Abstract
Dwarf stature is introduced to improve lodging resistance and harvest index in crop production. In many crops including maize, mining and application of novel dwarf genes are urgent to overcome genetic bottleneck and vulnerability during breeding improvement. Here we report the characterization and expression profiling analysis of a newly identified maize dwarf mutant Dwarf11 (D11). The D11 displays severely developmental abnormalities and is controlled by a dominant Mendelian factor. The D11 seedlings responds to both GA3 and paclobutrazol (PAC) application, suggesting that dwarf phenotype of D11 is caused by GA biosynthesis instead of GA signaling deficiency. In contrast, two well-characterized maize dominant dwarf plants D8 and D9 are all insensitive to exogenous GA3 stimulation. Additionally, sequence variation of D8 and D9 genes was not identified in the D11 mutant. Microarray and qRT-PCR analysis results demonstrated that transcripts encoding GA biosynthetic and catabolic enzymes ent-kaurenoic acid oxidase (KAO), GA 20-oxidase (GA20ox), and GA 2-oxidase (GA2ox) are up-regulated in D11. Our results lay a foundation for the following D11 gene cloning and functional characterization. Moreover, results presented here may aid in crops molecular improvement and breeding, especially breeding of crops with plant height ideotypes.
Introduction
The well-known ‘Green revolution’ highlighted in the late 1960s and increased food crops yield worldwide by incorporating semi-dwarf traits combined with other agricultural practices, such as fertilizer and pesticide utilization, irrigation equipment installation. The technical advance of ‘Green revolution’ mainly attributes to the development of ‘high-yielding varieties’ (HYVs) which show high capacity of nitrogen assimilation and tend to lodge. Dwarf traits were introduced to improve lodging resistance in this case. During the ‘Green revolution’, novel dwarf rice and wheat cultivars with increased yield were developed. Semi-dwarf variety IR8 (also known as ‘Miracle Rice’), the first widely used HYV of International Rice Research Institute (IRRI), was bred by a Chinese dwarf variety ‘Dee-geo-woo-gen’ crossed with an Indonesian high-yielding variety ‘Peta’. The Japanese dwarf wheat cultivar ‘Norin 10’ was adopted to breed wheat HYVs [1]. With the development of diverse omic-based technology, the genes of ‘Green revolution’ semidwarf1 (sd1) and Reduced height (Rht) were isolated and characterized. Rice sd1 encodes a defective gibberellin (GA) biosynthetic enzyme GA 20-oxidase (GA20ox). Wheat Rht has been proved to be involved in GA signaling cascade. Both sd1 and Rht genes participate in GA homeostasis [2].
Maize dwarf trait research could be traced back to Emmerson's work [3]. Over the past decades, a large amount of maize dwarf mutants have been identified and characterized. Detailed information of maize dwarf plants has been deposited in the MaizeGDB phenotypic database (http://www.maizegdb.org/phenotype.php). Maize dwarf mutants are primarily classified as GA-sensitive and GA-insensitive types based on their responses to exogenous GA application. Four recessive dwarf mutants dwarf-1 (d1), d2, d3, and d5, together with one dominant dwarf mutant D8-1023 belong to the GA-responsive group [4], [5]. Two well-characterized maize dominant dwarf plants Dwarf8 (D8) and D9 are insensitive to exogenous GA stimulation [6]. The change of DELLA domains in maize D8 and D9 proteins causes dwarf phenotype [7], [8]. D8-1023, allele of D8, produces dwarf plants by altering the VHYNP domain instead of DELLA domain [5]. A combined linkage and association analysis technique revealed that d8 functions not only in plant height control but flowering time determinant [9]. However, recent reanalysis of d8 locus by newly developed model indicated that d8 has minor effect on flowering time [10]. Besides, candidate genes regulating maize height were unraveled by genome wide association studies (GWAS) technique [11]. Recently, GA3ox2 was positionally cloned and proved to be responsible for maize height determinant [12].
Many plant height-regulating genes have been isolated in other cereal crops. Barley Sln1, wheat Rht, rice slender rice 1 (slr1), and maize d8 are orthologs of Arabidopsis Gibberellin Insensitive (GAI) and REPRESSOR OF ga1-3 (RGA) genes, which encode DELLA proteins and participate in GA signaling [7], [13], [14]. One member of APETALA2 (AP2)/Ethylene-Responsive Element Binding Factor (ERF) family significantly affects rice internode elongation by down-regulating of GA concentrations [15]. In addition to plant hormone GA, other phytohormones, such as auxin (Aux) and brassinosteroid (BR), modulate plant height. Maize dwarf plant with compact stems brachytic2 (br2) and sorghum counterpart dwarf3 (dw3) show polar auxin transport abnormalities in the stalk [16]. The BR metabolism pathway and signaling perception also relate to rice, maize, and wheat height control [17]–[20]. Besides, a recent research showed that an AP2-like gene mutation results in maize internode length decrease [21]. Plant cytochrome P450 member could also modulate rice height by influencing cell elongation [22].
Plant height, an important agronomic trait, is vital in shaping plant architecture and then ultimately affecting crop yield. Dissection of molecular bases underlying plant height is beneficial to both basic and applied research. During the past, a large number of plant height QTL/genes have been identified (http://www.gramene.org/qtl/), which is helpful for elucidating the genetic mechanisms of plant height determinant. Of note, dwarf mutants are informative for the plant height research. Herein, we report physiological and transcriptomic analysis of a maize dominant dwarf plant, which was temporarily termed Dwarf11 (D11) in our work. Results presented here provide insights into the biological network of plant height control and pave the way for the following D11 gene cloning and functional characterization.
Results
Characterization of maize D11 mutant
Maize inbred line Mo17 was pollinated with maize inbred line HN06 to generate F1 progeny. In F1 population, the D11 mutant was identified. Due to abnormal inflorescence, it failed to self-pollinate D11. To preserve mutant phenotype, we continuously backcrossed D11 (as donor parent) with three maize inbred lines Mo17, B73, and W22. Finally, three advanced backcross populations BC6F1 with different nuclear background were developed.
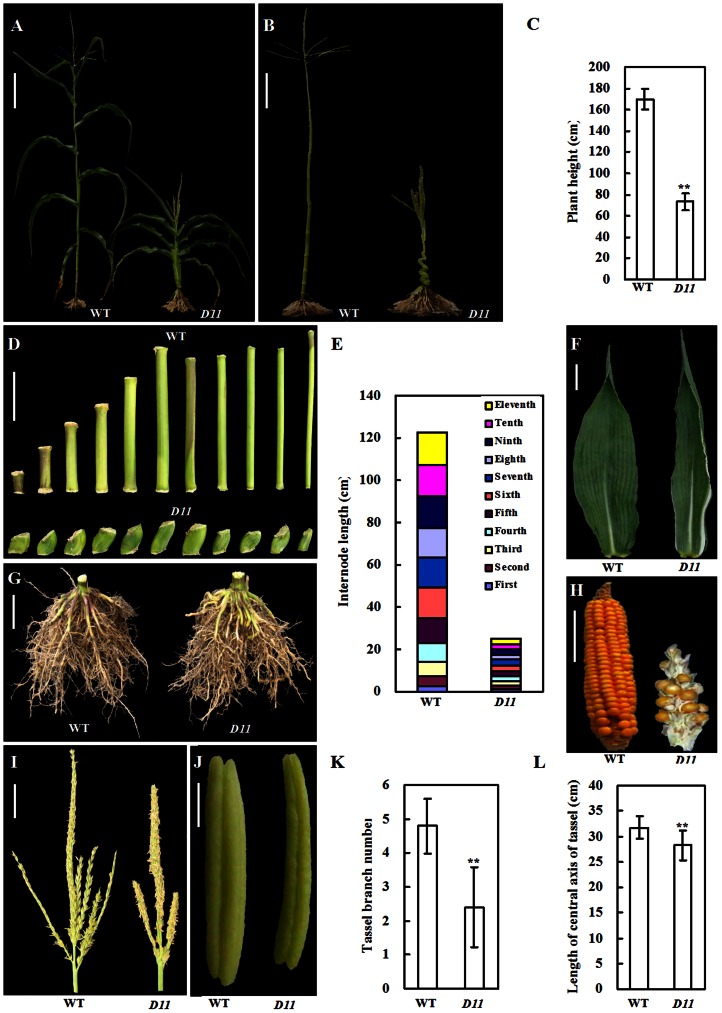
The D11 mutant displayed severely developmental abnormalities, such as shortened internodes, white leaf margins, multiple degenerated spikes. The D11 plant with Mo17 nuclear background was showed in Figure 1. The D11 mutant was about 73.5 cm tall, which was less than half of wild-type plant (169.8 cm) (Figures 1A–1C). Dwarf phenotype of D11 mainly attributed to shortened internodes, instead of internodes number decrease. Moreover, eleven internodes of D11 shortened evenly (Figures 1D and 1E). The D11 mutant had more leaves. Compared with wild-type plant, leaves of D11 were slender, dark green, and slightly-rolled. Additionally, leaves of D11 possessed white margins (Figures 1F and S1). Compared with wild-type, aerial roots of D11 were more sturdy (Figure 1G). As D11 displayed multiple degenerated spikes adjacent to the tassel, it was difficult to self-pollinate the mutant plant. In open-pollination conditions, some spikes were partly fertile, others were absolutely barren (Figure 1H). The fertility of D11 male inflorescence was normal, although anther size, branch number, and length of central spikes of D11 tassels were largely reduced (Figures 1I–1L). Besides, pollens shed from anthers in the central spikes of D11 were about two days earlier than shed in wild-type plant.
Figure 1. Gross morphology of maize D11 mutant.
(A) Phenotype of D11 and wild type (WT). Bar = 20 cm. (B) Whole plant. To get snapshot of internodes arrangement, leaves and spike were removed manually. Bar = 20 cm. (C) Plant height. (D) Internodes. Bar = 5 cm. (E) Internode length. (F) Leaf. Leaves of D11 are slender, dark green, slightly-rolled, and with white margins. Bar = 5 cm. (G) Roots. Aerial roots of D11 display more sturdy. Bar = 5 cm. (H) Spike. Spike of D11 degenerates severely. Bar = 5 cm. (I) Tassel. Bar = 5 cm. (J) Anther. Anthers of D11 are short and thin. Bar = 1 mm. (K) Tassel branch number. (L) Length of central axis of tassel. In figures (C), (E), (K), and (L), data are mean ±SD (n = 30). Double asterisks denote significant difference at P≤0.01 level compared with the wild type by Student's t test.
Genetic behavior of maize D11 gene
As the D11 mutant has abnormal female inflorescence, it was difficult to generate progeny derived from D11 by self-pollination. Thus, three advanced backcross populations were developed using D11 as donor parent. To investigate the genetic behavior of D11 gene, segregation of plant height in three backcross populations with different nuclear background (Mo17, B73, and W22) were analyzed in various years and regions. Results showed that the D11 mutant phenotype was stable in different years and growth conditions. Moreover, the D11 mutant phenotype appeared stably regardless of nuclear background. In backcross populations, the ratio of dwarf and normal height plants was 1∶1 which fit closely the expected segregation ratio. Progeny derived from self-pollination of normal height plants was all with normal height (Table S1). Evidence from segregation ratios in backcross and self-pollination populations, together with the phenomenon that D11 mutant phenotype was firstly observed in F1 generation, demonstrated that D11 inherits as a dominant Mendelian factor.
Sequence variation in maize D11 mutant
Two maize dominant dwarf genes D8 and D9 have been cloned. To test whether D11 is allelic to D8 or D9, we dissected sequence polymorphisms between wild-type and D11 plants. Two pairs of primers were designed on 5′ and 3′ untranslated regions (UTRs) of d8 and d9 (Table S2) and adopted to amplify wild-type and D11 templates (Figure S2). Sequencing of target PCR products showed that there were no sequence polymorphisms between wild-type and D11 plants, implying that D11 is not allelic to D8 and D9.
Response of maize D11 mutant to exogenous GA3 and PAC stimulation
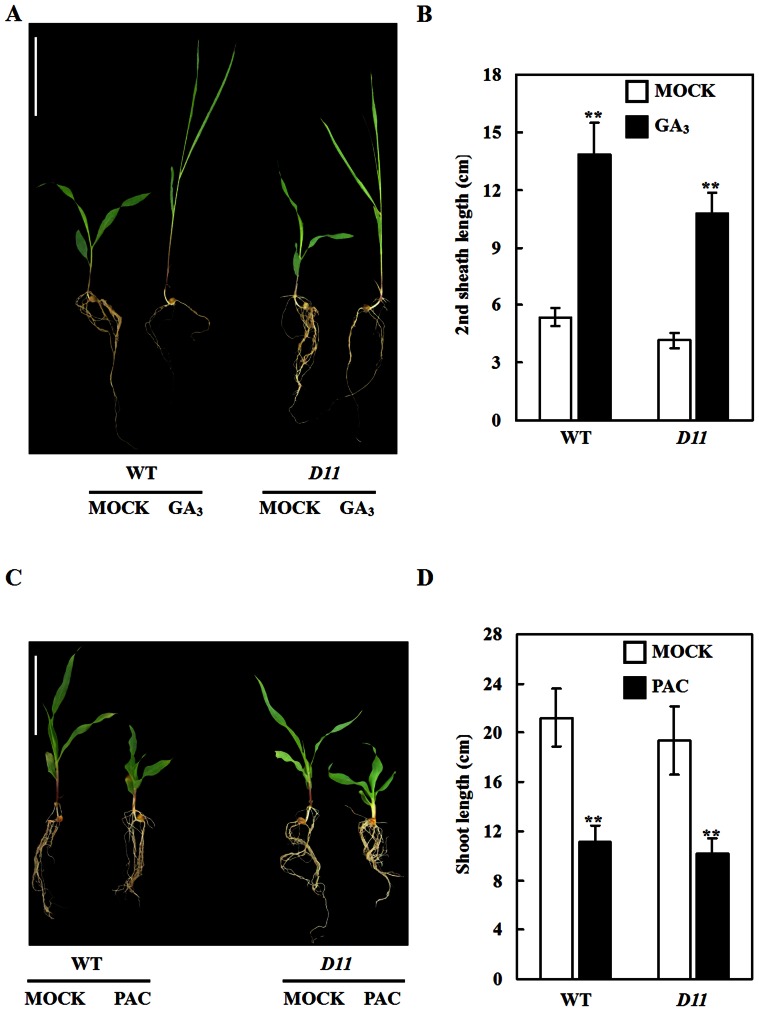
Two maize dominant dwarf plants D8 and D9 are all insensitive to exogenous GA3 application. We investigated responses of D11 to GA3 stimulation. When treated with a 10−4 M GA3 solution, shoot, coleoptile, the first leaf blade, the first leaf sheath, and the second leaf blade length were significantly increased in WT and D11 (Figures 2A, 2B, and S3). Moreover, GA biosynthesis inhibitor PAC was applied to WT and D11 seedlings. Results showed that shoot elongation in WT and D11 was significantly inhibited when treated with a 10−4 M PAC solution (Figures 2C and 2D). The D11 mutant responded to both GA3 and PAC stimulation, suggesting that dwarf phenotype of D11 is caused by GA biosynthesis instead of GA signaling deficiency.
Figure 2. Response of maize D11 mutant to GA3 and PAC application.
(A) Seedlings of WT and D11 when treated with a 10−4 M GA3 solution. Bar = 10 cm. (B) The second leaf sheath length of WT and D11 (n = 35) when treated with a 10−4 M GA3 solution. (C) Seedlings of WT and D11 when treated with a 10−4 M PAC solution. Bar = 10 cm. (D) Shoot length of WT and D11 (n = 40) when treated with a 10−4 M PAC solution. In figures (B) and (D), data are mean ±SD. Double asterisks indicate significant difference at P≤0.01 level compared with untreated samples by Student's t test.
To our knowledge, there are four maize dominant dwarf mutants (D8, D8-1023, D9, and Dt) which have been publicly reported. Features of these four maize dominant dwarf plants including D11 were described in Table 1.
Table 1. Snapshot of five maize dominant dwarf plants.
| Name | Leaf | Tillering | Floral organ | GA sensitivity | Chromosome | Gene cloning or not | Source |
| D8 | Narrow, dark green | √ | Andromonoecious | Insensitivity | 1 | √ | [9], [23] |
| D8-1023 | Broad, dark green, pursy | √ | Andromonoecious | Sensitivity | 1 | √ | [5] |
| D9 | Narrow, dark green | √ | Normal | Insensitivity | 5 | √ | [8], [23] |
| Dt | N.A. | N.A. | N.A. | Sensitivity | 10 | × | [24], [25] |
| D11 | Narrow, dark green, rolled | × | Abnormal | Sensitivity | 2 | × | [26], this study |
N.A. indicates not available.
Differentially expressed genes (DEGs) in response to D11 mutant phenotype
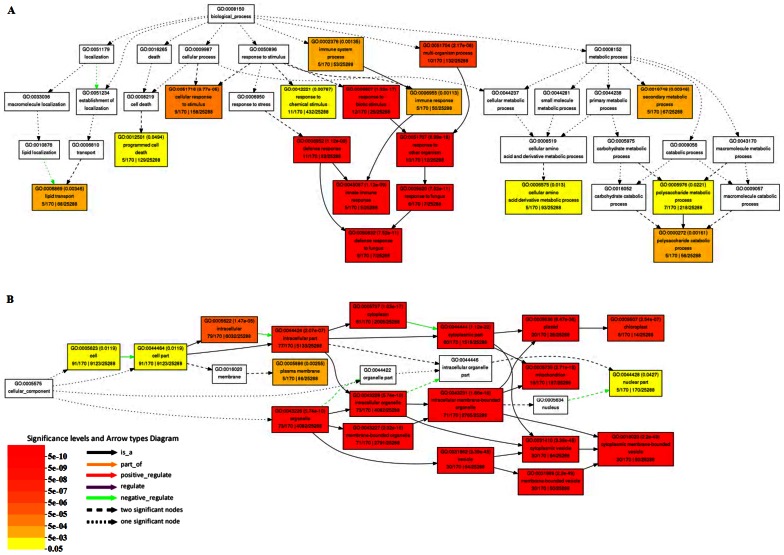
There were a total of 17,734 probes which could be detected both in WT and D11. In this study, only transcripts with fold change greater than five were considered as DEGs. Finally, 285 DEGs were identified, including 220 up-regulated genes and 65 down-regulated genes (Table S3). To investigate functional characterization of DEGs, up-regulated and down-regulated DEGs were separately subjected to GO enrichment analysis. Detailed information of GO clustering results was presented in Figure 3 and Figure S4.
Figure 3. GO clustering of up-regulated DEGs.
(A) GO enrichment analysis according to GO catalogue (GO:0008150 biological process). (B) GO enrichment analysis according to GO catalogue (GO:0005575 cellular component).
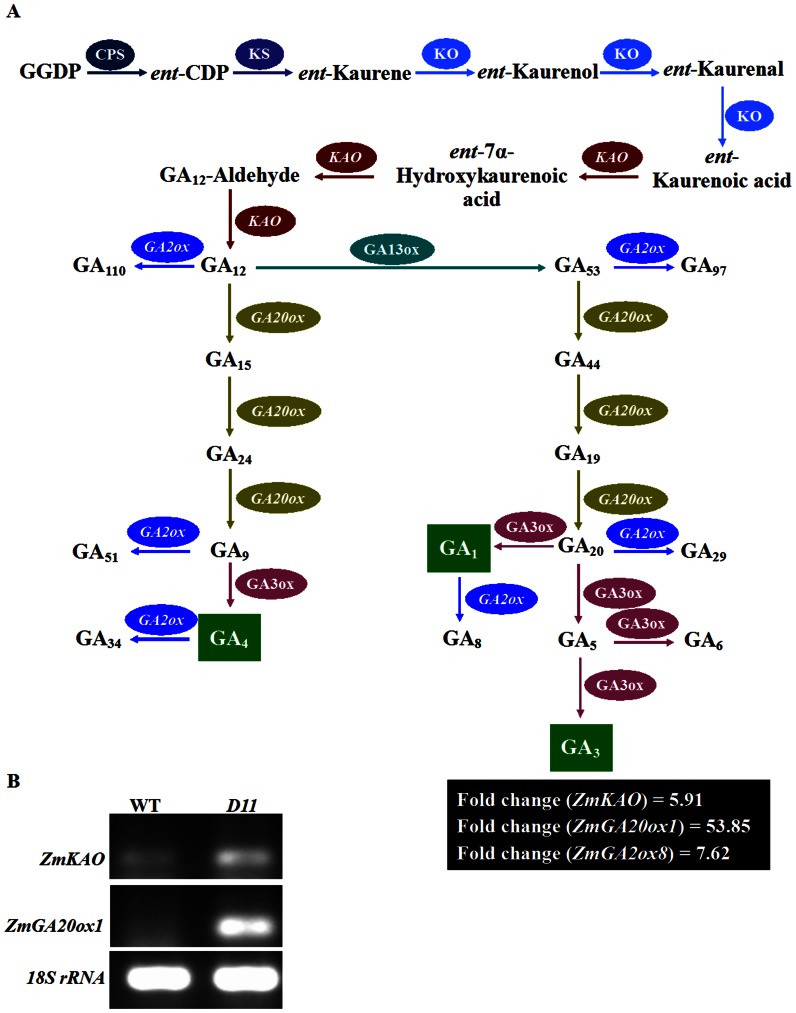
The D11 mutant is characteristic of dwarfism and responds to both GA3 and PAC stimulation. To this end, we specifically surveyed DEGs related to phytohormone GA biosynthesis and catabolism. Transcript encoding KAO which is responsible for precursor GA12 biosynthesis was up-regulated in D11. Expression of GA20ox1 which is required for the biosynthesis of active GAs from precursor GA12 was highly elevated in D11. Similarly, transcription of GA2ox8 which is involved in GA deactivation was relatively high in D11 (Figures 4A and 4B).
Figure 4. DEGs involved in GA biosynthesis and catabolism.
(A) GA biosynthesis and catabolism pathways were briefly diagramed. Transcripts encoding maize GA biosynthetic and catabolic enzymes ZmKAO, ZmGA20ox1, and ZmGA2ox8 are up-regulated in D11. (B) Semi-qRT-PCR validation of elevated transcripts ZmKAO and ZmGA20ox1. The 18S rRNA gene was used as an internal control.
Discussion
Possible relationship between plant hormone GA and D11 mutant phenotype
Plant hormones GA, BR, Aux together with other modulators form an elaborate network to finely trigger plant stature in nature [27]. Especially for phytohormone GA, there is ample evidence supporting its role in plant height control. Two well-characterized maize dominant dwarf plants D8 and D9 all belong to the GA-insensitive group, whose dwarf phenotype is caused by GA signaling instead of GA biosynthesis deficiency [6]. Contrarily, physiological analysis in our work demonstrated that maize dominant dwarf plant D11 responds to exogenous GA3 application and is a GA-sensitive mutant.
The GA metabolism pathway has been unraveled [28]. In this study, we investigated the expression patterns of GA metabolic enzyme genes in D11. Transcripts encoding GA metabolic enzymes KAO, GA20ox1, and GA2ox8 are all up-regulated in D11. Elevated transcripts of KAO and GA20ox1 were further validated by qRT-PCR method (Figure 4). Up-regulation of transcripts encoding GA metabolic enzymes in GA-deficient mutants has been reported previously. In Arabidopsis GA biosynthesis-deficient mutant dwarf and delayed-flowering 1 (ddf1), transcripts of three GA20ox genes AtGA20ox1, −2, and −3 are elevated [29]. Similarly, the expression levels of OsGA20ox2, −4, and OsGA2ox6 are increased in rice dominant dwarf and GA-deficient mutant H032 [30]. Up-regulation of transcripts encoding GA metabolic enzymes in GA-deficient mutants may be necessary for GA homeostasis throughout the life cycle of plants. Positive feedback mechanisms may account for this regulatory loop [29].
Plant hormone GA plays a vital role in flower organ development [31]. Systematic analysis of Arabidopsis GA20ox genes demonstrated that AtGA20ox1, −2, and −3 have significant effects on floral organ growth and anther development [32]. The D11 mutant displays severe inflorescence abnormalities, characteristic of slender tassel and degenerate spikes (Figures 1H–1L). Overdose of GA20ox1 gene revealed by microarray and qRT-PCR in our work (Figure 4) may be partly responsible for abnormal inflorescence development in D11. Undoubtedly, further physiological, genetic, and developmental data should be collected to test this hypothesis.
Phytohormone GA promotes chloroplast biogenesis through controlling leaf mesophyll cell expansion. In Arabidopsis and rice GA-deficient mutants, chloroplast division is largely inhibited. Transcript levels of genes related to chloroplast division are also significantly decreased in GA-deficient mutants [33]. Physiological and transcriptomic analysis in this study indicated that D11 has defects in GA metabolism. Additionally, D11 is with white leaf margins (Figures 1F and S1). The relationship between GA deficiency and albino phenotype of D11 need to be investigated in the future.
The D11 is a novel dominant dwarf mutant
Four maize dominant dwarf mutants, including D8, D8-1023, D9, and Dt, have been characterized (Table 1). The D8 and D9 all display narrow and dark green leaves. The D8-1023 is with broad, dark green, and pursy leaves. Leaves of D11 appear slender, dark green, and slightly-rolled. Remarkably, D11 possesses white leaf margins. Three maize dominant dwarf plants D8, D8-1023, and D9 show tillering ability. Floral organs of D9 are normal. Although D8 and D8-1023 display andromonoecious floral organs, they could be successfully self-pollinated. In contrast, D11 has severe defects in inflorescence development. Commonly, tassel of D11 is surrounded by multiple degenerate spikes which are almost barren. In extreme cases, female inflorescence dies aborning and no apparent ear is observed in D11. Thus, it is difficult to self-pollinate of D11. Compared with wild-type, D11 flowers earlier. Similar result was observed in D8 plant [9]. Intriguingly, maize transgenic experiment demonstrated that overexpression of D9-1 delays flowering, while overexpression of d9 promotes flowering [8]. Different dwarf genes may be recruited by distinct central modulators of complicated network to finely control flowering time in maize. Besides, D8 and D9 plants are insensitive to GA stimulation and belong to the GA-insensitive type [9], [23]. Conversely, D11 responds to exogenous GA stimulation (Figure 2). In this study, transcriptomic analysis provides further evidence of D11 as a GA-sensitive mutant (Figure 4).
Taken together, compared with other four maize dominant dwarf mutants (D8, D8-1023, D9, and Dt), D11 is a novel dominant dwarf mutant, characteristic of dwarf stature, slightly-rolled leaf with white margins, deformed inflorescence, and GA sensitivity. Further gene cloning and functional characterization are informative for dissecting D11-mediated network in the control of plant stature and other developmental programs.
Materials and Methods
Plant Materials and Traits Evaluation
Maize inbred line Mo17, as female plant, was crossed with maize inbred line HN06 with normal plant height and unknown pedigree to generate F1 progeny. In F1 population, the dwarf mutant D11 was identified. Then, D11 as donor parent was continuously backcrossed with three maize inbred lines Mo17, B73, and W22 to develop advanced backcross populations BC6F1 with different nuclear background.
All experimental materials were single-grain sowed and nursed with normal agricultural practice. Traits were evaluated with ten consecutive plants starting from the third plant in each row. Plant height was measured from the ground to tassel top. For leaf number count, the fifth, tenth, and fifteenth leaves were firstly marked in red paint. Leaf number was counted when the tassel achieved fixed-length. Leaf area was calculated with the following formula: Leaf area = Leaf length × leaf width ×0.75. Days to pollen shed were measured as number of days from sowing to emergence of the first pollen from anthers in the central axis of the tassel.
Sequence Polymorphisms Survey
Primers designed on 5′ and 3′ UTRs of d8 and d9 genes were used to amplify wild-type and D11 DNA templates. Target amplicons were purified from gel with the QIAquick® gel extraction kit (Qiagen, Germany) and ligated into pGEM-T vector (Promega, USA) for sequencing (BGI, China).
Physiological Characterization
Seeds of wild-type and D11 plants (n = 35) were surface sterilized with a 2% NaClO solution for 20 min, washed by distilled water for five times. For germination, sterilized seeds were placed on filter paper which was saturated with distilled water or 10−4 M GA3 (Sigma, USA) solution. Germinated seeds were transferred to roseite for growth. Seedlings of WT and D11 were sprayed with distilled water or 10−4 M GA3 solution per day. Ten days later, length of shoot, coleoptile, leaf blade, and leaf sheath was measured in WT and D11. For PAC treatment, sterilized seeds of wild-type and D11 (n = 40) were firstly soaked in a distilled water or 10−4 M PAC (Sigma, USA) solution for 24 h, then embedded in distilled water for 24 h. Treated seeds were transferred to roseite for growth. Fifteen days later, shoot length was determined in WT and D11.
Microarray Hybridization and DEGs Identification
The GeneChip® Maize Genome Array (Affymetrix, USA) was used for microarray analysis in this study. Firstly, total RNA was extracted from WT and D11 stems by TRIzol® Reagent (Invitrogen, USA), following the manufacturer's instructions. RNA integrity was assessed by the Agilent Bioanalyzer 2100 (Agilent technologies, USA). Qualified total RNA was further purified by RNeasy micro kit (QIAGEN, Germany) and RNase-Free DNase Set (QIAGEN, Germany). Then, qualified total RNA were amplified, labeled, and purified using GeneChip® 3′IVT Express Kit (Affymetrix, USA) with the guidance of the manufacturer's manual to obtain biotin labeled aRNA. Array hybridization and wash were implemented using GeneChip® Hybridization, Wash, and Stain Kit (Affymetrix, USA) in Hybridization Oven 645 (Affymetrix, USA) and Fluidics Station 450 (Affymetrix, USA) according to the manufacturer's instructions. Slides were scanned by GeneChip® Scanner 3000 (Affymetrix, USA).
Raw data were submitted to the Gene Expression Omnibus database (accession number: GSE46370). Raw data were normalized by MAS 5.0 algorithm implemented in the Gene Spring Software 11.0 (Agilent technologies, USA). Normalized data of D11 divided by those of WT obtained fold change. Genes whose transcripts were up-regulated or down-regulated at levels greater than a fivefold ratio were regards as DEGs in this study. Non-redundant DEGs were firstly annotated by the NCBI Entrez Gene resource (http://www.ncbi.nlm.nih.gov/gene). Then, DEGs were mapped on the B73 reference genome using the B73 sequencing database (http://www.maizesequence.org/index.html). Gene ontology (GO) enrichment analysis of DEGs was performed through the Gene Ontology (http://www.geneontology.org/) and agriGO (http://bioinfo.cau.edu.cn/agriGO/index.php) webservers [34].
qRT-PCR Validation of DEGs
Some DEGs involved in GA metabolism were validated by qRT-PCR technique. Purified RNA samples used for microarray analysis were firstly transcribed to obtain cDNA by M-MLV reverse transcriptase (Promega, USA). A total of 10 µl qRT-PCR reaction contained cDNA template, 1×PCR buffer, 200 µM dNTPs, 0.2 µM primers, 0.5 U Taq DNA polymerase (Invitrogen, USA), and appropriate amount of distilled water. The mixture was predenatured at 94°C for 2 min, followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 30 s. The final extension step was set at 72°C for 10 min. Primers reported by Song et al. [35] were used for qRT-PCR analysis.
Supporting Information
Characteristics of D11 leaf. (A) Phenotype of the first, second, and third leaves (from top to bottom) of D11 and wild type (WT). The D11 mutant has slender leaves with white margins. Bar = 5 cm. (B) Leaf number. The D11 mutant possesses more leaves. (C) Leaf length and width. Compared with wild-type, leaves of D11 are relatively long and narrow. (D) Ratio of leaf length to width. (E) Leaf area. Average values were calculated (n = 30). Data are mean ±SD. Single asterisk and double asterisks indicate significant difference at P≤0.05 and P≤0.01 levels compared with the wild type by Student's t test, respectively.
(TIF)
PCR products amplified by primers designed on UTRs of d8 and d9 . M: 1 kb marker; Lanes 1 and 2: amplicons from d8 UTRs primers; Lanes 3 and 4: amplicons from d9 UTRs primers.
(TIF)
Response of D11 to GA3 stimulation. (A) Shoot length. (B) Coleoptile length. (C) The first leaf blade length. (D) The first leaf sheath length. (E) The second leaf blade length. Data are mean ±SD (n = 35). Double asterisks denote significant difference at P≤0.01 level compared with untreated samples by Student's t test.
(TIF)
GO clustering of down-regulated DEGs.
(TIF)
Segregation in backcross and self-pollination populations from D11 and normal height plants.
(DOC)
Primers used in this study.
(DOC)
Detailed information of DEGs.
(XLS)
Acknowledgments
We thank Prof. Mingliang Xu for his valuable advices and assistance in experiments. And we also thank Prof. Jihua Tang and Dr. Xiaomin Lu for their help in material collection.
Funding Statement
This work was supported by grants from the National Natural Science Foundation of China (31201213), Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), and Innovative Foundation of Yangzhou University (2011CXJ046). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Khush GS (2001) Green revolution: the way forward. Nat Rev Genet 2: 815–822. [DOI] [PubMed] [Google Scholar]
- 2. Hedden P (2003) The genes of the Green Revolution. Trends Genet 19: 5–9. [DOI] [PubMed] [Google Scholar]
- 3. Emmerson RA (1912) The inheritance of certain “abnormalities” in maize. J Heredity os-8: 385–399. [Google Scholar]
- 4. Fujioka S, Yamane H, Spray CR, Gaskin P, Macmillan J, et al. (1988) Qualitative and quantitative analyses of gibberellins in vegetative shoots of normal, dwarf-1, dwarf-2, dwarf-3, and dwarf-5 seedlings of Zea mays L. Plant Physiol 88: 1367–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cassani E, Bertolini E, Badone FC, Landoni M, Gavina D, et al. (2009) Characterization of the first dominant dwarf maize mutant carrying a single amino acid insertion in the VHYNP domain of the dwarf8 gene. Mol Breeding 24: 375–385. [Google Scholar]
- 6. Winkler RG, Freeling M (1994) Physiological genetics of the dominant gibberellin-non responsive maize dwarf, Dwarf8 and Dwarf9 . Planta 193: 341–348. [Google Scholar]
- 7. Peng J, Richards DE, Hartley NM, Murphy GP, Devos KM, et al. (1999) ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 400: 256–261. [DOI] [PubMed] [Google Scholar]
- 8. Lawit SJ, Wych HM, Xu D, Kundu S, Tomes DT (2010) Maize DELLA proteins dwarf plant8 and dwarf plant9 as modulators of plant development. Plant Cell Physiol 51: 1854–1868. [DOI] [PubMed] [Google Scholar]
- 9. Thornsberry JM, Goodman MM, Doebley J, Kresovich S, Nielsen D, et al. (2001) Dwarf8 polymorphisms associate with variation in flowering time. Nat Genet 28: 286–289. [DOI] [PubMed] [Google Scholar]
- 10. Larsson SJ, Lipka AE, Buckler ES (2013) Lessons from dwarf8 on the strengths and weaknesses of structured association mapping. PLoS Genet 9: e1003246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Weng J, Xie C, Hao Z, Wang J, Liu C, et al. (2011) Genome-wide association study identifies candidate genes that affect plant height in Chinese elite maize (Zea mays L.) inbred lines. PLoS One 6: e29229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Teng F, Zhai L, Liu R, Bai W, Wang L, et al. (2013) ZmGA3ox2, a candidate gene for a major QTL, qPH 3.1, for plant height in maize. Plant J 73: 405–416. [DOI] [PubMed] [Google Scholar]
- 13. Chandler PM, Marion-Poll A, Ellis M, Gubler F (2002) Mutants at the Slender1 locus of barley cv Himalaya. Molecular and physiological characterization. Plant Physiol 129: 181–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ikeda A, Ueguchi-Tanaka M, Sonoda Y, Kitano H, Koshioka M, et al. (2001) slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8 . Plant Cell 13: 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Qi W, Sun F, Wang Q, Chen M, Huang Y, et al. (2011) Rice ethylene-response AP2/ERF factor OsEATB restricts internode elongation by down-regulating a gibberellin biosynthetic gene. Plant Physiol 157: 216–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Multani DS, Briggs SP, Chamberlin MA, Blakeslee JJ, Murphy AS, et al. (2003) Loss of an MDR transporter in compact stalks of maize br2 and sorghum dw3 mutants. Science 302: 81–84. [DOI] [PubMed] [Google Scholar]
- 17. Tong H, Jin Y, Liu W, Li F, Fang J, et al. (2009) DWARF AND LOW-TILLERING, a new member of the GRAS family, plays positive roles in brassinosteroid signaling in rice. Plant J 58: 803–816. [DOI] [PubMed] [Google Scholar]
- 18. Hartwig T, Chuck GS, Fujioka S, Klempien A, Weizbauer R, et al. (2011) Brassinosteroid control of sex determination in maize. Proc Natl Acad Sci USA 108: 19814–19819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Makarevitch I, Thompson A, Muehlbauer GJ, Springer NM (2012) Brd1 gene in maize encodes a brassinosteroid C-6 oxidase. PLoS One 7: e30798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gasperini D, Greenland A, Hedden P, Dreos R, Harwood W, et al. (2012) Genetic and physiological analysis of Rht8 in bread wheat: an alternative source of semi-dwarfism with a reduced sensitivity to brassinosteroids. J Exp Bot 63: 4419–4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jiang F, Guo M, Yang F, Duncan K, Jackson D, et al. (2012) Mutations in an AP2 transcription factor-like gene affect internode length and leaf shape in maize. PLoS One 7: e37040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ramamoorthy R, Jiang SY, Ramachandran S (2011) Oryza sativa cytochrome P450 family member OsCYP96B4 reduces plant height in a transcript dosage dependent manner. PLoS One 6: e28069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Harberd NP, Freeling M (1989) Genetics of dominant gibberellin-insensitive dwarfism in maize. Genetics 121: 827–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang S, Liu F, Liu B, Wang L, Dong S (2007) Discovery of a new dominant dwarf gene in maize and its preliminary study. J Maize Sci 15: 15–18. [Google Scholar]
- 25. Wang L, Ha L, Zhang S, Xu C, Liu B (2008) Identification and genetic analysis of a new dwarf mutant gene in maize. Acta Agriculturae Boreali-Sinica 23: 23–25. [Google Scholar]
- 26. Wang Y, Miao N, Shi Y, Deng D, Bian Y (2010) Genetic analysis of a dominant dwarf mutant in maize. Acta Agriculturae Boreali-Sinica 25: 90–93. [Google Scholar]
- 27. Wang Y, Li J (2008) Molecular basis of plant architecture. Annu Rev Plant Biol 59: 253–279. [DOI] [PubMed] [Google Scholar]
- 28. Yamaguchi S (2008) Gibberellin metabolism and its regulation. Annu Rev Plant Biol 59: 225–251. [DOI] [PubMed] [Google Scholar]
- 29. Magome H, Yamaguchi S, Hanada A, Kamiya Y, Oda K (2004) dwarf and delayed-flowering 1, a novel Arabidopsis mutant deficient in gibberellin biosynthesis because of overexpression of a putative AP2 transcription factor. Plant J 37: 720–729. [DOI] [PubMed] [Google Scholar]
- 30. Huang J, Tang D, Shen Y, Qin B, Hong L, et al. (2010) Activation of gibberellin 2-oxidase 6 decreases active gibberellin levels and creates a dominant semi-dwarf phenotype in rice (Oryza sativa L.). J Genet Genomics 37: 23–36. [DOI] [PubMed] [Google Scholar]
- 31. Plackett AR, Thomas SG, Wilson ZA, Hedden P (2011) Gibberellin control of stamen development: a fertile field. Trends Plant Sci 16: 568–578. [DOI] [PubMed] [Google Scholar]
- 32. Plackett AR, Powers SJ, Fernandez-Garcia N, Urbanova T, Takebayashi Y, et al. (2012) Analysis of the developmental roles of the Arabidopsis gibberellin 20-oxidases demonstrates that GA20ox1, −2, and −3 are the dominant paralogs. Plant Cell 24: 941–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jiang X, Li H, Wang T, Peng C, Wang H, et al. (2012) Gibberellin indirectly promotes chloroplast biogenesis as a means to maintain the chloroplast population of expanded cells. Plant J 72: 768–780. [DOI] [PubMed] [Google Scholar]
- 34. Du Z, Zhou X, Ling Y, Zhang Z, Su Z (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38: W64–W70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Song J, Guo B, Song F, Peng H, Yao Y, et al. (2011) Genome-wide identification of gibberellins metabolic enzyme genes and expression profiling analysis during seed germination in maize. Gene 482: 34–42. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Characteristics of D11 leaf. (A) Phenotype of the first, second, and third leaves (from top to bottom) of D11 and wild type (WT). The D11 mutant has slender leaves with white margins. Bar = 5 cm. (B) Leaf number. The D11 mutant possesses more leaves. (C) Leaf length and width. Compared with wild-type, leaves of D11 are relatively long and narrow. (D) Ratio of leaf length to width. (E) Leaf area. Average values were calculated (n = 30). Data are mean ±SD. Single asterisk and double asterisks indicate significant difference at P≤0.05 and P≤0.01 levels compared with the wild type by Student's t test, respectively.
(TIF)
PCR products amplified by primers designed on UTRs of d8 and d9 . M: 1 kb marker; Lanes 1 and 2: amplicons from d8 UTRs primers; Lanes 3 and 4: amplicons from d9 UTRs primers.
(TIF)
Response of D11 to GA3 stimulation. (A) Shoot length. (B) Coleoptile length. (C) The first leaf blade length. (D) The first leaf sheath length. (E) The second leaf blade length. Data are mean ±SD (n = 35). Double asterisks denote significant difference at P≤0.01 level compared with untreated samples by Student's t test.
(TIF)
GO clustering of down-regulated DEGs.
(TIF)
Segregation in backcross and self-pollination populations from D11 and normal height plants.
(DOC)
Primers used in this study.
(DOC)
Detailed information of DEGs.
(XLS)