Abstract
Limiting the levels of homologous recombination (HR) that occur at sites of DNA damage is a major role of BLM helicase. However, very little is known about the mechanisms dictating its relocalization to these sites. Here, we demonstrate that the ubiquitin/SUMO-dependent DNA damage response (UbS-DDR), controlled by the E3 ligases RNF8/RNF168, triggers BLM recruitment to sites of replication fork stalling via ubiquitylation in the N-terminal region of BLM and subsequent BLM binding to the ubiquitin-interacting motifs of RAP80. Furthermore, we show that this mechanism of BLM relocalization is essential for BLM’s ability to suppress excessive/uncontrolled HR at stalled replication forks. Unexpectedly, we also uncovered a requirement for RNF8-dependent ubiquitylation of BLM and PML for maintaining the integrity of PML-associated nuclear bodies and as a consequence the localization of BLM to these structures. Lastly, we identified a novel role for RAP80 in preventing proteasomal degradation of BLM in unstressed cells. Taken together, these data highlight an important biochemical link between the UbS-DDR and BLM-dependent pathways involved in maintaining genome stability.
Keywords: RAP80, RNF8, RNF168, K63-linked ubiquitylation, PML nuclear bodies
Introduction
The mammalian genome codes for five related RECQ-like 3′–5′ DNA helicases: RECQL1, BLM, WRN, RECQL4 and RECQL5, all of which have been demonstrated to act upon a number of topologically different DNA structures in vitro. The most extensively studied of these helicases is BLM, which has been shown to unwind a variety of DNA substrates that include 3’-tailed duplexes, bubble structures, forked duplexes, G-quadruplex structures, DNA displacement loops and four-way Holliday Junctions (HJs). In addition to its conventional helicase activity, BLM can also promote ATP-dependent branch migration of the HJs, a process that may be involved in replication fork restoration (reviewed in Monnat, 2010 and Hickson and Mankouri, 2011).
BLM is mutated in Bloom syndrome (BS); a rare autosomal recessive disorder that is typified by proportional dwarfism, sun-sensitive facial erythema, skin pigmentation abnormalities, immunodeficiency, infertility and an increased predilection to develop both lymphoid and epithelial-derived tumours (Ellis and German, 1996). Cells from BS patients characteristically exhibit increased chromosomal instability with increased numbers of chromatid gaps and breaks, as well as chromosome structural rearrangements, including symmetrical quadra-radials, telomere associations, anaphase bridges and lagging chromosomes being frequently observed. A unique cellular feature, commonly used in the molecular diagnosis of BS, is an ∼10-fold increase in the frequency of sister chromatid exchanges (SCEs), which is thought to arise as a consequence of uncontrolled homologous recombination (HR) during S and G2 phases of the cell cycle.
The exact role of BLM is complicated by the fact that it can act to both promote and suppress HR repair (HRR) depending on the cellular context and the type of DNA damage. Following exposure to ionizing radiation, cells that have incurred DSBs in late S or G2 phase of the cell cycle undergo MRN- and CtIP-dependent break end-resection to promote HR through the generation of ssDNA. While the MRN complex in association with CtIP initiates this resection, it was recently shown that the progressive nucleolytic degradation of the DNA end is coordinated by the actions of DNA2, Exo1 and BLM (Gravel et al, 2008; Nimonkar et al, 2008, 2011). In contrast, following the generation of stalled replication forks caused by the exposure of cells to hydroxyurea (HU), 53BP1 in association with BLM is recruited to sites of aberrant fork structures where it functions to suppress rather than promote HR. This may in part be achieved by the ability of 53BP1 and BLM to directly bind to pro-recombinogenic core HR proteins like RAD51 (Bischof et al, 2001; Sengupta et al, 2003; Tripathi et al, 2007) and RAD54 (Srivastava et al, 2009), and to disrupt RAD51 nucleoprotein filaments (Bugreev et al, 2007; Tripathi et al, 2007). Therefore, the role played by BLM in dictating the type of DNA repair depends on the nature of the DNA damage and the presence of critical key regulatory proteins such as 53BP1.
Despite our understanding of the enzymatic functions of BLM, very little is known about the mechanisms that trigger its relocalization to different DNA structures or how this is influenced by post-translational modifications. BLM seems to have a role both in sensing DNA damage and in transmission of the damage signal to downstream effector proteins (reviewed in Tikoo and Sengupta, 2010). BLM is phosphorylated via the Chk1/ATR pathway in response to replication stress (Davies et al, 2004; Sengupta et al, 2004). However, while ATR-dependent BLM phosphorylation is not required for its localization to sites of stalled replication forks, it is essential for S-phase checkpoint recovery (Davies et al, 2004). In contrast, dephosphorylation of Serine-646 has been shown to be essential for BLM to be recruited to the sites of damage (Kaur et al, 2010). Additionally, it has been suggested that modification of BLM by SUMO influences its cellular localization to PML-containing nuclear bodies (PML-NBs) as well as its ability to stimulate RAD51 recruitment to damaged replication forks (Eladad et al, 2005; Ouyang et al, 2009). Hence, phosphorylation, SUMOylation and possibly ubiquitylation of BLM may play important roles not only in regulating its dynamic movement between the PML-NBs and damaged DNA, but also its decision to promote or suppress RAD51-dependent HRR. Interestingly, following the induction of DSBs both the ubiquitin and SUMO conjugation pathways function in concert to stimulate repair of damaged DNA. This ubiquitin/SUMO-dependent DDR (UbS-DDR) is mediated by an E3 ubiquitin/SUMO ligase cascade composed of RNF8, HERC2, RNF168, PIAS1 and PIAS4, which promotes alterations in the local chromatin architecture surrounding the break to allow the efficient recruitment of essential repair factors such as the BRCA1-A complex and 53BP1 (Kolas et al, 2007; Mailand et al, 2007; Wang et al, 2007; Doil et al, 2009; Galanty et al, 2009; Stewart et al, 2009; Bekker-Jensen et al, 2010; Mattiroli et al, 2012). Given the link between BLM cellular localization regulation and the SUMO system (Eladad et al, 2005; Ouyang et al, 2009), it is conceivable that ubiquitin may also play a key role in regulating BLM recruitment to sites of DNA damage.
Here, we demonstrate that following replication stress, the RNF8/RNF168-dependent ubiquitylation of BLM directs its recruitment to sites of stalled replication forks via an interaction with the UIM domains of RAP80. In addition, we have identified a role for RNF8-dependent BLM ubiquitylation in undamaged cells that functions to maintain its localization to PML-NBs and prevents its sequestration into nucleoli. We also provide evidence that BLM binds constitutively to RAP80 in the absence of DNA damage in a ubiquitin-independent manner and this is required to maintain BLM stability within the cell. Lastly, we show that this ubiquitin-mediated relocalization of BLM is required for its ability to suppress excessive/uncontrolled HR.
Results
BLM is recruited to sites of replication stress in a ubiquitin-dependent manner
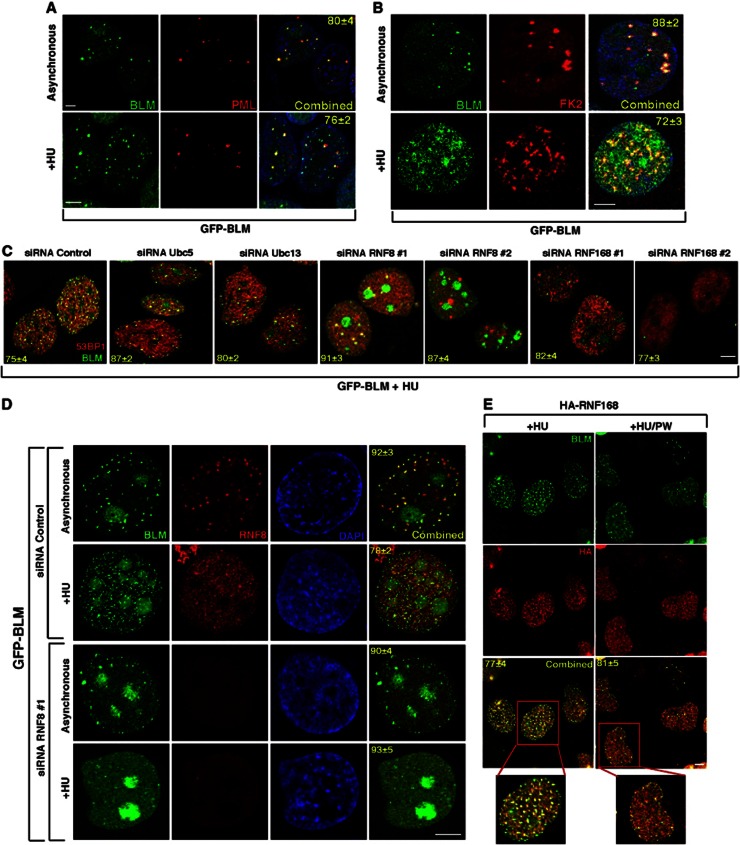
It has been demonstrated that the BLM helicase and 53BP1 together function to limit HR by suppressing the RAD51 response (Tripathi et al, 2007, 2008). Given the biochemical association between BLM, 53BP1 and the UbS-DDR, we hypothesized that the UbS-DDR may regulate the accumulation of BLM in response to replication stress. Hence, cells stably expressing GFP-tagged BLM were either mock treated or exposed to HU, fixed, stained with antibodies directed against PML or poly-ubiquitin chains (FK2). BLM is known to primarily reside in PML-NBs in undamaged cells (Yankiwski et al, 2000). Following treatment with HU, BLM relocalizes from PML-NBs to sites of stalled replication forks (Figure 1A). Consistent with the potential for the UbS-DDR to regulate the nuclear recruitment of BLM, HU-treated cells exhibited a significant colocalization of BLM with sites of ubiquitylation (Figure 1B).
Figure 1.
Recruitment of BLM to the sites of stalled replication depends on RNF8 and RNF168. (A, B) BLM colocalizes with PML-NBs and ubiquitin in the absence of DNA damage. GFP-BLM cells were either grown under asynchronous conditions or treated with HU (+HU). GFP-BLM cells were co-stained with (A) anti-PML antibody and (B) anti-ubiquitin FK2 antibody. Nuclei are stained by DAPI. Scale 5 μM. (C) Lack of E3 ligases leads to lack of BLM recruitment after HU treatment. GFP-BLM cells were transfected with either the control siRNA or siRNAs against Ubc5, Ubc13, RNF8 or RNF168. The cells were treated with HU. GFP-BLM was visualized along with 53BP1. Scale 5 μM. (D) BLM accumulates in the nucleolus in the absence of RNF8. Same as (C) except only siRNA Control or siRNA RNF8 #1 was used. GFP-BLM cells were stained with anti-RNF8 antibodies. Nuclei visualized by DAPI. Scale 5 μM. (E) RNF168 colocalizes with BLM after HU treatment. RIDDLE cells complemented with HA-tagged RNF168 were grown in the presence of HU (+HU) or in the postwash condition (+HU/PW). One of the cells under either conditions is zoomed. Immunofluorescence was carried out with anti-BLM (A300–110A) and anti-HA antibodies. Scale 5 μM.
To investigate whether the RNF8/RNF168-dependent E3 ubiquitin ligase cascade functioned during replication stress to promote the focal relocalization of BLM, cells expressing GFP-BLM were treated with either a control siRNA or siRNA directed against UbcH5a, Ubc13, RNF8 or RNF168. Following exposure to HU, immunofluorescence (IF) was used to determine the localization of BLM. Depletion of RNF8 or RNF168 (using two different siRNA sequences) resulted in a failure of cells to properly recruit BLM to sites of DNA damage (Figure 1C; Supplementary Figure 1A and C). These observations were also recapitulated in cells derived from a RIDDLE syndrome patient (Supplementary Figure 1B). Taken together, this indicates that BLM functions downstream of RNF8 and RNF168 during the DDR invoked by exposure to HU. Indeed we observed both RNF8/RNF168 relocate to sites of replication damage that contained BLM (Figure 1D and E) but not when the DNA damage has been repaired (compare +HU and +HU/PW conditions). Interestingly, BLM was predominantly localized to the nucleoli of cells lacking RNF8 (Figure 1D; Supplementary Figure 1C), suggesting that a basal level of RNF8- but not RNF168-dependent ubiquitylation is required for the normal localization of BLM within the nucleus even in undamaged cells. This is supported by the observation that BLM colocalized at sites of ubiquitylation in asynchronously growing cells (Figure 1B). Interestingly, depletion of either Ubc13 or UbcH5a alone did not affect BLM relocalization after HU treatment (Figure 1C; Supplementary Figure 1C). However, the combined loss of Ubc13 and UbcH5a completely abrogated the formation of BLM foci induced by replication stress comparable to a lack of RNF168 (Supplementary Figure 1C), possibly indicating functional redundancy between these two E2 enzymes.
BLM is targeted by RNF8 and RNF168 for ubiquitylation in vitro and in vivo
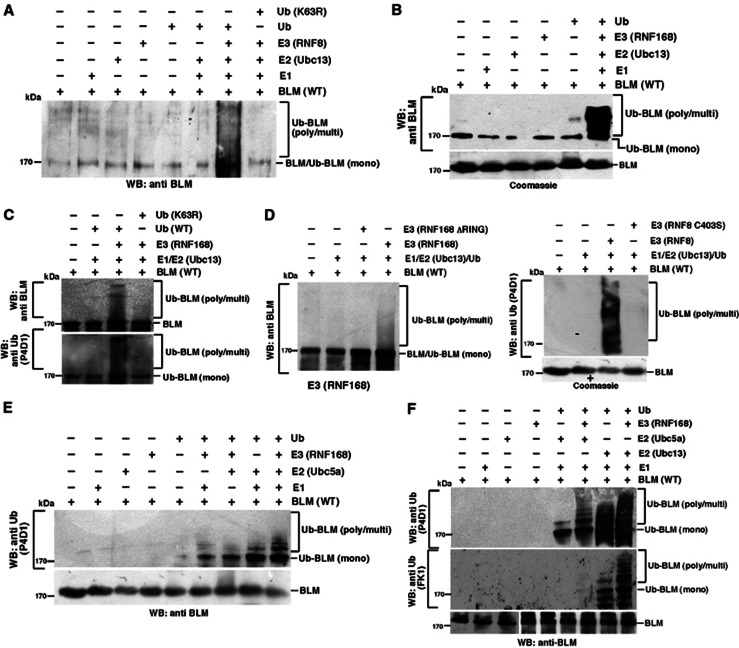
Based on the above observations, it is conceivable that the requirement for RNF8/RNF168 to facilitate BLM relocalization after HU treatment is mediated through their ability to target it for ubiquitylation. To investigate whether BLM is a substrate for RNF8/RNF168, in vitro ubiquitylation assays were carried out using recombinant BLM in combination with purified RNF8/RNF168 (Supplementary Figure 2A) in the presence of Ubc13/UbcH5a as the E2 conjugating enzyme. Both E3 ligases strongly stimulated the K63-dependent poly-ubiquitylation of BLM in vitro in the presence of Ubc13 (Figure 2A–C). Moreover, we demonstrated that this reaction was dependent on the active RING domain of the two E3 ligases (Figure 2D). Though UbcH5a could also be utilized as an E2 to catalyse BLM poly-ubiquitylation (Figure 2E), BLM ubiquitylation by Ubc13 was more robust (Figure 2F), indicating that the RNF8/RNF168 preferentially utilize Ubc13 to poly-ubiquitylate BLM in vitro.
Figure 2.
BLM is ubiquitylated by RNF8/RNF168 in vitro. (A) Ubc13/RNF8 leads to the poly-ubiquitylation of BLM. In vitro ubiquitylation reactions were carried out using recombinant full-length BLM and RNF8 as the E3 ligase. The ubiquitylated BLM was detected by an anti-BLM antibody (A300-120A). A parallel reaction was also carried out using K63R ubiquitin mutant. (B) BLM is ubiquitylated by RNF168/Ubc13. Ubiquitylation reactions were carried out with BLM using Ubc13 as the E2 and RNF168 as the E3 ligase. Westerns were carried out with antibody against BLM (A300-120A). The Coomassie stained gel for purified BLM used for ubiquitylation is shown at the bottom. (C) Poly-ubiquitylation of BLM mediated by Ubc13/RNF168 is K63 linked. In vitro ubiquitylation reactions using BLM as the substrate were carried out using either wild-type ubiquitin or ubiquitin mutant (K63R). Western analysis was carried out with antibody against (top) BLM (A300-120A) or (bottom) ubiquitin (P4D1). (D) The RING domain of RNF8 and RNF168 enhances the Ubc13-/RNF168-mediated ubiquitylation of BLM. In vitro BLM ubiquitylation reactions were carried out using either wild-type RNF8/RNF168 or their RING deleted/disrupted counterpart. The ubiquitylated forms of BLM were detected by using anti-BLM antibody (A300-120A) (for RNF168, left) or anti-Ubiquitin (P4D1) (for RNF8, right). For RNF8-dependent ubiquitylation, the Coomassie stained gel for purified BLM used for ubiquitylation is shown at the bottom. (E) BLM is ubiquitylated by RNF168/Ubc5a. Ubiquitylation reactions were carried out with BLM using Ubc5a as the E2 and RNF168 as the E3 ligase. Westerns were carried out with antibody against ubiquitin (P4D1). (F) Both Ubc5a and Ubc13 can act as the E2 conjugating enzyme for BLM. (F) Same as (A, E), except parallel ubiquitylation reactions were carried out using either Ubc5a or Ubc13. Western analysis was carried out with antibodies against ubiquitin-P4D1 (top) or FK1 (middle). The blots were further probed with anti-BLM antibody (A300-110A).
Source data for this figure is available on the online supplementary information page.
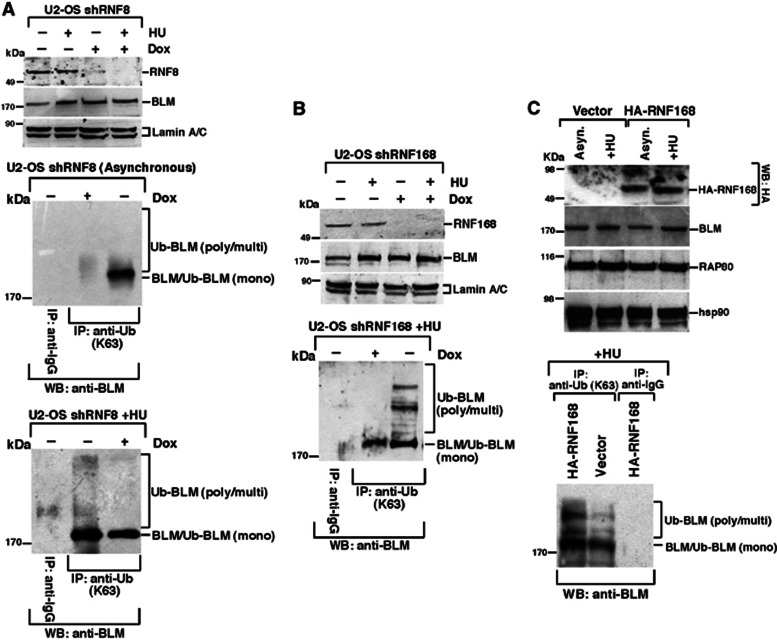
To ascertain whether BLM could also be ubiquitylated in vivo in a DNA damage-inducible manner, U2OS cells expressing doxycycline (Dox) inducible shRNA to either RNF8 or RNF168 were used. These cells were either left asynchronous or treated with HU in the presence or absence of Dox. Immunoprecipitations were carried out using an antibody to K63-linked poly-ubiquitin chains and western blotted for the presence of BLM. BLM poly-ubiquitylation was significantly induced following exposure to HU. However, the cells lacking either RNF8 or RNF168 failed to efficiently promote K63-linked ubiquitylation of BLM (Figure 3A and B). The dependency for RNF168 to mediate BLM ubiquitylation in vivo was confirmed in RIDDLE syndrome cells complemented with WT RNF168 but not an empty vector (Figure 3C). Interestingly BLM was mono-ubiquitylated in the absence of HU, which was compromised in the absence of RNF8 (Figure 3A). These data demonstrate that BLM is mono-ubiquitylated in the absence of DNA damage and poly-ubiquitylated following exposure to DNA damage in a RNF8- and RNF168-dependent manner.
Figure 3.
BLM is ubiquitylated by RNF8 and RNF168 in vivo. (A) Loss of RNF8 leads to decreased K63-linked ubiquitylation of BLM in vivo. U2OS shRNF8 cells were grown in the absence or presence of Doxycycline (Dox), without or with HU co-treatment. (Top) Nuclear extracts were probed with antibodies against RNF8, BLM (A300-110A) and Lamin A/C. (Middle and bottom) Immunoprecipitations were carried out with anti-Ubiquitin K63-linkage specific antibody (or the corresponding IgG). The immunoprecipitates were probed with antibodies against BLM (A300-120A). (B, C). Loss of RNF168 leads to decreased K63-linked ubiquitylation of BLM in vivo after HU treatment. Same as (A) except U2OS shRNF168 cells were used in (B) and RIDDLE syndrome cells complemented with either empty vector or HA-tagged RNF168 cells were used in (C). U2OS shRNF168 cells were grown in the absence or presence of Doxycycline (Dox). Both U2OS shRNF168 and RIDDLE syndrome cells were treated with HU. The direct westerns are on top while immunoprecipitation with anti-Ub (K63) antibodies (or the corresponding IgG) followed by anti-BLM (A300-120A) westerns is shown at the bottom.
Source data for this figure is available on the online supplementary information page.
Ubiquitylation of the BLM N-terminal region is required for its relocalization to sites of DNA damage
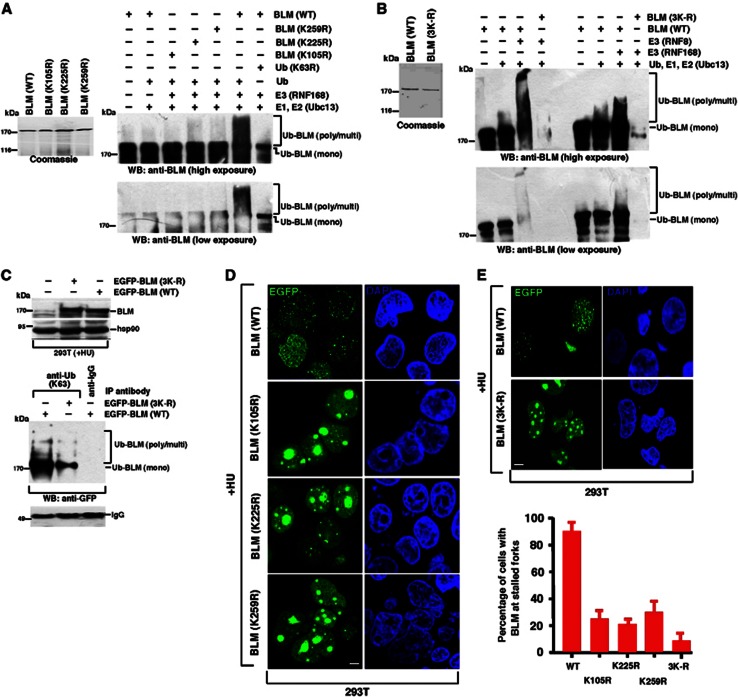
In order to ascribe a biological function to the RNF8-/RNF168-dependent ubiquitylation of BLM, initially the sites of ubiquitylation needed to be identified. To narrow down the lysine residues within BLM potentially targeted for ubiquitylation, two independent ubiquitylation site prediction programs (UbPred and UbiPred) were used, which both highlighted lysines at 105, 225 and 259 (K105, K225 and K259) as being high confidence target residues. To investigate whether these lysine residues were ubiquitylated by RNF8/RNF168, recombinant BLM containing each individual lysine mutated to arginine or all three sites mutated in combination (3K-R) were purified, tested for their ability to perform helicase activity to an equal extent (Supplementary Figure 2B) and subsequently used as the substrates during in vitro ubiquitylation reactions. Loss of any of the three predicted lysine residues individually resulted in a reduction in the level of in vitro BLM poly-ubiquitylation (Figure 4A). The RNF8/RNF168-dependent ubiquitylation was completely abrogated in 3K-R BLM mutant (Figure 4B), indicating that RNF8/RNF168 can target multiple lysine residues within BLM for ubiquitin chain conjugation in vitro.
Figure 4.
Ubiquitylation of BLM at 105, 225 and 259 is required for BLM recruitment to the sites of stalled replication. (A) BLM undergoes K63-linked ubiquitylation at lysines residues 105, 225 and 259. (Left) Coomassie gel demonstrating the expression of GST-tagged BLM (WT) or BLM (K105R), BLM (K225R), BLM (K259R). (Right) In vitro ubiquitylation reactions were carried out using equal amounts wild-type BLM or the three BLM mutants (K105R, K225R and K259R). Western blots were carried out with antibodies against BLM (A300-120A). Two different exposures are shown to demonstrate the differential ubiquitylation. (B) Complete abrogation of RNF8-/RNF168-mediated ubiquitylation in BLM (3K-R) mutant. Same as (A) except BLM (3K-R) mutant was used. Expression of wild-type GST-tagged BLM and BLM (3K-R) mutant via Coomassie staining is shown on the left. RNF8 or RNF168 was used as the E3 ligase in parallel reactions. Two different exposures are shown to demonstrate the differential ubiquitylation. (C) Mutation of lysines at 105, 225 and 259 on BLM leads to loss of BLM poly-ubiquitylation after DNA damage. Wild-type BLM or (3K-R) mutant was overexpressed in 293T cells and subsequently treated with HU. (Top) The expression levels were determined by western analysis using antibodies against BLM (A300-110A) and hsp90. (Bottom) Immunoprecipitations were carried out using anti-K63-linked ubiquitin antibody. Immunoprecipitates were probed with antibodies against GFP or the corresponding IgG. Equal amount of antibody used for immunoprecipitation is demonstrated by comparing the IgG level. (D) Ubiquitylation of BLM at lysines 105, 225 and 259 is required for its recruitment to the stalled replication forks. 293T cells were transfected with EGFP-tagged wild-type BLM or BLM (K105R), BLM (K225R), BLM (K259R). Post-transfection the cells were treated with HU for 24 h. Transfected cells were tracked. Nuclei are stained by DAPI. Scale 5 μM. (E) Mutation of lysines at 105, 225 and 259 on BLM leads to enhanced nucleolar accumulation of BLM after HU treatment. (Top) Same as (D) except after transfection with either BLM (WT) or BLM (3K-R), the transfected cells were tracked. Nuclei are stained by DAPI. Scale 5 μM. (Bottom) Quantitation of (D) and (E, top).
Source data for this figure is available on the online supplementary information page.
To determine whether the three sites of BLM ubiquitylation identified in vitro also mediated the conjugation of poly-ubiquitin chains in vivo, extracts derived from 293T cells transfected with either WT or mutant 3K-R BLM expression constructs were subjected to immunoprecipitation using an antibody specific for K63-linked poly-ubiquitin chains and western blotted for the presence of BLM. Loss of these three critical N-terminal lysine residues resulted in a significant decrease in the overall level of K63-linked BLM ubiquitylation after HU treatment (Figure 4C), supporting the notion that they represent the major sites of K63-linked BLM ubiquitylation in vivo.
It is conceivable that the RNF8-/RNF168-dependent ubiquitylation of BLM at specific residues is required for the relocalization of BLM to sites of replication stress following HU exposure. To test this hypothesis, cells transfected with either a WT or 3K-R mutant EGFP-tagged BLM expression construct were treated with HU and the focal recruitment of BLM monitored by fluorescence microscopy. In stark contrast to the WT BLM, the single mutants substantially compromised the ability of the exogenous BLM to form HU-induced foci (Figure 4D and E; Supplementary Figure 2C). This defect was exacerbated when all three sites of ubiquitylation on BLM were lost (Figure 4E). A similar lack of BLM 3K-R localization was also observed after ionizing radiation (Supplementary Figure 2D). Combined, these data support the concept that RNF8-/RNF168-dependent ubiquitylation of BLM promotes its recruitment to sites of DNA damage.
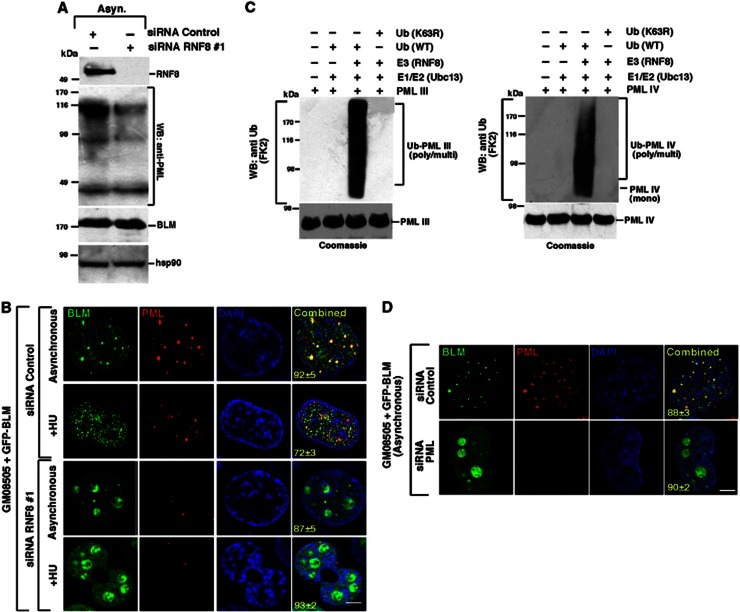
Each of the BLM lysine mutants exhibited a notable relocalization from PML-NBs into the nucleoli; a phenotype with striking similarity to that which we had previously observed for the nuclear localization of BLM in cells depleted of RNF8 (Figure 1C and D; Supplementary Figure 1C). Our previous observations have demonstrated that BLM was mono-ubiquitylated in undamaged cells in an RNF8-dependent manner (Figure 3A) and that in these cells BLM could be found colocalizing with sites of ubiquitin chain formation in PML-NBs (Figure 1A and B). This suggests that a basal level of mono-ubiquitylation of BLM by RNF8 may be required to maintain its normal nuclear localization and/or prevent its sequestration to the nucleoli. Interestingly, we noted that cells depleted of RNF8 exhibited a reduction in the number of PML-NBs as well as the overall level of PML expression (Figure 5A and B). This may be due to the K63-linked ubiquitylation-mediated stabilization of PML isoforms by Ubc13/RNF8 (Supplementary Figure 2A; Figure 5C). Hence, a lack of PML coupled with the loss of monoubiquitylation of BLM leads to the relocalization of BLM to the nucleolus in cells lacking RNF8 (Figure 5D).
Figure 5.
Destabilization of PML-NBs in the absence of RNF8 leads to the accumulation of BLM in nucleolus. (A, B) RNF8 knockdown destabilizes PML nuclear bodies. (A) GFP-BLM cells were transfected with either siRNA Control or siRNA RNF8 #1 and the cells were grown in asynchronous (Asyn.) conditions. Whole cell lysates were probed with antibodies against PML (PG-M3), BLM (A300-110A) and hsp90. (B) Same as (A), except GFP-BLM was tracked with PML-NBs by carrying out immunofluorescence with anti-PML (PG-M3) antibody. Nuclei are stained by DAPI. Scale 5 μM. (C) Ubc13/RNF8 leads to the poly-ubiquitylation of PML. In vitro ubiquitylation reaction was carried out using recombinant full-length PML III (left) or PML IV (right) and RNF8 as the E3 ligase. Poly-ubiquitylated forms of PML isoforms were detected by anti-ubiquitin antibody (FK2). Parallel reactions were also carried out using K63R ubiquitin mutant. The Coomassie-stained gels for purified PML III and IV used for ubiquitylation are shown. (D) PML knockdown relocated BLM to the nucleolus. Same as (B) except the GFP-BLM cells were transfected with either siRNA Control or siRNA PML. Post-transfection the cells were grown in asynchronous conditions.
Source data for this figure is available on the online supplementary information page.
Recruitment of ubiquitylated BLM is mediated by RAP80
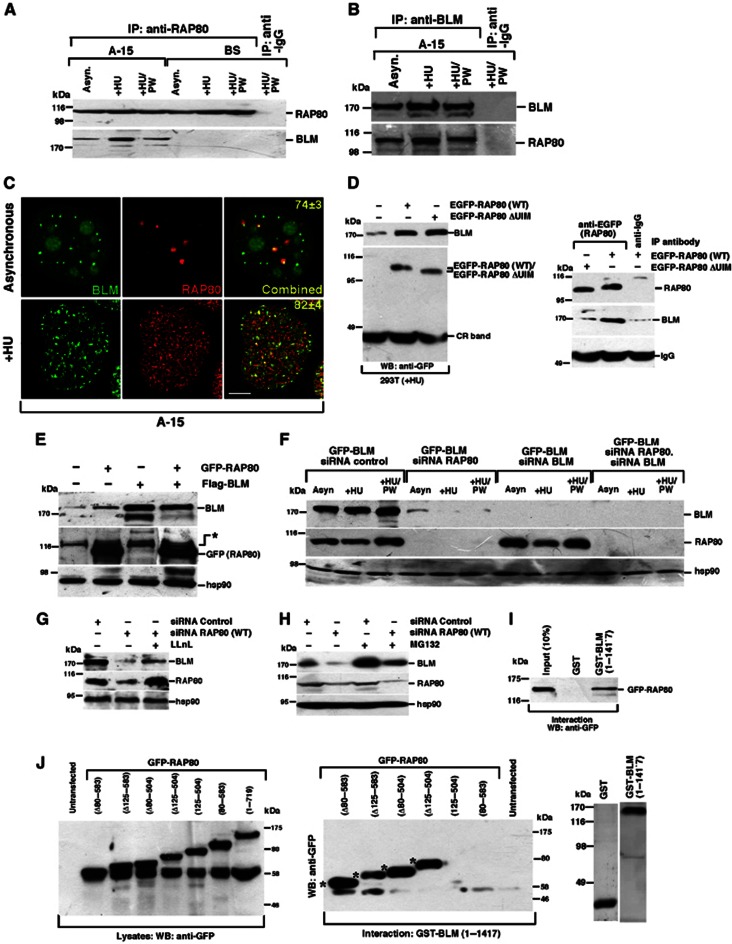
One of the primary functions of the DNA damage-linked RNF8/RNF168 E3 ubiquitin ligase cascade is to catalyse the formation of K63-linked poly-ubiquitin chains, which serve to direct/target protein binding (Kolas et al, 2007; Mailand et al, 2007; Doil et al, 2009; Stewart et al, 2009; Mattiroli et al, 2012). In the case of RNF8/RNF168, it has been shown that formation of poly-ubiquitin chains by these E3 ligases on H2A-type histones situated proximal to a DNA break promotes the recruitment of the BRCA1-A repair complex via their association with the ubiquitin-interacting motifs (UIMs) of the adaptor protein RAP80 (Kim et al, 2007; Sobhian et al, 2007; Wang et al, 2007). Since poly-ubiquitylation of BLM is required for its relocalization to sites of replication stress, it is plausible that this is also mediated via an interaction with the ubiquitin binding domains of RAP80. Hence, reciprocal co-immunoprecipitations were carried out using extracts derived from cells treated with and without HU. In keeping with the idea that RAP80 acts as a receptor for ubiquitylated BLM in response to DNA damage, we observed that BLM and RAP80 co-precipitated together in asynchronous cells or after the repair of the DNA damage (+HU/PW condition). Importantly, this interaction was increased following exposure of cells to HU (Figure 6A and B). These findings were supported by the increased colocalization of RAP80 and BLM in HU-treated cells (Figure 6C). To determine whether the DNA damage-inducible association of BLM with RAP80 required its ability to bind ubiquitin chains, cells were transfected with either an EGFP-tagged WT or mutant RAP80 expression construct lacking its UIM domains (ΔUIM), exposed to HU and then the tagged RAP80 was immunoprecipitated from cell extracts. In contrast to the robust interaction observed between BLM and WT EGFP-tagged RAP80, deletion of the RAP80 UIM domains completely abrogated the HU mediated enhancement of their association (Figure 6D).
Figure 6.
RAP80 regulates BLM stability. (A, B) BLM and RAP80 interact in vivo. Immunoprecipitation of BLM was carried out in (A) BS/A-15 cells or only in (B) A-15 cells using either (A) anti-RAP80 or (B) anti-BLM (NB 100–161) antibody or their corresponding IgG. Cells were grown in three conditions, namely Asynchronous (Asyn.), after HU treatment (+HU) and in postwash (+HU/PW) condition. Western blotting on the immunoprecipitates was carried out with antibodies against anti-BLM (NB 100-161) and anti-RAP80. (C) BLM and RAP80 colocalize after replication arrest. GFP-BLM cells were either grown under asynchronous conditions or treated with HU (+HU). GFP-BLM cells were co-stained with anti-RAP80 antibody. Scale 5 μM. (D) RAP80 interacts with BLM via its UIM after exposure to HU. 293T cells were transfected with either EGFP-tagged wild-type RAP80 or its ΔUIM mutant. (Left) Whole cell lysates were probed with antibodies against BLM (A300-110A) or GFP. CR band represents a cross-reactive band. (Right) Immunoprecipitations were carried out with antibodies against RAP80 or IgG control. Immunoprecipitates were probed with antibodies against BLM (A300-110A) and RAP80. IgG acted as a control for the usage of equal amount of antibody during immunoprecipitation. (E) Overexpression of RAP80 stabilizes BLM. 293T cells were either left untransfected or transfected with either Flag-BLM or GFP-RAP80 or both. Whole cell lysates were probed with antibodies against BLM (NB 100-161), GFP and hsp90. (*) is a cross contaminating band. (F) Ablation of RAP80 destabilizes BLM. GFP-BLM cells were transfected with siRNA control, siRNA BLM, siRNA RAP80 or siRNA BLM and siRNA RAP80. Whole cell lysates were probed with antibodies against BLM (A300-110A), RAP80 and hsp90. (G, H) Destabilization of BLM level after ablation of RAP80 is reversed after treatment with proteasome inhibitors. GFP-BLM cells were either transfected with either siRNA control, or siRNA RAP80 and grown in the absence or presence of (G) LLnL or (H) MG132. Whole cell lysates were probed with antibodies against BLM (A300-110A), RAP80 and hsp90. (I) BLM physically interacts with RAP80. (Top) Glutathione-Sepharose bound GST or GST-BLM (1–1417) was incubated with equal amounts of the lysates from 293T cells transfected with GFP-RAP80. The bound GFP-RAP80 was probed with antibody against GFP. (Bottom) Coomassie gel indicating purified GST and GST-BLM (1–1417). (J) Interaction between GST-BLM and the internal deletions and end fusion proteins of RAP80. (Left) Western blot depicting the expression of the wild-type RAP80 and the RAP80 internal deletions and the end fusion constructs as detected after probing with antibodies against GFP. (Right) Bead bound purified GST-BLM was incubated with lysates expressing EGFP-tagged RAP80 mutants. Bound RAP80 was detected by probing with anti-GFP antibody. (*) represents the EGFP-RAP80 fragments, which interact with GST-BLM.
Source data for this figure is available on the online supplementary information page.
Unexpectedly, we noticed that irrespective of its ability to bind ubiquitin, increasing expression levels of RAP80 stabilized endogenous BLM protein (Figure 6E), while depletion of RAP80 resulted in a dramatic reduction in the stability of BLM (Figure 6F). These results suggest that RAP80 is also required to maintain cellular pools of BLM protein independently of its function to facilitate its relocalization to sites of DNA damage. Furthermore, the expression of BLM in RAP80 siRNA-treated cells could be partially rescued with two proteasomal inhibitors, LLnL and MG132 (Figure 6G and H), indicating that BLM is targeted for destruction by the proteasome in the absence of RAP80. Loss of the UIM domains of RAP80 had little effect on BLM stability (Figure 6D). This suggests that BLM and RAP80 can bind directly in the absence of ubiquitin. To determine whether this was indeed the case, recombinant GST-tagged full-length BLM was incubated with overexpressed full-length GFP-RAP80. Consistent with our previous observations demonstrating that RAP80 and BLM associate together in undamaged cells (Figure 6A and B), we could detect a robust direct interaction between BLM and RAP80 in vitro (Figure 6I). Subsequently, a series of increasing N- and C-terminal deletion mutants of RAP80 were overexpressed and used in combination with FL-BLM in order to narrow down the binding site of BLM on RAP80. No single deletion mutant of RAP80 completely compromised its association with BLM possibly indicating the existence of multiple binding sites. Given that each RAP80 deletion mutant always contained either the N- or C-terminal ends, it seemed likely that BLM could independently bind to each of these regions. In keeping with this an RAP80 mutant protein lacking both N- and C-terminal ends (i.e., contains only amino acids 80–583) but retaining both its UIMs and ZnF domains failed to maintain its ability to bind BLM, whereas another RAP80 mutant protein with amino acids 80–583 deleted (Δ80–583) still robustly bound BLM (Figure 6J). Taken together, these observations suggest that RAP80 stabilizes BLM in undamaged cells and subsequently triggers its relocalization to sites of DNA damage.
RNF8-/RNF168-dependent ubiquitylation and recruitment of BLM is required to suppress HR at stalled replication forks
Since we have demonstrated that RNF8/RNF168 are required to localize BLM to sites of HU-induced DNA damage, it can be hypothesized that cells lacking these E3 ubiquitin ligases would display an elevated level of HR in response to HU, as observed in BS cells. In keeping with the proposed role for these enzymes in regulating BLM recruitment, cells depleted of either RNF8 or RNF168 exhibited an increased level of HR as judged by either a plasmid-based HR assay or SCEs (Figure 7B and C; Supplementary Figure 3A). The elevation in levels of SCEs induced by HU treatment was also evident in RIDDLE syndrome cells complemented with empty vector but not with WT RNF168 (Supplementary Figure 3B). Importantly, in keeping with an essential requirement for the RNF8-/RNF168-dependent ubiquitylation of BLM after DNA damage for its function, wild-type BLM but not the BLM 3K-R mutant was able to suppress HR in BS cells (Figure 7D).
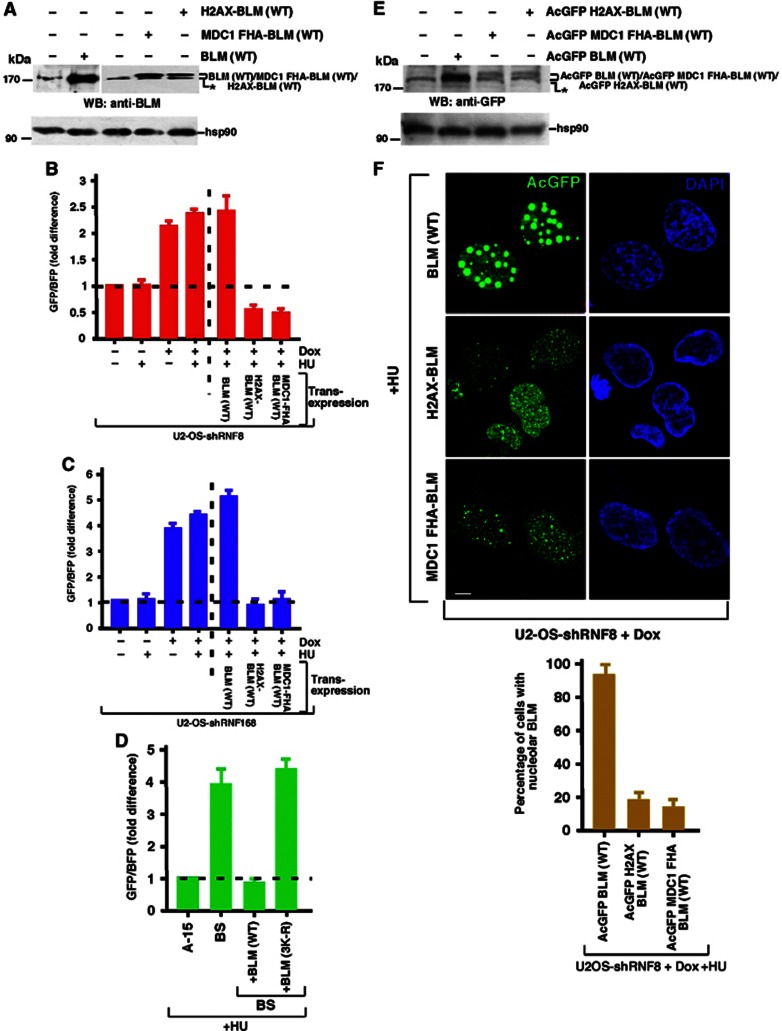
Figure 7.
Targeting of BLM to the chromatin in the absence of RNF8/RNF168 decreases HR. (A, E) Expression of chromatin targeted BLM fusions. (A) Untagged BLM (WT), H2AX-BLM (WT) or MDC1 FHA-BLM (WT) (cloned in pIRES vector) (E) AcGFP-tagged BLM (WT), H2AX-BLM (WT) or MDC1 FHA-BLM (WT) (cloned in AcGFP-N1 vector) were expressed in 293T cells. Western analysis was carried out with antibodies against (A) BLM (A300-110A) and hsp90 and (E) GFP and hsp90. (B, C) Chromatin targeted BLM rescues the hyper-recombinogenic phenotype of cells not expressing either RNF8 or RNF168. (B) U2OS shRNF8 or (C) U2OS shRNF168 cells were grown with or without Dox treatment. The cells were either transfected with pBHRF construct alone or along with BLM, MDC1 FHA-BLM or H2AX-BLM. Forty eight hours post transfection, the host cell reactivation assays were carried out after treatment with HU for the final 16 h. The GFP/BFP ratio was determined as the readout for the extent of HR. (D) Lack of BLM ubiquitylation prevents its anti-recombinogenic function. A-15/BS cells were transfected with pBHRF. In BS cells, co-transfection was carried out with either wild-type BLM or the BLM (3K-R) mutant. Forty eight hours post transfection, the host cell reactivation assays were carried out after treatment with HU for the final 16 h. The GFP/BFP ratio was determined as the readout for the extent of HR. (F) Direct chromatin targeting BLM rescues the loss of ubiquitylation machinery. (Top) BLM fusions (MDC1 FHA-EGFP-BLM and H2AX-EGFP-BLM) were transiently transfected into U2OS shRNF8 cells in the presence of Dox. Transfection was also carried out in parallel with BLM (WT). The cells were treated with HU. EGFP-BLM expression of the fusion proteins was tracked. Nuclei were stained by DAPI. Scale 5 μM. (Bottom) The percentage of cells with nucleolar BLM was determined from the above experiment.
Source data for this figure is available on the online supplementary information page.
Given that the underlying cause of the elevated HR in cells lacking RNF8/RNF168 could be in part explained by an inability of these cells to properly recruit BLM to regions of chromatin proximal to a stalled replication fork, it is conceivable that this could be corrected by forcing BLM onto chromatin and bypassing the need for the UbS-DDR. Based on this premise, two BLM fusion constructs were created, one that was fused to histone H2AX (H2AX-BLM) and the other that was fused to the FHA domain of MDC1 (MDC1-FHA-BLM). The rationale for the creation of these mutants is that H2AX-BLM should be constitutively bound to chromatin whereas the MDC1-FHA-BLM should only be relocalized to chromatin following the induction of DNA damage through its ability to bind to phosphorylated MDC1 located at sites of DNA damage. However, both these BLM fusion proteins circumvent the need for RNF8-/RNF168-dependent ubiquitylation to promote chromatin binding. Strikingly, both the H2AX-BLM or MDC1-FHA-BLM fusions completely suppressed the exacerbated levels of HR caused by depletion of either RNF8 or RNF168 (Figure 7A–C). Importantly, both the fusions, which cannot be ubiquitylated due to the lack of the E3 ligases, could however restore the nuclear relocalization of BLM to sites of DNA damage (Figure 7E and F). Taken together, these data strongly support a role of RNF8 and RNF168 in preventing excessive HR at sites of stalled or compromised replication forks through their ability to stimulate RAP80-dependent recruitment of ubiquitylated BLM.
Discussion
It is becoming clear that the ubiquitin-proteasome system (UPS) is a potent regulator of the cellular DNA damage response, in both its capacity to control protein degradation and also to stimulate the recruitment of repair factors to sites of genetic damage. While the protein degradation versus protein recruitment functions of the UPS are principally controlled at the level of ubiquitin chain linkage, it has been demonstrated that the DNA damage-associated E3 ubiquitin ligases RNF8/RNF168 can catalyse both K48- and K63-linked ubiquitin chains and therefore can mediate contrasting ubiquitin-dependent roles during the DDR. To highlight this, while it has been shown that K63-linked poly-ubiquitylation of H2A-type histones surrounding a DNA DSB is required to mediate recruitment of the BRCA1-A complex (Kolas et al, 2007; Mailand et al, 2007; Wang et al, 2007; Doil et al, 2009; Stewart et al, 2009; Mattiroli et al, 2012), RNF8-/RNF168-dependent degradation of the Jumonji/Tudor-domain containing proteins JMJD2A and JMJD2B is essential for exposure of the epigenetic mark on histone H4 that triggers 53BP1 relocalization (Mallette et al, 2012).
The role of these E3 ubiquitin ligases in regulating protein recruitment/degradation following the induction of replication stress is less well understood. Both RNF8 and RNF168 have been implicated in mediating the ubiquitylation of H2AX following HU exposure (Sy et al, 2011). However, only RNF8 has been proposed to have a functional role during the HU-induced DDR through its ability to promote the recruitment of RAD51 (Sy et al, 2011). While the underlying disparity between the requirement for RNF8 but not RNF168 during the cellular response to HU is unclear, the role of RNF8 is inconsistent with the observation that the recruitment of 53BP1, which is known to be RNF8/RNF168 dependent, functions to suppress HR.
In spite of these conflicting observations, it has been shown that the anti-recombinogenic activity of 53BP1 is important for preventing excessive/abnormal RAD51-dependent HR at sites of stalled replication forks (Tripathi et al, 2007; Bunting et al, 2010), an activity that is strikingly similar to that of BLM helicase. Consistent with a link between these repair factors, it has been demonstrated that following exposure to HU, BLM in conjunction with 53BP1 functions to inhibit RAD51 nucleofilament formation and SCE (Tripathi et al, 2007, 2008). Given the absolute requirement for the RNF8/RNF168 pathway to stimulate the relocalization of 53BP1 after the induction of DNA damage (Kolas et al, 2007; Mailand et al, 2007; Wang et al, 2007; Doil et al, 2009; Stewart et al, 2009; Mattiroli et al, 2012) and the functional and biochemical interaction between 53BP1 and BLM (Tripathi et al, 2007, 2008), it is hypothesized that the ubiquitin-dependent DDR may function to target BLM to damaged replication forks to control illegitimate recombination (summarized in Figure 8). In keeping with this hypothesis, we have demonstrated that BLM is poly-ubiquitylated in response to HU treatment by RNF8/ RNF168 (Figure 3) and that this promotes its targeting to sites of stalled replication forks via an interaction with the UIM domains of RAP80 (Figure 6D). Furthermore, we have shown that the RNF8-directed E3 ubiquitin ligase cascade as well as the ubiquitylation of BLM itself is essential to suppress RAD51-dependent HR after replication fork stalling/collapse (Figure 7). In light of these results, it raises the question as to whether the RNF8-/RNF168-dependent DNA damage response functions upstream of the BLM helicase to promote its recruitment to sites of DNA DSBs and if so, then is this function required to promote or suppress HR-mediated repair? This issue is complicated by the fact that it has been shown that BLM is required to stimulate end-resection of a DSB and thereby promote HR (Gravel et al, 2008; Nimonkar et al, 2008, 2011), whereas in contrast the RNF8-/RNF168-dependent relocalization of 53BP1 plays a role in limiting DNA end-resection and HR (Bunting et al, 2010; Bothmer et al, 2011). Moreover, given that BLM can function as both a pro-recombinogenic (Gravel et al, 2008; Nimonkar et al, 2008, 2011) and anti-recombinogenic factor during later stages of homology-directed DSB repair (Bischof et al, 2001; Sengupta et al, 2003; Bugreev et al, 2007; Tripathi et al, 2007, 2008; Srivastava et al, 2009), it is conceivable that it may also be required to moderate the levels of HR. In support of a role for RNF8 and RNF168 in facilitating the relocalization of BLM to DSBs, cells depleted of either E3 ligase fail to properly form BLM foci following exposure to laser-generated DSB tracks (Supplementary Figure 4).
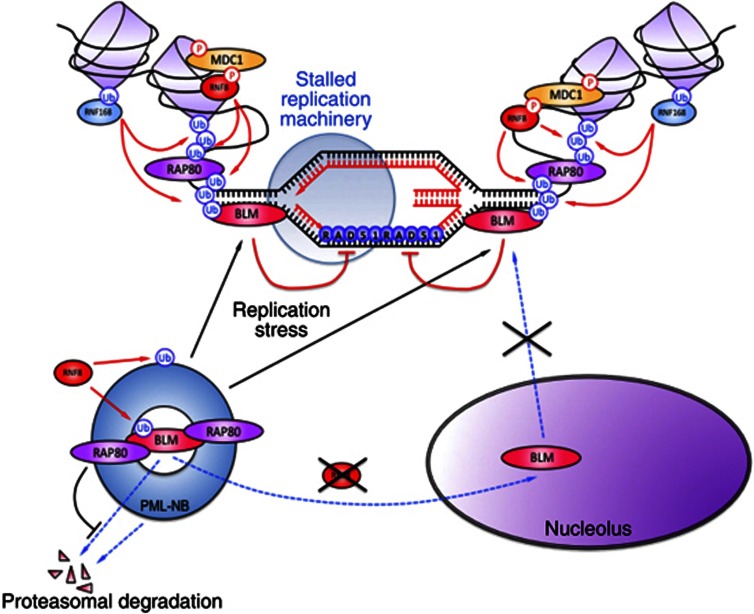
Figure 8.
Schematic diagram depicting the recruitment of BLM to the chromatin after DNA damage. In the absence of DNA damage, BLM remains in the PML nuclear bodies (PML-NBs) complexed with RAP80. Absence of RNF8 prevents the mono-ubiquitylation of BLM and poly-ubiquitylation of PML-NBs. These ubiquitylation events together result in the disruption of the PML-NBs and accumulation of BLM in the nucleolus. Ablation of RAP80 leads to proteasomal degradation of BLM. In the presence of replication stress, BLM is poly-ubiquitylated in the PML-NBs by RNF8. The poly-ubiquitylated BLM interacts with the ubiquitin-interacting motifs (UIMs) of RAP80, which recruits BLM to the site of damage on the chromatin. At the site of the lesion, BLM is acted on sequentially by RNF8 and RNF168. These two E3 ligases amplify the ubiquitylation signal and retain BLM at the site of the damage, so that BLM can subsequently carry out its functions during HRR.
Currently, it is unclear if and how lack of BLM-dependent DNA damage response contributes to clinical and cellular phenotype exhibited by patients with RNF168 dysfunction. However, one could postulate that a defect in the reversal of damaged DNA caused by a failure to recruit various repair proteins to the sites of the DNA lesion resulting from a loss of RNF168 may be offset by the elevated levels of HR attributed to an inability to properly relocalize BLM. As a consequence, cells lacking RNF168 will exhibit only a mild deficiency in DNA repair that is perhaps insufficient on its own to compromise genomic integrity and initiate tumourigenesis. Consistent with this idea, loss of RNF168 results in an extremely mild hypersensitivity to IR, UV and HU and unless combined with p53 dysfunction does not significantly predispose to the development of tumours (Bohgaki et al, 2011).
One unexpected finding from this work was that RAP80 has two separable roles in regulating BLM. Following exposure to DNA damage, the RNF8-/RNF168-dependent ubiquitylation of BLM potentiates its recruitment to sites of stalled replication forks by an increased association with the UIMs of RAP80 (Figure 6D). Interestingly, we uncovered a second role for RAP80 in maintaining BLM stability in the absence of genotoxic stress. Importantly, this function of RAP80 is independent of its ability to bind poly-ubiquitin residues. The ability of multiple proteasomal inhibitors to rescue the levels of BLM protein in RAP80-depleted cells (Figure 6E and H) indicates that in the absence of its binding to RAP80, BLM is targeted for degradation in a ubiquitin-/proteasome-dependent manner. Currently, the identity of the ubiquitin ligase(s) that mediates the destruction of BLM in the absence of RAP80 is unclear. However, it has been recently demonstrated that BLM is degraded during adenoviral infection by the E1B/E4orf6 proteins, which hijacks the cellular SCF E3 ubiquitin ligase complexes to facilitate removal of potentially deleterious host cell proteins (Orazio et al, 2011). This indicates that at least under certain circumstances BLM can be ubiquitylated by SCF-type ubiquitin ligases.
The identification that the stability of BLM is controlled by RAP80 would subsequently indicate that cells lacking RAP80 would resemble those in which the expression of BLM is compromised. Consistent with this premise, cells treated with RAP80-specific siRNA exhibit increased levels of HR and SCEs (Coleman and Greenberg, 2011), as also seen in BS cells. While it has been proposed that this is in part due to uncontrolled Mre11/CtIP-dependent DSB end-resection, it is conceivable that a loss of BLM’s anti-recombinogenic function also contributes to this phenotype, especially during the later stages of repair. Based on these observations, it is tempting to speculate that patients with deficiencies in RAP80 would exhibit both clinical and cellular features that overlap significantly with those resulting from loss of BLM. However, it should also be noted that the RAP80 knockout mice (Yin et al, 2012) have a much milder phenotype than BLM knockout mice (Luo et al, 2000). This may indicate that while RAP80 may stabilize BLM and be responsible for some of its functions, it is not essential for all BLM functions.
Another interesting aspect of our work is the observation that RNF8 but not RNF168 plays an additional role in regulating the nuclear localization of BLM in the absence of DNA damage. We demonstrated that loss of RNF8 results in BLM being predominantly relocalized from the nucleoplasm and PML-NBs to the nucleoli, indicating that RNF8 is required to properly localize BLM by ubiquitylating BLM (Figures 1C, D and 2A; Supplementary Figure 1C) and PML (Figure 5B, C). The observation that BLM lacking the sites of RNF8-/RNF168-dependent ubiquitylation also relocalized to the nucleoli (Figure 4D and E) suggests that a basal level of BLM mono-ubiquitylation mediated by RNF8 (Figure 3A) is a critical regulatory mechanism for controlling the intracellular distribution of the helicase. Hence, it is possible that the ubiquitylation of BLM on its N-terminus by RNF8 provides a docking site for a ubiquitin-binding protein located within the PML-NBs or that this post-translational modification masks a cryptic nucleolar targeting sequence that, unless RNF8 is absent, is otherwise hidden. It is conceivable that the ability of the N-terminal region of BLM to directly bind to RNF8 (Supplementary Figure 2E) may also contribute to this. However, we did note that cells lacking RNF8 also exhibited a reduction in the levels of endogenous PML and as a consequence also the numbers of PML-NBs (Figure 5A and B). It has been previously shown that the lack of N-terminal 133–237 amino acids in BLM (critical for BLM localization to PML NBs) leads to its sequestration in the nucleolus (Wang et al, 2001). This indicates that nucleolus is the preferred alternate storage site for BLM when it cannot localize to PML-NBs. Consistent with this premise, genetic ablation of PML in human fibroblasts also results in the nucleolar targeting of BLM (Figure 5D), similar to that which we observed in RNF8-depleted cells (Figure 1C and D). Hence, the post-translational modification of BLM as well as the integrity of the PML-NBs may together contribute for the redistribution of BLM to the nucleolus.
Taken together, it is clear that the regulation of BLM is intimately entwined with the UPS. As a consequence, it is likely that de-ubiquitylating enzymes (DUBs) will also play an important role. The demonstration that BLM stability is controlled by RAP80 may suggest that an RAP80-associated DUB, such as BRCC36 could be involved in removing unwanted K48-linked poly-ubiquitin chains conjugated to BLM that would target it for proteasomal degradation. Interestingly, loss of BRCC36, in a manner similar to that of RAP80 and BLM, also results in elevated levels of HR and the increased generation of SCEs (Coleman and Greenberg, 2011; Hu et al, 2011). It is also possible that BRCC36 alone or perhaps in combination with other DUBs implicated in dampening the UbS-DDR, such as USP3 and OTUB1, will also be involved in removing BLM from sites of DNA damage following the resolution of HR intermediates.
In conclusion, our work has identified a novel link between BLM and the ubiquitin system that is likely to reveal important regulatory mechanisms that are vital to the functions of BLM in maintaining genome stability and suppressing tumourigenesis. In future, we envisage the complete linkage of the various BLM post-translational modifications (like ubiquitylation, phosphorylation and SUMOylation) with the varied BLM functions will allow a better appreciation of the in vivo roles of this important caretaker tumour suppressor.
Materials and methods
Antibodies
Anti-BLM: rabbit polyclonal NB 100-161 (Novus Biologicals); rabbit polyclonal A300-110A (Bethyl Laboratories); goat polyclonal A300-120A (Bethyl Laboratories). Anti-Ub: K63 linkage-specific mouse monoclonal BML-PW0600 (Enzo Biosciences); FK1 mouse monoclonal BML-PW8805 (Enzo Biosciences); FK2 mouse monoclonal BML-PW8810 (Enzo Biosciences); mouse monoclonal (P4D1) sc-8017 (Santa Cruz Biotechnology). Anti-RAP80: rabbit polyclonal A300-763A (Bethyl Laboratories). Anti-GFP: mouse monoclonal 33-2600 (Invitrogen); mouse monoclonal 632375 (Clontech). Anti-hsp90 α/β: rabbit polyclonal (H-114) sc-7947 (Santa Cruz Biotechnology). Anti-Ubc5a: rabbit polyclonal A-615 (Boston Biochem). Anti-Ubc13: mouse monoclonal [4E11] (ab38795) (Abcam). Anti-RNF8: mouse monoclonal (B-2) sc-271462 (Santa Cruz Biotechnology). Anti-RNF168: mouse monoclonal (B-11) sc-101125 (Santa Cruz Biotechnology). Anti-Lamin A/C: mouse monoclonal (clone 14) #05-714 (Upstate). Anti-PML: mouse monoclonal (PG-M3) sc-966 (Santa Cruz Biotechnology); goat polyclonal (A-20) sc-9863 (Santa Cruz Biotechnology). Anti-HA: rabbit polyclonal 71-5500 (Invitrogen). Anti-His: rabbit polyclonal 552565 (BD Pharmingen). Anti 53BP1: mouse monoclonal 612523 (BD Pharmingen).
Recombinants
pGEX4T-1 BLM (1–212) (donated by Ian Hickson). pEGFP-C1 BLM (WT) (donated by Nathan Ellis). EGFP-RAP80 and EGFP RAP80 (ΔUIM) cloned in MSCV vector (donated by Steve Elledge and Bin Wang). pEGFP-C1-RAP80 (donated by Roger Greenberg). pBHRF (donated by Robbert Slebos). pGEX4T-1 BLM (191–660), pGEX4T-1 BLM (621–1041), pGEX4T-1 BLM (1001–1417) (Tripathi et al, 2008). pGEX4T-1 BLM (1–1417) (Srivastava et al, 2009). pLVX-AcGFP-N1-BLM (WT) was obtained by cloning the respective PCR fragment into the EcoR1 and BamH1 sites of the vector. For this purpose, a BLM cDNA was used in which the BamH1 site (at nucleotide 4132) and EcoR1 site (at nucleotide 1341) were destroyed by site-directed mutagenesis (using a kit by Stratagene), without any change in the amino-acid sequence. Both pLVX-AcGFP-N1-MDC1 FHA-BLM (WT) and pLVX-AcGFP-N1-H2AX BLM (WT) were obtained by a two-step cloning strategy: (A) PCR product encoding the MDC1 FHA or H2AX coding regions was cloned into pLVX-AcGFP-N1 at XhoI and EcoR1 sites to generate pLVX-AcGFP-N1-MDC1 FHA and pLVX-AcGFP-N1-H2AX recombinants, respectively; (B) PCR product for the full-length BLM (with destroyed EcoR1 and BamH1 sites) was sub-cloned in frame with either MDC1-FHA or H2AX at EcoR1 and BamH1 sites of pLVX-AcGFP-N1-MDC1 FHA and pLVX-AcGFP-N1-H2AX to generate pLVX-AcGFP-N1-MDC1 FHA-BLM (WT) and pLVX-AcGFP-N1-H2AX BLM (WT). pIRES-BLM (WT), pIRES-MDC1 FHA-BLM (WT) and pIRES-H2AX BLM (WT) were obtained by using pLVX-AcGFP-N1-BLM (WT), pLVX-AcGFP-N1-MDC1 FHA-BLM (WT) and pLVX-AcGFP-N1-H2AX BLM (WT) as the template and cloning the respective PCR fragments into the SpeI and NotI sites of the pIRES vector. pGEX4T-1 BLM (1–1417) (K105R), pGEX4T-1 BLM (1–1417) (K225R), pGEX4T-1 BLM (1–1417) (K259R), pGEX4T-1 BLM (1–1417) (K105R, K225R, K259R), that is, pGEX4T-1 BLM (1–1417) (3K-R), pEGFP-C1 BLM (K105R), pEGFP-C1 BLM (K225R), pEGFP-C1 BLM (K259R), pEGFP-C1 BLM (K105R, K225R, K259R), that is, pEGFP-C1 BLM (3K-R) were generated by site-directed mutagenesis using a Stratagene kit. All the pEGFP-BLM mutants were made on wild-type background. pEGFP C1-RAP80 (1–79), pEGFP C1-RAP80 (1–124), pEGFP C1-RAP80 (125–504) and pEGFP C1-RAP80 (80–583) were generated by cloning the respective PCR products into the EcoR1 and BamH1 sites of pEGFP-C1. For the internal deletions, pEGFP-C1 RAP80 (Δ80–583), pEGFP-C1 RAP80 (Δ125–583), pEGFP-C1 RAP80 (Δ80–504) and pEGFP-C1 RAP80 (Δ125–504) were generated by taking either the pEGFP C1-RAP80 (1–79) or pEGFP C1-RAP80 (1–124) as the cloning vector. These recombinants were digested with only BamH1 and respective PCR products coding the C-terminal regions of RAP80 were cloned in frame with the N-terminal regions at the BamH1 site of pEGFP-C1 vector (and checked for the right orientation). The His-tagged wild-type RNF8 and RNF8 C403S expression constructs were provided by Dan Durocher. The cloning of the GST-RNF168 expression vector has been previously described (Stewart et al, 2009). His-tagged PML III and PML IV were generated by cloning the respective PCR fragments into the EcoRI/NotI sites of pET28b.
IF and laser microirradiation
IF was done according to the published protocols (De et al, 2012). Fixations were carried out with either 100% ethanol (in case of a prelysis protocol that removes all the soluble nuclear proteins) or by 4% paraformaldehyde. Subsequent to fixation by either of the above reagents, the cells were rinsed three times with 1 × PBS, twice treated with 0.1%Triton X-100 for 5 min each, washed with 1 × PBS with 0.5% Tween-20 and blocked overnight with 10% normal chicken serum. Subsequently, the staining was carried out with the indicated primary antibodies at 37°C for 1 h in a humidified chamber. Incubation with secondary antibody was also carried out under similar conditions. Adequate controls like usage of the corresponding IgG or not using secondary antibody were included. At least 150 cells were analysed for all IF experiments. The slides were imaged in a Zeiss 510 Meta system with 63 × /1.4 oil immersion or 40 × /0.95 Corr objective. The laser lines used were Argon 458/477/488/514 nm (for FITC), DPSS 561 nm (for Texas Red and Mitotracker dyes) and a Chameleon Ultra autotunable femtosecond laser with a tuning range of 690–1050, nm (for DAPI). LSM5 software was used for image acquisition. Laser micro-irradiation of cells was carried out as previously described (Lukas et al, 2003). The numbers in the combined panel denote the percentage of the cells that show colocalization between the two proteins.
In vitro ubiquitylation assays
This method was adapted from Stewart et al (2009). Ubiquitin ligase assays were performed using 250 ng of soluble substrate (BLM or PML isoforms). Assays were set up at 37°C for 3 h in a total volume of 25 μl in 50 mM Tris–HCl (pH 8.0) and 1 mM DTT. Recombinant GST-RNF168/His-RNF8 (0.2 μM) was added to 0.4 μM of the indicated E2 enzymes Ubc5a or Ubc13 (Boston Biochem), 0.0125 μM E1 and 16 μM ubiquitin (Boston Biochem). Reactions were initiated by the addition of ATP (2 mM) and MgCl2 (5 mM). The reactions were stopped by boiling with 2 × SDS loading dye and samples were run on Nu-PAGE 4–12% gels in 1 × MOPS buffer (Invitrogen), transferred onto a nitrocellulose membrane and probed with the indicated antibodies. Ubiquitylated BLM was detected with goat polyclonal antibody A300-120A. The ubiquitylated forms of BLM or PML isoforms have been designated. Mono, poly and multi represent the ubiquitin linkages formed on BLM or PML isoforms. Multiple anti-ubiquitin antibodies (P4D1, FK1, FK2) were used to distinguish whether the high molecular weight species of BLM observed in the ubiquitylation reactions were due to multiple sites of mono-ubiquitylation versus poly-ubiquitylation.
Supplementary Material
Acknowledgments
We would like to acknowledge Ian Hickson, Nathan Ellis, Robbert Slebos, Jerry Shay, Steve Elledge, Bin Wang, Roger Greenberg, Dan Durocher and Jiri Lukas for plasmids and cells. SS would like to acknowledge National Institute of Immunology core funds, Department of Biotechnology (DBT), India (BT/PR11258/BRB/10/645/2008), Department of Science and Technology (DST), India (SR/SO/BB-08/2010), Indo-French Centre for the Promotion of Advanced Research (IFCPAR) (IFC/4603-A/2011/1250) and Council of Scientific and Industrial Research (CSIR), India [37(1541)/12/EMR-II] for financial assistance. GS, ES and KT are funded by a CR-UK Senior Fellowship (C17183/A13030). AZ is funded by a Lister Institute Prize and the University of Birmingham. We thank Siddharth De for help during confocal imaging and analysis.
Author contributions: ST, VM, MH, ESM, PA, AZ, PM and KT carried out the experiment work. GSS and SS analysed the data and wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bekker-Jensen S, Rendtlew Danielsen J, Fugger K, Gromova I, Nerstedt A, Lukas C, Bartek J, Lukas J, Mailand N (2010) HERC2 coordinates ubiquitin-dependent assembly of DNA repair factors on damaged chromosomes. Nat Cell Biol 12: 80–86,sup pp 1–12 [DOI] [PubMed] [Google Scholar]
- Bischof O, Kim SH, Irving J, Beresten S, Ellis NA, Campisi J (2001) Regulation and localization of the Bloom syndrome protein in response to DNA damage. J Cell Biol 153: 367–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohgaki T, Bohgaki M, Cardoso R, Panier S, Zeegers D, Li L, Stewart GS, Sanchez O, Hande MP, Durocher D, Hakem A, Hakem R (2011) Genomic instability, defective spermatogenesis, immunodeficiency, and cancer in a mouse model of the RIDDLE syndrome. PLoS Genet 7: e1001381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothmer A, Robbiani DF, Di Virgilio M, Bunting SF, Klein IA, Feldhahn N, Barlow J, Chen HT, Bosque D, Callen E, Nussenzweig A, Nussenzweig MC (2011) Regulation of DNA end joining, resection, and immunoglobulin class switch recombination by 53BP1. Mol Cell 42: 319–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugreev DV, Yu X, Egelman EH, Mazin AV (2007) Novel pro- and anti-recombination activities of the Bloom's syndrome helicase. Genes Dev 21: 3085–3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, Xu X, Deng CX, Finkel T, Nussenzweig M, Stark JM, Nussenzweig A (2010) 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141: 243–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman KA, Greenberg RA (2011) The BRCA1-RAP80 complex regulates DNA repair mechanism utilization by restricting end resection. J Biol Chem 286: 13669–13680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies SL, North PS, Dart A, Lakin ND, Hickson ID (2004) Phosphorylation of the Bloom's syndrome helicase and its role in recovery from S-phase arrest. Mol Cell Biol 24: 1279–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De S, Kumari J, Mudgal R, Modi P, Gupta S, Futami K, Goto H, Lindor NM, Furuichi Y, Mohanty D, Sengupta S (2012) RECQL4 is essential for the transport of p53 to mitochondria in normal human cells in the absence of exogenous stress. J Cell Sci 125: Pt 102509–2522 [DOI] [PubMed] [Google Scholar]
- Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, Ellenberg J, Panier S, Durocher D, Bartek J, Lukas J, Lukas C (2009) RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell 136: 435–446 [DOI] [PubMed] [Google Scholar]
- Eladad S, Ye TZ, Hu P, Leversha M, Beresten S, Matunis MJ, Ellis NA (2005) Intra-nuclear trafficking of the BLM helicase to DNA damage-induced foci is regulated by SUMO modification. Hum Mol Genet 14: 1351–1365 [DOI] [PubMed] [Google Scholar]
- Ellis NA, German J (1996) Molecular genetics of Bloom's syndrome. Hum Mol Genet 5(Spec No) 1457–1463 [DOI] [PubMed] [Google Scholar]
- Galanty Y, Belotserkovskaya R, Coates J, Polo S, Miller KM, Jackson SP (2009) Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature 462: 935–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel S, Chapman JR, Magill C, Jackson SP (2008) DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev 22: 2767–2772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickson ID, Mankouri HW (2011) Processing of homologous recombination repair intermediates by the Sgs1-Top3-Rmi1 and Mus81-Mms4 complexes. Cell Cycle 10: 3078–3085 [DOI] [PubMed] [Google Scholar]
- Hu Y, Scully R, Sobhian B, Xie A, Shestakova E, Livingston DM (2011) RAP80-directed tuning of BRCA1 homologous recombination function at ionizing radiation-induced nuclear foci. Genes Dev 25: 685–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur S, Modi P, Srivastava V, Mudgal R, Tikoo S, Arora P, Mohanty D, Sengupta S (2010) Chk1-dependent constitutive phosphorylation of BLM helicase at serine 646 decreases after DNA damage. Mol Cancer Res 8: 1234–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Chen J, Yu X (2007) Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science 316: 1202–1205 [DOI] [PubMed] [Google Scholar]
- Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, Panier S, Mendez M, Wildenhain J, Thomson TM, Pelletier L, Jackson SP, Durocher D (2007) Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 318: 1637–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas C, Falck J, Bartkova J, Bartek J, Lukas J (2003) Distinct spatiotemporal dynamics of mammalian checkpoint regulators induced by DNA damage. Nat Cell Biol 5: 255–260 [DOI] [PubMed] [Google Scholar]
- Luo G, Santoro IM, McDaniel LD, Nishijima I, Mills M, Youssoufian H, Vogel H, Schultz RA, Bradley A (2000) Cancer predisposition caused by elevated mitotic recombination in Bloom mice. Nat Genet 26: 424–429 [DOI] [PubMed] [Google Scholar]
- Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J (2007) RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 131: 887–900 [DOI] [PubMed] [Google Scholar]
- Mallette FA, Mattiroli F, Cui G, Young LC, Hendzel MJ, Mer G, Sixma TK, Richard S (2012) RNF8- and RNF168-dependent degradation of KDM4A/JMJD2A triggers 53BP1 recruitment to DNA damage sites. EMBO J 31: 1865–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattiroli F, Vissers JH, van Dijk WJ, Ikpa P, Citterio E, Vermeulen W, Marteijn JA, Sixma TK (2012) RNF168 Ubiquitinates K13-15 on H2A/H2AX to Drive DNA Damage Signaling. Cell 150: 1182–1195 [DOI] [PubMed] [Google Scholar]
- Monnat RJ Jr (2010) Human RECQ helicases: roles in DNA metabolism, mutagenesis and cancer biology. Semin Cancer Biol 20: 329–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimonkar AV, Genschel J, Kinoshita E, Polaczek P, Campbell JL, Wyman C, Modrich P, Kowalczykowski SC (2011) BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev 25: 350–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimonkar AV, Ozsoy AZ, Genschel J, Modrich P, Kowalczykowski SC (2008) Human exonuclease 1 and BLM helicase interact to resect DNA and initiate DNA repair. Proc Natl Acad Sci USA 105: 16906–16911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orazio NI, Naeger CM, Karlseder J, Weitzman MD (2011) The adenovirus E1b55K/E4orf6 complex induces degradation of the Bloom helicase during infection. J Virol 85: 1887–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang KJ, Woo LL, Zhu J, Huo D, Matunis MJ, Ellis NA (2009) SUMO modification regulates BLM and RAD51 interaction at damaged replication forks. PLoS Biol 7: e1000252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S, Linke SP, Pedeux R, Yang Q, Farnsworth J, Garfield SH, Valerie K, Shay JW, Ellis NA, Wasylyk B, Harris CC (2003) BLM helicase-dependent transport of p53 to sites of stalled DNA replication forks modulates homologous recombination. EMBO J 22: 1210–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S, Robles AI, Linke SP, Sinogeeva NI, Zhang R, Pedeux R, Ward IM, Celeste A, Nussenzweig A, Chen J, Halazonetis TD, Harris CC (2004) Functional interaction between BLM helicase and 53BP1 in a Chk1-mediated pathway during S-phase arrest. J Cell Biol 166: 801–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobhian B, Shao G, Lilli DR, Culhane AC, Moreau LA, Xia B, Livingston DM, Greenberg RA (2007) RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science 316: 1198–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava V, Modi P, Tripathi V, Mudgal R, De S, Sengupta S (2009) BLM helicase stimulates the ATPase and chromatin-remodeling activities of RAD54. J Cell Sci 122: Pt 173093–3103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, Nakada S, Ylanko J, Olivarius S, Mendez M, Oldreive C, Wildenhain J, Tagliaferro A, Pelletier L, Taubenheim N, Durandy A, Byrd PJ, Stankovic T, Taylor AM, Durocher D (2009) The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell 136: 420–434 [DOI] [PubMed] [Google Scholar]
- Sy SM, Jiang J, Dong SS, Lok GT, Wu J, Cai H, Yeung ES, Huang J, Chen J, Deng Y, Huen MS (2011) Critical roles of ring finger protein RNF8 in replication stress responses. J Biol Chem 286: 22355–22361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikoo S, Sengupta S (2010) Time to bloom. Genome Integr 1: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V, Kaur S, Sengupta S (2008) Phosphorylation-dependent interactions of BLM and 53BP1 are required for their anti-recombinogenic roles during homologous recombination. Carcinogenesis 29: 52–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V, Nagarjuna T, Sengupta S (2007) BLM helicase-dependent and -independent roles of 53BP1 during replication stress-mediated homologous recombination. J Cell Biol 178: 9–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Matsuoka S, Ballif BA, Zhang D, Smogorzewska A, Gygi SP, Elledge SJ (2007) Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science 316: 1194–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XW, Tseng A, Ellis NA, Spillare EA, Linke SP, Robles AI, Seker H, Yang Q, Hu P, Beresten S, Bemmels NA, Garfield S, Harris CC (2001) Functional interaction of p53 and BLM DNA helicase in apoptosis. J Biol Chem 276: 32948–32955 [DOI] [PubMed] [Google Scholar]
- Yankiwski V, Marciniak RA, Guarente L, Neff NF (2000) Nuclear structure in normal and Bloom syndrome cells. Proc Natl Acad Sci USA 97: 5214–5219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z, Menendez D, Resnick MA, French JE, Janardhan KS, Jetten AM (2012) RAP80 is critical in maintaining genomic stability and suppressing tumor development. Cancer Res 72: 5080–5090 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.