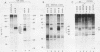Abstract
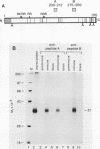
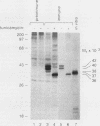
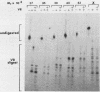
The proto-oncogene int-1 is activated by adjacent insertions of proviral DNA in mouse mammary tumor virus-induced tumors and has transforming activity in certain mammary epithelial cell lines. The gene is normally expressed in the central nervous system of mid-gestational embryos and in the adult testis. We raised antibodies against synthetic int-1 peptides and used these to identify protein products of the gene in cells transfected or infected with retroviral vectors expressing int-1. Four protein species of 36,000, 38,000, 40,000, and 42,000 Mr were immunoprecipitated by antibodies against two different int-1 peptides and were not present in control cells. Partial degradation with V8 protease showed the four species to be structurally related to each other and to int-1 polypeptide synthesized in vitro. Treatment of the cells with tunicamycin prevented the appearance of all but the 36,000-Mr species, suggesting that the slower-migrating forms are glycosylated derivatives. The unglycosylated 36,000-Mr species migrated faster in polyacrylamide gels than the in vitro translation product of int-1 and has probably undergone cleavage of an amino-terminal signal peptide.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brown A. M., Wildin R. S., Prendergast T. J., Varmus H. E. A retrovirus vector expressing the putative mammary oncogene int-1 causes partial transformation of a mammary epithelial cell line. Cell. 1986 Sep 26;46(7):1001–1009. doi: 10.1016/0092-8674(86)90699-9. [DOI] [PubMed] [Google Scholar]
- Cleveland D. W., Fischer S. G., Kirschner M. W., Laemmli U. K. Peptide mapping by limited proteolysis in sodium dodecyl sulfate and analysis by gel electrophoresis. J Biol Chem. 1977 Feb 10;252(3):1102–1106. [PubMed] [Google Scholar]
- Fung Y. K., Shackleford G. M., Brown A. M., Sanders G. S., Varmus H. E. Nucleotide sequence and expression in vitro of cDNA derived from mRNA of int-1, a provirally activated mouse mammary oncogene. Mol Cell Biol. 1985 Dec;5(12):3337–3344. doi: 10.1128/mcb.5.12.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobovits A., Shackleford G. M., Varmus H. E., Martin G. R. Two proto-oncogenes implicated in mammary carcinogenesis, int-1 and int-2, are independently regulated during mouse development. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7806–7810. doi: 10.1073/pnas.83.20.7806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg P. A., Melton D. A. Functional messenger RNAs are produced by SP6 in vitro transcription of cloned cDNAs. Nucleic Acids Res. 1984 Sep 25;12(18):7057–7070. doi: 10.1093/nar/12.18.7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Land H., Parada L. F., Weinberg R. A. Cellular oncogenes and multistep carcinogenesis. Science. 1983 Nov 18;222(4625):771–778. doi: 10.1126/science.6356358. [DOI] [PubMed] [Google Scholar]
- Lerner R. A., Sutcliffe J. G., Shinnick T. M. Antibodies to chemically synthesized peptides predicted from DNA sequences as probes of gene expression. Cell. 1981 Feb;23(2):309–310. doi: 10.1016/0092-8674(81)90126-4. [DOI] [PubMed] [Google Scholar]
- Mann R., Mulligan R. C., Baltimore D. Construction of a retrovirus packaging mutant and its use to produce helper-free defective retrovirus. Cell. 1983 May;33(1):153–159. doi: 10.1016/0092-8674(83)90344-6. [DOI] [PubMed] [Google Scholar]
- Nusse R., Varmus H. E. Many tumors induced by the mouse mammary tumor virus contain a provirus integrated in the same region of the host genome. Cell. 1982 Nov;31(1):99–109. doi: 10.1016/0092-8674(82)90409-3. [DOI] [PubMed] [Google Scholar]
- Nusse R., van Ooyen A., Cox D., Fung Y. K., Varmus H. Mode of proviral activation of a putative mammary oncogene (int-1) on mouse chromosome 15. Nature. 1984 Jan 12;307(5947):131–136. doi: 10.1038/307131a0. [DOI] [PubMed] [Google Scholar]
- Rijsewijk F., Schuermann M., Wagenaar E., Parren P., Weigel D., Nusse R. The Drosophila homolog of the mouse mammary oncogene int-1 is identical to the segment polarity gene wingless. Cell. 1987 Aug 14;50(4):649–657. doi: 10.1016/0092-8674(87)90038-9. [DOI] [PubMed] [Google Scholar]
- Rijsewijk F., van Deemter L., Wagenaar E., Sonnenberg A., Nusse R. Transfection of the int-1 mammary oncogene in cuboidal RAC mammary cell line results in morphological transformation and tumorigenicity. EMBO J. 1987 Jan;6(1):127–131. doi: 10.1002/j.1460-2075.1987.tb04729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackleford G. M., Varmus H. E. Expression of the proto-oncogene int-1 is restricted to postmeiotic male germ cells and the neural tube of mid-gestational embryos. Cell. 1987 Jul 3;50(1):89–95. doi: 10.1016/0092-8674(87)90665-9. [DOI] [PubMed] [Google Scholar]
- Vaidya A. B., Lasfargues E. Y., Sheffield J. B., Coutinho W. G. Murine mammary tumor virus (MuMTV) infection of an epithelial cell line established from C57BL/6 mouse mammary glands. Virology. 1978 Oct 1;90(1):12–22. doi: 10.1016/0042-6822(78)90328-8. [DOI] [PubMed] [Google Scholar]
- Wilkinson D. G., Bailes J. A., McMahon A. P. Expression of the proto-oncogene int-1 is restricted to specific neural cells in the developing mouse embryo. Cell. 1987 Jul 3;50(1):79–88. doi: 10.1016/0092-8674(87)90664-7. [DOI] [PubMed] [Google Scholar]
- van Ooyen A., Kwee V., Nusse R. The nucleotide sequence of the human int-1 mammary oncogene; evolutionary conservation of coding and non-coding sequences. EMBO J. 1985 Nov;4(11):2905–2909. doi: 10.1002/j.1460-2075.1985.tb04021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ooyen A., Nusse R. Structure and nucleotide sequence of the putative mammary oncogene int-1; proviral insertions leave the protein-encoding domain intact. Cell. 1984 Nov;39(1):233–240. doi: 10.1016/0092-8674(84)90209-5. [DOI] [PubMed] [Google Scholar]
- von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986 Jun 11;14(11):4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. Analysis of the distribution of charged residues in the N-terminal region of signal sequences: implications for protein export in prokaryotic and eukaryotic cells. EMBO J. 1984 Oct;3(10):2315–2318. doi: 10.1002/j.1460-2075.1984.tb02132.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. How signal sequences maintain cleavage specificity. J Mol Biol. 1984 Feb 25;173(2):243–251. doi: 10.1016/0022-2836(84)90192-x. [DOI] [PubMed] [Google Scholar]