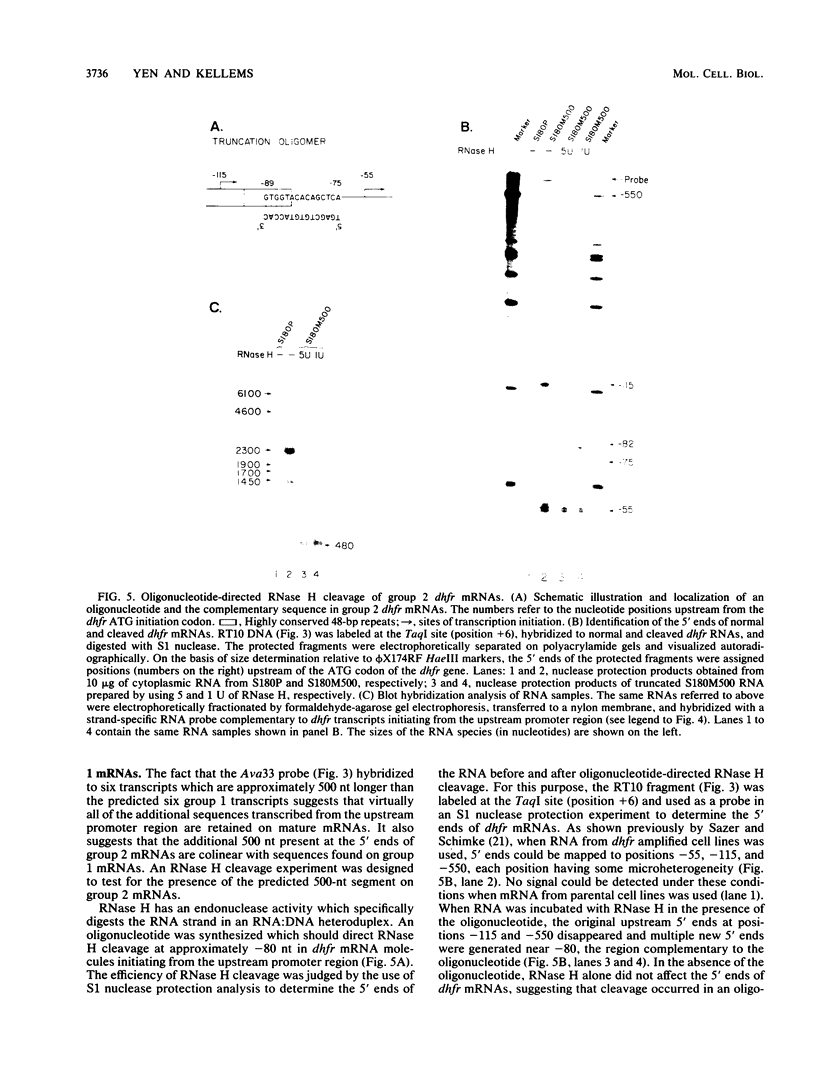
Multiple dihydrofolate reductase (dhfr) mRNAs, differing substantially in abundance, are produced as a result of the utilization of multiple transcription initiation sites and multiple polyadenylation sites. We have shown that dhfr mRNAs initiating from an upstream promoter region utilize the same collection of six polyadenylation sites and generate multiple dhfr mRNAs at the same relative abundance as do the mRNAs initiating from the major transcription promoter region. These results indicate that the 5' and 3' ends of dhfr mRNAs are independently determined. We show that the relative abundance of steady-state dhfr mRNAs was the same in nuclear and cytoplasmic RNA fractions. This finding makes it unlikely that differences in mRNA stability account for differences in the relative abundance of the multiple dhfr mRNAs in the cytoplasm. Our analysis of the dhfr promoter region revealed the existence of stable cytoplasmic polyadenylated transcripts complementary to the first 300 nucleotides of the dhfr transcripts initiating from the upstream promoter region. Therefore, the dhfr locus hosts two divergent and partially overlapping genes which share the same promoter region.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Bullock D. W., Woo S. L., O'Malley B. W. Uteroglobin messenger RNA: translation in vitro. Biol Reprod. 1976 Nov;15(4):435–443. doi: 10.1095/biolreprod15.4.435. [DOI] [PubMed] [Google Scholar]
- Chang A. C., Nunberg J. H., Kaufman R. J., Erlich H. A., Schimke R. T., Cohen S. N. Phenotypic expression in E. coli of a DNA sequence coding for mouse dihydrofolate reductase. Nature. 1978 Oct 19;275(5681):617–624. doi: 10.1038/275617a0. [DOI] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Crouse G. F., Leys E. J., McEwan R. N., Frayne E. G., Kellems R. E. Analysis of the mouse dhfr promoter region: existence of a divergently transcribed gene. Mol Cell Biol. 1985 Aug;5(8):1847–1858. doi: 10.1128/mcb.5.8.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouse G. F., Simonsen C. C., McEwan R. N., Schimke R. T. Structure of amplified normal and variant dihydrofolate reductase genes in mouse sarcoma S180 cells. J Biol Chem. 1982 Jul 10;257(13):7887–7897. [PubMed] [Google Scholar]
- Farnham P. J., Abrams J. M., Schimke R. T. Opposite-strand RNAs from the 5' flanking region of the mouse dihydrofolate reductase gene. Proc Natl Acad Sci U S A. 1985 Jun;82(12):3978–3982. doi: 10.1073/pnas.82.12.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Gasser C. S., Schimke R. T. Cell cycle regulation of transfected murine dihydrofolate reductase genes. J Biol Chem. 1986 May 25;261(15):6938–6946. [PubMed] [Google Scholar]
- Gudewicz T. M., Morhenn V. B., Kellems R. E. The effect of polyoma virus, serum factors, and dibutyryl cyclic AMP on dihydrofolate reductase synthesis, and the entry of quiescent cells into S phase. J Cell Physiol. 1981 Jul;108(1):1–8. doi: 10.1002/jcp.1041080102. [DOI] [PubMed] [Google Scholar]
- HAKALA M. T., ZAKRZEWSKI S. F., NICHOL C. A. Relation of folic acid reductase to amethopterin resistance in cultured mammalian cells. J Biol Chem. 1961 Mar;236:952–958. [PubMed] [Google Scholar]
- Johnson L. F., Fuhrman C. L., Wiedemann L. M. Regulation of dihydrofolate reductase gene expression in mouse fibroblasts during the transition from the resting to growing state. J Cell Physiol. 1978 Dec;97(3 Pt 2 Suppl 1):397–306. doi: 10.1002/jcp.1040970314. [DOI] [PubMed] [Google Scholar]
- Kaufman R. J., Sharp P. A. Construction of a modular dihydrofolate reductase cDNA gene: analysis of signals utilized for efficient expression. Mol Cell Biol. 1982 Nov;2(11):1304–1319. doi: 10.1128/mcb.2.11.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellems R. E., Morhenn V. B., Pfendt E. A., Alt F. W., Schimke R. T. Polyoma virus and cyclic AMP-mediated control of dihydrofolate reductase mRNA abundance in methotrexate-resistant mouse fibroblasts. J Biol Chem. 1979 Jan 25;254(2):309–318. [PubMed] [Google Scholar]
- Leff S. E., Rosenfeld M. G., Evans R. M. Complex transcriptional units: diversity in gene expression by alternative RNA processing. Annu Rev Biochem. 1986;55:1091–1117. doi: 10.1146/annurev.bi.55.070186.005303. [DOI] [PubMed] [Google Scholar]
- McGrogan M., Simonsen C. C., Smouse D. T., Farnham P. J., Schimke R. T. Heterogeneity at the 5' termini of mouse dihydrofolate reductase mRNAs. Evidence for multiple promoter regions. J Biol Chem. 1985 Feb 25;260(4):2307–2314. [PubMed] [Google Scholar]
- Mitchell P. J., Carothers A. M., Han J. H., Harding J. D., Kas E., Venolia L., Chasin L. A. Multiple transcription start sites, DNase I-hypersensitive sites, and an opposite-strand exon in the 5' region of the CHO dhfr gene. Mol Cell Biol. 1986 Feb;6(2):425–440. doi: 10.1128/mcb.6.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rave N., Crkvenjakov R., Boedtker H. Identification of procollagen mRNAs transferred to diazobenzyloxymethyl paper from formaldehyde agarose gels. Nucleic Acids Res. 1979 Aug 10;6(11):3559–3567. doi: 10.1093/nar/6.11.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed K. C., Mann D. A. Rapid transfer of DNA from agarose gels to nylon membranes. Nucleic Acids Res. 1985 Oct 25;13(20):7207–7221. doi: 10.1093/nar/13.20.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sazer S., Schimke R. T. A re-examination of the 5' termini of mouse dihydrofolate reductase RNA. J Biol Chem. 1986 Apr 5;261(10):4685–4690. [PubMed] [Google Scholar]
- Schimke R. T., Brown P. C., Kaufman R. J., McGrogan M., Slate D. L. Chromosomal and extrachromosomal localization of amplified dihydrofolate reductase genes in cultured mammalian cells. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 2):785–797. doi: 10.1101/sqb.1981.045.01.097. [DOI] [PubMed] [Google Scholar]
- Setzer D. R., McGrogan M., Nunberg J. H., Schimke R. T. Size heterogeneity in the 3' end of dihydrofolate reductase messenger RNAs in mouse cells. Cell. 1980 Nov;22(2 Pt 2):361–370. doi: 10.1016/0092-8674(80)90346-3. [DOI] [PubMed] [Google Scholar]
- Setzer D. R., McGrogan M., Schimke R. T. Nucleotide sequence surrounding multiple polyadenylation sites in the mouse dihydrofolate reductase gene. J Biol Chem. 1982 May 10;257(9):5143–5147. [PubMed] [Google Scholar]
- Yeung C. Y., Frayne E. G., Al-Ubaidi M. R., Hook A. G., Ingolia D. E., Wright D. A., Kellems R. E. Amplification and molecular cloning of murine adenosine deaminase gene sequences. J Biol Chem. 1983 Dec 25;258(24):15179–15185. [PubMed] [Google Scholar]
- Yoder S. S., Robberson B. L., Leys E. J., Hook A. G., Al-Ubaidi M., Yeung C. Y., Kellems R. E., Berget S. M. Control of cellular gene expression during adenovirus infection: induction and shut-off of dihydrofolate reductase gene expression by adenovirus type 2. Mol Cell Biol. 1983 May;3(5):819–828. doi: 10.1128/mcb.3.5.819. [DOI] [PMC free article] [PubMed] [Google Scholar]