Abstract
We describe a new supervised learning-based template matching approach for segmenting cell nuclei from microscopy images. The method uses examples selected by a user for building a statistical model which captures the texture and shape variations of the nuclear structures from a given dataset to be segmented. Segmentation of subsequent, unlabeled, images is then performed by finding the model instance that best matches (in the normalized cross correlation sense) local neighborhood in the input image. We demonstrate the application of our method to segmenting nuclei from a variety of imaging modalities, and quantitatively compare our results to several other methods. Quantitative results using both simulated and real image data show that, while certain methods may work well for certain imaging modalities, our software is able to obtain high accuracy across several imaging modalities studied. Results also demonstrate that, relative to several existing methods, the template-based method we propose presents increased robustness in the sense of better handling variations in illumination, variations in texture from different imaging modalities, providing more smooth and accurate segmentation borders, as well as handling better cluttered nuclei.
Keywords: Segmentation, nuclei, template matching, nonrigid registration, cross correlation
II. Introduction
Segmenting cell nuclei from microscopy images is an important image processing task necessary for many scientific and clinical applications due to the fundamentally important role of nuclei in cellular processes and diseases. Given a large variety of imaging modalities, staining procedures, experimental conditions, etc., many computational methods have been developed and applied to cell nuclei segmentation in 2D [1], [4]-[11] and 3D images [12]–[16]. Thresholding techniques [17], [18], followed by standard morphological operations, are amongst the simplest and most computationally efficient strategies. These techniques, however, are inadequate when the data contains strong intensity variations, noise, or when nuclei appear crowded in the field of view [9], [19] being imaged. The watershed method is able to segment touching or overlapping nuclei. Direct use of watershed algorithms, however, can often lead to over segmentation artifacts [8], [20]. Seeded or marker controlled watershed methods [4], [5], [7]–[9], [21], [22] can be utilized to overcome such limitations. We note that seed extraction is a decisive factor in the performance of seeded watershed algorithms. Missing or artificially added seeds can cause under or over segmentation. Different algorithms for extracting seeds have been proposed. In [22] for example, seeds are extracted using a gradient vector field (GVF) followed by Gaussian filtering. Jung and Kim [10] proposed to find optimal seeds by minimizing the residual between the segmented region boundaries and the fitted model. In addition, various of post-processing algorithms have been applied to improve the segmentation quality. For example, morphological algorithms (e.g. dilation and erosion) [9] can be used iteratively to overcome inaccuracies in segmentation. In [23] learning-based algorithms were used for discarding segmented regions deemed to be erroneous. Similar ideas using neural networks can be seen in [24].
When nuclei do not appear clearly in the images to be segmented (e.g. nuclear borders are not sharp enough or when a significant amount of noise is present), active contour-based methods [11], [13], [14], [25]–[29], especially those implicitly represented by level set [13], [14], [29], have been proposed to overcome some of these limitations successfully. As well known, the level set framework is well suited for accurate delineation of complicated borders and can be easily extended to higher dimensional datasets. Ortiz De Solorzano et al. [13], for example, proposed an edge-based deformable model that utilizes gradient information to capture nuclear surfaces. Considering that strong gradients at object boundaries may be blurred and the noise and intracellular structures may also show strong gradients, Mukherjee et al. [30] proposed a level set model that also incorporates a region term using the likelihood information for segmentation of leukocytes with homogeneous regions. In segmenting cells in culture or in tissue sections, Dufour et al. [14] proposed a multi-level deformable model incorporating both a gradient term and a region term, adopted from Chan and Vese model [31], to segment cells with ill-defined edges. In [32], Yan et al. also proposed a similar multilevel deformable model to segment RNAi fluorescence cellular images of drosophila. In [29], Cheng and Rajapakse utilized the Chan and Vese model [31] to obtain the outer contours of clustered nuclei, employing a watershed-like algorithm to separate clustered nuclei. Similarly, Nielsen et al [11] have described a method for segmenting Feulgen stained nuclei using a seeded watershed method, combined with a gradient vector flow-based deformable model method [33]. Considering that some nuclei may appear to overlap in 2D images, Plissiti and Nikou [34] proposed a deformable model driven by physical principles, helping to delineate the borders of overlapping nuclei. In [35], Dzyubachyk et al. proposed a modified region-based level set model, which addresses a number of shortcomings in [14] as well as speeds up computation. In order to reduce the large computational costs of variational deformable models, Dufour et al. [36] proposed a novel implementation of the piece-wise constant Mumford-Shah functional using 3D active meshes for 3D cell segmentation.
Besides the methods mentioned above, several other approaches for segmenting nuclei based on filter design [37], [38], multi-scale analysis [39], dynamic programming [40], Markov random fields [41], graph based methods [42]–[44] and learning based strategies [45]–[49] have been described. As new imaging modalities, staining techniques, etc., are developed, however, many existing methods specifically designed for current imaging modalities may not work well. Below we show that the application of some such methods can fail to detect adequate borders, or separate touching or overlapping nuclei, in several staining techniques. Therefore considerable resources have to be spent to modify existing methods (or developing entirely new segmentation methods) to better suit the new applications.
Here we describe a generic nuclear segmentation method based on the combination of template matching and supervised learning ideas. Our goal is to provide a method that can be used effectively for segmenting nuclei for many different types of cells imaged under a variety of staining or fluorescence techniques. We aim to guarantee robust performance by allowing the method to ‘calibrate’ itself automatically using training data, so that it will adapt itself to segmenting nuclei with different appearances (due to the staining techniques for example) and shapes. The method is also ‘constrained’ to produce smooth borders. Finally, given that the objective function used in the segmentation process is the normalized cross correlation (NCC), the method is also able to better handle variations in illumination within the same image, as well as across images. We note that template matching-based methods for image segmentation have long been used for segmenting biomedical images. One prominent example is the brain segmentation tool often used in the analysis of functional images [50]. When segmenting nuclei from microscopy images, contour templates have also been used [45], [46]. Here we utilize similar ideas with some adaptations. First, our approach is semi-automated in that it first seeks to learn a template and statistical model from images delineated by the user. The model is built based on estimating a ‘mean’ template, as well as the deformations from the template to all other nuclei provided in the training step. After this step, any image of the same modality can then be segmented via a template-based approach based on maximization of the normalized cross correlation between the template estimated from the input images and the image to be segmented. We describe the method in detail in the next section, and compare it to several other methods applied on different datasets in section IV. Finally, we note that our method is implemented in MATLAB computer language [3]. The necessary files can be obtained through contact with the corresponding author (GKR).
III. Materials and Methods
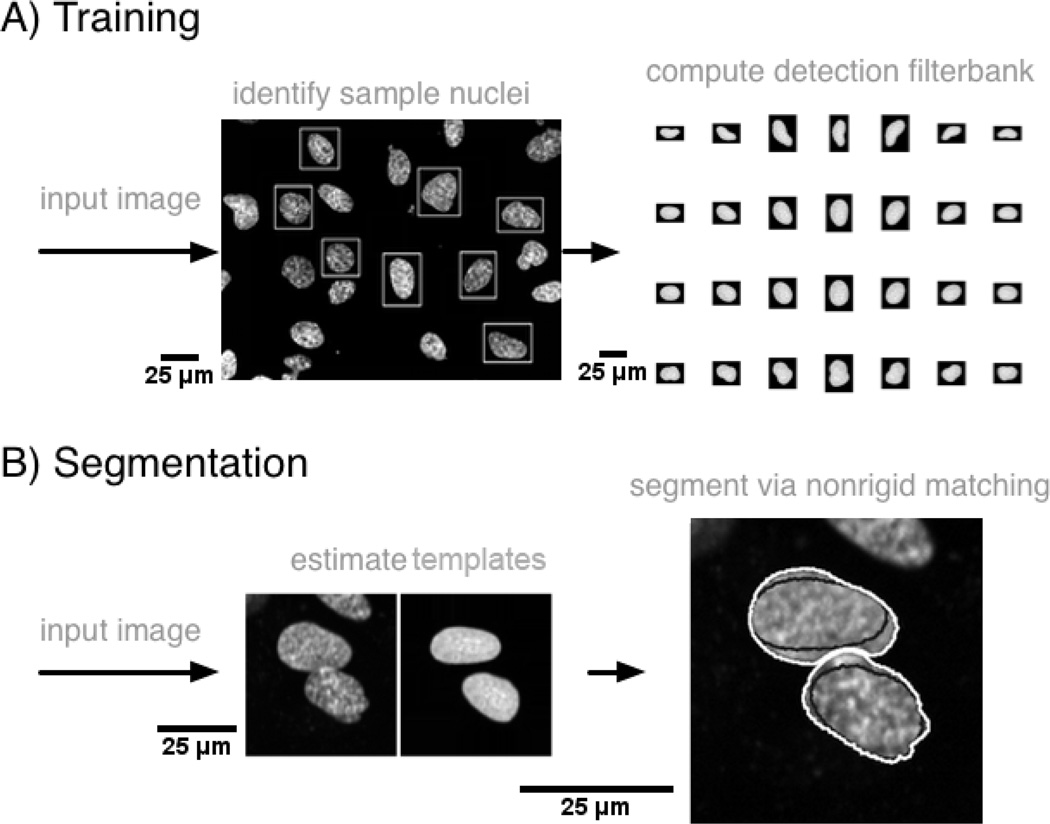
Given the large variation in appearance of nuclei in microscopy images, a completely automated (unsupervised) approach for segmenting nuclei from arbitrary images may be difficult to obtain. We therefore focus on a semi automated approach, depicted in Fig.1, where the idea is to first construct a statistical model for the mean texture and most likely variations of shape to be found in the dataset to be segmented from hand-delineated images. Segmentation of any image of similar type is then achieved by maximizing the normalized cross correlation (NCC) between the model and the local image region. Part A outlines the training procedure whereby the user utilizes a simple graphical user interface to isolate several nuclei samples, which are then used to build the statistical model. Part B outlines the actual segmentation procedure, which proceeds to first find an approximate segmentation (seed detection) of an input image by matching the statistical model with the given image, and then produces a final segmentation result via non-rigid registration.
Fig. 1.
Overview of nuclear segmentation approach. Part A outlines the training procedure, which utilizes sample nuclei manually identified by the user to build a statistical model for the texture and shape variations that could be present in the set of nuclei to be segmented. The model is then sampled to form a detection filter-bank. Part B outlines the actual segmentation procedure which utilizes the detection filter-bank to produce a rough segmentation, and then refines it using non-rigid registration based on the normalized cross correlation.
A. Training
As outlined in part A of Fig.1, we utilize a simple graphical user interface to enable an operator to manually delineate rectangular sub-windows each containing one nucleus sample from an image of the modality he or she wishes to segment. It is required by our system that each sub-window contains only one nucleus, and recommended that the set of sub-windows contain a variety of shapes and textures (small, large, bent, irregular shaped, hollow, etc.), since more variations present in the input images will translate into more variations being captured by the model. We note that it is not necessary for the user to provide the detailed outline for the nucleus present in each window. Rather, a rectangular bounding box suffices. In our implementation, given N such rectangular sub-windows, which can be of different sizes, each sub-window containing one nucleus from the training set, we first pad each sub-window image by replicating the border elements so as to render each sub-window of the same size (in terms of number of pixels in each dimension). The amount of padding applied to each sub-window is the amount necessary for that sub-window to match the size of the largest rectangular sub-window in the set. The set of sub-windows are then rigidly aligned to one sub-window image from the set (picked at random) via a procedure described in earlier work [51]. As a result, the major axis of nuclei samples are aligned to the same orientation. In this case, we choose the normalized cross correlation (NCC) as the optimization criterion for measuring how well two nuclei align and include coordinate inversions (image flips) in the optimization procedure.
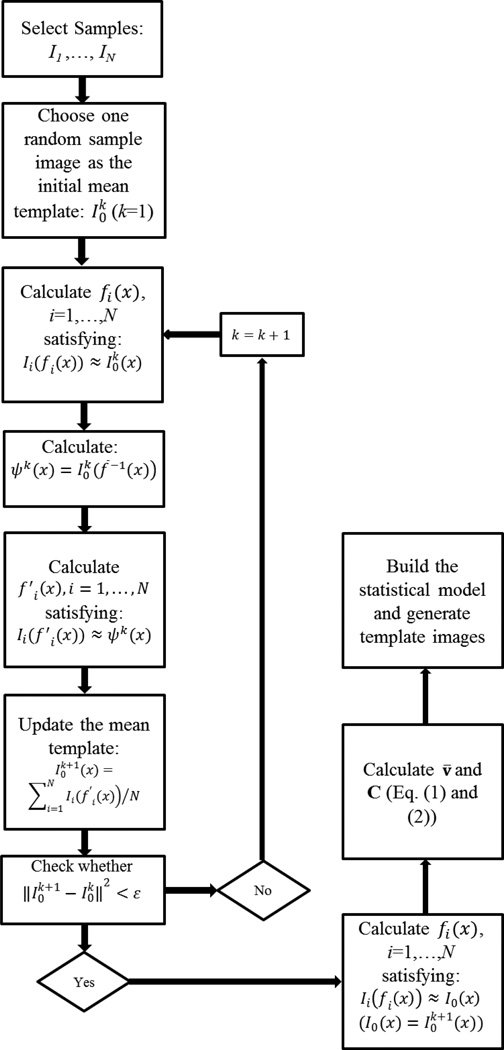
The set of N rigidly aligned sub-windows, denoted as I1, …,IN from now on, is then used to estimate a template that will represent an ”average” shape as well as texture for this set. Several procedures can be used for this purpose. In this work we choose the procedure outlined in Heitz et al. [52] where the idea is to iteratively deform all nuclear images (sub windows) towards a template image that is closest (in the sense of least deformation) to all other images in the set. Fig.2 contains a diagram depicting the procedure we use. The procedure depends on the computation of a nonrigid map that aligns two images Ii, and Ij, via Ii(fi(x)) ≈ Ij(x), with x an input coordinate in the image grid Ω, and a nonrigid mapping function fi(x) : Ω → ℛ2. In our approach, the nonrigid registration is computed via maximization of the normalized cross correlation cost function, which is described in detail in the appendix. Given the ability to non-rigidly align two nuclear images, the template estimation procedure consists of choosing a sub-window image from the set at random and denoting it . Then, starting with the iteration k = 1:
Non-rigidly register to each sub-window image Ii,i = 1, 2, …, N such that .
Calculate a temporary average shape template , with , and f−1 the inverse of the transformation function f (which we compute with Matlab’s ‘griddata’ function).
Compute the average texture on the same average shape template above by first registering each sub-window image in the set to Ψ(x) (i.e. ), and update the template via .
(sum of squared errors). If error < ε stop, otherwise set k = k + 1 and go to step 1.
Fig. 2.
Diagram depicting training procedure.
The end result is an image I0(x) that represents an average template (both in the sense of shape and texture), as well as a set of spatial transformations that map each sub-window image to the final template via Ii(fi(x)) ≈ I0(x),i = 1, … N. We next apply the principal component analysis (PCA) technique [53] to derive a statistical model for the possible variations in the shape of the sample nuclei. We encode each spatial transformation fi(x) as a vector of displacements via vi = [fi(x1), …, fi(xL)]T, with L the number of pixels in each image. Thus, the mean and the covariance of the set of spatial displacements v1, …, vN are:
| (1) |
| (2) |
Using the PCA method, the principal deformation modes are given by the eigenvectors qp, p = 1, 2, 3, … of the covariance matrix C satisfying Cqp = λpqp. A statistical model for the variations in shape is obtained by retaining the top eigenvalues and eigenvectors corresponding to 95% (this percentage chosen arbitrarily) of the variance in the dataset. This means that the number of eigenvectors used in each segmentation task (imaging modality) will depend on how much variability is present in the (training) dataset. In cases where variability is large, more eigenvectors will be necessary. In cases when variability is small, a small number of eigenvectors will be used. In all cases, the accuracy of the PCA reconstruction procedure is set to 95% (of the variance). The model can be evaluated by choosing an eigenvector qp and calculating vp, bp = v̄ + bpqp, where bp is a mode coefficient. The corresponding template is obtained by re-assembling vp,bp into a corresponding spatial transformation fp,bp, and computing .
In our approach, the statistical model is evaluated for in intervals of . The result of this operation is a set of images obtained by deforming the mean template and representing nuclear configurations likely to be encountered in data to be segmented. In addition, this set of images is augmented by including rotations (rotated every 30 degrees, totaling 7 orientations in our implementation) as well as variations in size (2 in our implementation). Finally, we discard the top 1% and bottom 1% (in the sense of area) of the templates to avoid potentially segmenting structures that would be too small or too large to be considered as nuclei. A data-dependent way of choosing this threshold is also described in the discussion section of this paper. The reason being that templates that are too small may cause over segmentation, while templates that are too large may merge nuclei that are close to each other. Fig.1 (top right) contains a few examples of template images generated in this way for a sample dataset. We denote the set of template images generated in this way as the ”detection filterbank” to be used as a starting point for the segmentation method described in the next subsection.
Segmenting the mean template
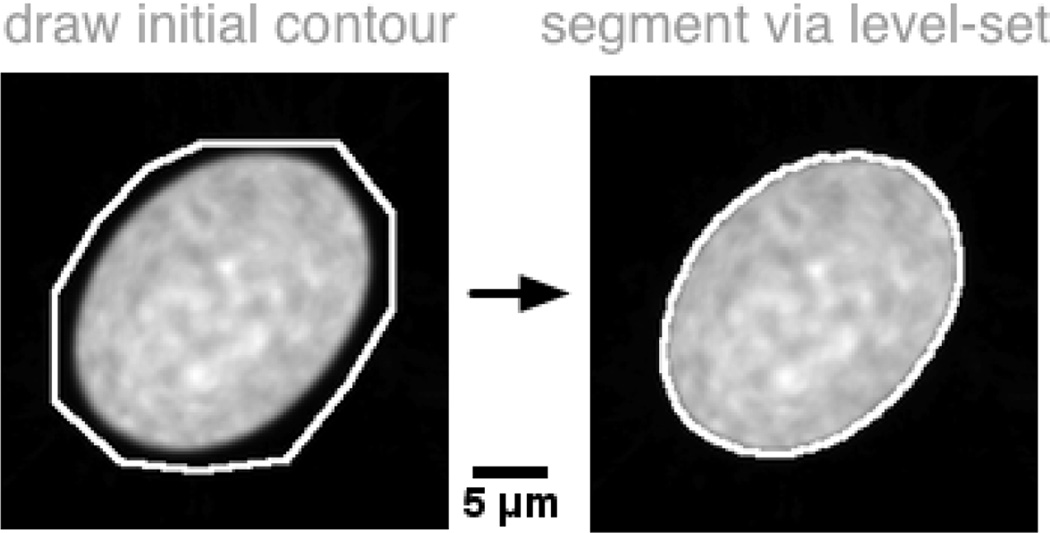
our procedure depends the mean template, computed as estimated above, on being segmented in the sense that pixels in the foreground and background are known. Though many automated methods can be considered for this step, we choose to utilize a rough contour manually provided by an user. The contour is then refined utilizing a level set approach [54]. The advantage is that such a process can be repeated until a satisfactory segmentation result has been made by the user. Fig.3 shows the outline of the procedure.
Fig. 3.
The mean template image must be segmented before the segmentation algorithm based on NCC maximization can be utilized. We utilize a semi automated approach wherein an user draws an initial contour, and a level set-based algorithm refines it to accurately match its borders.
B. Segmentation
Our segmentation algorithm is based on the idea of maximizing the NCC between the statistical model for a given dataset (its construction is described in the previous subsection) and local regions in an input image to be segmented. The first step in such a procedure is to obtain an approximate segmentation of an input image, here denoted as J(x), by computing the NCC of the input image against each filter (template image) in the detection filterbank to obtain an approximate segmentation. To that end, we compute the NCC between each filter Wp(x), p = 1, …, K and the image to be segmented via:
| (3) |
where , with Ωu denoting the neighborhood around u of the same size as filter Wp. We note that Nc is the number of channels in each image (e.g. 1 for scalar images and 3 for color images). A detection map denoted M is computed as M(u) = maxp γp(u). We note that the value of the cross correlation function γ above is bound to be in the range [−1, 1]. We also note that the index p that maximizes this equation also specifies the template Wp that best matches the region u and is used later as a starting point for the deformable model-based optimization.
The detection map M(u) is mined for potential locations of nuclei using the following two principles: (1) only pixels whose intensities in M are greater than a threshold μ are of interest. (2) The centers of detected nuclei must be at least a certain distance far away from each other. This helps to prevent, for example, two potential locations from being detected within one nucleus, causing over segmentation. These two principles can be implemented by first searching for the highest response in M. Subsequent detections must be at least a certain distance from the first. This is done by dilating the already detected nuclei (remember, the filtering step above not only defines regions where nuclei might be located, but also the rough shape of each). This process is able to detect nuclei of different shapes due to simulated templates of various shapes and orientations generated in the previous step, and is repeated until all pixels in the thresholded detection image M have been investigated. We note again that each detected pixel in M has its associated best matching template from the detection filterbank. Therefore, this part of the algorithm not only provides the location of a nucleus, but also a rough guess for its shape (see bottom middle of Fig.1) and texture.
Once an initial estimate for each nucleus in an input image is found via the procedure described above, the algorithm produces a spatially accurate segmentation by non-rigidly registering each approximate guess to the input image. The nonrigid registration nonlinearly adapts the borders of the detected template so as to accurately segment the borders of each nuclei in the input image. In addition, the nonrigid registration approach we use also is constrained to produce smooth borders. Details related to the nonrigid registration are provided in appendix. Rather than optimizing all guesses at once, which could lead to difficulties such as a large number of iterations in our gradient ascent-type strategy, each nucleus is segmented separately.
Segmenting touching nuclei
An important feature of our template matching approach is that it is capable of segmenting touching nuclei without difficulties with a small modification of the procedure described above. In our method, if two (or more) nuclei are detected to be close to each other (e.g. the closest distance between their best matching templates’ borders is smaller than 10 pixels), these nuclei are regarded as being potentially in close proximity to each other. If so, their best matching templates obtained from the filter bank procedure above are taken together under a sub-window and then non-rigidly registered to the same sub-window in the real image using the same optimization algorithm in the appendix. An example showing the segmentation of two nuclei in close proximity to each other is shown in Fig.1 (bottom row). The left part of this portion of the figure shows the initial estimates from the filterbank-based estimation of candidate locations. The result of the nonrigid registration-based estimation of the contours for each nucleus is shown at the bottom right corner of the same figure. The black contours indicate the borders of the best matching templates (the initial guesses) and the white lines delineate the final segmentation result after nonrigid registration.
C. Experiments Overview
1) Data Acquisition
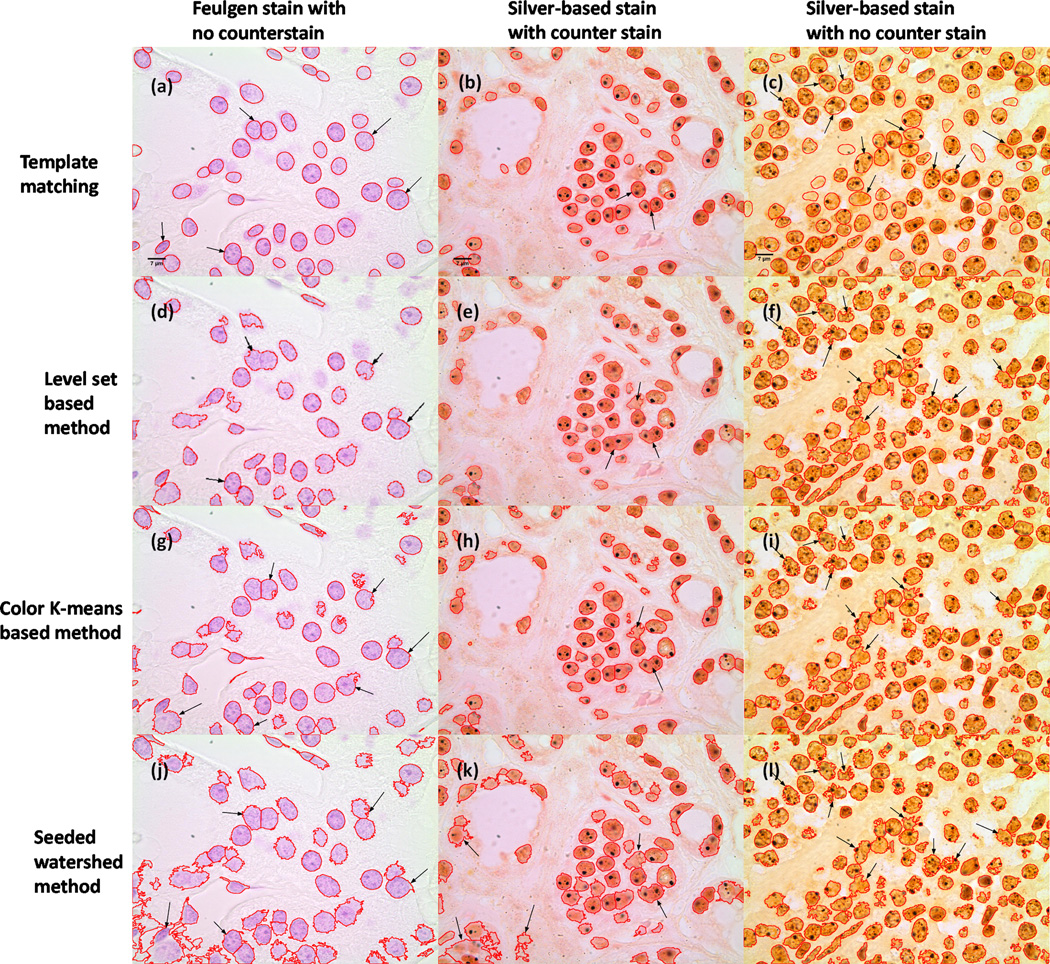
We demonstrate our system applied to several different cell nuclei datasets: (1) a synthetic dataset BBBC004v1 generated with the SIMCEP simulating platform for fluorescent cell population images [55], [56]; (2) two real cell datasets (U2OS cells and NIH3T3 cells) acquired with fluorescence imaging [57]; (3) and a histopathology dataset obtained using thyroid tissue specimens with several different staining techniques. The primary goal for the simulated dataset is to obtain an accurate count for the number of nuclei in each field of view. Each simulated image contains 300 objects with different degrees of overlap probability (ranging from 0.00 to 0.60). The U2OS (48 images, each containing multiple nuclei) and NIH3T3 (49 images) cells were obtained with the Hoechst 33342 fluorescence signal, and the ground truth (including accurately delineated borders) is provided by experts [57]. Of these, the U2OS dataset is more challenging, with nuclei tending to be more varied in shape and more clustered together. The intensity of the NIH3T3 images, however, is more nonuniform than the U2OS dataset. In addition we apply our method to segmenting nuclei from histopathology images taken from tissue sections of thyroid specimens. Tissue blocks were obtained from the archives of the University of Pittsburgh Medical Center (Institutional Review Board approval #PRO09020278). Briefly, tissue sections were cut at 5 µm thickness from the paraffin-embedded blocks and stained using three techniques. These include the Feulgen stain which stains deoxyribonucleic acids only. If no counterstaining is performed then only nuclei are visible demonstrating chromatin patterns as deep magenta hues shown in Fig. 6(a). The second is a silver-based technique that stains the intranuclear nucleolar organizing regions (NORs) (black intranuclear dots) and counterstained with nuclear fast red which uses kernechtrot that dyes nuclear chromatin red (Fig. 6(b)). The third is the same silver-based staining for NORs without counterstaining (Fig. 6(c)). All images used for analysis in this study were acquired using an Olympus BX51 microscope equipped with a 100X UIS2 objective (Olympus America, Central Valley, PA) and 2 mega pixel SPOT Insight camera (Diagnostic Instruments, Sterling Heights, MI). Image specifications were 24 bit RGB channels and 0.074 microns/pixel, 118 × 89 µm field of view. More details pertaining to the image acquisition process for this dataset are available in [58].
Fig. 6.
Nuclei segmentation from histopathology images with different staining. Note that the improvements are pointed out by black arrows. First row: results of our template matching approach. Second row: results of level set-based method. Third row: results of color K-means-based method. Fourth row: results of seeded watershed method.
2) Experimental Setup
We note that our system is able to work with grayscale (single color) images as well as with color images. Equation (4), in the Appendix section, allows color images to be used, while the method can also be used to segment 3D data by defining the inter products and convolutions utilized in equations (4) and (5) in three dimensions. In addition, we mention that for color images, each color channel (R, G and B) is equally weighted in the approach we described above. This allows for segmentation even in the case when the optimal color transformation for detecting nuclei is not known precisely (as is the case in many of the images shown). In cases where this information is known precisely, the approach we proposed can be used with only the color channel that targets nuclei, or with the image after optimal color transformation. In each experiment, k sample nuclei (k is arbitrarily chosen as 20 in our experiments) were chosen by the authors for the training process. All but one of the parameters remained constant for all experiments. The percent of variance retained in PCA analysis was set to (95%), ε = 0.01 for the calculation of the average template, the step size in the gradient ascent procedure κ was set as 5 × 104, the scale number in the multi-scale strategy s was set as 2. For smoothing parameter σ in the gradient ascent procedure, a higher σ value helps to smooth the contour, while a lower σ value helps to better capture the real border of nuclei. In this paper, σ was experimentally set as 1.5 (pixels). The only parameter that varied from dataset to dataset was the detection threshold μ. While a higher value of μ may miss some nuclei (e.g. out of focus), a lower value of μ may confuse noise and clutter for actual nuclear candidates. There are two ways to determine an appropriate value for detection threshold μ. When the ground truth (e.g. manual delineation of nuclei) for the training images is provided, μ value can be selected automatically by maximizing the dice metric [2] between the detections and provided ground truth. Here #(·) counts the number of nuclei in different results, GT corresponds to the ground truth result, while Detect(μ) corresponds to the nuclei detection result with respect to μ. When ground truth is not available, an appropriate μ value has to be empirically selected by the user in order to detect most nuclei in the training images for each application or dataset. In the experiments shown below, ground truth was not used for selecting μ. Rather μ was empirically determined for each dataset based on empirical experimentation with a given field of view (containing multiple nuclei) from the corresponding dataset.
For comparison, we choose several different types of algorithms commonly used for cell nuclei segmentation. These include the level set method (Chan and Vese model [31]), an unsupervised learning method (color K-means [59]), and the direct seeded watershed method, which uses a shape based method to separate clumped nuclei (implemented by CellProfiler [60]). Since the CellProfiler software [60] is only able to process 2D grayscale images, a typical choice is to convert the color histopathology image to grayscale image by forming a weighted sum of R, G, and B channels, which keeps the luminance channel (grayscale = 0.2989 × R + 0.5870 × G + 0.1140 × B) [61]. In addition, we prefer to take the general approach of normalizing all image data to fit the intensity range of [0, 1] by scaling the minimum and maximum of each image (discounting outliers set at 1% in our implementation). Since the level set method and the K-means method may not be able to separate clumped nuclei very well, a common solution is to apply seeded watershed algorithm on the binary masks segmented from level set method and K-means methods, in which seeds are defined as the local maxima in the distance transformed images of binary masks [62]. Note that H-dome maxima [62] are calculated on the distance transformed images in order to prevent over-segmentation, and for different dataset, the H value is arbitrarily selected for the best performance. These techniques were chosen since they are similar to several of the methods described in the literature for segmenting nuclei from microscopy images [14], [35]. In the following sections, we will show both qualitative and quantitative comparisons of these methods.
IV. Results
A. Qualitative Evaluation
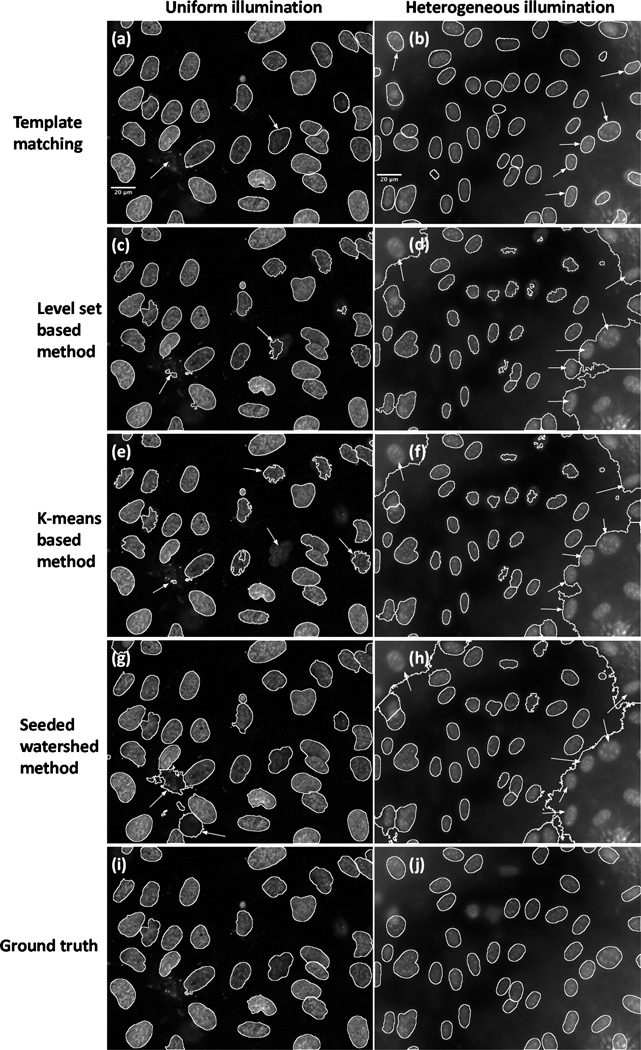
In Fig. 4, Fig. 5 and Fig. 6, we compare the results for different types of datasets using different methods. In Fig. 4, the results are obtained by different methods applied to the segmentation of synthetic nuclei with clustering probability set to 0.3. Note that we use green (in color), red, and yellow square dots to represent correct detections, missed detections, and spurious detections respectively. In Fig. 5, the first column shows the sample segmentations of the U2OS data (under uniform illumination), and the second column shows the sample segmentations of the NIH3T3 data (under heterogeneous illumination), in which the white contours delineate the borders of segmented nuclei. The first row of Fig. 5 corresponds to results computed using the approach we described in this paper, the second row corresponds to level set method-based method, the third row corresponds to color K-means-based method, and the fourth row corresponds to direct seeded watershed method. In addition, we show the hand-labeled images of U2OS data and NIH3T3 data as the ground truth separately in Fig. 5(i) and Fig. 5(j) in the final row. In Fig. 6, we show the segmentation results on sample histology images with different staining techniques, in which each column corresponds to a distinct staining technique (details have been described in the previous section), while each row corresponds to a distinct segmentation method (the row order is the same as Fig. 5).
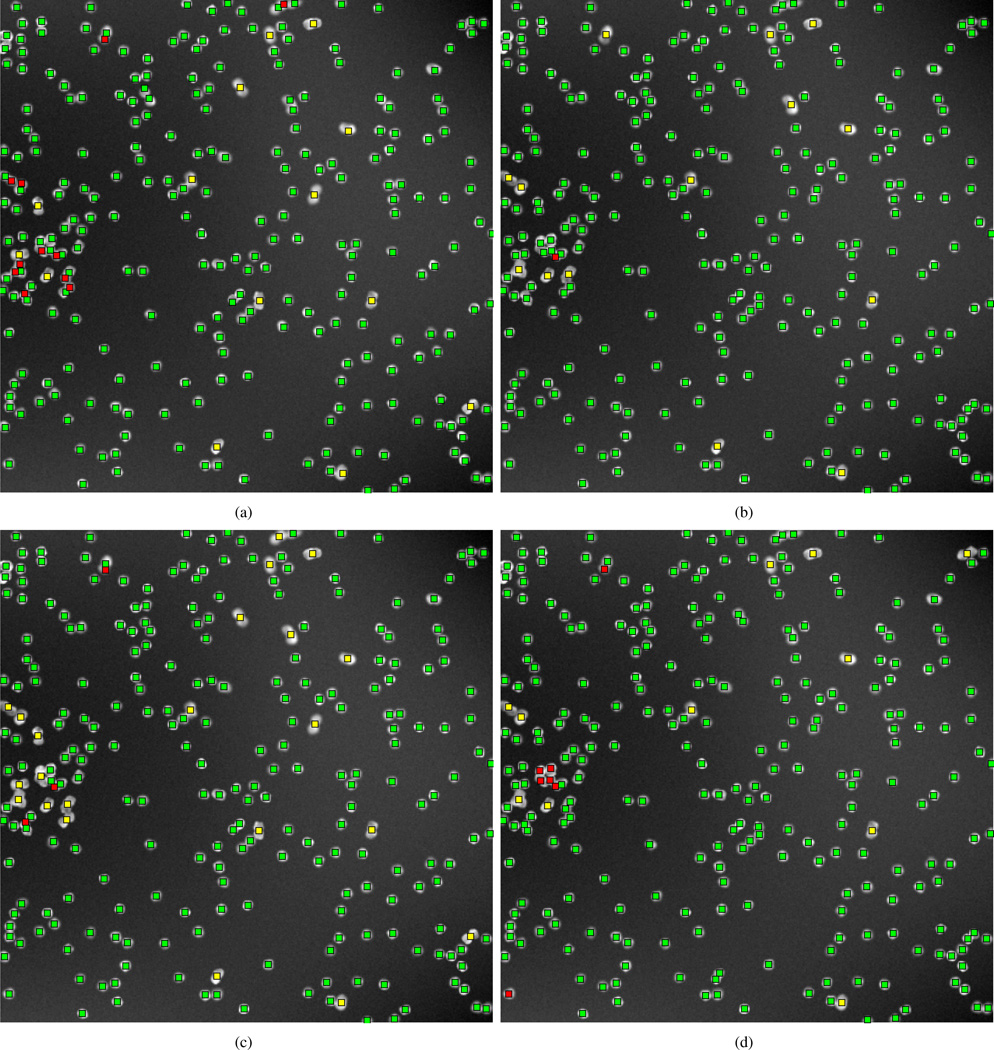
Fig. 4.
Nuclei counting in synthetic images. Upper left: results of our template matching approach. Upper right: result obtained with level set method. Bottom left: results obtained with color K-means-based method. Bottom right: results obtained with seeded watershed method. Note that green square dots represent correct detections, red square dots represent missed detections, and yellow square dots represent spurious detections.
Fig. 5.
Nuclei detection and segmentation from different fluorescence images. Note that the improvements are pointed out by white arrows. First row: results obtained with our template matching approach. Second row: results obtained with level set based method. Third row: results of color K-means-based method. Fourth row: results of seeded watershed method. Last row: hand-labeled results as the ground truth. First column: results of U2OS fluorescence image under uniform illumination. Second column: results of NIH3T3 fluorescence image under heterogeneous illumination.
From the comparison we can see that when nuclei samples are imaged clearly with distinct enough borders (for example, parts of image in Fig. 4(a) and Fig. 5(a)), all methods tested are able to achieve reasonable results. However, when noise or clutter is present, or when images are screened under uneven illumination (intensity inhomogeneities can be seen in Fig. 5(a) and Fig. 5(b)), most methods fail to segment nuclei very well (Fig. 5(d), (f) and (h)). Comparatively, our template matching approach still performs well on these images. The improvements are pointed out by white arrows in Fig. 5. In addition, our template matching approach can be naturally applied to higher dimensional data, as can other algorithms, such as RGB channel images (Fig. 6(a), (b) and (c)), and achieve what can be visually confirmed as better segmentation results over the existing methods tested. The improvements are pointed out by black arrows in Fig. 6. We note that in several locations (pointed out by arrows) our method performs better at segmenting cluttered nuclei. We also note that other methods often detect spurious locations as nuclei. Finally, we also note that our template matching approach is also much more likely (because it is constrained to do so) to produce contours that are more smooth and more realistic than the several other methods used for comparison.
B. Quantitative Evaluation
We used the synthetic dataset described above to calculate the average count produced by each method. We also studied the performance as a function of the clustering probability for this simulated dataset. The result is shown in Table I, where C.A. refers to ”Count Accuracy”, while O.P. refers to ”Overlap Probability” of data at each column. For the fluorescence microscopy data (U2OS and NIH3T3), we follow the same evaluation procedure as documented in [57], including: (1) Rand and Jaccard indices (RI and JI), which are used to measure the fraction of the pairs where the two clusterings agree (higher means better); (2) two spatially-aware evaluation metrics: Hausdorff metric and normalized sum of distances (NSD) (smaller means better); (3) counting errors: split, merged, added, and missing (smaller means better).
TABLE I.
Nuclei counting accuracy
| Algorithms | C. A. (O.P.: 0) | C. A. (O.P.: 0.15) | C. A. (O.P.: 0.30) | C. A. (O.P.: 0.45) | C. A. (O.P.: 0.60) |
|---|---|---|---|---|---|
| Template matching | 99.8% | 86.5% | 84.7% | 80.6% | 76.2% |
| Level Set [31] | 99.4% | 86.4% | 83.1% | 78.6% | 71.2% |
| K-means [59] | 100.0% | 87.7% | 84.3% | 80.2% | 72.5% |
| Seeded Watershed [21] | 99.9% | 91.0% | 88.1% | 84.7% | 78.4% |
Note that the bold face represents the best performance for each data (each column)
We also compare the results of the methods discussed above together with two other methods: active masks [63] and a merging based algorithm [5], as well as a manual delineation result. In Table II, for both U2OS and NIH3T3 data, we can see that although the Hausdorff metric values are quite high for our template matching approach, most segmentation metrics are comparable or better than many of the the existing algorithms. Our segmentation result also performs better than the manual delineation result (JI, Split for U2OS data, Split and Missing for NIH3T3 data) explored in [57]. More details pertaining to each method used in this comparison are available in [57]. The high Hausdorff metric can be explained by two reasons: (1) some bright noise regions are detected (especially in NIH3T3 dataset) and no morphological post-processing is used in our template matching approach and (2) we choose a relatively high threshold μ that discards some incomplete nuclei (small area) attached to the image border in some images. However, these incomplete nuclei are included in the manual delineated ground truth. The first reason may explain why the ”Added” error for NIH3T3 dataset is much higher than for U2OS dataset. In addition, the second reason may also explain why, excluding the active masks method [63], our algorithm misses more of the U2OS cells (last column of this table). On the other hand, for the NIH3T3 image data which contains intensity heterogeneities, the method we propose misses the fewest nuclei. We also note that, for the Rand and Jaccard indices, the normalized sum of distances (NSD) metric, and splitting errors, for both U2OS and NIH3T3 dataset, our results are similar to or better than the best results produced by other methods (excluding the manual delineation result).
TABLE II.
Quantitative Comparison of Nuclei Segmentation
| Algorithm (U2OS/NIH3T3) | RI | JI | Hausdorff | NSD(×10) | Split | Merged | Added | Missing |
|---|---|---|---|---|---|---|---|---|
| Manual | 95%/93% | 2.4/3.4 | 9.7/12.0 | 0.5/0.7 | 1.6/1.0 | 1.0/1.2 | 0.8/0.0 | 2.2/3.2 |
| Watershed (direct) [21] | 91%/78% | 1.9/1.6 | 34.9/19.3 | 3.6/3.7 | 13.8/2.9 | 1.2/2.4 | 2.0/11.6 | 3.0/5.5 |
| Active Masks [63] | 87%/72% | 2.1/2.0 | 148.3/98.0 | 5.5/5.0 | 10.5/1.9 | 2.1/1.5 | 0.4/3.9 | 10.8/31.1 |
| Merging Algorithm [5] | 96%/83% | 2.2/1.9 | 12.9/15.9 | 0.7/2.5 | 1.8/1.6 | 2.1/3.0 | 1.0/6.8 | 3.3/5.9 |
| Level Set [31] | 91%/81% | 2.39/2.30 | 96.6/122.8 | 0.85/5.0 | 1.1/1.4 | 0.35/1.4 | 2.75/4.2 | 0.85/8.2 |
| K-means [59] | 90%/78% | 2.36/2.35 | 94.6/100.6 | 1.05/6.15 | 1.56/0.45 | 0.3/0.9 | 2.6/2.75 | 1.6/17.4 |
| Template matching | 95%/91% | 2.50/2.72 | 77.8/131.2 | 0.64/2.65 | 0.58/0.51 | 1.45/2.49 | 0.9/3.7 | 3.48/2.8 |
Note that the bold face represents the best performance for each metric (each column)
Finally, we also studied how the number of nuclei used for training affects the performance of the proposed method. This was done for both U2OS and NIH3T3 dataset, by randomly selecting nuclei (of different sample sizes), and then implementing the method as described above. We found that the performance of several different quantitative metrics, such as the Rand index, NSD, etc., do not vary significantly when different amounts/types of nuclei samples are used (data omitted for brevity).
V. Summary and Discussion
We described a method for segmenting cell nuclei from several different modalities of images based on supervised learning and template matching. The method is suitable for a variety of imaging experiments given that it contains a training step that adapts the statistical model for the given type of data. In its simplest form, the method consists of building a statistical model for the texture and shape variations of the nuclei from the input of a user, and then segmenting arbitrary images by finding the instance in the model that best matches, in the sense of the NCC, local regions in the input images. We note that given an experimental setup, once the training operation is completed, the method is able to segment automatically any number of images from the same modality. We have demonstrated the application of the method to several types of images, and results showed that the method can achieve comparable, and often times better, performance compared with the existing specifically designed algorithms. Our main motivation was to design a method for segmenting nuclei from microscopy images of arbitrary types (scalar, color, fluorescence, different staining, etc.). To our knowledge ours is the first method to apply a template matching approach which includes texture and shape variations to accurately delineating nuclei from microscopy images. In addition, to our knowledge ours is the first method to utilize a supervised learning strategy to build such a statistical model, that includes texture and shape variations in multiple channels, for detecting nuclei from microscopy images.
In a practical sense, our method provides three main contributions. First, its overall performance is robust across different types of data with little tuning of parameters. We have demonstrated this here by applying the same exact software (with the only difference in each test being the value for μ) to a total of six different imaging modalities and showing the method performs as well or better than all other methods we were able to compare against. The performance was compared quantitatively and qualitatively, using both real and simulated data. Comparison results with a total of six alternative segmentation methods are shown here. Other, simpler, segmentation methods were also used for comparison, including several thresholding schemes followed by morphological operations. The results of these were not comparable to many of the methods shown here. Therefore, we have omitted them for brevity. Secondly, amongst the methods we have tested in this manuscript, we show that our method is the only method (besides manual segmentation) that is capable of handling significant intensity inhomogeneities. This is due to the fact that we utilize the NCC metric in the registration-based segmentation process. The NCC metric is independent of the overall intensity of the local region of the image being segmented. Finally, we also mention that, amongst all methods tried, the template matching method we described produced noticeably more smooth and accurate borders with fewer spurious contours. This can be seen, for example, by close observation of Fig. 6. The smoothness in the contours obtained by our method is primarily due to the fact that the statistical modeling we use includes only the main modes of variation in nuclear shape. These tend to be, topically, size, elongation, as well as bending (in addition to rotation). High order fluctuation in contours do occur in nuclei at times, but these do not occur as often as the ones already mentioned. We note that the method is still flexible enough to accurately segment nuclei that do not conform to these main modes of variation given the elastic matching procedure applied in the last step of the procedure.
We also note that our algorithm has several parameters including the percent of variance in PCA analysis, ε in the calculation of ”average” template, σ, s, κ in the non-rigid registration procedure, and μ in the approximate segmentation procedure. The algorithm is not unduly sensitive to these, as the same fixed parameters were utilized in all six experiments (datasets) used in this paper. The only parameter that was selected differently for each dataset was the detection threshold μ. When ground truth is available, we described a method to automatically choose the optimal μ for the given dataset. In addition, in our current implementation, we discard the top and bottom 1% (in size) of the generated templates, in an effort to reduce outlier detections. This percentage too could be made dataset dependent through a cross validation procedure, when the precise ground truth is present.
Finally, it is important to describe the computational cost of our template matching approach, which is also important in evaluating the performance of an algorithm. Our approach consists of a training stage and a testing stage, and is implemented in MATLAB 64 bit mode and tested on a PC laptop (CPU: Intel core i5 2.30GHz, memory: 8GB). The computational time for training a statistical model (560 simulated templates) from 20 nuclei samples (window size: 155 × 179), for example, is about 1.6 hours. Detecting and segmenting all cell nuclei (36 nuclei) from a fluorescence image (1030 × 1349) takes about 20 minutes (about half a minute per nucleus). We note however, that the computational time can often be significantly reduced by implementing the algorithm in a compiled language such as C, for example. In addition, we note that the computational time should be considered in context to alternative segmentation methods capable of producing results (albeit not as accurate) on similar datasets. The level set algorithm by Chan and Vese, which is used in a variety of other nuclear segmentation methods, takes even longer to compute on the same image (23 minutes) in our implementation (also in MATLAB). Finally, we note that the computational time of our algorithm can be decreased by utilizing a multi-scale framework. That is, instead of performing the filtering-based approach for detection in the original image space, we have also experimented with first reducing the size of the image (and templates) by two for the initial detection only (the remaining part of the method utilized the full resolution image). Thus we were able to reduce the total computation time for the same field of view to roughly 10.6 minutes. The accuracy of the final segmentation was not severely affected (data not shown for brevity). Future work will include improving the computational efficiency of this method by further investigation of multi-scale approaches, as well as faster optimization methods (e.g. conjugate gradient). Finally, we note again that the approach described above utilizes all color information contained in the training and test image. In cases where the nuclear stain color is known precisely, the approach can be easily modified to utilize only that color. In addition, many existing techniques for optimal color transformation [64] can also be combined with our proposed approach in the future for better performance.
Appendix
Here we describe the non-rigid registration algorithm used in both the training and segmentation steps outlined above. Let T(x) represent a target image (usually a raw image to be segmented) to which a source image (in our case the template) S(x) is to be deformed such that T(x) ≈ Su(x) = S(x − u(x)), with u(x) representing the warping function to be computed. We wish to maximize the square of the multichannel normalized cross correlation (NCC) between the two images:
| (4) |
where Nch is the number of channels, and ‖T‖2 = 〈T, T〉 = Σx T(x)T(x), where the sum is computed over the (fixed) image grid. We note that maximizing the squared NCC is equivalent to maximizing the NCC. We choose the squared NCC since it provides a more general framework, in which both positive and negative cross correlations can be optimized to the same effect. Equation (4) is maximized via steepest gradient ascent. The gradient of it is given by:
| (5) |
In practice we convolve ∇uΦ(u, x) with a radially symmetric Gaussian kernel of variance σ2 in order to regularize the problem. Optimization is conducted iteratively starting with u0(x) = 0, and uk+1(x) = uk + κGσ(x) * ∇uΦ(uk; x), where Gσ is the Gaussian kernel, * represents the digital convolution operation, and κ is a small step size. Optimization continues until the increase in the NCC value falls below a chosen threshold.
In addition, we perform the maximization above in a multi scale framework. That is, we utilize a sequence of images [T(x)]2 , [S(x)]2, [T(x)]1, [S(x)]1, and [T(x)]0 , [S(x)]0, where [T(x)]2 denotes the image T donwsampled by four (reduced to 1/8 of its size) after blurring, [T(x)]1 denotes the image T donwsampled by two (reduced to 1/4 of its size) after blurring, and [T(x)]0 denotes the original image being matched. The algorithm starts by obtaining an estimate for u(x) (using the gradient ascent algorithm described above) using images [T(x)]2, [S(x)]2. The estimate of the deformation map u is then used to initialized the same gradient ascent algorithm using images [T(x)]2 , [S(x)]2, and so on.
REFERENCES
- 1.Dow A, Shafer S, Kirkwood J, Mascari R, Waggoner A. Automatic multiparameter fluorescence imaging for determining lymphocyte phenotype and activation status in melanoma tissue sections. Cytometry. 1996;25:71–81. doi: 10.1002/(SICI)1097-0320(19960901)25:1<71::AID-CYTO8>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 2.Dice L. Measures of the amount of ecologic association between species. Ecology. 1945;26:297–302. [Google Scholar]
- 3.MATLAB. version 7.12.0 (R2011a) Natick, Massachusetts: The MathWorks Inc.; 2011. [Google Scholar]
- 4.Malpica N, Ortiz de Solorzano C, Vaquero J, Santos A, Vallcorba I, Garcia-Sagredo J, del Pozo F. Applying watershed algorithms to the segmentation of clustered nuclei. Cytometry. 1997;28:289–297. doi: 10.1002/(sici)1097-0320(19970801)28:4<289::aid-cyto3>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 5.Lin G, Adiga U, Olson K, Guzowski J, Barnes C, Roysam B. A hybrid 3d watershed algorithm incorporating gradient cues and object models for automatic segmentation of nuclei in confocal image stacks. Cytometry. 2003;56:23–36. doi: 10.1002/cyto.a.10079. [DOI] [PubMed] [Google Scholar]
- 6.Bengtsson E, Wahlby C, Lindblad J. Robust cell image segmentation methods. Pattern Recogn Image Analysis. 2004;14:157–167. [Google Scholar]
- 7.Chen X, Zhou X, Wong S. Automated segmentation, classification, and tracking of cancer cell nuclei in time-lapse microscopy. IEEE Trans Biomed Eng. 2006;53:762–766. doi: 10.1109/TBME.2006.870201. [DOI] [PubMed] [Google Scholar]
- 8.Yang X, Li H, Zhou X. Nuclei segmentation using marker-controlled watershed, tracking using mean-shift, and kalman filter in time-lapse microscopy. IEEE Trans Circuits Syst I. 2006;53:2405–2414. [Google Scholar]
- 9.Adiga U, Malladi R, Fernandez-Gonzalez R, de Solorzano C. High-throughput analysis of multispectral images of breast cancer tissue. IEEE Trans Image Process. 2006;15:2259–2268. doi: 10.1109/tip.2006.875205. [DOI] [PubMed] [Google Scholar]
- 10.Jung C, Kim C. Segmenting clustered nuclei using h-minima transform-based marker extraction and contour parameterization. IEEE Trans Biomed Eng. 2010;57:2600–2604. doi: 10.1109/TBME.2010.2060336. [DOI] [PubMed] [Google Scholar]
- 11.Nielsen B, Albregtsen F, Danielsen H. Automatic segmentation of cell nuclei in feulgen-stained histological sections of prostate cancer and quantitative evaluation of segmentation results. Cytometry. 2012;81:588–601. doi: 10.1002/cyto.a.22068. [DOI] [PubMed] [Google Scholar]
- 12.Lockett S, Sudar D, Thompson C, Pinkel D, Gray J. Efficient, interactive, and three-dimensional segmentation of cell nuclei in thick tissue sections. Cytometry. 1998;31:275–286. doi: 10.1002/(sici)1097-0320(19980401)31:4<275::aid-cyto7>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 13.Ortiz de Solorzano C, Malladi R, Lelievre S, Lockett S. Segmentation of nuclei and cells using membrane related protein markers. J Microsc. 2001;201:404–415. doi: 10.1046/j.1365-2818.2001.00854.x. [DOI] [PubMed] [Google Scholar]
- 14.Dufour A, Shinin V, Tajbakhsh S, Guillén-Aghion N, Olivo-Marin J, Zimmer C. Segmenting and tracking fluorescent cells in dynamic 3-d microscopy with coupled active surfaces. IEEE Trans Image Process. 2005;14:1396–1410. doi: 10.1109/tip.2005.852790. [DOI] [PubMed] [Google Scholar]
- 15.Lin G, Chawla M, Olson K, Barnes C, Guzowski J, Bjornsson C, Shain W, Roysam B. A multi-model approach to simultaneous segmentation and classification of heterogeneous populations of cell nuclei in 3d confocal microscope images. Cytometry. 2007;71:724–736. doi: 10.1002/cyto.a.20430. [DOI] [PubMed] [Google Scholar]
- 16.Hodneland E, Bukoreshtliev N, Eichler T, Tai X, Gurke S, Lundervold A, Gerdes H. A unified framework for automated 3-d segmentation of surfacestained living cells and a comprehensive segmentation evaluation. IEEE Trans Med Imag. 2009;28:720–738. doi: 10.1109/TMI.2008.2011522. [DOI] [PubMed] [Google Scholar]
- 17.Ridler T, Calvard S. Picture thresholding using an iterative selection method. IEEE Trans Syst, Man, Cybern. 1978;8:630–632. [Google Scholar]
- 18.Otsu N. A threshold selection method from gray-level histograms. IEEE Trans Syst, Man, Cybern. 1979;9:62–66. [Google Scholar]
- 19.Li G, Liu T, Tarokh A, Nie J, Guo L, Mara A, Holley S, Wong S. 3d cell nuclei segmentation based on gradient flow tracking. BMC cell biology. 2007;8:40–40. doi: 10.1186/1471-2121-8-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gonzalez R, Woods R. Digital image processing. 2nd ed. Prentice Hall Upper Saddle River; NJ: 2002. [Google Scholar]
- 21.Wählby C, Sintorn I, Erlandsson F, Borgefors G, Bengtsson E. Combining intensity, edge and shape information for 2d and 3d segmentation of cell nuclei in tissue sections. J Microsc. 2004;215:67–76. doi: 10.1111/j.0022-2720.2004.01338.x. [DOI] [PubMed] [Google Scholar]
- 22.Li F, Zhou X, Ma J, Wong S. Multiple nuclei tracking using integer programming for quantitative cancer cell cycle analysis. IEEE Trans Med Imag. 2010;29:96–105. doi: 10.1109/TMI.2009.2027813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Plissiti M, Nikou C, Charchanti A. Combining shape, texture and intensity features for cell nuclei extraction in pap smear images. Pattern Recog Lett. 2011;32:838–853. [Google Scholar]
- 24.Nandy K, Gudla P, Amundsen R, Meaburn K, Misteli T, Lockett S. Automatic segmentation and supervised learning-based selection of nuclei in cancer tissue images. Cytometry. 2012;81:743–754. doi: 10.1002/cyto.a.22097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leymarie F, Levine M. Tracking deformable objects in the plane using an active contour model. IEEE Trans Pattern Anal Mach Intell. 1993;15:617–634. [Google Scholar]
- 26.Bamford P, Lovell B. Unsupervised cell nucleus segmentation with active contours. Signal Pr. 1998;71:203–213. [Google Scholar]
- 27.Garrido A, Pérez de la Blanca N. Applying deformable templates for cell image segmentation. Pattern Recogn. 2000;33:821–832. [Google Scholar]
- 28.Yang L, Meer P, Foran D. Unsupervised segmentation based on robust estimation and color active contour models. IEEE Trans Inf Technol Biomed. 2005;9:475–486. doi: 10.1109/titb.2005.847515. [DOI] [PubMed] [Google Scholar]
- 29.Cheng J, Rajapakse J. Segmentation of clustered nuclei with shape markers and marking function. IEEE Trans Biomed Eng. 2009;56:741–748. doi: 10.1109/TBME.2008.2008635. [DOI] [PubMed] [Google Scholar]
- 30.Mukherjee D, Ray N, Acton S. Level set analysis for leukocyte detection and tracking. IEEE Trans Image Process. 2004;13:562–572. doi: 10.1109/tip.2003.819858. [DOI] [PubMed] [Google Scholar]
- 31.Chan T, Vese L. Active contours without edges. IEEE Trans Image Process. 2001;10:266–277. doi: 10.1109/83.902291. [DOI] [PubMed] [Google Scholar]
- 32.Yan P, Zhou X, Shah M, Wong S. Automatic segmentation of high-throughput rnai fluorescent cellular images. IEEE Trans Inf Technol Biomed. 2008;12:109–117. doi: 10.1109/TITB.2007.898006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu C, Prince J. Snakes, shapes, and gradient vector flow. IEEE Trans Image Process. 1998;7:359–369. doi: 10.1109/83.661186. [DOI] [PubMed] [Google Scholar]
- 34.Plissiti M, Nikou C. Overlapping cell nuclei segmentation using a spatially adaptive active physical model. IEEE Trans Image Process. 2012;21:4568–4580. doi: 10.1109/TIP.2012.2206041. [DOI] [PubMed] [Google Scholar]
- 35.Dzyubachyk O, van Cappellen W, Essers J, Niessen W, Meijering E. Advanced level-set-based cell tracking in time-lapse fluorescence microscopy. IEEE Trans Med Imag. 2010;29:852–867. doi: 10.1109/TMI.2009.2038693. [DOI] [PubMed] [Google Scholar]
- 36.Dufour A, Thibeaux R, Labruyere E, Guillen N, Olivo J. 3d active meshes: Fast discrete deformable models for cell tracking in 3d time-lapse microscopy. IEEE Trans Image Process. 2011;20:1925–1937. doi: 10.1109/TIP.2010.2099125. [DOI] [PubMed] [Google Scholar]
- 37.Quelhas P, Marcuzzo M, Mendonca AM, Campilho A. Cell nuclei and cytoplasm joint segmentation using the sliding band filter. IEEE Trans Med Imag. 2010;29:1463–1473. doi: 10.1109/TMI.2010.2048253. [DOI] [PubMed] [Google Scholar]
- 38.Esteves T, Quelhas P, Mendonça A, Campilho A. Gradient convergence filters and a phase congruency approach for in vivo cell nuclei detection. Mach Vision Appl. 2012;23:623–638. [Google Scholar]
- 39.Gudla P, Nandy K, Collins J, Meaburn K, Misteli T, Lockett S. A high-throughput system for segmenting nuclei using multiscale techniques. Cytometry. 2008;73:451–466. doi: 10.1002/cyto.a.20550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCullough D, Gudla P, Harris B, Collins J, Meaburn K, Nakaya M, Yamaguchi T, Misteli T, Lockett S. Segmentation of whole cells and cell nuclei from 3-d optical microscope images using dynamic programming. IEEE Trans Med Imag. 2008;27:723–734. doi: 10.1109/TMI.2007.913135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luck B, Carlson K, Bovik A, Richards-Kortum R. An image model and segmentation algorithm for reflectance confocal images of in vivo cervical tissue. IEEE Trans Image Process. 2005;14:1265–1276. doi: 10.1109/tip.2005.852460. [DOI] [PubMed] [Google Scholar]
- 42.Chen C, Li H, Zhou X, Wong S. Constraint factor graph cut–based active contour method for automated cellular image segmentation in rnai screening. J Microsc. 2008;230:177–191. doi: 10.1111/j.1365-2818.2008.01974.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ta V, Lézoray O, Elmoataz A, Schüpp S. Graph-based tools for microscopic cellular image segmentation. Pattern Recogn. 2009;42:1113–1125. [Google Scholar]
- 44.Al-Kofahi Y, Lassoued W, Lee W, Roysam B. Improved automatic detection and segmentation of cell nuclei in histopathology images. IEEE Trans Biomed Eng. 2010;57:841–852. doi: 10.1109/TBME.2009.2035102. [DOI] [PubMed] [Google Scholar]
- 45.Lee K, Street W. Model-based detection, segmentation, and classification for image analysis using on-line shape learning. Mach Vision Appl. 2003;13:222–233. [Google Scholar]
- 46.Lee K, Street W. An adaptive resource-allocating network for automated detection, segmentation, and classification of breast cancer nuclei topic area: image processing and recognition. IEEE Trans Neural Netw. 2003;14:680–687. doi: 10.1109/TNN.2003.810615. [DOI] [PubMed] [Google Scholar]
- 47.Fehr J, Ronneberger O, Kurz H, Burkhardt H. Self-learning segmentation and classification of cell-nuclei in 3d volumetric data using voxel-wise gray scale invariants. Pattern Recogn. 2005;3663:377–384. [Google Scholar]
- 48.Mao K, Zhao P, Tan P. Supervised learning-based cell image segmentation for p53 immunohistochemistry. IEEE Trans Biomed Eng. 2006;53:1153–1163. doi: 10.1109/TBME.2006.873538. [DOI] [PubMed] [Google Scholar]
- 49.Jung C, Kim C, Chae S, Oh S. Unsupervised segmentation of overlapped nuclei using bayesian classification. IEEE Trans Biomed Eng. 2010;57:2825–2832. doi: 10.1109/TBME.2010.2060486. [DOI] [PubMed] [Google Scholar]
- 50.Penny WD, Friston KJ, Ashburner JT, Kiebel SJ, Nichols TE, editors. Statistical Parametric Mapping: The Analysis of Functional Brain Images. Academic Press; 2006. [Google Scholar]
- 51.Rohde GK, Ribeiro AJS, Dahl KN, Murphy RF. Deformation-based nuclear morphometry: capturing nuclear shape variation in hela cells. Cytometry. 2008;73:341–350. doi: 10.1002/cyto.a.20506. [DOI] [PubMed] [Google Scholar]
- 52.Heitz G, Rohlfing T, Maurer Jr C. Statistical shape model generation using nonrigid deformation of a template mesh. Int P SPIE Med Imag. 2005;5747:1411–1421. [Google Scholar]
- 53.Shlens J. A tutorial on principal component analysis. Systems Neurobiology Laboratory, University of California at San Diego; 2005. [Google Scholar]
- 54.Li C, Xu C, Gui C, Fox M. Level set evolution without re-initialization: A new variational formulation. IEEE Int Proc Comput Vision Pattern Recog. 2005;1:430–436. [Google Scholar]
- 55.Lehmussola A, Ruusuvuori P, Selinummi J, Huttunen H, Yli-Harja O. Computational framework for simulating fluorescence microscope images with cell populations. IEEE Trans Med Imag. 2007;26:1010–1016. doi: 10.1109/TMI.2007.896925. [DOI] [PubMed] [Google Scholar]
- 56.Lehmussola A, Ruusuvuori P, Selinummi J, Rajala T, Yli-Harja O. Synthetic images of high-throughput microscopy for validation of image analysis methods. P IEEE. 2008;96:1348–1360. [Google Scholar]
- 57.Coelho L, Shariff A, Murphy R. Nuclear segmentation in microscope cell images: a hand-segmented dataset and comparison of algorithms. IEEE Int Sym Biomed Imag. 2009;1:518–521. doi: 10.1109/ISBI.2009.5193098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang W, Ozolek JA, Rohde GK. Detection and classification of thyroid follicular lesions based on nuclear structure from histopathology images. Cytometry. 2010;77:485–494. doi: 10.1002/cyto.a.20853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ravichandran K, Ananthi B. Color skin segmentation using k-means cluster. Int J Comput Appl Math. 2009;4:153–157. [Google Scholar]
- 60.Carpenter A, Jones T, Lamprecht M, Clarke C, Kang I, Friman O, Guertin D, Chang J, Lindquist R, Moffat J, et al. Cellprofiler: image analysis software for identifying and quantifying cell phenotypes. Genome biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pratt W. Digital Image Processing. John Wiley & Sons Inc.; 1991. [Google Scholar]
- 62.Raimondo F, Gavrielides M, Karayannopoulou G, Lyroudia K, Pitas I, Kostopoulos I. Automated evaluation of her-2/neu status in breast tissue from fluorescent in situ hybridization images. IEEE Trans Image Process. 2005;14:1288–1299. doi: 10.1109/tip.2005.852806. [DOI] [PubMed] [Google Scholar]
- 63.Srinivasa G, Fickus M, Guo Y, Linstedt A, Kovacevic J. Active mask segmentation of fluorescence microscope images. IEEE Trans Image Process. 2009;18:1817–1829. doi: 10.1109/TIP.2009.2021081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Deng J, Hu J, Wu J. A study of color space transformation method using nonuniform segmentation of color space source. Journal of Computers. 2011;6:288–296. [Google Scholar]