Abstract
In dividing animal cells, the centrosome, comprising centrioles and surrounding pericentriolar-material (PCM), is the major interphase microtubule-organizing center (MTOC), arranging a polarized array of microtubules (MTs) that controls cellular architecture. The mouse embryo is a unique setting for investigating the role of centrosomes in MT organization, since the early embryo is acentrosomal, and centrosomes emerge de novo during early cleavages. Here we use embryos from a GFP::CETN2 transgenic mouse to observe the emergence of centrosomes and centrioles in embryos, and show that unfocused acentriolar centrosomes first form in morulae (~16–32-cell stage) and become focused at the blastocyst stage (~64–128 cells) concomitant with the emergence of centrioles. We then used high-resolution microscopy and dynamic tracking of MT growth events in live embryos to examine the impact of centrosome emergence upon interphase MT dynamics. We report that pre-implantation mouse embryos of all stages employ a non-canonical mode of MT organization that generates a complex array of randomly oriented MTs that are preferentially nucleated adjacent to nuclear and plasmalemmal membranes and cell-cell interfaces. Surprisingly, however, cells of the early embryo continue to employ this mode of interphase MT organization even after the emergence of centrosomes. Centrosomes are found at MT-sparse sites and have no detectable impact upon interphase MT dynamics. To our knowledge, the early embryo is unique among proliferating cells in adopting an acentrosomal mode of MT organization despite the presence of centrosomes, revealing that the transition to a canonical mode of interphase MT organization remains incomplete prior to implantation.
Keywords: microtubule organization, pre-implantation embryo, centriole, centrosome, mouse
Introduction
The centrosome, consisting of a pair of perpendicular centrioles surrounded by a cloud of pericentriolar material (PCM), is the dominant microtubule organizing center (MTOC) in most animal cells. In interphase, the centrosome typically lies adjacent to the nucleus, organizing a polarized microtubule (MT) array with MT minus-ends proximal to the centrosome and plus-ends oriented toward the cell periphery. This MT network organizes the cytoplasmic architecture, controlling cell shape, polarity and motility.1-3
Early development poses a challenge in terms of centrosome number. Inheritance of a complete centrosome from each gamete would cause a centrosome excess after fertilization, with potentially damaging consequences.4,5 To circumvent this, oocytes of many species degrade their centrioles and the fertilizing sperm contributes the centrioles for the embryo.6-8 For reasons that remain poorly understood,8 in mouse the sperm also lacks centrioles,8-10 and embryonic development begins without centrioles or centrosomes. Immunofluorescence studies revealed that centrosomes containing the MT-nucleator γ-tubulin are first evident in interphase at morula-stage (~16–32 cells).11,12 These centrosomes are initially acentriolar, and classic electron micrographs first detect centrioles at the blastocyst stage (~64–128 cells).11,13,14 The absence of centrosomes and centrioles in early pre-implantation development suggests that the interphase MT network must be controlled in a centrosome-independent manner. How interphase MTs are organized in early embryos, and how embryos transition from acentrosomal to centrosomal MT organization is unknown.
Here we employ a GFP::CETN2-expressing transgenic mouse to observe the emergence of centrioles and centrosomes during early development and use high-resolution microscopy and tracking of MT growth events in live embryos to quantitatively assess the impact of centrosome emergence upon MT organization. Our experiments detail an acentrosomal mode of MT nucleation that functions in embryos and, unexpectedly, reveal that embryos continue to employ this pathway to govern MT layout in interphase despite the emergence of centrosomes in morulae and blastocysts.
Results
Centriole emergence occurs rapidly across both major cell lineages in late pre-implantation embryos
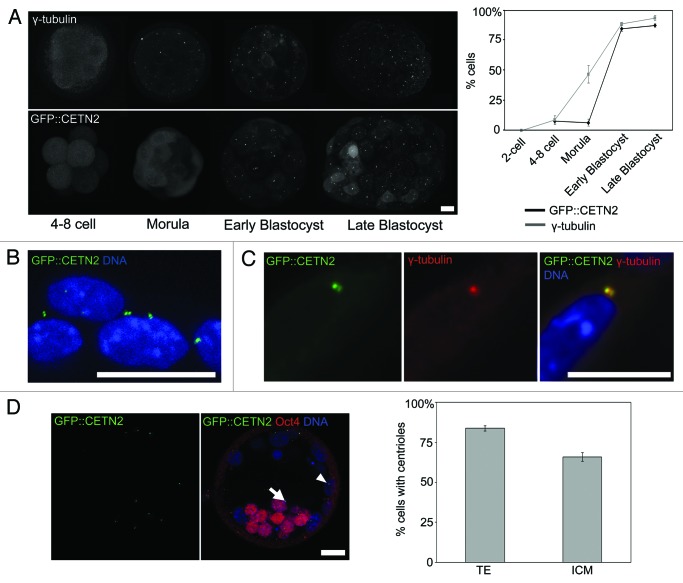
In mouse, centrioles are absent in sperm and oocytes and are first detectable by electron microscopy in blastocysts (~64–128 cells), revealing that centriole formation must occur de novo in late pre-implantation development.11,14 We first revisited this unusual scenario using a newly available transgenic mouse that constitutively expresses a GFP-tagged copy of the centriolar protein centrin-2 (GFP::CETN2). GFP::CETN2 mice display a pair of fluorescent centrioles in each somatic cell and have no distinguishable phenotype15 (and our unpublished data). GFP::CETN2 fluorescence was homogeneous in blastomeres of 4–8-cell stage embryos and in morulae (~16–32-cell stage, Fig. 1), confirming the absence of centrioles. In blastocysts, however, GFP::CETN2 was arranged into pairs of tightly focused spots associated with each nucleus in the majority of cells (Fig. 1), similar to somatic cells. GFP::CETN2-labeled centrioles co-labeled with centrosome markers, including the key MT nucleation factor γ-tubulin (Fig. 1C; Fig. S1). Previous studies suggested that interphase centrosomes, identified as accumulations of centrosomal markers such as γ-tubulin, can be detected in morulae (16–32-cell stage) despite the absence of centrioles.11-14 Accordingly we found that prominent interphase γ-tubulin foci were frequently evident in morulae (Fig. 1A). Thus, as previously suggested,11 centriole emergence occurs at the blastocyst stage of development, and is preceded by the formation of acentriolar centrosomes in ~16–32 cell morula stage embryos.
Figure 1. De novo emergence of centrosomes and centrioles in murine embryos. (A) Confocal Z-projections of GFP::CETN2-labeled and γ-tubulin immunolabeled embryos. Quantification of the proportion of nuclei with an associated GFP::CETN2 centriole pair (10–23 embryos/stage examined) or γ-tubulin focus (6–12/stage) is to the right. Note that the 2-cell embryo fluorescence pattern was identical to the 4-cells. (B) A zoomed image of a GFP::CETN2 blastocyst. Note pairs of GFP::CETN2-labeled centrioles associated with each nucleus. (C) Colocalization of γ-tubulin with GFP::CETN2-labeled centrioles. (D) Comparison of the proportion of Oct4-positive and Oct4-negative cells possessing centrioles in early cavitating blastocysts (n = 18). Note that although there is a greater proportion of centriole-containing cells in the trophoectoderm (arrowhead) compared with inner cell mass (arrow), the majority of cells possess centrioles in both lineages. Scale 20 µm.
The morula-blastocyst transition involves the establishment of two distinct cell lineages—the inner cell mass (ICM) and trophoectoderm (TE). To determine whether centrioles appear de novo in one lineage before the other, we examined GFP::CETN2-labeled centrioles in blastocysts co-stained for the ICM marker Oct4. In order to differentiate the emergence of centrioles in the 2-cell lineages, we examined GFP::CETN2 mice from newly forming blastocysts. At this stage, there was a significantly greater proportion of centriole-containing cells in the TE compared with in the ICM (p < 0.001). Nonetheless the majority of cells in both lineages possessed centrioles at this stage (83.74 +/− 1.67% in the TE vs. 65.98 +/−2.69% in the ICM; p < 0.001). Given that centrioles are almost never seen in morulae (16–32 cells), this indicates that centriole emergence at the morula-blastocyst transition is rapid and relatively synchronous across both cell types at the time of the morula-blastocyst transition (Fig. 1D). In summary, the early embryo lacks centrosomes, but acentriolar centrosomes are first detected in morulae (16–32 cells). Centrioles emerge 1–2 cell cycles later around the time of blastocyst formation (64–128 cells) within a relatively narrow window across both developing cell lineages.
Centrosome focusing at the morula-blastocyst transition
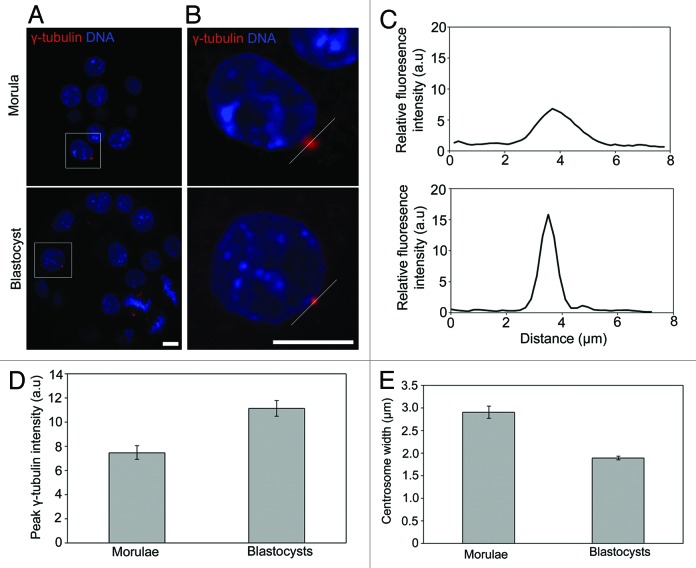
We next set out to examine the geometry of the γ-tubulin-containing centrosome in morulae and blastocysts. Given the key role of centrioles in centrosome organization,16,17 we wondered whether centriole emergence in blastocysts might coincide with a change in centrosome shape. To address this, we performed a quantitative comparison of the geometry of γ-tubulin-labeled centrosomes in morulae and blastocysts (Fig. 2). Centrosomes were immunolabeled using γ-tubulin antibodies (Fig. 2A and B), and linescan fluorescence intensity analysis of confocal images used to quantify centrosome geometry (Fig. 2B and C). Normalized peak fluorescence was used as a measure of γ-tubulin density at the centrosome, and the width of the linescan curve was used to measure centrosome width (Fig. 2C). Peak fluorescence intensity was significantly greater (p < 10−4, Fig. 2D) in blastocysts compared with morulae, and average centrosomal width was significantly smaller in blastocysts (p < 10−12, Fig. 2E), suggesting a focusing of the centrosome at the morula-blastocyst transition. Thus, preliminary centrosomes possessing γ-tubulin first emerge in morula, but then become focused in blastocysts coincident with the emergence of centrioles.
Figure 2. Centrosome focusing concomitant with centriole emergence. (A) Typical confocal sections of γ-tubulin foci in morulae and blastocysts. (B) Zoom of the area highlighted in (A). White lines indicate the linescan plotted in (C). (D) Quantification of peak γ-tubulin intensity and (E) centrosome width in morulae and blastocysts. 50–70 γ-tubulin foci examined from 9–10 embryos per stage. Scale 10 µm.
Centrosome emergence does not affect MT organization
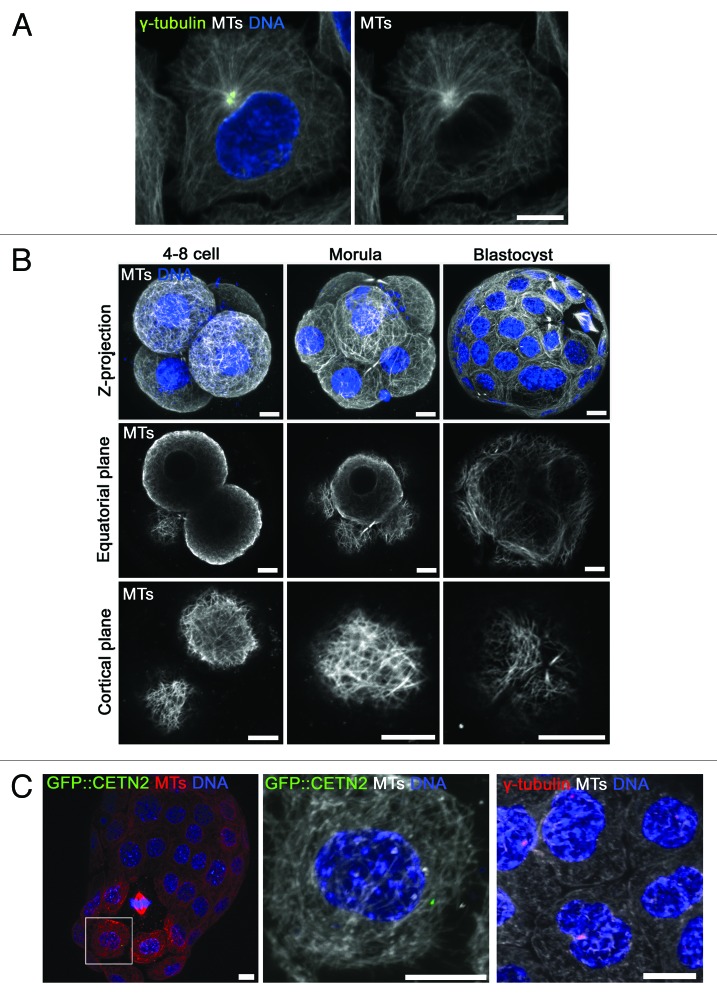
Having clarified the dynamics of centrosome and centriole emergence, we sought to explore the impact of centrosome emergence upon interphase MT organization. In typical dividing cells, the centrosome is juxta-nuclear and serves as a significant MTOC from which new MTs are nucleated.1,2,18,19 Accordingly, as is the case in many cultured cells, we found in HeLa cells that the centrosome is at the center of a significant thickening of MTs that radiate outwards toward the cortex (see Fig. 3A). To examine MT organization in embryos, we generated high-quality immunofluorescence images of interphase MTs. In 4–8-cell stage embryos, we observed an intricate network of MTs extending throughout the cytoplasm that was enriched adjacent to the nuclear membrane and plasmalemma, as reported for isolated early blastomeres.20,21 Consistent with the absence of centrosomes, we found no evidence of a thickening of MTs that would indicate a dominant site of MT nucleation (Fig. 3B).
Figure 3. Centrosomes are not major MTOCs in interphase embryos. (A) An interphase HeLa cell immunolabeled for MTs and γ-tubulin. (B) MT immunofluorescence in embryos. For each stage, a Z-projection is shown along with the equatorial and cortical plane from the same embryo. Note the intricate network of MTs observed in the cell cortex. However, no single major focus of MT nucleation was observed at any developmental stage. (C) Zoomed images of MT immunofluorescence in late (hatching; 6 d post-hCG) blastocysts from GFP::CETN2 mice (left) or labeled with γ-tubulin antibodies (right). Note that centrioles and centrosomes are not at sites of MT enrichment in the majority of blastomeres. 433 centrosomes were examined from 29 blastocysts. See also Figure S2. Scale 10 µm.
We next examined MTs in morulae and blastocysts. Notably, we were also unable to find a major thickening of MTs in these later embryos (Fig. 3B). This was unexpected, since these cells possess centrosomes. Examination of late blastocysts (day 6 post-hCG) expressing GFP::CETN2 or labeled with γ-tubulin antibodies revealed that centrioles and centrosomes were usually found at MT-sparse sites (Fig. 3C; Fig. S2A). Centrosomes were only occasionally found at sites of relative MT enrichment (< 15% of centrosomes; Fig. S2), and we never observed a radial array of MTs emanating from the centrosome. This suggested that centrosomes of the late pre-implantation embryo fail to serve as an interphase MTOC.
Examination of MT dynamics in interphase embryos
To better understand MT organization in embryos, we set out to perform more detailed analyses of interphase MT dynamics. For this we employed EB1::GFP, which labels growing MT plus-ends in live cells without affecting cellular function and so allows direct visualization of MT growth events and measurement of growth velocities and trajectories in live cells.18,19,22,23 EB1::GFP-expressing somatic cells are characterized by a major accumulation of EB1::GFP at the centrosome, with EB1::GFP comets radiating outwards from this central site toward the cell periphery18,19 (Fig. S3).
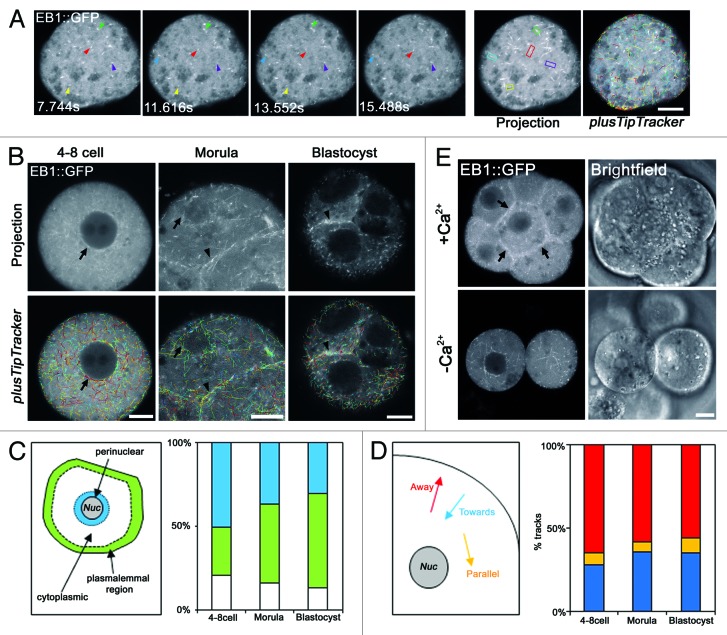
We expressed EB1::GFP in embryos by microinjection of mRNA at the 2-cell stage. Importantly, this approach had no effect upon embryo health as assessed by development to blastocyst in vitro. We first examined successive Z-sections of EB1::GFP-expressing embryos and found no evidence of a focal accumulation of EB1::GFP that might indicate a major MTOC at any stage, including morula and blastocysts which have centrosomes (Fig. S4). To obtain more detailed information, we generated rapid-acquisition (~2 sec intervals) movies of EB1::GFP-expressing embryos. At all developmental stages the movies revealed numerous comet-like structures moving in approximately linear trajectories, similar to those observed in other systems (Fig. 4A and B). To quantitatively assess the movement of the EB1::GFP comets, we used plusTipTracker software,24,25 which identifies EB1::GFP comets for automated assessment of location, velocity and directionality of MT growth events (Fig. 4A and B). We first analyzed EB1::GFP movie data sets made in 4–8-cell embryos. Consistent with our immunofluorescence observations of MT enrichment in the cortex and peri-nuclear region, EB1::GFP movies and automated analysis established that MT growth events are more frequent adjacent to nuclear and plasmalemmal membranes compared with the remainder of the cytoplasm (Fig. 4C). Analysis of trajectories of MT growth events in the remainder of the cytoplasm revealed them to be randomly oriented (Fig. 4D). In contrast to somatic cells, but consistent with the absence of centrosomes in 4–8-cell embryos, there was no single subcellular location acting as a source of EB1::GFP comets.
Figure 4. Dynamic tracking of MT growth events in early embryos. (A) Representative confocal time-series of EB1::GFP comets in a 4–8-cell stage blastomere. Each colored arrowhead tracks the movement of a single EB1::GFP comet. A maximum intensity projection of the entire time series is shown (right). Each MT growth event appears as a line of fluorescence. MT growth events identified by plusTipTracker software are shown far right. (B) Typical examples of MT growth events during development. (C) Analysis of relative density of MT growth events corrected for area in perinuclear, plasmalemmal and cytoplasmic regions. 1016–8309 tracks from 5–12 movies examined per developmental stage. (D) Analysis of cytoplasmic MT growth directionality in developing embryos. 49–125 tracks from 5–11 movies. Tracks were considered parallel to the nucleus within a ~10° range of the nuclear tangent. (E) Projections of morula-stage embryos experimentally de-compacted by removal of extracellular Ca2+. Note loss of cell-cell adhesion reduces EB1::GFP track abundance at cell-cell interfaces (arrows; n = 20). Scale 10 µm.
We next examined the MT growth events in morulae and blastocysts. Similar to earlier developmental stages, growth events were frequent in the perinuclear region and the sub-plasmalemmal region (Fig. 4C). In addition, MT growth events were enriched at sites of cell-cell contact (Fig. 4B and C). Experimentally reversing embryo compaction by removing extracellular Ca2+ reduced the density of MT growth events at the plasmalemmal region, confirming that cell-cell interfaces act as major sites of nucleation (Fig. 4E), as has been seen in epithelial cells.26,27 Strikingly, however, there was no evidence of any single subcellular location serving as a major source of MT nucleation in morulae and blastocysts. Similar to earlier developmental stages, cytoplasmic comets had random trajectories (Fig. 4B and D). Although there was a slight decrease in MT growth velocity (Fig. S5), analysis of MT growth trajectories revealed no change in MT orientation between 4-cell and blastocyst stages (CHI2 p = 0.64, Fig. 4D). The early embryo therefore employs a non-canonical acentrosomal mode of interphase MT organization consisting of apparently randomly oriented MTs enriched at nuclear and plasmalemmal surfaces and cell-cell boundaries. Unexpectedly, this persists as the major MT assembly pathway despite the presence of γ-tubulin-rich centrosomes at later developmental stages.
Discussion
Here we have examined MT dynamics in an unusual setting in which dividing cells transition from acentriolar to centriolar divisions within a relatively narrow developmental window. Our data detail an acentrosomal mechanism of interphase MT organization that operates throughout pre-implantation mouse development to generate a random non-radial cytoplasmic MT array enriched in the juxta-nuclear and plasmalemmal regions. Unexpectedly, however, centrosome formation and focusing has little or no effect upon the MT network, and embryo blastomeres continue to employ the acentrosomal MT assembly pathway despite the presence of centrosomes. The contrasting patterns of MT nucleation in the early embryo compared with the “textbook” dividing cell are illustrated by the cartoon in Figure 5. The discussion that follows will address the unusual scenario of de novo centrosome emergence in the mouse embryo, the mechanism of MT organization in embryos, and the physiological relevance of non-canonical MT dynamics in the early embryo.
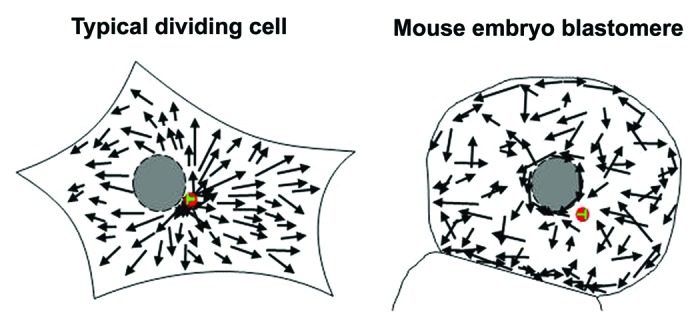
Figure 5. Comparison of MT nucleation in embryos and the typical dividing cell. Cartoon comparing MT nucleation in embryos and the typical dividing cell. In dividing cells the centrosome (red) nucleates MTs, which radiate toward the cell periphery. In embryo blastomeres, in contrast, the centrosome sits at MT sparse sites, with apparently random directionality of cytoplasmic MT growth events.
De novo centriole and centrosome formation in mouse embryos
Centrioles and centrosomes are absent in oocytes of many species, and the absence of centrioles in mouse sperm has been known for some time, classic studies revealing that centrioles are first detectable by electron microscopy in 64–128-cell blastocysts.11,13 The availability of a transgenic mouse expressing GFP::CETN2 allowed us to revisit this question, examining the presence of centrioles within each cell in relatively large numbers of embryos. We confirm that centriole emergence occurs in blastocysts, and show that this happens within a relatively narrow time-course across both major cell lineages and corresponds with a focusing of the previously acentriolar centrosomal material. The mechanism by which centrosomes and centrioles emerge in the embryo remains to be addressed. In somatic cells, a network of proteins has been identified that control centriole replication during mitotic S-phase.28,29 Whether the same network of proteins is involved in de novo assembly in mouse embryos remains to be seen. It is intriguing, however, that some members of this pathway including SAS6 and PLK4 can induce centriole-like structures when overexpressed in acentriolar Drosophila eggs,30-32 indicating that the same pathway may operate in centriole replication and de novo formation.
An interesting feature of centrosome emergence in the mouse embryo is that centriole formation is preceded by the emergence of an acentriolar accumulation of PCM in morulae. It might seem counterintuitive that acentriolar centrosomes should precede the emergence of bona fide centrioles, since centrioles are commonly viewed as the platform upon which the centrosome is assembled, and experimental disruption of the centrioles causes disassembly of the centrosome.16,17 A clue as to the role of the acentriolar centrosome in morulae may come from experiments in which overexpression of a core PCM protein generates extra sites for the assembly of extra centrioles.33 Similarly, the accumulation of PCM in morulae might be a prerequisite for the assembly of centrioles in blastocysts. The molecular underpinnings of centriole and centrosome emergence in the mouse embryo remain to be dissected.
Acentrosomal MT nucleation in mouse embryos
A focus of the current study was to use the mouse embryo to study how MTs are organized in dividing cells acquiring centrosomes and centrioles for the first time. We first detailed the mechanism of MT organization in acentriolar embryos, finding that MTs are randomly oriented and enriched adjacent to the nucleus and plasmalemma. We then examined MT dynamics in morulae and blastocysts, expecting the emergence of centrosomes and centrioles to cause a shift toward a canonical centrosome-dictated polarized MT array commonly seen in other cell types. Unexpectedly, however, two independent approaches failed to uncover a substantial impact of centrosome emergence. First, immunofluorescence images showed no evidence of a substantial thickening of MTs in blastocysts that might be expected to accompany the centrosome, and the centrosome was typically found at MT-sparse sites. Second, detailed analyses of MTs using EB1::GFP uncovered no shift in growth trajectories during morula and blastocyst formation when the centrosomes are forming. The centrosome-independence of MT dynamics is apparently specific to interphase, as GFP::CETN-2 and γ-tubulin are located at spindle poles in mitosis in blastocysts (refs. 11, 12 and 34 ; see Fig. S6), suggesting that centrosomes influence M-phase MT dynamics. Nonetheless, centrosomes in morulae and blastocysts apparently fail to dictate MT layout in interphase, and the acentrosomal mode persists as the dominant means of organizing MTs.
The absence of centrosomes in oocytes and (in some species) embryos is well known, and the mechanism of spindle assembly during M-phase in these settings has been intensely studied.35 However, less is known about how MTs can be organized in the absence of centrosomes in interphase. MTs can self-organize in experimentally generated acentrosomal cell fragments,36-39 or following laser ablation of centrosomes.40 Acentrosomal MT organization occurs normally in some terminally differentiated cells, where formation of linear MT arrays can accompany cellular specialization.41 However, examples of acentrosomal MT organization in normal dividing cells are rare. One clear example is dividing Drosophila cells, in which centrosomes disassemble in interphase,42 such that PCM dissociates from the centrioles. In this setting the MTs organize into a non-radial acentrosomal MT array morphologically similar to that which we see in mouse embryos,42 and the birth of live flies following centriole disruption reveals the functional sufficiency of the acentrosomal interphase array.43,44 The scenario in the mouse embryo is mechanistically distinct from Drosophila, however, in that centrosomes do not disassemble in interphase mouse embryos, as indicated by the continued presence of γ-tubulin at the centrioles in interphase. What nucleates MTs in interphase mouse embryos, and why the cytoplasm elects to persist with this pathway despite the presence of γ-tubulin-rich centrosomes in later pre-implantation embryos remain to be unravelled. To our knowledge, cells of the pre-implantation mouse embryo are unique among non-differentiated cells in employing an acentriolar mode of MT assembly to organize the cytoplasm despite the presence of γ-tubulin-rich centrosomes.
Transition toward canonical microtubule dynamics in the early embryo
Understanding the mechanisms of MT dynamics is of particular interest in the early mammalian embryo, since the early embryo is extraordinarily susceptible to chromosome segregation errors, which may be a contributing factor to infertility.45,46 Recent studies revealed that mitotic spindles in blastocyst are morphologically similar to somatic cell spindles, suggesting that mitotic MT organization transitions from an oocyte-mode to a somatic-mode by the end of pre-implantation development.34,47 In contrast, our detailed analysis of interphase MT dynamics provides clear evidence of non-canonical MT behavior throughout pre-implantation development. Further detailed analyses of spindle MT dynamics in blastocysts may yet uncover idiosyncrasies of mitotic MT dynamics that might underpin the apparent lack of chromosome segregation quality control in these cells. Interestingly, aneuploidy is also common in cultured human embryonic stem cells,48-50 which originate from the inner cell mass. We speculate that the completion of the journey toward canonical MT organization might coincide with a tightening of chromosome segregation fidelity.
Materials and Methods
Oocyte and embryo handling
To obtain embryos, MF1 female mice were administered with PMSG (7IU; Intervet) and hCG (5IU; Intervet) at 48 h intervals and mated with male mice at the time of hCG administration. Two-cell embryos were collected 48 h later. GFP::CETN2-expressing embryos were obtained by mating MF1 females with hemizygous transgenic males15 purchased from Jax Mice. Embryo handling was performed in Hepes-KSOM,51 and long-term embryo culture was performed in KSOM media (Millipore). Microinjection was performed on a Leica DMI4000 microscope using Narishige micromanipulators, as previously described.52
Fluorescent proteins and imaging
EB1::GFP19 was subcloned from pEGFP-N1 (Clontech) into pcDNA 3.1/myc-His(−) using NheI and NotI (NEB). Polyadenylated mRNA was manufactured using mMessage mMachine Ultra (Ambion), as previously.52 Oocyte and embryo processing for immunofluorescence was performed as previously.53 Antibodies used: mouse anti γ-tubulin (Sigma-T5326; 1:1,000), mouse anti-β-tubulin (Sigma-T4026; 1:1,000), rat anti-α-tubulin (YL1/2 abcam ab6160), with fluorescent secondary antibodies as appropriate (Invitrogen). Chromatin was labeled using 5 μg/ml Hoechst 33343 for 5 min for fixed samples, and 5 sec for live samples. Imaging was performed on a Zeiss-710 confocal microscope. Imaging of live embryos was performed at 37°C.
Image analysis
Analysis of centrosome focusing was performed using the “plot profile” function in ImageJ and normalized for each image.
Confocal images of live EB1::GFP-expressing embryos were obtained at 1.936 sec intervals. EB1::GFP movies were analyzed using plusTipTracker software, which identifies and quantitates comet motion. We first used plusTipParamSweepGUI to identify optimal tracking parameters for analysis of MT growth events. We tested a range of values for the following parameters: search radius, maximum gap length, maximum forward angle and fluctuation radius. Variations of each of these had a minimal effect on the number, speed and lifetime of growth tracks identified. We therefore used the standard parameters described in Applegate et al. (2011),24 which have previously been used to track MT growth events in mammalian cells, adjusted for pixel size and frame rate. Tracks that start in the first frame or finish in the last frame were automatically excluded from growth speed analysis. ROIs were defined using plusTipSeeTracks, which allowed identification of tracks in each of the three regions. Growth events were categorized as perinuclear or cortical if more than 50% of their lifetime was spent within 2 μm of the nuclear/plasma membrane, respectively. Analysis of cytoplasmic MT growth directionality was performed manually on ~20 plusTipTracker-identified tracks per movie.
Supplementary Material
Acknowledgments
Funded by an MRC Project Grant to GF. We thank Lynn Cassimeris and John Carroll for discussions and Zhi Yao, Rachel Moore and Jenny Bormann for advice and assistance.
Disclosure of Potential Conflicts
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/cc/article/24755
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/24755
References
- 1.Bornens M. Centrosome composition and microtubule anchoring mechanisms. Curr Opin Cell Biol. 2002;14:25–34. doi: 10.1016/S0955-0674(01)00290-3. [DOI] [PubMed] [Google Scholar]
- 2.Dammermann A, Desai A, Oegema K. The minus end in sight. Curr Biol. 2003;13:R614–24. doi: 10.1016/S0960-9822(03)00530-X. [DOI] [PubMed] [Google Scholar]
- 3.de Forges H, Bouissou A, Perez F. Interplay between microtubule dynamics and intracellular organization. Int J Biochem Cell Biol. 2012;44:266–74. doi: 10.1016/j.biocel.2011.11.009. [DOI] [PubMed] [Google Scholar]
- 4.Bettencourt-Dias M, Hildebrandt F, Pellman D, Woods G, Godinho SA. Centrosomes and cilia in human disease. Trends Genet. 2011;27:307–15. doi: 10.1016/j.tig.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nigg EA, Raff JW. Centrioles, centrosomes, and cilia in health and disease. Cell. 2009;139:663–78. doi: 10.1016/j.cell.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 6.Delattre M, Gönczy P. The arithmetic of centrosome biogenesis. J Cell Sci. 2004;117:1619–30. doi: 10.1242/jcs.01128. [DOI] [PubMed] [Google Scholar]
- 7.Manandhar G, Schatten H, Sutovsky P. Centrosome reduction during gametogenesis and its significance. Biol Reprod. 2005;72:2–13. doi: 10.1095/biolreprod.104.031245. [DOI] [PubMed] [Google Scholar]
- 8.Schatten G. The centrosome and its mode of inheritance: the reduction of the centrosome during gametogenesis and its restoration during fertilization. Dev Biol. 1994;165:299–335. doi: 10.1006/dbio.1994.1256. [DOI] [PubMed] [Google Scholar]
- 9.Manandhar G, Sutovsky P, Joshi HC, Stearns T, Schatten G. Centrosome reduction during mouse spermiogenesis. Dev Biol. 1998;203:424–34. doi: 10.1006/dbio.1998.8947. [DOI] [PubMed] [Google Scholar]
- 10.Manandhar G, Simerly C, Salisbury JL, Schatten G. Centriole and centrin degeneration during mouse spermiogenesis. Cell Motil Cytoskeleton. 1999;43:137–44. doi: 10.1002/(SICI)1097-0169(1999)43:2<137::AID-CM5>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 11.Gueth-Hallonet C, Antony C, Aghion J, Santa-Maria A, Lajoie-Mazenc I, Wright M, et al. gamma-Tubulin is present in acentriolar MTOCs during early mouse development. J Cell Sci. 1993;105:157–66. doi: 10.1242/jcs.105.1.157. [DOI] [PubMed] [Google Scholar]
- 12.Palacios MJ, Joshi HC, Simerly C, Schatten G. Gamma-tubulin reorganization during mouse fertilization and early development. J Cell Sci. 1993;104:383–9. doi: 10.1242/jcs.104.2.383. [DOI] [PubMed] [Google Scholar]
- 13.Szollosi D, Calarco P, Donahue RP. Absence of centrioles in the first and second meiotic spindles of mouse oocytes. J Cell Sci. 1972;11:521–41. doi: 10.1242/jcs.11.2.521. [DOI] [PubMed] [Google Scholar]
- 14.Abumuslimov SS, Nadezhdina ES, Chentsov IuS. [An electron microscopic study of centriole and centrosome morphogenesis in the early development of the mouse] Tsitologiia. 1994;36:1054–61. [PubMed] [Google Scholar]
- 15.Higginbotham H, Bielas S, Tanaka T, Gleeson JG. Transgenic mouse line with green-fluorescent protein-labeled Centrin 2 allows visualization of the centrosome in living cells. Transgenic Res. 2004;13:155–64. doi: 10.1023/B:TRAG.0000026071.41735.8e. [DOI] [PubMed] [Google Scholar]
- 16.Bobinnec Y, Khodjakov A, Mir LM, Rieder CL, Eddé B, Bornens M. Centriole disassembly in vivo and its effect on centrosome structure and function in vertebrate cells. J Cell Biol. 1998;143:1575–89. doi: 10.1083/jcb.143.6.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abal M, Keryer G, Bornens M. Centrioles resist forces applied on centrosomes during G2/M transition. Biol Cell. 2005;97:425–34. doi: 10.1042/BC20040112. [DOI] [PubMed] [Google Scholar]
- 18.Piehl M, Tulu US, Wadsworth P, Cassimeris L. Centrosome maturation: measurement of microtubule nucleation throughout the cell cycle by using GFP-tagged EB1. Proc Natl Acad Sci U S A. 2004;101:1584–8. doi: 10.1073/pnas.0308205100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piehl M, Cassimeris L. Organization and dynamics of growing microtubule plus ends during early mitosis. Mol Biol Cell. 2003;14:916–25. doi: 10.1091/mbc.E02-09-0607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Houliston E, Pickering SJ, Maro B. Redistribution of microtubules and pericentriolar material during the development of polarity in mouse blastomeres. J Cell Biol. 1987;104:1299–308. doi: 10.1083/jcb.104.5.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Houliston E, Maro B. Posttranslational modification of distinct microtubule subpopulations during cell polarization and differentiation in the mouse preimplantation embryo. J Cell Biol. 1989;108:543–51. doi: 10.1083/jcb.108.2.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tirnauer JS, O’Toole E, Berrueta L, Bierer BE, Pellman D. Yeast Bim1p promotes the G1-specific dynamics of microtubules. J Cell Biol. 1999;145:993–1007. doi: 10.1083/jcb.145.5.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mimori-Kiyosue Y, Shiina N, Tsukita S. The dynamic behavior of the APC-binding protein EB1 on the distal ends of microtubules. Curr Biol. 2000;10:865–8. doi: 10.1016/S0960-9822(00)00600-X. [DOI] [PubMed] [Google Scholar]
- 24.Applegate KT, Besson S, Matov A, Bagonis MH, Jaqaman K, Danuser G. plusTipTracker: Quantitative image analysis software for the measurement of microtubule dynamics. J Struct Biol. 2011;176:168–84. doi: 10.1016/j.jsb.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nishimura Y, Applegate K, Davidson MW, Danuser G, Waterman CM. Automated screening of microtubule growth dynamics identifies MARK2 as a regulator of leading edge microtubules downstream of Rac1 in migrating cells. PLoS One. 2012;7:e41413. doi: 10.1371/journal.pone.0041413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meng W, Mushika Y, Ichii T, Takeichi M. Anchorage of microtubule minus ends to adherens junctions regulates epithelial cell-cell contacts. Cell. 2008;135:948–59. doi: 10.1016/j.cell.2008.09.040. [DOI] [PubMed] [Google Scholar]
- 27.Chausovsky A, Bershadsky AD, Borisy GG. Cadherin-mediated regulation of microtubule dynamics. Nat Cell Biol. 2000;2:797–804. doi: 10.1038/35041037. [DOI] [PubMed] [Google Scholar]
- 28.Strnad P, Gönczy P. Mechanisms of procentriole formation. Trends Cell Biol. 2008;18:389–96. doi: 10.1016/j.tcb.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 29.Loncarek J, Khodjakov A. Ab ovo or de novo? Mechanisms of centriole duplication. Mol Cells. 2009;27:135–42. doi: 10.1007/s10059-009-0017-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peel N, Stevens NR, Basto R, Raff JW. Overexpressing centriole-replication proteins in vivo induces centriole overduplication and de novo formation. Curr Biol. 2007;17:834–43. doi: 10.1016/j.cub.2007.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodrigues-Martins A, Bettencourt-Dias M, Riparbelli M, Ferreira C, Ferreira I, Callaini G, et al. DSAS-6 organizes a tube-like centriole precursor, and its absence suggests modularity in centriole assembly. Curr Biol. 2007;17:1465–72. doi: 10.1016/j.cub.2007.07.034. [DOI] [PubMed] [Google Scholar]
- 32.Rodrigues-Martins A, Riparbelli M, Callaini G, Glover DM, Bettencourt-Dias M. Revisiting the role of the mother centriole in centriole biogenesis. Science. 2007;316:1046–50. doi: 10.1126/science.1142950. [DOI] [PubMed] [Google Scholar]
- 33.Loncarek J, Hergert P, Magidson V, Khodjakov A. Control of daughter centriole formation by the pericentriolar material. Nat Cell Biol. 2008;10:322–8. doi: 10.1038/ncb1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Courtois A, Schuh M, Ellenberg J, Hiiragi T. The transition from meiotic to mitotic spindle assembly is gradual during early mammalian development. J Cell Biol. 2012;198:357–70. doi: 10.1083/jcb.201202135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dumont J, Desai A. Acentrosomal spindle assembly and chromosome segregation during oocyte meiosis. Trends Cell Biol. 2012;22:241–9. doi: 10.1016/j.tcb.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carazo-Salas RE, Nurse P. Self-organization of interphase microtubule arrays in fission yeast. Nat Cell Biol. 2006;8:1102–7. doi: 10.1038/ncb1479. [DOI] [PubMed] [Google Scholar]
- 37.Rodionov V, Nadezhdina E, Borisy G. Centrosomal control of microtubule dynamics. Proc Natl Acad Sci U S A. 1999;96:115–20. doi: 10.1073/pnas.96.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karsenti E, Kobayashi S, Mitchison T, Kirschner M. Role of the centrosome in organizing the interphase microtubule array: properties of cytoplasts containing or lacking centrosomes. J Cell Biol. 1984;98:1763–76. doi: 10.1083/jcb.98.5.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Daga RR, Lee KG, Bratman S, Salas-Pino S, Chang F. Self-organization of microtubule bundles in anucleate fission yeast cells. Nat Cell Biol. 2006;8:1108–13. doi: 10.1038/ncb1480. [DOI] [PubMed] [Google Scholar]
- 40.Khodjakov A, Cole RW, Oakley BR, Rieder CL. Centrosome-independent mitotic spindle formation in vertebrates. Curr Biol. 2000;10:59–67. doi: 10.1016/S0960-9822(99)00276-6. [DOI] [PubMed] [Google Scholar]
- 41.Bartolini F, Gundersen GG. Generation of noncentrosomal microtubule arrays. J Cell Sci. 2006;119:4155–63. doi: 10.1242/jcs.03227. [DOI] [PubMed] [Google Scholar]
- 42.Rogers GC, Rusan NM, Peifer M, Rogers SL. A multicomponent assembly pathway contributes to the formation of acentrosomal microtubule arrays in interphase Drosophila cells. Mol Biol Cell. 2008;19:3163–78. doi: 10.1091/mbc.E07-10-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Basto R, Lau J, Vinogradova T, Gardiol A, Woods CG, Khodjakov A, et al. Flies without centrioles. Cell. 2006;125:1375–86. doi: 10.1016/j.cell.2006.05.025. [DOI] [PubMed] [Google Scholar]
- 44.Megraw TL, Kao LR, Kaufman TC. Zygotic development without functional mitotic centrosomes. Curr Biol. 2001;11:116–20. doi: 10.1016/S0960-9822(01)00017-3. [DOI] [PubMed] [Google Scholar]
- 45.Lightfoot DA, Kouznetsova A, Mahdy E, Wilbertz J, Höög C. The fate of mosaic aneuploid embryos during mouse development. Dev Biol. 2006;289:384–94. doi: 10.1016/j.ydbio.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 46.Vanneste E, Voet T, Le Caignec C, Ampe M, Konings P, Melotte C, et al. Chromosome instability is common in human cleavage-stage embryos. Nat Med. 2009;15:577–83. doi: 10.1038/nm.1924. [DOI] [PubMed] [Google Scholar]
- 47.Yamagata K, FitzHarris G. 4D imaging reveals a shift in chromosome segregation dynamics during mouse pre-implantation development. Cell Cycle. 2013;12:157–65. doi: 10.4161/cc.23052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Holubcová Z, Matula P, Sedláčková M, Vinarský V, Doležalová D, Bárta T, et al. Human embryonic stem cells suffer from centrosomal amplification. Stem Cells. 2011;29:46–56. doi: 10.1002/stem.549. [DOI] [PubMed] [Google Scholar]
- 49.Maitra A, Arking DE, Shivapurkar N, Ikeda M, Stastny V, Kassauei K, et al. Genomic alterations in cultured human embryonic stem cells. Nat Genet. 2005;37:1099–103. doi: 10.1038/ng1631. [DOI] [PubMed] [Google Scholar]
- 50.Draper JS, Smith K, Gokhale P, Moore HD, Maltby E, Johnson J, et al. Recurrent gain of chromosomes 17q and 12 in cultured human embryonic stem cells. Nat Biotechnol. 2004;22:53–4. doi: 10.1038/nbt922. [DOI] [PubMed] [Google Scholar]
- 51.Lawitts JA, Biggers JD. Culture of preimplantation embryos. Methods Enzymol. 1993;225:153–64. doi: 10.1016/0076-6879(93)25012-Q. [DOI] [PubMed] [Google Scholar]
- 52.Fitzharris G. A shift from kinesin 5-dependent metaphase spindle function during preimplantation development in mouse. Development. 2009;136:2111–9. doi: 10.1242/dev.035089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Illingworth C, Pirmadjid N, Serhal P, Howe K, Fitzharris G. MCAK regulates chromosome alignment but is not necessary for preventing aneuploidy in mouse oocyte meiosis I. Development. 2010;137:2133–8. doi: 10.1242/dev.048306. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.