Synopsis
The serine proteases of the trypsin-like (S1) family play critical roles in many key biological processes including digestion, blood coagulation, and immunity. Recent studies have identified members of this family which contain amino- or carboxy-terminal domains that serve to tether the serine protease catalytic domain directly at the plasma membrane. These membrane anchored serine proteases are proving to be key components of the cell machinery for activation of precursor molecules in the pericellular microenvironment, playing vital functions in the maintenance of homeostasis. Substrates activated by membrane anchored serine proteases include peptide hormones, growth and differentiation factors, receptors, enzymes, adhesion molecules and viral coat proteins. In addition, new insights into our understanding of the physiological functions of these proteases and their involvement in human pathology have come from animal models and patient studies. This review discusses emerging evidence for the diversity of this fascinating group of membrane serine proteases as potent modifiers of the pericellular microenvironment through proteolytic processing of diverse substrates. We also discuss the functional consequences of the activities of these proteases on mammalian physiology and disease.
Keywords: serine protease, TTSP, membrane serine protease, pericellular proteolysis, substrates
INTRODUCTION
The immediate context of a cell, or the cell microenvironment, is critically modulated by proteolytic activities that regulate a wide variety of developmental, physiological and disease-associated processes. Pericellular proteolysis via cell surface-localized proteases is recognized as an important pathway by which cells interact with the cell microenvironment [1]. Many proteins in the pericellular microenvironment, including growth factors, cytokines, receptors, enzymes and cell adhesion molecules, are present as inactive precursors, that require activation by endo-proteolytic cleavage of peptide bonds [2]. This activation may occur through the action of proteases that are cell surface-localized via protease receptors, but may also be mediated by proteases directly anchored on the cell surface via membrane anchoring domains (Figure 1). This latter proteolytic mechanism enables spatial and directional substrate processing directly at the cell membrane and can be mediated by at least 3 classes of enzymes including MT-MMPs, ADAMs and membrane-anchored serine proteases. Recent biochemical, cell biological and genetic advances have revealed the importance of the last enzyme class, the membrane anchored serine proteases, to mammalian health and survival, and have also provided important new insights into the mechanisms regulating their roles in pericellular activation of precursor proteins.
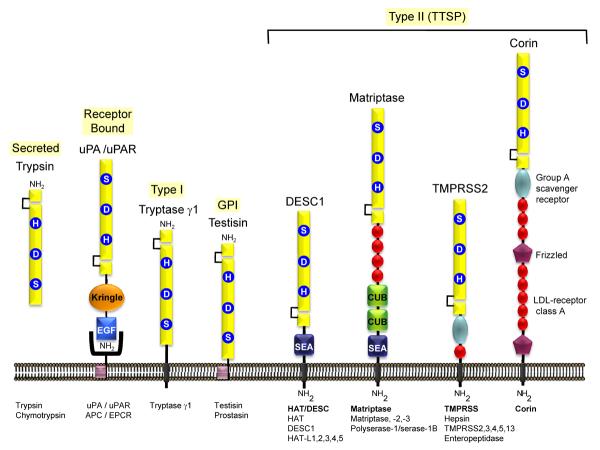
Figure 1. Membrane anchored serine proteases are linked directly to the plasma membrane.
Most of the well characterized S1A serine proteases, such as the prototype enzymes trypsin and chymotrypsin, are secreted enzymes, associated with extracellular proteolysis. Some of these enzymes (uPA, urinary-type plasminogen activator; APC, activated protein C) may participate in pericellular proteolysis by binding to specific cell surface receptors (uPAR, uPA receptor; EPCR, endothelial protein C receptor). Tryptase γ1 is the only known type I transmembrane serine protease. The human GPI anchored serine proteases are prostasin and testisin. The type II transmembrane serine proteases (TTSP) are the largest group of pericellular serine proteases and may be divided into four subfamilies [12,21]: (i) the Human Airway Trypsin-like protease/Differentially Expressed in Squamous cell Carcinoma (HAT/DESC) subfamily for which the stem regions are all composed of a single SEA domain, (ii) the Hepsin/Transmembrane Protease, Serine (TMPRSS) subfamily, each of which have a group A scavenger receptor domain (SRCR) in their stem region, preceded by a single LDL receptor class A-like (LDLRA) domain in TMPRSS2, 3, 4, and 13 or in enteropeptidase, an array of SEA, LDLRA, CUB, and MAM domains, (iii) the Matriptase subfamily, each containing a SEA domain, two CUB domains, and 3–4 LDLRA domains in their stem region. Polyserase-1 is unique comprising two active, and one catalytically inactive serine protease domains and a stem region containing an LDLRA domain, and (iv) the Corin subfamily, consisting of a single member, corin, which possesses a complex stem region composed of two frizzled domains, eight LDLRA domains, and one SRCR domain.
Membrane Anchored Serine Proteases
Serine proteases are defined by the nucleophilic serine (Ser) amino acid in the enzyme active site, which attacks the carbonyl moiety of the substrate peptide bond to form an acyl intermediate [3,4]. Completion of proteolysis is dependent on a catalytic triad of histidine (His), aspartate (Asp) and Ser amino acids, often referred to as a charge relay system. Over one third of all known proteolytic enzymes are serine proteases, and these have been grouped into families and clans based on common architectures and other functional attributes [4–6]. The serine proteases of clan PA, subfamily S1A, are a large group of enzymes that account for over 20% of the total known proteases. The most widely studied members of this group, which include trypsin, chymotrypsin, and thrombin, are produced as soluble, secreted proteins or are compartmentalized within intracellular granules and released in response to stress or inflammation (Figure 1). Some members, including the plasminogen activators, may be bound to specific cell surface receptors, enabling increased efficiency of plasminogen activation and subsequent plasmin-dependent proteolysis within the pericellular environment [7]. Only in the past decade has it been recognized that a unique sub-group of S1A serine proteases contain amino- or carboxy-terminal domains that serve to anchor the serine protease domains directly at the plasma membrane [8,9]. This review will focus on these enzymes as potent modifiers of the pericellular microenvironment through the diversity of their substrates and will discuss their contributions to mammalian physiology and disease. For historical perspectives, naming, classifications into subgroups, gene structure and chromosomal localization, and tissue- and cell-specific distribution of the membrane anchored serine proteases, the reader is referred to recent excellent reviews [8–12].
Membrane Localization
The membrane anchored serine proteases may be divided into sub-groups based on structural features (Figure 1). These proteases are tethered either by a carboxy-terminal transmembrane domain (Type I), through a glycosyl-phosphatidylinositol (GPI) linkage, or via an amino-terminal transmembrane domain with a cytoplasmic extension (Type II or TTSP) [8,9]. Type I serine proteases and the GPI anchored serine proteases are synthesized with a classical amino-terminal signal peptide and possess a hydrophobic domain at their carboxy-terminus. They are very similar in length, ranging from 310 to 370 amino acids. Tryptase γ1 is the only Type I transmembrane serine protease that has been identified to date [13,14]. The carboxy terminal domains of prostasin and testisin are post-transcriptionally modified with GPI-anchors [15–19]. Type II Transmembrane serine proteases or TTSPs [8] lack a classical signal peptide, but instead are synthesized with an N-terminal signal anchor. This signal anchor is not removed during synthesis, but serves as a transmembrane domain that positions the protease in the plasma membrane as an integral membrane protein with a cytoplasmic amino-terminus and an extracellular carboxy-terminus [8]. The first described TTSP was enteropeptidase, although its transmembrane molecular structure was only recognized when it was cloned [20]. The TTSPs share several structural features: an amino-terminal cytoplasmic domain of variable length (20 to 160 amino acids), a type II transmembrane sequence, a central stem region of variable length with modular structural domains, and an extracellular carboxy-terminal serine protease domain. A total of 19 TTSPs have been identified and divided into four subfamilies based on phylogenetic analyses of their serine protease domains and the domain structure of their extracellular stem regions [9,21] (Figure 1). These are the HAT/DESC, Hepsin/TMPRSS, Matriptase, and Corin subfamilies [10]. Polyserase-1 is an unusual member of the matriptase family that has a unique structure with three tandem serine protease domains and the ability to generate three independent serine proteases (i.e. Serase-1, -2 and -3) the third of which is predicted to be enzymatically inactive due to the lack of the catalytic Ser [22]. An alternatively spliced transcript encodes Serase-1B, a TTSP that has a SEA module in the stem region and a single protease domain with a mucin-like box at the C-terminus [23]. Differences in isoforms between humans and rodents have also been observed. For example, rodents express a secreted isoform of the human TTSP homologue HAT, that lacks the transmembrane and SEA domains [24]. In contrast, pancreasin/marapsin possesses a C-terminal GPI anchor in mice, which is absent in human and chimpanzee homologues where it is only found as a secreted enzyme [25].
The membrane anchoring domains of these enzymes contribute to their cellular trafficking and localization. Surface localization studies of the membrane anchored serine proteases for which antibodies are available show that they are localized at plasma membranes. An exception is the type I membrane anchored protease tryptase-γ, which is stored in the secretory granules of mast cells and does not reach the cell surface until degranulation [26]. The GPI anchored testisin and prostasin are found to be compartmentalized at plasma membranes within the dynamic microenvironment of specialized cholesterol-rich membrane microdomains or lipid rafts [17,18]. The cytoplasmic domains of the TTSPs, which are mosaic in nature and variable in length, are thought to contribute to the targeting of these enzymes to plasma membrane microdomains, based on the cellular sorting of other integral membrane proteins. Several of the TTSP cytoplasmic domains also contain consensus phosphorylation sites that may facilitate communication between the cell and the pericellular environment.
Membrane polarity and the epithelial to mesenchymal transition (EMT) impact the pericellular distribution of many of the membrane anchored serine proteases. In polarized epithelial monolayers, in which the plasma membranes are separated into apical and basolateral surfaces, some of these enzymes are targeted specifically to apical and others to the basolateral regions. In polarized epithelia, matriptase is present on basolateral membranes [27,28], and specifically localizes to the laterally located adherens junctions with E-cadherin in polarized epithelial cells [28,29]. Adherens junction formation and rearrangements of the actin cytoskeleton appear to be a prerequisite for sphingosine-1-phosphate (S1P)-induced matriptase localization at mammary epithelial cell-cell contacts [30]. Mutagenesis studies of the matriptase cytoplasmic domain has revealed that the juxtamembrane cytoplasmic amino acids (Lys45, Val47, and Arg50) are important for matriptase targeting to the basolateral surfaces of polarized Madin-Darby canine kidney (MDCK) epithelial monolayers [31]. The matriptase cytoplasmic domain has been shown to interact with actin-associated filamin [32], but whether this interaction is important for junctional localization in polarized epithelia has not been investigated. Hepsin also exhibits a junctional localization, colocalizing with desmosomal markers in OVCAR5 carcinoma cells, but not precisely with GAP junctions, adherens junctions, or tight junctions [33]. Desmosomal junctions appear to be specifically required for hepsin junctional localization, since hepsin localization at junctions does not occur in the related OVCAR5-TR cells, which lack the desomosomal junction protein desmoplakin, but retain normal adherens junctions. In contrast to these intercellular localizations, prostasin [16,34,35], TMPRSS2 [36,37], and enteropeptidase [38,39] localize to apical membranes in polarized epithelia. The mucin-like SEA domain of enteropeptidase directs its apical targeting in MDCK epithelial monolayers [38,39]. In some cases, loss of membrane polarity, such as occurs in tumor cells, is associated with mislocalization of the membrane serine proteases. TMPRSS2 for example localizes to the apical surfaces of renal tubular and airway epithelial cells [36] and along the apical membrane in normal prostate epithelium,[37,40,41], whereas in prostate carcinoma cells TMPRSS2 displays a more prominent cytoplasmic localization [41].
Ectodomain Shedding
Targeted release of extracellular domains from the cell surface, or ectodomain shedding, has been reported for several of the membrane anchored serine proteases, providing a means by which these enzymes may contribute to proteolytic activities in the extracellular space. Soluble forms of prostasin are found in human urine [42,43], and are elevated in hypertensive patients [44]. Aldosterone increases the expression and secretion of prostasin in a kidney cortical collecting duct cell line (M-1) and increases the urinary excretion of prostasin in rodents [45]. Prostasin is released apically from M-1 cells by cleavage of its GPI anchor via an endogenous GPI-specific phospholipase D1 [17]. Alternatively, there is evidence that prostasin may be shed via a tryptic-like proteolytic cleavage in its hydrophobic C-terminal domain [15,46]. The GPI anchored Testisin on the other hand, has not been found in a soluble shed form but may be released from cell membranes by addition of exogenous bacterial phosphatidylinositol-specific phospholipase C (PI-PLC)[18].
Among the TTSPs, naturally-occurring shed forms of matriptase, matriptase-2, enteropeptidase, and HAT have been detected in vivo. Shed matriptase was identified originally in complex with an inhibitor HAI-I in human milk [47] and additional shed forms have been reported in conditioned media of cultured breast epithelial lines, thymic epithelial lines and polarized intestinal epithelial cells [28,48–51]. Released enteropeptidase is found in mucosal fluid of the intestinal wall, where shedding is induced by hormones (secretin, cholecystokininpancreozymin) [52,53]. Soluble TMPRSS2 is a normal component of seminal fluid that accumulates within the lumen of the prostate, and in androgen-stimulated LNCaP cells, the TMPRSS2 ectodomain is released as a result of auto-activation [37,41]. Soluble forms of HAT are found in sputum from patients with chronic airway diseases [54].
The mechanisms by which the TTSPs are released are not yet fully understood and appear to vary depending on the specific protease. Porcine enteropeptidase, matriptase and matriptase-2 are shed as a result of proteolytic cleavage within their SEA domains [1,50,55], at sites that are distinct from the spontaneous processing known to occur in the SEA domain [50]. N-terminal sequencing of matriptase isofoms isolated from milk showed proteolytic cleavage at one of two sites either in the SEA domain (Lys188) or in the linker region between the SEA domain and CUB1 domain (Lys203), although the identity of the protease(s) that mediate these cleavages remains to be identified. The SEA domain may be critical for the shedding of these enzymes, since a mutation that disrupts the conformation of the SEA domain inhibits shedding of matriptase-2 in vitro [1]. Shedding of matriptase is thought to require its proteolytic activity [56], and can occur as a result of zymogen activation and HAI-I mediated inhibition [28,50]. Matriptase shedding may also be induced in response to PMA, causing local accumulation of the protease at the membrane, and interaction of the matriptase cytoplasmic domain with the cytoskeletal linker protein filamin, which was found to be essential for shedding [32]. PMA induced shedding could be inhibited by the MMP inhibitor GM6001 [32], implicating the involvement of a MMP in the shedding process. In contrast, Serase-1B is shed from HEK-293T cells in an inactive form following cleavage within the stem domain, which was not inhibited by GM6001 [23], suggesting specific mechanisms exist depending on the protease.
MODULATION OF MEMBRANE ANCHORED SERINE PROTEASE CATALYTIC ACTIVITY
The Serine Protease Catalytic Domain
The catalytic domain of the membrane anchored serine proteases are highly conserved and essential to the biological and physiological functions so far ascribed to these enzymes. Despite the high sequence homology of these domains, differences in amino acids that occupy key positions confer unique substrate specificities. All of the membrane anchored serine proteases share a serine protease tertiary domain structure with high sequence homology [9] that contains the catalytic triad of His, Asp and Ser amino acids necessary for S1 serine protease catalytic activity [8,9]. The catalytic domains are approximately 225–230 amino acids in size and are oriented in such a way that the domain is at the terminus of an extracellular region that is directly exposed to the pericellular environment. All of the membrane anchored serine proteases are synthesized as single-chain inactive pro-enzymes or zymogens, with an amino-terminal extension that acts as a propeptide, requiring proteolytic cleavage to generate the active enzyme. Activation results in a two-chain form with the pro- and catalytic domains linked by a disulfide bridge between two conserved cysteine residues [19]. Each active enzyme is defined by a binding pocket whose size, shape and charge are major determinants of substrate cleavage specificity. A specific nomenclature is used for the interaction of proteases with their substrates [57] where substrate amino acids (called P for peptide) are numbered P1 to Pn counting outward from the amino terminal side of the peptide bond that is cleaved during hydrolysis, and where those on the carboxy terminal side are numbered P1' through Pn'. Hydrolysis occurs between the P1 and P1' residues. The corresponding subsites of the enzymes are designated Sn through Sn'. All of the membrane serine proteases possess a conserved Asp amino acid at the bottom of the S1 substrate binding pocket in the activated serine protease domain which determines the preference for cleavage of substrates with a basic amino acid (Arg or Lys) in the P1 position (Table 1).
TABLE 1.
| Protease3 | Common Aliases | Gene Symbol | P4 | P3 | P2 | P1 | ↓ | P1' | P2' | P3' | P4' | Protease Source | Substrates | Ref |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GPI Anchored | ||||||||||||||
| Prostasin | PRSS8, CAP1 | PRSS8 | R | K | R | K | I | S | G | K | cDNA | ENAC(SCNN1γ) | [141] | |
| R/K | H/K/R | Y/W/R | R/K | X | X | X | A/S | SPD | Combinatorial peptide library. SPD | [85] | ||||
| Testisin | PRSS21, TESP, TEST1, ESP-1, tryptase 4 | PRSS21 | Boc | F | S | R | MCA | SPD | 4 peptide screen | [18] | ||||
| Type I | ||||||||||||||
| Tryptase γ1 | TMT/TPSG1, PRSS31 | TPSG1 | K | W/Y/F | R | AMC | ECD | Peptide library | [217] | |||||
| Type II | ||||||||||||||
| HAT | TMPRSS11D | TMPRSS11D | N | S | G | R | A | V | T | G | SPD | uPAR | [96] | |
| T | Y | S | R | S | R | Y | L | SPD | uPAR | [96] | ||||
| K | Q | T | R | G | L | F | G | cDNA | H3N2 HA protein, MDCK cells | [162] | ||||
| Boc | F | S | R | MCA | Purified human | peptide | [54] | |||||||
| DESC1 | TMPRSS11E | TMPRSS11E | R | R/Q | A/R | R | V | V | G | G | ECD | 18 peptide screen | [87] | |
| V | S | A | R | M | A | P | E | hECD, mSPD | hPAI-1 & RCL mutant | [86,87] | ||||
| F | T | F | R | S | A | R | L | mSPD | hPCI & RCL mutant | [86] | ||||
| HATL1 | TMPRSS11A, DESC3 | TMPRSS11A | S | L | L | R | S | T | S | Q | soluble protein | SARS S glycoprotein (triSpike) | [167] | |
| P | T | K | R | S | F | I | E | [167] | ||||||
| HATL3 | Neurobin, TMPRSS11C | Tmprss11c | V | G | R | pNA | SPD | 9 peptide screen | [102] | |||||
| R | G | H | K | V | A | G | G | cDNA | proHATL3 | [102] | ||||
| N | T | Y | R | S | R | K | Y | mSPD | FGF-2 fragment | [102] | ||||
| F | F | L | R | I | H | P | D | mSPD | FGF-2 fragment | [102] | ||||
| C | A | N | R | Y | L | A | M | mSPD | FGF-2 fragment | [102] | ||||
| V | R | E | K | S | D | P | H | mSPD | FGF-2 fragment | [102] | ||||
| P | H | I | K | L | Q | L | Q | mSPD | FGF-2 fragment | [102] | ||||
| Hepsin | TMPRSS1 | HPN | K/P | K/Q | T/N/L | R | ECD | Peptide library | [83] | |||||
| S | Q | L | R | L | Q | G | S | ECD | Laminin-332 β3 | [198] | ||||
| K | Q | L | R | V | V | N | ECD | proHGF | [121] | |||||
| P | Q | G | R | I | V | G | G | cDNA, ECD | FVII | [83,91] | ||||
| P | V | D | R | I | V | G | G | cDNA | m proHepsin | [101] | ||||
| P | R | F | K | I | I | G | G | ECD, cDNA | pro-uPA | [93] | ||||
| TMPRSS2 | epitheliasin | TMPRSS2 | R | Q | S | R | I | V | G | G | cDNA | proTMPRSS2 | [37] | |
| R | Q | S | R | F | V | L | G | cDNA | HMPV F protein Vero cells | [165] | ||||
| I | Q | S | R | G | L | F | G | cDNA | 1918 HA (South Carolina) 293T cells | [164] | ||||
| K | Q | T | R | G | L | F | G | cDNA | H3N2 HA protein MDCK cells | [162] | ||||
| S | K | G | R | S | L | I | G | purified | PAR2 | [112] | ||||
| Cbz | G | G | R | AMC | purified | Peptide assay | [112] | |||||||
| TMPRSS4 | MT-SP2, CAP2 | TMPRSS4 | I | Q | S | R | G | L | F | G | cDNA | 1918 HA (South Carolina) 293T cells | [164] | |
| R | K | R | R | E | A | G | S | cRNA | r γENAC xenopus oocytes | [145] | ||||
| L | N | Y | K | T | N | S | E | cRNA | r αENAC xenopus oocytes | [145] | ||||
| F | N | Y | R | T | I | E | E | cRNA | r βENAC xenopus oocytes | [145] | ||||
| L | N | Q | R | S | I | M | E | cRNA | r γENAC xenopus oocytes | [145] | ||||
| Boc | Q | A | R | AMC | ECD | Peptide assay | [187] | |||||||
| TMPRSS13 | MSPL | TMPRSS13 | Boc | Q/L | R/K | R | MCA | 19 peptide screen | [84] | |||||
| R/K | K | K | R | G | L | F | G | ECD | HA-based peptide | [161] | ||||
| TMPRSS5 | Spinesin | TMPRSS5 | Boc | Q | A | R | MCA | SPD | 10 peptide screen | [220] | ||||
| Enteropeptidase | PRSS7, Enterokinase | PRSS7 | D | D | D | K | I | V | G | G | purified porcine, bSPD | Trypsinogen 1 | [221,222] | |
| D/A | D | D | K/R | I | V | G | Purified bovine, porcine | 20 peptide screen | [223] | |||||
| Matriptase | MT-SP1, CAP3, TADG-15, PRSS14, ST14, SNC19, epithin (mouse) | ST14 | R | Q | AGL | R | V | V | G | G | SPD | 18 peptide screen | [87] | |
| R/K | X | S | R | A | SPD | Combinatorial peptide library | [81] | |||||||
| X | R/K | S | R | A | SPD | Combinatorial peptide library | [81] | |||||||
| R | Q | A | R | V | V | G | G | proMatriptase | [69] | |||||
| P/I | Q | A/P | R | I | T | G | G | KO, SPD | proProstasin (h/m) | [99,123] | ||||
| P | R | F | K | I | I | G | G | SPD | Pro-uPA | [81] | ||||
| S | K | G | R | S | L | I | G | SPD | hPAR2 xenopus oocytes | [81] | ||||
| K | Q | L | R | V | V | N | G | proHGF | [95,224,225] | |||||
| S | K | L | R | V | V | G | G | MSP-1 | [117] | |||||
| K | Q | S | R | K | F | V | P | SPD | ECD TRASK | [226] | ||||
| R | K | G | K | A | G | A | A | ECD, siRNA | IGFBP-rP1 | [227] | ||||
| R | R | V | R | K | E | D | E | VEGFR2 | [125] | |||||
| Matriptase-2 | TMPRSS6 | TMPRSS6 | boc | Q | A/G | R | AMC | GST-SPD | 5 peptide screen | [104] | ||||
| R | R | A | R | A/V | V | G | G | ECD | 18 peptide screen | [87] | ||||
| Matriptase-3 | TMPRSS7 | TMPRSS7 | A | A | p | R | pNA | mSPD | 7 peptide screen | [203] | ||||
| F | V | R | pNA | mSPD | 7 peptide screen | [203] | ||||||||
| V | S | A | R | M | A | P | E | mSPD | hPAI-1 & RCL mutant | [203] | ||||
| Polyserase-1/Serase-1B | TMPRSS9 | TMPRSS9 | Boc | Q | G | R | AMC | SPD | 4 peptide screen | [22] | ||||
| Boc | Q/L | S/T/A | R | MCA | ECD | 14 peptide screen | [23] | |||||||
| P | R | F | K | I | I | G | G | ECD | Pro-uPA | [23] | ||||
| Corin | LRP4, ATC2, TMPRSS10 | CORIN | Y | A | P | R | S | L | R | R | cDNA, ECD | proANP, HEK293 cells | [127,128] | |
| R | A | P | R | S | P | K | M | ECD | proBNP | [79] | ||||
| pyro | E | F | K | pNA | 5 peptide screen | [228] | ||||||||
Basic amino acids are highlighted in blue, acidic amino acids in red, and aromatic amino acids in green.
Abbreviations: AMC, 7-amino 4-methyl coumarin; Boc, t-butoxycarbonyl; ECD, extracellular domain; GST, Glutathione-S-Transferase; h, human; m, mouse; MCA, 4-methylcoumaryl-7-amide; pNA, p-nitroanilide; r, rat; RCL, reactive center loop; SPD, serine protease domain;
Substrate cleavage specificities for DESC4/HATL2, HATL4/TMPRSS11F, HATL5/TMPRSS11B, TMPRSS3, are not known.
Detailed comparative analyses of the amino acid sequences [8,9,21] combined with structural analyses of the serine protease catalytic domains of matriptase [58], hepsin [59], enteropeptidase [60], DESC1 [61] and prostasin [62–64] have revealed insights into the unique catalytic activities of the membrane serine proteases. In most but not all cases, crystal structures were obtained using truncated catalytic domains in complex with small molecular weight inhibitors. These structures reveal that each catalytic domain consists of two adjacent, six-stranded β-barrel domains that are connected by three trans-domain segments. The catalytic triad amino acids are located along the junction between the two barrels, whereas the active site cleft runs perpendicular to this junction. The specificity of these enzymes is determined by both the nature of the substrate-binding subsites (e.g. S4–S2'), and the differing loop regions that surround the active site. These loops can influence enzyme specificity both by `shaping' extended binding subsites and by providing secondary exosites for substrate or inhibitor binding. For example, the loop bordering the S2' pocket, varies markedly in length among the membrane serine proteases, and in matriptase, this loop is uniquely distorted due to a four-residue insertion, which leads to thrombin-like shielding of the prime site [58,61,65]. In contrast, DESC1 carries a one amino acid deletion in this loop compared with other TTSPs, resulting in a narrowing of the prime site [61]. The 99-loop which limits the space for the P2 and P4 residues of the substrate peptide at the top of the active site cleft, exhibits pronounced sequence heterogeneity among the membrane anchored serine proteases and contributes significantly to their respective specificities. The remarkably specific enteropeptidase is dependent on the Lys99 within its 99-loop for interaction with the P2-P4 aspartyl residues to facilitate cleavage of its physiological substrate, trypsinogen [60]. Ion interactions may also contribute to substrate specificities. The S1 subsite loop of prostasin directly binds the divalent cation Ca2+ and exhibits a large degree of conformational variation, able to move to block or to expose the S1 subsite [62]. This highlights a possible mechanism of regulation in pericellular spaces where ionic concentrations are variable.
In contrast to these crystal structures of truncated catalytic domains, the structure of the complete hepsin extracellular domain including the serine protease catalytic domain and the N-terminal scavenger receptor cysteine-rich (SRCR) domain contained within the stem region was determined [59]. Interestingly, structural analyses revealed that the SRCR domain was rigidly bound to the back of the catalytic serine protease domain [59]. Comparison of the surface charges of hepsin, DESC-1, matriptase and enteropeptidase suggest that this may be a common feature, with the stem region domains functioning as interaction partners with the exposed surfaces of the serine protease catalytic domains [61], potentially contributing to appropriate orientation of the active site cleft on the cell surface.
Extracellular Domains Affecting Serine Protease Catalytic Activities
There is now considerable evidence that the stem domains of the TTSPs not only contribute to surface orientation, but also modulate proteolysis by contributing to TTSP activation, the binding of substrates, and interactions with other proteins. N-terminal processing within a SEA domain, present in 11 members of this family, can regulate TTSP activation [66,67]. For matriptase, and potentially other SEA domain containing TTSPs, this domain undergoes spontaneous proteolytic processing at a conserved glycine residue [55,66], due to conformation driven non-enzymatic hydrolysis of the peptide bond [68]. After hydrolysis the matriptase extracellular domain remains non-covalently associated and this processing is required for matriptase zymogen activation. In addition to the contribution of the SEA domain, mutations that disrupt the domain structure of the LDL-receptor like (LDLRA) domain and deletions of either of the CUB domains of matriptase also inhibit zymogen activation [66,69]. Also, the extracellular stem domains of corin (Frizzled domain 1 and LDLRA1-4), and unidentified domains of the enteropeptidase stem region are important for recognition of macromolecular substrates, but are not required for cleavage of peptide substrates [70,71]. Furthermore, post-translational modifications such as N-linked glycosylation also affect biological activity of the membrane anchored serine proteases. For example, N-linked glycosylation at sites in the CUB1 domain and serine protease catalytic domain of matriptase are required for zymogen activation [66], and glycosylation in the serine protease domain of corin is required for both cell surface expression and zymogen activation [72,73]. The theme emerging from these studies is that for each TTSP its stem domain contributes uniquely to its cellular localization, activation and inhibition and that it is likely that further studies on these domains will identify important new binding partners that regulate TTSP mediated pericellular proteolysis.
Human Disease Mutations Affecting Membrane Serine Protease Functions
It is significant that mutations in several TTSP genes that impact protease activity have been causally linked to human diseases. For the interested reader a comprehensive list of TTSP mutations can be found in [12]. In summary, many of these mutations result in TTSPs with missing or truncated protease domains while others are point mutations predicted to influence enzyme expression or activity. Several of these point mutations have been investigated by cell transfection studies or through in vitro expression of the recombinant mutant proteases. These experiments have provided valuable insights into the functions of these TTSPs as well as mechanisms that regulate their proteolytic activities (summarized in Table 2). For example, missense mutations identified in the serine protease domains of matriptase, TMPRSS3 and TMPRSS5 result in catalytically inactive proteases, leading to human skin dysfunction in the case of matriptase and deafness in the case of TMPRSS3 and TMPRSS5. In contrast, missense mutations in the SEA or CUB-1 domains of matriptase-2 cause defective zymogen activation resulting in iron-refractory iron deficiency anemia (IRIDA) [1,74,75]. In addition, mutations in the LDLRA domains of this TTSP appear to be important for trafficking to the cell surface and subsequent zymogen activation [75]. Similarly a 30 amino acid deletion mutation in the LDLRA1/2 domains, and a point mutation in the CUB-1 domain of matriptase-2 are also associated with IRIDA [76], again showing the importance of non-serine protease domains for TTSP function. Further highlighting this point, a point mutation within the Frizzled-2 domain of corin, which is associated with systemic hypertension, also results in impaired zymogen activation [77–79]. Several other mutations in TMPRSS3 that are linked to congenital deafness are found in the LDLRA and SRCR domains and result in proteases that fail to become activated [80]. The impact of so many disease causing mutations in non-catalytic domains emphasizes the importance of these to the functional activities of the membrane anchored serine proteases.
Table 2.
Disease related human point mutations: effect on protease function
| Protease | Mutation | Location | Phenotype | Effect on function in vitro | Refs |
|---|---|---|---|---|---|
|
| |||||
| Type II | |||||
|
| |||||
| TMPRSS3 | R216L | SPD- zymogen activation site | Non-syndromic autosomal recessive deafness | Mutant does not undergo proteolytic processing representative of auto-activation. | [178] |
| P404L | SPD | Mutant does not undergo proteolytic processing representative of auto-activation. Catalytically inactive. | [80,177] | ||
| D103G,R109W | LDLRA | These mutants do not undergo proteolytic processing representative of auto-activation and are unable to activate ENaC substrate compared to WT. | [80] | ||
| C194F | SRCR | ||||
| W215C | SPD | ||||
|
| |||||
| TMPRSS5 | A317S | SPD | Nonsyndromic deafness | Serine protease domain is catalytically inactive | [40] |
|
| |||||
| Matriptase | G827R | SPD | Impaired epidermal barrier function, icthyosis, hypertrichosis, follicular atrophoderma, corneal opacity and photophobia | Mutation is in active site binding cleft. Serine protease domain is catalytically inactive and protease unable to auto-activate. | [103,170,172] |
|
| |||||
| Matriptase-2 | A118D | SEA | IRIDA | Mutant protein reaches cell surface and is shed, but is unable to become activated. | [1] |
| D521N | LDLRA | IRIDA | LDLRA mutants cannot not reach cell surface, are retained in Golgi and cannot activate. CUB mutant has reduced activation. Mutants are still able to interact with hemojuvelin and partially repressed hepcidin expression. | [75] | |
| E552K | LDLRA | ||||
| G442R | CUB-2 | ||||
|
| |||||
| Corin | T555I/Q568P | Frizzled-like domain-2 | Decreased processing of pro-atrial natriurectic peptide resulting in hypertension and cardiac hypertrophy. | Mutant reaches cell surface, but possesses impaired catalytic activity due to impaired zymogen activation. | [78,79] |
MEMBRANE SERINE PROTEASE SUBSTRATES
Substrate recognition is controlled in part by interactions between the active site of a protease and the amino acids spanning the cleavage site of its substrate. The following sections summarize the approaches that have been used to identify the peptide substrate specificities for membrane anchored serine proteases and experimentally identified protein cleavage sites (Table 1). Since additional factors will influence the activity and specificity of the membrane serine proteases for their substrates in vivo, we discuss the experimental evidence for cleavage of macromolecular substrates in various pericellular microenvironments. While it is clear from animal models and studies of human disease that the membrane anchored serine proteases are important components of specific physiological processes (Tables 2 and 3) much work remains to identify the relevant in vivo substrates for many of the membrane serine proteases.
Table 3.
Physiological functions defined in murine models
| Protease | Mouse model | Phenotype | Function | Refs |
|---|---|---|---|---|
| GPI Anchored | ||||
| Prostasin | Epidermal specific ablation: Prss8l0XΔ/K14-Cre mice | Death within 60 h due to severe dehydration, increased epidermal permeability, defective epidermal differentiation, defective profillagrin processing, absence of occludin | Maintenance of epidermal barrier integrity | [229] |
| Testisin | Prss21 Null mice | PRSS21 deficient spermatozoa show reduced oocyte fertilization, decreased motility, angulated and curled tails, fragile necks, increased susceptibility to decapitation and failed to mount an effective swelling response upon release into hypotonic media. These defects reflect aberrant maturation during passage through the epididymis. | Directs epididymal sperm cell maturation and sperm fertilizing ability | [180,181] |
| Type II | ||||
| Hepsin | Hpn Null mice | Deafness, abnormal cochlear development, thyroid hormone deficiency | Unknown | [40,179] |
| Over-expression in prostate epithelium: Hpn-probasin mice | Promotion of SV40 large T antigen induced prostate carcinoma and metastasis, basement membrane disorganization, reduced expression of laminin-322 | Tumor promotion | [230] | |
| TMPRSS2 | Tmprss2 Null mice | No phenotype | [231] | |
| Matriptase | St14 Null mice | Death within 48 h due to severe dehydration, increased epidermal permeability, defective epidermal differentiation, defective hair follicle development, increased thymocyte apoptosis. Defective pro-fillagrin processing in skin. Decreased activation of prostasin in epidermis. | Maintenance of epidermal barrier integrity | [99,168,169] |
| St14 hypomorphic mice | Epidermal hyperproliferative ichthyosis, impaired desquamation, sparse hair, tooth defects. Defective profillagrin processing. | Maintenance of epidermal barrier integrity | [172,232] | |
| Inducible ablation: B-actin-Cre-ERtm+/o;St14LoxP/−mice | Tamoxifen inducible ablation in adult mice. Dramatic weight loss, loss of fur, scaling of skin, edema of intestines, dissolution of colonic tissue architecture, inflammation of orofacial surfaces. Increased intestinal and colonic crypt cell proliferation. Abnormal expression/localization of tight junction markers in small intestine epithelium. Increased intestinal and epidermal permeability. | Global role in maintenance of epithelial homeostasis | [174] | |
| GI tract ablation: Villin-Cre+/o;St14LoxP/− mice | Gastrointestinal tract (GI) specific ablation. Death several weeks after birth, persistent diarrhea, low body weight, enlarged colon, increased proliferation of colonic crypt cells and loss of mucin production. Gross disruption of colonic tissue architecture, edema and inflammation. | Maintenance of intestinal epithelial barrier | [174] | |
| Salivary duct epithelium ablation: MMTV-Cre+/o;St14Loxp/− mice | Salivary ductal epithelium specific ablation. Loss of saliva production. No obvious abnormalities. | Maintenace of salivary epithelial function | [174] | |
| Expression in murine basal keratinocytes: St14 -cytokeratin 5 mice | Spontaneous squamous cell carcinoma, increased susceptibility to chemical carcinogenesis. | Tumor promotion | [185] | |
| Matriptase-2 | Tmprss6 Null mice | Alopecia and a severe iron deficiency anemia, upregulation of hepcidin expression, reduced ferroportin expression in enterocytes. | Regulation of hepcidin and iron homeostasis | [157,233] |
| Mask mice: Tmprss6msk/msk mice, chemically induced mutant phenotype | Lack expression of SPD of matriptase-2 due to splicing defect. Loss of body hair and microcytic anemia, reduced iron absorption caused by high levels of hepicidin. | Regulation of hepcidin and iron homeostasis | [153,233] | |
| Mask mice × hemojuvelin null mice: Tmprss6msk/msk; Hfe2tm1Nca/tm1Nca | Disruption of both matriptase-2 and the membrane receptor hemojuvelin, which is known to stimulate hepcidin expression in response to elevated iron levels. Results in low hepcidin expression, high serum and liver iron, suggesting hemojuvelin is a major matriptase-2 substrate that regulates hepcidin expression. | Regulation of membrane hemojuvelin to suppress hepcidin expression | [158] | |
| Corin | Corin Null mice | Defective pro-atrial natriuretic peptide processing leading to hypertension, cardiac hypertrophy, increased body weight, abnormal hair pigmentation. | Regulation of hypertension | [131,182] |
| Abnormal hair pigmentation `dirty blonde' phenotype | Role in specifying hair colour by suppression of the Agouti pathway | [182] | ||
Murine models are not yet available forTMPRSS3-5, TMPRSS13, HAT, HATL3-5, DESC1-4, Polyserase-1/Serase-1B.
Peptide Substrate Specificity
Biochemical and proteomics approaches have proven of considerable value as a first step for identification of substrate cleavage sites and for determining the amino acids that are preferred both side and proximal to these sites. In several cases, cell-based or synthetic libraries of peptide substrates have been applied to quantitatively determine the substrate specificity of the catalytic domains of the membrane anchored serine proteases [81–83]. Table 1 lists the known cleavage sequences of the membrane serine proteases against peptide substrates, macromolecular substrates and inhibitors. Of note, whereas the substrate specificities for hepsin, matriptase, prostasin, and tryptase γ1 have been extensively characterized using combinatorial peptide libraries, only limited or no information is available for other membrane anchored serine proteases. The listed cleavage sequences have been determined from in vitro studies using purified recombinant truncated serine protease domains (SPDs), recombinant extracellular TTSP domains including the stem region (ECD), or in some cases analyzed using cell based systems. The reader should be mindful that the pathophysiological relevance of substrate data obtained using SPDs alone may be imperfect, since truncated catalytic domains may not reveal secondary intramolecular interactions, protein and co-factor binding, or structural conformations that exist in vivo that can both positively and negatively impact substrate specificities.
These analyses demonstrate that all of the membrane anchored serine proteases show preference for cleavage of substrates with a basic amino acid (Arg or Lys) in the P1 position, although each enzyme displays different peptide substrate specificity, and recognition of diverse macromolecular substrates. Interestingly, several membrane anchored serine proteases, namely matriptase, prostasin, MSPL, tryptase γ1, and hepsin, prefer multibasic residues in the P4-P1 positions. In fact, the multibasic peptide specificity profiles of these membrane anchored serine proteases are similar to those of the proprotein convertases. Indeed, the furin inhibitor decanoyl-RVKR-CMK is a potent inhibitor of TMPRSS13 (Ki 2.9 nM) [84]. Then again, the furin selective mutant of α1-proteinase inhibitor (α1AT-PDX) is not an effective inhibitor of TMPRSS13 [84], or of prostasin [85]. As a group it could be generalized that the TTSPs select against acidic residues in the P4-P1 positions with the exception of enteropeptidase, which possesses a highly selective preference for acidic amino acids in P4-P2. In the P2 position of protein and peptide substrates, both prostasin and tryptase γ1 have a strong preference for aromatic residues, whereas TMPRSS2 and HAT have a strong preference for small polar amino acids or Gly at this position. Other TTSPs such as hepsin, HATL3, and TMPRSS4 can accommodate a variety of amino acid classes (e.g. aromatic, basic, hydrophobic) at the P2 position.
Overall the peptide substrate specificities of matriptase, hepsin, enteropeptidase, and prostasin determined by combinatorial peptide library screens strongly agree with the cleavage sites identified in protein substrates that have been characterized by amino acid sequencing. However, it is also interesting that in some instances, the substrate specificity predicted by active site geometry of the catalytic domain or by peptide screens, does not match with the preferred cleavage sites of biological substrates or inhibitors. For example, analysis of the crystal structure of DESC1 suggests that its substrate specificity differs markedly from that of other TTSPs, requiring large hydrophobic residues in P4 ∕ P3, small residues in P2, Arg or Lys in P1 and hydrophobic residues in P1' and P3' [61,86]. However, kinetic analyses show that DESC1 cleaves peptide substrates with Arg in P4, comparable to matriptase [87]. Additionally, the crystal structure of hepsin [83] predicts that large bulky side chains can be accommodated in the S2 subsite but a Gly residue would not provide strong interactions with the S2 pocket. The ability of hepsin to activate pro-uPA and pro-HGF, containing Phe and Leu respectively in the P2 position, are consistent with this observation. Conversely, the hepsin substrate FVII contains a Gly in the P2 position, suggesting other interactions may be compensating for the less favorable binding into the hepsin S2 pocket.
Zymogen Activation – Membrane Serine Proteases Activating other Serine Proteases
The involvement of serine proteases in zymogen cascades has been long recognized [2]. Zymogen cascades involve at least two consecutive proteolytic reactions with one protease zymogen being the substrate of another previously activated protease. This strategy confers the advantage that a signal can be specifically and irreversibly amplified each time a downstream zymogen is activated, unleashing a burst of proteolytic potential [88]. A range of key biological processes rely on protease zymogen activation, including blood coagulation, fibrinolysis, complement reaction, hormone production, metamorphosis, fertilization, and digestion [2]. While all of the membrane anchored serine proteases are synthesized as protease zymogens, for the most part, the specific endogenous proteases that catalyze the activation of their zymogen forms in vivo, are not known. In vitro, mild treatment with trypsin will convert recombinant serine protease pro-domains into active enzymes [51]; however, it is likely that specific proteases catalyze these cleavage reactions in vivo.
The substrate specificities of several of the membrane serine proteases are compatible with the cleavage and activation of serine protease zymogens, and a few have been demonstrated to participate in zymogen activation (Figure 2). For example, enteropeptidase produced on the brush border of the small intestine, activates pancreatic trypsinogen, which in turn, catalyzes the conversion of other pancreatic zymogens, chymotrypsinogen, proelastase, prolipases and procarboxypeptidases, to their active forms during digestion [89]. This processing is critical for normal digestion and nutritional well-being [90]. A further example is hepsin, which intersects the blood coagulation cascade by activating Factor VII to Factor VIIa, which in turn, is capable of initiating a coagulation pathway on the cell surface that leads to thrombin formation [91].
Figure 2. Membrane anchored serine proteases participate in zymogen cascades.
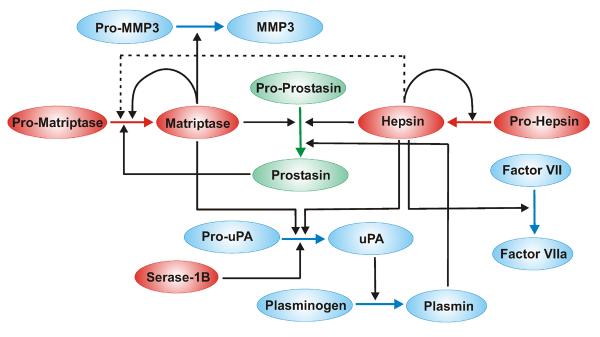
Pathways in which membrane anchored serine proteases have been shown to activate, or be activated by, serine proteases in vitro and in vivo. Proteases are color coded according to membrane localization sequences, (red) TTSPs, (green) GPI-anchored proteases, (blue) other secreted proteases. Lines indicate activation cleavages and loops indicate auto-activation. The dotted line indicates that hepsin is a weak activator of the matriptase zymogen. Membrane anchored serine proteases intersect the coagulation cascade (Factor VII activation), fibrinolysis (pro-uPA activation) and metalloproteinase pathways (pro-MMP-3 activation). Not shown is the activation of trypsinogen by enteropeptidase.
Several of the TTSPs intersect the plasminogen cascade, a key cascade critical for fibrinolysis, cell migration, extracellular matrix remodeling, and MMP activation [92], through the activation of pro-uPA to its catalytically active form. Pro-uPA is relatively promiscuous with respect to its activation in contrast to most serine protease zymogens, and a wide range of both serine proteases and enzymes from other proteolytic classes can catalyze pro-uPA activation in in vitro assays including the catalytic domains of hepsin, matriptase, and Serase-1B [23,81,93,94] (Figure 3A). Serase-1B mediates efficient conversion of pro-uPA into active uPA, which is negatively regulated by glycosaminoglycans [23]. Active uPA processes plasminogen on the cell surface with high efficiency when bound to its cellular receptor, uPAR. Kinetic analyses demonstrate that although matriptase is a relatively poor activator of pro-uPA in solution, it is an effective activator of uPAR-bound pro-uPA and initiates pericellular plasminogen activation on the monocyte cell surface [95]. The uPAR is also a substrate for HAT [96]. Recombinant HAT processes the full-length uPAR (D1D2D3) into a truncated (D2D3) species and shedding of the major ligand-binding D1 domain [97]. Recombinant truncated matriptase has also been shown to activate stromelysin (MMP-3) in several cell systems [98], implicating TTSPs in protease cascade networks associated with extracellular matrix degradation in tumor cell microenvironments.
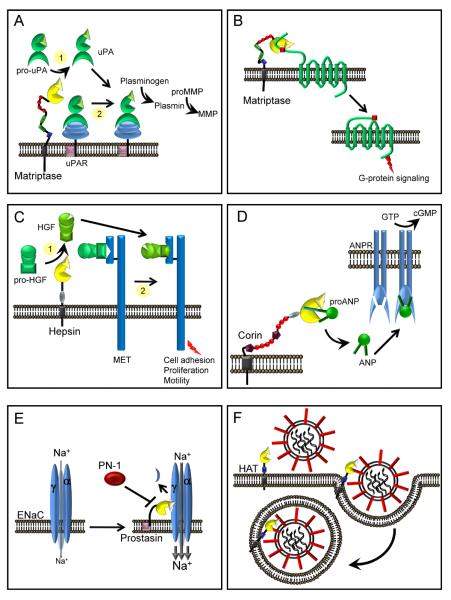
Figure 3. Proteolytic pathways associated with membrane anchored serine proteases in pericellular microenvironments.
(A) Fibrinolysis. The membrane anchored serine proteases, matriptase, hepsin and Serase-1B may participate in the initiation of the plasminogen activation cascade via conversion of pro-uPA to active uPA. Matriptase is a relatively poor activator of pro-uPA in solution (1), whereas it is an effective activator of uPAR-bound pro-uPA and initiates pericellular plasminogen activation on the monocyte cell surface (2). (B) Inflammation. Matriptase and possibly other membrane serine proteases, may participate in the activation of PARs triggering G-protein signaling pathways. (C) Growth factor activation. Hepsin and matriptase may participate in the activation of pro-HGF to initiate signaling via the HGF receptor, MET, to modulate cell adhesion, proliferation and cell motility. Whether these enzymes target soluble pro-HGF (1) or receptor bound pro-HGF (2) is not known. (D) Natriuretic peptides. Corin processes pro-ANP to an active form that bind ANP receptors (ANPR) in the kidney and vasculature, increasing guanylate cyclase activity to regulate blood pressure and blood volume. (E) Sodium homeostasis. The open probability of the ENaC is increased by prostasin and TMPRSS4, resulting in increased cellular uptake of sodium (Na+) which in turn can regulate homeostasis of extracellular fluid volume, blood pressure and Na+ reabsorption. Processing of the α or γ subunits of ENaC is predicted to release an inhibitory fragment that leads to full activation of the channel. The serpin protease nexin-1 (PN-1, serpinE2) can inhibit prostasin mediated activation of ENaC. (F) Viral Pathogenicity. Several TTSPs, including HAT, TMPRSS2, 4, and 13 can process precursor viral proteins that facilitate virus entry and fusion into host cells.
In addition to activating other serine protease zymogens, several of the membrane anchored serine proteases are able to activate other membrane anchored serine proteases or to self-activate (Figure 2). The prostasin zymogen was shown to be converted to its active enzyme form by matriptase in vitro and in vivo in skin, where the matriptase/prostasin axis is a physiological activator of terminal epidermal differentiation [99]. Intriguingly, in in vitro assays the zymogen form of matriptase can also be converted to the active form by the addition of soluble active prostasin or hepsin, suggesting that, in certain cellular contexts, prostasin might function both upstream and downstream of matriptase, thus constituting a positive-feedback loop similar to the amplification cascades associated with coagulation [100]. Self or autocatalytic activation of some of the purified TTSPs in vitro has been reported. In these cases, the specificity preference of the TTSP is compatible with the activation motif of the TTSP, and inactivation of the active site catalytic triad residues of the biochemically purified catalytic domain can be shown to prevent autocatalytic activation [10,66,101,102]. In vitro autoactivation has been reported for the TTSPs matriptase [66,81,103], matriptase-2 [104], hepsin [105], TMPRSS2 [37], TMPRSS3 [80], TMPRSS4 [106] and TMPRSS11C [102], although the ability to undergo an intermolecular activating cleavage requires more rigorous demonstration for several of these enzymes.
Cleavage of Protease Activated Receptors (PARs)
G-protein coupled signaling receptors on the cell surface known as protease activated receptors or PARs are activated by proteolytic cleavage [107,108], and several membrane anchored serine proteases have been investigated either as direct or indirect activators of PAR signaling. PARs are cleaved at specific amino acid sequences within the extracellular N-terminus of the receptor, exposing a new N-terminus that serves as a tethered ligand domain, initiating varying intracellular signal transduction pathways [109]. The four members of the PAR family (PAR1–4) may be processed by different activating or inactivating proteases. PAR1, PAR3, and PAR4 are activated by thrombin and PAR3 has been reported to be a co-factor for the activation of PAR4 by thrombin. PAR2 is a target of several trypsin-like serine proteases including trypsin, mast cell tryptase and kallikrein 4 [110,111], and is activated by the soluble catalytic domains of matriptase [81,100] and TMPRSS2 [112], resulting in protease-triggered signaling, indicative of G-protein-coupled-receptor activation (Figure 3B). Matriptase was also found to induce release of proinflammatory cytokines including IL-6 and IL-8 in endothelial cells through activation of PAR2 [113]. Truncated HAT has been shown to induce amphiregulin release through PAR2-mediated ERK activation and TNFα-converting enzyme (TACE) activity in airway epithelial cells [114]. While it has not been shown that prostasin proteolytically activates PAR2, the expression of prostasin was found to play a modulatory role in PAR2 signaling in prostate epithelial cells [115], and more recent studies show that soluble recombinant prostasin can activate PAR2 indirectly through a matriptase-dependent mechanism [100]. Whether membrane-tethered serine proteases are direct physiological activators of PARs juxtapositioned on plasma membranes in the pericellular environment will be important to investigate. Indeed, recent studies suggest this may be the case, since full length matriptase co-expressed in a lung fibroblast cell line in the presence of HAI-1 was shown to activate PAR2 after activation of the matriptase zymogen by soluble matriptase or prostasin [100].
Growth Factor Processing
Proteolytic cleavage governs many multi-component signaling events including growth factor activation and availability. In vitro matriptase is a very efficient activator of pro-hepatocyte growth factor/scatter factor (HGF) (Figure 3C) and pro-macrophage stimulating-1 (MSP/MST-1), growth factors that serve as ligands for two receptor tyrosine kinases associated with epithelial cell motility, migration and proliferation, MET/HGF receptor and RON/MST1 receptor, respectively, [116–118]. Interestingly, HGF and MSP are inactive serine protease analogues, with substitutions in two of the three residues of the catalytic triad, but require processing by conversion of a single chain precursor to the bioactive two-chain form [94]. At the cellular level, expression of matriptase by transfection activates proHGF on the surface of colon and prostate carcinoma cell lines [119]. Matriptase activation of proHGF may further contribute to mammary gland morphogenesis [120], since RNAi mediated suppression of murine matriptase could inhibit induction of complex ductal structures induced by pro-HGF in a three-dimensional mammary epithelial model. MSP was shown to be activated by matriptase expressed on the surface of primary peritoneal macrophages, where activation of pro-MSP could be inhibited by the presence of HAI-I or an anti-matriptase antibody [117]. Similarly, the hepsin serine protease domain was also shown to activate proHGF into its bioactive form in vitro, with an activity comparable to HGF-activator (HGFA) [83,121]. The colocalization of hepsin with HGF at the cell junctions of ovarian tumor cells [33] is supportive of a potential role for hepsin in proHGF activation, however the physiological relevance of this activity remains to be demonstrated. Fibroblast growth factor-2 (FGF-2) is another growth factor reported to be cleaved into several fragments by the catalytic domain of HAT-L3 [102].
In addition to activation of growth factors, several of the membrane anchored serine proteases have been reported to modulate the epidermal growth factor receptor (EGFR). In PC3 prostate cells, prostasin expression induced down-regulation of EGFR at the mRNA level, an activity that required protease catalytic activity [122]. At a post-translational level, both matriptase and hepsin were found to cleave the extracellular domain of the EGFR in co-transfection studies [123,124]. Although they cleaved the extracellular domain of the EGFR at different sites, only matriptase processing resulted in a constituively active membrane anchored form of the receptor that was capable of initiating intracellular signalling. The physiological relevance of EGFR modifications by membrane anchored serine proteases warrants further investigation. Interestingly, matriptase has also been shown to cleave the VEGFR-2 in vitro at a site that is predicted to induce its shedding from the cell surface, and addition of recombinant matriptase to HUVECs inhibited the signaling response to VEGF [125].
Activation of Natriuretic Peptides
Processing of pro-atrial natriuretic peptide (pro-ANP) to the cardiac hormone ANP is catalyzed by corin (Figure 3D). ANP is synthesized as a cell associated inactive pro-hormone that requires proteolytic processing for release and activity. In vitro studies show that corin, which is highly expressed in cardiac tissue [126], is able to convert the cell-associated inactive pro-ANP into its active form on the surface of cardiomyocytes [127,128]. ANP plays a key role in regulating blood pressure by regulating systemic salt and water balance [129,130]. Consistent with an in vivo role in this conversion, corin deficient mice show defects in the conversion of pro-ANP to ANP, and development of spontaneous hypertension and cardiac hypertrophy that is exacerbated by a high salt diet [131]. The hypertensive phenotype of corin deficient mice mimics that of ANP deficient mice [132], and emphasizes that pro-ANP is an important in vivo corin substrate. Several corin single nucleotide polymorphisms identified in African American populations and associated with high blood pressure and cardiac hypertrophy [77,78], result in production of corin mutants with impaired zymogen activation, leading to reduced zymogen activity [79]. Corin has also been shown to activate the pro-form of brain or B-type natriuretic peptide (BNP) [79,127], a marker of congestive heart failure [133]. Mice deficient in BNP develop cardiac fibrosis, indicating that it is involved in ventricular remodeling rather than blood pressure regulation [134]. Accordingly, a recent study has shown that patients with the corin I555(P568) allele exhibit impaired BNP processing and are at increased risk for adverse outcomes associated with heart failure [135].
Epithelial Sodium Channel Activation
The epithelial sodium channel (ENaC) regulates sodium and water flux across high-resistance epithelia, such as the airway, bladder, kidney, colon and skin. During biosynthesis, ENaCα and ENaCγ subunits are proteolytically processed by the proprotein convertase furin, but are maximally activated by further processing by cellular proteases [34]. Several membrane anchored serine proteases referred to as channel-activating proteases (CAPs) activate ENaC when coexpressed in heterologous expression systems [34,136,137]. Prostasin activation of ENaC has been the most well characterized (Figure 3E). Studies in Xenopus oocytes showed that co-expression of prostasin and ENaC subunits increases the amiloride-sensitive sodium current [136,138] and expression of prostasin in mammalian kidney epithelial cells [139] and in a cystic fibrosis epithelial line [140] induces increased ENaC activity. At a molecular level, prostasin cleaves the ENaCγ subunit in the extracellular loop at a site (K186) distal to the furin cleavage site which releases a 43 amino acid inhibitory peptide shown to inhibit ENaC activity, resulting in increased open probability of the channel and full activation [141,142]. Recent studies also show that low levels of plasmin found in urine from patients with nephritic syndrome can activate ENaCs via activation of membrane anchored prostasin [143]. On the other hand, there is also evidence that noncatalytic protease mechanisms may be involved in ENaC activation [106,144]. For example, in the Xenopus oocyte expression system, catalytically inactive prostasin mutants were able to fully activate ENaC, with no evidence of prostasin-mediated intramolecular cleavage, although cell surface expression via the prostasin GPI anchor was essential for this activity. In addition to prostasin, TMPRSS3, TMPRSS4 and matriptase are able to activate ENaC when co-expressed in Xenopus oocytes [80,136]. Missense TMPRSS3 mutations associated with deafness fail to activate ENaC [80]. Direct cleavage of ENaC subunits by matriptase and TMPRSS3 has not been demonstrated, but TMPRSS4 was recently shown to cleave in the inhibitory ENaCγ subunit at a site distinct from that of prostasin [145]. Interestingly, expression of TMPRSS2 has been shown to decrease sodium currents and ENaC protein levels [146], suggesting that the membrane anchored proteases may both positively and negatively regulate ENaC function. The channel-activating activity of prostasin is likely to be pathophysiologically important. Mice lacking the prostasin gene in lung epithelium demonstrate impaired ENaC-mediated alveolar fluid clearance [147]. In addition, recent findings suggest that increased ENaC activation in the airway of cystic fibrosis patients may be linked to excessive prostasin activity on the surface of lung epithelium [148,149]. Increased ENaC activation [150] is associated with hypertension and a recent independent study identified prostasin as a candidate gene implicated in the development of hypertension in youths [151]. Hypertensive patients exhibit increased levels of urinary prostasin [42], and in a rat model of hypertension, treatment with a synthetic inhibitor of prostasin was able to significantly reduce blood pressure and increase the urinary Na/K ratio [152].
Regulation of Iron Homeostasis
Recently, matriptase-2 has emerged as an essential regulator of systemic iron levels. Mutations in this TTSP are causal for suppression of hepcidin [1] an hepatic peptide hormone that controls body iron levels by limiting iron egress from duodenal enterocytes and macrophages to serum transferin through its degradation of the iron transporter, ferroportin [74,153]. Insight into the involvement of matriptase-2 in iron regulation has come from the identification of frame-shift, splice junction and missense mutations in the encoding gene, TMPRSS6, which cause IRIDA in humans [154–156]. These observations are supported by studies in mice. A chemically induced Tmprss6 mutation, eliminating the splice acceptor site of intron 14 and resulting in loss of the matriptase-2 serine protease domain (Mask mice), results in anemia, low plasma iron and depleted iron stores [156]. In addition, levels of transcription from the hepcidin encoding gene hamp were ~10 fold higher in homozygous Tmprss6 mutant mice following iron deprivation than in wildtype mice. Investigation of Tmprss6 knockout mice revealed a severe iron deficiency anemic phenotype accompanied by a marked up-regulation of hamp transcription, reduced ferroportin expression in the basolateral membrane of enterocytes and accumulation of iron in these cells [157]. Further studies on the mechanism of matriptase-2 activity suggest that this protease may proteolytically process hemojuvelin [153,158], a GPI-anchored protein that is a key activator of Hamp transcription [159].
Viral Proteins and Promotion of Viral Pathogenicity
Increasing evidence suggests that influenza and other respiratory viruses exploit membrane anchored proteases to promote viral spread [160]. For example, the TTSPs TMPRSS2, HAT, TMPRSS4, TMPRSS13 and TMPRSS11A support activation and replication of human influenza viruses. Influenza entry and membrane fusion with host cells is mediated by hemagglutinin (HA), which is synthesized as a precursor protein HA0 that must be cleaved by a host cellular protease into the covalently linked subunits HA1 and HA2. Mutational evolution of the HA proteolytic cleavage site is a determinant of viral pathogenicity and impacts multiplication of the virus in vivo [161]. Highly pathogenic avian influenza viruses possess multibasic HA cleavage motifs, e.g. R-X-K/R-R and K-X-K/R-R, which are processed intracellularly and on the cell surface by the ubiquitously expressed, calcium-dependent subtilisin-like proteases, furin and proprotein convertases 5/6, enabling rapid systemic spread of viral infection. Recently TMPRSS13 was shown to recognize and process HA multibasic motifs in a calcium-independent manner. TMPRSS13 was able to process HA of a highly virulent recombinant strain of pathogenic influenza virus in vitro, that is poorly susceptible to furin, and to facilitate cell fusion [161]. In contrast to these high virulence strains, HA of low pathogenic viruses usually contain monobasic cleavage sites, and cleavage by trypsin is required to support efficient replication of these viruses in cell culture. HAT and TMPRSS2 were found to process monobasic cleavage of HA and to support viral replication in vitro [162,163] (Figure 3F). Interestingly, TMPRSS2 and TMPRSS4 but not matriptase-3 were able to cleave and activate HA of the genetically reconstituted 1918 influenza virus [164]. The presence of these enzymes in human respiratory epithelium may suggest a pathogenic role in promoting viral spread in human airways [164]. TTSPs have also been associated with the propagation of other viruses. TMPRSS2 supports multicycle replication of human metapneumovirus in vitro by proteolytic cleavage of the fusion protein F [165]. In addition, hepsin interacts with the × protein of human hepatitis B virus, although proteolytic activity was not reported [166]. Recently, the spike (S) protein of the severe acute respiratory syndrome coronavirus (SARS-CoV) was found to be susceptible to cleavage by TMPRSS11A, implicating protease-mediated enhancement of SARS-CoV entry in a fashion dependent on previous receptor binding [167].
Maintenance of Epithelial Barriers
Several membrane serine proteases are found to be essential for epithelial barrier integrity, although the biochemical basis for this activity is not yet defined. A role for matriptase in epithelial barrier function, was first identified by examining the phenotype of mice null for the St14 gene encoding matriptase [168]. Although these mice develop to term, they die shortly after birth due to severe dehydration caused by defective epidermal barrier function [168,169]. In humans, mutations in the matriptase gene underlie autosomal recessive ichthyosis with hypotrichosis syndrome (ARIH), a rare human skin disease characterized by congenital ichthyosis associated with abnormal hair [170] [171]. One characterized missense mutation, G827R, affects access to the binding/catalytic cleft of the enzyme, thereby preventing autocatalysis of the zymogen form resulting in a catalytically inactive protease [103,172]. Affected individuals and matriptase deficient mice show defective epidermal differentiation processes, including altered extrusion of extracellular lipids that form the lipid lamellae, defective shedding of the stratum corneum, and defective processing of the epidermal structural polyprotein pro-fillagrin [169,172,173].
The recent generation of murine models of inducible matriptase ablation in the whole animal, and tissue specific ablation in the GI tract and salivary epithelium, has revealed that matriptase is essential for the maintenance of epithelial barrier integrity [12,174]. Ablation of matriptase in intestinal and salivary gland epithelium disrupts organ function, increases epithelial permeability, and in tissues colonized by microbial flora such as the large intestine, leads to major architectural breakdown of tissue integrity [174]. Matriptase hypomorphic mice, which have a 100-fold reduction in intestinal matriptase mRNA levels, display a 35% reduction in intestinal transepithelial electrical resistance (TEER), but retain histologically normal intestinal epithelium. Reduced barrier integrity is mediated at least in part by an inability to regulate the expression and localization of the permeability associated, `leaky' tight junction protein claudin-2 both in vivo and in Caco-2 polarized epithelial monolayers [29]. The direct molecular target(s) of matriptase during the maintenance of epithelial integrity in multiple diverse epithelial tissues however, remains to be established.
Prostasin has also been implicated in the regulation of epithelial barriers and tight junction function [17]. Over-expression of prostasin in M-1 epithelial monolayers decreases transepithelial electrical resistance (TEER), a sensitive measure of epithelial barrier function, and enhances paracellular permeability to inulin. Interestingly, over-expression of a form of prostasin in which the GPI-anchor was replaced with the transmembrane domain of tryptase-γ1 resulted in enhanced TEER reflecting increased M-1 barrier integrity. It is interesting to note that these effects on TEER appeared unrelated to ENaC activity, suggesting that prostasin may target an alternate substrate involved in regulation of epithelial barrier permeability, although the nature of the physiological matriptase-prostasin pathway substrate(s) mediating this process remains elusive.
Roles in Hearing
The TTSPs TMPRSS3, TMPRSS5 and hepsin are implicated in hearing and their dysregulation is associated with deafness [175], although the biochemical mechanisms by which these enzymes are involved in normal hearing have not been elucidated. The TMPRSS3 gene was first identified as a gene located on a chromosomal locus associated with human familial congenital deafness [176]. Mutations in the TMPRSS3 and TMPRSS5 genes that contribute to hearing loss render the respective proteases inactive [80,177,178]. Hepsin null mice are also associated with a markedly increased hearing threshold with abnormal cochlear structures, reduced myelin protein expression in the auditory nerve by histological analysis, and reduced level of the large conductance voltage- and Ca2+-activated K+ channel [40,179]. Hearing deficiency in hepsin null mice has been proposed to be due to a defect in thyroid hormone metabolism since these mice demonstrate low plasma levels of thyroxine, a hormone required for inner ear development [179].
Membrane Anchored Serine Proteases in Development
Several of the membrane anchored serine proteases are associated with mammalian development. For example, testisin is required for epididymal sperm maturation and fertilizing ability, since mouse sperm lacking testisin display several functional abnormalities [180,181]. Ablation of the gene encoding corin in mice revealed a function in the regulation of hair shaft pigmentation, acting downstream of agouti gene expression as a suppressor of the agouti pathway [182]. Inhibition of matriptase is essential for formation of the placental labyrinth in mice, since genetic ablation of the matriptase inhibitor HAI-1, leads to deregulation of matriptase activity and disruption of the epithelial integrity of chorionic trophoblasts [183]. In a series of elegant studies, it was shown that genetic ablation of matriptase activity in HAI-1-deficient embryos restores trophoblast integrity and enables placental labyrinth formation and development to term [183]. The specific substrates targeted by these enzymes are important areas for future study.
Dysregulation in Cancer
Many membrane anchored serine proteases are aberrantly expressed in human cancers and have been suggested as biomarkers indicative of disease state. Evidence comes from expression studies in human cancers and in some cases, from mouse models of carcinogenesis. As several recent reviews have extensively discussed membrane anchored serine proteases in cancer [9,10], here we focus on the key findings in this area.
Several of the membrane anchored serine proteases are overexpressed by tumor cells and are implicated in promoting tumor development and progression [184,185]. A modest increase in matriptase activity on the epidermis of a transgenic mouse model was sufficient for the induction of spontaneous squamous cell carcinomas [185,186]. In addition, matriptase overexpression in a several cell lines leads to more aggressive tumors [98,185,186]. Hepsin overexpression also induces tumor growth in mice, that was shown to be dependent on hepsin catalytic activity [33]. The expression of TMPRSS4 in human tumor cells was shown to promote invasion, migration, and metastasis by inducing the loss of E-cadherin-mediated cell-cell adhesion and facilitating the epithelial-to-mesenchymal-transition (EMT), a step in the metastatic process [187]. Interestingly, chromosomal rearrangements in the human TMPRSS2 gene (21q22.3) result in gene fusions of the 5'-untranslated region of TMPRSS2 with ETS transcription factor family members (e.g. TMPRSS2-ERG), where ERG expression is driven by the TMPRSS2 gene promoter, and associated with high rate of prostate cancer recurrence, metastasis, and death after prostatectomy [188,189].
In contrast to promotion of tumor progression, several membrane anchored serine proteases are present in normal tissues and found to be down-regulated or absent in corresponding malignant carcinomas. When expressed in prostate and breast tumor cell lines, matriptase-2 acts as a tumor suppressor, leading to the suppression of tumor migration and metastasis, and its expression has been correlated with a better disease prognosis in breast cancer patients [190,191]. DESC1 is expressed in normal epithelial cells of prostate, skin, testes, head and neck, whereas it is down-regulated in squamous cell carcinomas [192]. Prostasin expression is frequently down-regulated in gastric cancer [193] and loss of prostasin expression in human bladder transitional cell carcinoma cell lines has been shown to be associated with the EMT [115]. Testisin expression was reported to be lost in testicular germ cell tumors [19], and it has been shown that epigenetic gene silencing contributes to the down-regulation of both prostasin and testisin expression in tumors [193–197].
The truncated serine protease domains of the membrane anchored serine proteases are found to degrade in vitro extracellular matrix protein components fibronectin, fibrinogen, and denatured type-1 collagen, gelatin and casein to varying extents [48,104,186,198]. In addition, hepsin expressed in prostate cancer cells was shown to cleave basement membrane laminin-322, leading to increased tumor cell migration in vitro [198]. The physiological significance of these activities is not known, although they may well have particular relevance for cancer and inflammation, where protease dysregulation may result in encounters with substrates not normally seen.
INHIBITION OF MEMBRANE SERINE PROTEASE ACTIVITIES
The importance of regulated membrane anchored serine protease expression to normal homeostasis and the frequent associations between these enzymes and cancer and other diseases, indicates that these enzymes must be tightly regulated in normal settings. This regulation is partly mediated by endogenous protease inhibitors, including members of the serpin family of serine protease inhibitors and Kunitz-domain containing inhibitors (Table 4). In addition, a number of small molecule inhibitors that specifically modulate the activities of these enzymes have recently been reported.
TABLE 4.
Membrane Serine Protease Inhibitors1
| Endogenous Protein Inhibitors | Other Protein Inhibitors | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Proteases2 | Kunitz | Serpin3 | Assay | Refs | SLPI4 | SBTI5 | Aprotinin | Leupeptin | Ecotin | Refs | ||||||||
| HAI-1 | HAI-2 | A1 | A3 | A5 | C1 | D1 | E1 | E2 | F2 | |||||||||
| GPI Anchored | ||||||||||||||||||
| Prostasin | + | + | − | − | + | CA, GE | [85,205,234] | − | + | + | [15] [85] | |||||||
| Testisin | + | + | Upub6 | |||||||||||||||
| Type I | ||||||||||||||||||
| Tryptase γ1 | + | CA, GE | [26] | +/− | + | + | [26] | |||||||||||
| Type II | ||||||||||||||||||
| HAT | − | + | + | + | [54] | |||||||||||||
| DESC1 | − | − | + | + | − | + | + | CA,GE | [86,87,235] | + | + | [86,235] | ||||||
| HATL3 | + | + | [102] | |||||||||||||||
| Hepsin | + | + | +/− | + | + | + | CA | [87,91,121] | +/− | + | [91,214] | |||||||
| TMPRSS13 | − | + | CA | [84] | + | + | − | [84] | ||||||||||
| TMPRSS5 | − | [220] | ||||||||||||||||
| Enteropeptidase | + | + | + | [236,237] | ||||||||||||||
| Matriptase | + | + | + | + | + | + | CA, GE | [87,202] [238,239] | + | + | + | + | [240] [48] | |||||
| Matriptase-2 | − | + | + | + | CA | [87] | + | + | [104] | |||||||||
| Matriptase-3 | − | + | + | − | + | + | CA,GE | [203] | + | + | [203] | |||||||
| Polyserase-1 | + | + | CA | [22] | ||||||||||||||
| Serase-1B | − | +/− | + | + | CA, GE | [23] | + | +/− | + | + | [23] | |||||||
| Corin | +/− | + | + | [228] | ||||||||||||||
Inhibitory activity as assayed by catalytic assay (CA) or complex formation by gel electrophoresis (GE). (−) Inhibitory activity less than 10%; (+/−) Inhibitory activity less than 50%; (+) Inhibitory activity greater than 50%.
Inhibitory profiles for HATL1/DESC3/TMPRSS11A, HATL2/DESC4, HATL4/TMPRSS11F, HATL5/TMPRSS11B, TMPRSS2, TMPRSS3, and TMPRSS4, are not known.
Abbreviations: Serpins: A1, α1-proteinase inhibitor/α1-antitrypsin; A3, α1-antichymotrypsin; A5, protein C inhibitor (PCI); C1, antithrombin III; D1, heparin cofactor II; E1, plasminogen activator inhibitor-1 (PAI-1); E2, protease nexin 1; F2, α2-antiplasmin.
SLPI, secretory leukocyte proteinase inhibitor
SBTI, soybean Kunitz trypsin inhibitor
published data, S. Netzel-Arnett
Serpins
Serpins interact with proteases via a `pseudo substrate' exposed binding loop known as the reactive center loop (RCL), that upon cleavage, covalently traps the protease by undergoing an irreversible conformational rearrangement [199,200]. The suicide nature of serpin inhibition is an effective strategy for regulation of proteolytic activity by direct removal of unwanted proteases via membrane bound endocytic receptors [201]. Matriptase has been isolated from human milk in inhibitory complexes with the secreted serpins, antithrombin III (serpinC1), α1-proteinase inhibitor (serpinA1) and α2-antiplasmin (serpinF2) [202]. In addition, inhibitory complexes form in vitro between mouse DESC1 and the serpins PAI-1 (serpinE1) and protein C inhibitor (PCI, serpinA5) [86], matriptase-3 and PAI-1, PCI, α1-proteinase inhibitor, α2-antiplasmin and antithrombin III [203], and serase-1B and PAI-1 and α2-antiplasmin [23].
The selectivity of a serpin towards a particular serine protease is not always readily evident from the sequence of the protease sensitive RCL sequence because of the flexible nature of the reactive segment of the serpin and possible exosite binding interactions. For example, the RCL sequences of the serpins PAI-1 and PCI do not match the optimal docking sequence of DESC1 predicted by the crystal structure [61], but these serpins form effective inhibitory complexes with DESC1 [86]. Additionally, α1-proteinase inhibitor (serpinA1) possesses a Met in the P1 substrate position, yet can inhibit trypsin-like proteases including tryptase γ1, matriptase, hepsin and polyserase-1 [22,26,87]. Conversely, serpinA1 is a poor inhibitor of the matriptase catalytic domain in peptide assays, however endogenous complexes of matriptase-serpinA1 have been isolated from breast milk [87,202]. While most studies have been performed with recombinant soluble forms of the enzymes and inhibitors, anti-thrombin III (serpinC1) and protease nexin I (serpinE2) have also been shown to inhibit cell surface associated activities of hepsin [91] and prostasin [204](Figure 3E), respectively. In general, inhibition studies have provided information on the ability of the membrane anchored serine proteases to form SDS-stable protease-inhibitor complexes and have determined IC50 values in peptide based assays, however, detailed kinetic analyses of these protease-serpin interactions is lacking for the most part.
Kunitz-type Inhibitors
Kunitz-type inhibitors are plasma inhibitors that contain domains that inhibit protease activity by forming very tight, but reversible complexes with target proteases. In contrast to serpins, reversible Kunitz inhibitors may simply compete with physiological substrates (such as ECM components), to reduce the availability of the protease. The transmembrane Kunitz-type inhibitor HAI-1/SPINT1 was originally isolated as an inhibitor of HGFA [67,83,205], but has also been implicated in the expression, zymogen activation and inhibition of matriptase [206,207,207,208], hepsin [121] and prostasin [67,83,121,205]. Indeed, HAI-1/Spint1 deficient mice display severe growth retardation and early postnatal lethality as well as skin and hair defects [209,210], phenotypes that are rescued by matriptase deficiency [209], demonstrating that HAI-1 maintains tissue homeostasis in vivo through inhibition of matriptase. The closely related Kunitz-type transmembrane serine protease inhibitor, HAI-2 (SPINT2/Placental bikunin), also displays potent inhibitory activity towards matriptase [211], hepsin [121] and prostasin [85,212]. Genetic inactivation of the mouse HAI-2/Spint2 gene leads to defects in neural tube closure, and abnormal placental labyrinth development associated with embryonic lethality. These developmental defects are also caused by unregulated matriptase activity, as both placental development and embryonic survival in HAI2-deficient embryos were completely restored by the simultaneous genetic inactivation of matriptase [213].
Exogenous Inhibitors
The activities of recombinant membrane anchored serine proteases are inhibited to varying extents by small molecular weight serine protease inhibitors, e.g. PMSF, AEBSF, benzamidine, leupeptin, and aprotinin (Table 4). SBTI appears to be an effective inhibitor of hepsin in peptide substrate assays, although it was shown to have minimal effect on hepsin's ability to activate FVII on the cell surface [91,214]. Ecotin, a macromolecular inhibitor of trypsin-like serine proteases isolated from Escherichia coli [215], was demonstrated to be a potent subnanomolar inhibitor of matriptase [215], but its selectivity and inhibitory profile against other membrane anchored serine proteases is not known.
Interestingly, a number of specific, small molecular weight inhibitors have been developed based upon the distinct structural features of individual membrane anchored serine proteases. The unique fine structures of the binding pockets of the membrane serine proteases have been exploited using peptidic inhibitor library screens arrayed in a positional scanning format [61,63]. This has led to the development of nanomolar inhibitors to DESC1 [61], prostasin [63] and hepsin [216], and a submicromolar inhibitor for tryptase γ1 [217]. A second strategy has been the design and synthesis of synthetic small molecule inhibitors based on preferred cleavage sequences of the enzymes. One of these, a selective matriptase inhibitor (CVS3893, Ki 3.3nM) was shown to suppress the growth of prostate tumor xenografts [218]. In addition, an inhibitor of uPA was optimized to selectively inhibit matriptase with Ki values below 5 nM and this inhibitor also showed tumor inhibitory activity in a mouse model [219]. Thus far however the selectivity of the reported inhibitors for individual membrane anchored serine proteases relative to other family members has not been extensively characterized.
Compounds that are already in clinical use have also shown efficacy as inhibitors of membrane anchored serine proteases. An inhibitor of trypsin-like serine proteases, camostat, is in clinical use for the treatment of chronic pancreatitis and two of the TTSPs, matriptase and prostasin are inhibited by camostat in peptide and cell based assays [152,212]. Camostat inhibits prostasin-dependent regulation of ENaC activity in animal models of airway epithelium [212] as well as sodium transport in the kidney [152]. The inhibition of prostasin-dependent activation of ENaCs is being developed as a therapeutic strategy for cystic fibrosis and salt-sensitive hypertension.
FUTURE PERSPECTIVES
The membrane anchored serine proteases are present in a wide range of tissues and biological fluids, and play key roles as potent modifiers of the cellular microenvironment. These enzymes catalyze pericellular proteolytic processing of a range of substrates that include peptide hormones, growth and differentiation factors, receptors, enzymes, adhesion molecules and viral proteins. While it is clear that these proteases have roles in homeostasis, increasing evidence suggests that their aberrant expression is a hallmark of several cancers and other diseases, and their activities are hijacked by viruses to facilitate infection and propagation. Elucidation of the endogenous substrate repertoire of the membrane anchored proteases will facilitate our understanding of their roles in normal physiology and disease while development of specific inhibitors may provide new avenues for the treatment of a range of ailments.
Acknowledgements/Funding
This work was supported in part by grants from the National Institutes of Health (CA098369, HL084387, DK081376)(TMA), the Mary Kay Ash Charitable Foundation 075-07 (TMA), the Maryland Stem Cell Research Fund 0145-00 (SNA) and by Fellowships from the National Health and Medical Research Council of Australia (CJ Martin Training Fellowship 384359 to MSB and RD Wright Fellowship 339732 to JDH).
Abbreviations
- ADAM
A Disintegrin And Metalloprotease
- CAP
channel activating protease
- CUB
complement C1r/C1s, Uegf, Bmp1
- ECD
extracellular domain
- GPI
glycosyl-phosphatidyl inositol
- HGF
hepatocyte growth factor
- HGFA
hepatocyte growth factor activator
- IRIDA
iron-refractory iron deficiency anemia
- LDLRA
low density lipoprotein-receptor class A-like
- MT-MMP
membrane-type matrix metalloproteinase
- PMA
phorbol myristate acetate
- SEA
sea urchin sperm protein, enteropeptidase, agrin
- SPD
serine protease domain
- SRCR
group A scavenger receptor cysteine-rich domain
- TEER
transepithelial electrical resistance
- TTSP
type II transmembrane serine protease
REFERENCES
- 1.Ramsay AJ, Quesada V, Sanchez M, Garabaya C, Sarda MP, Baiget M, Remacha A, Velasco G, Lopez-Otin C. Matriptase-2 mutations in iron-refractory iron deficiency anemia patients provide new insights into protease activation mechanisms. Hum. Mol. Genet. 2009;18:3673–3683. doi: 10.1093/hmg/ddp315. [DOI] [PubMed] [Google Scholar]
- 2.Neurath H, Walsh KA. Role of proteolytic enzymes in biological regulation (a review) Proc. Natl. Acad. Sci. U.S.A. 1976;73:3825–3832. doi: 10.1073/pnas.73.11.3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hedstrom L. Serine protease mechanism and specificity. Chem. Rev. 2002;102:4501–4524. doi: 10.1021/cr000033x. [DOI] [PubMed] [Google Scholar]
- 4.Rawlings ND, Barrett AJ. Introduction: serine peptidases and their clans. In: Barrett AJ, Rawlings ND, Woessner JF, editors. In Handbook of Proteolytic Enzymes. 2nd Edition Vol. 2. Elsevier Ltd; London UK: 2004. pp. 1417–1439. [Google Scholar]
- 5.Rawlings ND, Morton FR, Kok CY, Kong J, Barrett AJ. MEROPS: the peptidase database. Nucleic Acids Res. 2008;36:D320–D325. doi: 10.1093/nar/gkm954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rawlings ND, Morton FR. The MEROPS batch BLAST: a tool to detect peptidases and their non-peptidase homologues in a genome. Biochimie. 2008;90:243–259. doi: 10.1016/j.biochi.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 7.Plow EF, Miles LA. Plasminogen receptors in the mediation of pericellular proteolysis. Cell Differ. Dev. 1990;32:293–298. doi: 10.1016/0922-3371(90)90042-u. [DOI] [PubMed] [Google Scholar]
- 8.Hooper JD, Clements JA, Quigley JP, Antalis TM. Type II transmembrane serine proteases. Insights into an emerging class of cell surface proteolytic enzymes. J. Biol. Chem. 2001;276:857–860. doi: 10.1074/jbc.R000020200. [DOI] [PubMed] [Google Scholar]
- 9.Netzel-Arnett S, Hooper JD, Szabo R, Madison EL, Quigley JP, Bugge TH, Antalis TM. Membrane anchored serine proteases: a rapidly expanding group of cell surface proteolytic enzymes with potential roles in cancer. Cancer Metastasis Rev. 2003;22:237–258. doi: 10.1023/a:1023003616848. [DOI] [PubMed] [Google Scholar]
- 10.Szabo R, Bugge TH. Type II transmembrane serine proteases in development and disease. Int. J. Biochem. Cell Biol. 2008;40:1297–1316. doi: 10.1016/j.biocel.2007.11.013. [DOI] [PubMed] [Google Scholar]
- 11.Puente XS, Sanchez LM, Overall CM, Lopez-Otin C. Human and mouse proteases: a comparative genomic approach. Nat. Rev. Genet. 2003;4:544–558. doi: 10.1038/nrg1111. [DOI] [PubMed] [Google Scholar]
- 12.Bugge TH, Antalis TM, Wu Q. Type II transmembrane serine proteases. J. Biol. Chem. 2009;284:23177–23181. doi: 10.1074/jbc.R109.021006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caughey GH, Raymond WW, Blount JL, Hau LW, Pallaoro M, Wolters PJ, Verghese GM. Characterization of human gamma-tryptases, novel members of the chromosome 16p mast cell tryptase and prostasin gene families. J. Immunol. 2000;164:6566–6575. doi: 10.4049/jimmunol.164.12.6566. [DOI] [PubMed] [Google Scholar]
- 14.Wong GW, Tang Y, Feyfant E, Sali A, Li L, Li Y, Huang C, Friend DS, Krilis SA, Stevens RL. Identification of a new member of the tryptase family of mouse and human mast cell proteases which possesses a novel COOH-terminal hydrophobic extension. J. Biol. Chem. 1999;274:30784–30793. doi: 10.1074/jbc.274.43.30784. [DOI] [PubMed] [Google Scholar]
- 15.Yu JX, Chao L, Chao J. Prostasin is a novel human serine proteinase from seminal fluid. Purification, tissue distribution, and localization in prostate gland. J. Biol. Chem. 1994;269:18843–18848. [PubMed] [Google Scholar]
- 16.Chen LM, Skinner ML, Kauffman SW, Chao J, Chao L, Thaler CD, Chai KX. Prostasin is a glycosylphosphatidylinositol-anchored active serine protease. J. Biol. Chem. 2001;276:21434–21442. doi: 10.1074/jbc.M011423200. [DOI] [PubMed] [Google Scholar]
- 17.Verghese GM, Gutknecht MF, Caughey GH. Prostasin regulates epithelial monolayer function: cell-specific Gpld1-mediated secretion and functional role for GPI anchor. Am. J. Physiol Cell Physiol. 2006;291:C1258–C1270. doi: 10.1152/ajpcell.00637.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Honda A, Yamagata K, Sugiura S, Watanabe K, Baba T. A mouse serine protease TESP5 is selectively included into lipid rafts of sperm membrane presumably as a glycosylphosphatidylinositol-anchored protein. J. Biol. Chem. 2002;277:16976–16984. doi: 10.1074/jbc.M112470200. [DOI] [PubMed] [Google Scholar]
- 19.Hooper JD, Nicol DL, Dickinson JL, Eyre HJ, Scarman AL, Normyle JF, Stuttgen MA, Douglas ML, Loveland KA, Sutherland GR, Antalis TM. Testisin, a new human serine proteinase expressed by premeiotic testicular germ cells and lost in testicular germ cell tumors. Cancer Res. 1999;59:3199–3205. [PubMed] [Google Scholar]
- 20.Kitamoto Y, Yuan X, Wu Q, McCourt DW, Sadler JE. Enterokinase, the initiator of intestinal digestion, is a mosaic protease composed of a distinctive assortment of domains. Proc. Natl. Acad. Sci. U.S.A. 1994;91:7588–7592. doi: 10.1073/pnas.91.16.7588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szabo R, Wu Q, Dickson RB, Netzel-Arnett S, Antalis TM, Bugge TH. Type II transmembrane serine proteases. Thromb. Haemost. 2003;90:185–193. doi: 10.1160/TH03-02-0071. [DOI] [PubMed] [Google Scholar]
- 22.Cal S, Quesada V, Garabaya C, Lopez-Otin C. Polyserase-I, a human polyprotease with the ability to generate independent serine protease domains from a single translation product. Proc. Natl. Acad. Sci. U.S.A. 2003;100:9185–9190. doi: 10.1073/pnas.1633392100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okumura Y, Hayama M, Takahashi E, Fujiuchi M, Shimabukuro A, Yano M, Kido H. Serase-1B, a new splice variant of polyserase-1/TMPRSS9, activates urokinase-type plasminogen activator and the proteolytic activation is negatively regulated by glycosaminoglycans. Biochem. J. 2006;400:551–561. doi: 10.1042/BJ20060212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hansen IA, Fassnacht M, Hahner S, Hammer F, Schammann M, Meyer SR, Bicknell AB, Allolio B. The adrenal secretory serine protease AsP is a short secretory isoform of the transmembrane airway trypsin-like protease. Endocrinology. 2004;145:1898–1905. doi: 10.1210/en.2003-0930. [DOI] [PubMed] [Google Scholar]
- 25.Bhagwandin VJ, Hau LW, Mallen-St Clair J, Wolters PJ, Caughey GH. Structure and activity of human pancreasin, a novel tryptic serine peptidase expressed primarily by the pancreas. J. Biol. Chem. 2003;278:3363–3371. doi: 10.1074/jbc.M209353200. [DOI] [PubMed] [Google Scholar]
- 26.Wong GW, Foster PS, Yasuda S, Qi JC, Mahalingam S, Mellor EA, Katsoulotos G, Li L, Boyce JA, Krilis SA, Stevens RL. Biochemical and functional characterization of human transmembrane tryptase (TMT)/tryptase gamma. TMT is an exocytosed mast cell protease that induces airway hyperresponsiveness in vivo via an interleukin-13/interleukin-4 receptor alpha/signal transducer and activator of transcription (STAT) 6-dependent pathway. J. Biol. Chem. 2002;277:41906–41915. doi: 10.1074/jbc.M205868200. [DOI] [PubMed] [Google Scholar]
- 27.Tsuzuki S, Murai N, Miyake Y, Inouye K, Hirayasu H, Iwanaga T, Fushiki T. Evidence for the occurrence of membrane-type serine protease 1/matriptase on the basolateral sides of enterocytes. Biochem. J. 2005;388:679–687. doi: 10.1042/BJ20041639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang JK, Lee MS, Tseng IC, Chou FP, Chen YW, Fulton A, Lee HS, Chen CJ, Johnson MD, Lin CY. Polarized epithelial cells secrete matriptase as a consequence of zymogen activation and HAI-1-mediated inhibition. Am. J. Physiol Cell Physiol. 2009;297:C459–C470. doi: 10.1152/ajpcell.00201.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buzza MS, Netzel-Arnett S, Shea-Donohue T, Zhao A, Lin CY, List K, Szabo R, Fasano A, Bugge TH, Antalis TM. Membrane-anchored serine protease matriptase regulates epithelial barrier formation and permeability in the intestine. Proc. Natl. Acad. Sci. U.S.A. 2010;107:4200–4205. doi: 10.1073/pnas.0903923107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hung RJ, Hsu I, Dreiling JL, Lee MJ, Williams CA, Oberst MD, Dickson RB, Lin CY. Assembly of adherens junctions is required for sphingosine 1-phosphate-induced matriptase accumulation and activation at mammary epithelial cell-cell contacts. Am. J. Physiol Cell Physiol. 2004;286:C1159–C1169. doi: 10.1152/ajpcell.00400.2003. [DOI] [PubMed] [Google Scholar]
- 31.Murai N, Miyake Y, Tsuzuki S, Inouye K, Fushiki T. Involvement of the cytoplasmic juxtamembrane region of matriptase in its exclusive localization to the basolateral membrane domain of Madin-Darby canine kidney epithelial cells. Cytotechnology. 2009;59:169–176. doi: 10.1007/s10616-009-9205-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim C, Cho Y, Kang CH, Kim MG, Lee H, Cho EG, Park D. Filamin is essential for shedding of the transmembrane serine protease, epithin. EMBO Rep. 2005;6:1045–1051. doi: 10.1038/sj.embor.7400534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miao J, Mu D, Ergel B, Singavarapu R, Duan Z, Powers S, Oliva E, Orsulic S. Hepsin colocalizes with desmosomes and induces progression of ovarian cancer in a mouse model. Int. J. Cancer. 2008;123:2041–2047. doi: 10.1002/ijc.23726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vallet V, Chraibi A, Gaeggeler HP, Horisberger JD, Rossier BC. An epithelial serine protease activates the amiloride-sensitive sodium channel. Nature. 1997;389:607–610. doi: 10.1038/39329. [DOI] [PubMed] [Google Scholar]
- 35.Selzer-Plon J, Bornholdt J, Friis S, Bisgaard HC, Lothe IM, Tveit KM, Kure EH, Vogel U, Vogel LK. Expression of prostasin and its inhibitors during colorectal cancer carcinogenesis. BMC. Cancer. 2009;9:201. doi: 10.1186/1471-2407-9-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jacquinet E, Rao NV, Rao GV, Hoidal JR. Cloning, genomic organization, chromosomal assignment and expression of a novel mosaic serine proteinase: epitheliasin. FEBS Lett. 2000;468:93–100. doi: 10.1016/s0014-5793(00)01196-0. [DOI] [PubMed] [Google Scholar]
- 37.Afar DE, Vivanco I, Hubert RS, Kuo J, Chen E, Saffran DC, Raitano AB, Jakobovits A. Catalytic cleavage of the androgen-regulated TMPRSS2 protease results in its secretion by prostate and prostate cancer epithelia. Cancer Res. 2001;61:1686–1692. [PubMed] [Google Scholar]
- 38.Zheng X, Lu D, Sadler JE. Apical sorting of bovine enteropeptidase does not involve detergent- resistant association with sphingolipid-cholesterol rafts. J. Biol. Chem. 1999;274:1596–1605. doi: 10.1074/jbc.274.3.1596. [DOI] [PubMed] [Google Scholar]
- 39.Zheng X, Sadler JE. Mucin-like domain of enteropeptidase directs apical targeting in Madin- Darby canine kidney cells. J. Biol. Chem. 2002;277:6858–6863. doi: 10.1074/jbc.m109857200. [DOI] [PubMed] [Google Scholar]
- 40.Guipponi M, Toh MY, Tan J, Park D, Hanson K, Ballana E, Kwong D, Cannon PZ, Wu Q, Gout A, Delorenzi M, Speed TP, Smith RJ, Dahl HH, Petersen M, Teasdale RD, Estivill X, Park WJ, Scott HS. An integrated genetic and functional analysis of the role of type II transmembrane serine proteases (TMPRSSs) in hearing loss. Hum. Mutat. 2008;29:130–141. doi: 10.1002/humu.20617. [DOI] [PubMed] [Google Scholar]
- 41.Lucas JM, True L, Hawley S, Matsumura M, Morrissey C, Vessella R, Nelson PS. The androgen-regulated type II serine protease TMPRSS2 is differentially expressed and mislocalized in prostate adenocarcinoma. J. Pathol. 2008;215:118–125. doi: 10.1002/path.2330. [DOI] [PubMed] [Google Scholar]
- 42.Koda A, Wakida N, Toriyama K, Yamamoto K, Iijima H, Tomita K, Kitamura K. Urinary prostasin in humans: relationships among prostasin, aldosterone and epithelial sodium channel activity. Hypertens. Res. 2009;32:276–281. doi: 10.1038/hr.2009.6. [DOI] [PubMed] [Google Scholar]
- 43.Zhu H, Chao J, Guo D, Li K, Huang Y, Hawkins K, Wright N, Stallmann-Jorgensen I, Yan W, Harshfield GA, Dong Y. Urinary prostasin: a possible biomarker for renal pressure natriuresis in black adolescents. Pediatr. Res. 2009;65:443–446. doi: 10.1203/PDR.0b013e3181994b85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Olivieri O, Castagna A, Guarini P, Chiecchi L, Sabaini G, Pizzolo F, Corrocher R, Righetti PG. Urinary prostasin: a candidate marker of epithelial sodium channel activation in humans. Hypertension. 2005;46:683–688. doi: 10.1161/01.HYP.0000184108.12155.6b. [DOI] [PubMed] [Google Scholar]
- 45.Narikiyo T, Kitamura K, Adachi M, Miyoshi T, Iwashita K, Shiraishi N, Nonoguchi H, Chen LM, Chai KX, Chao J, Tomita K. Regulation of prostasin by aldosterone in the kidney. J. Clin. Invest. 2002;109:401–408. doi: 10.1172/JCI13229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iwashita K, Kitamura K, Narikiyo T, Adachi M, Shiraishi N, Miyoshi T, Nagano J, Tuyen DG, Nonoguchi H, Tomita K. Inhibition of prostasin secretion by serine protease inhibitors in the kidney. J. Am. Soc. Nephrol. 2003;14:11–16. doi: 10.1097/01.asn.0000043900.39397.48. [DOI] [PubMed] [Google Scholar]
- 47.Lin CY, Anders J, Johnson M, Dickson RB. Purification and characterization of a complex containing matriptase and a Kunitz-type serine protease inhibitor from human milk. J. Biol. Chem. 1999;274:18237–18242. doi: 10.1074/jbc.274.26.18237. [DOI] [PubMed] [Google Scholar]
- 48.Shi YE, Torri J, Yieh L, Wellstein A, Lippman ME, Dickson RB. Identification and characterization of a novel matrix-degrading protease from hormone-dependent human breast cancer cells. Cancer Res. 1993;53:1409–1415. [PubMed] [Google Scholar]
- 49.Cho EG, Kim MG, Kim C, Kim SR, Seong IS, Chung C, Schwartz RH, Park D. N-terminal processing is essential for release of epithin, a mouse type II membrane serine protease. J. Biol. Chem. 2001;276:44581–44589. doi: 10.1074/jbc.M107059200. [DOI] [PubMed] [Google Scholar]
- 50.Benaud C, Dickson RB, Lin CY. Regulation of the activity of matriptase on epithelial cell surfaces by a blood-derived factor. Eur. J. Biochem. 2001;268:1439–1447. doi: 10.1046/j.1432-1327.2001.02016.x. [DOI] [PubMed] [Google Scholar]
- 51.Jin X, Hirosaki T, Lin CY, Dickson RB, Higashi S, Kitamura H, Miyazaki K. Production of soluble matriptase by human cancer cell lines and cell surface activation of its zymogen by trypsin. J. Cell Biochem. 2005;95:632–647. doi: 10.1002/jcb.20418. [DOI] [PubMed] [Google Scholar]
- 52.Moss S, Birch GR, Lobley RW, Holmes R. The release of enterokinase following secretin and cholecystokinin-pancreozymin in man. Scand. J. Gastroenterol. 1979;14:1001–1007. [PubMed] [Google Scholar]
- 53.Lebenthal E, Lee PC. Effect of pancreozymin and secretin on intraluminal enterokinase, trypsin, and chymotrypsin activities of cystic fibrosis and control children. Digestion. 1982;23:39–47. doi: 10.1159/000198708. [DOI] [PubMed] [Google Scholar]
- 54.Yasuoka S, Ohnishi T, Kawano S, Tsuchihashi S, Ogawara M, Masuda K, Yamaoka K, Takahashi M, Sano T. Purification, characterization, and localization of a novel trypsin-like protease found in the human airway. Am. J. Respir. Cell Mol. Biol. 1997;16:300–308. doi: 10.1165/ajrcmb.16.3.9070615. [DOI] [PubMed] [Google Scholar]
- 55.Matsushima M, Ichinose M, Yahagi N, Kakei N, Tsukada S, Miki K, Kurokawa K, Tashiro K, Shiokawa K, Shinomiya K. Structural characterization of porcine enteropeptidase. J. Biol. Chem. 1994;269:19976–19982. [PubMed] [Google Scholar]
- 56.Cho EG, Schwartz RH, Kim MG. Shedding of membrane epithin is blocked without LDLRA4 and its protease activation site. Biochem. Biophys. Res. Commun. 2005;327:328–334. doi: 10.1016/j.bbrc.2004.12.014. [DOI] [PubMed] [Google Scholar]
- 57.Schechter I, Berger A. On the size of the active site in proteases. I. Papain. Biochem. Biophys. Res. Commun. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- 58.Friedrich R, Fuentes-Prior P, Ong E, Coombs G, Hunter M, Oehler R, Pierson D, Gonzalez R, Huber R, Bode W, Madison EL. Catalytic domain structures of MT-SP1/matriptase, a matrix-degrading transmembrane serine proteinase. J. Biol. Chem. 2002;277:2160–2168. doi: 10.1074/jbc.M109830200. [DOI] [PubMed] [Google Scholar]
- 59.Somoza JR, Ho JD, Luong C, Ghate M, Sprengeler PA, Mortara K, Shrader WD, Sperandio D, Chan H, McGrath ME, Katz BA. The structure of the extracellular region of human hepsin reveals a serine protease domain and a novel scavenger receptor cysteine-rich (SRCR) domain. Structure. 2003;11:1123–1131. doi: 10.1016/s0969-2126(03)00148-5. [DOI] [PubMed] [Google Scholar]
- 60.Lu D, Futterer K, Korolev S, Zheng X, Tan K, Waksman G, Sadler JE. Crystal structure of enteropeptidase light chain complexed with an analog of the trypsinogen activation peptide. J. Mol. Biol. 1999;292:361–373. doi: 10.1006/jmbi.1999.3089. [DOI] [PubMed] [Google Scholar]
- 61.Kyrieleis OJ, Huber R, Ong E, Oehler R, Hunter M, Madison EL, Jacob U. Crystal structure of the catalytic domain of DESC1, a new member of the type II transmembrane serine proteinase family. FEBS J. 2007;274:2148–2160. doi: 10.1111/j.1742-4658.2007.05756.x. [DOI] [PubMed] [Google Scholar]
- 62.Spraggon G, Hornsby M, Shipway A, Tully DC, Bursulaya B, Danahay H, Harris JL, Lesley SA. Active site conformational changes of prostasin provide a new mechanism of protease regulation by divalent cations. Protein Sci. 2009;18:1081–1094. doi: 10.1002/pro.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tully DC, Vidal A, Chatterjee AK, Williams JA, Roberts MJ, Petrassi HM, Spraggon G, Bursulaya B, Pacoma R, Shipway A, Schumacher AM, Danahay H, Harris JL. Discovery of inhibitors of the channel-activating protease prostasin (CAP1/PRSS8) utilizing structure-based design. Bioorg. Med. Chem. Lett. 2008;18:5895–5899. doi: 10.1016/j.bmcl.2008.08.029. [DOI] [PubMed] [Google Scholar]
- 64.Rickert KW, Kelley P, Byrne NJ, Diehl RE, Hall DL, Montalvo AM, Reid JC, Shipman JM, Thomas BW, Munshi SK, Darke PL, Su HP. Structure of human prostasin, a target for the regulation of hypertension. J. Biol. Chem. 2008;283:34864–34872. doi: 10.1074/jbc.M805262200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Friedrich R, Steinmetzer T, Huber R, Sturzebecher J, Bode W. The methyl group of N(alpha)(Me)Arg-containing peptides disturbs the active-site geometry of thrombin, impairing efficient cleavage. J. Mol. Biol. 2002;316:869–874. doi: 10.1006/jmbi.2001.5394. [DOI] [PubMed] [Google Scholar]
- 66.Oberst MD, Williams CA, Dickson RB, Johnson MD, Lin CY. The activation of matriptase requires its noncatalytic domains, serine protease domain, and its cognate inhibitor. J. Biol. Chem. 2003;278:26773–26779. doi: 10.1074/jbc.M304282200. [DOI] [PubMed] [Google Scholar]
- 67.Lin CY, Tseng IC, Chou FP, Su SF, Chen YW, Johnson MD, Dickson RB. Zymogen activation, inhibition, and ectodomain shedding of matriptase. Front Biosci. 2008;13:621–635. doi: 10.2741/2707. [DOI] [PubMed] [Google Scholar]
- 68.Macao B, Johansson DG, Hansson GC, Hard T. Autoproteolysis coupled to protein folding in the SEA domain of the membrane-bound MUC1 mucin. Nat. Struct. Mol. Biol. 2006;13:71–76. doi: 10.1038/nsmb1035. [DOI] [PubMed] [Google Scholar]
- 69.Lee MS, Tseng IC, Wang Y, Kiyomiya K, Johnson MD, Dickson RB, Lin CY. Autoactivation of matriptase in vitro: requirement for biomembrane and LDL receptor domain. Am. J. Physiol. Cell Physiol. 2007;293:C95–105. doi: 10.1152/ajpcell.00611.2006. [DOI] [PubMed] [Google Scholar]
- 70.Knappe S, Wu F, Madlansacay MR, Wu Q. Identification of domain structures in the propeptide of corin essential for the processing of proatrial natriuretic peptide. J. Biol. Chem. 2004;279:34464–34471. doi: 10.1074/jbc.M405041200. [DOI] [PubMed] [Google Scholar]
- 71.Lu D, Yuan X, Zheng X, Sadler JE. Bovine proenteropeptidase is activated by trypsin, and the specificity of enteropeptidase depends on the heavy chain. J. Biol. Chem. 1997;272:31293–31300. doi: 10.1074/jbc.272.50.31293. [DOI] [PubMed] [Google Scholar]
- 72.Gladysheva IP, King SM, Houng AK. N-glycosylation modulates the cell-surface expression and catalytic activity of corin. Biochem. Biophys. Res. Commun. 2008;373:130–135. doi: 10.1016/j.bbrc.2008.05.181. [DOI] [PubMed] [Google Scholar]
- 73.Liao X, Wang W, Chen S, Wu Q. Role of glycosylation in corin zymogen activation. J. Biol. Chem. 2007;282:27728–27735. doi: 10.1074/jbc.M703687200. [DOI] [PubMed] [Google Scholar]
- 74.Ramsay AJ, Reid JC, Velasco G, Quigley JP, Hooper JD. The type II transmembrane serine protease matriptase-2--identification, structural features, enzymology, expression pattern and potential roles. Front Biosci. 2008;13:569–579. doi: 10.2741/2702. [DOI] [PubMed] [Google Scholar]
- 75.Silvestri L, Guillem F, Pagani A, Nai A, Oudin C, Silva M, Toutain F, Kannengiesser C, Beaumont C, Camaschella C, Grandchamp B. Molecular mechanisms of the defective hepcidin inhibition in TMPRSS6 mutations associated with iron-refractory iron deficiency anemia. Blood. 2009;113:5605–5608. doi: 10.1182/blood-2008-12-195594. [DOI] [PubMed] [Google Scholar]
- 76.Tchou I, Diepold M, Pilotto PA, Swinkels D, Neerman-Arbez M, Beris P. Haematologic data, iron parameters and molecular findings in two new cases of iron-refractory iron deficiency anaemia. Eur. J. Haematol. 2009;83:595–602. doi: 10.1111/j.1600-0609.2009.01340.x. [DOI] [PubMed] [Google Scholar]
- 77.Rame JE, Drazner MH, Post W, Peshock R, Lima J, Cooper RS, Dries DL. Corin I555(P568) allele is associated with enhanced cardiac hypertrophic response to increased systemic afterload. Hypertension. 2007;49:857–864. doi: 10.1161/01.HYP.0000258566.95867.9e. [DOI] [PubMed] [Google Scholar]
- 78.Dries DL, Victor RG, Rame JE, Cooper RS, Wu X, Zhu X, Leonard D, Ho SI, Wu Q, Post W, Drazner MH. Corin gene minor allele defined by 2 missense mutations is common in blacks and associated with high blood pressure and hypertension. Circulation. 2005;112:2403–2410. doi: 10.1161/CIRCULATIONAHA.105.568881. [DOI] [PubMed] [Google Scholar]
- 79.Wang W, Liao X, Fukuda K, Knappe S, Wu F, Dries DL, Qin J, Wu Q. Corin variant associated with hypertension and cardiac hypertrophy exhibits impaired zymogen activation and natriuretic peptide processing activity. Circ. Res. 2008;103:502–508. doi: 10.1161/CIRCRESAHA.108.177352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Guipponi M, Vuagniaux G, Wattenhofer M, Shibuya K, Vazquez M, Dougherty L, Scamuffa N, Guida E, Okui M, Rossier C, Hancock M, Buchet K, Reymond A, Hummler E, Marzella PL, Kudoh J, Shimizu N, Scott HS, Antonarakis SE, Rossier BC. The transmembrane serine protease (TMPRSS3) mutated in deafness DFNB8/10 activates the epithelial sodium channel (ENaC) in vitro. Hum. Mol. Genet. 2002;11:2829–2836. doi: 10.1093/hmg/11.23.2829. [DOI] [PubMed] [Google Scholar]
- 81.Takeuchi T, Harris JL, Huang W, Yan KW, Coughlin SR, Craik CS. Cellular localization of membrane-type serine protease 1 and identification of protease-activated receptor-2 and single-chain urokinase-type plasminogen activator as substrates. J. Biol. Chem. 2000;275:26333–26342. doi: 10.1074/jbc.M002941200. [DOI] [PubMed] [Google Scholar]
- 82.Boulware KT, Daugherty PS. Protease specificity determination by using cellular libraries of peptide substrates (CLiPS) Proc. Natl. Acad. Sci. U.S.A. 2006;103:7583–7588. doi: 10.1073/pnas.0511108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Herter S, Piper DE, Aaron W, Gabriele T, Cutler G, Cao P, Bhatt AS, Choe Y, Craik CS, Walker N, Meininger D, Hoey T, Austin RJ. Hepatocyte growth factor is a preferred in vitro substrate for human hepsin, a membrane-anchored serine protease implicated in prostate and ovarian cancers. Biochem. J. 2005;390:125–136. doi: 10.1042/BJ20041955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kido H, Okumura Y. MSPL/TMPRSS13. Front. Biosci. 2008;13:754–758. doi: 10.2741/2717. [DOI] [PubMed] [Google Scholar]
- 85.Shipway A, Danahay H, Williams JA, Tully DC, Backes BJ, Harris JL. Biochemical characterization of prostasin, a channel activating protease. Biochem. Biophys. Res. Commun. 2004;324:953–963. doi: 10.1016/j.bbrc.2004.09.123. [DOI] [PubMed] [Google Scholar]
- 86.Hobson JP, Netzel-Arnett S, Szabo R, Rehault SM, Church FC, Strickland DK, Lawrence DA, Antalis TM, Bugge TH. Mouse DESC1 is located within a cluster of seven DESC1-like genes and encodes a type II transmembrane serine protease that forms serpin inhibitory complexes. J. Biol. Chem. 2004;279:46981–46994. doi: 10.1074/jbc.M403299200. [DOI] [PubMed] [Google Scholar]
- 87.Beliveau F, Desilets A, Leduc R. Probing the substrate specificities of matriptase, matriptase-2, hepsin and DESC1 with internally quenched fluorescent peptides. FEBS J. 2009;276:2213–2226. doi: 10.1111/j.1742-4658.2009.06950.x. [DOI] [PubMed] [Google Scholar]
- 88.Antalis TM, Shea-Donohue T, Vogel SN, Sears C, Fasano A. Mechanisms of disease: protease functions in intestinal mucosal pathobiology. Nat. Clin. Pract. Gastroenterol. Hepatol. 2007;4:393–402. doi: 10.1038/ncpgasthep0846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Neurath H. Proteolytic enzymes, past and present. Fed. Proc. 1985;44:2907–2913. [PubMed] [Google Scholar]
- 90.Holzinger A, Maier EM, Buck C, Mayerhofer PU, Kappler M, Haworth JC, Moroz SP, Hadorn HB, Sadler JE, Roscher AA. Mutations in the proenteropeptidase gene are the molecular cause of congenital enteropeptidase deficiency. Am. J. Hum. Genet. 2002;70:20–25. doi: 10.1086/338456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kazama Y, Hamamoto T, Foster DC, Kisiel W. Hepsin, a putative membrane-associated serine protease, activates human factor VII and initiates a pathway of blood coagulation on the cell surface leading to thrombin formation. J. Biol. Chem. 1995;270:66–72. doi: 10.1074/jbc.270.1.66. [DOI] [PubMed] [Google Scholar]
- 92.Cao C, Lawrence DA, Li Y, Von Arnim CA, Herz J, Su EJ, Makarova A, Hyman BT, Strickland DK, Zhang L. Endocytic receptor LRP together with tPA and PAI-1 coordinates Mac-1-dependent macrophage migration. EMBO J. 2006;25:1860–1870. doi: 10.1038/sj.emboj.7601082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Moran P, Li W, Fan B, Vij R, Eigenbrot C, Kirchhofer D. Pro-urokinase-type plasminogen activator is a substrate for hepsin. J. Biol. Chem. 2006;281:30439–30446. doi: 10.1074/jbc.M605440200. [DOI] [PubMed] [Google Scholar]
- 94.Lee SL, Dickson RB, Lin CY. Activation of hepatocyte growth factor and urokinase/plasminogen activator by matriptase, an epithelial membrane serine protease. J. Biol. Chem. 2000;275:36720–36725. doi: 10.1074/jbc.M007802200. [DOI] [PubMed] [Google Scholar]
- 95.Kilpatrick LM, Harris RL, Owen KA, Bass R, Ghorayeb C, Bar-Or A, Ellis V. Initiation of plasminogen activation on the surface of monocytes expressing the type II transmembrane serine protease matriptase. Blood. 2006;108:2616–2623. doi: 10.1182/blood-2006-02-001073. [DOI] [PubMed] [Google Scholar]
- 96.Beaufort N, Leduc D, Eguchi H, Mengele K, Hellmann D, Masegi T, Kamimura T, Yasuoka S, Fend F, Chignard M, Pidard D. The human airway trypsin-like protease modulates the urokinase receptor (uPAR, CD87) structure and functions. Am. J. Physiol. Lung Cell Mol. Physiol. 2007;292:L1263–L1272. doi: 10.1152/ajplung.00191.2006. [DOI] [PubMed] [Google Scholar]
- 97.Preissner KT, Kanse SM, May AE. Urokinase receptor: a molecular organizer in cellular communication. Curr. Opin. Cell Biol. 2000;12:621–628. doi: 10.1016/s0955-0674(00)00141-1. [DOI] [PubMed] [Google Scholar]
- 98.Jin X, Yagi M, Akiyama N, Hirosaki T, Higashi S, Lin CY, Dickson RB, Kitamura H, Miyazaki K. Matriptase activates stromelysin (MMP-3) and promotes tumor growth and angiogenesis. Cancer Sci. 2006;97:1327–1334. doi: 10.1111/j.1349-7006.2006.00328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Netzel-Arnett S, Currie BM, Szabo R, Lin CY, Chen LM, Chai KX, Antalis TM, Bugge TH, List K. Evidence for a matriptase-prostasin proteolytic cascade regulating terminal epidermal differentiation. J. Biol. Chem. 2006;281:32941–32945. doi: 10.1074/jbc.C600208200. [DOI] [PubMed] [Google Scholar]
- 100.Camerer E, Barker A, Duong DN, Ganesan R, Kataoka H, Cornelissen I, Darragh MR, Hussain A, Zheng YW, Srinivasan Y, Brown C, Xu SM, Regard JB, Lin CY, Craik CS, Kirchhofer D, Coughlin SR. Local protease signaling contributes to neural tube closure in the mouse embryo. Dev. Cell. 2010;18:25–38. doi: 10.1016/j.devcel.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Vu TK, Liu RW, Haaksma CJ, Tomasek JJ, Howard EW. Identification and cloning of the membrane-associated serine protease, hepsin, from mouse preimplantation embryos. J. Biol. Chem. 1997;272:31315–31320. doi: 10.1074/jbc.272.50.31315. [DOI] [PubMed] [Google Scholar]
- 102.Stallmach R, Gloor SM. Neurobin/TMPRSS11c, a novel type II transmembrane serine protease that cleaves fibroblast growth factor-2 in vitro. Biochem. J. 2008;412:81–91. doi: 10.1042/BJ20071432. [DOI] [PubMed] [Google Scholar]
- 103.Desilets A, Beliveau F, Vandal G, McDuff FO, Lavigne P, Leduc R. Mutation G827R in matriptase causing autosomal recessive ichthyosis with hypotrichosis yields an inactive protease. J. Biol. Chem. 2008;283:10535–10542. doi: 10.1074/jbc.M707012200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Velasco G, Cal S, Quesada V, Sanchez LM, Lopez-Otin C. Matriptase-2, a membrane-bound mosaic serine proteinase predominantly expressed in human liver and showing degrading activity against extracellular matrix proteins. J. Biol. Chem. 2002;277:37637–37646. doi: 10.1074/jbc.M203007200. [DOI] [PubMed] [Google Scholar]
- 105.Qiu D, Owen K, Gray K, Bass R, Ellis V. Roles and regulation of membrane-associated serine proteases. Biochem. Soc. Trans. 2007;35:583–587. doi: 10.1042/BST0350583. [DOI] [PubMed] [Google Scholar]
- 106.Andreasen D, Vuagniaux G, Fowler-Jaeger N, Hummler E, Rossier BC. Activation of epithelial sodium channels by mouse channel activating proteases (mCAP) expressed in Xenopus oocytes requires catalytic activity of mCAP3 and mCAP2 but not mCAP1. J. Am. Soc. Nephrol. 2006;17:968–976. doi: 10.1681/ASN.2005060637. [DOI] [PubMed] [Google Scholar]
- 107.Coughlin SR. How the protease thrombin talks to cells. Proc. Natl. Acad. Sci. U.S.A. 1999;96:11023–11027. doi: 10.1073/pnas.96.20.11023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Arora P, Ricks TK, Trejo J. Protease-activated receptor signalling, endocytic sorting and dysregulation in cancer. J Cell Sci. 2007;120:921–928. doi: 10.1242/jcs.03409. [DOI] [PubMed] [Google Scholar]
- 109.Ossovskaya VS, Bunnett NW. Protease-activated receptors: contribution to physiology and disease. Physiol Rev. 2004;84:579–621. doi: 10.1152/physrev.00028.2003. [DOI] [PubMed] [Google Scholar]
- 110.Cottrell GS, Amadesi S, Schmidlin F, Bunnett N. Protease-activated receptor 2: activation, signalling and function. Biochem. Soc.Trans. 2003;31:1191–1197. doi: 10.1042/bst0311191. [DOI] [PubMed] [Google Scholar]
- 111.Ramsay AJ, Reid JC, Adams MN, Samaratunga H, Dong Y, Clements JA, Hooper JD. Prostatic trypsin-like kallikrein-related peptidases (KLKs) and other prostate-expressed tryptic proteinases as regulators of signalling via proteinase-activated receptors (PARs) Biol. Chem. 2008;389:653–668. doi: 10.1515/BC.2008.078. [DOI] [PubMed] [Google Scholar]
- 112.Wilson S, Greer B, Hooper J, Zijlstra A, Walker B, Quigley J, Hawthorne S. The membrane-anchored serine protease, TMPRSS2, activates PAR-2 in prostate cancer cells. Biochem. J. 2005;388:967–972. doi: 10.1042/BJ20041066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Seitz I, Hess S, Schulz H, Eckl R, Busch G, Montens HP, Brandl R, Seidl S, Schomig A, Ott I. Membrane-type serine protease-1/matriptase induces interleukin-6 and -8 in endothelial cells by activation of protease-activated receptor-2: potential implications in atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2007;27:769–775. doi: 10.1161/01.ATV.0000258862.61067.14. [DOI] [PubMed] [Google Scholar]
- 114.Chokki M, Eguchi H, Hamamura I, Mitsuhashi H, Kamimura T. Human airway trypsin-like protease induces amphiregulin release through a mechanism involving protease-activated receptor-2-mediated ERK activation and TNF alpha-converting enzyme activity in airway epithelial cells. FEBS J. 2005;272:6387–6399. doi: 10.1111/j.1742-4658.2005.05035.x. [DOI] [PubMed] [Google Scholar]
- 115.Chen LM, Hatfield ML, Fu YY, Chai KX. Prostasin regulates iNOS and cyclin D1 expression by modulating protease-activated receptor-2 signaling in prostate epithelial cells. Prostate. 2009;69:1790–1801. doi: 10.1002/pros.21030. [DOI] [PubMed] [Google Scholar]
- 116.Nusrat A, Parkos CA, Bacarra AE, Godowski PJ, Delp-Archer C, Rosen EM, Madara JL. Hepatocyte growth factor/scatter factor effects on epithelia. Regulation of intercellular junctions in transformed and nontransformed cell lines, basolateral polarization of c-met receptor in transformed and natural intestinal epithelia, and induction of rapid wound repair in a transformed model epithelium. J. Clin. Invest. 1994;93:2056–2065. doi: 10.1172/JCI117200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bhatt AS, Welm A, Farady CJ, Vasquez M, Wilson K, Craik CS. Coordinate expression and functional profiling identify an extracellular proteolytic signaling pathway. Proc. Natl. Acad. Sci. U.S.A. 2007;104:5771–5776. doi: 10.1073/pnas.0606514104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sakamoto O, Iwama A, Amitani R, Takehara T, Yamaguchi N, Yamamoto T, Masuyama K, Yamanaka T, Ando M, Suda T. Role of macrophage-stimulating protein and its receptor, RON tyrosine kinase, in ciliary motility. J. Clin. Invest. 1997;99:701–709. doi: 10.1172/JCI119214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Forbs D, Thiel S, Stella MC, Sturzebecher A, Schweinitz A, Steinmetzer T, Sturzebecher J, Uhland K. In vitro inhibition of matriptase prevents invasive growth of cell lines of prostate and colon carcinoma. Int. J. Oncol. 2005;27:1061–1070. [PubMed] [Google Scholar]
- 120.Lee SL, Huang PY, Roller P, Cho EG, Park D, Dickson RB. Matriptase/epithin participates in mammary epithelial cell growth and morphogenesis through HGF activation. Mech. Dev. 2010;127:82–95. doi: 10.1016/j.mod.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 121.Kirchhofer D, Peek M, Lipari MT, Billeci K, Fan B, Moran P. Hepsin activates pro-hepatocyte growth factor and is inhibited by hepatocyte growth factor activator inhibitor-1B (HAI-1B) and HAI-2. FEBS Lett. 2005;579:1945–1950. doi: 10.1016/j.febslet.2005.01.085. [DOI] [PubMed] [Google Scholar]
- 122.Chen M, Fu YY, Lin CY, Chen LM, Chai KX. Prostasin induces protease-dependent and independent molecular changes in the human prostate carcinoma cell line PC-3. Biochim. Biophys. Acta. 2007;1773:1133–1140. doi: 10.1016/j.bbamcr.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chen M, Chen LM, Lin CY, Chai KX. The epidermal growth factor receptor (EGFR) is proteolytically modified by the Matriptase-Prostasin serine protease cascade in cultured epithelial cells. Biochim. Biophys. Acta. 2008;1783:896–903. doi: 10.1016/j.bbamcr.2007.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chen M, Chen LM, Lin CY, Chai KX. Hepsin activates prostasin and cleaves the extracellular domain of the epidermal growth factor receptor. Mol. Cell Biochem. 2009 doi: 10.1007/s11010-009-0307-y. [DOI] [PubMed] [Google Scholar]
- 125.Darragh MR, Bhatt AS, Craik CS. MT-SP1 proteolysis and regulation of cell-microenvironment interactions. Front. Biosci. 2008;13:528–539. doi: 10.2741/2698. [DOI] [PubMed] [Google Scholar]
- 126.Gladysheva IP, Robinson BR, Houng AK, Kovats T, King SM. Corin is co-expressed with pro-ANP and localized on the cardiomyocyte surface in both zymogen and catalytically active forms. J. Mol. Cell Cardiol. 2008;44:131–142. doi: 10.1016/j.yjmcc.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 127.Yan W, Wu F, Morser J, Wu Q. Corin, a transmembrane cardiac serine protease, acts as a pro-atrial natriuretic peptide-converting enzyme. Proc. Natl. Acad. Sci. U.S.A. 2000;97:8525–8529. doi: 10.1073/pnas.150149097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wu F, Yan W, Pan J, Morser J, Wu Q. Processing of pro-atrial natriuretic peptide by corin in cardiac myocytes. J. Biol. Chem. 2002;277:16900–16905. doi: 10.1074/jbc.M201503200. [DOI] [PubMed] [Google Scholar]
- 129.Das BB, Solinger R. Role of natriuretic peptide family in cardiovascular medicine. Cardiovasc. Hematol. Agents Med. Chem. 2009;7:29–42. doi: 10.2174/187152509787047630. [DOI] [PubMed] [Google Scholar]
- 130.Lee CY, Burnett JC., Jr Natriuretic peptides and therapeutic applications. Heart Fail. Rev. 2007;12:131–142. doi: 10.1007/s10741-007-9016-3. [DOI] [PubMed] [Google Scholar]
- 131.Chan JC, Knudson O, Wu F, Morser J, Dole WP, Wu Q. Hypertension in mice lacking the proatrial natriuretic peptide convertase corin. Proc. Natl. Acad. Sci. U.S.A. 2005;102:785–790. doi: 10.1073/pnas.0407234102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.John SW, Krege JH, Oliver PM, Hagaman JR, Hodgin JB, Pang SC, Flynn TG, Smithies O. Genetic decreases in atrial natriuretic peptide and salt-sensitive hypertension. Science. 1995;267:679–681. doi: 10.1126/science.7839143. [DOI] [PubMed] [Google Scholar]
- 133.Berger R, Huelsman M, Strecker K, Bojic A, Moser P, Stanek B, Pacher R. B-type natriuretic peptide predicts sudden death in patients with chronic heart failure. Circulation. 2002;105:2392–2397. doi: 10.1161/01.cir.0000016642.15031.34. [DOI] [PubMed] [Google Scholar]
- 134.Tamura N, Ogawa Y, Chusho H, Nakamura K, Nakao K, Suda M, Kasahara M, Hashimoto R, Katsuura G, Mukoyama M, Itoh H, Saito Y, Tanaka I, Otani H, Katsuki M. Cardiac fibrosis in mice lacking brain natriuretic peptide. Proc. Natl. Acad. Sci. U.S.A. 2000;97:4239–4244. doi: 10.1073/pnas.070371497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Rame JE, Tam SW, McNamara D, Worcel M, Sabolinski ML, Wu AH, Dries DL. Dysfunctional Corin I555(P568) Allele Is Associated With Impaired Brain Natriuretic Peptide Processing and Adverse Outcomes in Blacks With Systolic Heart Failure: Results From the Genetic Risk Assessment in Heart Failure Substudy. Circ. Heart Fail. 2009;2:541–548. doi: 10.1161/CIRCHEARTFAILURE.109.866822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Vuagniaux G, Vallet V, Jaeger NF, Hummler E, Rossier BC. Synergistic Activation of ENaC by Three Membrane-bound Channel- activating Serine Proteases (mCAP1, mCAP2, and mCAP3) and S. J. Gen. Physiol. 2002;120:191–201. doi: 10.1085/jgp.20028598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kleyman TR, Carattino MD, Hughey RP. ENaC at the cutting edge: regulation of epithelial sodium channels by proteases. J. Biol. Chem. 2009;284:20447–20451. doi: 10.1074/jbc.R800083200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Adachi M, Kitamura K, Miyoshi T, Narikiyo T, Iwashita K, Shiraishi N, Nonoguchi H, Tomita K. Activation of epithelial sodium channels by prostasin in Xenopus oocytes. J. Am. Soc. Nephrol. 2001;12:1114–1121. doi: 10.1681/ASN.V1261114. [DOI] [PubMed] [Google Scholar]
- 139.Vuagniaux G, Vallet V, Jaeger NF, Pfister C, Bens M, Farman N, Courtois-Coutry N, Vandewalle A, Rossier BC, Hummler E. Activation of the amiloride-sensitive epithelial sodium channel by the serine protease mCAP1 expressed in a mouse cortical collecting duct cell line. J. Am. Soc. Nephrol. 2000;11:828–834. doi: 10.1681/ASN.V115828. [DOI] [PubMed] [Google Scholar]
- 140.Tong Z, Illek B, Bhagwandin VJ, Verghese GM, Caughey GH. Prostasin, a membrane-anchored serine peptidase, regulates sodium currents in JME/CF15 cells, a cystic fibrosis airway epithelial cell line. Am. J. Physiol. Lung Cell Mol. Physiol. 2004;287:L928–L935. doi: 10.1152/ajplung.00160.2004. [DOI] [PubMed] [Google Scholar]
- 141.Bruns JB, Carattino MD, Sheng S, Maarouf AB, Weisz OA, Pilewski JM, Hughey RP, Kleyman TR. Epithelial Na+ channels are fully activated by furin- and prostasin-dependent release of an inhibitory peptide from the gamma-subunit. J. Biol. Chem. 2007;282:6153–6160. doi: 10.1074/jbc.M610636200. [DOI] [PubMed] [Google Scholar]
- 142.Carattino MD, Hughey RP, Kleyman TR. Proteolytic processing of the epithelial sodium channel gamma subunit has a dominant role in channel activation. J. Biol. Chem. 2008;283:25290–25295. doi: 10.1074/jbc.M803931200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Svenningsen P, Uhrenholt TR, Palarasah Y, Skjoedt K, Jensen BL, Skott O. Prostasin-dependent activation of epithelial Na+ channels by low plasmin concentrations. Am. J. Physiol Regul. Integr. Comp. Physiol. 2009;297:R1733–R1741. doi: 10.1152/ajpregu.00321.2009. [DOI] [PubMed] [Google Scholar]
- 144.Vallet V, Pfister C, Loffing J, Rossier BC. Cell-surface expression of the channel activating protease xCAP-1 is required for activation of ENaC in the Xenopus oocyte. J. Am. Soc. Nephrol. 2002;13:588–594. doi: 10.1681/ASN.V133588. [DOI] [PubMed] [Google Scholar]
- 145.Garcia-Caballero A, Dang Y, He H, Stutts MJ. ENaC proteolytic regulation by channel-activating protease 2. J. Gen. Physiol. 2008;132:521–535. doi: 10.1085/jgp.200810030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Donaldson SH, Hirsh A, Li DC, Holloway G, Chao J, Boucher RC, Gabriel SE. Regulation of the epithelial sodium channel by serine proteases in human airways. J. Biol. Chem. 2002;277:8338–8345. doi: 10.1074/jbc.M105044200. [DOI] [PubMed] [Google Scholar]
- 147.Planes C, Randrianarison NH, Charles RP, Frateschi S, Cluzeaud F, Vuagniaux G, Soler P, Clerici C, Rossier BC, Hummler E. ENaC-mediated alveolar fluid clearance and lung fluid balance depend on the channel-activating protease 1. EMBO Mol. Med. 2010;2:26–37. doi: 10.1002/emmm.200900050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Donaldson SH, Boucher RC. Sodium channels and cystic fibrosis. Chest. 2007;132:1631–1636. doi: 10.1378/chest.07-0288. [DOI] [PubMed] [Google Scholar]
- 149.Myerburg MM, McKenna EE, Luke CJ, Frizzell RA, Kleyman TR, Pilewski JM. Prostasin expression is regulated by airway surface liquid volume and is increased in cystic fibrosis. Am. J. Physiol. Lung Cell Mol. Physiol. 2008;294:L932–L941. doi: 10.1152/ajplung.00437.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hummler E. Implication of ENaC in salt-sensitive hypertension. J. Steroid Biochem. Mol. Biol. 1999;69:385–390. doi: 10.1016/s0960-0760(99)00073-4. [DOI] [PubMed] [Google Scholar]
- 151.Zhu H, Guo D, Li K, Yan W, Tan Y, Wang X, Treiber FA, Chao J, Snieder H, Dong Y. Prostasin: a possible candidate gene for human hypertension. Am. J. Hypertens. 2008;21:1028–1033. doi: 10.1038/ajh.2008.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Maekawa A, Kakizoe Y, Miyoshi T, Wakida N, Ko T, Shiraishi N, Adachi M, Tomita K, Kitamura K. Camostat mesilate inhibits prostasin activity and reduces blood pressure and renal injury in salt-sensitive hypertension. J. Hypertens. 2009;27:181–189. doi: 10.1097/hjh.0b013e328317a762. [DOI] [PubMed] [Google Scholar]
- 153.Silvestri L, Pagani A, Nai A, De D, I, Kaplan J, Camaschella C. The serine protease matriptase-2 (TMPRSS6) inhibits hepcidin activation by cleaving membrane hemojuvelin. Cell Metab. 2008;8:502–511. doi: 10.1016/j.cmet.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Finberg KE, Heeney MM, Campagna DR, Aydinok Y, Pearson HA, Hartman KR, Mayo MM, Samuel SM, Strouse JJ, Markianos K, Andrews NC, Fleming MD. Mutations in TMPRSS6 cause iron-refractory iron deficiency anemia (IRIDA) Nat. Genet. 2008;40:569–571. doi: 10.1038/ng.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Guillem F, Lawson S, Kannengiesser C, Westerman M, Beaumont C, Grandchamp B. Two nonsense mutations in the TMPRSS6 gene in a patient with microcytic anemia and iron deficiency. Blood. 2008;112:2089–2091. doi: 10.1182/blood-2008-05-154740. [DOI] [PubMed] [Google Scholar]
- 156.Melis MA, Cau M, Congiu R, Sole G, Barella S, Cao A, Westerman M, Cazzola M, Galanello R. A mutation in the TMPRSS6 gene, encoding a transmembrane serine protease that suppresses hepcidin production, in familial iron deficiency anemia refractory to oral iron. Haematologica. 2008;93:1473–1479. doi: 10.3324/haematol.13342. [DOI] [PubMed] [Google Scholar]
- 157.Folgueras AR, de Lara FM, Pendas AM, Garabaya C, Rodriguez F, Astudillo A, Bernal T, Cabanillas R, Lopez-Otin C, Velasco G. Membrane-bound serine protease matriptase-2 (Tmprss6) is an essential regulator of iron homeostasis. Blood. 2008;112:2539–2545. doi: 10.1182/blood-2008-04-149773. [DOI] [PubMed] [Google Scholar]
- 158.Truksa J, Gelbart T, Peng H, Beutler E, Beutler B, Lee P. Suppression of the hepcidin-encoding gene Hamp permits iron overload in mice lacking both hemojuvelin and matriptase-2/TMPRSS6. Br. J. Haematol. 2009;147:571–581. doi: 10.1111/j.1365-2141.2009.07873.x. [DOI] [PubMed] [Google Scholar]
- 159.Babitt JL, Huang FW, Xia Y, Sidis Y, Andrews NC, Lin HY. Modulation of bone morphogenetic protein signaling in vivo regulates systemic iron balance. J. Clin. Invest. 2007;117:1933–1939. doi: 10.1172/JCI31342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Choi SY, Bertram S, Glowacka I, Park YW, Pohlmann S. Type II transmembrane serine proteases in cancer and viral infections. Trends Mol. Med. 2009;15:303–312. doi: 10.1016/j.molmed.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 161.Kido H, Okumura Y, Takahashi E, Pan HY, Wang S, Chida J, Le TQ, Yano M. Host envelope glycoprotein processing proteases are indispensable for entry into human cells by seasonal and highly pathogenic avian influenza viruses. J. Mol. Genet. Med. 2008;3:167–175. [PMC free article] [PubMed] [Google Scholar]
- 162.Bottcher E, Matrosovich T, Beyerle M, Klenk HD, Garten W, Matrosovich M. Proteolytic activation of influenza viruses by serine proteases TMPRSS2 and HAT from human airway epithelium. J. Virol. 2006;80:9896–9898. doi: 10.1128/JVI.01118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Bottcher E, Freuer C, Steinmetzer T, Klenk HD, Garten W. MDCK cells that express proteases TMPRSS2 and HAT provide a cell system to propagate influenza viruses in the absence of trypsin and to study cleavage of HA and its inhibition. Vaccine. 2009;27:6324–6329. doi: 10.1016/j.vaccine.2009.03.029. [DOI] [PubMed] [Google Scholar]
- 164.Chaipan C, Kobasa D, Bertram S, Glowacka I, Steffen I, Solomon TT, Takeda M, Bugge T, Kim S, Park Y, Marzi A, Pohlmann S. Proteolytic activation of the 1918 influenza virus hemagglutinin. J. Virol. 2009;83:3200–3211. doi: 10.1128/JVI.02205-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Shirogane Y, Takeda M, Iwasaki M, Ishiguro N, Takeuchi H, Nakatsu Y, Tahara M, Kikuta H, Yanagi Y. Efficient multiplication of human metapneumovirus in Vero cells expressing the transmembrane serine protease TMPRSS2. J. Virol. 2008;82:8942–8946. doi: 10.1128/JVI.00676-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Zhang JL, Zhao WG, Wu KL, Wang K, Zhang X, Gu CF, Li Y, Zhu Y, Wu JG. Human hepatitis B virus × protein promotes cell proliferation and inhibits cell apoptosis through interacting with a serine protease Hepsin. Arch. Virol. 2005;150:721–741. doi: 10.1007/s00705-004-0446-0. [DOI] [PubMed] [Google Scholar]
- 167.Kam YW, Okumura Y, Kido H, Ng LF, Bruzzone R, Altmeyer R. Cleavage of the SARS coronavirus spike glycoprotein by airway proteases enhances virus entry into human bronchial epithelial cells in vitro. PLoS. One. 2009;4:e7870. doi: 10.1371/journal.pone.0007870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.List K, Haudenschild CC, Szabo R, Chen W, Wahl SM, Swaim W, Engelholm LH, Behrendt N, Bugge TH. Matriptase/MT-SP1 is required for postnatal survival, epidermal barrier function, hair follicle development, and thymic homeostasis. Oncogene. 2002;21:3765–3779. doi: 10.1038/sj.onc.1205502. [DOI] [PubMed] [Google Scholar]
- 169.List K, Szabo R, Wertz PW, Segre J, Haudenschild CC, Kim SY, Bugge TH. Loss of proteolytically processed filaggrin caused by epidermal deletion of Matriptase/MT-SP1. J. Cell Biol. 2003;163:901–910. doi: 10.1083/jcb.200304161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Basel-Vanagaite L, Attia R, Ishida-Yamamoto A, Rainshtein L, Ben Amitai D, Lurie R, Pasmanik-Chor M, Indelman M, Zvulunov A, Saban S, Magal N, Sprecher E, Shohat M. Autosomal recessive ichthyosis with hypotrichosis caused by a mutation in ST14, encoding type II transmembrane serine protease matriptase. Am. J. Hum. Genet. 2007;80:467–477. doi: 10.1086/512487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Avrahami L, Maas S, Pasmanik-Chor M, Rainshtein L, Magal N, Smitt J, van Marle J, Shohat M, Basel-Vanagaite L. Autosomal recessive ichthyosis with hypotrichosis syndrome: further delineation of the phenotype. Clin. Genet. 2008;74:47–53. doi: 10.1111/j.1399-0004.2008.01006.x. [DOI] [PubMed] [Google Scholar]
- 172.List K, Currie B, Scharschmidt TC, Szabo R, Shireman J, Molinolo A, Cravatt BF, Segre J, Bugge TH. Autosomal ichthyosis with hypotrichosis syndrome displays low matriptase proteolytic activity and is phenocopied in ST14 hypomorphic mice. J. Biol. Chem. 2007;282:36714–36723. doi: 10.1074/jbc.M705521200. [DOI] [PubMed] [Google Scholar]
- 173.Alef T, Torres S, Hausser I, Metze D, Tursen U, Lestringant GG, Hennies HC. Ichthyosis, Follicular Atrophoderma, and Hypotrichosis Caused by Mutations in ST14 Is Associated with Impaired Profilaggrin Processing. J. Invest Dermatol. 2009;129:862–869. doi: 10.1038/jid.2008.311. [DOI] [PubMed] [Google Scholar]
- 174.List K, Kosa P, Szabo R, Bey AL, Wang CB, Molinolo A, Bugge TH. Epithelial integrity is maintained by a matriptase-dependent proteolytic pathway. Am. J. Pathol. 2009;175:1453–1463. doi: 10.2353/ajpath.2009.090240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Guipponi M, Antonarakis SE, Scott HS. TMPRSS3, a type II transmembrane serine protease mutated in non-syndromic autosomal recessive deafness. Front. Biosci. 2008;13:1557–1567. doi: 10.2741/2780. [DOI] [PubMed] [Google Scholar]
- 176.Scott HS, Kudoh J, Wattenhofer M, Shibuya K, Berry A, Chrast R, Guipponi M, Wang J, Kawasaki K, Asakawa S, Minoshima S, Younus F, Mehdi SQ, Radhakrishna U, Papasavvas MP, Gehrig C, Rossier C, Korostishevsky M, Gal A, Shimizu N, Bonne-Tamir B, Antonarakis SE. Insertion of beta-satellite repeats identifies a transmembrane protease causing both congenital and childhood onset autosomal recessive deafness. Nat. Genet. 2001;27:59–63. doi: 10.1038/83768. [DOI] [PubMed] [Google Scholar]
- 177.Lee YJ, Park D, Kim SY, Park WJ. Pathogenic mutations but not polymorphisms in congenital and childhood onset autosomal recessive deafness disrupt the proteolytic activity of TMPRSS3. J. Med. Genet. 2003;40:629–631. doi: 10.1136/jmg.40.8.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Wattenhofer M, Di I, V, Rabionet R, Dougherty L, Pampanos A, Schwede T, Montserrat-Sentis B, Arbones L, Iliades T, Pasquadibisceglie A, D'Amelio M, Alwan S, Rossier C, Dahl HH, Petersen MB, Estivill X, Gasparini P, Scott HS, Antonarakis SE. Mutations in the TMPRSS3 gene are a rare cause of childhood nonsyndromic deafness in Caucasian patients. J. Mol. Med. 2002;80:124–131. doi: 10.1007/s00109-001-0310-6. [DOI] [PubMed] [Google Scholar]
- 179.Guipponi M, Tan J, Cannon PZ, Donley L, Crewther P, Clarke M, Wu Q, Shepherd RK, Scott HS. Mice deficient for the type II transmembrane serine protease, TMPRSS1/hepsin, exhibit profound hearing loss. Am. J. Pathol. 2007;171:608–616. doi: 10.2353/ajpath.2007.070068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Netzel-Arnett S, Bugge TH, Hess RA, Carnes K, Stringer BW, Scarman AL, Hooper JD, Tonks ID, Kay GF, Antalis TM. The glycosylphosphatidylinositol-anchored serine protease PRSS21 (testisin) imparts murine epididymal sperm cell maturation and fertilizing ability. Biol. Reprod. 2009;81:921–932. doi: 10.1095/biolreprod.109.076273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Yamashita M, Honda A, Ogura A, Kashiwabara S, Fukami K, Baba T. Reduced fertility of mouse epididymal sperm lacking Prss21/Tesp5 is rescued by sperm exposure to uterine microenvironment. Genes Cells. 2008;13:1001–1013. doi: 10.1111/j.1365-2443.2008.01222.x. [DOI] [PubMed] [Google Scholar]
- 182.Enshell-Seijffers D, Lindon C, Morgan BA. The serine protease Corin is a novel modifier of the Agouti pathway. Development. 2008;135:217–225. doi: 10.1242/dev.011031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Szabo R, Molinolo A, List K, Bugge TH. Matriptase inhibition by hepatocyte growth factor activator inhibitor-1 is essential for placental development. Oncogene. 2007;26:1546–1556. doi: 10.1038/sj.onc.1209966. [DOI] [PubMed] [Google Scholar]
- 184.Lee JW, Yong SS, Choi JJ, Lee SJ, Kim BG, Park CS, Lee JH, Lin CY, Dickson RB, Bae DS. Increased expression of matriptase is associated with histopathologic grades of cervical neoplasia. Hum. Pathol. 2005;36:626–633. doi: 10.1016/j.humpath.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 185.List K, Szabo R, Molinolo A, Sriuranpong V, Redeye V, Murdock T, Burke B, Nielsen BS, Gutkind JS, Bugge TH. Deregulated matriptase causes ras-independent multistage carcinogenesis and promotes ras-mediated malignant transformation. Genes Dev. 2005;19:1934–1950. doi: 10.1101/gad.1300705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Ihara S, Miyoshi E, Ko JH, Murata K, Nakahara S, Honke K, Dickson RB, Lin CY, Taniguchi N. Prometastatic effect of N-acetylglucosaminyltransferase V is due to modification and stabilization of active matriptase by adding beta 1–6 GlcNAc branching. J. Biol. Chem. 2002;277:16960–16967. doi: 10.1074/jbc.M200673200. [DOI] [PubMed] [Google Scholar]
- 187.Jung H, Lee KP, Park SJ, Park JH, Jang YS, Choi SY, Jung JG, Jo K, Park DY, Yoon JH, Park JH, Lim DS, Hong GR, Choi C, Park YK, Lee JW, Hong HJ, Kim S, Park YW. TMPRSS4 promotes invasion, migration and metastasis of human tumor cells by facilitating an epithelial-mesenchymal transition. Oncogene. 2008;27:2635–2647. doi: 10.1038/sj.onc.1210914. [DOI] [PubMed] [Google Scholar]
- 188.Tomlins SA, Laxman B, Dhanasekaran SM, Helgeson BE, Cao X, Morris DS, Menon A, Jing X, Cao Q, Han B, Yu J, Wang L, Montie JE, Rubin MA, Pienta KJ, Roulston D, Shah RB, Varambally S, Mehra R, Chinnaiyan AM. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448:595–599. doi: 10.1038/nature06024. [DOI] [PubMed] [Google Scholar]
- 189.Nam RK, Sugar L, Yang W, Srivastava S, Klotz LH, Yang LY, Stanimirovic A, Encioiu E, Neill M, Loblaw DA, Trachtenberg J, Narod SA, Seth A. Expression of the TMPRSS2:ERG fusion gene predicts cancer recurrence after surgery for localised prostate cancer. Br. J. Cancer. 2007;97:1690–1695. doi: 10.1038/sj.bjc.6604054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Sanders AJ, Parr C, Martin TA, Lane J, Mason MD, Jiang WG. Genetic upregulation of matriptase-2 reduces the aggressiveness of prostate cancer cells in vitro and in vivo and affects FAK and paxillin localisation. J. Cell Physiol. 2008;216:780–789. doi: 10.1002/jcp.21460. [DOI] [PubMed] [Google Scholar]
- 191.Parr C, Sanders AJ, Davies G, Martin T, Lane J, Mason MD, Mansel RE, Jiang WG. Matriptase-2 inhibits breast tumor growth and invasion and correlates with favorable prognosis for breast cancer patients. Clin. Cancer Res. 2007;13:3568–3576. doi: 10.1158/1078-0432.CCR-06-2357. [DOI] [PubMed] [Google Scholar]
- 192.Lang JC, Schuller DE. Differential expression of a novel serine protease homologue in squamous cell carcinoma of the head and neck. Br. J. Cancer. 2001;84:237–243. doi: 10.1054/bjoc.2000.1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Sakashita K, Mimori K, Tanaka F, Tahara K, Inoue H, Sawada T, Ohira M, Hirakawa K, Mori M. Clinical significance of low expression of Prostasin mRNA in human gastric cancer. J. Surg. Oncol. 2008;98:559–564. doi: 10.1002/jso.21158. [DOI] [PubMed] [Google Scholar]
- 194.Tang T, Kmet M, Corral L, Vartanian S, Tobler A, Papkoff J. Testisin, a glycosylphosphatidylinositol-linked serine protease, promotes malignant transformation in vitro and in vivo. Cancer Res. 2005;65:868–878. [PubMed] [Google Scholar]
- 195.Manton KJ, Douglas ML, Netzel-Arnett S, Fitzpatrick DR, Nicol DL, Boyd AW, Clements JA, Antalis TM. Hypermethylation of the 5' CpG island of the gene encoding the serine protease Testisin promotes its loss in testicular tumorigenesis. Br. J. Cancer. 2005;92:760–769. doi: 10.1038/sj.bjc.6602373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Kempkensteffen C, Christoph F, Weikert S, Krause H, Kollermann J, Schostak M, Miller K, Schrader M. Epigenetic silencing of the putative tumor suppressor gene testisin in testicular germ cell tumors. J. Cancer Res. Clin. Oncol. 2006;132:765–770. doi: 10.1007/s00432-006-0124-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Shigemasa K, Underwood LJ, Beard J, Tanimoto H, Ohama K, Parmley TH, O'Brien TJ. Overexpression of testisin, a serine protease expressed by testicular germ cells, in epithelial ovarian tumor cells. J. Soc. Gynecol. Investig. 2000;7:358–362. [PubMed] [Google Scholar]
- 198.Tripathi M, Nandana S, Yamashita H, Ganesan R, Kirchhofer D, Quaranta V. Laminin-332 is a substrate for hepsin, a protease associated with prostate cancer progression. J. Biol. Chem. 2008;283:30576–30584. doi: 10.1074/jbc.M802312200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Silverman GA, Bird PI, Carrell RW, Church FC, Coughlin PB, Gettins PG, Irving JA, Lomas DA, Luke CJ, Moyer RW, Pemberton PA, Remold-O'Donnell E, Salvesen GS, Travis J, Whisstock JC. The serpins are an expanding superfamily of structurally similar but functionally diverse proteins. Evolution, mechanism of inhibition, novel functions, and a revised nomenclature. J. Biol. Chem. 2001;276:33293–33296. doi: 10.1074/jbc.R100016200. [DOI] [PubMed] [Google Scholar]
- 200.Gettins PG. Serpin structure, mechanism, and function. Chem. Rev. 2002;102:4751–4804. doi: 10.1021/cr010170+. [DOI] [PubMed] [Google Scholar]
- 201.Herz J, Strickland DK. LRP: a multifunctional scavenger and signaling receptor. J. Clin. Invest. 2001;108:779–784. doi: 10.1172/JCI13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Tseng IC, Chou FP, Su SF, Oberst M, Madayiputhiya N, Lee MS, Wang JK, Sloane DE, Johnson M, Lin CY. Purification from human milk of matriptase complexes with secreted serpins: mechanism for inhibition of matriptase other than HAI-1. Am. J. Physiol. Cell Physiol. 2008;295:C423–C431. doi: 10.1152/ajpcell.00164.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Szabo R, Netzel-Arnett S, Hobson JP, Antalis TM, Bugge TH. Matriptase-3 is a novel phylogenetically preserved membrane-anchored serine protease with broad serpin reactivity. Biochem. J. 2005;390:231–242. doi: 10.1042/BJ20050299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Wakida N, Kitamura K, Tuyen DG, Maekawa A, Miyoshi T, Adachi M, Shiraishi N, Ko T, Ha V, Nonoguchi H, Tomita K. Inhibition of prostasin-induced ENaC activities by PN-1 and regulation of PN-1 expression by TGF-beta1 and aldosterone. Kidney Int. 2006;70:1432–1438. doi: 10.1038/sj.ki.5001787. [DOI] [PubMed] [Google Scholar]
- 205.Fan B, Wu TD, Li W, Kirchhofer D. Identification of hepatocyte growth factor activator inhibitor-1B as a potential physiological inhibitor of prostasin. J. Biol. Chem. 2005;280:34513–34520. doi: 10.1074/jbc.M502119200. [DOI] [PubMed] [Google Scholar]
- 206.Lin CY, Anders J, Johnson M, Sang QA, Dickson RB. Molecular cloning of cDNA for matriptase, a matrix-degrading serine protease with trypsin-like activity. J. Biol. Chem. 1999;274:18231–18236. doi: 10.1074/jbc.274.26.18231. [DOI] [PubMed] [Google Scholar]
- 207.Oberst MD, Chen LY, Kiyomiya K, Williams CA, Lee MS, Johnson MD, Dickson RB, Lin CY. HAI-1 regulates activation and expression of matriptase, a membrane-bound serine protease. Am. J. Physiol. Cell Physiol. 2005;289:C462–C470. doi: 10.1152/ajpcell.00076.2005. [DOI] [PubMed] [Google Scholar]
- 208.Kojima K, Tsuzuki S, Fushiki T, Inouye K. Roles of functional and structural domains of hepatocyte growth factor activator inhibitor type 1 in the inhibition of matriptase. J. Biol. Chem. 2008;283:2478–2487. doi: 10.1074/jbc.M709073200. [DOI] [PubMed] [Google Scholar]
- 209.Szabo R, Kosa P, List K, Bugge TH. Loss of matriptase suppression underlies spint1 mutation-associated ichthyosis and postnatal lethality. Am. J. Pathol. 2009;174:2015–2022. doi: 10.2353/ajpath.2009.090053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Nagaike K, Kawaguchi M, Takeda N, Fukushima T, Sawaguchi A, Kohama K, Setoyama M, Kataoka H. Defect of hepatocyte growth factor activator inhibitor type 1/serine protease inhibitor, Kunitz type 1 (Hai-1/Spint1) leads to ichthyosis-like condition and abnormal hair development in mice. Am. J. Pathol. 2008;173:1464–1475. doi: 10.2353/ajpath.2008.071142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Szabo R, Hobson JP, List K, Molinolo A, Lin CY, Bugge TH. Potent inhibition and global co-localization implicate the transmembrane Kunitz-type serine protease inhibitor hepatocyte growth factor activator inhibitor-2 in the regulation of epithelial matriptase activity. J. Biol. Chem. 2008;283:29495–29504. doi: 10.1074/jbc.M801970200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Coote K, Atherton-Watson HC, Sugar R, Young A, MacKenzie-Beevor A, Gosling M, Bhalay G, Bloomfield G, Dunstan A, Bridges RJ, Sabater JR, Abraham WM, Tully D, Pacoma R, Schumacher A, Harris J, Danahay H. Camostat attenuates airway epithelial sodium channel function in vivo through the inhibition of a channel-activating protease. J. Pharmacol. Exp. Ther. 2009;329:764–774. doi: 10.1124/jpet.108.148155. [DOI] [PubMed] [Google Scholar]
- 213.Szabo R, Hobson JP, Christoph K, Kosa P, List K, Bugge TH. Regulation of cell surface protease matriptase by HAI2 is essential for placental development, neural tube closure and embryonic survival in mice. Development. 2009;136:2653–2663. doi: 10.1242/dev.038430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Zhukov A, Hellman U, Ingelman-Sundberg M. Purification and characterization of hepsin from rat liver microsomes. Biochim. Biophys. Acta. 1997;1337:85–95. doi: 10.1016/s0167-4838(96)00152-5. [DOI] [PubMed] [Google Scholar]
- 215.Stoop AA, Craik CS. Engineering of a macromolecular scaffold to develop specific protease inhibitors. Nat. Biotechnol. 2003;21:1063–1068. doi: 10.1038/nbt860. [DOI] [PubMed] [Google Scholar]
- 216.Katz BA, Luong C, Ho JD, Somoza JR, Gjerstad E, Tang J, Williams SR, Verner E, Mackman RL, Young WB, Sprengeler PA, Chan H, Mortara K, Janc JW, McGrath ME. Dissecting and designing inhibitor selectivity determinants at the S1 site using an artificial Ala190 protease (Ala190 uPA) J. Mol. Biol. 2004;344:527–547. doi: 10.1016/j.jmb.2004.09.032. [DOI] [PubMed] [Google Scholar]
- 217.Yuan J, Beltman J, Gjerstad E, Nguyen MT, Sampang J, Chan H, Janc JW, Clark JM. Expression and characterization of recombinant gamma-tryptase. Protein Expr. Purif. 2006;49:47–54. doi: 10.1016/j.pep.2006.04.018. [DOI] [PubMed] [Google Scholar]
- 218.Galkin AV, Mullen L, Fox WD, Brown J, Duncan D, Moreno O, Madison EL, Agus DB. CVS-3983, a selective matriptase inhibitor, suppresses the growth of androgen independent prostate tumor xenografts. Prostate. 2004;61:228–235. doi: 10.1002/pros.20094. [DOI] [PubMed] [Google Scholar]
- 219.Steinmetzer T, Schweinitz A, Sturzebecher A, Donnecke D, Uhland K, Schuster O, Steinmetzer P, Muller F, Friedrich R, Than ME, Bode W, Sturzebecher J. Secondary amides of sulfonylated 3-amidinophenylalanine. New potent and selective inhibitors of matriptase. J. Med. Chem. 2006;49:4116–4126. doi: 10.1021/jm051272l. [DOI] [PubMed] [Google Scholar]
- 220.Yamaguchi N, Okui A, Yamada T, Nakazato H, Mitsui S. Spinesin/TMPRSS5, a novel transmembrane serine protease, cloned from human spinal cord. J. Biol. Chem. 2002;277:6806–6812. doi: 10.1074/jbc.M103645200. [DOI] [PubMed] [Google Scholar]
- 221.Knecht W, Cottrell GS, Amadesi S, Mohlin J, Skaregarde A, Gedda K, Peterson A, Chapman K, Hollenberg MD, Vergnolle N, Bunnett NW. Trypsin IV or mesotrypsin and p23 cleave protease-activated receptors 1 and 2 to induce inflammation and hyperalgesia. J. Biol. Chem. 2007;282:26089–26100. doi: 10.1074/jbc.M703840200. [DOI] [PubMed] [Google Scholar]
- 222.Kunitz M. FORMATION OF TRYPSIN FROM CRYSTALLINE TRYPSINOGEN BY MEANS OF ENTEROKINASE. J. Gen. Physiol. 1939;22:429–446. doi: 10.1085/jgp.22.4.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Kim YT, Nishii W, Matsushima M, Inoue H, Ito H, Park SJ, Takahashi K. Substrate specificities of porcine and bovine enteropeptidases toward the peptide Val-(Asp)4-Lys-Ile-Val-Gly and its analogs. Biosci. Biotechnol. Biochem. 2008;72:905–908. doi: 10.1271/bbb.70732. [DOI] [PubMed] [Google Scholar]
- 224.Kang JY, Dolled-Filhart M, Ocal IT, Singh B, Lin CY, Dickson RB, Rimm DL, Camp RL. Tissue microarray analysis of hepatocyte growth factor/Met pathway components reveals a role for Met, matriptase, and hepatocyte growth factor activator inhibitor 1 in the progression of node-negative breast cancer. Cancer Res. 2003;63:1101–1105. [PubMed] [Google Scholar]
- 225.Parr C, Watkins G, Mansel RE, Jiang WG. The hepatocyte growth factor regulatory factors in human breast cancer. Clin. Cancer Res. 2004;10:202–211. doi: 10.1158/1078-0432.ccr-0553-3. [DOI] [PubMed] [Google Scholar]
- 226.Bhatt AS, Erdjument-Bromage H, Tempst P, Craik CS, Moasser MM. Adhesion signaling by a novel mitotic substrate of src kinases. Oncogene. 2005;24:5333–5343. doi: 10.1038/sj.onc.1208582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Ahmed S, Jin X, Yagi M, Yasuda C, Sato Y, Higashi S, Lin CY, Dickson RB, Miyazaki K. Identification of membrane-bound serine proteinase matriptase as processing enzyme of insulin-like growth factor binding protein-related protein-1 (IGFBP-rP1/angiomodulin/mac25) FEBS J. 2006;273:615–627. doi: 10.1111/j.1742-4658.2005.05094.x. [DOI] [PubMed] [Google Scholar]
- 228.Knappe S, Wu F, Masikat MR, Morser J, Wu Q. Functional analysis of the transmembrane domain and activation cleavage of human corin: design and characterization of a soluble corin. J. Biol. Chem. 2003;278:52363–52370. doi: 10.1074/jbc.M309991200. [DOI] [PubMed] [Google Scholar]
- 229.Leyvraz C, Charles RP, Rubera I, Guitard M, Rotman S, Breiden B, Sandhoff K, Hummler E. The epidermal barrier function is dependent on the serine protease CAP1/Prss8. J. Cell Biol. 2005;170:487–496. doi: 10.1083/jcb.200501038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Klezovitch O, Chevillet J, Mirosevich J, Roberts RL, Matusik RJ, Vasioukhin V. Hepsin promotes prostate cancer progression and metastasis. Cancer Cell. 2004;6:185–195. doi: 10.1016/j.ccr.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 231.Kim TS, Heinlein C, Hackman RC, Nelson PS. Phenotypic analysis of mice lacking the Tmprss2-encoded protease. Mol. Cell Biol. 2006;26:965–975. doi: 10.1128/MCB.26.3.965-975.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.List K, Szabo R, Molinolo A, Nielsen BS, Bugge TH. Delineation of matriptase protein expression by enzymatic gene trapping suggests diverging roles in barrier function, hair formation, and squamous cell carcinogenesis. Am. J. Pathol. 2006;168:1513–1525. doi: 10.2353/ajpath.2006.051071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Du X, She E, Gelbart T, Truksa J, Lee P, Xia Y, Khovananth K, Mudd S, Mann N, Moresco EM, Beutler E, Beutler B. The serine protease TMPRSS6 is required to sense iron deficiency. Science. 2008;320:1088–1092. doi: 10.1126/science.1157121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Chen LM, Zhang X, Chai KX. Regulation of prostasin expression and function in the prostate. Prostate. 2004;59:1–12. doi: 10.1002/pros.10346. [DOI] [PubMed] [Google Scholar]
- 235.Viloria CG, Peinado JR, Astudillo A, Garcia-Suarez O, Gonzalez MV, Suarez C, Cal S. Human DESC1 serine protease confers tumorigenic properties to MDCK cells and it is upregulated in tumours of different origin. Br. J. Cancer. 2007;97:201–209. doi: 10.1038/sj.bjc.6603856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Gasparian ME, Ostapchenko VG, Dolgikh DA, Kirpichnikov MP. Biochemical characterization of human enteropeptidase light chain. Biochemistry (Mosc.) 2006;71:113–119. doi: 10.1134/s0006297906020015. [DOI] [PubMed] [Google Scholar]
- 237.Liepnieks JJ, Light A. The preparation and properties of bovine enterokinase. J. Biol. Chem. 1979;254:1677–1683. [PubMed] [Google Scholar]
- 238.Janciauskiene S, Nita I, Subramaniyam D, Li Q, Lancaster JR, Jr., Matalon S. Alpha1-antitrypsin inhibits the activity of the matriptase catalytic domain in vitro. Am. J. Respir. Cell Mol. Biol. 2008;39:631–637. doi: 10.1165/rcmb.2008-0015RC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Kirchhofer D, Peek M, Li W, Stamos J, Eigenbrot C, Kadkhodayan S, Elliott JM, Corpuz RT, Lazarus RA, Moran P. Tissue expression, protease specificity, and Kunitz domain functions of hepatocyte growth factor activator inhibitor-1B (HAI-1B), a new splice variant of HAI-1. J. Biol. Chem. 2003;278:36341–36349. doi: 10.1074/jbc.M304643200. [DOI] [PubMed] [Google Scholar]
- 240.Yamasaki Y, Satomi S, Murai N, Tsuzuki S, Fushiki T. Inhibition of membrane-type serine protease 1/matriptase by natural and synthetic protease inhibitors. J. Nutr. Sci. Vitaminol. (Tokyo) 2003;49:27–32. doi: 10.3177/jnsv.49.27. [DOI] [PubMed] [Google Scholar]