Abstract
LEAFY COTYLEDON1 (LEC1) is a central regulator of seed development that plays a key role in controlling the maturation phase during which storage macromolecules accumulate and the embryo becomes tolerant of desiccation. We queried the genomes of seedless plants and identified a LEC1 homolog in the lycophyte, Selaginella moellendorffii , but not in the bryophyte, Physcomitrella patens . Genetic suppression experiments indicated that Selaginella LEC1 is the functional ortholog of Arabidopsis LEC1. Together, these results suggest that LEC1 originated at least 30 million years before the first seed plants appeared in the fossil record. The accumulation of Selaginella LEC1 RNA primarily in sexual and asexual reproductive structures suggests its involvement in cellular processes similar to those that occur during the maturation phase of seed development.
Introduction
The seed is a complex structure that is comprised of three distinct regions: embryo, endosperm, and seed coat. Seed development can be divided conceptually into two temporal phases. During the early morphogenesis phase, the single-celled zygote develops into a multicellular embryo with a shoot-root axis and the embryonic tissue and organ systems [1]. The endosperm develops initially as a syncytium in which nuclei become regionalized to form distinct morphological domains, and it later cellularizes [2]. It is also during this period that ovule integuments differentiate into the seed coat [3]. The maturation phase follows morphogenesis and is characterized by the accumulation of storage macromolecules, such as lipids and proteins, within the embryo and endosperm. The embryo also becomes tolerant of the stresses of desiccation that occur during seed drying [4,5]. At the end of maturation, the embryo and endosperm are quiescent metabolically and arrested developmentally. Under permissive conditions, the mature seed germinates, and growth and differentiation of the seedling begins.
Seed plants constitute the largest and most species-rich group of extant land plants. However, the evolutionary origin of the seed 385 to 365 million years ago (mya) [6] is poorly understood, because early seed plants radiated rapidly and the earliest seed plant lineages are known only from the fossil record [7–9]. Evolution of the seed habit required a number of developmental innovations, including the advent of heterospory, retention and encasement of the megasporangium in maternally-derived integuments to form the ovule, and incorporation of the maturation phase into sporophyte development [10]. The maturation phase represents an intrusive pause in the morphogenesis of the sporophyte that enables the mature embryo to remain quiescent until conditions are favorable for germination [4,5]. By contrast, the life cycle of basal, non-seed bearing land plants is uninterrupted. Despite the importance of the maturation phase, little is known of how it has become integrated into the seed plant life cycle.
LEAFY COTYLEDON1 (LEC1) is a central regulator of seed development with a critical role in regulating the maturation phase (reviewed in 11–13). Loss-of-function mutations in LEC1 cause defects in storage macromolecule accumulation and the loss of desiccation tolerance, whereas ectopic expression of LEC1 confers embryonic characteristics to seedlings, including the expression of genes encoding storage macromolecules [14–16]. LEC1 is a novel HAP3 (NF–YB) subunit of the CCAAT-binding (NF–Y) transcription factor [14]. The HAP3 gene family has undergone a dramatic radiation and specialization in land plants as compared with yeast and animals that have single HAP3 genes [17–20]. Arabidopsis possesses 13 HAP3 subunits that can be grouped broadly into two classes, the LEC1-type and the non-LEC1-type, based on phylogenetic and functional criteria [19,21]. HAP3 subunits share a highly conserved, central B domain that is flanked by divergent A and C regions. LEC1 and another LEC1-type HAP3 subunit, LEC1-LIKE (L1L), share 16 signature B-domain amino acid residues that differ from residues conserved in non-LEC1-type HAP3 subunits. Moreover, overexpression of LEC1 and L1L confers embryonic characteristics to vegetatively growing plants, whereas ectopic expression of non-LEC1-type HAP3 genes does not induce these traits [19]. Thus, LEC1 represents a functionally specialized HAP3 subunit required for the maturation phase.
Because of the radiation and functional specialization of HAP3 subunits and the central role of LEC1 in seed development, we investigated the origin of LEC1 in basal land plants. We examined two lower plant lineages, Physcomitrella patens and Selaginella moellendorffii , representing bryophytes and lycophytes, respectively, to identify the most basal LEC1-type HAP3 subunit and to obtain clues about its role in the evolution of the seed.
Materials and Methods
Adult Selaginella moellendorffii plants were purchased (Plant Delights Nursery Inc., Raleigh, NC) or grown from bulbils (generously provided by Jo Ann Banks, Purdue University). Bulbils were surface sterilized (2 min in 70% ethanol, 5 min in 20% bleach, followed by five rinses with sterile water) and grown on one-half strength MS salts agar medium [22] in growth chambers under 56 µmol m-2 s-1, 16 hour light, 8 hour dark at 21°C until they had produced a shoot and root system. At this point, they were transferred to soil and grown in the greenhouse at approximately 26°C, 80% relative humidity, and 57 µmol m-2 s-1 light intensity.
Total RNA was isolated from S . moellendorffii organs with RNEasy Mini-Kit (Qiagen) and treated twice with DNase (DNA-free kit, Ambion). First-strand cDNA was synthesized with Thermoscript reverse transcriptase (Invitrogen) and used as a template for cloning and qRT-PCR.
DNA sequences for HAP3 genes were originally identified with BLAST searches against EST databases and trace archives of genomic DNA sequences from the moss, Physcomitrella patens (http://genome.jgi-psf.org/euk_cur1.html), and the lycophyte, S . moellendorffii (http://selaginella.genomics.purdue.edu/), to find ancient homologs of LEC1. The DNA sequences were confirmed with current genome assemblies (www.phyotozome.net) [23,24]. Coordinates for putative S . moellendorffii and P . patens HAP3 genes are given in Table S1 along with corresponding cDNA accession number. Because the S . mollendorffii genome contains two nearly identical haplotypes (98.5% identical at the nucleotide level) [23] that have not been organized into chromosome-scale pseudomolecules, we checked ~100 kb of genome sequence flanking HAP3 candidate loci for synteny using BLAST2. This analysis allowed us to identify and remove duplicate loci corresponding to the same chromosome position from the two haplotypes (Figure S1). Only unique S . mollendorffii loci were retained for analysis. Protein sequences for Saccharomyces cerevisiae, Arabidopsis thaliana, S. mollendorffii and P . patens HAP3 subunits were aligned using CLUSTALX [25] and trimmed to the B domains. PHYLIP [26] was used to construct maximum parsimony trees with 1000 bootstrap replicates, and the consensus majority rule tree is displayed. The alignment was formatted using BoxShade (http://www.ch.embnet.org/software/BOX_form.html).
SmLEC1 cDNA was amplified using 5' and 3' SMART-RACE (Clontech). Nested 3' RACE primers were as follows: 5'-CGAGTTCATCAGCTTCATCACCAG-3', 5'-CTCTCGCTCTTCCTCCACAAGTACC-3', 5'-GTACTTGTGGAGGAAGAGCGAGAGC-3' and 5'-CTGGTGATGAAGCTGATGAACTCG-3'. cDNA products were cloned into pCR-2.1 and sequenced.
The SmLEC1 cDNA clone was inserted into the LEC1 expression cassette [19]. This construct was transformed into Agrobacterium tumefaciens strain GV3101 and transferred into both wild-type Ws-0 and lec1-1 mutant Arabidopsis plants. Suppression of the lec1 mutation was assessed by the ability of seeds to germinate [14], and genotypes were confirmed by PCR amplification.
Quantitative RT-PCR was done essentially as described previously [27]. Primers for the control RNA, S . moellendorffii ubiquitin EST (DN839032) were: Sm_Ubi_1F 5'-ATACCATCGGCGATTTGAAG-3',
Sm_Ubi_1R 5'-CGCTTACAAGGAAAGCACCT-3'. Gene-specific primers for SmLEC1 were: Sm_B_Contig_2F 5'-CAGGACCGCTTCATGCCCAT-3' Sm_B_Contig_2R 5'-GGGTCGGCGTAATCGTCGAA-3'. The statistical significance of mRNA level differences was determined by subjecting all pairwise comparisons of delta-Ct values to Bonferroni-corrected Student’s t-tests [28].
To prepare thin histological sections, bulbils were fixed in triple fixative [29] and dehydrated using a graded water/ethanol series. The samples were then put through a graded ethanol/xylenes series and embedded in paraffin. Bulbils were sectioned at 7 µm on a rotary microtome, mounted on slides, deparaffinized with xylenes and stained with toluidine blue.
Staining for lipid contents was performed using fresh plant material sectioned at ~30 µm using a vibratome. The slides containing sections were then transferred to 50% ethanol for a few seconds and stained with 0.07% Sudan Black B in 70% ethanol for five minutes. Excess stain was removed by immersing slides in 50% ethanol for 1 minute, followed by mounting in glycerol and imaging.
Results
We identified several DNA sequences corresponding to the B domain of HAP3 subunits in both S . moellendorffii and P . patens (see Materials and Methods). As shown in Figure 1, phylogenetic analysis of these HAP3 B domain amino acid sequences from Arabidopsis and S. cerevisiae suggested that S . moellendorffii and P . patens possessed non-LEC1-type HAP3 subunits, although much of the phylogeny of the non-LEC1-type HAP3 subunits was not fully resolved. By contrast, a LEC1-type HAP3 subunit that grouped in a well-supported, monophyletic clade with Arabidopsis LEC1 and L1L was detected only S . moellendorffii and not P . patens . Thus, a LEC1-type HAP3 subunit was detected in S . moellendorffii but not in P . patens .
Figure 1. Phylogenetic tree based on the B domains of HAP3 subunits from yeast, Arabidopsis , S. moellendorffii, and P . patens .
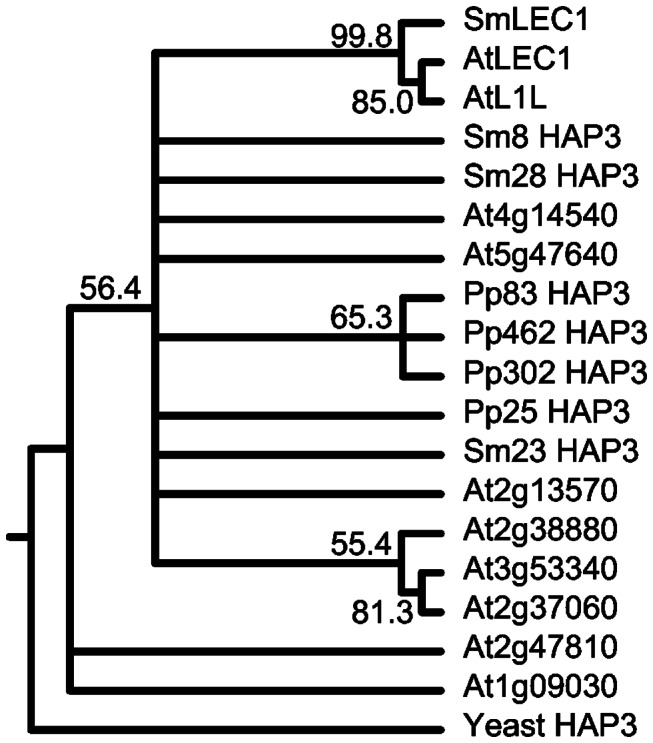
Nodes with bootstrap values below 50 were collapsed.
A single gene, designated SmLEC1, encoding the S . moellendorffii LEC1-type HAP3 subunit was identified. We showed that the gene was expressed by cloning its corresponding cDNA. The putative SmLEC1 polypeptide consists of 175 amino acids, with a 90-residue B domain that shares 83% amino acid identity with the Arabidopsis LEC1 B domain (Figure 2). More significantly, 12 of the 16 signature amino acid residues characteristic of angiosperm LEC1-type B domains [19] were conserved in SmLEC1, including the essential Asp residue at position 28 that is required for LEC1 function [21]. The A and C regions of SmLEC1, flanking the LEC1-type B domain, were not conserved between S . moellendorffii and Arabidopsis , consistent with the reports that Arabidopsis LEC1 and L1L do not share similarity in A and C regions [19] and that the B domain is responsible for specifying LEC1-type function [21].
Figure 2. Amino acid sequence alignment of the B domains of yeast, Arabidopsis and putative S . moellendorffii and P . patens HAP3 subunits.

Positions of residues diagnostic for the LEC1-type B domain are marked with arrows. Red arrow indicates the key Asp residue required for LEC1 activity. Grey and black arrows, respectively, show residues that are different or identical between S . moellendorffii and Arabidopsis LEC1-type HAP3 subunits. Amino acids identical to the consensus (>50%) sequence are shown with a black background, whereas similar amino acids are shown with a grey background.
To determine if SmLEC1 and Arabidopsis LEC1 have orthologous functions, we conducted genetic suppression assays using the Arabidopsis lec1-1 mutant. lec1-1 mutant embryos are intolerant of seed desiccation, and mutant seeds do not germinate after drying [16,30]. The ability of transgenic mutant seeds to germinate is a indication that the transgene can genetically suppress the lec1 mutation [14]. We fused the SmLEC1 cDNA clone with Arabidopsis DNA sequences 5' and 3' of the LEC1 protein coding region [19] and transferred the transgene into lec1-1 mutants. lec1-1 mutant plants transformed with SmLEC1, Arabidopsis LEC1, and an empty vector control vector generated 0.73% (51 of 7,015), 0.40% (56 of 13,950), and 0.062% (5 of 8,064) viable seedlings, respectively. The germination percentages reflected both the transformation efficiency and ability of the transgenes to suppress the lec1 mutation. This result suggests strongly that SmLEC1 is orthologous and functionally conserved with Arabidopsis LEC1.
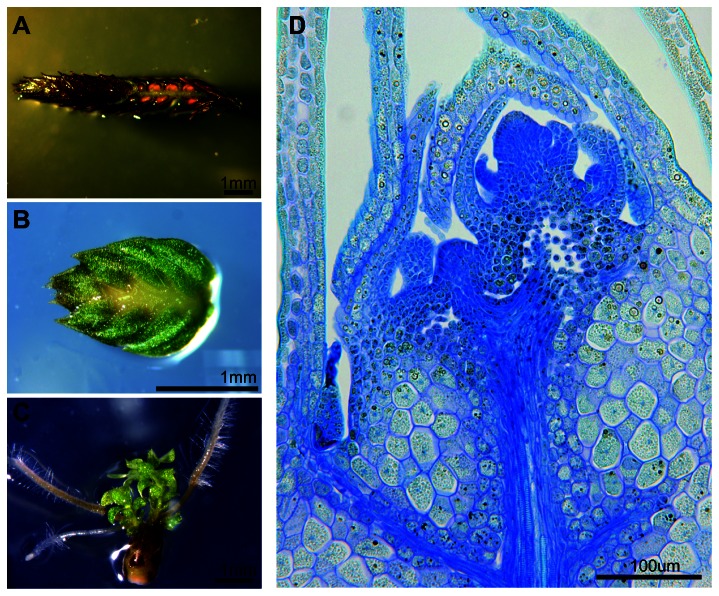
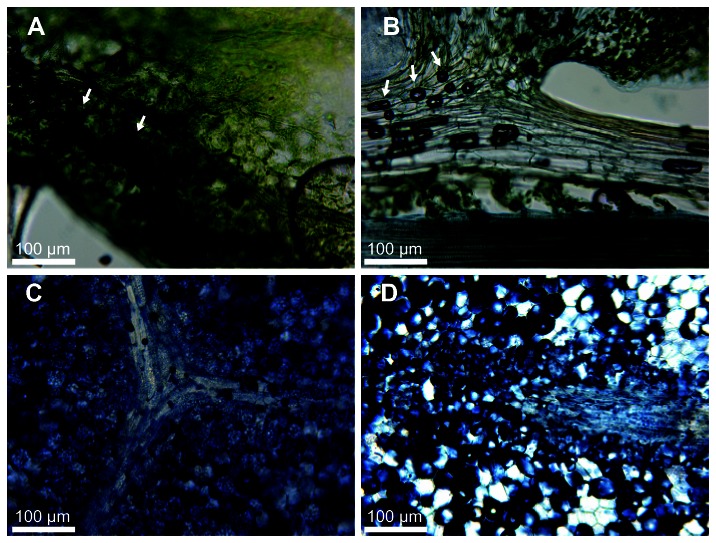
To obtain insight into the role of LEC1 in a basal land plant, we analyzed SmLEC1 RNA levels in S . moellendorffii shoots, shoots with apices removed to avoid incipient bulbils, rhizomes, roots, bulbils and strobili. Figure 3 shows that SmLEC1 RNA was present at highest levels in strobili and bulbils. Strobili, as shown in Figure 4A, contain both the female and male reproductive organs, megasporangia and microsporangia, that are located in the axils of microphylls, although microsporangia are far more abundant [31]. Bulbils are a loose class of vegetative propagules that are produced from various organs. S . moellendorffii bulbils are enlarged shoot apices that have ceased elongation and microphyll production (Figure 4B). They are quiescent developmentally and can be dispersed after being detached from the plant, retaining viability for approximately one year. Under appropriate conditions, bulbils can regenerate the adult plant through reactivation of shoot growth at the bulbil tip along with root formation from angle meristems (Figure 4C) [32]. Figure 4D shows that bulbils possessed enlarged cells reminiscent of those containing storage macromolecules. Consistent with this interpretation, we showed that sections through bulbils stained strongly for lipids (Figure 5D) similar to those obtained from an oil-storing angiosperm seed (Figure 5C). By contrast, sections from stems and leaves of S . moellendorffii and angiosperm leaves (Figure 5B and 5A, respectively) did not stain substantially for lipids. We conclude that S . moellendorffii bulbils expressed LEC1-type genes and accumulated lipids, similar to what occurs in seeds.
Figure 3. SmLEC1 is expressed in vegetative and reproductive propagules.
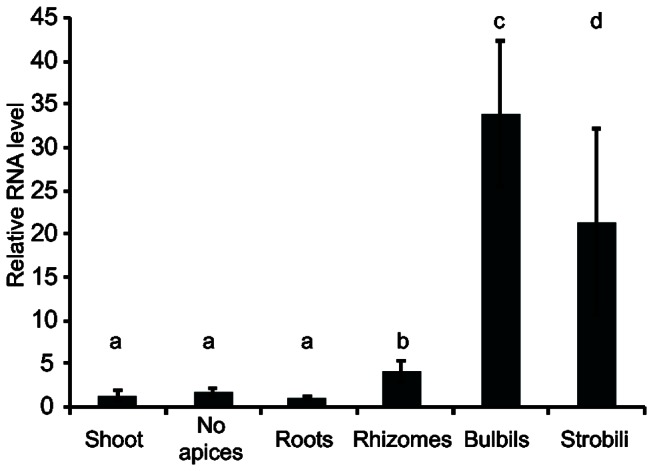
qRT-PCR survey of SmLEC1 mRNA levels in various S . moellendorffii organs. No apices are shoot samples with growing tip removed to avoid sampling incipient propagules. Relative quantification of RNA levels was calculated using ubiquitin as a control. Error bars indicate standard deviations. The following comparisons were determined to be significantly different (p < 0.05): a vs c, b vs c, and a vs d.
Figure 4. Vegetative and reproductive propagules of S . moellendorffii .
(A) Strobilus with basal microphylls removed to reveal axillary sporangia. (B) Mature, dormant bulbil. (C) Mature bulbil showing new shoot and root growth from apical and angle meristems respectively. (D) Longitudinal section of dormant bulbil stained with toluidine blue. showing the shoot apex and enlarged cells in the main body of the bulbil.
Figure 5. Lipid storage in bulbils.
Vibratome-cut sections were stained with Sudan Black B to identify cells with neutral lipids. (A) Section through an Arabidopsis leaf with a low storage lipid content. (B) Stem and leaf section from S . moellendorffii . (C) Section through a pumpkin oilseed with high storage lipid content. (D) S . moellendorffii bulbil section. Stained lipids appears as dark blue or black. Air bubbles in A and B (arrows) appear dark and do not indicate staining. The images are representative of material from at least three different plants.
Discussion
Our results demonstrate that a gene encoding a functional LEC1-type HAP3 subunit is present in a lycophyte but is not detected in a bryophyte. These findings place the origin of LEC1 at approximately 400 to 430 mya [33] which is approximately 20 to 30 million years before the appearance of the first seed plants. Although the sequence of the P . patens genome is largely complete [24], we cannot formally exclude the possibility that the LEC1 gene is present in an unsequenced region of the P . patens genome. It is also possible that a LEC1-type gene is present in other bryophytes and its absence in P . patens is due to a lineage-specific loss. Similar results have been reported by Xie et al. [34] who identified LEC1-type HAP3 genes in other lycophyte and fern genomes. The mode of origin of the LEC1-type B domain, however, remains uncertain. The sudden appearance of the LEC1-type gene in evolutionary time and a lack of evidence of recent duplication among other HAP3 genes make it unlikely that the appearance of LEC1 was the result of genome duplication. Others have postulated that the absence of introns in SmLEC1 relative to another class of basal land plant HAP3 genes may support an origin via a retrotransposition event [34].
Because LEC1-type HAP3 genes appeared prior to the origin of seed plants, we were interested to learn their physiological roles in Sellaginella and whether these roles relate to LEC1 function in seed development. In higher plants, LEC1 is a major regulator of the maturation phase, a developmental innovation critical for the seed habit [15,16]. For example, overexpression of LEC1 causes the induction of genes encoding storage proteins and fatty acid biosynthetic enzymes and the accumulation of fatty acids [14,35,36]. Features of the maturation phase that are critical for the seed habit include the acquisition of desiccation tolerance that permits the embryo to withstand the stresses of drying and become metabolically quiescent and the accumulation of macromolecular reserves, such as storage proteins and lipids that are used as a nutrient source by the developing embryo and/or seedling. Thus, integration of the maturation phase into the sporophytic generation enables a period of developmental and metabolic quiescence in which the embryo remains arrested until conditions are appropriate for subsequent vegetative and reproductive growth [4,5].
Lycophytes are basal land plants that represent the first extant group that diverged after the lineages that make up the bryophyte grade. They are the earliest diverging vascular plant lineage possessing a dominant sporophytic generation and true vasculature. Although some lycophyte genera, including Selaginella , are heterosporous, these instances of heterospory were derived independently from that of seed plants [37]. Because lycophytes are seedless plants that have not integrated the maturation phase into their life cycles, the LEC1-type gene must have evolved in lycophytes to fulfill other physiological processes.
Two sets of physiological processes that characterize lycophytes and some other basal land plants overlap with those that occur in maturation-phase seeds. First, several structures, such as the bulbils of lycophytes and monilophytes and gemmae of bryophytes, serve as specialized propagules that accumulate storage macromolecules [38,39]. For example, we showed that S . molenlendorffii bulbils possess substantial quantities of lipid (Figure 5). Second, lycophytes acquire the ability to withstand desiccation at specialized developmental stages or environmental conditions. Spores, particularly the megaspores of heterosporous plants such as S . moellendorffii , undergo desiccation prior to giving rise to viable gametophytes [40]. Furthermore, gametophytes and sporophytes of several lower plants, including certain species of Selaginella known as resurrection plants, desiccate and become quiescent upon drying, yet they remain viable [41].
Based on the finding that SmLEC1 RNA is present at high levels in strobili and bulbils and at low levels elsewhere in the plant (Figure 3), we hypothesize that SmLEC1 may function to induce storage lipid accumulation in S . mollendorffi bulbils and confer desiccation tolerance to megaspores. The findings that ectopic expression of Arabidopsis LEC1 induces storage lipid accumulation in vegetative Arabidopsis organs [35] and that LEC1-type HAP3 RNA levels are upregulated in desiccating foliage leaves of the resurrection plant, Selaginella sinensis [34] are consistent with this hypothesis. A role for SmLEC1 in the accumulation of storage reserves and the acquisition of desiccation tolerance would suggest that LEC1 was coopted in angiosperms for its role in seed development. Alternatively, because RNA encoding a LEC1-type HAP3 subunit is detected in S . mollendorffii strobili and pine pollen cones [19], LEC1-type HAP3 subunits may have evolved with an original role in gametophyte development. While functional characterization of SmLEC1 in S . moellendorffii remains difficult in the absence of a gene transfer system, further exploration of the role of the LEC1-type HAP3 subunits in other basal land plants should provide insight into ancient processes regulated by LEC1 that were coopted for seed development.
Supporting Information
Identification of S. moellendorffii HAP3 haplotypes.
The sequenced S . moellendorffii genome contains two haplotypes. Approximately 100 kb regions surrounding HAP3 alleles were compared to identify haplotypes. Dot Matrix View of BLAST2 alignments show exceptionally high sequence alignment (slanted lines) of HAP3 alleles, indicating that the pairs occur in nearly identical scaffolds and not likely to be distinct loci.
(EPS)
Genomic locations of all putative genes encoding HAP3 subunits identified in this study.
Scaffold and coordinate identifiers correspond to the Physcomitrella patens 1.6 genome release and the Selaginella moellendorffii 1.0 genome release.
(DOCX)
Acknowledgments
We thank Bob Goldberg for his thoughtful comments about the manuscript.
Funding Statement
Department of Energy http://energy.gov/ DE-FG02-03ER15392 and National Science Foundation http://www.nsf.gov/ IOS-1027494 The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript
References
- 1. Capron A, Chatfield S, Provart N, Berleth T (2009) Embryogenesis: pattern formation from a single cell. Arabidopsis Book 7: e0126 . PubMed: 22303250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Li J, Berger F (2012) Endosperm: food for humankind and fodder for scientific discoveries. New Phytol 195: 290-305 . doi:10.1111/j.1469-8137.2012.04182.x. PubMed: 22642307. [DOI] [PubMed] [Google Scholar]
- 3. Haughn G, Chaudhury A (2005) Genetic analysis of seed coat development in Arabidopsis. Trends Plant Sci 10: 472-477 . doi:10.1016/j.tplants.2005.08.005. PubMed: 16153880. [DOI] [PubMed] [Google Scholar]
- 4. Harada JJ (1997) Seed maturation and control of germination. In: Larkins BA, Vasi IK. Advances in Cellular and Molecular Biology of Plants, Volume 4, Cellular and Molecular Biology of Seed Development. Dordrecht: Kluwer Publishing House Academic Publishers; pp. 545-592. [Google Scholar]
- 5. Vicente-Carbajosa J, Carbonero P (2005) Seed maturation: developing an intrusive phase to accomplish a quiescent state. Int J Dev Biol 49: 645-651 . doi:10.1387/ijdb.052046jc. PubMed: 16096971. [DOI] [PubMed] [Google Scholar]
- 6. Gerrienne P, Meyer-Berthaud B, Fairon-Demaret M, Streel M, Steemans P (2004) Runcaria, a middle Devonian seed plant precursor. Science 306: 856-858 . doi:10.1126/science.1102491. PubMed: 15514154. [DOI] [PubMed] [Google Scholar]
- 7. Dimichele WA, Davis JI, Olmstead RG (1989) Origins of heterospory and the seed habit the role of heterochrony. Taxon 38: 1-11 . doi:10.2307/1220881. [Google Scholar]
- 8. Hilton J, Edwards D (1996) A new Late Devonian acupulate preovule from the Taff Gorge, South Wales. Rev Paleobot Palynol 93: 235-252 . doi:10.1016/0034-6667(95)00128-X. [Google Scholar]
- 9. Rothwell GW, Scheckler SE, Gillespie WH (1989) Elkinsia new-genus a late devonian gymnosperm with cupulate ovules. Bot Gaz 150: 170-189 . doi:10.1086/337763. [Google Scholar]
- 10. Steeves TA (1983) The evolution and biological significance of seeds. Can J Bot 61: 3550-3560 . doi:10.1139/b83-404. [Google Scholar]
- 11. Braybrook SA, Harada JJ (2008) LECs go crazy in embryo development. Trends Plant Sci 13: 624-630 . doi:10.1016/j.tplants.2008.09.008. PubMed: 19010711. [DOI] [PubMed] [Google Scholar]
- 12. Gutierrez L, Van Wuytswinkel O, Castelain M, Bellini C (2007) Combined networks regulating seed maturation. Trends Plant Sci 12: 294-300 . doi:10.1016/j.tplants.2007.06.003. PubMed: 17588801. [DOI] [PubMed] [Google Scholar]
- 13. Santos-Mendoza M, Dubreucq B, Baud S, Parcy F, Caboche M et al. (2008) Deciphering gene regulatory networks that control seed development and maturation in Arabidopsis. Plant J 54: 608-620 . doi:10.1111/j.1365-313X.2008.03461.x. PubMed: 18476867. [DOI] [PubMed] [Google Scholar]
- 14. Lotan T, Ohto M, Yee KM, West, MAL, Lo R, et al. (1998) Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93: 1195-1205. [DOI] [PubMed] [Google Scholar]
- 15. Meinke DW, Franzmann LH, Nickle TC, Yeung EC (1994) Leafy Cotyledon Mutants of Arabidopsis. Plant Cell 6: 1049-1064 . doi:10.2307/3869884. PubMed: 12244265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. West M, Yee KM, Danao J, Zimmerman JL, Fischer RL et al. (1994) LEAFY COTYLEDON1 Is an Essential Regulator of Late Embryogenesis and Cotyledon Identity in Arabidopsis. Plant Cell 6: 1731-1745 . doi:10.1105/tpc.6.12.1731. PubMed: 12244233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Edwards D, Murray JAH, Smith AG (1998) Multiple genes encoding the conserved CCAAT-box transcription factor complex are expressed in Arabidopsis. Plant Physiol 117: 1015-1022 . doi:10.1104/pp.117.3.1015. PubMed: 9662544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gusmaroli G, Tonelli C, Mantovani R (2001) Regulation of the CCAAT-Binding NF-Y subunits in Arabidopsis thaliana. Gene 264: 173-185 . doi:10.1016/S0378-1119(01)00323-7. PubMed: 11250072. [DOI] [PubMed] [Google Scholar]
- 19. Kwong RW, Bui AQ, Lee H, Kwong LW, Fischer RL et al. (2003) LEAFY COTYLEDON1-LIKE defines a class of regulators essential for embryo development. Plant Cell 15: 5-18 . doi:10.1105/tpc.006973. PubMed: 12509518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Siefers N, Dang KK, Kumimoto RW, Bynum WE, Tayrose G et al. (2009) Tissue-Specific Expression Patterns of Arabidopsis NF-Y Transcription Factors Suggest Potential for Extensive Combinatorial Complexity. Plant Physiol 149: 625-641 . PubMed: 19019982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee H, Fischer RL, Goldberg RB, Harada JJ (2003) Arabidopsis LEAFY COTYLEDON1 represents a functionally specialized subunit of the CCAAT binding transcription factor. Proc Natl Acad Sci U S A 100: 2152-2156 . doi:10.1073/pnas.0437909100. PubMed: 12578989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Olsen LJ, Ettinger WF, Damsz B, Matsudaira K, Webb MA et al. (1993) Targeting of glyoxysomal proteins to peroxisomes in leaves and roots of a higher plant. Plant Cell 5: 941-952 . doi:10.2307/3869662. PubMed: 8400872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Banks JA, Nishiyama T, Hasebe M, Bowman JL, Gribskov M et al. (2011) The Selaginella Genome Identifies Genetic Changes Associated with the Evolution of Vascular Plants. Science 332: 960-963 . doi:10.1126/science.1203810. PubMed: 21551031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A et al. (2008) The Physcomitrella Genome Reveals Evolutionary Insights into the Conquest of Land by Plants. Science 319: 64-69 . doi:10.1126/science.1150646. PubMed: 18079367. [DOI] [PubMed] [Google Scholar]
- 25. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876-4882 . doi:10.1093/nar/25.24.4876. PubMed: 9396791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Felsenstein J (1989) PHYLIP - Phylogeny Inference Package (Version 3.2). Cladistics 5: 164-166. [Google Scholar]
- 27. Yamagishi K, Nagata N, Yee KM, Braybrook SA, Pelletier J et al. (2005) TANMEI/EMB2757 encodes a WD repeat protein required for embryo development in Arabidopsis. Plant Physiol 139: 163-173 . doi:10.1104/pp.105.060467. PubMed: 16113228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yuan JS, Reed A, Chen F, Stewart CN Jr. (2006) Statistical analysis of real-time PCR data. BMC Bioinformatics 7: 85 . doi:10.1186/1471-2105-7-85. PubMed: 16504059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ohto MA, Floyd SK, Fischer RL, Goldberg RB, Harada JJ (2009) Effects of APETALA2 on embryo, endosperm, and seed coat development determine seed size in Arabidopsis. Sex Plant Reprod 22: 277-289 . doi:10.1007/s00497-009-0116-1. PubMed: 20033449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Meinke DW (1992) A Homoeotic Mutant of Arabidopsis thaliana with Leafy Cotyledons. Science 258: 1647-1650 . doi:10.1126/science.258.5088.1647. PubMed: 17742538. [DOI] [PubMed] [Google Scholar]
- 31. Little DP, Moran RC, Brenner ED, Stevenson DW (2007) Nuclear genome size in Selaginella. Genome 50: 351-356 . doi:10.1139/G06-138. PubMed: 17546093. [DOI] [PubMed] [Google Scholar]
- 32. Jernstedt JA, Cutter EG, Gifford EM, Lu P (1992) Angle Meristem Origin and Development in Selaginella martensii. Ann Bot 69: 351-363. [Google Scholar]
- 33. Kenrick P, Crane PR (1997) The origin and early evolution of plants on land. Nature 389: 33-39 . doi:10.1038/37918. [Google Scholar]
- 34. Xie Z, Li X, Glover BJ, Bai S, Rao GY et al. (2008) Duplication and functional diversification of HAP3 genes leading to the origin of the seed-developmental regulatory gene, LEAFY COTYLEDON1 (LEC1), in nonseed plant genomes. Mol Biol Evol 25: 1581-1592 . doi:10.1093/molbev/msn105. PubMed: 18453547. [DOI] [PubMed] [Google Scholar]
- 35. Mu J, Tan H, Zheng Q, Fu F, Liang Y et al. (2008) LEAFY COTYLEDON1 is a key regulator of fatty acid biosynthesis in Arabidopsis. Plant Physiol 148: 1042-1054 . doi:10.1104/pp.108.126342. PubMed: 18689444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tan H, Yang X, Zhang F, Zheng X, Qu C et al. (2011) Enhanced seed oil production in canola by conditional expression of Brassica napus LEAFY COTYLEDON1 and LEC1-LIKE in developing seeds. Plant Physiol 156: 1577-1588 . doi:10.1104/pp.111.175000. PubMed: 21562329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bateman RM, Dimichele WA (2008) Heterospory: the most iterative key innovation in the evolutionary history of the plant kingdom. Biol Rev Camb Philos Soc 69: 345-417. [Google Scholar]
- 38. Duckett J (1995) The Formation of Catenate Foliar Gemmae and the Origin of Oil Bodies in the Liverwort Odontoschisma denudatum (Mart.) Dum. (Jungermanniales): a Light and Electron Microscope Study. Ann Bot 76: 405-419 . doi:10.1006/anbo.1995.1114. [Google Scholar]
- 39. Smith RW (1920) Bulbils of Lycopodium lucidulum. Bot Gaz 69: 426-437 . doi:10.1086/332675. [Google Scholar]
- 40. Hoekstra FA (2005) Differential longevities in desiccated anhydrobiotic plant systems. Integr Comp Biol 45: 725-733 . doi:10.1093/icb/45.5.725. PubMed: 21676823. [DOI] [PubMed] [Google Scholar]
- 41. Proctor MCF, Pence VC (2002) Vegetative tissues: bryophytes vascular resurrection plants and vegetative propagules. In: Black M, Pritchard HW. Desiccation and Survival in Plants: Drying Without Dying. Wallingford CABI Publishing; pp. 207-238. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Identification of S. moellendorffii HAP3 haplotypes.
The sequenced S . moellendorffii genome contains two haplotypes. Approximately 100 kb regions surrounding HAP3 alleles were compared to identify haplotypes. Dot Matrix View of BLAST2 alignments show exceptionally high sequence alignment (slanted lines) of HAP3 alleles, indicating that the pairs occur in nearly identical scaffolds and not likely to be distinct loci.
(EPS)
Genomic locations of all putative genes encoding HAP3 subunits identified in this study.
Scaffold and coordinate identifiers correspond to the Physcomitrella patens 1.6 genome release and the Selaginella moellendorffii 1.0 genome release.
(DOCX)