Abstract
Telomeres are repetitive DNA structures that, together with the shelterin and the CST complex, protect the ends of chromosomes. Telomere shortening is mitigated in stem and cancer cells through the de novo addition of telomeric repeats by telomerase. Telomere elongation requires the delivery of the telomerase complex to telomeres through a not yet fully understood mechanism. Factors promoting telomerase–telomere interaction are expected to directly bind telomeres and physically interact with the telomerase complex. In search for such a factor we carried out a SILAC-based DNA–protein interaction screen and identified HMBOX1, hereafter referred to as homeobox telomere-binding protein 1 (HOT1). HOT1 directly and specifically binds double-stranded telomere repeats, with the in vivo association correlating with binding to actively processed telomeres. Depletion and overexpression experiments classify HOT1 as a positive regulator of telomere length. Furthermore, immunoprecipitation and cell fractionation analyses show that HOT1 associates with the active telomerase complex and promotes chromatin association of telomerase. Collectively, these findings suggest that HOT1 supports telomerase-dependent telomere elongation.
Keywords: DNA–protein interaction, HOT1, mass spectrometry, telomeres, telomere length
Introduction
Telomeres, the nucleoprotein structures at the ends of chromosomes, consist of 5′-TTAGGG-3′ repeats bound by a dedicated set of proteins forming the shelterin complex (Palm and de Lange, 2008). Three members of the six protein complex directly bind telomeric DNA and were the only direct telomere-specific binding proteins known so far: TRF1 and TRF2 bind double-stranded DNA (dsDNA; Zhong et al, 1992; Bilaud et al, 1997; Broccoli et al, 1997), whereas POT1 binds single-stranded 5′-TTAGGG-3′ repeats (Baumann, 2001). This complex constitutively associates with telomeres and shields the ends of linear chromosomes from being recognized as a double-stranded break, thus protecting telomeres from end-to-end fusions (Palm and de Lange, 2008). While this solves the end-protection problem (de Lange, 2009), maintaining telomere integrity itself is of outstanding importance and major factors may have remained elusive.
Telomere length homeostasis, a crucial process in stem cell biology, aging and cancer, depends on the equilibrium between telomere lengthening (in most cases due to telomerase activity) and shortening reactions (generally due to replication and controlled processing) (Jain and Cooper, 2010). Telomerase is capable of adding telomeric repeats to chromosome ends de novo. The enzyme works as a ribonucleoprotein complex, which consists of a catalytic subunit with reverse-transcriptase activity (called TERT), and an RNA serving as the elongation matrix for telomeres (called TR or TERC) (Greider and Blackburn, 1989). While these two core elements are sufficient for telomerase activity in vitro, biochemical analyses have shown that in vivo telomerase resides in a large complex of about 1 MDa (Schnapp et al, 1998). Some additional components of this large multi-subunit holoenzyme complex have been identified. In particular, the core components of box H/ACA small nucleolar ribonucleoprotein particles (snoRNPs), DKC1 (dyskerin), GAR1, NHP2 and NOP10, are part of the active telomerase complex, and are necessary for proper RNP assembly as well as for TERC stability (Mitchell et al, 1999; Wang and Meier, 2004). More recently, the ATPases RUVBL1 and RUVBL2 have been identified as factors essential for holoenzyme assembly (Venteicher et al, 2008), and TCAB1 (WDR79/WRAP53), identified as a DKC1 interaction partner, was shown to be required for proper localization of CAB box containing small Cajal body (CB)-specific RNPs (scaRNPs) to CBs, including TERC, and is part of the active telomerase complex (Tycowski et al, 2009; Venteicher et al, 2009).
The presence of major scaRNA processing and trafficking factors in the telomerase complex hints to an important aspect of telomerase cell biology: the orchestrated maturation of telomerase and interaction with telomeres in the CB. Telomere maintenance by telomerase requires that both TERT and TERC are recruited from distinct subnuclear sites to telomeres during S phase (synthesis phase) (Tomlinson et al, 2008). Like other scaRNAs, TERC contains a common CB-specific localization signal and accumulates in CBs (Jády et al, 2004; Zhu et al, 2004), where it is found together with TERT (Tomlinson et al, 2008). In a cell cycle-dependent manner, telomerase-containing CBs are then recruited to telomeres, suggesting that CBs represent an enzymatic hub in which telomere elongation by telomerase takes place (Jády et al, 2006; Tomlinson et al, 2006; Cristofari et al, 2007). This trafficking model is further supported by telomere elongation defects in the absence of TCAB1 or presence of dysfunctional TCAB1, disrupting TERC accumulation in the CB (Venteicher et al, 2009; Zhong et al, 2011). Nevertheless, so far it remains elusive how telomeres are recruited to CBs, how this selective interaction is regulated and what drives the conversion from telomeres in a closed state, in which telomerase has little or no access, to telomeres in an open, accessible state.
Telomerase is usually limiting and, under physiological conditions, acts preferentially on short telomeres (Hemann et al, 2001; Britt-Compton et al, 2009), due to a well-established negative feedback loop mediated in cis by TRF1 and POT1, likely by hiding the 3′-overhang, which serves as a template for telomerase (Loayza and de Lange, 2003). Indeed, diminished loading of POT1 or expression of a dominant-negative version lacking DNA-binding activity leads to telomere elongation by telomerase, and in vitro experiments have shown that POT1 is competing with telomerase for its substrate (Loayza and de Lange, 2003; Ye et al, 2004; Kelleher et al, 2005; Lei et al, 2005). However, POT1 also interacts with TPP1, and both proteins together promote telomerase activity in vitro (Latrick and Cech, 2010). Furthermore, TPP1 has been shown to be required for the recruitment of telomerase to its substrate in vitro and to telomeric chromatin in vivo (Xin et al, 2007; Abreu et al, 2010; Tejera et al, 2010; Zaug et al, 2010; Zhong et al, 2012). While TPP1 has been proposed as a telomerase recruiter, it does not completely fit the definition, since it has initially been described as a negative regulator of telomere length in telomerase-positive cells (Liu et al, 2004; Ye et al, 2004), although these results may in part be attributed to secondary effects, such as the lack of POT1 tethering to telomeres in the absence of TPP1. Based on these bivalent results, a ying-yang model for telomerase recruitment and activity control has been proposed for TPP1–POT1 (Xin et al, 2007). Indeed, recently, a specific patch of amino acids on the surface of TPP1, the TEL patch (TPP1 glutamate (E)- and leucine (L)-rich patch), has been identified as crucial for the TPP1 function in telomerase recruitment and regulation (Nandakumar et al, 2012). Analysis of point mutations within the TEL patch demonstrated that the TEL patch is physically and functionally distant from the portion of TPP1 engaged in end protection, separating these two functions (Nandakumar et al, 2012).
Regulating the amount of telomeric repeats added by telomerase also involves how long telomerase can act on a given chromosome end. Therefore, turning off telomerase needs to be regulated in addition to the recruitment step and processivity control. In a recent study, Chen et al (2012) described the human CST (CTC1, STN1 and TEN1) complex as a terminator of telomerase activity (Chen et al, 2012). CST competes with POT1–TPP1 for telomeric DNA and is increasingly enriched on telomeric DNA during late S/G2 phase, correlating with the period in the cell cycle when telomerase action is terminated. In agreement with a suppression of telomerase action, depletion of any of the three CST complex members led to a steady increase in telomere length in a telomerase-dependent manner. The authors suggested that CST binds to telomerase-extended 3′-ends and thereby suppresses telomerase access and further elongation (Chen et al, 2012). However, depletion of STN1 has been reported not to affect telomere length in various telomerase-positive cellular contexts (Wang et al, 2012) and murine CTC1-null cells do not exhibit the reported telomere lengthening phenotype (Gu et al, 2012). The proposed model is nonetheless appealing, as human CST subunits stimulate DNA polymerase α-primase (Casteel et al, 2009) and therefore CST binding to telomerase-extended 3′-ends could initiate a switch from telomere elongation to fill-in synthesis. This mechanism could provide an autonomous end point to telomerase action at single telomeres, ensuring that every telomere is extended by telomerase once and only once during every cell cycle (Chen et al, 2012).
We reasoned that the identification of novel telomere-binding proteins would be a first step to the identification of additional factors implicated in telomere biology. SILAC-based quantitative mass spectrometry (MS) has been adapted for DNA–protein interactions (Mittler et al, 2009) and has been used by us and others successfully to identify factors binding to particular functional DNA fragments (Markljung et al, 2009; Butter et al, 2010; Bartels et al, 2011; Butter et al, 2012). For our purpose, we applied this approach to telomeric DNA in order to screen for telomere repeat-binding proteins and identified the protein HOT1 (HMBOX1; homeobox telomere-binding protein 1). HOT1 had previously been described as a putative transcriptional repressor based on reporter gene assays (Chen et al, 2006) and had been identified as a telomere-associated protein by the proteomics of isolated chromatin segments (PICh) approach (Déjardin and Kingston, 2009). Here we demonstrate that HOT1 directly binds to telomeric DNA, and characterize this binding in atomic detail by resolving a crystal structure of the HOT1 homeobox domain in a cocrystal with telomeric DNA. In vivo, HOT1 localizes to a subset of telomeres with a higher degree of HOT1–telomere association in cellular contexts of elevated telomere processing. In addition, we show that HOT1 associates with the active telomerase complex and that HOT1 is required for telomerase chromatin binding. These findings suggest that HOT1 contributes to the association of telomerase with telomeres and telomere length maintenance in various cellular settings, and classify HOT1 as the first direct telomere-binding protein that acts as a positive regulator of telomere length.
Results
Identification of HOT1 as a direct telomere repeat-binding protein
To identify telomere-binding proteins we used polymerized biotinylated double-stranded oligonucleotides of the telomeric sequence (5′-TTAGGG-3′) and a scrambled control sequence (5′-GTGAGT-3′), separately immobilized on paramagnetic streptavidin beads and incubated with heavy and light SILAC-labelled nuclear extracts from HeLa cells, respectively. Specific binding of proteins is detected by incubation of cell lysates encoded by ‘heavy’ amino acids (15N- and 13C-labelled Lys and Arg) with the bait sequence, while a control sequence is incubated with ‘light’, nonlabelled amino acids. Specific binders display a differential SILAC ratio, whereas background binders have a 1:1 ratio. After mild washing, bead fractions were combined and captured proteins were analysed by quantitative, high-resolution MS (Cox and Mann, 2008) (Figure 1A).
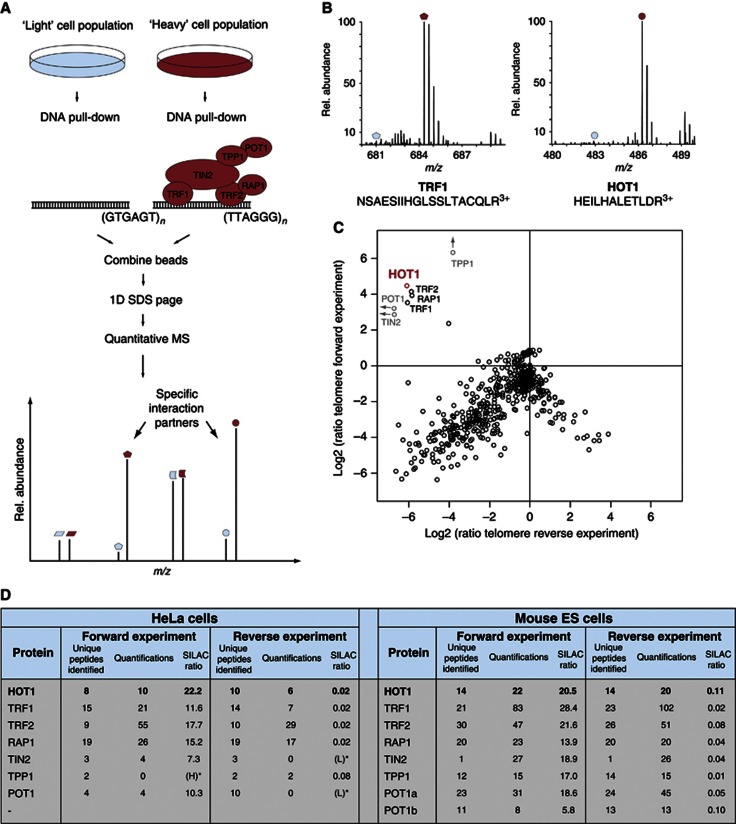
Figure 1.
Detection of specific telomere-interacting proteins. (A) A schematic of the quantitative SILAC-based DNA interaction screen with DNA oligonucleotides containing either the telomeric repeat or a control sequence. Specific interaction partners are differentiated from background binders by a SILAC ratio other than 1:1. (B) MS spectra of representative peptides from the ‘forward’ pull-down experiment. The heavy peptide partners are easily detected (red dots), while the light partner is barely observable (blue dots) in the mass spectrum. (C) Two-dimensional interaction plot: known shelterin components cluster together with HOT1, demonstrating enrichment at the telomere sequence compared to the control sequence. (D) Summary of the MS data for HOT1 and the core shelterin components from the SILAC-based DNA–protein interaction screens carried out with nuclear extracts derived from HeLa and murine ES cells.
We identified all the six core shelterin components with a SILAC ratio of about 10 or higher in the ‘forward’ and about 0.1 in the ‘reverse’ experiment, in which we had switched the labels (Figure 1B–D, Supplementary Figure S1 and Supplementary Table S1). In contrast, none of the proteins known to interact with shelterin were identified with SILAC ratios sufficiently high to be consistent with telomere binding, demonstrating that this approach was very stringent and exclusively detected telomere repeat-binding proteins and their strong interaction partners (Figure 1C). In addition to the shelterin components, we found the protein HOT1 with a high SILAC ratio that clustered with those of the shelterin components (Figure 1B–D, Supplementary Figure S1 and Supplementary Table S1). This indicates that HOT1 must either strongly associate with the shelterin complex or directly bind to the 5′-TTAGGG-3′ repeats.
To verify that the HOT1 identification was not cancer-, cell- or species-specific, we repeated our telomere-binding assay with SILAC-labelled nuclear extracts derived from mouse embryonic stem cells (ES cells). Again, all components of the shelterin complex and HOT1 were identified with SILAC ratios, indicating specific binding to the telomere repeats (Figure 1D and Supplementary Table S2). Here we also identified the two paralogues, POT1a and POT1b, which result from a gene duplication of the Pot1 gene in the rodent lineage (Hockemeyer et al, 2006), underscoring the specificity of our assay for direct telomere-binding proteins. Hence, HOT1 is a putative telomere repeat-binding protein conserved in mammalian cells.
HOT1 contains a homeobox domain (Chen et al, 2006), suggesting that it may bind DNA directly. To determine whether HOT1 was detected in our assay due to direct binding to the 5′-TTAGGG-3′ repeats, we performed DNA-binding assays with HOT1 in vitro. Recombinant HOT1 bound specifically to telomeric repeats, whereas no binding to the negative control repeat fragments (5′-GTGAGT-3′) was detected (Figure 2A). Exhibiting similar binding behaviour as TRF1, HOT1 was not enriched on any of the subtelomeric variant repeats 5′-TCAGGG-3′, 5′-TGAGGG-3′ and 5′-TTGGGG-3′, nor on the C. elegans telomere 5′-TTAGGC-3′ repeat sequence (Wicky et al, 1996; Figure 2A). To test whether HOT1 also associates with telomeres in vivo, we performed chromatin immunoprecipitation (ChIP) experiments with extracts from HeLa cells using an antibody directed against HOT1. Similar to TRF2, HOT1 IPs showed enrichment of telomeric DNA in comparison to two negative controls (anti-GFP antibody and IgG; Figure 2B). Thus, HOT1 is a direct and specific telomere repeat-binding protein.
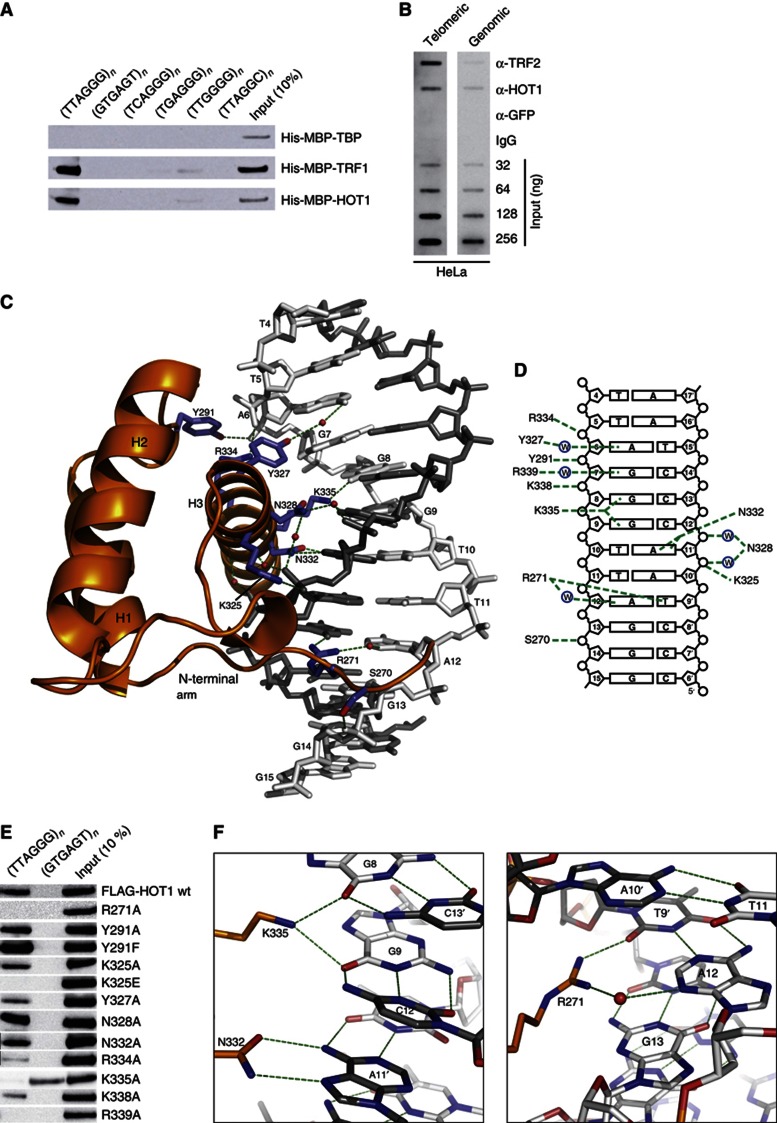
Figure 2.
The DBD of human HOT1 recognizes telomeric DNA in a sequence-specific manner. (A) Sequence-specific pull-down of recombinant HOT1, TRF1 (positive control) and TBP (TATA-binding protein, negative control). Proteins were incubated with dsDNA of telomeric repeats (5′-TTAGGG-3′), the control sequence (5′-GTGAGT-3′), the subtelomeric repeat variants (5′-TCAGGG-3′, 5′-TGAGGG-3′ and 5′-TTGGGG-3′, as well as the C. elegans telomere repeat 5′-TTAGGC-3′). All DNA substrates were concatemerized from 60 bp oligonucleotides to larger DNA fragments (on average at least 1 kb). (B) ChIP of telomeric DNA using antibodies against HOT1, TRF2 (positive control), GFP and IgG (negative controls). Representative slotblot images are shown for ChIP from HeLa extracts after hybridization with a telomeric and genomic control. Input dilutions demonstrate the linearity of the signals acquired. (C) Structure of the DBD of HOT1 bound to double-stranded telomeric DNA. The protein is shown as a cartoon representation (orange), whereas DNA is shown as a stick model (grey). The interacting amino acid residues in HOT1 are shown as blue sticks, water molecules as red spheres and protein–DNA contacts are visualized as green dashed lines. (D) Schematic representation of all protein–DNA contacts in the complex. (E) Sequence-specific pull-down of FLAG–HOT1 and selected single mutations to investigate binding specificity. Proteins were incubated with either telomeric repeats (5′-TTAGGG-3′) or a control oligonucleotide (5′-GTGAGT-3′). (F) Atomic details of DNA sequence recognition by HOT1. K335 of helix 3 is involved in direct hydrogen bonding to O6 of G8 and O6 of G9. N332 of helix 3 specifically recognizes A11′ of the complementary strand by forming two direct H-bonds with the bicyclic ring system of A11′ (N6 and N7) (left panel). R271 of the N-terminal arm binds two bases of an AT base pair, directly to T9′ and via a water-mediated H-bond to A12 (right panel).
Source data for this figure is available on the online supplementary information page.
HOT1 recognizes telomeric DNA by means of its homeodomain
Driven by these findings and with the aim to fully understand the molecular interactions between HOT1 and telomeric DNA, we crystallized the DNA-binding domain (DBD) of HOT1 with telomeric DNA. In order to identify a construct suitable for crystallization, we initially tested six different HOT1 fragments for their DNA-binding ability (Supplementary Figure S2). The three longer constructs Q144–A345, L156–A345 and G233–A345 all bound to immobilized telomeric dsDNA baits, demonstrating that the homeodomain of HOT1 is sufficient for recognizing telomeric DNA and that integrity of the predicted N-terminal POU-specific (POUs) domain (Chi et al, 2002) is not required. The three shorter constructs (P242–A345, P254–A345 and R271–A345) were not able to bind to the bait DNA (Supplementary Figure S2). For crystallization, we reconstituted and purified telomeric DNA complexes with all three binding constructs, but only one (G233–A345) yielded crystals when reconstituted with a duplex telomeric DNA (5′-cTGTTAGGGTTAGGGTTAG-3′ and 3′-ACAATCCCAATCCCAATCt-5′) similar to the one present in the crystal structures of the TRF1 and TRF2 homeodomains bound to telomeric DNA (Court et al, 2005).
The optimized crystals diffracted to 2.9 Å resolution and we could solve the structure by molecular replacement using the NMR model of the human HOT1 homeodomain (residues 268–343, PDB entry 2CUF, unpublished data from RIKEN Structural Genomics/Proteomics Initiative). In the orthorhombic crystals, the DNA forms an infinite double helix via a C–T nonWatson–Crick base pairing of the single-base overhangs (not shown). Two copies of the HOT1 DBD are bound to one duplex DNA that comprises two and a half 5′-TTAGGG-3′ repeats in a regular and undistorted B-form conformation (Supplementary Figure S3). As expected, and in accordance with the NMR model, the homeodomain of HOT1 folds into a small structure of three consecutive helices, α1 (res. 276–288), α2 (res. 293–309) and α3 (res. 322–342), separated by a loop (α1–α2) and a turn (α2–α3; Figures 2C and 3A, and Supplementary Figure S3). The N-terminal residues 233–266 and the two C-terminal residues were not defined by electron density and were, thus, not built.
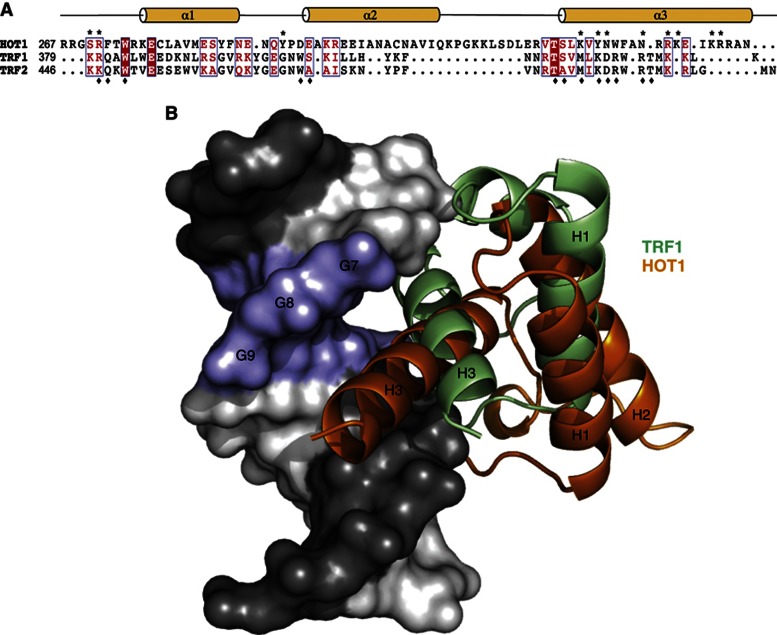
Figure 3.
Comparison of the molecular recognition of telomeric DNA by HOT1 and TRFs. (A) Schematic representation of the domain structure of the homeobox domains of TRF1, TRF2 and HOT1. Residues involved in DNA binding are marked with an asterisk (HOT1) or diamond (TRF1). Strictly conserved residues are shown with white font on red background and conserved residues are written in red font. (B) Superposition of structures of the HOT1 DBD and TRF1 DBD bound to telomeric DNA. Both binding domains recognize a different set of DNA bases, resulting in a different positioning relative to the 5′-TTAGGG-3′ motif.
As reported for TRF1 and TRF2, each copy of the HOT1 homeodomain binds to the major groove of the DNA double helix around a 5′-TTAGGG-3′ motif (Figure 2C and Supplementary Figure S3). Typical for homeobox domains, binding to DNA is mainly mediated by an N-terminal unstructured arm (267–276), the loop between α1 and α2, and the C-terminal α3 (Figure 2C). Binding and sequence recognition is achieved through a combination of either water-mediated or direct contacts with the phosphate backbone (on both sides of the major groove) and the DNA nucleobases in the major (α3) and minor groove (N-terminal arm; Figures 2C and D; the structure is deposited under PBD entry 4J19; for crystallographic statistics please see Table I).
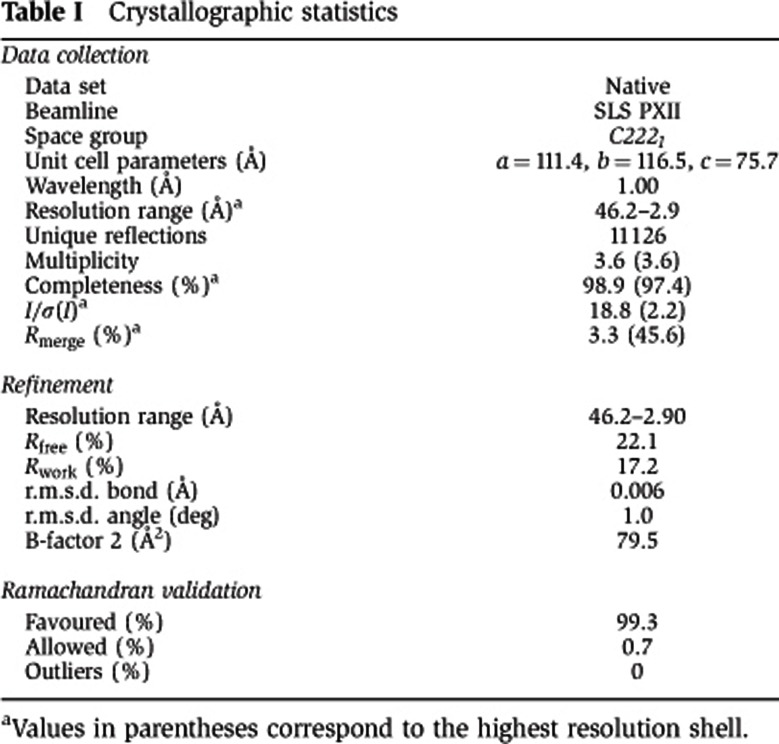
Table 1. Crystallographic statistics.
aValues in parentheses correspond to the highest resolution shell.
Lysine 335 is a key residue for telomere sequence specificity of HOT1
In order to bind a specific sequence, the HOT1 homeodomain must recognize and directly contact individual bases of telomeric DNA. We could identify five such direct interactions (R271, Y327, N332, K335, R339; Figures 2C, D and F) in our structure. Four of these residues reside on helix 3, which is generally referred to as the recognition helix and one, R271, is part of the unstructured N-terminal arm.
In detail, Y327 binds to N6 of the adenine base of A6 via a water molecule (Figure 2F). K338 and R339 are in close proximity and oriented towards the phosphate of G8 and the nucleobase of G7, respectively, but their electron density was not as clearly defined, probably due to conformational flexibility. However, in our mutant analysis the single mutants K338A and R339A no longer bind DNA, and Y327A shows weakened binding—a clear indication that these residues are important for binding (Figure 2E). In addition, N332 makes two direct hydrogen bonds with the adenine base of A11′ of the complementary strand, but mutating N332 to A had no or little effect in our binding assay.
The interaction of a lysine, K335, of this helix might be the most noteworthy. The ε-amino group of K335 makes a bifurcated hydrogen bond with carbonyl oxygens of two adjacent guanine bases (G8 and G9) in the major groove. Mutation of this lysine to alanine (K335A) not only completely abrogates binding of HOT1 to the 5′-TTAGGG-3′ motif, suggesting that this residue is essential for binding (Figures 2E and F), but this mutation also results in the ‘gained’ ability to bind a nontelomeric control sequence (5′-GTGAGT-3′) in our pull-down experiments. To our understanding, this ‘gain-of-function’ is a strong indication that K335 is a critical determinant for recognizing and binding to the telomeric sequence and, thus, for the specificity of HOT1.
Cooperatively, residues in helix 3 recognize a 5′-AGGGT-3′ (=A′) motif in the telomeric DNA. Furthermore, residues K325, N328 and R334 of this helix bind to the ribose phosphate backbone either directly (K325 to A11′, and R334 to A6) or through water (N328 to A11′ and C12′). Apart from helix 3, we observe sequence-specific interactions from residues in the α1–α2 loop (Y291 binding to the phosphate O of G7) and the N-terminal arm. In full-length HOT1, this arm is predicted to be part of an unstructured stretch of ∼65 residues between the homeodomain and the preceding putative POUs domain. Notably, in the structures of TRF1 and TRF2 this arm was discussed to be important for extending the DNA-binding interface by adding specificity and affinity (Court et al, 2005). In our structure, the hydroxyl group of S270 of this arm binds to the G14 backbone phosphate, and R271 specifically contacts both base moieties of the A12–T9′ base pair, either directly (T9′), or through a water molecule (A12; Figure 2F). The importance of R271 was also confirmed by the fact that mutating this residue to alanine (R271A) completely abolishes DNA binding (Figure 2E). As outlined in Figure 2D, the homeodomain of HOT1 docks onto telomeric DNA by specifically interacting with nucleobases and backbone groups of more than one telomeric repeat. In particular, R271 binds to the A12–T9′ base pair of the following repeat. This overlapping binding, which was also seen in the structures of TRF1 and TRF2, accounts for the repetitive nature of telomeric DNA and probably provides an extra level of selectivity.
While TRF1 and TRF2 also recognize telomeric dsDNA via their respective homeobox domains, they only share low sequence similarity with the homeodomain of HOT1 (13 and 5%, respectively, Figure 3A) and DNA binding is different in both cases. TRF1 and TRF2 both bind DNA via highly conserved interactions, involving direct interactions of α3 to TTxGG (direct contacts with phosphate (x) or bases), or xAGGGTx when water-mediated contacts are included. In HOT1, α3 of the homeodomain recognizes AxGGT directly (or AxGGTxAx with all resolved contacts; Court et al, 2005). When aligning and superimposing the DNA moieties of the TRF1 and HOT1 crystal structures, the difference in binding is easily visualized (Figure 3B). With respect to TRF1, HOT1 is shifted ‘down’ (5′→3′) the major groove by about one base and towards the subsequent telomeric repeat.
Our crystal structure reveals in atomic detail how HOT1 specifically recognizes and binds 5′-TTAGGG-3′ repeats by means of its homeodomain. We could underpin these findings with mutational analyses and DNA pull-down assays, and we conclude that K335 is a key residue for telomere recognition and binding in HOT1.
HOT1–telomere associations correlate with the degree of telomere processing
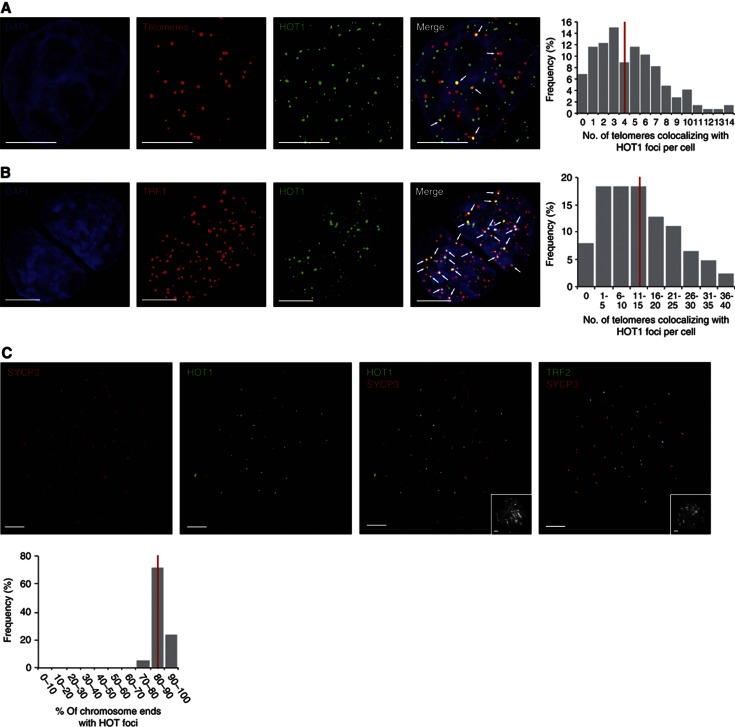
To investigate the interaction between HOT1 and telomeres in vivo, we analysed HOT1 intracellular localization by immunoFISH (immunohistochemistry combined with fluorescence in situ hybridization) microscopy in HeLa cells. In agreement with previous observations (Déjardin and Kingston, 2009), HOT1 showed a nuclear-punctuated localization pattern, but in contrast to TRF1 and TRF2 only associated with a subset of telomeres. Colocalization between HOT1 and telomeric DNA was observed in about 90% of all cells, with on average 4.5 HOT1 and telomeric foci colocalizing per cell (Figure 4A, Supplementary Figure S4a). To study the HOT1 localization in another cell type, we performed IF stainings in mouse ES cells (Figure 1D). Interestingly, in this setting we observed a higher degree of colocalization of HOT1 with telomeres, with an average of 13.5 HOT1 and TRF1 foci colocalizing (Figure 4B). In telomerase-positive cancer cells, such as HeLa, only a small fraction of telomeres is actively processed, as telomerase is only associated with a few telomeres at any given time (Jády et al, 2006; Tomlinson et al, 2006). Here most telomeres are believed to exist in a ‘closed’ state, while only some telomeres are in an ‘open’, actively processed state. In contrast, ES cells have high telomerase activity and maintain long telomeres (Varela et al, 2011). We reasoned that more colocalization events of HOT1 with telomeres might reflect the degree of active telomere extension in these cells. If this was true, then cells that very actively extend their telomeres should have the highest frequency of HOT1–telomere associations. To test this hypothesis, we carried out IF stainings for HOT1 on pachytene chromosome spreads from mouse spermatocytes, in which telomerase very actively elongates telomeres (Eisenhauer et al, 1997; Kozik et al, 1998; Yashima et al, 1998; Riou et al, 2005; Tanemura et al, 2005), using SYCP3 as a marker for the synaptonemal complex/the chromosome axis (Adelfalk et al, 2009). In agreement with a correlation between telomere processing and HOT1–telomere association, HOT1 robustly localized to telomeres on all spreads analysed with on average 87% of chromosome ends being HOT1 positive (Figure 4C). We also confirmed this frequent association of HOT1 with telomeres in mouse testes sections using the same IF set-up. Again, we could confirm the prominent localization of HOT1 to telomeres in SYCP3-positive pachytene cells, highlighting the in vivo relevance of our discovery (Supplementary Figure S5). Together, this data validates the in vivo localization of HOT1 to telomeres and suggests that HOT1 is a dynamic telomere-binding protein, putatively associated with actively processed telomeres.
Figure 4.
The degree of HOT1–telomere association varies between cell types. (A) Colocalization analysis of telomeres and HOT1 in HeLa cells by immunoFISH staining. A representative image illustrating the colocalization between several HOT1 foci (green) and telomeres (red) is shown. DAPI (blue) is used as nuclear counterstain. Colocalization events are indicated by arrows. Scale bars represent 5 μm. The quantification of the frequency of colocalization events was done after a 3D reconstruction of the acquired Z-stacks (n=147). The average value is indicated by a red bar. (B) Colocalization analysis of TRF1 and HOT1 in mouse ES cells by IF staining. To visualize TRF1 a LAP cell line (Poser et al, 2008) was used, expressing GFP-tagged TRF1 at endogenous expression levels. A representative image illustrating the colocalization between several HOT1 foci (green) and TRF1 (red) is shown. DAPI (blue) is used as a nuclear counterstain. Colocalization events are indicated by arrows. Scale bars represent 5 μm. The quantification of the frequency of colocalization events was done after a 3D reconstruction of the acquired Z-stacks (n=126). The average value is indicated by a red bar. (C) IF stainings of HOT1 at chromosome ends of mouse pachytene chromosome spreads. Representative images illustrating the localization of HOT1 and TRF2 (in green) to chromosome ends are shown. The synaptonemal complex/chromosome axis is marked by SYCP3 (red). The same field of view for the DNA counterstained by DAPI (greyscale) is shown in the bottom right corners. Scale bars represent 5 μm. The quantification of HOT1 foci at chromosome ends was done after a 3D reconstruction of the acquired Z-stacks (n=21). The average value is indicated by a red bar.
HOT1 interacts with the active telomerase complex and associates with CBs
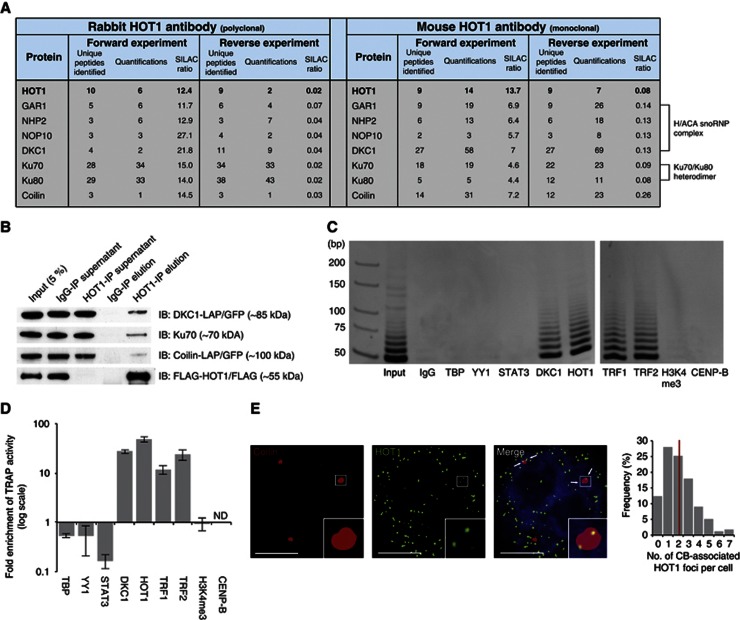
To gain further insights about HOT1 function at telomeres, we performed HOT1 immunoprecipitation assays combined with quantitative MS (Vermeulen et al, 2008), using both a polyclonal rabbit and a monoclonal mouse HOT1 antibody and SILAC-labelled nuclear protein extracts from HeLa cells. HOT1 itself was recovered with SILAC ratios indicative of specific binding to the antibody (Figure 5A and Supplementary Figure S6a). We also identified several proteins relevant to telomere homeostasis: the four core components of box H/ACA snoRNPs (DKC1, GAR1, NHP2 and NOP10; all part of the active telomerase RNP (Wang and Meier, 2004)) and the Ku70–Ku80 heterodimer proteins. We validated several of these interactions by co-IP experiments, confirming a physical interaction of endogenous HOT1 with components of the active telomerase complex (Figure 5B, Supplementary Figure S6 and Supplementary Tables S3–S6). In addition, CB proteins, notably the CB-scaffolding and -marker protein Coilin (Cioce and Lamond, 2005), were strongly enriched in our IP experiments (Figures 5A, B, Supplementary Figure S6a and Supplementary Tables S3–S6), indicating that HOT1 functions in the CB, where telomerase is assembled and potentially brought together with telomeres (Cioce and Lamond, 2005; Jády et al, 2006; Tomlinson et al, 2006), or in association with CB components in the nucleoplasm. We could further validate these interactions with analogous immunoprecipitations using extracts from mouse ES cells (Supplementary Figure S6b and Supplementary Table S7). It is noteworthy that in none of our HOT1 IPs any of the shelterin components were enriched and that in agreement with this HOT1 was not identified in a reciprocal IP of the shelterin component POT1 (Supplementary Figure S6c and Supplementary Table S8), suggesting that HOT1 does not directly interact with the shelterin complex.
Figure 5.
HOT1 associates with telomerase and CB complex components. (A) Summary of SILAC-based protein–protein interactions. Identification and normalized SILAC ratios are indicated for HOT1 (bait) and the identified interaction partner relevant for telomere biology from immunoprecipitation using both a rabbit and a mouse anti-HOT1 antibody. (B) Validation of the MS identifications by conventional immunoprecipitation. Nuclear HeLa extracts were subject to immunoprecipitation with either a polyclonal rabbit anti-HOT1 or an IgG antibody, and were immunoblotted for DKC1, Ku70 and Coilin. HOT1 IPs for the coprecipitation of DKC1 and Coilin were carried out in corresponding LAP cell lines (Poser et al, 2008) and both proteins were detected with anti-GFP antibody. FLAG–HOT1 was used to monitor the efficiency of the IP and a representative blot is shown. (C) Visualization of telomerase activity enrichment by gel electrophoresis in immunoprecipitations using antibodies against HOT1, DKC1 (positive control), TBP, YY1, STAT3, H3K4me3 and CENP-B (negative controls), as well as TRF1 and TRF2, using extracts from HeLa cells. All antibodies are rabbit polyclonal. A representative gel image of quantitative TRAP reaction products is shown. Samples were loaded on two gels and run in parallel represented by a gap between gel pictures. (D) Quantification of telomerase activity enrichment from the immunoprecipitations in panel C. Enrichments are normalized to immunoprecipitations using an IgG control. Error bars represent the s.d. of three independent experiments. Enrichments for DKC1, HOT1, TRF1 and TRF2 are statistically significant with P<0.05 (Student’s t-test). (E) Colocalization analysis of Coilin and HOT1 in HeLa cells by immunofluorescence staining. A representative image illustrating the colocalization between several HOT1 foci (green) and CBs (red; staining for Coilin) is shown. DAPI (blue) is used as nuclear counterstain. Colocalization events are indicated by arrows. An enhanced magnification of the boxed area is shown in the bottom right corners. Scale bars represent 5 μm. The quantification of the frequency of colocalization events was done after a 3D reconstruction of the acquired Z-stacks (n=179). The average value is indicated by a red bar.
Source data for this figure is available on the online supplementary information page.
The interaction of HOT1 with the active telomerase complex components box H/ACA snoRNPs subunits (Wang and Meier, 2004) raises the possibility that HOT1 binds to active telomerase. To substantiate this hypothesis, we performed immunoprecipitation experiments followed by telomerase activity measurements using the quantitative TRAP (telomere repeat amplification protocol) assay for immunoprecipitates obtained from IPs for both HOT1 and the positive control DKC1. Indeed, we detected telomerase activity in both cases (Figures 5C and 5D), while five other nuclear DNA-binding proteins (TBP, YY1, STAT3, Histone3[K4me3] and CENP-B) did not show any enrichment of telomerase activity compared to an IgG control. Antibodies against TRF1 and TRF2 immunoprecitated telomerase activity, putatively dependent on the previously established link between the shelterin component TPP1 and telomerase (Xin et al, 2007). In sum, we conclude that HOT1 associates with the active telomerase complex.
To corroborate the physical interaction between HOT1 and Coilin, we performed IF stainings for HOT1 and Coilin in nonsynchronized HeLa cells. After deconvolution and 3D reconstruction of the IF images the colocalization between both proteins was analysed. In about 85% of all cells analysed, we observed colocalization of one to seven HOT1 foci with CBs (Figure 5E). Remarkably, HOT1 foci preferentially localized to the periphery of Coilin, reminiscent of previous findings on the association of telomerase RNA and telomeres with CBs (Jády et al, 2006). This data underscores the idea that HOT1 might be associated with actively processed telomeres in a telomerase-positive context.
HOT1 is a positive regulator of telomere length
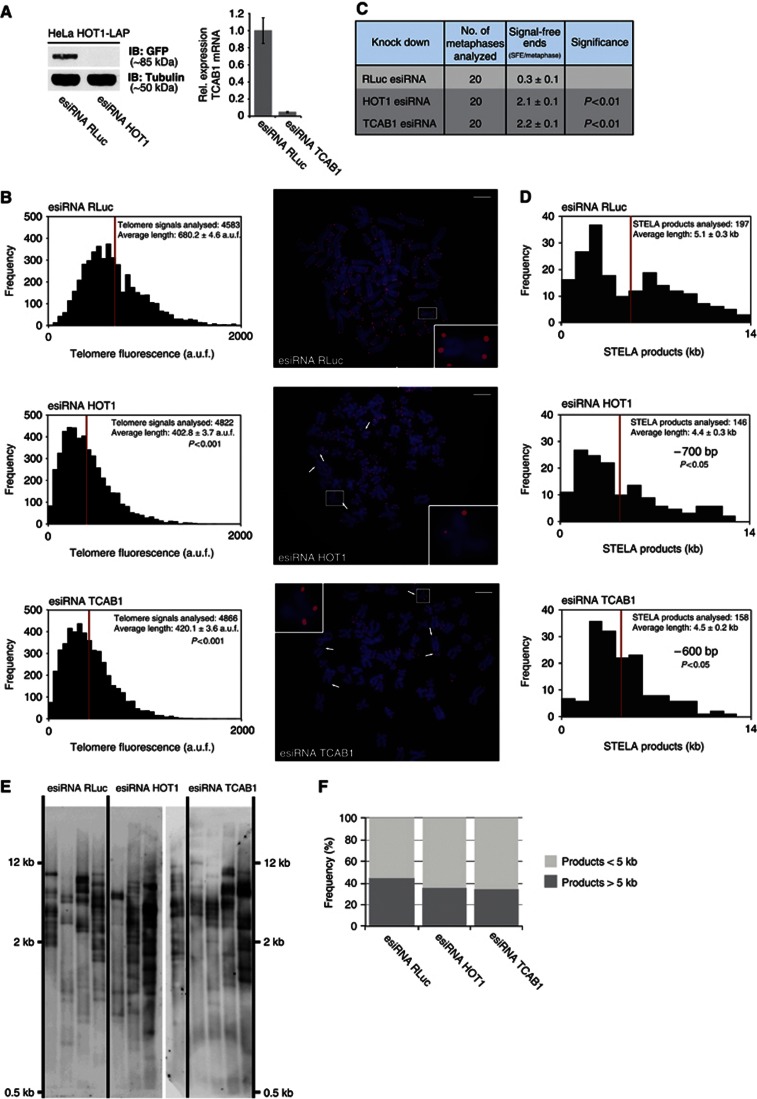
The differential degree of the HOT1–telomere colocalization, and the association with the active telomerase complex as well as the reminiscent localization to the periphery of CBs, suggested that HOT1 might be involved in telomere maintenance. To investigate this, we depleted both HOT1 and TCAB1 in HeLa cells with endoribonuclease-prepared siRNA (esiRNA; Kittler et al, 2005; Figure 6A and Supplementary Figure S7). Here TCAB1, as a recently described DKC1 interaction partner necessary for proper trafficking of TERC (Venteicher et al, 2009; Zhong et al, 2011), serves as a reference for the extent of telomere shortening that can be assigned as telomerase dependent. From HOT1- and TCAB1-depleted, as well as control-transfected cells, metaphase spreads were prepared 3 days post transfection and telomere length was determined using quantitative telomeric FISH (Londoño-Vallejo et al, 2001). Knockdown of both HOT1 and TCAB1 resulted in significant telomere shortening (Figure 6B and Supplementary Figure S7) as indicated by reduced FISH signals. Here the entire populations of FISH signals shifted towards weaker signals, indicating that telomeres globally shortened in the absence of HOT1. In these experiments the distributions after HOT1 and TCAB1 knockdown were indistinguishable from each other, indicating that their effect on telomere homeostasis is similar. Consistent with this finding we also observed a significant increase in the appearance of signal-free chromosome ends both upon HOT1 and TCAB1 knockdown (Figures 6B and C, and Supplementary Figure S7). We also obtained qualitatively similar results by measuring telomere length after HOT1 and TCAB1 knockdowns 3 days post transfection by a universal, single telomere-elongation length analysis (STELA; Figure 6D–F). Again, depletion of both HOT1 and TCAB1 led to shorter telomere length with a shortening of on average 700 and 600 bp, respectively. Given the short nature of telomeres in this cell line (on average, 5 kb measured by STELA) and the substantial contribution of variant repeats and subtelomeric DNA to this value (Aubert et al, 2012), our quantitative FISH and STELA data mutually confirm HOT1 as a positive regulator of telomere length that quantitatively behaves similar to the established telomerase pathway member TCAB1.
Figure 6.
HOT1 regulates telomere length similar to the telomerase pathway member TCAB1. (A) Verification of HOT1 knockdown efficiency by western blot 48 h post transfection using the corresponding LAP cell line (Poser et al, 2008) as a reporter for protein expression. The TCAB1 knockdown was evaluated by quantitative PCR 24 h post transfection. (B) Quantification of telomere length by quantitative telomeric FISH after transient knockdown of HOT1 and TCAB1. The distributions of fluorescence intensities, in arbitrary units of fluorescence (a.u.f.), of individual telomeres from a total of 20 metaphases per treatment are displayed; the average intensity is indicated in red. For the gene-specific knockdowns, changes of average telomere signal intensity relative to the RLuc (Renilla Luciferase) control are shown (left). Representative FISH images are shown for each treatment and signal-free ends are indicated by arrows (right). Examples of individual chromosomes are magnified and the respective chromosomes are marked by rectangles (right). Scale bars represent 5 μm. (C) Summary of the quantification of signal-free ends per metaphase after gene-specific knockdown. (D) Quantification of telomere length by universal STELA after transient knockdown of HOT1 and TCAB1. The distributions of telomere length groups in kb of individual telomeres are displayed; the average length is indicated in red and averages are stated with the respective s.e.m. Changes of average telomere length are shown relative to the RLuc (Renilla Luciferase) control. (E) Raw data of STELA reactions for the quantification of telomere length. STELA products after gel electrophoresis, transfer and hybridization from 12 individual reactions (lanes) per treatment are shown. Samples were loaded on two gels and were run in parallel, represented by a gap within HOT1 esiRNA lanes. (F) Quantification of the frequency of short STELA products (<5 kb) relative to long STELA products (>5 kb). Five kilobases were used as a cut-off value based on this being the average telomere length as determined in the RLuc control.
Source data for this figure is available on the online supplementary information page.
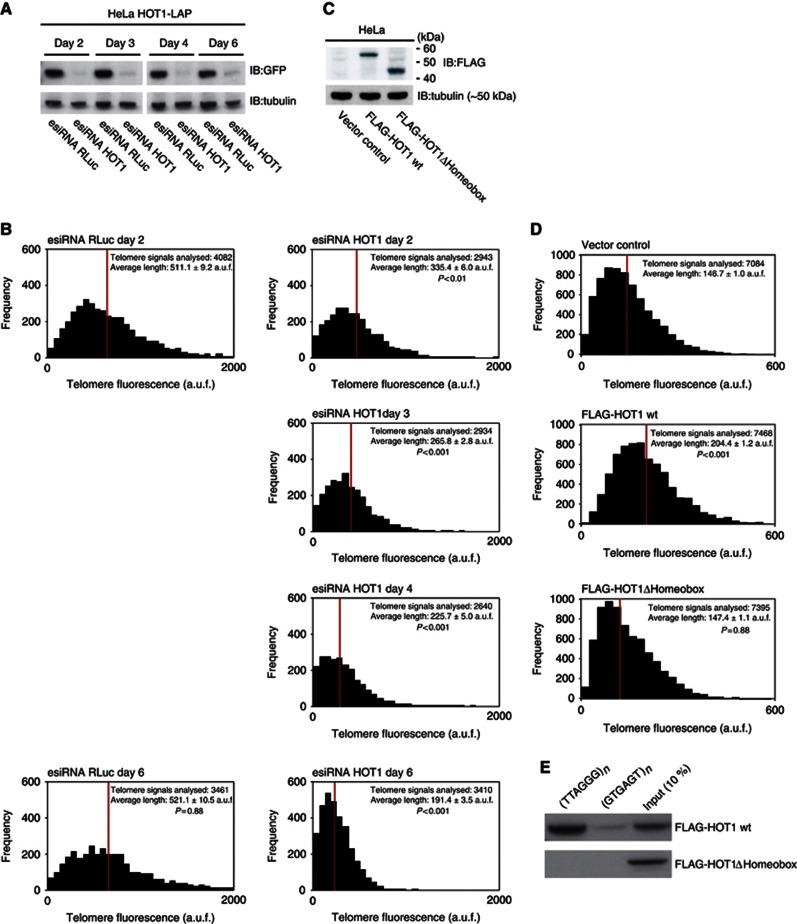
Next we performed telomere length measurements after HOT1 depletion at different time points. We reasoned that if HOT1 is influencing telomere length in a mode dependent on telomerase activity, its depletion should lead to a gradual loss of telomeric tracts. To test this, we performed telomere length measurements upon HOT1 depletion as a time course over several days. Indeed, the telomeric signal was gradually reduced in the absence of HOT1 (Figures 7A and B). Hence, the telomere length changes observed due to manipulations of HOT1 levels are in agreement with telomerase-dependent changes.
Figure 7.
HOT1 acts as a positive regulator of telomere length. (A) Verification of HOT1 esiRNA knockdown efficiency using a HOT1-LAP cell line (Poser et al, 2008) as a reporter for HOT1 expression by western blot after 2, 3, 4 and 6 days. Cells analysed after 4 and 6 days were transfected twice: After the initial transfection, cells were transfected a second time on day 3 (72 h post transfection). All samples were run on the same gel, irrelevant lanes were spliced out. (B) Quantification of telomere length by quantitative telomeric FISH after transient knockdown of HOT1. The distributions of fluorescence intensities, in arbitrary units of fluorescence (a.u.f.), of individual telomeres from a total of 15–20 metaphases per treatment are displayed; the average intensity is indicated in red. Changes of average telomere signal intensity are shown relative to the RLuc (Renilla Luciferase) control. (C) Verification of FLAG–HOT1 and FLAG–HOT1ΔHomeobox expression by western blot 48 h post transfection. (D) Results of telomere length measurements after transient overexpression of FLAG–HOT1 and FLAG–HOT1ΔHomeobox. The distributions of fluorescence intensities, in a.u.f., of individual telomeres from a total of 30 metaphases per treatment are displayed; the average intensity is indicated in red. (E) Sequence-specific pull-down of FLAG–HOT1 and FLAG–HOT1ΔHomeobox. Proteins were incubated with either telomeric repeats (5′-TTAGGG-3′) or a control oligonucleotide (5′-GTGAGT-3′).
Source data for this figure is available on the online supplementary information page.
In a complementary experiment, we transiently overexpressed FLAG–HOT1 in HeLa cells and analysed telomere length 3 days after transfection. Consistent with the observed telomere shortening upon HOT1 depletion, telomeres were elongated upon its overexpression as indicated by an increase of the entire population of FISH signals (Figures 7C and D). The qFISH data suggests that the telomere length regulation exerted by HOT1 is dependent on its telomere-binding capacity. To test whether HOT1-dependent telomere elongation indeed requires DNA binding, we used a HOT1ΔHomeobox variant. In vitro-binding studies with recombinant HOT1ΔHomeobox demonstrated that the deletion of this domain indeed abolished binding to telomeric 5′-TTAGGG-3′ repeats (Figure 7E) and the lack of binding also coincides with loss of telomeric localization in vivo (Supplementary Figure S4b). To examine functional consequences of DNA-binding-deficient HOT1 on telomere length regulation, we overexpressed FLAG–HOT1ΔHomeobox in HeLa cells. In contrast to telomere elongation upon expression of full-length FLAG–HOT1, the expression of FLAG–HOT1ΔHomeobox did not lead to an effect on telomere length in comparison to the control (Figures 7C and D). Thus, the HOT1–telomere interaction is essential for telomere elongation. Collectively, these findings demonstrate that HOT1 acts as a positive regulator of telomere length. The fact that overexpression of HOT1 can stimulate telomere lengthening further suggests that similar to TERT and TERC (Cristofari and Lingner, 2006), HOT1 is a limiting factor, at least in this cellular context.
TERT binding to chromatin is dependent on HOT1
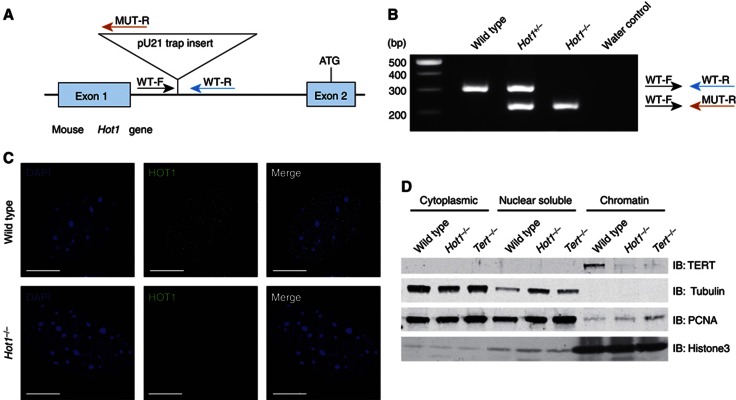
The combined features of HOT1—direct binding to telomeric dsDNA, preferential localization to telomeres in settings of active processing, association with active telomerase, localization to the periphery of CBs and a positive effect on telomere length—suggest a putative role of HOT1 in telomerase recruitment. To test this possibility we assayed TERT binding to chromatin in the absence of HOT1, based on a previous experimental set-up that contributed to the identification of TPP1 as a telomerase recruiter (Tejera et al, 2010). To avoid potential convolutions of experimental results by the presence of residual HOT1 protein, we established a HOT1 genetrap mouse. In Hot1Gt(pU-21T)346Card/(Gt(pU-21T)346Card mice, the genetrap construct is inserted in the first intron of the Hot1 gene. As the start codon resides in exon 2, we expected that protein synthesis would be entirely abrogated (Figure 8A and B). To verify this, we established mouse embryonic fibroblasts (MEFs) of the different genotypes from littermate embryos and performed immunofluorescence stainings with our HOT1 antibody. While wild-type cells showed the typical nuclear-punctuated pattern, HOT1 signals were absent in Hot1Gt(pU-21T)346Card/(Gt(pU-21T)346Card MEFs (Figure 8C). Based on this data and for the sake of simplicity, we will refer to this as Hot1−/− genotype. We then performed the chromatin fractionation assay and could, as expected, detect TERT on the chromatin fraction of wild-type MEFs, but not on chromatin fractions from Tert−/− MEFs. Notably, the TERT signal was also absent when extracts of Hot1−/− MEFs were used (Figure 8D), reminiscent of previous data on TPP1 (Tejera et al, 2010). This data indicate that HOT1 indeed contributes to telomerase recruitment in vivo. Thus, HOT1 as a dynamic and differential telomere-binding protein contributes to telomerase–telomere association, mechanistically underlying its function as a positive regulator of telomere length.
Figure 8.
TERT binding to chromatin is dependent on HOT1. (A) Schematic of the genetrap insertion in the Hot1 gene. (B) PCR genotyping. Genomic DNA from wild-type, heterozygous and homozygous mice were analysed by PCR. Genotypes were confirmed using three primers WT-F, WT-R and MUT-R. WT-F and WT-R amplify the wild-type allele (upper band), and WT-F and MUT-R amplify the genetrap allele (lower band). (C) Immunofluorescence stainings of HOT1 on wild-type and Hot1Gt(pU-21T)346Card/(Gt(pU-21T)346Card MEFs (referred to as Hot1−/− MEFs). HOT1 foci (green) are absent in Hot1−/− MEFs. DAPI (blue) is used as a nuclear counterstain. Scale bars represent 5 μm. (D) Subcellular fractionation analysis of wild-type, Hot1−/− and Tert−/− MEFs. Tubulin, PCNA and Histone3 serve as loading controls and to monitor the cell fractionation.
Source data for this figure is available on the online supplementary information page.
Discussion
We have adapted our SILAC-based DNA–protein interaction screening technique to repetitive sequences as a straightforward and robust approach to identify novel interactions. In addition to the established telomere-binding proteins and their tight interaction partner, we have identified HOT1 as the first direct telomere-binder that positively regulates telomere length. HOT1 has previously been found as a telomere-associated factor that localizes to some telomeres in HeLa cells using the PICh approach (Déjardin and Kingston, 2009). Interestingly, this is the only one of several recent large scale studies searching for telomere-associated factors, which has identified HOT1 (Déjardin and Kingston, 2009; Giannone et al, 2010; Nittis et al, 2010; Lee et al, 2011), possibly because also in this approach telomeric DNA was used as bait, rather than using the shelterin components. While by the PICh approach several hundreds of putative telomere-associated factors were identified and the PICh technique is, in general, addressing the global composition of (telomeric) chromatin, our approach is more apt to rapidly identify the direct DNA binders and their tight interaction partners as illustrated by the identification of HOT1. Thus, our in vitro reconstitution assay seems particularly suited to discover novel direct telomere binders.
In contrast to the other direct telomere-binding proteins, TRF1 and TRF2, HOT1 does not localize to all telomeres and the degree of telomere association varies between cell types. While in situ telomerase activity is limited in HeLa cells to a simple maintenance of short telomeres, it is increased in mouse ES cells and is very high in mouse spermatogonia and spermatocytes, where telomere length is reset to the very long telomeres observed in mature sperm (Eisenhauer et al, 1997; Kozik et al, 1998; Yashima et al, 1998; Riou et al, 2005; Tanemura et al, 2005). The fact that the nature of the HOT1 association with telomeres seems to be more transient in normally cycling telomerase-positive cancer cells compared to an intermediate degree of association in mouse ES cells, and to a prominent and robust association with telomeres in mouse spermatocytes, is consistent with promoting telomerase-dependent telomere lengthening. This raises the possibility that HOT1 selectively interacts with actively processed telomeres. How exactly this interaction is regulated and which precise step in telomere lengthening HOT1 promotes will have to be addressed in the future.
While HOT1 localizes to a subset of telomeres together with shelterin components, our HOT1 immunoprecipitation experiments failed to establish any putative association between HOT1 and shelterin complex members. Likewise, we could not retrieve HOT1 after a POT1–IP, which is in agreement with several other studies that searched for shelterin-associated factors either by immunoprecipitation or by bimolecular fluorescence complementation (Giannone et al, 2010; Nittis et al, 2010; Lee et al, 2011). Our comparison of the cocrystal structures of telomeric DNA with the HOT1, TRF1 and TRF2 homeodomains indicates that while HOT1 is shifted ‘down’ in 5′→3′ direction by one base towards the following telomeric repeat, the binding sites are largely overlapping. Hence, it is intriguing how these proteins coexist at telomeres, how they compete for binding sites and whether the shelterin proteins are found interspersed on telomeres or whether there are discrete, mutually exclusive patches along the telomeric tracts. This might further contribute to answering the question how HOT1 is selectively restricted to a subset of telomeres.
How does HOT1 fit into the current view of telomere homeostasis? TCAB1 (WDR79/WRAP53) has been shown to be required for proper localization of CAB box containing scaRNPs to CBs, including TERC (Tycowski et al, 2009; Venteicher et al, 2009). Similar to HOT1, TCAB1 acts as a positive regulator of telomere length by recruiting TERC to CBs (Venteicher et al, 2009). The telomere-binding properties of HOT1 and its functional consequences, which are quantitatively similar to those of TCAB1, suggest that HOT1 may act downstream of TCAB1. The putative mechanism may involve recruiting telomerase-containing CBs to telomeres, and thus promoting telomerase association with telomeres, perhaps through additional interactions involving TPP1 (Abreu et al, 2010; Tejera et al, 2010; Zaug et al, 2010). The fact that overexpression of HOT1 further stimulates telomere lengthening suggests that HOT1, similar to TERT and TERC, is also a limiting factor. Therefore, the shelterin and CST complexes, and the controlled levels of TERT, TERC and HOT1, may all act cooperatively to define a mean telomere length in the cell.
Our chromatin fractionation analysis indicates that HOT1 is required for TERT recruitment to chromatin and, therefore, qualifies as a telomerase recruitment factor. Similarly, TPP1 has been established as a telomerase recruiter likely by (direct or indirect) interaction with TERT via its TEL patch (Nandakumar et al, 2012), but based on our immunoprecipitation data and previous studies there is no indication for an interaction between HOT1 and TPP1. This discrepancy may be explained by a step-wise telomerase recruitment model, in which HOT1 simply acts before TPP1. Previous reports have shown that POT1–TPP1 act as a processivity factor for telomerase (Wang et al, 2007; Latrick and Cech, 2010; Zaug et al, 2010). Enhanced in vivo processivity would also result in an increased residence time and a higher affinity to telomeres, thus explaining the reduction of telomerase–telomere association in TPP1 knockdown/knockout conditions. Given that HOT1 preferentially localizes to telomeres in settings of high in situ telomerase activity, one possibility is that HOT1 contributes to an ‘open’ telomere state that allows/promotes telomere elongation. In such a model, HOT1 might contribute to rendering telomeres accessible and/or contributes to the delivery of telomerase to ‘open’ telomeres. In this scenario, TPP1 would contribute more prominently to maintaining telomerase at telomeres and, thus, to processivity stimulation. Undoubtedly, the mechanistic contribution of both factors and potential genetic interactions are exciting avenues for future research.
Materials and methods
Cell culture
HeLa cells (Epitheloid carcinoma, cervix) and MEFs were cultivated in 4.5 g/l glucose Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum, 100 U/ml penicillin and 100 μg/ml streptomycin (Gibco) at 37°C and 5% CO2. For SILAC labelling, HeLa cells were incubated in RPMI 1640 (–Arg, –Lys) medium containing 10% dialysed fetal bovine serum (Gibco) supplemented with 84 mg/l 13C615N4 L-arginine and 50 mg/l 13C615N2 L-lysine (Sigma Isotec or Euriso-top) or the corresponding nonlabelled amino acids, respectively. R1/E murine ES cells were also grown in 4.5 g/l GlutaMAX DMEM containing sodium pyruvate, but supplemented with 20% FBS, 50 μM 2-mercaptoethanol (Invitrogen), LIF (MPI-CBG protein expression facility), as well as 100 U/ml penicillin and 100 μg/ml streptomycin (Gibco). For SILAC labelling of R1/E cells, DMEM (–Arg, –Lys) medium containing 20% dialysed FBS (Gibco) supplemented with 40 mg/l 13C615N4 L-arginine and 80 mg/l 13C615N2 L-lysine (Sigma Isotec or Euris-top) or the corresponding nonlabelled amino acids, respectively, was used. Cells were collected and nuclear extracts were prepared as described (Butter et al, 2010).
Telomere pull-down
Chemically synthesized oligonucleotides (Table II; Metabion) were annealed and polymerized by T4 ligase (Fermentas) and biotinylated with biotin–dATP (Invitrogen) by Klenow fragment (Fermentas) following the manufacturer’s instructions. Twenty-five microgram baits were immobilized on 750 μg paramagnetic streptavidin beads (Dynabeads MyOne C1, Invitrogen) and subsequently incubated with either 400 μg of SILAC-labelled nuclear extract or 20 μl of supernatants of crude E. coli lysates (3 μg/ml) in PBB buffer (150 mM NaCl, 50 mM Tris–HCl pH 8.0, 10 mM MgCl2, 0.5% NP-40, Complete Protease Inhibitor without EDTA (Roche)) for 2 h at 4°C on a rotation wheel. After three washes with PBB buffer bead fractions were pooled, bound proteins boiled at 80°C in 1 × LDS buffer (Invitrogen) and separated on a 4–12% gradient gel (Novex, Invitrogen; Butter et al, 2010).
Table 2. Oligonucleotides used for pull-down experiments.
| No | Sequence motif (5′→3′) | Primer sequence (5′→3′) |
|---|---|---|
| 1a | TTAGGG for | TTAGGGTTAGGGTTAGGGTTAGGGTTAGGGTTAGGGTTAGGGTTAGGGTTAGGGTTAGGG |
| 1b | TTAGGG rev | AACCCTAACCCTAACCCTAACCCTAACCCTAACCCTAACCCTAACCCTAACCCTAACCCT |
| 2a | GTGAGT for | TTGACAGTGAGTGTGAGTGTGAGTGTGAGTGTGAGTGTGAGTGTGAGTGTGAGTGTGAGT |
| 2b | GTGAGT rev | AAACTCACACTCACACTCACACTCACACTCACACTCACACTCACACTCACACTCACTGTC |
| 3a | TTAGGC for | TTAGGCTTAGGCTTAGGCTTAGGCTTAGGCTTAGGCTTAGGCTTAGGCTTAGGCTTAGGC |
| 3b | TTAGGC rev | AAGCCTAAGCCTAAGCCTAAGCCTAAGCCTAAGCCTAAGCCTAAGCCTAAGCCTAAGCCT |
| 4a | TTGGGG for | TTGGGGTTGGGGTTGGGGTTGGGGTTGGGGTTGGGGTTGGGGTTGGGGTTGGGGTTGGGG |
| 4b | TTGGGG rev | AACCCCAACCCCAACCCCAACCCCAACCCCAACCCCAACCCCAACCCCAACCCCAACCCC |
| 5a | TCAGGG for | GTCAGGGTCAGGGTCAGGGTCAGGGTCAGGGTCAGGGTCAGGGTCAGGGTCAGGGTCAGG |
| 5b | TCAGGG rev | ACCCTGACCCTGACCCTGACCCTGACCCTGACCCTGACCCTGACCCTGACCCTGACCCTG |
| 6a | TGAGGG for | GTGAGGGTGAGGGTGAGGGTGAGGGTGAGGGTGAGGGTGAGGGTGAGGGTGAGGGTGAGG |
| 6b | TGAGGG rev | ACCCTCACCCTCACCCTCACCCTCACCCTCACCCTCACCCTCACCCTCACCCTCACCCTC |
MS data acquisition
In-gel digestion and MS analysis was performed essentially as previously described by Butter et al, 2010. Peptides were desalted on StageTips and analysed by nanoflow liquid chromatography on an EASY-nLC system from Proxeon Biosystems coupled to a LTQ-Orbitrap XL or a Orbitrap-Velos (Thermo Electron). Peptides were separated on a C18-reversed phase column (15 cm long, 75 μm inner diameter, packed in-house with ReproSil-Pur C18-AQ 3 μm resin (provided by Dr Maisch)) directly mounted on the electrospray ion source. We used a 107-min gradient from 2 to 60% acetonitrile in 0.5% acetic acid at a flow of 200 nl/min. The LTQ-Orbitrap XL was operated with a Top5 MS/MS spectra acquisition method in the linear ion trap per MS full scan in the Orbitrap, while for the Orbitrap-Velos a Top10 acquisition method with HCD fragmentation was used.
MS spectrum and data analysis
The raw files were processed with MaxQuant (Cox and Mann, 2008) and searched with the Mascot search engine (Matrix Science) against IPI human v3.37 protein database concatenated with a decoy of the reversed sequences. Carbamidomethylation was set as fixed modification, while methionine oxidation and protein N-acetylation were considered as variable modifications. The search was performed with an initial mass tolerance of 7 p.p.m. mass accuracy for the precursor ion and 0.5 Da for the MS/MS spectra obtained with CID fragmentation and 20 p.p.m. for the MS/MS spectra in the HCD fragmentation mode. Search results were processed with MaxQuant filtered with a false-discovery rate of 0.01. Before statistical analysis, known contaminants and reverse hits were removed. The protein ratios of a ‘forward’ experiment and the ‘reverse’ experiment were plotted in R (prerelease version 2.8.0). Only proteins identified with at least two unique peptides and two quantitation events were plotted for the telomere pull-down (quality filter).
Recombinant protein expression and deletion variant construction
The HOT1 clone was obtained from the ORFeome collection (IOH40784, Invitrogen). The sequence was subcloned into SLIC-compatible pETM44 or pETM14 vector via SLIC cloning (Li and Elledge, 2007; Scholz et al, 2013) and was expressed in E. coli Rosetta at 18°C. E. coli extracts with overexpressed recombinant proteins were used for binding studies on variant DNA motives. For crystallization, protein was purified under nondenaturing conditions using Ni2+ sepharose high-performance columns (GE Healthcare) followed by size-exclusion chromatography on Superdex 75 (GE Healthcare), using 150 mM NaCl and 20 mM Tris–HCl (pH 7.5) as a running buffer.
The HOT1 ORFeome clone was LR recombined into a gateway compatible pcDNA3.1 vector with N-terminal FLAG-tag (gift of Christian Brandts). A FLAG-tagged HOT1 homeobox deletion variant was constructed by PCR amplification of the pcDNA3.1-FLAG vector using primers with site-specific overhangs (Table III; Metabion) following the QuickChange II Site-directed mutagenesis kit protocol (Stratagene; Zheng et al, 2004). Point mutants were generated similarly using the QuickChange protocol. For expression in ES cells, constructs were subcloned into a pCAGGS vector. The constructs were sequence verified using an Applied Biosystems 3730 Genetic Analyzer (Applied Biosystems), according to the manufacturer’s instructions.
Table 3. Oligonucleotides used for QuickChange.
| No | Cloning/BAC engineering purpose | Primer sequence (5′→3′) |
|---|---|---|
| 1 | HOT1 homeobox domain deletion, forward primer | CTGCGACGAGGGAGTGCCAATATTGAAGCAGCAATCCTGG |
| 2 | HOT1 homeobox domain deletion, reverse primer | TGCTTCAATATTGGCACTCCCTCGTCGCAGTCGGAAAGTA |
Crystallization and structure determination
The complex of human HOT1 DBD and double-stranded telomeric DNA (5′-TTAGGGTTAGGGTTA-3′) was obtained as follows: synthetic single-strand complementary oligonucleotides (Table IV; Eurofins MWG) were dissolved in water and mixed to yield a 5-mM stock solution. For double-strand annealing, this solution was heated to 96°C for 10 min and allowed to cool down to room temperature (RT) in the switched-off heat block within 2–3 h. This solution was mixed in a 1–1.2 M excess with a solution of purified HOT1 DBD at 10 mg/ml and incubated on ice for 10 min. Crystals of the complex were grown at 4°C by sitting-drop vapour diffusion from drops formed by mixing equal volumes of the complex solution and the crystallization condition 24% PEG 3350, 100 mM Na Acetate (pH 4.5) and 100 mM KSO4. For data collection, crystals were cryoprotected by soaking in a mother liquor supplemented with 20% glycerol and was flash-frozen in liquid nitrogen. Diffraction data were collected at the PXII beam line of the Swiss Light Source (SLS; Villigen, Switzerland) and processed using XDS (Kabsch, 1993). The structure was solved by molecular replacement with the programme PHASER (McCoy et al, 2007), using the NMR structure of the HOT1 DBD as a search model (PDB entry 2CUF). Model building and refinement was carried out with Phenix (Afonine et al, 2005) and COOT (Emsley and Cowtan, 2004), and models were validated with the programme Molprobity (Davis et al, 2007).
Table 4. Oligonucleotides used for crystallization.
| No | Telomeric DNA oligos for crystallization | Primer sequence (5′→3′) |
|---|---|---|
| 1 | T1; forward oligo | CTGTTAGGGTTAGGGTTAG |
| 2 | T2; reverse oligo | TCTAACCCTAACCCTAACA |
The alignment of the HOT1, TRF1 and TRF2 homeobox domain sequences was done in two steps. First, a secondary structure-based alignment was generated using the programme Chimera (www.cgl.ucsf.edu/chimera) followed by feeding it into the web application Espript (espript.ibcp.fr) for a simple sequence alignment scoring and colouring.
Antibody production
His-MBP-tagged HOT1 (pETM44 vector construct) was expressed in E. coli Rosetta at 18°C in a fermentation tank. Cell pellet was lysed with Avestin and the soluble fraction subjected to affinity purification using Ni-sepharose. The elution fraction was concentrated with an Amicon Ultra 15 concentrator column and dialysed into buffer containing 50 mM K2PO3, 20 mM NaCl, 10% glycerol, 1 mM TCEP and protein inhibitors. Purified MBP-HOT1 was injected into rabbits for immunization and the rabbits were ultimately sacrificed. The antibody was affinity purified using His-MBP-HOT1 and MBP immobilized on HiTRAP desalting columns (GE Healthcare). First, the serum was applied to the His-MBP-HOT1 column, eluted and then applied to the MBP column. The flow trough was quantified and used for subsequent experiments.
Purified MBP-HOT1 was injected into mice for immunization and the mice were ultimately sacrificed. Immortalized hybrid cells (hybridomas) were obtained by the fusion of B cells from the spleen of an immunized mouse with a myeloma cell line, which itself does not produce antibodies, using PEG and AH selection (Sigma Aldrich). Hybridoma clones were generated and screened using the Meso Scale Discovery platform (Meso Scale Diagnostics) by comparing affinity to His-MBP-HOT1 and His-MBP-Katanin as an unspecific negative control. Positive clones were subcloned by limiting dilution and retested using the MSD platform. Based on the subcloned hybridoma cell lines, antibodies were purified using HiTRAP protein G columns (GE Healthcare) followed by acid elution.
Chromatin immunoprecipitation
Cells were crosslinked with 1% formaldehyde (Thermo Scientific) for 10 min at 37°C, and the reaction was stopped by adding glycine to a final concentration of 0.125 M for 5 min at RT. Nuclei were prepared from fixed and washed cells by homogenization in cell lysis buffer (5 mM PIPES pH 8, 85 mM KCl, 0.5% NP-40) and centrifugation at 1800 g for 10 min. Finally, nuclei were lysed in 900 μl nuclei lysis buffer (50 mM Tris–HCl pH 8, 10 mM EDTA pH 8, 1% SDS) and lysates were sonicated for 30 min (30 s on/30 s off) in a Diagenode water-bath sonicator at speed 5. Following a centrifugation at 14 000 r.p.m. for 10 min, the cleared supernatants were snap-frozen in liquid nitrogen and stored at −80°C. Sonication efficiency was routinely monitored by DNA gel electrophoresis to ensure that the bulk of DNA fragments was between 100 and 500 bp.
Sonicated chromatin containing 50 μg DNA was diluted 10 times in ChIP dilution buffer (16.7 mM Tris–HCl pH 8, 167 mM NaCl, 1.2 mM EDTA pH 8, 1.1% Triton X-100, 0.01% SDS) and precleared for 2 h, rotating at 4°C, with 20 μl blocked beads (Dynabeads Protein A beads (Invitrogen) incubated for 2 h with 5 mg/ml BSA) before the overnight incubation with 5 μg of antibody. The following antibodies were used: mouse anti-HOT1 (MPI-CBG Antibody Facility), rabbit anti-TRF2 (NB110-57130, Novus), rabbit IgG (ab37415, Abcam) or mouse anti-GFP (ab1218, Abcam)). The bound material was recovered after a 2 h incubation, rotating at 4°C, with 30 μl blocked beads. The beads were washed for 10 min in each of the following wash buffers: low-salt buffer (20 mM Tris–HCl pH 8, 150 mM NaCl, 2 mM EDTA pH 8, 1% Triton X-100, 0.1% SDS), high-salt buffer (20 mM Tris–HCl pH 8, 500 mM NaCl, 2 mM EDTA pH 8, 1% Triton X-100, 0.1% SDS), LiCl buffer (10 mM Tris–HCl pH 8, 0.25 M LiCl, 1 mM EDTA pH 8, 1% NP-40, 1% Na deoxycholate) and twice, 5 min each, in TE. ChIPed material was eluted by two 15 min incubations at RT with 225 μl elution buffer (0.1 M NaHCO3, 1% SDS). Chromatin was reverse-crosslinked by adding 18 μl of 5 M NaCl and incubated overnight at 65°C, and DNA was submitted to RNase and proteinase K digestion and extracted by phenol–chloroform.
Purified DNA recovered by ChIP was denatured in 0.2 M NaOH by heating to 100°C for 10 min and spotted onto a positively charged Biodyne B nylon membrane (Pall, VWR). Membranes were hybridized at 42°C in 6 × SSC, 0.01% SDS, 0.1% milk with 20 pmol of DIG-labelled telomeric C-rich LNA probe (36 bp, Exiqon). Following hybridization washes (twice 5 min in 2 × SSC, 0.01% SDS, and once 2 min in 0.1 × SSC, 0.01% SDS), the signal was revealed using the anti-DIG-AP antibodies (Roche) and CDP-Star (Roche) following the manufacturer’s instructions. Images were obtained using the Luminescent image analyser LAS-4000 mini (GE Healthcare). Following the detection of a telomeric signal, membranes were stripped twice for 15 min each in 0.5% SDS at 60°C and twice for 15 min each in 0.2 N NaOH, 0.1% SDS at 37°C. Stripped membranes were then hybridized at 65°C in 6 × SSC, 0.01% SDS, 0.1% milk with P32-random-primed labelled total genomic DNA from HeLa cells. Following hybridization washes (10 min in 2 × SSC, 0.01% SDS, 10 min in 0.5 × SSC, 0.01% SDS and 10 min in 0.1 × SSC, 0.01% SDS) the signal was revealed using the phosphorimager and an image obtained by the Typhoon scanner.
Immunofluorescence and immunoFISH stainings on cycling cells
For immunofluorescence stainings, cells were seeded 24 h before the treatment on either glass coverslips (0.17 mm, assorted glass, Thermo Scientific) or in LabTek II chambered coverglass chambers (Labtek). After a brief wash with 1x PBS cells were fixed in 10% formalin solution (Sigma Aldrich) for 10 min at RT, followed by two washes with 1 × PBS+30 mM glycine. Cells were then permeabilized with 1 × PBS+0.5% Triton X-100 for 5 min at 4°C, followed by again two washes with 1 × PBS+30 mM glycine. Afterwards, cells were blocked in blocking solution (1 × PBS, 0.2% fish-skin gelatine (Sigma Aldrich)) for 15 min at RT. Primary antibodies were diluted in blocking solution and incubated for 1 h at RT. The following primary antibodies were used: mouse anti-HOT1 (MPI-CBG Antibody Facility, 1:1000), rabbit anti-Coilin (sc-32860, Santa Cruz, 1:500), and goat anti-GFP (MPI-CBG Antibody Facility, 1:2000). Cells were washed three times for 3 min each in blocking solution followed by a 30-min incubation at RT with secondary antibodies, which were diluted in blocking solution. As secondary antibodies, fluorescent-labelled donkey anti-rabbit-IgG or donkey anti-mouse-IgG antibodies with either Alexa488, Alexa555, Alexa594 or Alexa647 as fluorochromes (1:500, Invitrogen) were used. After three final washes for 3 min each in blocking solution, slides were briefly rinsed in distilled water and mounted using DAPI Prolong Gold Antifade Reagent (Invitrogen). In the case of Labtek chambers, samples were incubated for 5 min with blocking solution containing 1 μg/ml DAPI, followed by one wash in blocking solution.
If FISH stainings were combined with immunofluorescence stainings (immunoFISH), the FISH labelling was carried out directly after the IF protocol. Cells were post-fixed in 10% formalin solution (Sigma Aldrich) for 10 min at RT, followed by three washes in 70, 90 and 100% ethanol for 5 min each, followed by a standard telomeric FISH protocol as previously described.
All images were acquired with a DeltaVision Core Microscope (Applied Precision, Olympus IX71 microscope) using a 100 × /1.4 UPlanSApo oil-immersion objective. Z-stacks (0.2 μm optical sections) were collected and deconvolved using softWoRx (Applied Precision). Z-stacks were reconstructed in 3D using Imaris (Bitplane), and colocalization events were determined for signals above the background using the colocalization function.
esiRNA synthesis
The detailed protocol of esiRNA production has been previously published (Kittler et al, 2005). Briefly, optimal regions for designing esiRNAs were chosen using the Deqor design algorithm (Henschel et al, 2004) in order to fulfill two criteria: to obtain the most efficient silencing trigger in terms of silencing efficiency, and to get lowest chances to cross-silence other genes. The most favourable fragments were used to design gene-specific primers (Table V) using the Primer3 algorithm (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). Two esiRNAs for HOT1 and one each for TRF1 and TCAB1 were designed and synthesized. PCR products for the esiRNA production were sequenced using an Applied Biosystems 3730 Genetic Analyzer (Applied Biosystems) according to the manufacturer’s instructions. All positions of sequence trace files were confirmed by manual inspection.
Table 5. Oligonucleotides used for esiRNA production.
Please note that esiRNA–template production consists of two consecutive PCRs. In the table only the transcript-specific primers for the first PCR round are listed. Universal tags have to be added to all primers that allow amplification with universal T7 primers in the second PCR round. The universal tags are 5′-TGTAAAACGACGGCCAGT-3′ (forward) and 5′-AGGAAACAGCTATGACCAT-3′ (reverse). The universal T7 primers are 5′-GCTAATACGACTCACTATAGGGAGAG-3′ (forward) and 5′-GCTAATACGACTCACTATAGGGAGAC-3′ (reverse).
esiRNA and plasmid transfection
For all transfections, HeLa cells were seeded in six-well plates (40 000 cells per well corresponding to 9.6 cm2) dish and incubated overnight before transfection. For esiRNA transfection, 30 μl Oligofectamine (Invitrogen) were diluted in 250 μl OptiMEM (Invitrogen) and incubated for 5 min at RT. In a separate tube, 2 μg esiRNA were diluted in 250 μl OptiMEM. Solutions were combined, mixed and incubated for 20 min at RT after which the transfection mix was evenly distributed over the dish.
The FLAG–HOT1 and the homeobox deletion variant construct were transfected using either Effectene (Qiagen) or Lipofectamine 2000 (Invitrogen) as the transfection reagent according to the manufacturers’ instructions.
Telomeric quantitative FISH
For metaphase preparation, cells were incubated for 4 h with 200 nM nocodazole in order to induce mitotic arrest. A hypotonic shock was achieved in 0.03 M sodium citrate at 37°C for 40 min. Cells were fixed in an ethanol/acetic acid solution (3:1) and washed three times in this fixing reagent. Metaphase spreads were obtained by dropping suspensions of fixed cells onto clean glass slides.
The QFISH procedure was carried out as described, using an Alexa488-O-O-(CCCTAA)3 or Cy3-O-O-(CCCTAA)3 PNA probe (Panagene) (Londoño-Vallejo et al, 2001).
Telomeric signals were quantified using the iVision software (Chromaphor). Telomere signals were segmented manually and average pixel intensities from every segment were quantified. For each metaphase, the average background intensity was determined and subtracted from individual telomere signals. Statistical analyses were done using Student’s t-test.
The number of signal-free ends per metaphase was determined by manual inspection of the same metaphase images that were used for telomere signal-intensity quantification. Statistical analyses were done using Student’s t-test.
Universal STELA
One hundred picogram of NdeI/MseI (NEB)-digested DNA were amplified according to the previously published Universal STELA PCR amplification protocol (Bendix et al, 2010) with the following cycling conditions: 68°C for 5 min, 95°C for 2 min, 26 cycles of 95°C for 15 s, 58°C for 30 s and 72°C for 12 min, and 72°C for 15 min. In brief, 10 ng of DNA were digested with 0.4 units of NsdeI and 0.4 units of MseI, annealed at 16°C overnight to 4.2 μM of both 42-mer (5′-TGTAGCGTGA-AGACGACAGAAAGGGCGTGGTGCGGACGCGGG-3′)and 11+2-mer (5′-TACCCGCGTCCGC-3′) panhandle-oligos with 1.3 units of T4 ligase (NEB) and ligated to 1 nM of telorette3 (5′-TGCTCCGTGCATCTGGCATCCCTAACC-3′) at 35°C over night. One hundred picogram of ligated DNA was then used for PCR amplification with 0.1 μM of Adapter (5′-TGTAGC-GTGAAGACGACAGAA-3′) and of Teltail (5′-TGCTCCGTGCATCTGGCATC-3′) primers by the Failsafe Enzyme with the FailSafe buffer premix H (Epicentre). The PCR products were resolved on a 1.2% agarose gel, blotted onto a nylon membrane by capillary osmosis and hybridized at 42°C over night to a DIG-labelled telomeric C-rich LNA probe (36 bp, Exiqon). The blots were then washed with Maleic Acid/Tween solution and DIG signal was revealed with anti-DIG-AP antibody (1:20 000, Roche) and CDP-star solution (Roche) according to the manufacturer’s instructions. Statistical analyses were done using Student’s t-test.
Protein immunoprecipation
Immunoprecipitations were carried out using the Dynabeads Protein G or A immunoprecipitation kit (Invitrogen). Fifty microlitre of beads were treated with 10 μg rabbit anti-HOT1, mouse anti-HOT1 (both MPI-CBG Antibody Facility), rabbit IgG (sc-66931, Santa Cruz; 2729s, Biolabs) or mouse IgG (ChromPure, Jackson ImmunoResearch) in PBB buffer (150 mM NaCl, 50 mM Tris–HCl pH 8.0, 10 mM MgCl2, 0.5% NP-40, Complete Protease Inhibitor–EDTA (Roche)) and subsequently incubated with 400 μg HeLa nuclear extract (SILAC-labelled if followed by MS analysis) for 2 h at 4°C on a rotation wheel, followed by three washes with PBB buffer. For MS, bead fractions were pooled, bound proteins were eluted and separated on a 4–12% gradient gel (Novex, Invitrogen). For co-IP experiments followed by western blot, bound proteins were simply eluted and subjected to western blot analysis.
Western blot
For western blot samples were boiled in Laemmli buffer (Sigma Aldrich) and subjected to SDS–PAGE (NuPage 4–12% Bis-Tris gels; Invitrogen). Gels were blotted to nitrocellulose (Protran; Schleicher & Schuell), blocked in 5% nonfat milk in PBST (PBS containing 0.1% Tween-20) for 1 h at RT and incubated over night at 4°C with primary antibody. The following primary antibodies were used: mouse anti-GFP (Roche Diagnostics, 1:4000 dilution), mouse anti-DM1alpha tubulin (MPI-CBG Antibody Facility, 1:50 000 dilution), mouse anti-Ku70 (sc17789, Santa Cruz, 1:1000 dilution), mouse anti-FLAG (M2, Sigma Aldrich, 1:5000) mouse anti-PCNA (sc-9847, Santa Cruz, 1:1000), mouse anti Histone3 (ab, 1791, Abcam, 1:10 000) and rabbit anti-TERT (gift from Madalena Tarsounas, 1:2000). The next day, membranes were washed three times for 10 min each in 5% milk PBST and were incubated for 1 h at RT with secondary antibody (goat anti-mouse antibody conjugated to horseradish peroxidase, Bio-Rad, 1:4000, or donkey anti-mouse IRDye 800CW or donkey anti-rabbit IRDye 800CW, both LI-COR Odyssey, 1:15 000). Membranes were washed three times for 10 min each in PBST followed by one PBS wash. Bands were visualized with enhanced chemiluminescence Western Blotting Detection Reagents (GE Healthcare) or with the LI-COR Odyssey imaging system. For detection of His-tagged HOT1, TRF1 and TBP, the Penta-His HRP Conjugate Kit (Qiagen) was used according to the manufacturer’s instructions. As a molecular weight standard, Spectra Multicolor Broad Range Protein Ladder (Fermentas), Seablue 2 (Invitrogen) and MagicMark XP Western Protein Standard (Invitrogen) were used.
Immunoprecipitation of telomerase activity and quantitative TRAP assay
For the immunoprecipitation of telomerase activity HeLa cells were lysed in lysis buffer (50 mM Tris–HCl (pH 8.0), 150 mM NaCl and 1% NP-40 supplemented with Complete Protease Inhibitor–EDTA (Roche)) for 30 min on ice followed by a 30 min centrifugation step in a table-top centrifuge at 4°C and 20 000 g. Per IP assay, 25 μl of magnetic protein G beads (Invitrogen) were used. Beads were washed three times with 1 × PBS before use and incubated with 5 mg/ml BSA (in 1 × PBS) for 1 h at 4°C on a rotating wheel, while lysates were precleared with uncoated beads for 1 h at 4°C on a rotating wheel. Per IP assay, 1 mg lysate was incubated with 5 μg rabbit anti-HOT1 (MPI-CBG Antibody Facility), rabbit anti-DKC1 (ab64667, Abcam), rabbit anti-TRF1 (ab1423, Abcam), rabbit anti-TRF2 (NB110-57130, Novus), rabbit anti-TBP (sc-273, Santa Cruz), rabbit anti-YY1 (ab12132, Abcam), rabbit anti-STAT3 (9132, Cell Signaling), rabbit anti-Histone3K4tri-methylated (07-473, Upstate Antibodies), rabbit anti-CENP-B (ab25734, Abcam) or rabbit IgG (sc-66931, Santa Cruz) in PBS for 2 h at 4°C on a rotating wheel. BSA-coated beads were added, followed by a second incubation for 2 h at 4°C on a rotating wheel. Beads were then washed once with PBS, twice with lysis buffer and again once with PBS and finally recovered in 75 μl Chaps buffer (Chemicon).
The quantitative TRAP assay was carried out using GoTaq qPCR Master Mix (Promega) and both the TS (5′-AATCCGTCGAGCAGAGTT-3′) and ACX primer (5′-GCGCGGCTTACCCTTACCCTTACCCTAACC-3′) at 200 nM. The reaction was run on a Mx3000p real-time PCR system (Stratagene) with the following protocol: 25°C for 20 min, 95°C for 10 min and 32 cycles with 95°C for 30 s, 60°C for 30 s and 72°C for 1 min. Statistical analyses were done using Student’s t-test.
For verification that signals were due to the presence of the characteristic TRAP ladder, samples were run on a 20% precast TBE gel (Invitrogen) for 2 h at 200 V. For size reference, the GeneRuler Ultra Low Range DNA Ladder (Fermentas) was used. The gel was stained with EtBr for visualization.
Quantitative real-time PCR
For quantification of TCAB1 mRNA levels RNA was extracted with the RNeasy kit (Qiagen), including DNaseI digestion and from the eluted RNA cDNA was synthesized using the SuperScript III first-strand synthesis kit with oligodT according to manufacturer’s instructions. qPCR primers (Table VI) were used at 70 nM concentration together with the Absolute qPCR SYBR green mix (Abgene) on a Mx3000p real-time PCR system (Stratagene). Target gene mRNA levels were normalized against quantification of GAPDH mRNA levels for housekeeping.
Table 6. Oligonucleotides used for quantitative PCRs.
| No | Gene name | ENSEMBL ID | Forward primer (5′→3′) | Reverse primer (5′→3′) |
|---|---|---|---|---|
| 1 | GAPDH | ENSG00000111640 | CAGCCTCAAGATCATCAGCA | TGTGGTCATGAGTCCTTCCA |
| 2 | TCAB1 | ENSG00000141499 | AGCCAGACACCTCCTACGTG | GGTTGAAGCCACAGAAGAGC |
Statistical analyses were done using student’s t-test.
BAC TransgeneOmics
The following BACs (bacterial artificial chromosomes) were used in this study: human HOT1 RP11-789B24 (Invitrogen), DKC1 RP11-107C18, Coilin RP23-375L19 and POT1 CTD-3053M7 and mouse TRF1 RP24-402F23 (BACPAC Resource Center). A LAP (localization and affinity purification) cassette was inserted as a C-terminal fusion using recombineering. Isolated BAC DNA was transfected and selected for stable integration as described (Poser et al, 2008). The BAC RP11-789B24 does not cover the entire HOT1 gene and was complemented by insertion of a cDNA fragment covering the missing coding and 3′-UTR sequence (Table VII).
Table 7. Oligonucleotides used for cloning and BAC engineering.
| No | Cloning/BAC engineering purpose | Primer sequence (5′→3′) |
|---|---|---|
| 1a | HOT1 BAC stitching primers to add missing CDS and 3′-UTR, forward primer | CGATTTACCTGGAGAAAGGAGTGCCTGGCTGTTATGGAAAGT-TACTTCAATGAGAATCAATACCCAGATG |
| 1b | HOT1 BAC stitching primers to add missing CDS and 3′-UTR, reverse primer | TATAATACAGCATTTATGATATTCTAAAGTACTTTTAGAGAT-AGAACCAACCCTGTGCTGCTACATTGAA |
Immunofluorescence stainings on testes chromosome spreads and testes sections
Testes were isolated from wild-type 129T2/SvEms male mice, between 4–6 weeks old. To prepare chromosome spreads, the tunica albuginea was detached and the seminiferous tubules were incubated in 500 μl of 1 mg/ml Collagenase Type I (Gibco) in PBS for 10 min at 32°C. The tubules were agitated slightly, and the liquid was removed in order to remove a proportion of interstitial cells. A fresh 500 μl aliquot of 1 mg/ml collagenase was added and incubated at 32°C for 30 min, disaggregating by pipetting every 10 min. The cells were spun at 2000, r.p.m. for 5 min at 4°C. The pellet was resuspended in 1 ml cold PBS and filtered through a 40 μm nylon membrane to create a single-cell suspension.
Fifteen microlitre of fixation and permeabilization solution (1% PFA, 5 mM sodium borate pH 8.5, 0.2% Triton X-100) were added to each well (Ø 7 μm) on a 10-well glass slide (StarFrost coating, Engelbrecht). An ImmEdge pen (Vector Laboratories) was used to circle the wells and prevent leakage. The cells were diluted 1:5 into cold 100 mM sucrose in 1 × PBS for 2–3 min to allow hypertonic swelling. One to two microlitre of cell suspension were added to each well. The slides were incubated in a humidified chamber for 30 min, followed by 2–3 h of drying in a laminar hood or on the bench at 22°C. The slides were washed briefly three times in 0.5% Photo-Flo (Kodak) then once in distilled water.
To prepare testes tissue sections, whole testes were immersed in O.C.T Compound (Tissue-Tek 4583) in specimen molds (Tissue-Tek 4566 Cyromold 15 mm × 15 mm × 5 mm) and frozen at −80°C. Seven-micrometer sections were cut using a Leica CM1900 and placed on microscope slides (StarFrost K078; 76 × 26 mm). Sections were fixed using 4% formaldehyde (Sigma F8775) in 1 × PBS for 15 min at 22°C and permeabilized using 0.15% Triton X-100 for 10 min at 22°C and washed twice in 1 × PBS.
To perform immunofluorescence for both chromosome spreads and tissue sections, blocking solution (0.2% fish gelatin (Sigma) in PBS−0.1% Tween-20) was added for 1 h at RT. Primary antibodies were added for 16 h at RT. The following primary antibodies were used: rabbit anti-HOT1 (MPI-CBG Antibody Facility, 1:1000), rabbit anti-TRF2 (sc-9143, Santa Cruz, 1:100) and mouse anti-SYCP3 (as previously described (Adelfalk et al, 2009)). The cells were washed 3 × in blocking solution for 10 min each and secondary antibodies (goat anti-rabbit Alexa555 or goat anti-mouse Alexa488 at 1:500, Invitrogen) were added for 1 h at RT. After 3 × washing in blocking solution for 10 min each, Vectashield (Vecta Laboratories) containing 1 μg/ml DAPI was added and a glass cover slip was placed on top of the wells and sealed closed. Slides were stored at −20°C.
Images were acquired with a DeltaVision Core Microscope (Applied Precision, Olympus IX71 microscope) using a 100 × /1.4 UPlanSApo oil-immersion objective. Z-stacks (0.2–0.5 μm optical sections) were collected and deconvolved using softWoRx (Applied Precision). Z-stacks were reconstructed in 3D using Imaris (Bitplane) and colocalization events were determined for signals above the background using the colocalization function. For imaging of entire tubules, separate images were acquired as one panel and stitched together using the stitch function in softWoRx for visualization.
Hot1Gt(pU-21T)346Card/(Gt(pU-21T)346Card mice and generation of MEFs
C57BL/6-CBA-Hot1Gt(pU-21T)346Card/+ embryos were retrieved from the Center for Animal Resources and Development and are based on the Exchangeable Gene Trap Clones system using the exchangeable pU21 trap vector (Araki et al, 2009). Live animals were retrieved by embryo transfer and were outcrossed for several generations against C57BL/6 wild-type animals. MEFs were generated from littermate embryos at E13.5 by crossing C57BL/6-Hot1Gt(pU-21T)346Card/+ animals with each other.
Genotyping PCR
For genotyping of Hot1Gt(pU-21T)346Card/(Gt(pU-21T)346Card mice, primers were designed to amplify the wild-type and mutant allele in one PCR reaction, both sharing the same forward primer. The PCR was carried out with a reaction containing 2.5 μl 10 × PCR buffer, 0.8 μl 50 mM MgCl2, 0.2 μl 25 mM dNTP mix, 2 μl 10 μM WT-F primer, each 1 μl 10 μM WT-R and MUT-R primers, 1 μl TaqRed polymerase (Bioloine) and 2 μl crude genomic DNA extracts, filled up with HPLC-grade H2O to a total volume of 25 μl (for primer sequences see Table VIII). PCR conditions were as follows: initial hotstart denaturation at 94°C for 3 min followed by 10 cycles with 94°C for 30 s, 62°C for 30 s with a 0.5°C touchdown decline per cycle and 72°C for 60 s, followed by 25 cycles with 94°C for 30 s, 57°C for 30 s and 72°C for 60 s, finished by a final elongation at 72°C for 5 min. The reaction was run on a DNA Engine Thermocycler (Bio-Rad). After the reaction the PCR products were checked by standard gel electrophoresis
Table 8. Oligonucleotides used for genotyping PCRs.
| No | Primer name | Reverse primer (5′→3′) |
|---|---|---|
| 1 | WT-F | CCTCCTGCTCTGTCTCCTTG |
| 2 | WT-R | CCCATTCAATCCCATCAAGG |
| 3 | MUT-R | CCCTTCAGTCTTCCTGTCCA |
Cell fractionation
Cell fractionation analysis for TERT binding to chromatin was carried out as previously described, with minor modifications (Tejera et al, 2010). Per sample one million cells were washed once in cold PBS and then resuspended in 200 μl ice-cold buffer A+ (10 mM HEPES pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, Complete Protease Inhibitor–EDTA (Roche)). Samples were incubated for 5 min on ice and centrifuged for 5 min at 1300 g and 4°C. The supernatant was further cleaned by centrifugation for 25 min at 20 000 g and 4°C, and represents the cytoplasmic fraction. The pellet was washed twice in 500 μl buffer A+ and resuspended in 100 μl ice-cold buffer B (3 mM EDTA, 0.2 mM EGTA, Complete Protease Inhibitor–EDTA (Roche)). Samples were incubated for 30 min on ice and centrifuged for 5 min at 1700 g and 4°C. The supernatant was further cleaned by centrifugation for 25 min at 20 000 g and 4°C, and represents the nuclear soluble fraction. The pellet was washed twice in 500 μl buffer B, resuspended in 100 μl Laemmli buffer (Sigma Aldrich) and sonicated twice for 10 min each in a water-bath sonicator. This sample represents the chromatin fraction. The other fractions were equally mixed with Laemmli buffer, and all samples were boiled at 95°C for 5 min and separated on a 4–12% gradient gel (Novex, Invitrogen).
Supplementary Material
Acknowledgments
We thank members from the Buchholz, Mann and Londoño laboratories for helpful comments on the manuscript, Charlott-Amélie Teutsch, an internship student selected by the Förderverein der Biologieolympiade e.V., for performing DNA pull-down experiments, Sebastian Rose for technical assistance and Karla Neugebauer and Karl Lenhard Rudolph for critical reading of an earlier version of the manuscript. We are indebted to Sabine Suppmann and Claudia Franke of the MPI-B core facility for help with recombinant protein expression, the MPI-CBG antibody facility for raising anti-HOT1 antibodies, the MPI-CBG light microscopy facility for assistance with imaging and the MPI-B crystallization facility for help with crystallization screening. We thank Anthony Hyman for providing reagents, and Ina Poser, Martina Augsburg, Marit Leuschner and Andrea Ssykor for help with generating LAP cell lines. We also thank Anja Wehner and Bianca Splettstoesser for excellent technical assistance. The anti-SYCP3 antibody was a kind gift from Christa Heyting. Tert−/− MEFs and the anti-TERT antibody were kindly provided by Madalena Tarsounas. Work in the Buchholz laboratory is supported by the Max Planck Society, the University of Technology Dresden, the DFG-Center for Regenerative Therapies Dresden (CRTD), the Bundesministerium für Bildung und Forschung grants Go-Bio (0315105) and NGFN-Plus (01GS1102), and the Deutsche Forschungsgemeinschaft (SFB 655 and SPP 1356). Work in the Mann laboratory is supported by the Max Planck Society, NGFN-Plus (01GS0859) and the 7th Framework Program PROSPECTS (HEALTH-F4-2008-021648). Work in the Londoño laboratory was supported by grants from ARC, LCC, ANR and INCa, and is an ‘Equipe Labellisée Ligue’.
Author contributions: DK and F Butter initiated the research, and planned and performed most experiments, with assistance from C Benda, M Scheibe, ID, M Stevense, CLN, C Basquin, DBK and RK; MA and KA generated the Hot1Gt(pU-21T)346Card/(Gt(pU-21T)346Card strain. JAL and RJ contributed to the planning of the experiments; F Buchholz and MM supervised the research and contributed to the planning of the experiments; all authors contributed to the writing of the manuscript.
Footnotes
DK, F Butter, MM and F Buchholz declare a patent application of this work as patent PCT/EP2011/065943. The remaining authors declare that they have no conflict of interest.
References
- Abreu E, Aritonovska E, Reichenbach P, Cristofari G, Culp B, Terns RM, Lingner J, Terns MP (2010) TIN2-tethered TPP1 recruits human telomerase to telomeres in vivo. Mol Cell Biol 30: 2971–2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adelfalk C, Janschek J, Revenkova E, Blei C, Liebe B, Göb E, Alsheimer M, Benavente R, de Boer E, Novak I, Höög C, Scherthan H, Jessberger R (2009) Cohesin SMC1beta protects telomeres in meiocytes. J Cell Biol 187: 185–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonine PV, Grosse-Kunstleve RW, Adams PD (2005) The Phenix refinement framework. CCP4 Newslett 42: [Google Scholar]
- Araki M, Araki K, Yamamura K-I (2009) International Gene Trap Project: towards gene-driven saturation mutagenesis in mice. Curr Pharm Biotechnol 10: 221–229 [DOI] [PubMed] [Google Scholar]
- Aubert G, Hills M, Lansdorp PM (2012) Telomere length measurement-caveats and a critical assessment of the available technologies and tools. Mutat Res 730: 59–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels SJJ, Spruijt CG, Brinkman AB, Jansen PWTC, Vermeulen M, Stunnenberg HG (2011) A SILAC-based screen for Methyl-CpG binding proteins identifies RBP-J as a DNA methylation and sequence-specific binding protein. PLoS One 6: e25884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P (2001) Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 292: 1171–1175 [DOI] [PubMed] [Google Scholar]
- Bendix L, Horn PB, Jensen UB, Rubelj I, Kolvraa S (2010) The load of short telomeres, estimated by a new method, Universal STELA, correlates with number of senescent cells. Aging Cell 9: 383–397 [DOI] [PubMed] [Google Scholar]
- Bilaud T, Brun C, Ancelin K, Koering CE, Laroche T, Gilson E (1997) Telomeric localization of TRF2, a novel human telobox protein. Nat Genet 17: 236–239 [DOI] [PubMed] [Google Scholar]
- Britt-Compton B, Capper R, Rowson J, Baird DM (2009) Short telomeres are preferentially elongated by telomerase in human cells. FEBS Lett 583: 3076–3080 [DOI] [PubMed] [Google Scholar]
- Broccoli D, Smogorzewska A, Chong L, de Lange T (1997) Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat Genet 17: 231–235 [DOI] [PubMed] [Google Scholar]
- Butter F, Davison L, Viturawong T, Scheibe M, Vermeulen M, Todd JA, Mann M (2012) Proteome-wide analysis of disease-associated SNPs that show allele-specific transcription factor binding. PLoS Genet 8: e1002982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butter F, Kappei D, Buchholz F, Vermeulen M, Mann M (2010) A domesticated transposon mediates the effects of a single-nucleotide polymorphism responsible for enhanced muscle growth. EMBO Rep 11: 305–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casteel DE, Zhuang S, Zeng Y, Perrino FW, Boss GR, Goulian M, Pilz RB (2009) A DNA polymerase-alpha primase cofactor with homology to replication protein A-32 regulates DNA replication in mammalian cells. J Biol Chem 284: 5807–5818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L-Y, Redon S, Lingner J (2012) The human CST complex is a terminator of telomerase activity. Nature 488: 540–544 [DOI] [PubMed] [Google Scholar]
- Chen S, Saiyin H, Zeng X, Xi J, Liu X, Li X, Yu L (2006) Isolation and functional analysis of human HMBOX1, a homeobox containing protein with transcriptional repressor activity. Cytogenet Genome Res 114: 131–136 [DOI] [PubMed] [Google Scholar]
- Chi Y-I, Frantz JD, Oh B-C, Hansen L, Dhe-Paganon S, Shoelson SE (2002) Diabetes mutations delineate an atypical POU domain in HNF-1alpha. Mol Cell 10: 1129–1137 [DOI] [PubMed] [Google Scholar]
- Cioce M, Lamond AI (2005) Cajal bodies: a long history of discovery. Annu Rev Cell Dev Biol 21: 105–131 [DOI] [PubMed] [Google Scholar]
- Court R, Chapman L, Fairall L, Rhodes D (2005) How the human telomeric proteins TRF1 and TRF2 recognize telomeric DNA: a view from high-resolution crystal structures. EMBO Rep 6: 39–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26: 1367–1372 [DOI] [PubMed] [Google Scholar]
- Cristofari G, Adolf E, Reichenbach P, Sikora K, Terns RM, Terns MP, Lingner J (2007) Human telomerase RNA accumulation in Cajal bodies facilitates telomerase recruitment to telomeres and telomere elongation. Mol Cell 27: 882–889 [DOI] [PubMed] [Google Scholar]
- Cristofari G, Lingner J (2006) Telomere length homeostasis requires that telomerase levels are limiting. EMBO J 25: 565–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, Snoeyink J, Richardson JS, Richardson DC (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res 35: W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T (2009) How telomeres solve the end-protection problem. Science 326: 948–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Déjardin J, Kingston RE (2009) Purification of proteins associated with specific genomic loci. Cell 136: 175–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhauer KM, Gerstein RM, Chiu CP, Conti M, Hsueh AJ (1997) Telomerase activity in female and male rat germ cells undergoing meiosis and in early embryos. Biol Reprod 56: 1120–1125 [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Giannone RJ, McDonald HW, Hurst GB, Shen R-F, Wang Y, Liu Y (2010) The protein network surrounding the human telomere repeat binding factors TRF1, TRF2, and POT1. PLoS One 5: e12407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH (1989) A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature 337: 331–337 [DOI] [PubMed] [Google Scholar]
- Gu P, Min J-N, Wang Y, Huang C, Peng T, Chai W, Chang S (2012) CTC1 deletion results in defective telomere replication, leading to catastrophic telomere loss and stem cell exhaustion. EMBO J 31: 2309–2321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemann MT, Strong MA, Hao LY, Greider CW (2001) The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell 107: 67–77 [DOI] [PubMed] [Google Scholar]
- Henschel A, Buchholz F, Habermann B (2004) DEQOR: a web-based tool for the design and quality control of siRNAs. Nucleic Acids Res 32: W113–W120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Daniels J-P, Takai H, de Lange T (2006) Recent expansion of the telomeric complex in rodents: two distinct POT1 proteins protect mouse telomeres. Cell 126: 63–77 [DOI] [PubMed] [Google Scholar]
- Jain D, Cooper JP (2010) Telomeric strategies: means to an end. Annu Rev Genet 44: 243–269 [DOI] [PubMed] [Google Scholar]
- Jády BE, Bertrand E, Kiss T (2004) Human telomerase RNA and box H/ACA scaRNAs share a common Cajal body-specific localization signal. J Cell Biol 164: 647–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jády BE, Richard P, Bertrand E, Kiss T (2006) Cell cycle-dependent recruitment of telomerase RNA and Cajal bodies to human telomeres. Mol Biol Cell 17: 944–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch WJ (1993) Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Cryst 26: 795–800 [Google Scholar]
- Kelleher C, Kurth I, Lingner J (2005) Human protection of telomeres 1 (POT1) is a negative regulator of telomerase activity in vitro. Mol Cell Biol 25: 808–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittler R, Heninger A-K, Franke K, Habermann B, Buchholz F (2005) Production of endoribonuclease-prepared short interfering RNAs for gene silencing in mammalian cells. Nat Methods 2: 779–784 [DOI] [PubMed] [Google Scholar]
- Kozik A, Bradbury EM, Zalensky A (1998) Increased telomere size in sperm cells of mammals with long terminal (TTAGGG)n arrays. Mol Reprod Dev 51: 98–104 [DOI] [PubMed] [Google Scholar]
- Latrick CM, Cech TR (2010) POT1-TPP1 enhances telomerase processivity by slowing primer dissociation and aiding translocation. EMBO J 29: 924–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee O-H, Kim H, He Q, Baek HJ, Yang D, Chen L-Y, Liang J, Chae HK, Safari A, Liu D, Songyang Z (2011) Genome-wide YFP fluorescence complementation screen identifies new regulators for telomere signaling in human cells. Mol Cell Proteomics 10: M110.001628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M, Zaug AJ, Podell ER, Cech TR (2005) Switching human telomerase on and off with hPOT1 protein in vitro. J Biol Chem 280: 20449–20456 [DOI] [PubMed] [Google Scholar]
- Li MZ, Elledge SJ (2007) Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat Methods 4: 251–256 [DOI] [PubMed] [Google Scholar]
- Liu D, Safari A, O'Connor MS, Chan DW, Laegeler A, Qin J, Songyang Z (2004) PTOP interacts with POT1 and regulates its localization to telomeres. Nat Cell Biol 6: 673–680 [DOI] [PubMed] [Google Scholar]
- Loayza D, de Lange T (2003) POT1 as a terminal transducer of TRF1 telomere length control. Nature 423: 1013–1018 [DOI] [PubMed] [Google Scholar]
- Londoño-Vallejo JA, DerSarkissian H, Cazes L, Thomas G (2001) Differences in telomere length between homologous chromosomes in humans. Nucleic Acids Res 29: 3164–3171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markljung E, Jiang L, Jaffe JD, Mikkelsen TS, Wallerman O, Larhammar M, Zhang X, Wang L, Saenz-Vash V, Gnirke A, Lindroth AM, Barrés R, Yan J, Strömberg S, De S, Pontén F, Lander ES, Carr SA, Zierath JR, Kullander K et al. (2009) ZBED6, a novel transcription factor derived from a domesticated DNA transposon regulates IGF2 expression and muscle growth. PLoS Biol 7: e1000256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ (2007) Phaser crystallographic software. J Appl Crystallogr 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell JR, Wood E, Collins K (1999) A telomerase component is defective in the human disease dyskeratosis congenita. Nature 402: 551–555 [DOI] [PubMed] [Google Scholar]
- Mittler G, Butter F, Mann M (2009) A SILAC-based DNA protein interaction screen that identifies candidate binding proteins to functional DNA elements. Genome Res 19: 284–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandakumar J, Bell CF, Weidenfeld I, Zaug AJ, Leinwand LA, Cech TR (2012) The TEL patch of telomere protein TPP1 mediates telomerase recruitment and processivity. Nature 492: 285–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nittis T, Guittat L, LeDuc RD, Dao B, Duxin JP, Rohrs H, Townsend RR, Stewart SA (2010) Revealing novel telomere proteins using in vivo cross-linking, tandem affinity purification, and label-free quantitative LC-FTICR-MS. Mol Cell Proteomics 9: 1144–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W, de Lange T (2008) How shelterin protects mammalian telomeres. Annu Rev Genet 42: 301–334 [DOI] [PubMed] [Google Scholar]
- Poser I, Sarov M, Hutchins JRA, Hériché J-K, Toyoda Y, Pozniakovsky A, Weigl D, Nitzsche A, Hegemann B, Bird AW, Pelletier L, Kittler R, Hua S, Naumann R, Augsburg M, Sykora MM, Hofemeister H, Zhang Y, Nasmyth K, White KP et al. (2008) BAC TransgeneOmics: a high-throughput method for exploration of protein function in mammals. Nat Methods 5: 409–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riou L, Bastos H, Lassalle B, Coureuil M, Testart J, Boussin FD, Allemand I, Fouchet P (2005) The telomerase activity of adult mouse testis resides in the spermatogonial alpha6-integrin-positive side population enriched in germinal stem cells. Endocrinology 146: 3926–3932 [DOI] [PubMed] [Google Scholar]
- Schnapp G, Rodi HP, Rettig WJ, Schnapp A, Damm K (1998) One-step affinity purification protocol for human telomerase. Nucleic Acids Res 26: 3311–3313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz J, Besir HS, Strasser C, Suppmann S (2013) A new method to customize protein expression vectors for fast, efficient and background free parallel cloning. BMC Biotechnol 13: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanemura K, Ogura A, Cheong C, Gotoh H, Matsumoto K, Sato E, Hayashi Y, Lee H-W, Kondo T (2005) Dynamic rearrangement of telomeres during spermatogenesis in mice. Dev Biol 281: 196–207 [DOI] [PubMed] [Google Scholar]
- Tejera AM, Alcontres MSD, Thanasoula M, Marion RM, Martinez P, Liao C, Flores JM, Tarsounas M, Blasco MA (2010) TPP1 Is Required for TERT Recruitment, Telomere Elongation during Nuclear Reprogramming, and Normal Skin Development in Mice. Dev Cell 18: 775–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson RL, Abreu EB, Ziegler T, Ly H, Counter CM, Terns RM, Terns MP (2008) Telomerase reverse transcriptase is required for the localization of telomerase RNA to cajal bodies and telomeres in human cancer cells. Mol Biol Cell 19: 3793–3800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson RL, Ziegler TD, Supakorndej T, Terns RM, Terns MP (2006) Cell cycle-regulated trafficking of human telomerase to telomeres. Mol Biol Cell 17: 955–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycowski KT, Shu M-D, Kukoyi A, Steitz JA (2009) A conserved WD40 protein binds the Cajal body localization signal of scaRNP particles. Mol Cell 34: 47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela E, Schneider RP, Ortega S, Blasco MA (2011) Different telomere-length dynamics at the inner cell mass versus established embryonic stem (ES) cells. Proc Natl Acad Sci USA 108: 15207–15212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venteicher AS, Abreu EB, Meng Z, McCann KE, Terns RM, Veenstra TD, Terns MP, Artandi SE (2009) A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science 323: 644–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venteicher AS, Meng Z, Mason PJ, Veenstra TD, Artandi SE (2008) Identification of ATPases pontin and reptin as telomerase components essential for holoenzyme assembly. Cell 132: 945–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen M, Hubner NC, Mann M (2008) High confidence determination of specific protein–protein interactions using quantitative mass spectrometry. Curr Opin Biotechnol 19: 331–337 [DOI] [PubMed] [Google Scholar]
- Wang C, Meier UT (2004) Architecture and assembly of mammalian H/ACA small nucleolar and telomerase ribonucleoproteins. EMBO J 23: 1857–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Podell ER, Zaug AJ, Yang Y, Baciu P, Cech TR, Lei M (2007) The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature 445: 506–510 [DOI] [PubMed] [Google Scholar]
- Wang F, Stewart JA, Kasbek C, Zhao Y, Wright WE, Price CM (2012) Human CST has independent functions during telomere duplex replication and C-strand fill-in. Cell Rep 2: 1096–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicky C, Villeneuve AM, Lauper N, Codourey L, Tobler H, Müller F (1996) Telomeric repeats (TTAGGC)n are sufficient for chromosome capping function in Caenorhabditis elegans. Proc Natl Acad Sci USA 93: 8983–8988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin H, Liu D, Wan M, Safari A, Kim H, Sun W, O'Connor MS, Songyang Z (2007) TPP1 is a homologue of ciliate TEBP-beta and interacts with POT1 to recruit telomerase. Nature 445: 559–562 [DOI] [PubMed] [Google Scholar]
- Yashima K, Maitra A, Rogers BB, Timmons CF, Rathi A, Pinar H, Wright WE, Shay JW, Gazdar AF (1998) Expression of the RNA component of telomerase during human development and differentiation. Cell Growth Differ 9: 805–813 [PubMed] [Google Scholar]
- Ye JZ-S, Hockemeyer D, Krutchinsky AN, Loayza D, Hooper SM, Chait BT, de Lange T (2004) POT1-interacting protein PIP1: a telomere length regulator that recruits POT1 to the TIN2/TRF1 complex. Genes Dev 18: 1649–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaug AJ, Podell ER, Nandakumar J, Cech TR (2010) Functional interaction between telomere protein TPP1 and telomerase. Genes Dev 24: 613–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Baumann U, Reymond J-L (2004) An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res 32: e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong F, Savage SA, Shkreli M, Giri N, Jessop L, Myers T, Chen R, Alter BP, Artandi SE (2011) Disruption of telomerase trafficking by TCAB1 mutation causes dyskeratosis congenita. Genes Dev 25: 11–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong FL, Batista LFZ, Freund A, Pech MF, Venteicher AS, Artandi SE (2012) TPP1 OB-fold domain controls telomere maintenance by recruiting telomerase to chromosome ends. Cell 150: 481–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Z, Shiue L, Kaplan S, de Lange T (1992) A mammalian factor that binds telomeric TTAGGG repeats in vitro. Mol Cell Biol 12: 4834–4843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Tomlinson RL, Lukowiak AA, Terns RM, Terns MP (2004) Telomerase RNA accumulates in Cajal bodies in human cancer cells. Mol Biol Cell 15: 81–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.