Summary
Dimerization-driven activation of the intracellular kinase domains of the epidermal growth factor receptor (EGFR) upon extracellular ligand binding is a key step in cellular pathways regulating proliferation, migration, and differentiation. Inactive EGFR can exist as both monomers and dimers, suggesting that the mechanism regulating EGFR activity may be subtle. The membrane itself may play a role, but creates substantial difficulties for structural studies. Our molecular dynamics simulations of membrane-embedded EGFR suggest that, in ligand-bound dimers, the extracellular domains assume conformations favoring dimerization of the transmembrane helices near their N-termini, dimerization of the juxtamembrane segments, and formation of asymmetric (active) kinase dimers. In ligand-free dimers, by holding apart the N-termini of the transmembrane helices, the extracellular domains instead favor C-terminal dimerization of the transmembrane helices, juxtamembrane segment dissociation and membrane burial, and formation of symmetric (inactive) kinase dimers. Electrostatic interactions of EGFR’s intracellular module with the membrane were found to be critical in maintaining this coupling.
Introduction
The Epidermal growth factor receptor (EGFR, or Her1/ErbB1) is one of the four members of the Her (ErbB) family of receptor tyrosine kinases, which also includes Her2 (ErbB2/Neu), Her3 (ErbB3), and Her4 (ErbB4). These proteins serve as cell-surface receptors for the peptide ligands of the epidermal growth factor (EGF) family and play crucial roles in regulating cell proliferation, migration, and differentiation (Citri and Yarden, 2006); their aberrant activity is implicated in a variety of cancers (Riese et al., 2007). Consequently, Her proteins, and EGFR and Her2 in particular, are among the most intensely pursued drug targets.
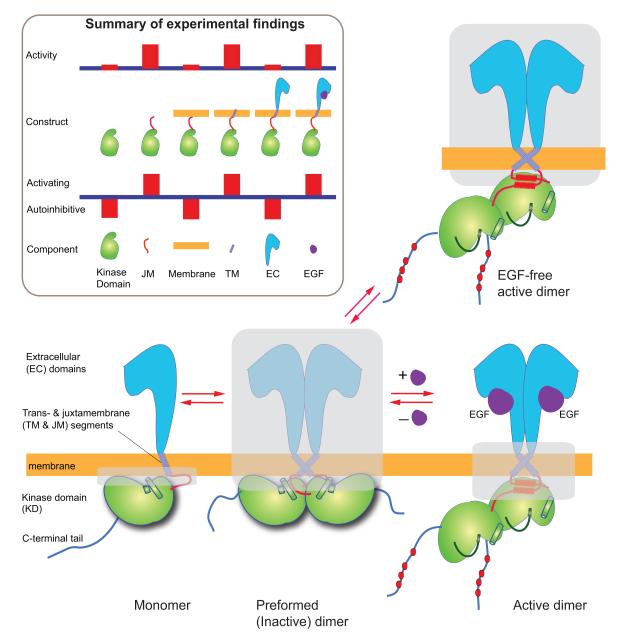
The EGF receptor consists of an extracellular module (comprising domains I, II, III, and IV) and an intracellular kinase domain (with a long regulatory C-terminal tail), which are connected by a single-helix transmembrane segment and a juxtamembrane segment (Figure 1). EGFR activation is dimerization dependent (Schlessinger, 2002). Ligand binding elicits a “back-to-back” dimer of the extracellular domains (Ogiso et al., 2002; Garrett et al., 2002), which leads the intracellular kinase domains to form enzymatically active (asymmetric) dimers (Zhang et al., 2006). Crystal structures of monomeric extracellular domains (Ferguson et al., 2003) and of an inactive, symmetric kinase dimer (Jura et al., 2009) have also been resolved. At low resolution, detergent-solubilized dimers of nearly full-length receptors have been visualized (Mi et al., 2011). A number of studies (e.g., Low-Nam et al., 2011; Chung et al., 2010; Clayton et al., 2005) have shown that EGFR can also exist as preformed dimers in the absence of ligands.
Figure 1. Schematic view of three EGFR states and summary of some key experimental results.
Cartoon of the monomer, ligand-free, and ligand-bound dimers of EGFR. The structurally unresolved portions are shaded. The inset is a summary of experiments (referenced in the main text) that measured the activity of various EGFR constructs, and the inferred contribution of EGFR components to the balance between EGFR activation and autoinhibition.
A body of experimental evidence shows that the EGFR components that promote the dimerization and activation of the receptors are intertwined with the components that inhibit these processes (Figure 1). While isolated kinase domains are predominantly monomeric in solution (Zhang et al., 2006), they dimerize and activate strongly when the juxtamembrane segments are included (Jura et al., 2009). When such EGFR constructs are localized to cell membranes, however, their activities are abrogated. This surprising finding is reported in the accompanying paper (Endres et al., 2013), which also shows that the activity may be recovered by the addition of the transmembrane segments and abrogated at low expression levels by the further addition of the extracellular domains. These findings suggest that the transmembrane and juxtamembrane segments on balance favor EGFR activation, whereas the kinase domain, the extracellular domains, and the EGFR interaction with the cell membrane contribute to EGFR autoinhibition. Although ligand binding enhances dimerization of the extracellular domains only modestly in solution (Odaka et al., 1997), at the cell surface it can tip the balance decisively toward activation.
The structural basis of these findings is uncertain, however, since the architecture of intact EGFR remains obscure. Various experiments have suggested the need to take the cell membranes into account (Bessman and Lemmon, 2012), but this presents a formidable challenge to structural analysis. One key problem is the relatively poor understanding of the crucial middle sections of EGFR that are embedded in, or adjacent to, the cell membrane. It has been proposed that in EGFR dimers the transmembrane helices alternate between two dimer forms, one associated with inactive and the other with active EGFR dimers (Fleishman et al., 2002), although direct evidence supporting this hypothesis is lacking. While the juxtamembrane segments promote the dimerization and activation of EGFR kinases (Jura et al., 2009; Red Brewer et al., 2009), the structural mechanism that couples the trans- and juxtamembrane segments is not understood.
The preformed dimers present another key problem. Even though inactive EGFR is usually present as monomers in normal cells, at higher levels of expression, inactive, preformed dimers are commonly observed, and knowledge of their structure could potentially shed light on the mechanisms by which EGFR activity is controlled. Their structure, however, remains unknown, although the observation that preformed dimers are primed for ligand binding (Chung et al., 2010) hints at a potentially close structural relationship to the active dimer in its extracellular domains. The existence of inactive dimers suggests a tight conformational coupling between the extra- and intracellular domains to prevent kinase activity in the absence of ligand, but puzzlingly, the dimerization of the extracellular domains and the dimerization of the intracellular kinase domains are not necessarily correlated, as shown by experiments on intact EGFRs in detergent micelles (Mi et al., 2011; Wang et al., 2011). One possibility is that embedding EGFR in detergent micelles alters the receptor’s behavior in important ways (Bessman and Lemmon, 2012), and that a structural understanding of signal transduction may need to take the membrane environment into account.
Here, we use molecular dynamics (MD) simulations to elucidate the overall architecture of EGFR and its interaction with the membrane to understand the cross-membrane coupling in these receptors. We adopted a divide-and-conquer strategy, initiating our model building with simulations of individual EGFR components in various contexts. Guided by observations from these simulations and by insights gained from coordinated experimental work described in a companion paper (Endres et al., 2013), the components were then assembled into larger models, the simulations of which helped motivate further experimental work. We ultimately constructed and characterized nearly full-length models for the monomer, the ligand-free, inactive dimer, and the ligand-bound, active dimer. In simulations of the extracellular portion of a ligand-bound EGFR dimer, we found that the extracellular domains remained close to the (active) crystal structure, but that this structure was no longer stable when the bound EGF ligands were removed. In particular, the two domain IVs underwent a conformational change that substantially increased the distance between their C-termini. Since in intact EGFR the domain IVs are directly linked to the transmembrane domains, these extracellular domain simulations shed light on how ligand binding may be coupled to the arrangement of the transmembrane helices. Further simulations, together with nuclear magnetic resonance (NMR) and mutagenesis data (Endres et al., 2013), demonstrated that the transmembrane helices may dimerize either near their N- or C-termini. The data further showed that antiparallel juxtamembrane helix dimers coexist with N-terminal transmembrane dimers, but are incompatible with the C-terminal transmembrane dimers. Our simulations showed that, when the juxtamembrane dimers are disrupted, the juxtamembrane segments become embedded in the membrane. Consistent with earlier findings (Jura et al., 2009), we show that a juxtamembrane helix dimer is compatible with the asymmetric kinase dimer, and hence with EGFR activation. We find membrane-embedded juxtamembrane segments, on the other hand, to be compatible with both the symmetric kinase dimer and EGFR monomers, and thus with EGFR autoinhibition. Combining these findings, we propose a structural mechanism that enables the receptors to relay signals across the membrane. Notably, the simulations indicate that membrane lipids, especially anionic lipids, interact extensively with the receptors and are integral to signal transduction. A detailed description of EGFR–membrane interaction from these simulations provides a mechanistic understanding of a dual role for anionic lipids in EGFR regulation, in both inhibiting EGFR in the absence of ligand stimulus and in accentuating EGFR response to a stimulus.
Results
Monomeric extracellular domains are conformationally highly flexible
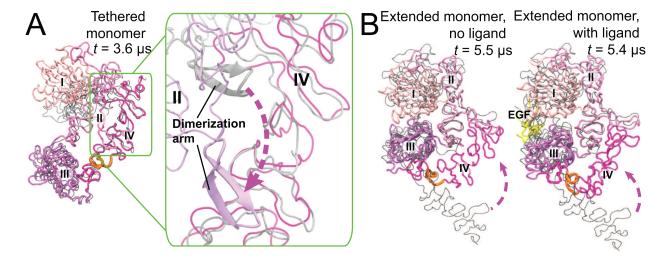
In the ligand-bound extracellular EGFR dimer, each subunit binds one ligand between domains I and III and adopts an extended conformation (Figure 2; Garrett et al., 2002; Ogiso et al., 2002; Lu et al., 2010). In the absence of ligands, monomeric extracellular domains may assume a “tethered” conformation (Cho and Leahy, 2002; Ferguson et al., 2003) that precludes dimerization. This conformation is characterized by a wide separation of domains I and III and the tether interaction between domain IV and the dimerization arm of domain II (Figure 2). EGFR, however, can dimerize prior to EGF binding, suggesting that the extracellular domains in monomeric EGFR may adopt conformations other than tethered.
Figure 2. Conformational diversity of the monomeric extracellular module.
(A) Simulation of the tethered monomeric extracellular module (starting from PDB entry 1NQL). The overall conformation is maintained, but the “tether” contacts are lost (inset). (B) Simulation of the extended monomeric extracellular module (one subunit taken from the ligand-bound extracellular dimer in PDB entry 3NJP). Domain IV undergoes a large conformational change and reaches the dimerization arm of domain II, while domains I, II, and III largely remain stable. In both (A) and (B), the “hinge” in domain IV (residues 502–514) is highlighted in orange, and the starting conformations are shown in gray.
To investigate these conformations, we first simulated the monomer starting from the tethered conformation, in which, on a timescale of microseconds, the “tether” readily disengaged, but the two domains II and IV remained in extensive contact, shielding the dimerization arm (Figure 2A). Thus, our simulations suggest a limited role of the tether in EGFR autoinhibition, which is consistent with experimental findings (Mattoon et al., 2004; Dawson et al., 2007; Liu et al., 2012a).Simulations of monomeric extracellular domains were also initiated from the extended conformation (both with and without EGF bound). These simulations demonstrated a significant conformational change occurring within 1–5 μs, in which the C-terminus of the extracellular domains traveled a distance of ~80 Å (Figure 2B). This happened largely as a result of the bending of domain IV around a “hinge” (residues 502–514), which produced a compact conformation of the extracellular domains not yet captured by crystallography, which resembled the extended conformation in domains I–III, and the tethered conformation in domain IV. The instability of the extended conformation for EGFR monomers confirms that the extracellular domains in monomers generally adopt compact conformations (Du et al., 2012).
The dimer conformation of ligand-free extracellular domains
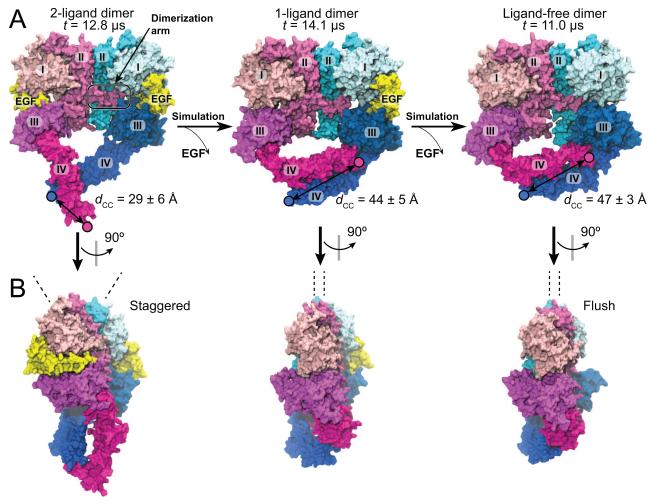
In contrast to the instability of the extended conformation of extracellular monomers, the extended conformation of the two subunits in a ligand-bound dimer (the “2-ligand dimer”) remained stable in our simulations, consistent with the stabilizing effect of ligand binding to EGFR active dimers (Figure S1). EGFR can also form ligand-free dimers that presumably need to be autoinhibited in normal cells. Since the crystal structure of the ligand-free extracellular dimer is not yet available for human EGFR, we attempted to investigate the structure using MD simulations. From the notion that the inactive dimers are primed for ligand binding (Chung et al., 2010; Sako et al., 2000), we hypothesized that their extracellular domains bear structural resemblance to the active structure. We thus performed simulations based on the crystal structure of the 2-ligand extracellular dimer. First, the crystal structure was simulated after one ligand was removed. In the simulation, the gap left by the removed ligand (Figure 3) was filled by domains I and III, which came into contact with each other. The resultant conformation of the ligand-free subunit bears significant similarity in domains I–III (Figure 3) to the ligand-free structures of Her2 (Cho et al., 2003; Garrett et al., 2003) and Drosophila EGFR (Alvarado et al., 2009; Alvarado et al., 2010). Moreover, we observed a rotation of one subunit with respect to the dimerization arm of the other (Figures 3B and S1E), reflecting a transition between the “staggered” and the “flush” conformations (Liu et al., 2012b).
Figure 3. Conformations of the ligand-bound and ligand-free EGFR extracellular dimer.
(A) The dimer is simulated starting from the crystal structure (PDB entry 3NJP), retaining either both ligands (2-ligand dimer, left) or one ligand (1-ligand dimer, center). The final conformation of the 1-ligand dimer simulation is used to initiate the simulation of the ligand-free dimer (right). The two domain IVs undergo a significant movement in the 1-ligand dimer simulation, but not in the 2-ligand or ligand-free dimer simulations. This results in a greater distance between the C-termini (dCC) in the 1-ligand and ligand-free dimers than in the 2-ligand dimer. (B) Side views of the dimer conformations. Transitions from the “staggered” toward the “flush” conformation (Liu et al., 2012b) are observed in our simulations of the 1-ligand and ligand-free dimers. The dashed lines here indicate the principal axes of the domain IIs. See also Figure S1.
Perhaps more importantly, the rearrangement of domains I and III led to a “bending” motion at domain IV around the hinge region in the ligand-free subunit of the “1-ligand dimer” (where only one EGFR subunit is ligand-bound), reminiscent of the “bending” in monomeric extracellular domains (Figure 2B). The bent domain IV was observed to occasionally return to its initial conformation (Figure S1), likely reflecting a fluctuation between the two conformations in a 1-ligand dimer. To further investigate the conformation of the ligand-free extracellular dimer, we removed the remaining ligand from the 1-ligand extracellular dimer. Starting with a bent domain IV, subsequent simulations did not exhibit significant conformational changes, suggesting that a bent domain IV is stable in absence of bound ligands in the dimer (Figure 3).
Our results thus suggest that the removal of bound ligands results in significant rearrangement of the domain IVs. Instead of a V-shape arrangement of domain IVs in the 2-ligand dimer, in the new arrangement they adopt an antiparallel arrangement with a distance of ~45 Å between the two C-terminal ends (dCC), which is significantly greater than the distance in the 2-ligand dimers (10 Å in crystal structures and ~30 Å on average in our simulations; Figure S1E). The distance dCC is important, because by altering it the extracellular domains may communicate with the one-pass transmembrane helices and ultimately the cytoplasmic portion of the EGFR dimer. The basic observation concerning dCC was found to be independent of the choice of force field in our simulations (Supplemental Information).
The transmembrane helices favor an N-terminal dimer
The transmembrane segment connects the extra- and intracellular domains. We expect that the transmembrane conformations in active and inactive EGFR dimers differ in ways that place substantial constraints on the possible structures of the intra- and extracellular modules. An EGFR transmembrane helix contains two GxxxG-like motifs (where the G represents a glycine or other small amino acid), one close to the N-terminus (TGMVGA, residues 624–629, which itself comprises two overlapping GxxxG-like motifs) and the other to the C-terminus (ALGIG, residues 637–641). Since GxxxG motifs often serve as dimerization interfaces of transmembrane helices (Lemmon et al., 1994; Russ and Engelman, 2000), dimerization of the EGFR transmembrane helices at the N- or the C-terminal motifs has been suggested (Mendrola et al., 2002; Fleishman et al., 2002). Crosslinking experiments (Lu et al., 2010) confirmed that the ligand-bound active EGFR dimer contains dimer contacts at the N-terminal motif. The C-terminal transmembrane dimer, on the other hand, has been proposed to be part of the ligand-free inactive EGFR dimer (Landau and Ben-Tal, 2008).
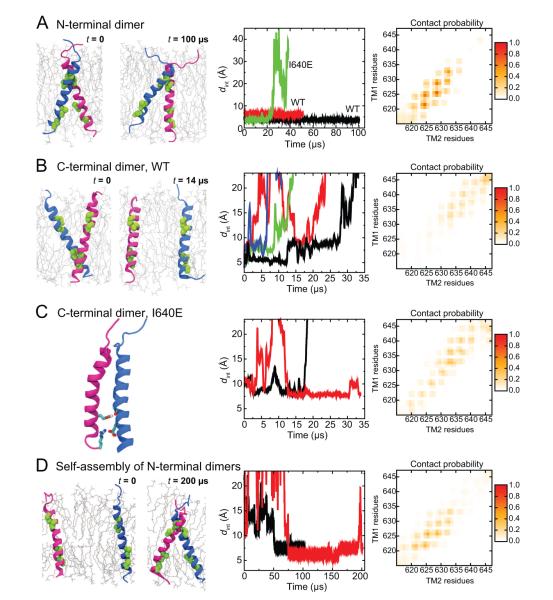
To assess the relevance of these two potentially competing transmembrane-dimer forms for EGFR signaling, we investigated their stability using MD simulations. We constructed a model of the N-terminal transmembrane dimer using the resolved Her2 N-terminal dimer (Bocharov et al., 2008) as a template. The C-terminal dimer model was constructed so that the two helices were in contact at the C-terminal GxxxG-like motifs and the angle between these two helices was similar to that in the Her2 N-terminal dimer. In a POPC/POPS lipid bilayer (see Experimental Procedures), the N-terminal transmembrane dimer was found to be stable by itself, as it remained intact in simulations up to 100 μs long (Figure 4A), whereas the C-terminal dimer dissociated on a timescale of 100 ns to 10 μs (Figure 4B).
Figure 4. N-terminal and C-terminal transmembrane dimers.
(A) Simulations of the N-terminal transmembrane dimer. The starting and ending conformations in one of the simulations are shown in the left panel, where the GxxxG-like motifs are highlighted in green. In the middle panel, the (center-of-mass) distance between the motifs of the two helices is plotted. The right panel shows the residue–residue contacts between the two helices, computed over the two simulations, where the intensity represents the fraction of simulation time in which a contact is maintained. (B) Simulations of the C-terminal transmembrane dimers. The distance between the dimer interfaces (middle panel) shows the instability of these dimers. The residue–residue contacts (right panel) are averaged over all four simulations. (C) Simulations of the I640E C-terminal dimer. The contact between the Glu640 and the backbone of the other helix is highlighted in the left panel. The I640E mutation stabilized the C-terminal transmembrane dimer (middle panel). Note that the residue contact map (right panel) differs from that of the C-terminal dimer of the wild type (B). (D) Self-assembly simulation of EGFR transmembrane helices. The self-assembly was observed in two independent simulations (middle panel). The residue contacts (right panel) are similar to those observed in simulations of the modeled N-terminal transmembrane dimers in (A). See also Figure S2.
Consistent with the crosslinking experiments (Lu et al., 2010), although the wild-type N-terminal dimer remained intact in simulations, the dimer interfaces were variable, including the GxxxG-like motif, as well as the adjacent residues. This is reflected in the distance between the interfacing residues, dint, which fluctuated around slightly different averages in the simulations, and in the varied residue contacts (Figure 4A).
The different stability of the N- and C-terminal transmembrane dimers may be in part attributed to the fact that the N-terminal dimerization interface is more extensive, consisting of two overlapping GxxxG-like motifs. Moreover, the glycine (Gly625) in the N-terminal motif is more favorable for dimerization than its counterpart (Ala637) at the C-terminal motif. Consistent with this observation, Her2 features glycine in its C-terminal motif, and in our simulations the C-terminal Her2 transmembrane dimer was stable for as long as 100 μs (Figure S2).
The relative stability of the N-terminal EGFR transmembrane dimer was corroborated by further self-assembly simulations of the transmembrane helices initially placed distant from one other in a POPC/POPS lipid bilayer. In both such simulations, the transmembrane helices dimerized at the N-terminal GxxxG-like motifs, and resultant dimers remained stable throughout the simulations—50 μs or longer (Figure 4D).
The model of the N-terminal transmembrane dimer is consistent with NMR measurements
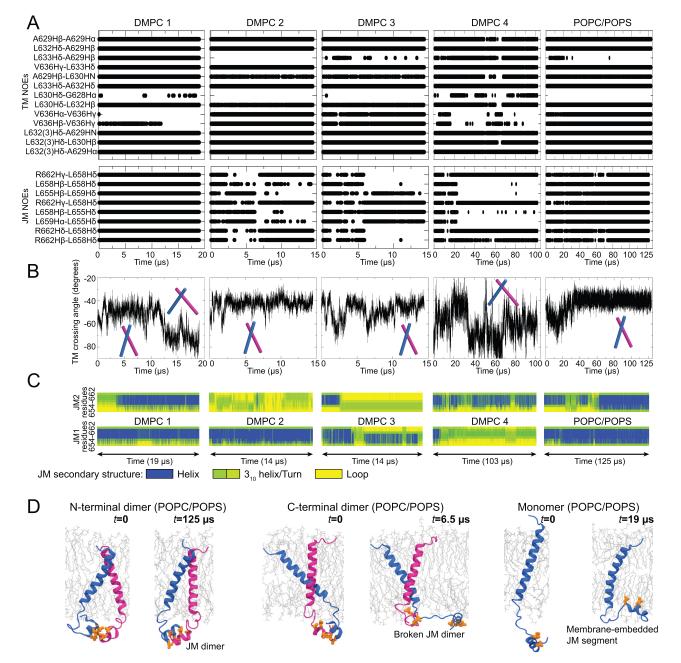
From the nuclear Overhauser effect (NOE) spectroscopy of EGFR trans- and juxtamembrane segments (TM–JM) embedded in DMPC lipids, 21 pairs of adjacent residues from a TM–JM dimer (21 inter-subunit “NOEs,” including 13 for the transmembrane and 8 for the juxtamembrane segments) were identified (Endres et al., 2013). Almost all (up to 11 out of 13) of the transmembrane NOEs were satisfied in the above-discussed simulations of the N-terminal transmembrane dimer in POPC/POPS membrane bilayers, where the structural model was constructed independently from the NMR data, and the membrane lipids were different from those in the NMR experiments.
To further compare our model against the NMR data, we simulated the same system probed by the NMR experiments, where the transmembrane dimer was embedded in neutral-charged DMPC lipid bilayers with the juxtamembrane segments attached (Figure 5A). In the first of these simulations (“DMPC 1”), the initial conformation of the transmembrane dimer was taken from the last snapshot of the 100-μs simulation of the N-terminal transmembrane dimer in POPC/POPS lipids (Figure 4A), which satisfied 9 of the 13 transmembrane NOEs. The dimer reached a new conformation with a wider (approximately −70°) angle between the two helices 12 μs into the simulation (Figure 5B). Similarly, in simulation “DMPC 4” an ensemble of conformations with angles ranging from −30° to −80° was observed. The alternative, “narrow” conformations also appeared to be accessible in DMPC, as the conformation remained stable in two other simulations (“DMPC 2” and “DMPC 3”). Each of these two sets of conformations satisfies up to 11 of the 13 transmembrane NOEs (Figures 5A and S3); the wide conformations do not satisfy the two NOEs involving Val636, which are mostly satisfied in the narrow conformations (Figures 5A and S3).
Figure 5. Properties of the TM–JM-A dimers.
(A) NOEs (Endres et al., 2013) satisfied by simulations. Each dot indicates a satisfied NOE at a given time in the simulation. The five columns correspond to four TM–JM-A simulations with DMPC and one with POPC/POPS lipids. Note that the overlapping dots may appear a straight line in the figure. (B) The angle between the two transmembrane helices in the simulated TM–JM-A dimers. The narrow and wide conformations are marked schematically. (C) JM-A helicity. (D) JM-A conformations. The JM-A dimer is stable when connected to the N-terminal transmembrane dimer (left), but not when connected to the C-terminal dimer. A JM-A embedded in the membrane, with its hydrophobic residues (orange) placed into the hydrophobic membrane interior. See also Figure S3.
Based on these observations, we suggest that an N-terminal transmembrane dimer exists in a dynamic equilibrium in DMPC bilayers. Indeed, we confirmed that an ensemble consisting of one wide and one narrow transmembrane conformation can satisfy all of the 13 transmembrane NOEs (Figures 5A and S3). It should be noted that the wide transmembrane-dimer conformations observed in the DMPC bilayers may not be viable in the thicker POPC/POPS bilayers or typical biological membranes. Transmembrane helices tend to be more tilted in thinner bilayers (Holt and Killian, 2010), leading to larger inter-helix angles in a dimer in the DMPC bilayers. It is noteworthy that simulated annealing, a standard method in NMR analysis, generated a model of a left-handed N-terminal transmembrane dimer. This model was unstable in simulations (Figure S3) and is likely not adopted by EGFR. This indicates that the relatively small number of NOEs themselves may be insufficient to distinguish different models.
The N- and the C-terminal transmembrane dimers represent an active and an inactive conformation respectively
The N-terminal transmembrane dimer is geometrically compatible with the 2-ligand active extracellular dimer, in that the short distance between the N-termini of the transmembrane helices matches that of the C-termini of the two domain IVs in the active extracellular dimer. This is not the case for the C-terminal transmembrane dimer. We infer that the N-terminal transmembrane dimer is integral to an active EGFR dimer, a conclusion consistent with experiments (Lu et al., 2010) showing elevated levels of crosslinking at the N-terminal GxxxG-like motif upon EGF stimulus. The same study, however, also showed that mutations (T624L, G625L, G628L, A629L) at the N-terminal motif do not disrupt EGFR activation, thus suggesting that N-terminal dimerization is not essential. We performed simulations of these mutants and found that the N-terminal transmembrane dimer in fact is resilient to these mutations (Figure S2). (This observation is likely connected to Lu et al.’s finding that the N-terminal dimerization interface is structurally variable, a finding directly supported by our simulations, as discussed above.) The mutagenesis findings of Lu et al. thus are not necessarily inconsistent with an essential role for N-terminal dimerization of the transmembrane helices in EGFR activation. Additional support for this conclusion comes from investigations of the T624I/G625I/G628I/A629I quadruple mutation, which impairs EGFR activation (Endres et al., 2013); our simulations of this mutant show that the N-terminal dimerization is indeed disrupted (Figure S2).
The C-terminal transmembrane dimer has been hypothesized to be part of an inactive EGFR dimer (Landau and Ben-Tal, 2008). While our simulations suggest that the C-terminal dimers are relatively unstable on their own (Figure S3D), further simulations showed that they can be maintained if other EGFR components are present. That the C-terminal transmembrane dimer matches the inferred structure of the ligand-free extracellular dimer is consistent with this hypothesis.
This hypothesis is further supported by the finding that an I640E mutation near the C-terminal GxxxG-like motif strongly inhibits EGFR activity in cells (Endres et al., 2013). This mutation broke the N-terminal transmembrane dimer in simulation (Figure 4A). The glutamate side chain tended to interact with lipid head groups rather than stay inside the membrane, reducing the effective hydrophobic length of the transmembrane helices, changing their orientation with respect to the membrane, and disrupting the dimer interface. This mechanism resembles that by which a similar transmembrane mutation of integrin β leads to the dissociation of its transmembrane domains and integrin activation (Kim et al., 2012). Moreover, glutamate side chains are known to form inter-chain hydrogen bonds in membranes (Sternberg and Gullick, 1989) and the I640E mutation may strengthen the dimerization at the C-terminus (Figure 4C). The contact in the mutant transmembrane helices was mostly by interactions of Glu640 and the backbone groups near the C-terminus of the partner helix. The I640E dimer is nevertheless similar to the GxxxG-mediated C-terminal dimer, in that the C-termini of the helices are closer to each other than the N-termini are.
The JM-A helix dimer is induced by the N-terminal transmembrane dimer and is stabilized by anionic lipids
It has been previously shown that the juxtamembrane segments linking the transmembrane helices and the kinase domains are critical in EGFR activation (Jura et al., 2009; Red Brewer et al., 2009; Thiel and Carpenter, 2007). In the active state, the N-terminal portion of the juxtamembrane segments (JM-A) forms antiparallel helix dimers (Scheck et al., 2012; Jura et al., 2009) and the C-terminal portion (JM-B) interacts with the kinase domains. The JM-A helix dimer thus couples the N-terminal transmembrane dimer at one end with the (active) asymmetric kinase dimer at the other. The new NMR measurements of the TM–JM segments (Endres et al., 2013) identified eight JM-A NOEs, all consistent with an antiparallel helix dimer.
We modeled the active TM–JM-A dimer consisting of an antiparallel JM-A helix dimer attached to an N-terminal transmembrane dimer. The JM-A dimer was so constructed that the hydrophobic sides of the amphipathic JM-A helices were in contact with each other. Notably, simulations of this model showed that the JM-A helices were less stable with DMPC lipids than with POPS/POPC lipids: the JM-A helices melted in three of the four DMPC simulations, and the JM-A dimers fell apart (Figure 5C). This is consistent with the NMR finding that JM-A helicity is maintained only ~30% of the time with DMPC (Endres et al., 2013). Moreover, when the JM-A helix dimer was maintained, the model satisfied the NOEs remarkably well (Figure 5A), although neither the construction of the model nor the simulation used any information from NMR.
It is especially noteworthy that, compared to DMPC, in a POPC/POPS bilayer the JM-A helices and the helix dimer were better stabilized (Figure 5C) and the NOEs were better satisfied throughout the simulation. The JM-A is rich in positively charged basic residues and the simulation showed that they interacted extensively with the anionic POPS lipids (Figure S3). The stabilizing effect of the POPS/POPC bilayer most likely arose from these electrostatic interactions, which are absent in the neutral DMPC lipids. Additionally, in simulations where the TM–JM dimer was connected to the asymmetric kinase dimer, we observed further stabilization of the JM-A helices (Figure S3F), suggesting a cooperative interaction between these domains.
The JM-A segments of inactive EGFRs are embedded in the membrane
To communicate signals across the membrane, the trans- and juxtamembrane segments presumably adopt distinct conformations in the active or inactive EGFR states. Having argued that an active dimer of EGFR comprises an N-terminal transmembrane dimer and an antiparallel JM-A helix dimer, we now seek to identify the JM-A conformations in a monomeric receptor or an inactive dimer. We first simulated monomeric TM–JM-A. Notably, the JM-A, which was initially placed in solution away from the POPC/POPS membrane, became embedded in the membrane with the JM-A hydrophobic residues buried in the bilayer interior and the basic residues paired with the head groups of the charged lipid (Figure 5D, S3). The membrane embedding of the juxtamembrane segments is consistent with the NMR data of JM-A in detergent micelles (Choowongkomon et al., 2005). We then performed simulations (with a POPC/POPS bilayer) starting from a C-terminal transmembrane dimer combined with a JM-A dimer. In these simulations the JM-A dimer either dissociated or significantly deformed (Figure 5D, S3), suggesting that the C-terminal transmembrane dimer, representative of an inactive state, and the JM-A antiparallel helix dimer, representative of an active state, may not coexist. Structurally, this may be because residues (645–653) connecting the trans- and juxtamembrane segments are rich in positive charges, and when the C-terminal transmembrane dimer brings them close to one another, the resultant repulsion destabilizes the JM-A dimer. By contrast, the N-terminal transmembrane dimer separates these residues and thus stabilizes the JM-A dimer. The same effect may contribute to the relative weakness of the C-terminal transmembrane dimer in comparison with the N-terminal one. The C-terminal dimer, while not stable by itself in simulations, was stable in simulations of inactive dimer embedded in membrane of 15% POPS concentration (Figure S3D). Taken together, these results support a scenario in which the JM-A conformations alternate between the antiparallel JM-A helix dimer and separated JM-A embedded in the membrane, the former corresponding to the active and the latter to the inactive state.
Assembly of complete EGFRs
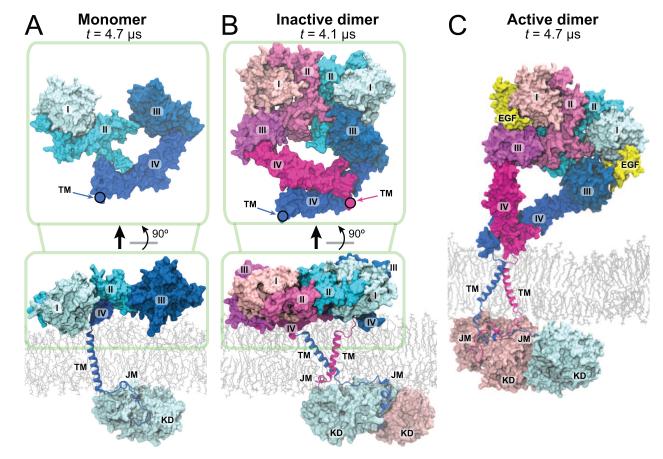
Having characterized the main EGFR components individually, we proceeded to assemble these components into models of near-complete EGFRs (missing the natively unstructured tails C-terminal to the kinase domains). A model of monomeric EGFR (Figure 6A) includes the EGFR extracellular domains adopting a non-extended conformation (Figure 2), a transmembrane helix, a JM-A embedded in POPC/POPS membrane, and the JM-B connected to the kinase domain (KD) in its inactive kinase conformation (Wood et al., 2004; Jura et al., 2009). The kinase domain was placed relative to the membrane so that its two positively charged patches (Figure 7A; Lys689, Lys690, Lys692, Lys715 in one and Arg779, Arg817, Lys851, Lys889 in the other) were in contact with the membrane and interacting with the anionic lipids. Such an arrangement potentially constitutes another layer of inhibition by occluding the substrate binding site of the kinase by the membrane (Figure 7A).
Figure 6. Models of the near-complete EGFR monomer and dimers.
The models are taken from simulations of the EGFR monomer (A), inactive dimer (B), and active dimer (C), at the noted simulation time. The connecting points between the extracellular and the transmembrane helices are marked by circles.
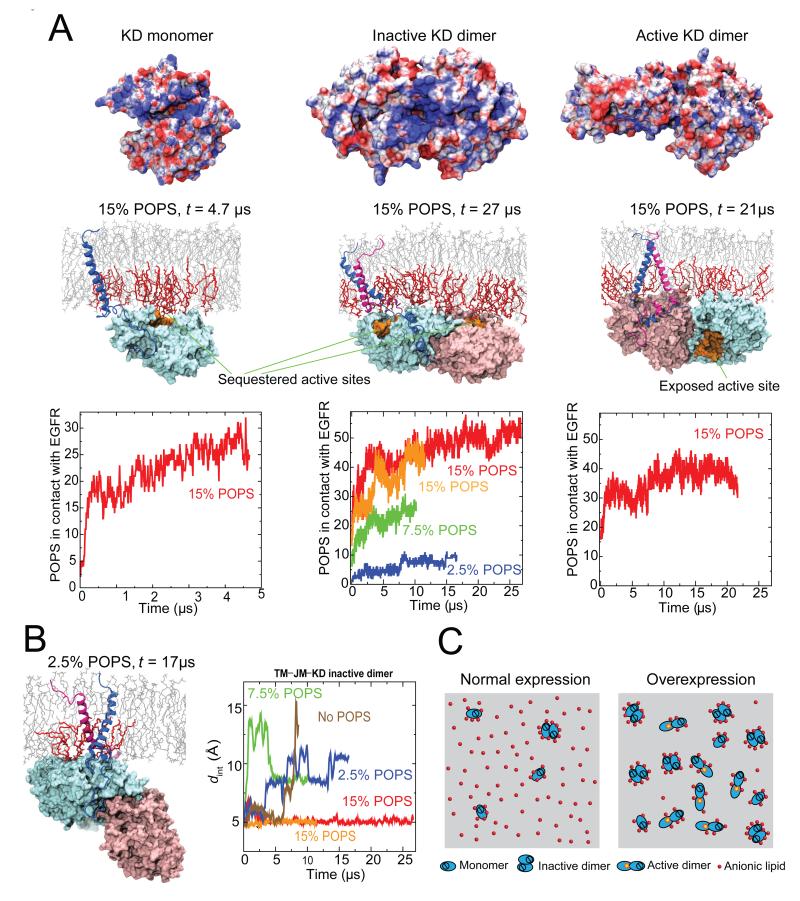
Figure 7. EGFR interaction with the intracellular leaflet of the membrane.
(A) Electrostatic potentials of EGFR kinases on the surface in contact with the membrane (first row), kinase interactions with the inner leaflet (second row), and aggregation of anionic (POPS) lipids around EGFR in simulations (third row). The anionic lipids are shown in red, and the other lipids in gray. The fractions indicate the relative concentration of POPS lipids in the membrane bilayer. The electrostatic potential is shown on a scale from −5 to 5 kBT/∣e∣ (red to blue). Note that the kinase domains are attached to the membrane and their active sites (shown in orange in Row 2) are sequestered by the membrane except in the active dimer. (B) Instability of the inactive dimer at low concentrations of POPS lipids. With low POPS concentrations, the kinase domains detached from the membrane (left) and the C-terminal transmembrane dimer dissociated (right); here dint denotes the separation between the two C-terminal GxxxG-like motifs. (C) A model of overexpression-induced EGFR activation due to reduced availability of the anionic lipids. At normal expression levels (left), extensive interaction with the anionic lipids favors inactive EGFR monomers and dimers over active dimers. At high expression levels (right), a relative scarcity of the anionic lipids leads to EGFR activation. See also Figure S4.
Similarly, we assembled a model of an EGFR inactive dimer (Figure 6B), which included the simulation-generated, ligand-free extracellular dimer (Figure 3), a C-terminal transmembrane dimer, and membrane-embedded JM-A connected via the extended JM-B to the (inactive) symmetric kinase dimer (Jura et al., 2009). The involvement of the symmetric dimer in the inactive dimer of EGFR is consistent with the low-resolution visualization of a globular kinase dimer that differs from the rod-like asymmetric active kinase dimer (Mi et al., 2010). The symmetric kinase dimer, with the large distance between its N-termini, may be readily connected to a pair of separated juxtamembrane segments embedded in the membrane.
Previously it has been noted that the positively charged patches of the subunits in the symmetric kinase dimer face the same direction (Figure 7A; Jura et al., 2009). Given that these patches may interact favorably with anionic lipids, in the inactive dimer model we placed the kinase dimer so that these patches faced the membrane. As in the monomer model, here the substrate binding sites of the kinase domains were again occluded by the membrane (Figure 7A).
Our model of the active dimer (Figure 6C) consisted of the 2-ligand active extracellular dimer, the N-terminal transmembrane dimer connected with the antiparallel JM-A dimer, and an (active) asymmetric JM-B–KD dimer. The asymmetric kinase dimer (PDB entry 2GS6; Zhang et al., 2006) was placed relative to the JM-A dimer according to the orientation seen in the crystal structure of the JM–KD construct (PDB entry 3GOP; Red Brewer et al., 2009). Unlike the inactive EGFR dimer, where the substrate binding sites of the kinase domains are occluded by the membrane, in the active EGFR dimer, the site of the enzymatically activated receiver kinase faces the interior of the cell (Figure 7A). While the key components of EGFR dimers were stable in our simulations of the EGFR models, some flexibility was observed between these components (Figure 6). For instance, the extracellular module of the active dimer may undergo significant motions relative to the transmembrane segments and the membrane but itself maintains a small distance between its C-termini (Figure S1E). Rather than standing upright on the membrane, the extracellular modules are flexible in orientation and often align on the membrane surface, in agreement with previous FRET measurements (Kästner et al., 2009).
EGFR interactions with anionic membrane lipids
Our models highlight the extensive electrostatic interaction between EGFR and the intracellular leaflet of the membrane. Such interaction is reflected in the clustering of anionic POPS lipids around EGFR in our simulations. The simulations show that the EGFR monomer, and inactive and active dimers, respectively, are in contact (within 5 Å) with 25 ± 3, 52 ± 2, and 37 ± 3 POPS lipid molecules on average (at 15% POPS; see Figures 7A and S4). In all three cases the POPS in contact with EGFR accounted for ~50% of all the lipids in contact, while POPS accounted for only 30% (15% overall for the bilayer) of the inner-leaflet lipids (see Experimental Procedures; Figure S4B and S4C). We have also observed similar trends in simulations with lower (7.5% and 2.5% of the overall lipid number) POPS content. The JM-A, which is rich in basic residues, interacts extensively with the anionic lipids both in inactive EGFRs, where it is embedded in the membrane, and in the active EGFR dimers, where it is part of an antiparallel helix dimer (Figure S3). As discussed above, such interactions in the active dimer presumably stabilize the JM-A dimer and strengthen the coupling between the extra- and intracellular modules.
We found that inactive EGFR interacted with the anionic lipids more extensively than did active EGFR. For example, on average ~26 POPS molecules were in contact with each receptor in the inactive monomer or dimer, versus ~18–19 in the active dimer. This is because in inactive EGFR the basic residues of the kinase domains are exposed and in contact with the membrane (Figure 7A), whereas in the active dimer the patches are shielded by the C-terminal tails, as shown in the crystal structure (PDB entry 2GS6; Zhang et al., 2006).
The interaction of EGFR with anionic lipids thus is likely more energetically favorable for inactive than for active EGFR. If so, the interaction of the membrane with the intracellular portion of EGFR would give a net contribution to EGFR inhibition. This is consistent with our finding that lowering POPS concentration leads to destabilization of the inactive dimer conformation and detachment of the kinase domains from the membrane (Figure 7B and S4D). While the C-terminal transmembrane dimer associated with EGFR autoinhibition was stable in the inactive TM–JM–KD dimer at 15% (overall) POPS, it dissociated in simulations with lower POPS content (Figure 7B), presumably because the interaction of the anionic lipids with the juxtamembrane segments is needed to stabilize the inactive EGFR dimer. Remarkably, simulations with reduced POPS concentration (0–2.5% ) also show that the kinase domains of the inactive EGFR dimer may detach from the membrane when there is a lack of negatively charged lipids (Figure 7B). Such detachment may favor activation, as the intracellular EGFR module has been found to be active in solution, but inhibited when attached to the cell membrane (Endres et al., 2013).
Discussion
The simulation studies and the experimental findings (Endres et al., 2013) together shed light on the overall architecture of intact EGFRs in the membrane environment and on the structural mechanism of autoinhibition and activation. The extracellular module apparently plays an inhibitory role in the absence of ligands, as its deletion leads to ligand-independent activation. Our simulations show that, in addition to impeding receptor dimerization, ligand-free extracellular dimers (especially the two domain IVs) disfavor activation in preformed dimers by assuming conformations inconsistent with the formation of the N-terminal transmembrane dimers. This explains why the insertion of a flexible linker between the extracellular and the transmembrane segments, which presumably decouples the former from the rest of EGFR, causes enhanced activity in the absence of ligands.
In addition, our studies highlight the previously largely overlooked role of the membrane in maintaining the coupling of EGFR extracellular domains with the rest of the receptors. The anionic lipids were found to extensively interact with the basic residues of the juxtamembrane segments, helping stabilize the juxtamembrane helix dimer and, indirectly, the active kinase dimer. These interactions led to a stronger conformational coupling between the trans- and juxtamembrane segments in the POPC/POPS membrane than in the DMPC one, as reflected in the greater stability of the juxtamembrane helix dimer in the former membrane. This may explain why EGFR activity in response to EGF stimulus is reduced at lower levels of anionic PIP2 lipids in the cell membrane (Michailidis et al., 2011). It is likewise not entirely surprising that, for EGFRs immersed in detergent micelles rather than embedded in a cell membrane, the dimerization of their extracellular modules is not necessarily coupled to the dimerization of their intracellular modules (Mi et al., 2011; Wang et al., 2011).
While the membrane helps ensure that ligand binding leads to robust EGFR activation, our studies show that it also plays a crucial role in the autoinhibition. The surface of an EGFR kinase domain features extensive patches of basic residues (Jura et al., 2009), which are shielded by the C-terminal tails only in an active dimer. Our simulations show that inactive EGFR kinases, whether monomers or dimers, attach to the membrane, and the basic residues interact extensively with the anionic lipids. As a result, the active sites of the kinase domains are obstructed. In comparison, the asymmetric kinase dimer in an active EGFR dimer has less interactions with the anionic lipids, and the active site of the enzymatically active “receiver” kinase is exposed (Figure 7A).
On balance, the anionic lipids of the membrane favor the inactive state of EGFR. This is an important feature of the “electrostatic engine” model, in which EGFR activation is postulated to involve the breaking of electrostatic interactions and the release of the intracellular module from the membrane (McLaughlin et al., 2005). Our observation that the inactive dimer interacts more favorably with the membrane (Figure 7A) may be of importance to EGFR autoinhibition. In normal cells the effective EGFR concentration on the cell surface can reach 20 μM (~150 molecules per μm2 in surface density), but the dissociation constant for active dimers of EGFR kinase domain is approximately 6 μM without stabilization by the juxtamembrane segment (Shan et al., 2012). Since the juxtamembrane domains are long and flexible, these numbers suggest that active kinase domain dimers would form in the absence of ligand stimulation if the formation does not require breaking favorable interactions with the membrane.
The important role of anionic lipids in EGFR inhibition led us to speculate that a relative shortage of anionic lipids may underlie the aberrant activity associated with EGFR overexpression that has been observed in many cancer cells. Although overexpression presumably causes an enhanced level of EGFR dimerization, it remains unclear how overexpression overwhelms the autoinhibitory mechanism that ordinarily ensures that ligand-free dimers are inactive. It has been shown (Endres et al., 2013) that in the absence of ligands EGFR activity depends on EGFR density at the cell surface. Notably, EGFR activity appears to grow strongly only in the density range beyond 800 μm−2, suggesting a possible autoinhibitory mechanism that breaks down at high EGFR density. Our analysis shows that a dimerization model with a density-independent dissociation constant of the active dimer fits the data less well than a similar model with a dissociation constant that decreases with higher densities (Figure S4E). The latter model implies that the active dimer is favored at high densities beyond the effect of high concentration. We conjecture that EGFR overexpression, and a consequent relative shortage of anionic lipids, may weaken the coupling between EGFR extra- and intracellular modules, and shift the balance toward active kinase dimers (Figure 7C).
Experimental Procedures
The simulations were performed on the special-purpose supercomputer Anton (Shaw et al., 2009). The simulated systems ranged in size from ~35,000 to ~554,000 atoms. All proteins, water molecules, and membrane lipids were represented in full atomic detail. See the Supplemental Information for details of the protocols of the MD simulation and system setup, and the force fields used in the simulations.
Supplementary Material
Research Highlights.
Full-length EGF receptors are modeled in a realistic membrane environment
The models show how EGF binding controls the extracellular domains in EGFR dimers
The trans- and juxtamembrane segments alternate between two dimer forms as a result
Anionic lipids in the membrane are critical to the regulation of the kinase domains
Acknowledgements
We thank Morten Jensen, Stefano Piana, and Kresten Lindorff-Larsen for helpful discussions, Ansgar Philippsen for assistance with graphics, and Mollie Kirk and Berkman Frank for editorial assistance. N.E. was a Leukemia and Lymphoma Society Fellow. R.D. was supported by post-doctoral fellowships from the Natural Sciences and Engineering Research Council of Canada (2009-2011) and the Canadian Institutes of Health Research (2011-2012). This work was supported by a grant from the National Cancer Institute (NCI) to J.K. (2-R01-CA096504-06).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alvarado D, Klein DE, Lemmon MA. ErbB2 resembles an autoinhibited invertebrate epidermal growth factor receptor. Nature. 2009;461:287–291. doi: 10.1038/nature08297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarado D, Klein DE, Lemmon MA. Structural basis for negative cooperativity in growth factor binding to an EGF receptor. Cell. 2010;142:568–579. doi: 10.1016/j.cell.2010.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessman NJ, Lemmon MA. Finding the missing links in EGFR. Nat. Struct. and Mol. Biol. 2012;19:1–3. doi: 10.1038/nsmb.2221. [DOI] [PubMed] [Google Scholar]
- Bocharov EV, Mineev KS, Volynsky PE, Ermolyuk YS, Tkach EN, Sobol AG, Chupin VV, Kirpichnikov MP, Efremov RG, Arseniev AS. Spatial structure of the dimeric transmembrane domain of the growth factor receptor ErbB2 presumably corresponding to the receptor active state. J. Biol. Chem. 2008;283:6950–6956. doi: 10.1074/jbc.M709202200. [DOI] [PubMed] [Google Scholar]
- Cho HS, Leahy DJ. Structure of the extracellular region of HER3 reveals an interdomain tether. Science. 2002;297:1330–1333. doi: 10.1126/science.1074611. [DOI] [PubMed] [Google Scholar]
- Cho HS, Mason K, Ramyar KX, Stanley AM, Gabelli SB, Denney DW, Jr., Leahy DJ. Structure of the extracellular region of HER2 alone and in complex with the Herceptin Fab. Nature. 2003;421:756–760. doi: 10.1038/nature01392. [DOI] [PubMed] [Google Scholar]
- Choowongkomon K, Carlin CR, Sonnichsen FD. A structural model for the membrane-bound form of the juxtamembrane domain of the epidermal growth factor receptor. J. Biol. Chem. 2005;280:24043–24052. doi: 10.1074/jbc.M502698200. [DOI] [PubMed] [Google Scholar]
- Chung I, Akita R, Vandlen R, Toomre D, Schlessinger J, Mellman I. Spatial control of EGF receptor activation by reversible dimerization on living cells. Nature. 2010;464:783–787. doi: 10.1038/nature08827. [DOI] [PubMed] [Google Scholar]
- Citri A, Yarden Y. EGF–ERBB signalling: towards the systems level. Nat. Rev. Mol. Cell Biol. 2006;7:505–516. doi: 10.1038/nrm1962. [DOI] [PubMed] [Google Scholar]
- Clayton AHA, Walker F, Orchard SG, Henderson C, Fuchs D, Rothacker J, Nice EC, Burgess AW. Ligand-induced dimer-tetramer transition during the activation of the cell surface epidermal growth factor receptor-A multidimensional microscopy analysis. J. Biol. Chem. 2005;280:30392–30399. doi: 10.1074/jbc.M504770200. [DOI] [PubMed] [Google Scholar]
- Dawson JP, Bu Z, Lemmon MA. Ligand-induced structural transitions in ErbB receptor extracellular domains. Structure. 2007;15:942–954. doi: 10.1016/j.str.2007.06.013. [DOI] [PubMed] [Google Scholar]
- Du Y, Yang H, Xu Y, Cang X, Luo C, Mao C, Wang C, Qin G, Luo X, Jiang J. Conformational transition and energy landscape of ErbB4 activated by neuregulin1β: one microsecond molecular dynamics simulations. J. Am. Chem. Soc. 2012;134:6720–6731. doi: 10.1021/ja211941d. [DOI] [PubMed] [Google Scholar]
- Endres NF, Das R, Smith A, Arkhipov A, Kovacs E, Huang Y, Pelton JG, Shan YB, Shaw DE, Wemmer DE, Groves JT, Kuriyan J. Conformational coupling across the plasma membrane in activation of the EGF receptor. Cell. 2013 doi: 10.1016/j.cell.2012.12.032. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson KM, Berger MB, Mendrola JM, Cho HS, Leahy DJ, Lemmon MA. EGF activates its receptor by removing interactions that autoinhibit ectodomain dimerization. Mol. Cell. 2003;11:507–517. doi: 10.1016/s1097-2765(03)00047-9. [DOI] [PubMed] [Google Scholar]
- Fleishman SJ, Schlessinger J, Ben-Tal N. A putative molecular-activation switch in the transmembrane domain of erbB2. Proc. Natl. Acad. Sci. U.S.A. 2002;99:15937–15940. doi: 10.1073/pnas.252640799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett TPJ, McKern NM, Lou M, Elleman TC, Adams TE, Lovrecz GO, Zhu HJ, Walker F, Frenkel MJ, Hoyne PA, et al. Crystal structure of a truncated epidermal growth factor receptor extracellular domain bound to transforming growth factor α. Cell. 2002;110:763–773. doi: 10.1016/s0092-8674(02)00940-6. [DOI] [PubMed] [Google Scholar]
- Garrett TP, McKern NM, Lou M, Elleman TC, Adams TE, Lovrecz GO, Kofler M, Jorissen RN, Nice EC, Burgess AW, et al. The crystal structure of a truncated ErbB2 ectodomain reveals an active conformation, poised to interact with other ErbB receptors. Mol. Cell. 2003;11:495–505. doi: 10.1016/s1097-2765(03)00048-0. [DOI] [PubMed] [Google Scholar]
- Holt A, Killian JA. Orientation and dynamics of transmembrane peptides: the power of simple models. Eur. Biophys. J. 2010;39:609–621. doi: 10.1007/s00249-009-0567-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jura N, Endres NF, Engel K, Deindl S, Das R, Lamers MH, Wemmer DE, Zhang X, Kuriyan J. Mechanism for activation of the EGF receptor catalytic domain by the juxtamembrane segment. Cell. 2009;137:1293–1307. doi: 10.1016/j.cell.2009.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kästner J, Loeffler HH, Roberts SK, Martin-Fernandez ML, Winn MD. Ectodomain orientation, conformational plasticity and oligomerization of ErbB1 receptors investigated by molecular dynamics. J. Struct. Biol. 2009;167:117–128. doi: 10.1016/j.jsb.2009.04.007. [DOI] [PubMed] [Google Scholar]
- Kim C, Schmidt T, Cho EG, Ye F, Ulmer TS, Ginsberg MH. Basic amino-acid side chains regulate transmembrane integrin signaling. Nature. 2012;481:209–213. doi: 10.1038/nature10697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau M, Ben-Tal N. Dynamic equilibrium between multiple active and inactive conformations explains regulation and oncogenic mutations in ErbB receptors. Biochim. Biophys. Acta. 2008;1785:12–31. doi: 10.1016/j.bbcan.2007.08.001. [DOI] [PubMed] [Google Scholar]
- Lemmon MA, Treutlein HR, Adams PD, Brünger AT, Engelman DM. A dimerization motif for transmembrane alpha-helices. Nat. Struct. Biol. 1994;1:157–63. doi: 10.1038/nsb0394-157. [DOI] [PubMed] [Google Scholar]
- Liu P, Bouyain S, Eigenbrot C, Leahy DJ. The ErbB4 extracellular region retains a tethered-like conformation in the absence of the tether. Protein Sci. 2012a;21:152–155. doi: 10.1002/pro.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Cleveland TE, Bouyain S, Byrne PO, Longo PA, Leahy DJ. A single ligand is sufficient to activate EGFR dimers. Proc. Natl. Acad. Sci. U.S.A. 2012b;109:10861–10866. doi: 10.1073/pnas.1201114109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low-Nam ST, Lidke KA, Cutler PJ, Roovers RC, van Bergen en Henegouwen PMP, Wilson BS, Lidke DS. ErbB1 dimerization is promoted by domain co-confinement and stabilized by ligand binding. Nat. Struct. Mol. Biol. 2011;18:1244–1249. doi: 10.1038/nsmb.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Mi LZ, Grey MJ, Zhu J, Graef E, Yokoyama S, Springer TA. Structural evidence for loose linkage between ligand binding and kinase activation in the epidermal growth factor receptor. Mol. Cel. Biol. 2010;30:5432–5443. doi: 10.1128/MCB.00742-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattoon D, Klein P, Lemmon MA, Lax I, Schlessinger J. The tethered configuration of the EGF receptor extracellular domain exerts only a limited control of receptor function. Proc. Natl. Acad. Sci. U.S.A. 2004;101:923–928. doi: 10.1073/pnas.0307286101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin S, Smith SO, Hayman MJ, Murray D. An electrostatic engine model for autoinhibition and activation of the epidermal growth factor receptor (EGFR/ErbB) family. J. Gen. Physiol. 2005;126:41–53. doi: 10.1085/jgp.200509274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendrola JM, Berger MB, King MC, Lemmon MA. The single transmembrane domains of ErbB receptors self-associate in cell membranes. J. Biol. Chem. 2002;277:4704–4712. doi: 10.1074/jbc.M108681200. [DOI] [PubMed] [Google Scholar]
- Mi LZ, Lu C, Li Z, Nishida N, Walz T, Springer TA. Simultaneous visualization of the extracellular and cytoplasmic domains of the epidermal growth factor receptor. Nat. Struct. Mol. Biol. 2011;18:984–989. doi: 10.1038/nsmb.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michailidis IE, Rusinov R, Georgakopoulos A, Chen Y, Iyengar R, Robakis NK, Logothetis DE, Baki L. Phosphatidylinositol-4,5-bisphosphate regulates epidermal growth factor receptor activation. Pflugers Arch. 2011;461:387–397. doi: 10.1007/s00424-010-0904-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogiso H, Ishitani R, Nureki O, Fukai S, Yamanaka M, Kim J-H, Saito K, Sakamoto A, Inoue M, Shirouzu M, Yokoyama S. Crystal structure of the complex of human epidermal growth factor and receptor extracellular domains. Cell. 2002;110:775–787. doi: 10.1016/s0092-8674(02)00963-7. [DOI] [PubMed] [Google Scholar]
- Odaka M, Kohda D, Lax I, Schlessinger J, Inagaki F. Ligand binding enhances the affinity of dimerization of the extracellular domain of the epidermal growth factor receptor. J. Biochem. 1997;122:116–121. doi: 10.1093/oxfordjournals.jbchem.a021718. [DOI] [PubMed] [Google Scholar]
- Red Brewer M, Choi SH, Alvarado D, Moravcevic K, Pozzi A, Lemmon MA, Carpenter G. The juxtamembrane region of the EGF receptor functions as an activation domain. Mol. Cell. 2009;34:641–651. doi: 10.1016/j.molcel.2009.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riese DJ, Gallo RM, Settleman J. Mutational activation of ErbB family receptor tyrosine kinases: insights into mechanisms of signal transduction and tumorigenesis. BioEssays. 2007;29:1521–1878. doi: 10.1002/bies.20582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russ WP, Engelman DM. The GxxxG motif: a framework for transmembrane helix-helix association. J. Mol. Biol. 2000;296:911–919. doi: 10.1006/jmbi.1999.3489. [DOI] [PubMed] [Google Scholar]
- Sako Y, Minoghchi S, Yanagida T. Single-molecule imaging of EGFR signaling on the surface of living cells. Nat. Cell Biol. 2000;2:168–172. doi: 10.1038/35004044. [DOI] [PubMed] [Google Scholar]
- Scheck RA, Lowder MA, Appelbaum JS, Schepartz A. Bipartite tetracysteine display reveals allosteric control of ligand-specific EGFR activation. ACS Chem. Biol. 2012;7:1367–76. doi: 10.1021/cb300216f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlessinger J. Ligand-induced, receptor-mediated dimerization and activation of EGF receptor. Cell. 2002;110:669–672. doi: 10.1016/s0092-8674(02)00966-2. [DOI] [PubMed] [Google Scholar]
- Shan Y, Eastwood MP, Zhang X, Kim TE, Arkhipov A, Dror RO, Jumper J, Kuriyan J, Shaw DE. Oncogenic mutations counteract intrinsic disorder in the EGFR kinase and promote receptor dimerization. Cell. 2012;19:860–870. doi: 10.1016/j.cell.2012.02.063. [DOI] [PubMed] [Google Scholar]
- Shaw DE, Dror RO, Salmon JK, Grossman JP, Mackenzie KM, Bank JA, Young C, Deneroff MM, Batson B, Bowers KJ, et al. Millisecond-scale molecular dynamics simulations on Anton. Proceedings of the Conference on High Performance Computing, Networking, Storage and Analysis (SC09).2009. [Google Scholar]
- Sternberg MJ, Gullick WJ. Neu receptor dimerization. Nature. 1989;339:587. doi: 10.1038/339587a0. [DOI] [PubMed] [Google Scholar]
- Thiel KW, Carpenter G. Epidermal growth factor receptor juxtamembrane region regulates allosteric tyrosine kinase activation. Proc. Natl. Acad. Sci. U.S.A. 2007;104:19238–19243. doi: 10.1073/pnas.0703854104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Longo PA, Tarrant MK, Kim K, Head S, Leahy DJ, Cole PA. Mechanistic insights into the activation of oncogenic forms of EGF receptor. Nat. Struct. and Mol. Biol. 2011;18:1388–1393. doi: 10.1038/nsmb.2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood ER, Truesdale AT, McDonald OB, Yuan D, Hassell A, Dickerson SH, Ellis B, Pennisi C, Horne E, Lackey K, et al. A unique structure for epidermal growth factor receptor bound to GW572016 (lapatinib): relationships among protein conformation, inhibitor off-rate, and receptor activity in tumor cells. Cancer Res. 2004;64:6652–6659. doi: 10.1158/0008-5472.CAN-04-1168. [DOI] [PubMed] [Google Scholar]
- Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.