Abstract
DREF was first characterized for its role in the regulation of transcription of genes encoding proteins involved in DNA replication and found to interact with sequences similar to the DNA recognition motif of the BEAF-32 insulator protein. Insulators are DNA-protein complexes that mediate intra- and inter-chromosome interactions. Several DNA-binding insulator proteins have been described in Drosophila, including BEAF-32, dCTCF and Su(Hw). Here we find that DREF and BEAF-32 co-localize at the same genomic sites, but their enrichment shows an inverse correlation. Furthermore, DREF co-localizes in the genome with other insulator proteins, suggesting that the function of this protein may require components of Drosophila insulators. This is supported by the finding that mutations in insulator proteins modulate DREF-induced cell proliferation. DREF persists bound to chromatin during mitosis at a subset of sites where it also co-localizes with dCTCF, BEAF-32 and CP190. These sites are highly enriched for sites where Orc2 and Mcm2 are present during interphase and at the borders of topological domains of chromosomes defined by Hi-C. The results suggest that DREF and insulator proteins may help maintain chromosome organization during the cell cycle and mark a subset of genomic sites for the assembly of pre-replication complexes and gene bookmarking during the M/G1 transition.
Keywords: transcription, chromatin, epigenetics, replication, cell cycle, mitosis
Introduction
Drosophila DNA replication-related element binding factor (DREF) is a homodimeric transcription factor that binds to the DNA replication-related element (DRE) of many genes involved in DNA replication and cell proliferation.1 DREF has been found in a complex with TRF2,2 which is present at more than a thousand sites in the genome, where it is responsible for the expression of TATA-less promoters.3 Like the insulator protein boundary element-associated factor 32 (BEAF-32), the N-terminal end of DREF has one BED finger that binds the DRE recognition sequence.4 The DNA recognition motif of DREF (TATCGATA) is similar to that of BEAF-32 (CGATA), and binding of DREF to DNA has been shown to antagonize BEAF-32 binding in vitro.5 It is currently unclear whether DREF and BEAF-32 bind the same or different sites, and whether these sites are close or in different regions of the genome.
Genes containing DREs include those involved in DNA replication (DNA polymerase α, PCNA, E2f, CycA),6-9 chromatin regulators (Moira, Osa),10 cell proliferation (p38a)11 and apoptosis (p53).12 Overexpression of DREF in the eye imaginal disc causes excessive cell proliferation, resulting in a rough eye phenotype. Expression of a dominant-negative form of DREF protein in larval salivary glands shows defective endo-reduplication. Depletion of the DREF protein in developing tissues results in failure of normal cell cycle progression (G1/S), which affects growth of the imaginal discs and derived adult organs.5,13 These effects of DREF on DNA synthesis and cell cycle progression may result from its role in the transcription of genes involved in these processes. In addition, given its ability to bind similar recognition sequences as the BEAF-32 insulator protein, it is also possible that the role of DREF depends on, or it is related to, that of classical insulators.
Insulators were originally identified based on their ability to regulate interactions between enhancers and promoters and to stop the spreading of heterochromatic silencing in transgene assays. More recent results suggest that these properties may not reflect the main role of insulators. Instead, insulators may have diverse functions that are context-dependent and derived from their ability to mediate inter- and intra-chromosomal contacts.14 Insulators are found not only in intergenic regions, but also at promoters, 5′UTRs and introns, where they appear to play such diverse roles as targeting distal enhancers to the appropriate promoter, regulating RNAPII pausing, alternative splicing and V(D)J recombination.15
Interactions among different loci in the genome appear to be a major organizing principle underlying nuclear architecture.16,17 Such interactions can take place at a local level, across large distances within a chromosome,18 or between chromosomes.19,20 In some cases these interactions are required for activation of gene expression,19 whereas in other cases they may facilitate silencing.21 Recent results using 5C and Hi-C suggest that chromosomes are organized into topologically associating domains (TADs) during interphase, and that insulator proteins are enriched at the boundaries between these domains.22-25 Therefore, insulator-mediated interactions may contribute to the three-dimensional organization of the chromosomes during interphase. How this 3D structure is maintained or altered during the cell cycle and the role of chromosome architecture in gene expression are issues currently not well understood.
Five different DNA-binding insulator proteins have been characterized in Drosophila, including BEAF-32, Drosophila CCCTC binding factor (dCTCF), suppressor of Hairy-wing [Su(Hw)], GAGA factor (GAF) and Zeste-white 5 (Zw5).26-31 Two additional proteins, centrosomal protein 190 (CP190) and Mod(mdg4), do not bind DNA; instead, they interact with the insulator DNA-binding proteins and with themselves to mediate inter/intra-chromosomal interactions.32-35 Analysis of the genome-wide distribution of these proteins has given some insights into the issue of whether the different Drosophila insulators play distinct or overlapping roles in genome organization and function.36-38 For example, BEAF-32 is found more often close to gene promoters, where it appears to confer independent regulation to closely adjacent, divergently transcribed genes.39 On the other hand, Su(Hw) tends to be present in intergenic regions next to genes transcribed at low levels.36 In addition, insulators appear to play overlapping roles at some genomic locations, where several different insulator DNA-binding proteins cluster together. These “aligned” insulators are preferentially located at borders of functional chromosome domains, such as those defined by the presence of H3K27me3 and Polycomb (Pc),40 and TADs defined by Hi-C.25,41
In this study, we have examined the genome-wide distribution of DREF in Drosophila Kc167 cells in interphase and mitosis in order to gain further insights into its function in gene expression and the possibility that the role of this protein depends on or is related to chromosome architecture during the cell cycle. We find that DREF overlaps with BEAF-32 and other insulator proteins in the Drosophila genome. This and results from genetic analysis of the effect of mutations in insulator proteins on DREF-induced over-proliferation phenotypes suggest that DREF may cooperate with classical insulator proteins to accomplish some of its functions. Sites where DREF co-localizes with insulator proteins are maintained during mitosis. Interestingly, sites of DREF and other insulator proteins in mitotic chromosomes are highly enriched at the boundaries of TADs and correspond to sites of Orc2 and Mcm2 binding during interphase. We suggest that sites of DREF and aligned insulators may mark sites of pre-replication complex assembly during mitosis and bookmark genes involved in DNA replication and cell proliferation to ensure their expression in early G1.
Results
DREF is present at insulator sites throughout the Drosophila genome
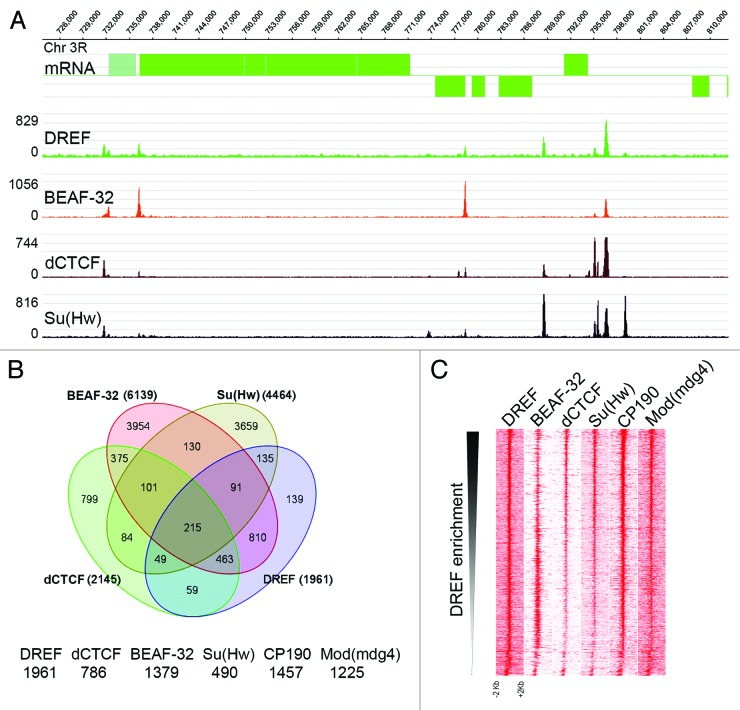
DREF has been extensively studied in the context of its ability to bind DRE elements located in the promoter and regulatory regions of genes encoding proteins involved in DNA replication and cell cycle control.1 As a consequence, it has been shown to be a classical transcription factor involved in the expression of this class of genes both in Drosophila and humans. The DREF DNA recognition sequence overlaps extensively with that of the Drosophila BEAF-32 insulator protein, and in vitro experiments suggest the two proteins compete for binding to this sequence.42 This observation opens the possibility of a role for DREF related to insulator function. To explore the possibility that the role of DREF in transcription is related to that of insulators, we analyzed the genome-wide distribution of DREF in Drosophila Kc167 cells using Chip-seq. Results indicate that DREF is present at 1961 sites in the genome that show significant level of enrichment over the input. Earlier studies have shown that DREF overlaps with BEAF-32 at several hundred sites on polytene chromosome.42 Results from ChIP-seq confirm this observation, with most sites of DREF overlapping those of BEAF-32 (Fig. 1A). However, some DREF sites appear to lack BEAF-32 and, instead, overlap with sites of dCTCF and/or Su(Hw) (Fig. 1A). In order to obtain a quantitative estimate of the overlap between DREF and other insulator proteins, we intersected DREF peaks with those previously described for other insulator DNA-binding proteins35,40 using Galaxy tools to generate different subgroups of co-localizing proteins. The majority of DREF binding regions (1822/1961) also overlap with one or more insulator proteins. DREF shows highest overlap with BEAF-32 among the DNA-binding insulator proteins (Fig. 1B), something unexpected in view of their apparent ability to recognize the same DNA sequence. DREF is alone at 139 sites and overlaps with other DNA-binding insulator proteins in the absence of BEAF-32, including 135 sites where it is present in combination with Su(Hw), 59 sites where it overlaps with dCTCF and 49 sites where it overlaps with both. In addition, DREF overlaps extensively with the two accessory proteins required for inter-insulator interactions, CP190 (1457 sites) and Mod(mdg4) (1225) (Fig. 1B). However, since BEAF-32 and DREF are present at the same genomic locations, it is unclear whether CP190 and Mod(mdg4) interact with both or just one of these two proteins. In order to compare the relative tag enrichments between DREF and other insulator proteins, we plotted tag densities by descending intensity of DREF summits. We observe an inverse tag density correlation between DREF and BEAF-32, whereas other insulator proteins show a direct correlation in tag density with respect to DREF (Fig. 1C).These data are in agreement with earlier results suggesting an antagonism between DREF and BEAF-32 binding.42 Nevertheless, the results indicate that BEAF32 and DREF do not appear to exclude each other from sites in the genome.
Figure 1. DREF associates with insulator proteins. (A) A 100 kb region of the genome is shown. Genes from the UCSC Genome Browser are shown in green. Enrichment regions from wig files are shown using SignalMap (NimbleGene) for DREF and other insulator proteins. (B) Venn diagram showing the overlap of genomic sites for DREF and other insulator proteins. (C) Heatmaps of the distribution of ChIP-seq data sets using DREF as an anchor. The total number of reads is plotted for each ChIP-seq data set in 20-bp bins covering 1 kb upstream of and downstream from DREF sites and viewed in Java Treeview. The data are arranged in decreasing intensities of DREF enrichment.
DREF and BEAF-32 bind similar sequences in the genome
Earlier studies determined the DNA binding motifs of BEAF-32 and DREF using DNA footprinting experiments in the scs’ insulator and the E2F promoter, respectively. The recognition motif for both proteins shares the sequence 5` CGATA 3′ but the reported DREF binding site contains three additional nucleotides to form the motif 5′ TATCGATA 3′.8,43 Although DREF and BEAF-32 contain a BED Zn finger DNA binding domain in the N-terminal region, the actual sequence of the BED finger is not the same in the two proteins. Therefore, it is possible that the two proteins bind similar but not identical sequences in the genome. We thus used the genome-wide binding data for DREF and BEAF-32 to derive consensus sequences for sites co-occupied by the two proteins and sites where BEAF-32 is present alone (Fig. 2A). The recognition sequence for BEAF-32 alone contains the previously found core motif, but this protein appears to have a preference for the sequence 5′ TATCGATA 3′ (Fig. 2A). Sites containing both BEAF-32 and DREF are further enriched in sequences containing the TAT trinucleotide 5′ to the core motif, suggesting that the two proteins share a core motif 5′CGAT3′, but the adjacent nucleotides are either different, or the two proteins have different preferences for the same nucleotides (Fig. 2A). These results suggest that, rather than competing for the same sequence, DREF and BEAF-32 may bind slightly different sequences located close to each other at specific genomic sites. To test this possibility, we determined the number of consensus sequences present at genomic sites found to contain DREF and BEAF-32 by ChIP-seq. We find that, on average, sites binding DREF and BEAF-32 contain 2.5 copies of the recognition motifs for these two proteins, suggesting that both of them could bind simultaneously to the same genomic regions. In fact, most DREF sites obtained by ChIP-seq contain 2–11 recognition motifs within +/−150 bp of the summits (Fig. 2C), supporting the idea that both DREF and BEAF-32 can bind simultaneously in the same genomic sites. Interestingly, sites containing DREF without BEAF-32 are enriched for the Su(Hw) or dCTCF recognition sequences instead. The presence of DREF at genomic sites lacking its consensus sequence but containing binding motifs for Su(Hw) or dCTCF may suggest that DREF does not directly bind to these sites and is, instead, recruited through direct or indirect interactions with other insulator proteins.
Figure 2. Characterization of DREF binding sequences. (A) Consensus sequences predicted from peaks where the DREF and BEAF-32 proteins overlap and peaks having only BEAF-32. (B) Three separate regions of chromosome 3 containing the genes encoding RNA polymerase I, p53 and Warts proteins. Genes from the UCSC Genome Browser are shown in purple. Enrichment regions for DREF from wig files are shown in green. The locations of predicted DREF consensus sequences are shown in red. (C) Histogram representing the average number of DREF or BEAF-32 motifs at sites in the genome where the BEAF-32 protein is present alone or together with DREF. (D) Histogram representing number of occurrences of DREF consensus sequences in each DREF-bound gene promoter. (E) Analysis of DREF-bound genes based on molecular function (GO terms).
Earlier studies have shown binding of DREF upstream of TSSs of genes involved in DNA replication, cell proliferation and apoptosis.4,6-12 ChIP-seq data confirms enrichment of DREF at these loci, including DNA polymerase I, p53 and warts (Fig. 2B). The peaks with DREF enrichment contain the DREF consensus sequence corresponding to DREs (Fig. 2A). Nearly 82% of DREF peaks contain this consensus sequence. DREF peaks in the genome contain one or more copies of the DRE motif within 150 bp of the summit, but not all DREs show a corresponding enrichment of DREF, suggesting that some of these sites are not functional (Fig. 2B and 2D). We then defined DREF-bound genes as those containing a DREF peak within 200 bp upstream of the TSS, and these genes were analyzed for molecular function. The highest enriched group corresponds to genes involved in cell cycle, M phase and cell cycle processes (Fig. 2E), which are known to be expressed during early G1.44,45 Other groups include metabolism, ubiquitin protein ligation, centrosome cycle, oogenesis and development or metamorphosis (Fig. 2E).
DREF is present near the TSSs of active genes
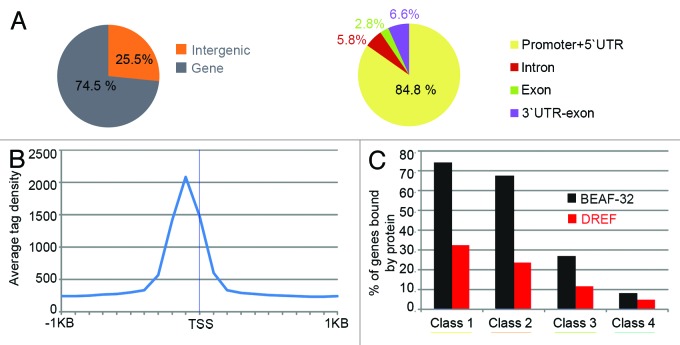
DREF is known to bind to DRE sequences upstream of the TSS of several genes.1 We have used the genome-wide DREF mapping data to examine the general distribution of this protein with respect to various gene features. Most DREF sites (75%) are located in gene regions (defined from −200 bp from the TSS to +200 bp from the TTS), with 25% located in inter-genic regions (Fig. 3A). Most gene-associated DREF sites are present in the promoter/5′UTR region. The highest tag enrichment of DREF is located around 100 bp upstream from TSSs, indicating that, as is the case for BEAF-32, DREF preferentially binds upstream of the promoter region (Fig. 3B). Also like BEAF-32 in wing imaginal disc cells,46 DREF associates preferentially with highly transcribed genes (Fig. 3C). These characteristics of the distribution of DREF protein in the genome are in agreement with a direct role for DREF in transcription. However, BEAF-32 displays a similar distribution pattern, but has been recently shown to act as an insulator to allow independent regulation of close adjacent divergently transcribed genes.39
Figure 3. Distribution of DREF binding regions in the genome. (A) Distribution of DREF binding sites with respect to gene features. (B) DREF tag enrichment around transcription start sites (TSSs) of DREF-bound genes. (C) Histogram indicating the fraction of DREF and BEAF-32 bound genes and their gene expression levels in wing imaginal tissue. The percentages refer to the fraction of BEAF-32-bound genes within each class. Class I corresponds to the 25% most highly transcribed genes, whereas class 4 corresponds to the lowest 25%.
A subset of DREF sites is maintained during mitosis
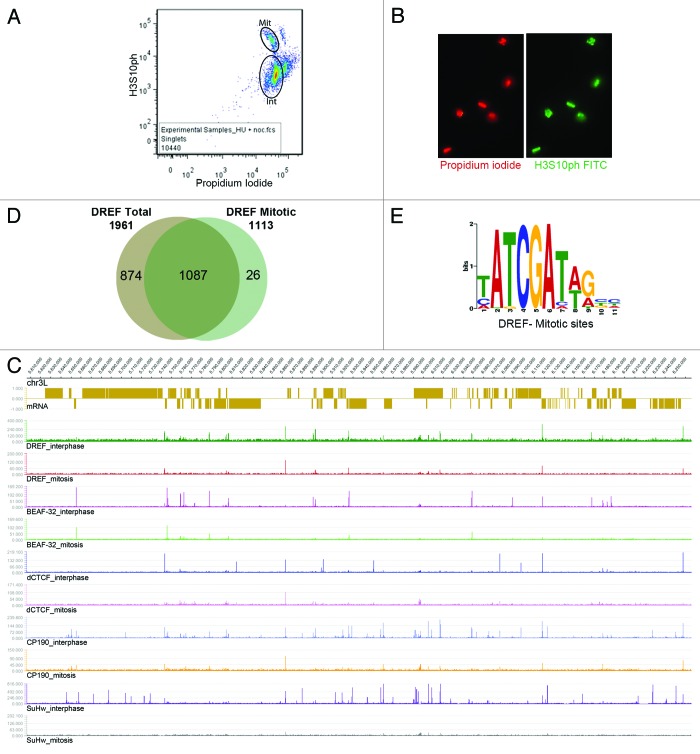
Insulator proteins, with the exception of Su(Hw), remain bound to chromosomes at a subset of sites during mitosis.47 On the other hand, most transcription factors do not stay bound to mitotic chromosomes. To further distinguish between the roles of DREF as a transcription factor and its location at insulator sites throughout the genome, we tested the possibility that DREF stays bound to chromatin during mitosis, perhaps with other insulator proteins, using ChIP-seq. Since Drosophila cells are difficult to synchronize by standard procedures, cycling cells were partially synchronized, fixed and labeled with antibodies to lamin Dm0, and mitotic cells were separated by FACS (Fig. 4A). The purity of the mitotic cell population was confirmed by immunofluorescence microscopy with antibodies to H3S10ph and ranged between 97 and 99% (Fig. 4B). Mitotic cells were then used for ChIP-seq analyses using DREF antibodies. Figure 4C displays a snapshot of a ca. 600 kb region of chromosome 3L showing that a subset of DREF sites indeed persists during mitosis and co-localizes with other insulator proteins. To determine the functional significance of these sites, we first performed a detailed analysis of cell cycle changes in the localization of DREF and found 1,113 sites of this protein in mitotic chromosomes with significant enrichment over input. Of these, 1,087 overlap with peaks found in cycling cells (Fig. 4D). Since approximately 90–95% of cells present in a cycling population are in interphase (Fig. 4A), we will consider the peaks of DREF, as well as previously published data for other insulator proteins,35,40 found in cycling cells as interphase sites. Therefore, out of a total of 1,961 sites of DREF present in interphase, 1,087 are retained during mitosis, whereas 874 are not (Fig. 4D). The 26 sites found de novo in mitotic cells are probably not significant. Prediction of a DREF binding motif for mitotic sites using MEME-ChIP48 results in the same sequence found for interphase sites (Fig. 4E), suggesting that the different site occupancy of the protein during the two phases of the cell cycle is not determined by differences in the recognition sequence. The DREF binding sites predicted using the mitotic peaks also have the 5′ATCGA3′ core sequence and are enriched for genes involved in cell cycle processes (GO terms). The reason for the presence of DREF at these sites during mitosis is unclear, but DREF may function with insulator proteins to organize chromosome structure during the cell cycle.
Figure 4. Analysis of DREF bound regions in mitotic chromosomes. (A) Selection of interphase and mitotic cells using FACS. Approximate gates used to select interphase and mitotic populations are indicated as ellipses. (B) Immunofluorescence microscopy of cells in the mitotic population stained with propidium iodide and antibodies to H3S10ph indicate that all cells are in mitosis. (C) Snapshot of a 650 kb region of chromosome 3L showing the location of genes (top) and distribution of DREF and insulator protein sites in asynchronous (interphase) and mitotic cells. (D) Extent of overlap between DREF sites obtained in asynchronous (total) and mitotic cells. (E) DNA consensus obtained for DREF bound regions during mitosis.
Sites of DREF in mitotic chromosomes also contain other insulator proteins
To test whether the presence of DREF in mitotic chromosomes is related to insulator function, we compared the distribution of DREF during mitosis with that of other insulator proteins. Su(Hw) does not remain bound to mitotic chromosomes, whereas a subpopulation of sites for dCTCF, BEAF-32 and CP190 persists in mitotic chromatin.47 We therefore compared the distribution of regions bound by DREF and other insulator proteins in mitotic chromosomes. There is a significant enrichment of insulator proteins dCTCF, BEAF-32 and CP190 around mitotic DREF regions (Fig. 5A). These results suggest that mitotic sites of DREF also contain other insulator proteins. Given the extensive co-localization of DREF with other insulator proteins throughout the cell cycle, the role of DREF in mitosis and interphase may depend in part, or be related to, that of classical insulators.
Figure 5. (A) Heatmaps of the distribution of ChIP-seq data sets using DREF as an anchor. The total number of reads is plotted for each ChIP-seq data set in 20-bp bins covering 2 kb upstream of and downstream from DREF sites and viewed in Java Treeview. The data are arranged in decreasing intensities of enrichment of DREF mitotic peaks. (B) Distribution of mitotic DREF sites with respect to borders of physical chromosome domain. (C) Heatmaps of the distribution of ChIP-seq data sets using DREF as an anchor. The total number of reads is plotted for each ChIP-seq data set in 20-bp bins covering 2 kb upstream of and downstream from DREF sites and viewed in Java Treeview. The data are arranged in decreasing intensities of enrichment of DREF mitotic peaks. (D) Box plot showing distribution of the DREF bound regions with respect to replication timing. (E) Histogram showing enrichment of Orc2 (gray) and Mcm2 (red) at sites of DREF present in both interphase and mitosis and sites of DREF present only in interphase.
It has been recently shown that insulators are enriched at the boundaries of topologically associating domains in interphase.22-25 Specifically, aligned insulators, which contain several insulator DNA binding proteins within a small 100–200 bp region,40 are enriched at the borders of TADs.22 We thus examined whether DREF mitotic sites are enriched at interphase chromosome domains borders. Figure 5B shows that this is indeed the case. Mitotic sites of DREF, which are also present in interphase, are enriched at interphase TAD borders compared with interphase-only sites (Fisher’s exact test < 0.0001), whereas interphase DREF sites appear to be enriched inside of domains. These results suggest that a subset of DREF sites persists during mitosis co-localized with other insulator proteins at aligned insulator sites. Given the enrichment of these sites at the borders of TADs, DREF may contribute to the maintenance of these domains during mitosis, or its function may depend on the properties of TAD borders.
Mitotic sites of DREF may mark sites of replication complex assembly
One possible interpretation of the finding of mitotic DREF sites at boundaries of topologically associating domains is that these regions of chromosomes are more accessible and allow transcription of DREF-inducible genes early at the M/G1 transition. A second, related and not mutually exclusive, possibility is that these open regions of chromatin allow the assembly of the replication complex in late M phase. This possibility is supported by the location of DREF close to promoters of actively transcribed genes, which is reminiscent of that of components of the pre-replication complex such as Orc2.49 To explore the possibility of a more direct role for DREF in replication events, we first compared its distribution with that of Orc2 and Mcm2.49Figure 5C shows that the distribution of DREF closely associates with that of these two proteins. The association of the Mcm2 helicase and Orc2 with chromatin represents an initial step during the formation of the pre-replication complex. Orc2 binding is highly enriched in early replicating compared with late replicating regions.49 We therefore tested whether DREF sites are preferentially represented at early replication regions. We found that DREF-bound sites are significantly enriched for early replication regions (Fig. 5D). Furthermore, Orc2 and Mcm2 are more enriched in DREF mitotic regions than in non-mitotic DREF regions: over 70% of DREF mitotic peaks overlap with Orc2 or Mcm2, whereas 50–60% of DREF interphase sites do (Fisher’s exact test < 0.0001) (Fig. 5E). Since Orc2 is not present at sites of replication during S phase and only binds to replication origins at the end of M,50,51 it is possible that DREF marks replication origins to allow Orc2 binding at the end of mitosis.
DREF-induced cell proliferation and growth is dependent on insulator proteins
Overexpression of DREF in cells of eye imaginal tissue results in increased cell proliferation and growth. This phenotype could result from alterations in the localization of DREF in mitotic chromosomes, which could cause misregulation of bookmarked genes or changes in replication timing. In the background of mutations in the head involution defective (hid) gene, which inhibits cell death, the proliferating cells do not undergo apoptosis, and a characteristic overgrowth phenotype can be observed in the eye. The increased proliferation causes increased disarray in the ommatidia and the formation of bulges in the eye due to increased number and growth of the ommatidia (Fig. 6A).5 When the dosage of DREF is reduced by genetic crosses that introduce a DREF loss of function allele, the phenotype caused by DREF overexpression is suppressed (Fig. 6B).The same effect is observed when a BEAF-32-null allele is introduced (Fig. 6C). The reduction in levels of BEAF-32 suggests that both proteins are required for the observed phenotypic effect,5 although it is possible that a second mutation in the chromosome containing the BEAF-32 allele is responsible for the observed phenotype. Surprisingly, mutations in dCTCF and CP190 show an enhanced phenotype with increased proliferation and larger ommatidia (Fig. 6E–H), while mutations in su(Hw) did not show significant changes with respect to the control (Fig. 6D). The genetic interaction between alleles of insulator proteins and DREF suggests that the effect of DREF on cell proliferation, which may depend on its presence in mitotic chromosomes to bookmark genes for early G1 expression or mark early replication origins, is contingent upon proper insulator function. The lack of an effect of Su(Hw) could be due to its absence from mitotic chromosomes, suggesting that the presence of insulator proteins during mitosis is required for DREF-induced overgrowth phenotypes.
Figure 6. Insulator protein alleles show genetic interaction with DREF-induced eye phenotypes. DREF overexpression in a hid(W1) background shows increased growth and proliferation. Each panel displays pictures of adult eyes from flies of the genotypes shown above each photograph.
Discussion
Proper cellular homeostasis requires the expression of specific sets of genes, which takes place mostly during interphase and requires a complex interplay between chromatin, the transcription complex and binding of transcription factors to regulatory sequences. Interaction of transcription factors with the chromatin substrate involves modifications of the histone octamer to allow access of the transcription machinery to the DNA substrate. However, chromatin states established during interphase must be profoundly altered when cells divide, since the chromatin fiber undergoes condensation, and RNA polymerase II and most transcription factors dissociate from the DNA during mitosis.52 How do cells remember the patterns of transcription present in the previous G1 after the conclusion of mitosis? It has been suggested that cells use a combination of specific proteins, histone modifications and histone variants to bookmark genes and facilitate their expression at the M/G1 interface. As a consequence, whereas most transcription factors are evicted from mitotic chromatin, proteins such as Brd4 and MLL persist at the promoters of specific genes to facilitate their expression upon re-entry in G1.53,54
DREF has been extensively characterized as a transcription factor responsible for the expression of many genes, including those involved in various aspects of DNA replication and cell cycle control, which are transcribed early during G1. Here we show that, contrary to typical transcription factors, DREF persists bound to chromatin during mitosis. Furthermore, sites where DREF binds in mitotic chromosomes also contain a specific subset of insulator proteins, and these sites are occupied during G1 by the Orc2 and Mcm2 components of the pre-replication complex. Although these observations are only correlative, they suggest interesting models for further testing.
Drosophila has at least five different insulators that share a common core of accessory proteins but use different DNA binding proteins to target the accessory proteins to different regions of the genome.55,56 The different subclasses of Drosophila insulators show distinct localization patterns with respect to gene features, suggesting different roles in nuclear biology. However, the different insulator subclasses also cluster at specific genomic locations, where they synergize to create stronger insulators.40 These aligned insulators are enriched at the borders of TADs22 and are maintained during mitosis.47
The existence of TADs in interphase chromosomes raises the question of how these structures change during the cell cycle, and whether aligned insulators play a role in the regulation of the dynamic changes that occur to chromosomes as the cells cycle between interphase and mitosis. The presence of aligned insulator sites in mitotic chromosomes suggests that these insulator clusters may contribute to the maintenance of certain chromosome architecture during the cell cycle. Interestingly, borders of TADs have been shown to be more accessible than the surrounding chromatin, independent of the types of histone modifications present in the chromatin fiber.22 Results presented here suggest that DREF associates with insulator proteins during interphase and mitosis. DREF is enriched at the borders of topological domains, and sites of DREF in mitotic chromosomes correspond to aligned insulators, suggesting that genomic locations where DREF persists in mitotic chromosomes may be more accessible than the rest of the condensed mitotic chromosomes. These regions may become accessible to the transcription machinery earlier than the rest of the chromosome, when these decondense at the end of M phase. This would explain the role of DREF in the transcription of genes involved in cell cycle regulation and DNA replication, which are transcribed at the M/G1 interphase.
Mutations in the DREF gene result in cell cycle arrest in G1 and lower levels of expression of genes involved in DNA synthesis and cell growth.5 The promoter regions of affected genes are occupied by the DREF protein, suggesting a direct effect on their expression. The most obvious explanation for this observation is that DREF is a classic transcription factor that controls the expression of these genes. Nevertheless, ChIP-seq analysis of DREF localization in the genome suggests that DREF is located at insulator sites. Furthermore, DREF clusters with dCTCF, BEAF-32 and CP190 to form aligned insulators at specific genomic locations during interphase, and a subset of these aligned insulator sites is maintained in mitotic chromosomes. This is in contrast to what happens to most transcription factors, which are displaced from chromatin as the cells enter M phase. Interestingly, Su(Hw) is also displaced from the chromosomes during mitosis, suggesting that the persistence of other insulator proteins during this stage of the cell cycle serves a specific functional role. This role could be simply structural, i.e., the maintenance of a subset of chromosome physical domains between interphase and mitosis. Nevertheless, the phenotypes of DREF mutations suggest that the role of DREF in mitosis and, by extension, the role of insulator proteins with which it co-localizes during this phase of the cell cycle, is more complex.
Genes that encode proteins involved in DNA synthesis and cell growth, which represent many of those bound by DREF, are expressed at the M/G1 interface,44,45,57 perhaps to allow sufficient time for the proteins to perform their function in S phase. This process has been termed bookmarking,52,58 and it is possible that the persistence of DREF in mitotic chromosomes is related to its role in the expression of genes that need to be transcribed at the M/G1 interface. In addition, DREF co-localizes extensively with Orc2 and Mcm2, two proteins responsible for the assembly of the pre-replication complex at M/G1 that are displaced from the chromosomes during S phase to only bind again late in M phase. It is possible that DREF and aligned insulators play a role in these two processes: gene bookmarking to allow early transcription of cell cycle and cell growth genes and targeting of Orc2 to early replication origins at the end of mitosis to regulate replication timing.
The mechanisms by which DREF and insulator proteins allow access to RNAPII and Orc2 to chromatin at the end of mitosis is unknown, but their presence at TAD borders may suggest some plausible explanation. TADs are separated by boundaries enriched in aligned insulator sites.22 These are the same sites that are maintained during mitosis and co-localize with DREF. Interestingly, it has been shown that regions adjacent to TAD borders have a more open chromatin, independent of the type of histone modifications present, that allows higher insertion rates of transposable elements and higher expression of transgenes.22 It is possible that the persistence of DREF and other insulator proteins in mitosis allows the maintenance of these domain borders of open chromatin, which then facilitate the recruitment of RNAPII and Orc2 at the end of mitosis.
Materials and Methods
Cell culture and flow cytometry
Drosophila Kc167 cells were grown at 25°C in CCM3 media (Hyclone) to a density of 2 × 10.6 To synchronize the cells, the culture was treated with hydroxyurea (1 mg/ml in ethanol, to a final concentration of 15 ng/ml) for 16 h, then pelleted at 300 g and rinsed with fresh medium. Cells were then incubated for 8 h with nocodazole (5 mg/ml in DMSO, to a final concentration of 2 ng/ml) and harvested. Isolation of mitotic and interphase cells by flow cytometry was performed as previously described.47 Briefly, cells were fixed for 10 min by adding formaldehyde (37% stock, to a final concentration of 1%) directly to the medium. Cells were then blocked in suspension for 30 min in blocking buffer and incubated overnight with mouse α-Lamin Dm0 at 1:500. Cells were washed 3 × 15 min in blocking buffer then incubated overnight with secondary antibody Alexa Fluor 488 α-mouse at 1:5,000. Cells were washed as before and incubated 30 min in blocking buffer plus propidium iodide (0.1 mg/ml). Samples were passed several times through a 25-gauge syringe to reduce clumping and then sorted on a FACSAria II cell sorter.
Immunofluorescence
Kc167 cells were harvested while in log phase growth, washed once and then resuspended in 1× PBS. Cell suspension was applied to slides treated with 0.01% poly-L-lysine, and allowed to settle for 10 min. Slides were fixed for 30 min (1× PBS plus 0.1% Triton-X100 and 4% paraformaldehyde), then blocked for 30 min (1× PBS plus 0.1% Triton-X100 and 5% BSA). Mouse α-H3S10ph (Millipore 05-806) at 1:4,000 was diluted in blocking buffer, and slides were incubated overnight at 4°C. Slides were washed 3 × 15 min (1× PBS plus 0.1% Triton-X100), incubated 2 h with secondary antibody in blocking buffer (1:250 dilution) and then washed again as before. Slides were treated with DAPI and mounted in Vectashield. Images were taken using a Zeiss Axiophot fluorescence microscope.
Chromatin immunoprecipitation and Illumina sequencing
Antibodies to DREF were prepared by expression in E. coli of the region of the protein containing amino acids 16–608, followed by purification and immunization of rabbits. Chromatin immunoprecipitation was performed as described.36 Approximately 40 × 106 cells in log phase were pelleted and suspended in fresh CCM3 media. They were then fixed using formaldehyde in CCM3 to a final concentration of 1% at room temperature with gentle shaking. The reaction was stopped by adding glycine to a final concentration of 0.125 mM. Chromatin was prepared by shearing the chromatin using sonication. Chromatin was immunoprecipitated using antibodies to DREF,35,40 and the DNA was used for library preparation using the Illumina TruSeq kit. ChIP Seq libraries were sequenced at the HudsonAlpha Institute for Biotechnology using an Illumina HiSeq system. Sequences were mapped to the dm3 genome with Bowtie 0.12.359 using default settings. Peaks were then called using MACS 1.4.0alpha2, with a p value of 10−10.60
Bioinformatics and software used for analysis
Overlapping intervals of insulator proteins and DREF were obtained using GALAXY tools61 by intersecting peak data sets obtained with MACS. The Venn diagram shown in Figure 1 was prepared using a universal set of all peak regions from insulator proteins and DREF. The tag enrichment around peak summits using wig data files were extracted using in-house R scripts. Heat maps were generated using JAVA Treeview. SignalMap software (NimbleGen) was used to generate the graphical output for genome tracks (dm3 build). Gene ontology analysis for DREF-bound genes was done using DAVID (http://david.abcc.ncifcrf.gov) functional annotation with high stringency.62
Fly crosses and genetic interaction analysis
Drosophila strains were grown at 25°C under standard culture conditions. The fly strain GMR-GAL-UAS DREF ;hidP1/Cyo-Tb was obtained from Dr Dirk Bohmann at the University of Rochester Medical Center.5 The DREF allele DrefKG09294 was obtained from the Bloomington stock center, and BEAF-32np6377 from the National Institute of Genetics, Kyoto, Japan. The following strains have been previously published: CP190,1,4 CP190H32-1 32, su(Hw) ,2,63 CTCFy+2 and CTCFy+.1,26 Images of adult eyes were obtained using a Zeiss stereo scope with white light illumination, with the same magnification settings, and processed using Adobe Photoshop.
Acknowledgments
We would like to thank members of the lab for helpful discussions and suggestions during this study. We also thank The Genomic Services Lab at the HudsonAlpha Institute for Biotechnology for their help in performing Illumina sequencing of ChIP-Seq samples. Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number R01GM035463. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Accession Numbers
ChIP-seq data are deposited in NCBI’s Gene Expression Omnibus (GEO) (www.ncbi.nlm.nih.gov/geo) under accession number GSE39664.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/24742
References
- 1.Matsukage A, Hirose F, Yoo MA, Yamaguchi M. The DRE/DREF transcriptional regulatory system: a master key for cell proliferation. Biochim Biophys Acta. 2008;1779:81–9. doi: 10.1016/j.bbagrm.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 2.Hochheimer A, Tjian R. Diversified transcription initiation complexes expand promoter selectivity and tissue-specific gene expression. Genes Dev. 2003;17:1309–20. doi: 10.1101/gad.1099903. [DOI] [PubMed] [Google Scholar]
- 3.Isogai Y, Keles S, Prestel M, Hochheimer A, Tjian R. Transcription of histone gene cluster by differential core-promoter factors. Genes Dev. 2007;21:2936–49. doi: 10.1101/gad.1608807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hirose F, Yamaguchi M, Matsukage A. Targeted expression of the DNA binding domain of DRE-binding factor, a Drosophila transcription factor, attenuates DNA replication of the salivary gland and eye imaginal disc. Mol Cell Biol. 1999;19:6020–8. doi: 10.1128/mcb.19.9.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hyun J, Jasper H, Bohmann D. DREF is required for efficient growth and cell cycle progression in Drosophila imaginal discs. Mol Cell Biol. 2005;25:5590–8. doi: 10.1128/MCB.25.13.5590-5598.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hirose F, Yamaguchi M, Handa H, Inomata Y, Matsukage A. Novel 8-base pair sequence (Drosophila DNA replication-related element) and specific binding factor involved in the expression of Drosophila genes for DNA polymerase alpha and proliferating cell nuclear antigen. J Biol Chem. 1993;268:2092–9. [PubMed] [Google Scholar]
- 7.Yamaguchi M, Hayashi Y, Nishimoto Y, Hirose F, Matsukage A. A nucleotide sequence essential for the function of DRE, a common promoter element for Drosophila DNa replication-related genes. J Biol Chem. 1995;270:15808–14. doi: 10.1074/jbc.270.26.15808. [DOI] [PubMed] [Google Scholar]
- 8.Sawado T, Hirose F, Takahashi Y, Sasaki T, Shinomiya T, Sakaguchi K, et al. The DNA replication-related element (DRE)/DRE-binding factor system is a transcriptional regulator of the Drosophila E2F gene. J Biol Chem. 1998;273:26042–51. doi: 10.1074/jbc.273.40.26042. [DOI] [PubMed] [Google Scholar]
- 9.Ohno K, Hirose F, Sakaguchi K, Nishida Y, Matsukage A. Transcriptional regulation of the Drosophila CycA gene by the DNA replication-related element (DRE) and DRE binding factor (DREF) Nucleic Acids Res. 1996;24:3942–6. doi: 10.1093/nar/24.20.3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakamura K, Ida H, Yamaguchi M. Transcriptional regulation of the Drosophila moira and osa genes by the DREF pathway. Nucleic Acids Res. 2008;36:3905–15. doi: 10.1093/nar/gkn291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park JS, Kim YS, Kim JG, Lee SH, Park SY, Yamaguchi M, et al. Regulation of the Drosophila p38b gene by transcription factor DREF in the adult midgut. Biochim Biophys Acta. 2010;1799:510–9. doi: 10.1016/j.bbagrm.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 12.Trong-Tue N, Thao DT, Yamaguchi M. Role of DREF in transcriptional regulation of the Drosophila p53 gene. Oncogene. 2010;29:2060–9. doi: 10.1038/onc.2009.483. [DOI] [PubMed] [Google Scholar]
- 13.Yoshida H, Kwon E, Hirose F, Otsuki K, Yamada M, Yamaguchi M. DREF is required for EGFR signalling during Drosophila wing vein development. Genes Cells. 2004;9:935–44. doi: 10.1111/j.1365-2443.2004.00775.x. [DOI] [PubMed] [Google Scholar]
- 14.Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Phillips-Cremins JE, Corces VG. CTCF: Linking genome organization to cellular function. Mol Cell. 2013 doi: 10.1016/j.molcel.2013.04.018. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cremer T, Cremer M, Dietzel S, Müller S, Solovei I, Fakan S. Chromosome territories--a functional nuclear landscape. Curr Opin Cell Biol. 2006;18:307–16. doi: 10.1016/j.ceb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 17.van Driel R, Fransz PF, Verschure PJ. The eukaryotic genome: a system regulated at different hierarchical levels. J Cell Sci. 2003;116:4067–75. doi: 10.1242/jcs.00779. [DOI] [PubMed] [Google Scholar]
- 18.Chambeyron S, Bickmore WA. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 2004;18:1119–30. doi: 10.1101/gad.292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126:403–13. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- 20.Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435:637–45. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- 21.Li T, Hu JF, Qiu X, Ling J, Chen H, Wang S, et al. CTCF regulates allelic expression of Igf2 by orchestrating a promoter-polycomb repressive complex 2 intrachromosomal loop. Mol Cell Biol. 2008;28:6473–82. doi: 10.1128/MCB.00204-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hou C, Li L, Qin ZS, Corces VG. Gene density, transcription, and insulators contribute to the partition of the Drosophila genome into physical domains. Mol Cell. 2012;48:471–84. doi: 10.1016/j.molcel.2012.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–80. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–5. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, et al. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–72. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 26.Gerasimova TI, Lei EP, Bushey AM, Corces VG. Coordinated control of dCTCF and gypsy chromatin insulators in Drosophila. Mol Cell. 2007;28:761–72. doi: 10.1016/j.molcel.2007.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moon H, Filippova G, Loukinov D, Pugacheva E, Chen Q, Smith ST, et al. CTCF is conserved from Drosophila to humans and confers enhancer blocking of the Fab-8 insulator. EMBO Rep. 2005;6:165–70. doi: 10.1038/sj.embor.7400334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parkhurst SM, Harrison DA, Remington MP, Spana C, Kelley RL, Coyne RS, et al. The Drosophila su(Hw) gene, which controls the phenotypic effect of the gypsy transposable element, encodes a putative DNA-binding protein. Genes Dev. 1988;2:1205–15. doi: 10.1101/gad.2.10.1205. [DOI] [PubMed] [Google Scholar]
- 29.Zhao K, Hart CM, Laemmli UK. Visualization of chromosomal domains with boundary element-associated factor BEAF-32. Cell. 1995;81:879–89. doi: 10.1016/0092-8674(95)90008-X. [DOI] [PubMed] [Google Scholar]
- 30.Belozerov VE, Majumder P, Shen P, Cai HN. A novel boundary element may facilitate independent gene regulation in the Antennapedia complex of Drosophila. EMBO J. 2003;22:3113–21. doi: 10.1093/emboj/cdg297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gaszner M, Vazquez J, Schedl P. The Zw5 protein, a component of the scs chromatin domain boundary, is able to block enhancer-promoter interaction. Genes Dev. 1999;13:2098–107. doi: 10.1101/gad.13.16.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pai CY, Lei EP, Ghosh D, Corces VG. The centrosomal protein CP190 is a component of the gypsy chromatin insulator. Mol Cell. 2004;16:737–48. doi: 10.1016/j.molcel.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 33.Gause M, Morcillo P, Dorsett D. Insulation of enhancer-promoter communication by a gypsy transposon insert in the Drosophila cut gene: cooperation between suppressor of hairy-wing and modifier of mdg4 proteins. Mol Cell Biol. 2001;21:4807–17. doi: 10.1128/MCB.21.14.4807-4817.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ghosh D, Gerasimova TI, Corces VG. Interactions between the Su(Hw) and Mod(mdg4) proteins required for gypsy insulator function. EMBO J. 2001;20:2518–27. doi: 10.1093/emboj/20.10.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wood AM, Van Bortle K, Ramos E, Takenaka N, Rohrbaugh M, Jones BC, et al. Regulation of chromatin organization and inducible gene expression by a Drosophila insulator. Mol Cell. 2011;44:29–38. doi: 10.1016/j.molcel.2011.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bushey AM, Ramos E, Corces VG. Three subclasses of a Drosophila insulator show distinct and cell type-specific genomic distributions. Genes Dev. 2009;23:1338–50. doi: 10.1101/gad.1798209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nègre N, Brown CD, Shah PK, Kheradpour P, Morrison CA, Henikoff JG, et al. A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet. 2010;6:e1000814. doi: 10.1371/journal.pgen.1000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang N, Emberly E, Cuvier O, Hart CM. Genome-wide mapping of boundary element-associated factor (BEAF) binding sites in Drosophila melanogaster links BEAF to transcription. Mol Cell Biol. 2009;29:3556–68. doi: 10.1128/MCB.01748-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang J, Ramos E, Corces VG. The BEAF-32 insulator coordinates genome organization and function during the evolution of Drosophila species. Genome Res. 2012;22:2199–207. doi: 10.1101/gr.142125.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Bortle K, Ramos E, Takenaka N, Yang J, Wahi JE, Corces VG. Drosophila CTCF tandemly aligns with other insulator proteins at the borders of H3K27me3 domains. Genome Res. 2012;22:2176–87. doi: 10.1101/gr.136788.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hou C, Corces VG. Throwing transcription for a loop: expression of the genome in the 3D nucleus. Chromosoma. 2012;121:107–16. doi: 10.1007/s00412-011-0352-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hart CM, Cuvier O, Laemmli UK. Evidence for an antagonistic relationship between the boundary element-associated factor BEAF and the transcription factor DREF. Chromosoma. 1999;108:375–83. doi: 10.1007/s004120050389. [DOI] [PubMed] [Google Scholar]
- 43.Hart CM, Zhao K, Laemmli UK. The scs’ boundary element: characterization of boundary element-associated factors. Mol Cell Biol. 1997;17:999–1009. doi: 10.1128/mcb.17.2.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whitfield ML, Sherlock G, Saldanha AJ, Murray JI, Ball CA, Alexander KE, et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol Biol Cell. 2002;13:1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Young DW, Hassan MQ, Pratap J, Galindo M, Zaidi SK, Lee SH, et al. Mitotic occupancy and lineage-specific transcriptional control of rRNA genes by Runx2. Nature. 2007;445:442–6. doi: 10.1038/nature05473. [DOI] [PubMed] [Google Scholar]
- 46.Gurudatta BV, Ramos E, Corces VG. The BEAF insulator regulates genes involved in cell polarity and neoplastic growth. Dev Biol. 2012;369:124–32. doi: 10.1016/j.ydbio.2012.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang J, Sung E, Donlin-Asp PG, Corces VG. A subset of Drosophila Myc sites remain associated with mitotic chromosomes co-localized with insulator proteins. Nat Commun. 2013;4:1464. doi: 10.1038/ncomms2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Machanick P, Bailey TL. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics. 2011;27:1696–7. doi: 10.1093/bioinformatics/btr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.MacAlpine HK, Gordân R, Powell SK, Hartemink AJ, MacAlpine DM. Drosophila ORC localizes to open chromatin and marks sites of cohesin complex loading. Genome Res. 2010;20:201–11. doi: 10.1101/gr.097873.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Araki M, Yu H, Asano M. A novel motif governs APC-dependent degradation of Drosophila ORC1 in vivo. Genes Dev. 2005;19:2458–65. doi: 10.1101/gad.1361905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baldinger T, Gossen M. Binding of Drosophila ORC proteins to anaphase chromosomes requires cessation of mitotic cyclin-dependent kinase activity. Mol Cell Biol. 2009;29:140–9. doi: 10.1128/MCB.00981-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zaidi SK, Young DW, Montecino MA, Lian JB, van Wijnen AJ, Stein JL, et al. Mitotic bookmarking of genes: a novel dimension to epigenetic control. Nat Rev Genet. 2010;11:583–9. doi: 10.1038/nrg2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Blobel GA, Kadauke S, Wang E, Lau AW, Zuber J, Chou MM, et al. A reconfigured pattern of MLL occupancy within mitotic chromatin promotes rapid transcriptional reactivation following mitotic exit. Mol Cell. 2009;36:970–83. doi: 10.1016/j.molcel.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dey A, Nishiyama A, Karpova T, McNally J, Ozato K. Brd4 marks select genes on mitotic chromatin and directs postmitotic transcription. Mol Biol Cell. 2009;20:4899–909. doi: 10.1091/mbc.E09-05-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gurudatta BV, Corces VG. Chromatin insulators: lessons from the fly. Brief Funct Genomic Proteomic. 2009;8:276–82. doi: 10.1093/bfgp/elp032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Van Bortle K, Corces VG. Nuclear organization and genome function. Annu Rev Cell Dev Biol. 2012;28:163–87. doi: 10.1146/annurev-cellbio-101011-155824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Young DW, Hassan MQ, Yang XQ, Galindo M, Javed A, Zaidi SK, et al. Mitotic retention of gene expression patterns by the cell fate-determining transcription factor Runx2. Proc Natl Acad Sci USA. 2007;104:3189–94. doi: 10.1073/pnas.0611419104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sarge KD, Park-Sarge OK. Mitotic bookmarking of formerly active genes: keeping epigenetic memories from fading. Cell Cycle. 2009;8:818–23. doi: 10.4161/cc.8.6.7849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Goecks J, Nekrutenko A, Taylor J, Galaxy Team Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang W, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, et al. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8:R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hoover KK, Chien AJ, Corces VG. Effects of transposable elements on the expression of the forked gene of Drosophila melanogaster. Genetics. 1993;135:507–26. doi: 10.1093/genetics/135.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]