Abstract
Overexpression of ABCG2 has been reported in cell lines selected for drug resistance and it is widely believed to be important in the clinical pharmacology of anticancer drugs. We and others have previously identified and validated two microRNAs (hsa-miR-519c and hsa-miR-520h) targeting ABCG2. In this study, the shortening of the ABCG2 3′ untranslated region (3′UTR) was found to be a common phenomenon in a number of ABCG2-overexpressing resistant cell lines, which as a result removes the hsa-miR-519c binding site and its repressive effects on mRNA stability and translation blockade, thereby contributing to drug resistance. On the other hand, reduced expression of hsa-miR-520h, previously thought to have allowed ABCG2 overexpression, was found to be caused by the sequestering of the microRNA by the highly expressed ABCG2. In drug sensitive cells, inhibitors against hsa-miR-519c and hsa-miR-520h could augment the cytotoxic effect of mitoxantrone, suggesting a substantial role for both microRNAs in controlling ABCG2 level and thereby anticancer drug response. However, in drug resistant cells, altering the levels of the two microRNAs did not have any effect on sensitivity to mitoxantrone. Taken together, these studies suggest that in ABCG2 overexpressing drug resistant cells, hsa-miR-519c, is unable to impact ABCG2 expression because the mRNA lacks its binding site; whereas hsa-miR-520h is sequestered and unable to limit ABCG2 expression. Given the recent observation that a truncated 3′UTR is also observed in ABCG2-overexpressing human embryonic stem cells, our results in drug resistant cell lines suggest that 3′UTR truncation is a relatively common mechanism of ABCG2 regulation.
Keywords: ABCG2, microRNA, multidrug resistance, gene translocation, mRNA stability
Introduction
ABCG2 is an efflux transporter important in clinical pharmacology and in normal tissue protection (1). ABCG2 was first identified and is often observed in drug resistant cancer cells (2–6). However, relatively little is known about the mechanisms underlying its upregulation. Most studies of this question have focused on the ABCG2 promoter (7–14). Less is known about its regulation at the 3′ untranslated region (3′UTR) of the gene. MicroRNAs (miRNAs) represent a large class of endogenous 19–25 nucleotide-long non-coding RNA molecules that are capable of inducing mRNA degradation and/or translational repression by forming imperfect hybrids with the 3′UTR sequences of their target mRNAs (15–17). We demonstrated that ABCG2 mRNA adopts a longer 3′UTR in a parental S1 colon cancer cell line than in its drug-resistant S1M180 subline and that a miRNA (hsa-miR-519c; hereafter referred to as miR-519c) decreases endogenous ABCG2 mRNA and protein levels by acting through a putative miR-519c binding site located within the longer 3′UTR region found only in the parental cells (18). These findings suggest that escape from miRNA-mediated translational repression and mRNA degradation could contribute to overexpression of ABCG2 in drug resistant cancer cells.
miRNAs play important roles in the regulation of basic cell functions, including proliferation, differentiation and apoptosis (16,19). Recently, attention has been paid to the involvement of miRNA in the acquisition of drug resistance by cancer cells (20–24). In this study, we sought to examine the ABCG2 3′UTR in various ABCG2-overexpressing resistant cell lines and to assess the functional role of the two previously validated miRNAs (hsa-miR-519c and hsa-miR-520h, hereafter referred to as miR-520h) targeting ABCG2.
Materials and Methods
Tissue Culture
Several pairs of parental and resistant cell lines were used. The human colon cancer cell line S1 and its resistant sublines (S1M1 0.4, S1M1 0.8, S1M1 1.6, S1M1 3.2, S1M1 80 and S1M1 200), obtained by selection in mitoxantrone in a stepwise manner, have been previously described (4,25). A549, H460, MCF-7, SF295 and SW620 cells were studied, along with their drug resistant counterparts developed by stepwise selection of parental cells in increasing concentrations of anticancer agent. A549 Beca250 (26), H460 MX20 (27), MCF-7 FLV1000 (5), SF295 MX2000, and SW620 Ad300 (28) were maintained in 250 nM becatecarin, 20 nM mitoxantrone, 1000 nM flavopiridol, 2000 nM mitoxantrone, and 300 ng/ml doxorubicin, respectively. ABCG2 is overexpressed in A549 Beca250, H460 MX20, MCF-7 FLV1000, and SF295 MX2000, contributing to drug resistance; no MRP1 or P-glycoprotein was detected in these resistant cell lines (5,27). The SW620 Ad300 subline does not express ABCG2 but has high levels of P-glycoprotein as the mechanism of drug resistance (28). The cell lines were maintained in IMEM (S1, S1M1 series of resistant sublines, MCF-7, MCF-7 FLV1000) or RPMI medium (A549, A549 Beca250, H460, H460 MX20, SF295, SF295 MX2000, SW620, SW620 Ad300) supplemented with 10% fetal bovine serum, 100 units/mL streptomycin sulfate and 100 units/mL penicillin G sulfate, and incubated at 37°C in 5% CO2.
RNA Degradation Analysis
Parental and resistant cells in 10-cm tissue culture dishes were grown until subconfluent, and then actinomycin D (Sigma-Aldrich, St Louis, MO) was added to a final concentration of 5μg/mL to arrest de novo RNA synthesis. At 0, 4, 8 and 16 hr after actinomycin D treatment, the cells were harvested and ABCG2 mRNA was quantified by RT-PCR as previously described (18). c-myc and glyceraldehyde-3-phosphate (GAPDH) mRNA levels were also monitored as controls. The value recorded was the percentage of mRNA remaining compared with the amount before the addition of actinomycin D, after normalization with GAPDH. All experiments were repeated three times.
Semiquantitative and real-time RT-PCR
Semiquantitative RT-PCR was performed as previously described (18). Amplification of cDNA was done using primers specific for:
ABCA8 (5′-GTGTGTGTCAACAAACTTGGGCCT-3′ [forward] and
5′-AGCTGTATGGTCCTTGTGCTCCTT-3′ [reverse]),
SMAD6 (5′-AGGAGAAACTCGCTCCAAGTGCAT-3′ [forward] and
5′-TCTGGAAAGCCCTGCCTTTACCTT-3′ [reverse]),
ID1a (5′-TCGCTTTGCCCATTCTGTTTCAGC-3′ [forward] and
5′-TGTCGTAGAGCAGCACGTTTACCT-3′ [reverse]) and
BRCA2 (5′-AGGAAGTTGTACCGTCTTTGGCCT-3′ [forward] and
5′-TCCTGCAGGCATGACAGAGAATCA-3′ [reverse]).
GAPDH was used as the internal control.
ABCG2 mRNA expression in the S1M1 series of resistant cell lines was measured by real-time PCR using a Light Cycler Fast Start DNA Master Taqman kit (Universal Probe Library; Roche Molecular Biochemicals, Indianapolis, IN) as previously described (18).
For the detection of miR-519c and miR-520h, 2 μg total RNA was used in the reverse transcription (RT) reaction by using the QuantMir RT Kit (System Biosciences, Mountain View, CA). Quantitative real-time PCR was performed using the KAPA SYBR FAST qPCR Kit (KapaBiosystems, Woburn, MA) in a LightCycler 480 Instrument I (Roche Applied Science). The mature miR-519c and miR-520h DNA sequence was used as the forward primer and the 3′ universal primer provided from the QuantiMiR RT Kit was used as the reverse primer. The human U6 RNA was amplified in parallel as the internal control. PCRs were performed at 95°C for 10 min, followed by 40 cycles of 95°C for 10s and 60°C for 10s. Fluorescence signal was acquired at the end of the elongation step of every PCR cycle (72°C for 10s) to monitor the increasing amount of amplified DNA. ΔCt was calculated by subtracting the Ct of U6 from the Ct of miR-519c or miR-520h. ΔΔCt was then calculated by subtracting the ΔCt of the parental cells from the ΔCt of the corresponding resistant cells. Fold change of miRNAs was calculated by the equation 2−ΔΔCt.
3′RACE Assay
Total RNA was extracted from parental cells and resistant sublines using Trizol reagent (Invitrogen, Carlsbad, CA). 3′RACE assay was performed as described previously (18; a detailed scheme is shown in Supplementary Figure 1). DNA as a mixture from the PCR reaction was subcloned into a pCR2.1 vector (Invitrogen). Multiple clones were then sequenced by the DNA sequencing core facility at the Laboratory of Experimental Carcinogenesis, NCI. All sequences were aligned against reported mRNA sequence for ABCG2 (GenBank accession no. NM_0004872). For S1 and SW620 cells, 3′RACE analysis was performed with total RNA harvested from cells after transfection with a miR-519c specific inhibitor (Dharmacon, Lafayette, CO).
Cell Transfection with miRNA Inhibitors or Mimics
miRNA (miRIDIAN) inhibitors or mimics against miR-519c or miR-520h were purchased from Dharmacon. Cells were transfected with up to 100 nM of the specific inhibitors or mimics in a total volume of 6 mL by using DharmaFECT Duo Transfection Reagent (Dharmacon). The transfection efficiency was monitored by measuring the expression of the cotransfected GFP protein (pEGFP-C1; BD Bioscience Clontech). To evaluate the role of the two miRNAs in the sensitivity of cells to ABCG2 substrate drugs, cells were seeded into 96-well plate for growth inhibitory assays at 48 h after transfection with the miRIDIAN inhibitors or mimics.
Stem-loop RT-PCR for miRNAs
Stem-loop RT-PCR for evaluating expression of mature miR-519c and miR-520h was performed as previously described (18).
Growth Inhibition Assays
Mitoxantrone was obtained from Sigma-Aldrich. The growth inhibitory effect of mitoxantrone under the various miRIDIAN inhibitor/mimic transfection conditions as indicated was tested using the sulforhodamine B proliferation assay (29). At 48 h after transfection with either the miRNA inhibitors or mimics, cells were inoculated into 96 well microtitre plates in 100μL at plating densities ranging from 5,000 to 20,000 cells/well depending on the doubling time of individual cell lines. The cells were then treated with mitoxantrone at a range of concentrations and allowed to incubate at 37°C in 5% CO2 for 96 h. Each drug concentration was tested in quadruplicate and controls were tested in replicates of eight. Dose modifying factors were calculated for the miRNA inhibitor or mimic with mitoxantrone by dividing the IC50 for mitoxantrone without the miRNA inhibitor/mimic by the IC50 for mitoxantrone in the presence of the miRNA inhibitor/mimic. Each experiment was carried out independently at least three times. To determine whether differences between IC50 values were significant, a Student’s t-test was performed with p < 0.05 being considered significant.
Western Blot Analysis
Whole cell lysates prepared from S1 and its resistant sublines were separated by SDS-PAGE and subjected to immunoblot analysis with the respective antibodies (ABCG2, Kamiya Biomedical Company, Seattle, WA; GAPDH, American Research Products, Belmont, MA). The blot was analyzed in the Odyssey (LI-COR, Licolin, NE) infrared imaging system after incubation with a 1:10000 dilution of the goat anti-mouse (IRDye800CW) or anti-rabbit (IRDye680) secondary antibody (LI-COR).
Flow cytometry
Detection of ABCG2-mediated efflux was performed by flow cytometry assays as previously described (30). Briefly, cells were trypsinized and incubated in 10 μM pheophorbide A (PhA) with or without 10 μM of the ABCG2 specific inhibitor fumitremorgin C (FTC) in complete medium (phenol red-free RPMI medium with 10% fetal calf serum) at 37°C in 5% CO2 for 30 minutes. Subsequently, the cells were washed once with cold complete medium and then incubated for 1 h at 37°C in PhA-free complete medium continuing in 10 μM FTC to generate the FTC/Efflux histogram, or without FTC to generate the Efflux histogram. Cells were then washed with cold DPBS and placed on ice in the dark until analyzed.
To determine cell surface expression of ABCG2, cells were trypsinized and resuspended in 2% BSA/DPBS to which was added the phycoerythrin-conjugated anti-ABCG2 antibody 5D3 (eBioscience, San Diego, CA) or phycoerythrin-conjugated mouse IgG2b negative control antibody (eBioscience). The cells were incubated with antibody for 30 min at room temperature and washed with DPBS. Surface expression of ABCG2 was calculated as the difference in mean channel number between the 5D3 antibody histogram and the negative control antibody histograms.
Samples were analyzed on a FACSort flow cytometer (Becton Dickinson, San Jose, CA). Phycoerythrin fluorescence was detected with a 488-nm argon laser and a 585-nm bandpass filter, whereas PhA was detected using a 635-nm red diode laser and a 561-nm filter. At least 10,000 events were collected for all flow cytometry studies. By gating on forward versus side scatter, debris was eliminated, and dead cells were excluded based on propidium iodide staining.
Fluorescence in situ hybridization analysis (FISH)
Fluorescence in situ hybridization (FISH) analysis was performed using whole chromosome paint kits for chromosome 4 in Spectrum Orange and chromosome 17 in Spectrum Green (Vysis, Downers Grove, IL). Earlier studies had been performed using an ABCG2 bacterial artificial chromosome (BAC) probe (>50kb and containing the 3′ end of the gene; NCBI Clone Registry ID RP11-368G2, Invitrogen, Carlsbad, CA) to localize the ABCG2 gene to normal and rearranged chromosomes. The ABCG2 BAC probe localizes to chromosome position 4q22 and has previously been used to study ABCG2 overexpression (31). Standard FISH hybridization procedure was used. All slides were counterstained with 4′,6-diamidino-2-phenylindole (DAPI).
Results
Stability and 3′UTR length of ABCG2 mRNA in parental S1 cells and its ABCG2-overexpressing mitoxantrone-resistant sublines
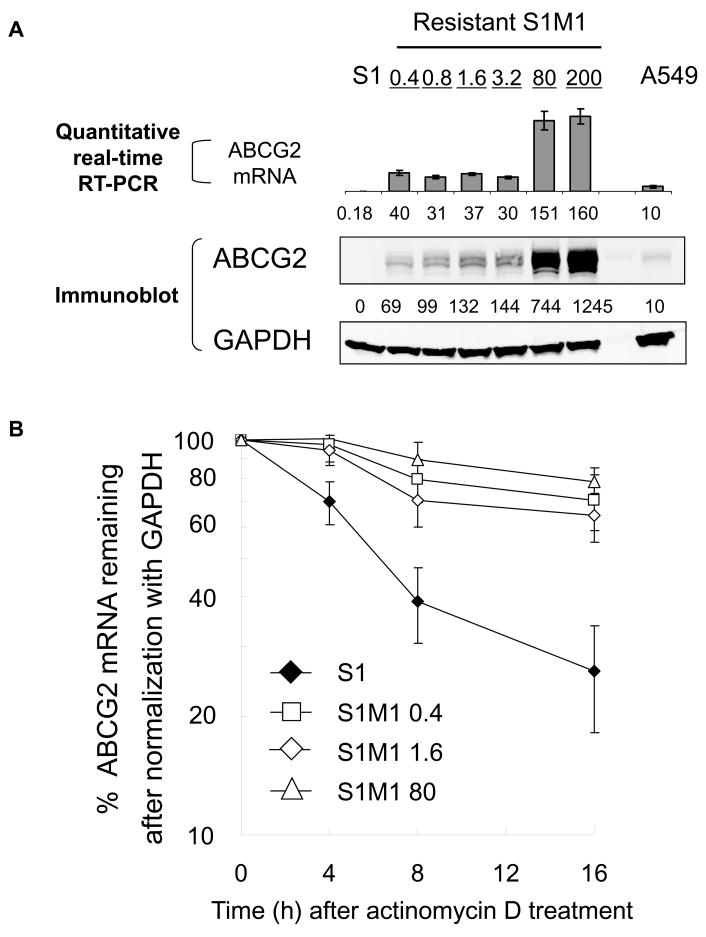
We previously observed that ABCG2 mRNA in the parental S1 colon cancer cell line was found to adopt different lengths at the 3′UTR, with a long (about 2 kb) and multiple shorter (200 – 400 bp) 3′UTRs, whereas only the shorter ones exist in the resistant S1M1 80 subline (18). As the parental S1 cells underwent stepwise selection with increasing concentrations of mitoxantrone, individual selection steps were maintained at 0.4, 0.8, 1.6, 3.2, 80 and 200 μM of mitoxantrone and named accordingly (4,25). Overexpression of ABCG2 was observed in the whole series with detection of upregulated mRNA and protein expression in as early as the first resistant step (i.e. S1M1 0.4) (Figure 1A). We asked whether an increase in mRNA stability and shortening of 3′UTR could contribute to the overexpression of ABCG2 in the early and low-resistance steps. Degradation of mRNA was followed by RT-PCR (18) after the cells were treated with actinomycin D to inhibit RNA synthesis. While ABCG2 mRNA in S1M1 0.4, S1M1 1.6 and S1M1 80 was fairly stable up to 16 h after actinomycin D treatment, ABCG2 in the parental cells had a half-life of about 6 h (Figure 1B). GAPDH and c-myc, controls with long (more than 24 h) and short (15–40 min) mRNA half-lives, respectively, were found to yield opposite degradation profiles (data not shown), confirming that actinomycin D efficiently stopped transcription in our experiments. These results suggested a reduced degradation rate for ABCG2 mRNA, even in the early selection steps.
Figure 1.
(A) The relative expression of ABCG2 mRNA and protein in the S1M1 series of resistant cell lines was examined by quantitative real-time RT-PCR and immunoblot analysis, respectively. ABCG2 expression was also measured in A549 non-small cell lung cancer cells because endogenous ABCG2 protein expression is readily detectable in these cells for comparison purpose. The relative ABCG2 mRNA levels were shown after normalization with GAPDH (mean values from three independent RNA preparations were shown). The numbers under the gel image (Western) represent the relative expression of ABCG2 after normalization with GAPDH; A549 level is set as 10 for comparison. (B) ABCG2 mRNA is more stable in resistant S1M1 0.4, S1M1 1.6 and S1M1 80 cells than in parental S1 cells. Nascent RNA synthesis was inhibited with actinomycin D (5 μg/mL) and RNA harvested at 0, 4, 8, and 16 h post treatment. RT-PCR analysis of ABCG2 mRNA was carried out to trace the remaining amount of ABCG2 mRNA with time. c-myc and GAPDH mRNA levels were also monitored as controls for fast-degrading and stable mRNA, respectively (not shown). The results presented were the percentage of mRNA remaining compared with the amount before the addition of actinomycin D after normalization with GAPDH. All experiments were repeated three times.
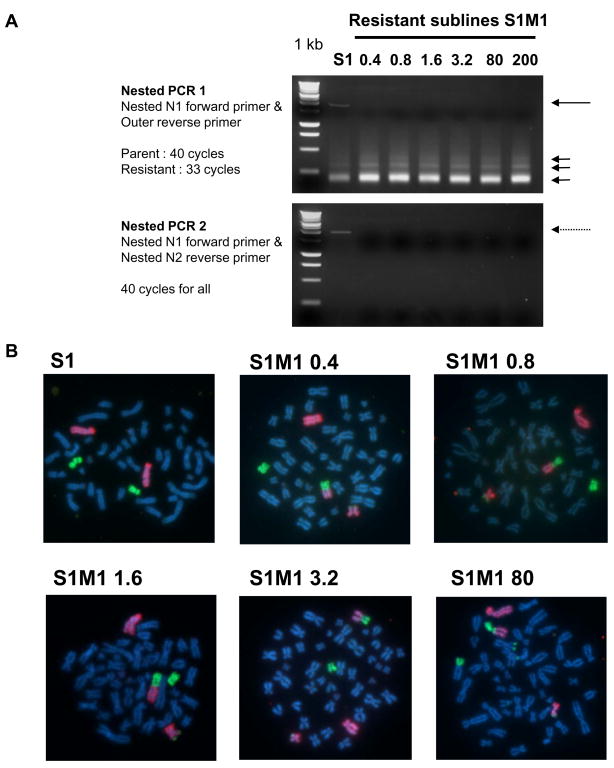
3′RACE reactions were performed to examine the 3′ ends of the ABCG2 mRNA and to define the location of poly(A) sites in the S1M1 series of resistant sublines (18). The nested PCR 1 gave rise to a long fragment and multiple shorter fragments in the parental S1 cells but only to short fragments in the resistant sublines (Figure 2A). Nested PCR 2 further confirmed the existence of the long ABCG2 3′UTR in the S1 cells but not in the resistant sublines. There was no significant reaction with samples primed without reverse transcription (data not shown).
Figure 2.
(A) 3′RACE analysis of ABCG2 mRNA in the S1M1 series of resistant cell lines. The schematics for the 3′RACE assay can be found in Supplementary Figure 1. The first PCR and the nested PCR 1 gave the same pattern of PCR products. Only the nested PCR 1 is shown. In miR-519c inhibitor-transfected S1 parental cells, a long PCR product (about 2 kb, long arrow) and three much smaller fragments (~200, 300 and 400 bp, shorter arrows) were obtained. In all resistant S1M1 sublines, only the smaller fragments were seen. Nested PCR 2 was performed to further confirm the existence of the long ABCG2 3′UTR fragment in the parental S1 cells only. (B) FISH analysis of chromosome metaphase spreads with whole chromosome paints. Chromosome 4 is painted with WCP 4 Spectrum Orange and chromosome 17 is painted with WCP 17 Spectrum Green; DAPI counterstain. S1 parent line: two normal chromosomes 4; S1M1 0.4, S1M1 0.8, S1M1 1.6, S1M1 3.2, S1M1 80: one normal chromosome 4, one normal 17, and t(4;17)(q21-q22; p13). Earlier studies had shown that the ABCG2 signal is split at the breakpoint (31).
Chromosomal rearrangement in the early steps of the S1M1 series and its coincidence with the shortening of ABCG2 3′UTR
The chromosomal rearrangements of the S1 parental and S1M1 80 resistant cells have been characterized (31). A balanced translocation t(4;17)(q21-q22; p13), presumably at the 3′ end of the ABCG2 gene, was found in the resistant S1M1 80 subline using FISH and spectral karyotyping (SKY) analysis. It was then hypothesized that the sequences juxtaposed to ABCG2 after the translocation conferred the increased ABCG2 expression in S1M1 80 cells, although the breakpoint has never been characterized. In the present study, the early selection steps (S1M1 0.4, 0.8, 1.6, 3.2) were assessed for chromosomal translocations as a possible initial event for induction of ABCG2 and the subsequent multidrug resistance phenotype. FISH analysis using whole chromosome paints was performed to identify normal and rearranged chromosomes in cytogenetic preparations from the parent S1 and the S1M1 resistant sublines. Interestingly, the same translocation reported in S1M1 80, involving chromosome 4 downstream of the ABCG2 gene, was also observed in all selection steps (Figure 2B). The parental S1 cells had no chromosome 4 rearrangement and no copy number gain or amplification of chromosome 4. It is notable that the S1M1 80 cells have this gene rearrangement and, despite intense selection pressure in very high concentrations of mitoxantrone, ABCG2 has never undergone gene amplification.
Shortening of the ABCG2 mRNA 3′UTR in other ABCG2-overexpressing resistant cell lines
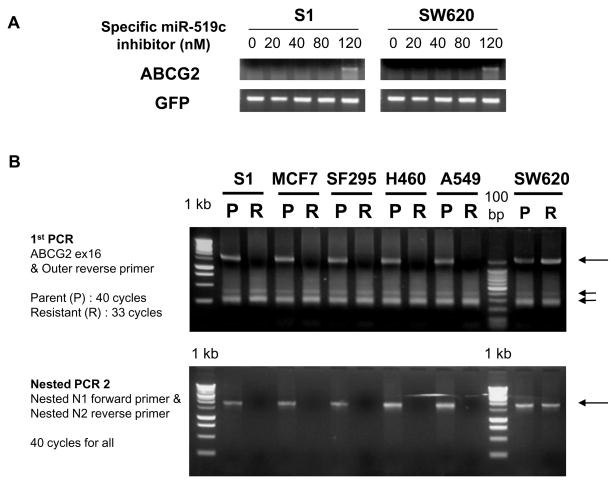
To determine whether 3′UTR truncation was unique to the S1M1 80 cell line, the length of the ABCG2 mRNA 3′UTR in other ABCG2-overexpressing resistant cell lines was examined by 3′RACE assay. Parental S1 and SW620 cells were transfected with specific miR-519c inhibitor to provide higher ABCG2 expression levels for the 3′RACE assay (Figure 3A). For all parental cell lines tested (S1, MCF-7, SF295, H460, A549 and SW620), 3′RACE assay gave a long 3′UTR fragment and multiple shorter fragments (Figure 3B, upper panel). However, in the ABCG2-overexpressing resistant sublines (S1M1 80, MCF-7 FLV1000, SF295 MX2000, H460 MX20 and A549 Beca250), only the shorter fragments were obtained. Interestingly, in Pgp-overexpressing SW620 Ad300 cells, where there is no ABCG2 overexpression, the long 3′UTR fragments persist together with the other shorter fragments observed in other resistant cell lines (Figure 3B). The existence of the long ABCG2 3′UTR in the parental cells and in the SW620 resistant subline without ABCG2 overexpression was further confirmed by nested PCR2 (Figure 3B, Lower Panel).
Figure 3.
(A) ABCG2 mRNA expression become detectable in S1 and SW620 cells transfected with miR-519c inhibitor. Total RNA was harvested for RT-PCR analysis. Co-transfected GFP was also measured to ensure equal transfection efficiency. (B) 3′RACE analysis of the ABCG2 mRNA was performed in pairs of parental and ABCG2-overexpressing resistant cell lines (S1, MCF-7, SF295, H460 and A549) as in Figure 2A above. The parental cells (P) gave both the long (long arrow) and short (short arrows) ABCG2 3′UTR fragments but the corresponding resistant cells (R) (S1M1 80, MCF-7 FLV1000, SF295 MX2000, H460 MX20, A549 Beca250, respectively) gave only the shorter ones. SW620 Ad300 (the resistant subline of SW620 cells) do not express ABCG2 and they gave both long and short fragments from the 3′RACE assay as with the parental SW620 cells. The specific detection of the long 3′UTR fragment in the parental S1, MCF-7, SF295, H460, A549, SW620 and the resistant SW620 Ad300 was confirmed by the nested PCR 2.
Since the 3′RACE assay may have missed the long 3′UTR PCR products because of their lower PCR efficiency, we also conducted RT-PCR experiments by using total RNA from the pairs of parental and resistant cell lines, and primers complementary to different regions in the ABCG2 3′UTR cDNA. Six separate and overlapping PCR fragments (A–F) that covered the reported ABCG2 3′UTR sequence (NM_004872), were amplified (Supplementary Figure 2A) (18). All six PCR fragments were obtained with the predicted size in all RT samples tested, eliminating the possibility of alternate splicing at the ABCG2 3′UTR. Calibration curves were constructed using a range of concentrations of input genomic DNA from S1 cells. The relative abundance of different 3′UTR fragments in each cell line, compared to the most upstream region A, was then estimated by reading from the calibration curves. For all parental cell lines tested, as well as the SW620 Ad300 subline without ABCG2 overexpression, the relative abundance of the six 3′UTR regions was found to be similar (Supplementary Figure 2B). However, for the ABCG2-overexpressing resistant cell lines, the relative abundance of the more downstream 3′UTR regions decreased from region A to F, suggesting a higher proportion of truncated 3′UTR in the resistant cells (Supplementary Figure 2B).
Stability of ABCG2 mRNA in various parental and resistant cell lines
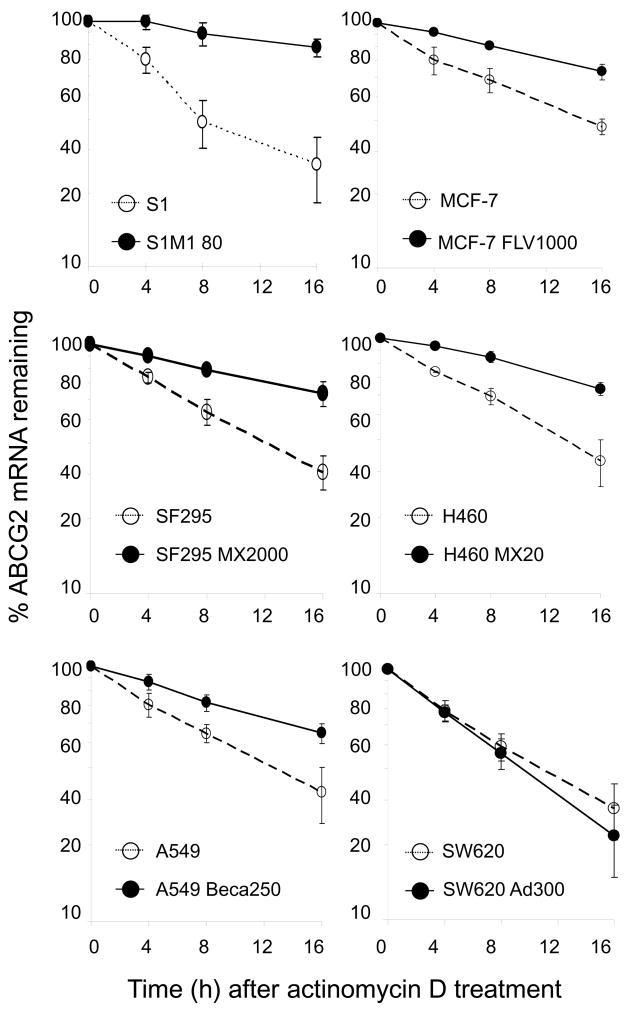
Since the various resistant sublines (except the Pgp-expressing SW620 Ad300 subline) lack the long ABCG2 3′UTR fragment, as reported for S1M1 80 cells (18), the ABCG2 mRNA in these resistant cells may also be more stable than that in the parental counterparts. The half-life of ABCG2 mRNA in this panel of parental and drug-resistant cells was measured after treatment with actinomycin D to inhibit RNAsynthesis. While ABCG2 mRNA in the resistant sublines is fairly stable up to 24 h after actinomycin D treatment, ABCG2 in the parental cells has a half-life of about 12 h (except in S1 where the half-life is about 8 h), which is comparable to the number reported by Nakanishi for MCF-7 cells (12). Thus, the reduced degradation rate of ABCG2 mRNA may have contributed to the upregulation of ABCG2 in these resistant cells. In resistant Pgp-overexpressing SW620 Ad300, ABCG2 mRNA stability was found to be similar to that in the parental cells (Figure 4).
Figure 4.
ABCG2 mRNA degradation detected by RT-PCR. Nascent RNA synthesis was inhibited with actinomycin D (5 μg/mL), and RNAs were harvested at 0, 4, 8, and 16 h posttreatment. RT-PCR analysis of ABCG2 mRNA was carried out to trace the remaining amount of ABCG2 mRNA with time. For S1, MCF-7, SF295, H460 and A549 cells, ABCG2 mRNA is more stable in their corresponding ABGC2-overexpressing resistant counterparts. SW620 Ad300 cells do not over-express ABCG2. ABCG2 mRNA stability in the parental SW620 cells is similar to that in the resistant sublines.
Sequence analysis of ABCG2 3′UTR with different lengths
We cloned the 3′RACE PCR reaction mixture as a whole (1st PCR) into the TOPO vector and subjected the resulting clones to DNA sequencing. ABCG2 3′UTR transcripts of five different lengths were found. Consistent with the 3′RACE assay, while we found a long (~2 kb) and a few much shorter (100 – 400bp) 3′UTR transcripts in the parental cells, only the shorter ones were found in the resistant cells. A summary of the 3′UTR ends found for the various pairs of parental and resistant cell lines is tabulated in Supplementary Figure 3. The numbering of nucleotides at the 3′UTR is relative to the first nucleotide in the ABCG2 transcript designated in NM_004827. The sequence of the 3′UTR was identical to the full-length sequence except for the early termination, indicating that multiple site polyadenylation rather than alternate splicing was involved.
Reduced levels of miR-520h found in ABCG2-overexpressing resistant cells
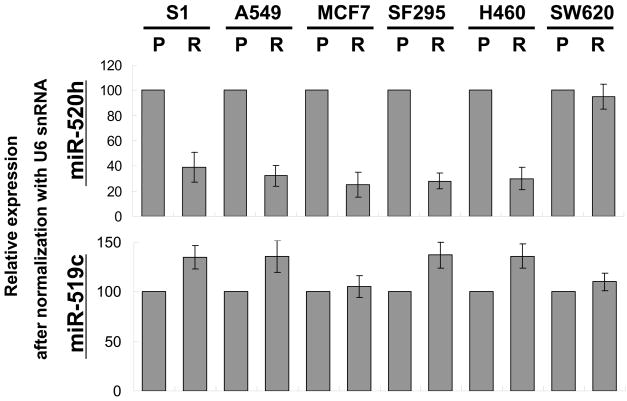
As discussed previously, miR-520h binds to both long and short ABCG2 3′UTR in parental S1 and resistant S1M1 80 cells while miR-519c binds to only the long ABCG2 3′UTR in the parental S1 cells (18). We therefore examined the expression level of these two miRNAs by quantitative real time PCR in the various parental and resistant cells to try to elucidate their role in regulating ABCG2. Interestingly, the levels of miR520h in the resistant cells (except SW620 Ad300) were reduced 2–3 fold relative to those in the respective parental cells (Figure 5). In contrast, miR-519c levels in the resistant cells were found to be slightly higher in the resistant cells than in the parental cells.
Figure 5.
Differential miR-519c and miR-520h expression levels between the parental and ABCG2-overexpressing resistant cells. Expression of the two miRNAs was measured by quantitative real time PCR. U6 small nuclear RNA was used as the internal control. The results shown represent means ± SD from three independent and reproducible experiments.
High levels of ABCG2 in resistant cells serve as a sink for miR-520h in the resistant cells
Although the reduction in miR-520h expression in the resistant cells may mediate the overexpression of ABCG2, we also considered an alternate possibility – that the high levels of ABCG2 could serve as a sink for miR-520h in the resistant cells, yielding the same observation. ABCG2 was thus knocked down by transfecting parental MCF-7 and resistant MCF-7 FLV1000 cells with a pool of four siRNA duplexes specifically designed against ABCG2 (ONTARGETplus siRNA, Dharmacon). ABCG2 was knocked down significantly by 90% and 60% in MCF-7 and MCF-7 FLV1000, respectively (Supplementary Figure 4). While the miR-519c level was not affected by ABCG2 siRNA transfection, the miR-520h level increased more than 2-fold in the ABCG2-silenced MCF-7 FLV1000 cells. The effect of lower endogenous miR-520h levels in the resistant cells on four of its putative target genes (BRCA2, SMAD6, ABCA8 and ID1a), predicted by the miRDB miRNA target prediction database (22), were also evaluated by RT-PCR (Supplementary Figure 5). Compared with the parental MCF-7 cells, no consistent increase in these putative miR-520h target genes was observed in the resistant MCF-7 FLV1000 cells, despite the low levels of miR-520h in these cells.
Effects of miRNA silencing and forced expression on sensitivity of cells to the ABCG2 substrate mitoxantrone
We previously demonstrated that resistant cells could have overexpressed ABCG2 by escaping repression mediated by miR-519c. To further assess the intrinsic effects of miR-519c and miR-520h on sensitivity towardsABCG2 substrate drugs, we performed cytotoxicity assays using sulforhodamine B to compare the growth of S1 and MCF-7 cells in which the endogenous pool of miR-519c or miR-520h was altered by transfection with synthetic miRIDIAN inhibitor or mimic (Dharmacon). Cells were seeded into 96-well plates for cytotoxicity assay at 48 h after transfection. The cells were then exposed to mitoxantrone for another 96 h and cell proliferation was assessed (29). The data from quadruplicate samples were analyzed for significant differences by the paired Student’s t-test (Supplementary Figure 6). The miRNA inhibitor used in our study is a chemically modified antisense oligonucleotide perfectly complementary to the mature miRNA, which binds tightly to the endogenous miRNA, resulting in its sequestration (33). The specific miR-519c and miR-520h inhibitor (100 nM) increased the IC50 of mitoxantrone by 13.5-fold and 10.1-fold in the parental S1, and 7.0-fold and 4.3-fold in the parental MCF-7 cells, respectively. On the other hand, miR-519c and miR-520h mimics are double stranded RNAs that contain the mature miRNA sequence. When added to cells, the mimic functions to reduce ABCG2 levels in the same way as the endogenous miRNA. When the specific miR-519c or miR-520h mimic was transfected into S1 or MCF-7 cells, minimal change in the IC50 of mitoxantrone was observed. In resistant S1M1 80 and MCF-7 FLV1000 cells, the antiproliferative effect of mitoxantrone was not affected by either miR519c or miR520h inhibitors or mimics. The universal negative controls for miRNA mimic and inhibitor based on C. elegans miRNA sequences (cel-miR-67; Dharmacon) did not have any effect on the antiproliferative effect of mitoxantrone in both S1 and MCF-7 cells (Supplementary Figure 6).
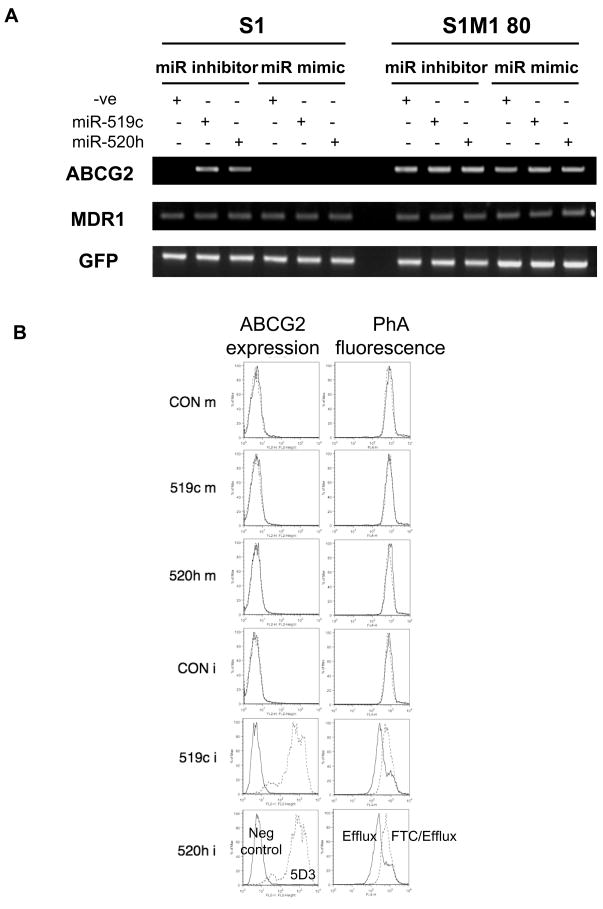
The effect of the microRNAs on ABCG2 mRNA expression (Figure 6A), cell surface expression and pheophorbide A efflux (Figure 6B) was also studied. Consistent with the result from the drug cytotoxicity assay, cell surface ABCG2 expression and PhA efflux were induced remarkably in parental S1 cells (and less dramatic in MCF-7) upon miR-519c and miR-520h inhibitor transfection (Figure 6B & Supplementary Figure 7). On the other hand, miRNA mimics against either miR-519c or miR-520h did not have any appreciable effect on the expression or function of ABCG2 in either S1 or MCF-7 cells (Figure 6 & Supplementary Figure 7).
Figure 6.
(A) RT-PCR analysis of ABCG2 and MDR1 mRNA expression in S1 and S1M1 80 cells transfected with specific inhibitor/mimic against miR-519c or miR-520h. GFP from cotransfection was also measured to assure sufficient transfection efficiency. ABCG2 expression was increased by miR-519c and miR-520h inhibitor in the parental S1 cells but no change was recorded in the resistant S1M1 80 cells. (B) ABCG2 surface expression and FTC-inhibitable PhA efflux in miRNA inhibitor or mimic-transfected S1 cells. Left column: Cells were incubated for 30 min with either phycoerythrin-labeled negative control antibody (solid lines) or the phycoerythrin-labeled anti-ABCG2 antibody 5D3 (dashed lines) according to the manufacturer’s instructions. Right column: Transfected cells were incubated in PhA with or without 10 μM FTC for 30 min at 37°C, washed, then allowed to efflux for 1 h at 37°C in substrate-free medium continuing with (dashed lines, FTC/Efflux histogram) or without (solid lines, Efflux histogram) FTC. Representative histograms are shown. No change in ABCG2 expression or PhA efflux was observed in S1M1 80 cells upon the miRNA inhibitor or mimic transfection (not shown).
ABCG2 mRNA expression and efflux function were also induced, to varying extent, in other parental cell lines upon transfection with miR-519c or miR-520h inhibitors (Supplementary Figure 8A & 8B).
Discussion
High levels of ABCG2 expression are found in normal tissues involved in xenobiotic protection, in stem cells, and in drug resistant cancer cell lines (2–6). However, little is known about the mechanisms underlying its upregulation, especially at the 3′UTR. Previously, we reported that the ABCG2 mRNA is more stable in the mitoxantrone-selected and ABCG2-overexpressing resistant subline S1M1 80, compared to the ABCG2 mRNA in S1 parental cancer cell line (18). Our results suggested that miR-519c binds to the 3′UTR of ABCG2 in the S1 cells, thereby suppressing ABCG2 expression by either mRNA degradation and/or protein translation repression, but that the binding site for this miRNA is lost in the shorter 3′UTR present in the resistant S1M1 80 cells. Similar findings have recently been reported in an undifferentiated human embryonic stem (HuES) cell line where its high ABCG2 expression was associated with only the short 3′ UTR variant forms (34). In contrast, another differentiated HuES cell line with lower ABCG2 levels possesses the longer 3′UTR variant (34). Sandberg et al. also found that rapidly proliferating cells express ABCG2 mRNAs with shorter 3′UTRs, presumably to escape miRNA regulation (35). In the present study, we examined this phenomenon in additional cancer cell lines to gain further insight into this mechanism of ABCG2 gene overexpression.
The importance of ABCG2 and other drug transporters in oncology has not yet been clearly established. Certainly overexpression of ABCG2 and P-glycoprotein in clinical samples has been linked with poor prognosis in multiple settings (36,37). However, the addition of inhibitors to therapeutic regimens did not improve outcome in multiple studies. This left transporters as therapeutic targets an unanswered question that has been renewed in recent years with the discovery that most of the tyrosine kinase inhibitors (TKIs) currently in clinical use are substrates or inhibitors of the transporters (1). Since the TKIs are being tested in combination with chemotherapy, a method to confirm whether ABCG2 confers resistance to them, or is inhibited by them, or both, is exceedingly relevant.
Earlier steps of selection in the S1M1 series were examined, and stabilization of ABCG2 mRNA and shortening of the 3′UTR were observed in all the earlier steps (Figure 1B & 2A). We previously reported that the effect of miR-519c on inhibiting ABCG2 protein translation is more pronounced than that promoting mRNA degradation, probably because of imperfect binding of miR-519c to its binding site on ABCG2 3′UTR (18). When we review the S1M1 series of resistant sublines, the increase in ABCG2 protein expression is more dramatic than that in ABCG2 mRNA levels (Figure 1A). This lack of proportionality is consistent with a differential effect of the miRNA on mRNA stability and protein translation. Taken together, our results suggest that loss of miR-519c binding may be involved in the initial step of selection when these sublines acquired multidrug resistance, prompting a marked overexpression of ABCG2.
miR-520h has been reported to target ABCG2 in hematopoietic stem cells during their differentiation into progenitor cells (38). Unlike miR-519c, miR-520h binds to ABCG2 3′UTR in both parental and resistant cells. We found that ABCG2-overexpressing resistant cells have lower levels of miR-520h than their parental counterparts (Figure 5). Interestingly, this is likely caused by the sequestration of miR-520h by the highly expressed ABCG2 in the resistant cells (Supplementary Figure 4).
A balanced t(4;7) translocation downstream of the ABCG2 gene has been reported for S1M1 80 cells (31). Coincident with the shortening of the ABCG2 3′UTR in the early S1M1 resistant sublines, the same translocation was also found in these sublines starting from S1M1 0.4. Since the breakpoint for this translocation is believed to be 3′ to the ABCG2 gene, it remains to be determined if there is any correlation between the translocation and the differential 3′UTR length adopted in resistant cells. Although this chromosomal rearrangement was not identified in several other drug resistant cell lines examined and was considered to be a unique event (31), the possibility that this rearrangement could be found in other tumors cannot be excluded. Indeed, there is increasing evidence that fusion genes are important, even in non-hematologic tumor types such as the EML4-Alk fusion gene recently identified as an important target in a subset of lung cancer (39,40). Further investigation is warranted in other ABCG2-overexpressing resistant cancer cell lines and in primary clinical samples.
The concurrence of the 3′ translocation, distinct histone modifications at the promoter (14), and the shortening of 3′UTR of ABCG2 in the resistant sublines, although possibly coincidental, suggests an interaction of multiple mechanisms of gene regulation. Intriguingly, besides the S1M1 series of resistant cells, all the ABCG2-overexpressing resistant human cancer cell lines tested in this study exhibited the absence of the long ABCG2 3′UTR, effectively removing miR-519c-mediated repression on ABCG2 (Figure 3B) and stabilizing the ABCG2 mRNA (Figure 4). In contrast, the long ABCG2 3′UTR is retained in the doxorubicin resistant cell line derived from the SW620 colon cancer cell line (Ad300) without the overexpression of ABCG2 (Figure 3B).
Although our study has focused on ABCG2 overexpression due to drug selection, it is possible that the same regulatory mechanism identified (i.e. stabilization of mRNA due to truncation at the 3′UTR) could apply to other cells with high endogenous ABCG2 levels, as observed in the undifferentiated HuES cell line (34). To date, however, we have not confirmed ABCG2 3′UTR and mRNA stabilization as a generalized mechanism contributing to intrinsic drug resistance due to high endogenous ABCG2 expression. None of the unselected cell lines with relatively high ABCG2 expression that we have examined to date (MCF-7, SF295, A549, H460, RPMI-8226 or KM12) (41) has had the truncated phenotype. Rather, ABCG2 adopts both a long and several short 3′UTR fragments (Supplementary Figure 1B). However, 3′UTR length has not been examined in tumor samples with ABCG2 overexpression, nor in putative cancer stem cells.
During the review of this manuscript, Pan et al identified another miRNA (miR-328) targeting ABCG2 3′UTR (42). They also observed an inverse relation between the levels of miR-328 and ABCG2 in the parental MCF-7 and the resistant MCF-7/MX100 cells. Interestingly, the binding site for miR-328 on ABCG2 3′UTR is also located in the long 3′UTR species identified only in parental cells in our study. Although Pan et al did not evaluate the ABCG2 3′UTR length in the parental and resistant cells, we speculate that ABCG2 3′UTR also adopts a long form in their resistant cell line model. Therefore, the interplay between differential miRNA expression and ABCG2 3′UTR length could determine the precise mechanism by which ABCG2 is overexpressed in a particular resistant cell line.
Evidence pointing to the role of miRNAs in determining drug sensitivity and resistance is steadily emerging. Blower et al. reported significant correlations in the NCI-60 tumor drug screen panel between microRNA expression patterns and potency patterns for a large set of compounds (43). It has been shown that a genetic polymorphism of the miR-24 miRNA binding site in dihydrofolate reductase gene could result in resistance to methotrexate (44). The upregulation of miR-214 has been shown to promote the survival of ovarian cancer cells and to induce cisplatin resistance through targeting the 3′UTR of PTEN, which leads to down regulation of PTEN protein and activation of Akt pathway (45). More recently, it has been demonstrated that the increased expression of miR-27a and miR-451 contributes to the multidrug resistance phenotype in A2780 human ovarian cancer cells (21), though the precise mechanism is not clear. In another study, miR-451 was validated as a true miRNA targeting MDR1 in MCF-7 cells (22).
We evaluated the role of miR-519c and miR-520h in affecting ABCG2 expression, function, and resistance to mitoxantrone. Unselected cells transfected with inhibitors against both miR-519c or miR-520h demonstrated increased ABCG2, pheophorbide A efflux, and mitoxantrone resistance upon removal of the repressive effect by the two miRNAs (Figure 6 and Supplementary Figures 6 and 8). Although the studies raise the possibility of miRNA manipulation as a novel strategy to alter drug sensitivity, the experiments do show the limitations of such a strategy. The miRNA mimics do not reduce ABCG2 expression or function where levels are already low. Furthermore, neither miRNA inhibitor nor mimic affect expression in resistant S1M1 80 and MCF-7 FLV1000 cells. This may indicate that while the microRNAs are able to control low levels of ABCG2 mRNA, they cannot have a measurable impact on the very high levels found in drug resistant cell lines.
While reducing drug resistance through increasing microRNA expression is not achievable at this writing, the possibility that it could opens new avenues of investigation. For example, we need to know whether the 3′UTR truncation is observed in any drug resistance setting. Drug-selected cell lines, with exceedingly high expression of an ABC transporter, do not faithfully reproduce clinical drug resistance. We need to evaluate the 3′UTR in clinical samples. Given the observation that both undifferentiated stem cells and proliferating cells have truncated 3′UTR as part of their phenotype, it seems rational that tumors with ABCG2 overexpression may have truncated 3′UTR as well. Identifying truncation would be convincing evidence of activation of the transporter in that tumor type. In the event that ABCG2 3′UTR truncation can be confirmed in specific tumor types, methods of reversing the truncation will need to be developed. Since a genetic change is very unlikely as a cause, the shortening must be epigenetic in origin, and this leads to the possibility that an epigenetic therapy could be identified to modulate the length of the 3′UTR. With the increasing numbers of epigenetic therapies on the horizon, the possibility is an intriguing one (46).
Supplementary Material
Supplementary Figure 1. (A) 3′RACE (Rapid Amplification of cDNA ends) Assay – Overview (18). First strand cDNA synthesis was carried out with total RNA, using a 3′RACE adaptor and Bioscript reverse transcriptase (Bioline). PCR was then performed on the cDNA template using a 3′RACE outer reverse primer (from the assay kit), BIO-X-ACT Long and a forward ABCG2 specific primer (annealing to exon 16). A first nested PCR 1 was performed using the nested N1 ABCG2 specific forward primer and the outer reverse primer to increase the specificity and intensity of the signal. A second nested PCR 2 was carried out by using the nested N1 ABCG2-specific forward primer and a nested N2 reverse primer, complementary to nt 4240–4263 of ABCG2, to ascertain the specificity of the long ABCG2 3′UTR in parental cell lines. (B) 3′RACE analysis of ABCG2 mRNA in four cancer cell lines with high endogenous ABCG2 expression (A549, H460, RPMI-8226 & KM12). Among the NCI Anticancer Drug Screen cancer cell lines, these four cell lines have the highest ABCG2 expression and transport activity (41). Both the long and short ABCG2 3′UTR species were found in these cell lines.
Supplementary Figure 2. RT-PCR analysis investigating the relative abundance of different fragments of ABCG2 3′UTR in pairs of parental and resistant cells.
(A) Schematic showing the approximate position of the six different and overlapping fragments within ABCG2 3′UTR analyzed by RT-PCR. Each fragment is about 500 bp in length.
(B) Relative abundance of different 3′UTR fragments was plotted relative to the amount in region A. Open bar: parental cells; Closed bar: resistant cells.
Supplementary Figure 3. Sequence analysis of cloned 3′UTR PCR products from pairs of parental and resistant cells. Ten clones were sequenced from each parental or resistant cell line. Five ABCG2 3′UTR transcripts of different lengths were found (with the 3′ end at 2471, 2525, 2644, 3013, and 4419, respectively). The numbering is relative to the first nucleotide in the ABCG2 transcript designated in NM_004827.
Supplementary Figure 4. High expression of ABCG2 mops up miR-520h in the resistant MCF-7 FLV1000 cells. Expression levels of ABCG2 and miR-519c/miR-520h were assayed by conventional RT-PCR and stem-loop RT-PCR (18), respectively. GAPDH and U6 small RNA were used as the internal control. ABCG2 mRNA levels were knocked down effectively by a pool of four siRNA duplexes (Dharmacon) by 90% and 60% in the parental and resistant cell lines, respectively. The endogenous level of miR-520h is lower in the resistant MCF-7 FLV1000 than in the parental MCF-7 cells as already shown in Figure 5. When ABCG2 was knocked down in MCF-7 FLV1000, miR-520h level was increased by about 2 fold whereas miR-519c level was not changed. A representative gel image from three reproducible experiments is shown. * PCR reaction for ABCG2 was performed for 30 cycles from MCF-7 cDNA but for 28 cycles from a 1/20 dilution of MCF-7 FLV1000 cDNA.
Supplementary Figure 5. mRNA expression levels of four putative miR-520h target genes (BRCA2, SMAD6, ABCA8 and ID1a) were examined in pairs of parental and ABCG2-overexpressing resistant cells by RT-PCR. ID1a was not detectable in the cells after 40 cycles of PCR reactions. GAPDH was used as the loading control.
Supplementary Figure 6. Antiproliferative activity of mitoxantrone in the parental and resistant S1 and MCF-7 cells after transfection with the two miRNA mimic/inhibitor were evaluated by sulforhodamine D assay (29). The numbers in the parenthesis represent the dose modifying factor caused by the miRNA mimic/inhibitor when compared with the mock transfected cells. Statistical analysis was performed by the Student’s t-test.
Supplementary Figure 7. (A) RT-PCR analysis of ABCG2 mRNA expression in MCF-7 and MCF-7 FLV1000 cells transfected with specific inhibitor/mimic against miR-519c or miR-520h. Cotransfected GFP was also measured to demonstrate equal transfection efficiency. ABCG2 expression was slightly increased by mi-R519c and miR-520h inhibitor in the parental MCF-7 cells but no change was recorded in the resistant MCF-7 FLV1000 cells. (B) ABCG2 surface expression and FTC-inhibitable PhA efflux in miRNA inhibitor or mimic-transfected MCF-7 cells as described in Figure 6. A slight increase in ABCG2 surface expression and FTC-inhibitable PhA efflux was observed in miR-519c or miR-520h inhibitors-transfected MCF-7 cells.
Supplementary Figure 8. Specific miR-519c and miR-520h inhibitors also increased ABCG2 expression and transport activity in other parental cell lines
(A) ABCG2 mRNA expression was measured by RT-PCR in the indicated cell lines after transfection with the miRNA inhibitors (1 = Mock transfection; 2 = miR-519c inhibitor; 3 = miR-520h inhibitor). To indicate the transfection efficiency of the miRNA inhibitors, the level of the cotransfected GFP mRNA was also assessed.
(B) FTC-inhibitable PhA efflux was assessed as a functional assay (as in Figure 6B) for the upregulation of ABCG2 levels in the miRNA inhibitors-transfected cells. Solid line: PhA efflux; Dashed line: FTC-inhibitable PhA efflux.
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, Medical Oncology Branch.
Footnotes
Disclosure of Potential Conflict of Interest
The authors reported no potential conflicts of interest.
References
- 1.Polgar O, Robey RW, Bates SE. ABCG2: structure, function and role in drug response. Expert Opin Drug Metab Toxicol. 2008;4:1–15. doi: 10.1517/17425255.4.1.1. [DOI] [PubMed] [Google Scholar]
- 2.Doyle LA, Yang W, Abruzzo LV, et al. A multidrug resistance transporter from human MCF-7 breast cancer cells. Proc Natl Acad Sci USA. 1998;95:15665–70. doi: 10.1073/pnas.95.26.15665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maliepaard M, van Gastelen MA, de Jang LA, et al. Overexpression of the BCRP/MXR/ABCP gene in a topotecan-selected tumor cell line. Cancer Res. 1999;59:4559–63. [PubMed] [Google Scholar]
- 4.Miyake K, Mickley L, Litman T, et al. Molecular cloning of cDNAs which are highly overexpressed in mitoxantrone-resistance cells: demonstration of homology to ABC transport genes. Cancer Res. 1999;59:8–13. [PubMed] [Google Scholar]
- 5.Robey R, Medina-Perez WY, Nishiyama K, et al. Overexpression of the ATP-binding cassette half transporter, ABCG2 (Mxr/BCrp/ABCP1) in flavopiridol-resistant human breast cancer cells. Clin Cancer Res. 2001;7:145–52. [PubMed] [Google Scholar]
- 6.Volk EL, Farley KM, Wu Y, Li F, Robey RW, Schneider E. Overexpression of wild-type breast cancer resistance protein mediates methotrexate resistance. Cancer Res. 2002;62:5035–40. [PubMed] [Google Scholar]
- 7.Bailey-Dell KJ, Hassel B, Doyle LA, Ross DD. Promoter characterization and genomic organization of the human breast cancer resistance protein (ATP-binding cassette transporter G2) gene. Biochim Biophys Acta. 2001;1520:234–41. doi: 10.1016/s0167-4781(01)00270-6. [DOI] [PubMed] [Google Scholar]
- 8.Ee PL, Kamalakaran S, Tonetti D, He X, Ross DD, Beck WT. Identification of a novel estrogen response element in the breast cancer resistance protein (ABCG2) gene. Cancer Res. 2004;64:1247–51. doi: 10.1158/0008-5472.can-03-3583. [DOI] [PubMed] [Google Scholar]
- 9.Krishnamurthy P, Ross DD, Nakanishi T, et al. The stem cell marker Bcrp/ABCG2 enhances hypoxic cell survival through interactions with heme. J Biol Chem. 2004;279:24218–25. doi: 10.1074/jbc.M313599200. [DOI] [PubMed] [Google Scholar]
- 10.Szatmari I, Vamosi G, Brazda P, et al. Peroxisome proliferator-activated receptor gamma-regulated ABCG2 expression confers cytoprotection to human dendritic cells. J Biol Chem. 2006;281:23812–23. doi: 10.1074/jbc.M604890200. [DOI] [PubMed] [Google Scholar]
- 11.Ebert B, Seidel A, Lampen A. Identification of BCRP as transporter of benz[a]pyrene conjugates metabolically formed in Caco-2 cells and its induction by Ah-receptor agonists. Carcinogenesis. 2005;26:1754–63. doi: 10.1093/carcin/bgi139. [DOI] [PubMed] [Google Scholar]
- 12.Nakanishi T, Bailer-Dell KJ, Hassel BA, et al. Novel 5′ untranslated region variants of BCRP mRNA are differentially expressed in drug-selected cancer cells and in normal human tissues: implications for drug resistance, tissue-specific expression, and alternative promoter usage. Cancer Res. 2006;66:5007–11. doi: 10.1158/0008-5472.CAN-05-4572. [DOI] [PubMed] [Google Scholar]
- 13.To KKW, Zhan Z, Bates SE. Aberrant promoter methylation of the ABCG2 gene in renal carcinoma. Mol Cell Biol. 2006;26:8572–85. doi: 10.1128/MCB.00650-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.To KKW, Polgar O, Huff L, Kuniaki M, Bates SE. Histone modifications at the ABCG2 promoter following treatment with HDAC inhibitor mirror those in multidrug-resistant cells. Mol Cancer Res. 2008;6:151–64. doi: 10.1158/1541-7786.MCR-07-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farh KK, Grimson A, Jan C, et al. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817–21. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 16.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 17.Doench JG, Sharp PA. Specificity of microRNA target selection in translation repression. Genes Dev. 2004;18:504–11. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.To KKW, Zhan Z, Litman T, Bates SE. Regulation of ABCG2 expression at the 3′ untranslated region of its mRNA through modulation of transcript stability and protein translation by a putative microRNA in the S1 colon cancer cell line. Mol Cell Biol. 2008;28:5147–61. doi: 10.1128/MCB.00331-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–5. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 20.Weidhaas JB, Babar I, Nallur SM, et al. MicroRNAs as potential agents to alter resistance to cytotoxic anticancer therapy. Cancer Res. 2007;67:11111–6. doi: 10.1158/0008-5472.CAN-07-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu H, Wu H, Liu X, et al. Role of microRNA miR-27a and miR-451 in the regulation of MDR1/P-glycoprotein expression in human cancer cells. Biochem Pharmacol. 2008;76:582–8. doi: 10.1016/j.bcp.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olga K, Filkowski J, Meservy J, et al. Involvement of microRNA-451 in resistance of the MCF-7 breast cancer cells to chemotherapeutic drug doxorubicin. Mol Cancer Ther. 2008;7:2152–9. doi: 10.1158/1535-7163.MCT-08-0021. [DOI] [PubMed] [Google Scholar]
- 23.Sorrentino A, Liu CG, Addario A, Peschle C, Scambia G, Ferlini C. Role of microRNAs in drug-resistant ovarian cancer cells. Gynecol Oncol. 2008;111:478–86. doi: 10.1016/j.ygyno.2008.08.017. [DOI] [PubMed] [Google Scholar]
- 24.Xia L, Zhang D, Du R, et al. miR-15b and miR-16 modulate multidrug resistance by targeting BCL2 in human gastric cancer cells. Int J Cancer. 2008;123:372–9. doi: 10.1002/ijc.23501. [DOI] [PubMed] [Google Scholar]
- 25.Rabindran SK, He H, Singh M, et al. Reversal of a novel multidrug resistance mechanism in human colon carcinoma cells by fumitremorgin C. Cancer Res. 1998;58:5850–8. [PubMed] [Google Scholar]
- 26.Robey RW, Obrzut T, Shukla S, et al. Becatecarin (rebeccamycin analog, NSC 655649) is a transport substrate and induces expression of the ATP-binding cassette transporter, ABCG2, in lung carcinoma cells. Cancer Chemother Pharmacol. 2009;64:575–83. doi: 10.1007/s00280-008-0908-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang S, Yang Z, Morris ME. Flavonoids are inhibitors of breast cancer resistance protein (ABCG2)-mediated transport. Mol Pharm. 2004;65:1208–16. doi: 10.1124/mol.65.5.1208. [DOI] [PubMed] [Google Scholar]
- 28.Lai GM, Chen YN, Mickley LA, Fojo AT, Bates SE. P-glycoprotein expression and schedule dependence of adriamycin cytotoxicity in human colon carcinoma cell lines. Int J Cancer. 1991;49:696–703. doi: 10.1002/ijc.2910490512. [DOI] [PubMed] [Google Scholar]
- 29.Skehan P, Stornet R, Scudiero D, et al. New colorimetric cytotoxicity assay for anticancer-drug screening. J Natl Cancer Inst. 1990;82:1107–12. doi: 10.1093/jnci/82.13.1107. [DOI] [PubMed] [Google Scholar]
- 30.Robey RW, Steadman K, Polgar O, et al. Pheophorbide A is a specific probe for ABCG2 function and inhibition. Cancer Res. 2004;64:1242–6. doi: 10.1158/0008-5472.can-03-3298. [DOI] [PubMed] [Google Scholar]
- 31.Knutsen T, Rao VK, Ried T, et al. Amplification of 4q21-q22 and the MXR gene in independently derived mitoxantrone-resistant cell lines. Genes Chromosomes Cancer. 2000;27:110–6. [PubMed] [Google Scholar]
- 32.Wang X. miRDB: a microRNA target prediction and functional annotation database with a wiki interface. RNA. 2008;14:1012–7. doi: 10.1261/rna.965408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davis S, Lollo B, Freier S, Esau C. Improved targeting of miRNA with antisense oligonucleotides. Nucleic Acids Res. 2006;34:2294–304. doi: 10.1093/nar/gkl183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Apati A, Orban TI, Varga N, et al. High level functional expression of the ABCG2 multidrug transporter in undifferentiated human embryonic stem cells. Biochim Biophys Acta. 2008;1778:2700–9. doi: 10.1016/j.bbamem.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 35.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–7. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leonard GD, Fojo T, Bates SE. The role of ABC transporters in clinical practice. Oncologist. 2003;8:411–24. doi: 10.1634/theoncologist.8-5-411. [DOI] [PubMed] [Google Scholar]
- 37.Benderra Z, Faussat AM, Sayada L, et al. MRP3, BCRP, and P-glycoprotein activities are prognostic factors in adult acute myeloid leukemia. Clin Cancer Res. 2005;11:7764–72. doi: 10.1158/1078-0432.CCR-04-1895. [DOI] [PubMed] [Google Scholar]
- 38.Liao R, Sun J, Zhang L, et al. MicroRNAs play a role in the development of human hematopoietic stem cells. J Cell Biochem. 2008;104:805–17. doi: 10.1002/jcb.21668. [DOI] [PubMed] [Google Scholar]
- 39.Choi YL, Takeuchi K, Soda M, et al. Identification of novel isoforms of the EML4-ALK transforming gene in non-small cell lung cancer. Cancer Res. 2008;68:4971–6. doi: 10.1158/0008-5472.CAN-07-6158. [DOI] [PubMed] [Google Scholar]
- 40.Kaye FJ. Mutation-associated fusion cancer genes in solid tumors. Mol Cancer Ther. 2009;8:1399–408. doi: 10.1158/1535-7163.MCT-09-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deeken JF, Robey RW, Shukla S, et al. Identification of compounds that correlate with ABCG2 transporter function in the National Cancer Institute Anticancer Drug Screen. Mol Pharmacol. doi: 10.1124/mol.109.056192. Published online on Jul 24, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pan YZ, Morris ME, Yu AM. MicroRNA-328 negatively regulates the expression of breast cancer resistance protein (BCRP/ABCG2) in human cancer cells. Mol Pharmacol. 2009;75:1374–9. doi: 10.1124/mol.108.054163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blower PE, Chung JH, Verducci JS, et al. MicroRNAs modulate the chemosensitivity of tumor cells. Mol Cancer Ther. 2008;7:1–9. doi: 10.1158/1535-7163.MCT-07-0573. [DOI] [PubMed] [Google Scholar]
- 44.Mishra PJ, Humeniuk R, Mishra PJ, et al. A miR-24 microRNA binding-site polymorphism in dihydrofolate reductase gene leads to methotrexate resistance. Proc Natl Acad Sci USA. 2007;104:13513–8. doi: 10.1073/pnas.0706217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang H, Kong W, He L, et al. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 2008;68:425–33. doi: 10.1158/0008-5472.CAN-07-2488. [DOI] [PubMed] [Google Scholar]
- 46.Piekarz RL, Bates SE. Epigenetic modifiers: basic understanding and clinical development. Clin Cancer Res. 2009;15:3918–26. doi: 10.1158/1078-0432.CCR-08-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. (A) 3′RACE (Rapid Amplification of cDNA ends) Assay – Overview (18). First strand cDNA synthesis was carried out with total RNA, using a 3′RACE adaptor and Bioscript reverse transcriptase (Bioline). PCR was then performed on the cDNA template using a 3′RACE outer reverse primer (from the assay kit), BIO-X-ACT Long and a forward ABCG2 specific primer (annealing to exon 16). A first nested PCR 1 was performed using the nested N1 ABCG2 specific forward primer and the outer reverse primer to increase the specificity and intensity of the signal. A second nested PCR 2 was carried out by using the nested N1 ABCG2-specific forward primer and a nested N2 reverse primer, complementary to nt 4240–4263 of ABCG2, to ascertain the specificity of the long ABCG2 3′UTR in parental cell lines. (B) 3′RACE analysis of ABCG2 mRNA in four cancer cell lines with high endogenous ABCG2 expression (A549, H460, RPMI-8226 & KM12). Among the NCI Anticancer Drug Screen cancer cell lines, these four cell lines have the highest ABCG2 expression and transport activity (41). Both the long and short ABCG2 3′UTR species were found in these cell lines.
Supplementary Figure 2. RT-PCR analysis investigating the relative abundance of different fragments of ABCG2 3′UTR in pairs of parental and resistant cells.
(A) Schematic showing the approximate position of the six different and overlapping fragments within ABCG2 3′UTR analyzed by RT-PCR. Each fragment is about 500 bp in length.
(B) Relative abundance of different 3′UTR fragments was plotted relative to the amount in region A. Open bar: parental cells; Closed bar: resistant cells.
Supplementary Figure 3. Sequence analysis of cloned 3′UTR PCR products from pairs of parental and resistant cells. Ten clones were sequenced from each parental or resistant cell line. Five ABCG2 3′UTR transcripts of different lengths were found (with the 3′ end at 2471, 2525, 2644, 3013, and 4419, respectively). The numbering is relative to the first nucleotide in the ABCG2 transcript designated in NM_004827.
Supplementary Figure 4. High expression of ABCG2 mops up miR-520h in the resistant MCF-7 FLV1000 cells. Expression levels of ABCG2 and miR-519c/miR-520h were assayed by conventional RT-PCR and stem-loop RT-PCR (18), respectively. GAPDH and U6 small RNA were used as the internal control. ABCG2 mRNA levels were knocked down effectively by a pool of four siRNA duplexes (Dharmacon) by 90% and 60% in the parental and resistant cell lines, respectively. The endogenous level of miR-520h is lower in the resistant MCF-7 FLV1000 than in the parental MCF-7 cells as already shown in Figure 5. When ABCG2 was knocked down in MCF-7 FLV1000, miR-520h level was increased by about 2 fold whereas miR-519c level was not changed. A representative gel image from three reproducible experiments is shown. * PCR reaction for ABCG2 was performed for 30 cycles from MCF-7 cDNA but for 28 cycles from a 1/20 dilution of MCF-7 FLV1000 cDNA.
Supplementary Figure 5. mRNA expression levels of four putative miR-520h target genes (BRCA2, SMAD6, ABCA8 and ID1a) were examined in pairs of parental and ABCG2-overexpressing resistant cells by RT-PCR. ID1a was not detectable in the cells after 40 cycles of PCR reactions. GAPDH was used as the loading control.
Supplementary Figure 6. Antiproliferative activity of mitoxantrone in the parental and resistant S1 and MCF-7 cells after transfection with the two miRNA mimic/inhibitor were evaluated by sulforhodamine D assay (29). The numbers in the parenthesis represent the dose modifying factor caused by the miRNA mimic/inhibitor when compared with the mock transfected cells. Statistical analysis was performed by the Student’s t-test.
Supplementary Figure 7. (A) RT-PCR analysis of ABCG2 mRNA expression in MCF-7 and MCF-7 FLV1000 cells transfected with specific inhibitor/mimic against miR-519c or miR-520h. Cotransfected GFP was also measured to demonstrate equal transfection efficiency. ABCG2 expression was slightly increased by mi-R519c and miR-520h inhibitor in the parental MCF-7 cells but no change was recorded in the resistant MCF-7 FLV1000 cells. (B) ABCG2 surface expression and FTC-inhibitable PhA efflux in miRNA inhibitor or mimic-transfected MCF-7 cells as described in Figure 6. A slight increase in ABCG2 surface expression and FTC-inhibitable PhA efflux was observed in miR-519c or miR-520h inhibitors-transfected MCF-7 cells.
Supplementary Figure 8. Specific miR-519c and miR-520h inhibitors also increased ABCG2 expression and transport activity in other parental cell lines
(A) ABCG2 mRNA expression was measured by RT-PCR in the indicated cell lines after transfection with the miRNA inhibitors (1 = Mock transfection; 2 = miR-519c inhibitor; 3 = miR-520h inhibitor). To indicate the transfection efficiency of the miRNA inhibitors, the level of the cotransfected GFP mRNA was also assessed.
(B) FTC-inhibitable PhA efflux was assessed as a functional assay (as in Figure 6B) for the upregulation of ABCG2 levels in the miRNA inhibitors-transfected cells. Solid line: PhA efflux; Dashed line: FTC-inhibitable PhA efflux.