Abstract
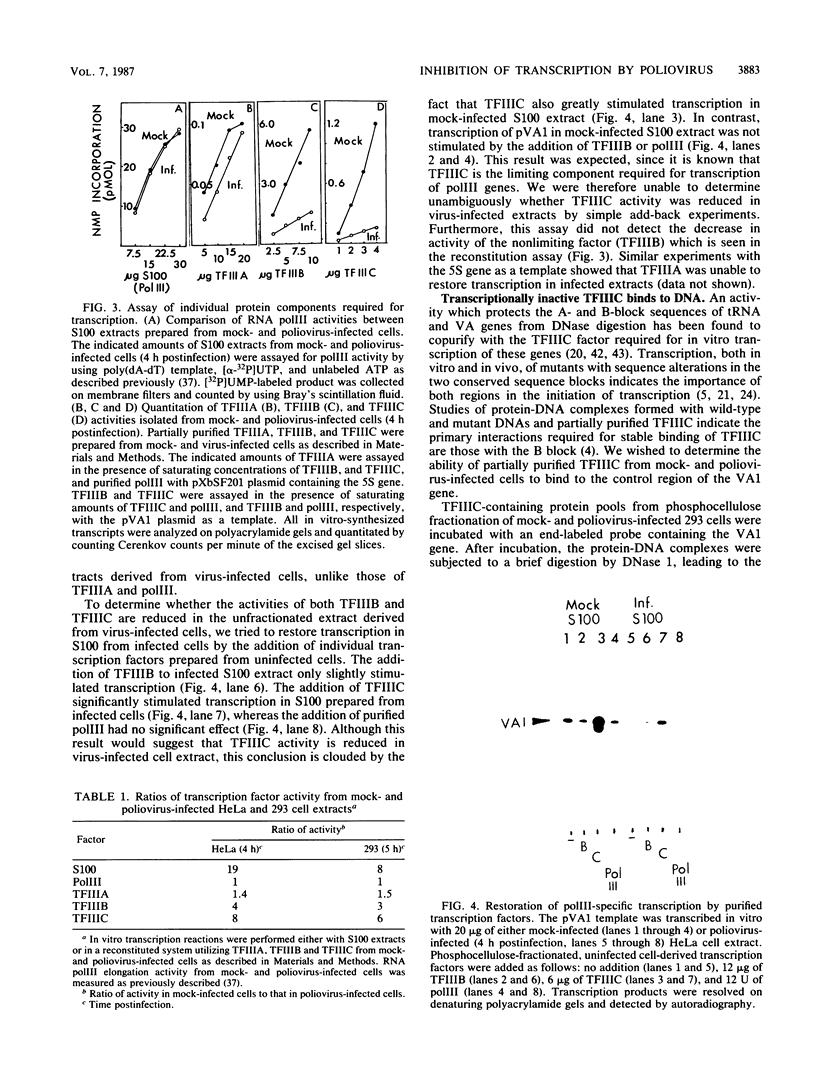
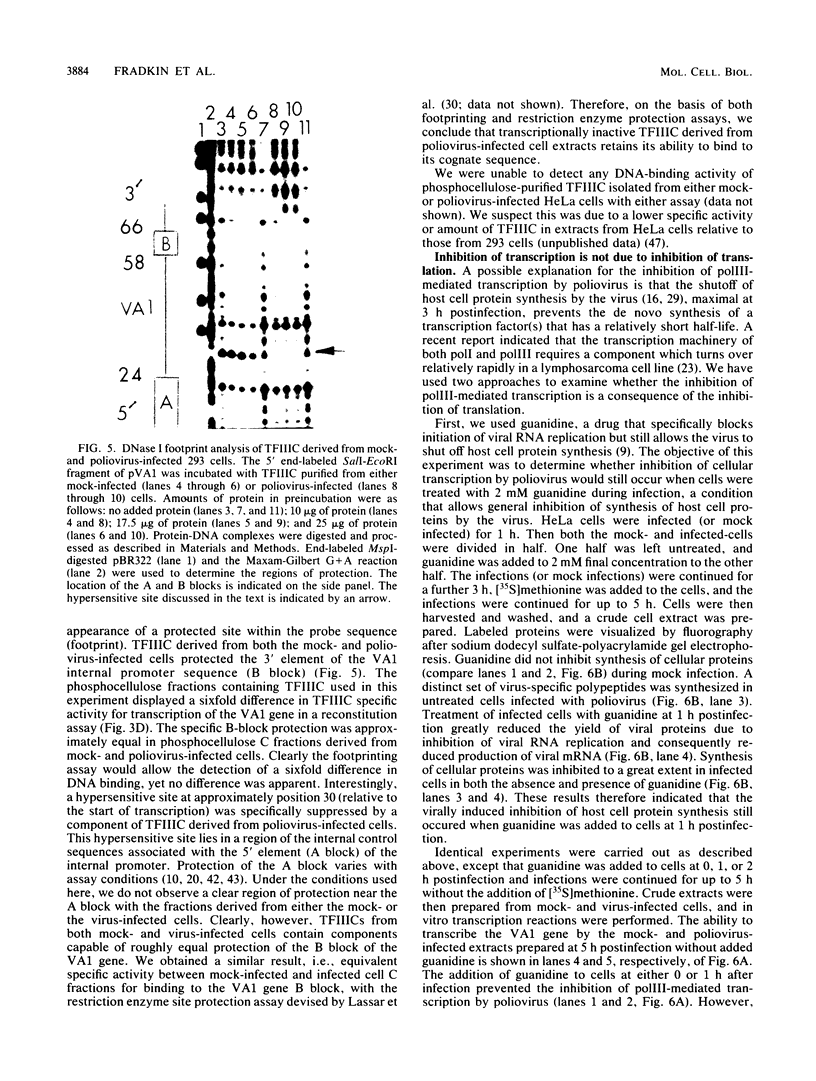
The inhibition of transcription by RNA polymerase III in poliovirus-infected cells was studied. Experiments utilizing two different cell lines showed that the initiation step of transcription by RNA polymerase III was impaired by infection of these cells with the virus. The observed inhibition of transcription was not due to shut-off of host cell protein synthesis by poliovirus. Among four distinct components required for accurate transcription in vitro from cloned DNA templates, activities of RNA polymerase III and transcription factor TFIIIA were not significantly affected by virus infection. The activity of transcription factor TFIIIC, the limiting component required for transcription of RNA polymerase III genes, was severely inhibited in infected cells, whereas that of transcription factor TFIIIB was inhibited to a lesser extent. The sequence-specific DNA-binding of TFIIIC to the adenovirus VA1 gene internal promoter, however, was not altered by infection of cells with the virus. We conclude that (i) at least two transcription factors, TFIIIB and TFIIIC, are inhibited by infection of cells with poliovirus, (ii) inactivation of TFIIIC does not involve destruction of its DNA-binding domain, and (iii) sequence-specific DNA binding by TFIIIC may be necessary but is not sufficient for the formation of productive transcription complexes.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Apriletti J. W., Penhoet E. E. Cellular RNA synthesis in normal and mengovirus-infected L-929 cells. J Biol Chem. 1978 Jan 25;253(2):603–611. [PubMed] [Google Scholar]
- Apriletti J. W., Penhoet E. E. Recovery of DNA-dependent RNA polymerase activities from L cells after mengovirus infection. Virology. 1974 Oct;61(2):597–601. doi: 10.1016/0042-6822(74)90294-3. [DOI] [PubMed] [Google Scholar]
- Baker R. E., Gabrielsen O., Hall B. D. Effects of tRNATyr point mutations on the binding of yeast RNA polymerase III transcription factor C. J Biol Chem. 1986 Apr 25;261(12):5275–5282. [PubMed] [Google Scholar]
- Baker R. E., Hall B. D. Structural features of yeast tRNA genes which affect transcription factor binding. EMBO J. 1984 Dec 1;3(12):2793–2800. doi: 10.1002/j.1460-2075.1984.tb02211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balanian R. Structural and functional alterations in cultured cells infected with cytocidal viruses. Prog Med Virol. 1975;19:40–83. [PubMed] [Google Scholar]
- Bieker J. J., Martin P. L., Roeder R. G. Formation of a rate-limiting intermediate in 5S RNA gene transcription. Cell. 1985 Jan;40(1):119–127. doi: 10.1016/0092-8674(85)90315-0. [DOI] [PubMed] [Google Scholar]
- Bogenhagen D. F., Sakonju S., Brown D. D. A control region in the center of the 5S RNA gene directs specific initiation of transcription: II. The 3' border of the region. Cell. 1980 Jan;19(1):27–35. doi: 10.1016/0092-8674(80)90385-2. [DOI] [PubMed] [Google Scholar]
- Brown D. D. The role of stable complexes that repress and activate eucaryotic genes. Cell. 1984 Jun;37(2):359–365. doi: 10.1016/0092-8674(84)90366-0. [DOI] [PubMed] [Google Scholar]
- Camier S., Gabrielsen O., Baker R., Sentenac A. A split binding site for transcription factor tau on the tRNA3Glu gene. EMBO J. 1985 Feb;4(2):491–500. doi: 10.1002/j.1460-2075.1985.tb03655.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciliberto G., Castagnoli L., Cortese R. Transcription by RNA polymerase III. Curr Top Dev Biol. 1983;18:59–88. doi: 10.1016/s0070-2153(08)60579-7. [DOI] [PubMed] [Google Scholar]
- Contreras G., Summers D. F., Maizel J. V., Ehrenfeld E. HeLa cell nucleolar RNA synthesis after poliovirus infection. Virology. 1973 May;53(1):120–129. doi: 10.1016/0042-6822(73)90471-6. [DOI] [PubMed] [Google Scholar]
- Crawford N., Fire A., Samuels M., Sharp P. A., Baltimore D. Inhibition of transcription factor activity by poliovirus. Cell. 1981 Dec;27(3 Pt 2):555–561. doi: 10.1016/0092-8674(81)90397-4. [DOI] [PubMed] [Google Scholar]
- Dasgupta A., Baron M. H., Baltimore D. Poliovirus replicase: a soluble enzyme able to initiate copying of poliovirus RNA. Proc Natl Acad Sci U S A. 1979 Jun;76(6):2679–2683. doi: 10.1073/pnas.76.6.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam J. D., Martin P. L., Shastry B. S., Roeder R. G. Eukaryotic gene transcription with purified components. Methods Enzymol. 1983;101:582–598. doi: 10.1016/0076-6879(83)01039-3. [DOI] [PubMed] [Google Scholar]
- Engelke D. R., Ng S. Y., Shastry B. S., Roeder R. G. Specific interaction of a purified transcription factor with an internal control region of 5S RNA genes. Cell. 1980 Mar;19(3):717–728. doi: 10.1016/s0092-8674(80)80048-1. [DOI] [PubMed] [Google Scholar]
- Fernández-Tomás C. The presence of viral-induced proteins in nuclei from poliovirus-infected HeLa cells. Virology. 1982 Jan 30;116(2):629–634. doi: 10.1016/0042-6822(82)90154-4. [DOI] [PubMed] [Google Scholar]
- Flores-Otero G., Fernández-Tomás C., Gariglio-Vidal P. DNA-bound RNA polymerases during poliovirus infection: reduction in the number of form II enzyme molecules. Virology. 1982 Jan 30;116(2):619–628. doi: 10.1016/0042-6822(82)90153-2. [DOI] [PubMed] [Google Scholar]
- Fuhrman S. A., Engelke D. R., Geiduschek E. P. HeLa cell RNA polymerase III transcription factors. Functional characterization of a fraction identified by its activity in a second template rescue assay. J Biol Chem. 1984 Feb 10;259(3):1934–1943. [PubMed] [Google Scholar]
- Galli G., Hofstetter H., Birnstiel M. L. Two conserved sequence blocks within eukaryotic tRNA genes are major promoter elements. Nature. 1981 Dec 17;294(5842):626–631. doi: 10.1038/294626a0. [DOI] [PubMed] [Google Scholar]
- Ginsberg A. M., King B. O., Roeder R. G. Xenopus 5S gene transcription factor, TFIIIA: characterization of a cDNA clone and measurement of RNA levels throughout development. Cell. 1984 Dec;39(3 Pt 2):479–489. doi: 10.1016/0092-8674(84)90455-0. [DOI] [PubMed] [Google Scholar]
- Gokal P. K., Cavanaugh A. H., Thompson E. A., Jr The effects of cycloheximide upon transcription of rRNA, 5 S RNA, and tRNA genes. J Biol Chem. 1986 Feb 25;261(6):2536–2541. [PubMed] [Google Scholar]
- Guilfoyle R., Weinmann R. Control region for adenovirus VA RNA transcription. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3378–3382. doi: 10.1073/pnas.78.6.3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffler W. K., Roeder R. G. Enhancement of RNA polymerase III transcription by the E1A gene product of adenovirus. Cell. 1985 Jul;41(3):955–963. doi: 10.1016/s0092-8674(85)80076-3. [DOI] [PubMed] [Google Scholar]
- Keegan L., Gill G., Ptashne M. Separation of DNA binding from the transcription-activating function of a eukaryotic regulatory protein. Science. 1986 Feb 14;231(4739):699–704. doi: 10.1126/science.3080805. [DOI] [PubMed] [Google Scholar]
- Kingston R. E., Baldwin A. S., Sharp P. A. Transcription control by oncogenes. Cell. 1985 May;41(1):3–5. doi: 10.1016/0092-8674(85)90049-2. [DOI] [PubMed] [Google Scholar]
- Kozak M. Regulation of protein synthesis in virus-infected animal cells. Adv Virus Res. 1986;31:229–292. doi: 10.1016/S0065-3527(08)60265-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käriäinen L., Ranki M. Inhibition of cell functions by RNA-virus infections. Annu Rev Microbiol. 1984;38:91–109. doi: 10.1146/annurev.mi.38.100184.000515. [DOI] [PubMed] [Google Scholar]
- Lassar A. B., Martin P. L., Roeder R. G. Transcription of class III genes: formation of preinitiation complexes. Science. 1983 Nov 18;222(4625):740–748. doi: 10.1126/science.6356356. [DOI] [PubMed] [Google Scholar]
- McCormick W., Penman S. Inhibition of RNA synthesis in HeLa and L cells by Mengovirus. Virology. 1967 Jan;31(1):135–141. doi: 10.1016/0042-6822(67)90017-7. [DOI] [PubMed] [Google Scholar]
- Pelham H. R., Brown D. D. A specific transcription factor that can bind either the 5S RNA gene or 5S RNA. Proc Natl Acad Sci U S A. 1980 Jul;77(7):4170–4174. doi: 10.1073/pnas.77.7.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razvi F., Gargiulo G., Worcel A. A simple procedure for parallel sequence analysis of both strands of 5'-labeled DNA. Gene. 1983 Aug;23(2):175–183. doi: 10.1016/0378-1119(83)90049-5. [DOI] [PubMed] [Google Scholar]
- Ruet A., Camier S., Smagowicz W., Sentenac A., Fromageot P. Isolation of a class C transcription factor which forms a stable complex with tRNA genes. EMBO J. 1984 Feb;3(2):343–350. doi: 10.1002/j.1460-2075.1984.tb01809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakonju S., Bogenhagen D. F., Brown D. D. A control region in the center of the 5S RNA gene directs specific initiation of transcription: I. The 5' border of the region. Cell. 1980 Jan;19(1):13–25. doi: 10.1016/0092-8674(80)90384-0. [DOI] [PubMed] [Google Scholar]
- Schwartz L. B., Lawrence C., Thach R. E., Roeder R. G. Encephalomyocarditis virus infection of mouse plasmacytoma cells. II. Effect on host RNA synthesis and RNA polymerases. J Virol. 1974 Sep;14(3):611–619. doi: 10.1128/jvi.14.3.611-619.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz L. B., Sklar V. E., Jaehning J. A., Weinmann R., Roeder R. G. Isolation and partial characterization of the multiple forms of deoxyribonucleic acid-dependent ribonucleic acid polymerase in the mouse myeloma, MOPC 315. J Biol Chem. 1974 Sep 25;249(18):5889–5897. [PubMed] [Google Scholar]
- Segall J. Assembly of a yeast 5 S RNA gene transcription complex. J Biol Chem. 1986 Sep 5;261(25):11578–11584. [PubMed] [Google Scholar]
- Segall J., Matsui T., Roeder R. G. Multiple factors are required for the accurate transcription of purified genes by RNA polymerase III. J Biol Chem. 1980 Dec 25;255(24):11986–11991. [PubMed] [Google Scholar]
- Setzer D. R., Brown D. D. Formation and stability of the 5 S RNA transcription complex. J Biol Chem. 1985 Feb 25;260(4):2483–2492. [PubMed] [Google Scholar]
- Shastry B. S., Ng S. Y., Roeder R. G. Multiple factors involved in the transcription of class III genes in Xenopus laevis. J Biol Chem. 1982 Nov 10;257(21):12979–12986. [PubMed] [Google Scholar]
- Stillman D. J., Sivertsen A. L., Zentner P. G., Geiduschek E. P. Correlations between transcription of a yeast tRNA gene and transcription factor-DNA interactions. J Biol Chem. 1984 Jun 25;259(12):7955–7962. [PubMed] [Google Scholar]
- Taylor W., Jackson I. J., Siegel N., Kumar A., Brown D. D. The developmental expression of the gene for TFIIIA in Xenopus laevis. Nucleic Acids Res. 1986 Aug 11;14(15):6185–6195. doi: 10.1093/nar/14.15.6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinaga S. K., Boulanger P. A., Berk A. J. Resolution of human transcription factor TFIIIC into two functional components. Proc Natl Acad Sci U S A. 1987 Jun;84(11):3585–3589. doi: 10.1073/pnas.84.11.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinaga S., Dean N., Han M., Berk A. J. Adenovirus stimulation of transcription by RNA polymerase III: evidence for an E1A-dependent increase in transcription factor IIIC concentration. EMBO J. 1986 Feb;5(2):343–354. doi: 10.1002/j.1460-2075.1986.tb04218.x. [DOI] [PMC free article] [PubMed] [Google Scholar]