Abstract
High-throughput techniques have identified numerous antisense (AS) transcripts and long non-coding RNAs (ncRNAs). However, their significance in cancer biology remains largely unknown. Here, we report an androgen-responsive long ncRNA, CTBP1-AS, located in the AS region of C-terminal binding protein 1 (CTBP1), which is a corepressor for androgen receptor. CTBP1-AS is predominantly localized in the nucleus and its expression is generally upregulated in prostate cancer. CTBP1-AS promotes both hormone-dependent and castration-resistant tumour growth. Mechanistically, CTBP1-AS directly represses CTBP1 expression by recruiting the RNA-binding transcriptional repressor PSF together with histone deacetylases. CTBP1-AS also exhibits global androgen-dependent functions by inhibiting tumour-suppressor genes via the PSF-dependent mechanism thus promoting cell cycle progression. Our findings provide new insights into the functions of ncRNAs that directly contribute to prostate cancer progression.
Keywords: androgen, non-coding RNA, prostate cancer
Introduction
Emerging evidence has shown that long noncoding RNAs (ncRNAs) are widely transcribed across the entire genome (FANTOM Consortium, 2005; Guttman et al, 2009; Kurokawa et al, 2009). Interestingly, antisense (AS) transcripts link adjacent genes in complex loci into chains of linked transcriptional units (Yelin et al, 2003; Katayama et al, 2005; Yu et al, 2008). AS ncRNAs are expected to modulate transcription of the human genome in a similar fashion, but how they are regulated and what biological significance ncRNAs may have remain controversial. The actions of androgen and its cognate nuclear receptor, the androgen receptor (AR), are essential for the development and proliferation of prostate cancer and its subsequent progression to castration-resistant prostate cancer (CRPC) (Chen et al, 2004; Debes and Tindall, 2004; Wang et al, 2009). We employed a combination of AR transcriptional network analysis using cap analysis gene expression (CAGE), mapping of transcriptional start sites (TSSs) regulated by androgen, and chromatin immunoprecipitation with subsequent analysis by whole-genome tiling array using an anti-AR specific antibody (ChIP-chip) to demonstrate that AR-regulated transcripts of unknown function are transcribed from intergenic or AS regions of genes in prostate cancer (Takayama et al, 2007, 2009, 2011). These results indicated that androgen-regulated transcripts may include diverse ncRNAs of unknown function. Moreover, a recent comprehensive analysis of transcripts expressed in prostate cancer tissue samples by RNA sequence discovered that long ncRNAs are dysregulated during prostate cancer development (Prensner et al, 2011).
Here we report a novel function of androgen-responsive ncRNA (designated as CTBP1-AS) located in the AS region of the CTBP1 gene, a transcriptional coregulator. Functional analyses revealed a novel sense–antisense mechanism for CTBP1 repression by CTBP1-AS. We also found that CTBP1-AS promoted AR transcriptional activity and cell cycle for prostate cancer. Moreover, CTBP1-AS promotes both hormone-dependent and castration-resistant tumour growth. We assume that the present findings will reveal the new pathophysiological relevance of ncRNAs that directly contributes to the prostate cancer.
Results
Regulation of a functional AS ncRNA CTBP1-AS by androgen
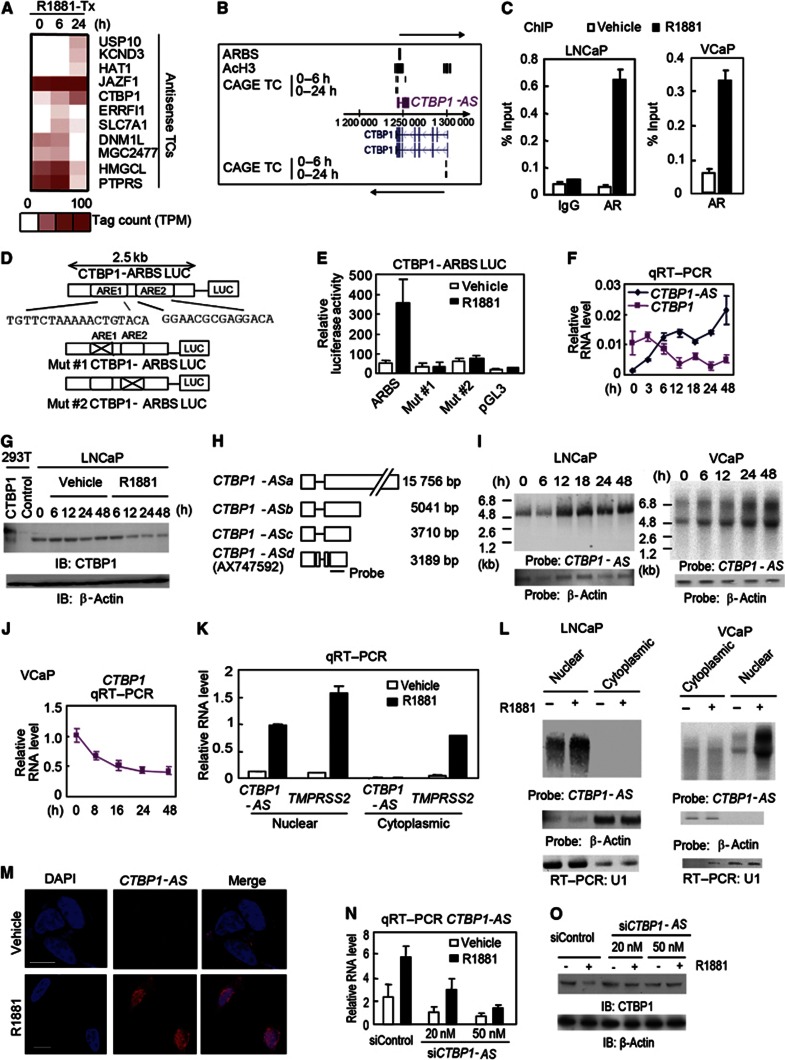
To analyse the roles of AS transcripts in prostate cancer proliferation and progression, we searched for androgen-regulated tag clusters (TCs) situated in the AS regions of the AR-binding RefSeq genes identified by our previous CAGE analysis (Takayama et al, 2011). We identified pairs of sense–antisense-regulated TCs with AR-binding sites (ARBSs) (Figure 1A). Interestingly, we found CTBP1 among the androgen-regulated genes potentially governed by AS TCs. CTBP1 has been reported to function as a corepressor in the nucleus (Shi et al, 2003; Chinnadurai, 2007, 2009) and to be involved in tumour invasion (Bergman et al, 2009). We identified androgen-repressed TCs at the TSS of CTBP1 (Figure 1B). In contrast, an ARBS combined with histone H3 acetylation chromatin status was detected at the 3′-untranslated region (UTR) (Figure 1B). This CTBP1–ARBS genomic region including androgen response element (ARE) sequences showed strong ligand-dependent transcriptional activity (Figure 1C–E). We found that another TC highly upregulated by androgen was detected just downstream of ARBS on the AS strand. By GenBank searching, we found that this TC was located in the exon of AX747592, suggesting that a transcriptional variant of this transcript started from the TC. We confirmed upregulation of CTBP1-AS together with downregulation of CTBP1 by androgen (Figure 1F and G). Although, 3′ RACE (rapid amplification of cDNA ends) PCR analysis revealed that this AS transcript has multiple transcriptional termination sites (Figure 1H), our northern blot analysis revealed that an ∼5-kb transcript is the predominant isoform in LNCaP cells (Figure 1I). In addition, reciprocal regulation of CTBP1 and CTBP1-AS was confirmed in AR-positive VCaP prostate cancer cells (Figure 1I and J). We also found enhanced expression of CTBP1-AS in the nucleus relative to the cytoplasm (Figure 1K and L). Moreover, RNA fluorescence in situ hybridization (FISH) analysis revealed that CTBP1-AS is expressed diffusely throughout the nucleus induced by R1881 treatment (Figure 1M). We investigated the biological significance of CTBP1-AS in prostate cancer cells by short interference RNA (siRNA) targeting CTBP1-AS (Figure 1N). We confirmed that the androgen-dependent decrease in CTBP1 protein levels is abolished by siCTBP1-AS (Figure 1O). In contrast, CTBP1 repression and CTBP1-AS induction were not observed in AR-negative DU145 cells (Supplementary Figure S1). Taken together, these results indicate that CTBP1-AS is an AS long ncRNA regulated by AR in the nucleus repressing the sense CTBP1.
Figure 1.
Identification of a functional AR-regulated AS noncoding RNA in the CTBP1 locus. (A) CAGE TCs significantly regulated by androgen were extracted (Takayama et al, 2011). Among 125 Refseq genes with androgen-regulated AS TCs, 39 genes also had androgen-regulated TCs in the sense direction. Of these sense-AS pairs, 11 included the genes adjacent to ChIP-chip-determined ARBSs (Takayama et al, 2011). The heat map represents CAGE tag counts. (B) Mapping of ChIP-chip and CAGE data in the CTBP1 locus. (C) Chromatin immunoprecipitation (ChIP) analysis of AR. LNCaP or VCaP cells were treated with 1 nM R1881 or vehicle. Cell lysates were immunoprecipitated with rabbit IgG or anti-AR antibody. Bar: s.d. (D, E) CTBP1 ARBS mediates transcriptional activity. A 2-kb fragment that included the CTBP1 ARBS was cloned into the pGL3 luciferase vector (D). Two noncanonical AREs were mutated in the Mut #1 and Mut #2 vectors. A luciferase assay was performed in LNCaP cells with the indicated treatments for 24 h (E). Bar: s.d. (F) Reciprocal CTBP1 and CTBP1-AS expression at the indicated time points after androgen treatment (n=3). (G) Protein level regulation of CTBP1 in LNCaP cells treated with R1881 or vehicle. (H) Schematic view of the CTBP1-AS transcripts obtained by our RACE PCR analysis. The location of the RNA probe is indicated. (I) Northern blot analysis of CTBP1-AS or β-actin by using total RNA from cells treated with androgen. (J) qRT–PCR analysis for transcriptional regulation of CTBP1 in VCaP cells after androgen treatment. Bar: s.d. (K, L) Total RNAs from nuclear and cytoplasmic cell fractions were used for qRT–PCR (n=2) (K) and northern blot (L) analysis. TMPRSS2 is an androgen-induced protein-coding gene used as a positive control. (M) RNA FISH of CTBP1-AS in LNCaP cells. Bar=10 μm. (N) siRNA-mediated knockdown of CTBP1-AS (N=3). (O) siCTBP1-AS-dependent changes in androgen-stimulated expression of CTBP1 protein in LNCaP cells.
Source data for this figure is available on the online supplementary information page.
CTBP1-AS is upregulated in prostate cancer
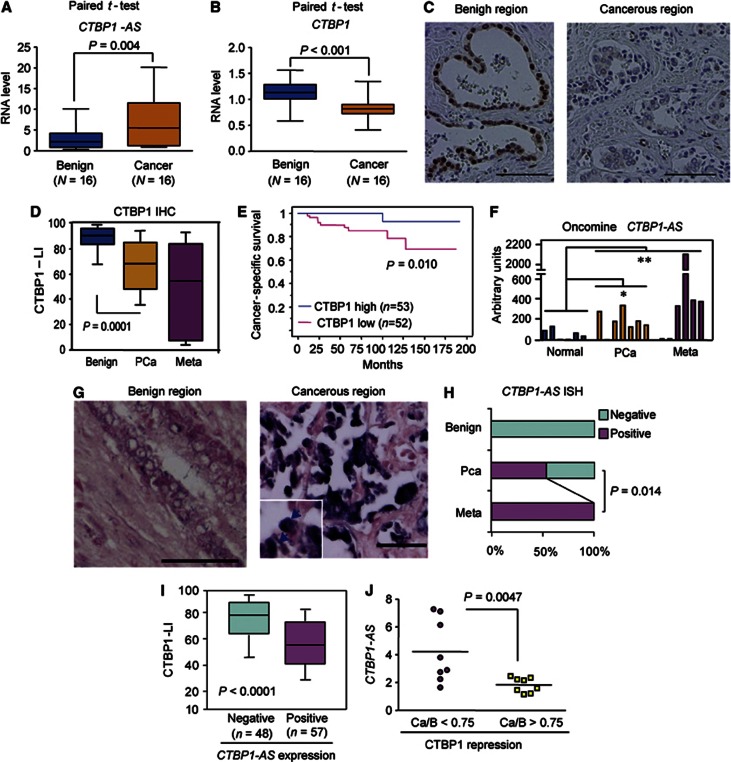
To investigate whether sense–antisense transcriptional regulation by CTBP1-AS might be associated with the biology of prostate cancer, we performed laser capture microdissection (LCM) and quantitative reverse transcriptase–PCR (qRT–PCR) analysis to compare the expression levels of both CTBP1 and CTBP1-AS in benign and cancerous regions. We observed the upregulation of CTBP1-AS and downregulation of CTBP1 in cancer (Figure 2A and B). Next, we further performed an immunohistochemical analysis of CTBP1 expression in prostate cancer clinical specimens (Figure 2C and Supplementary Figure S2A–D). CTBP1 expression was downregulated in cancer samples, particularly in metastatic cancer samples, compared with benign prostate samples (Figure 2D). Kaplan–Meier analysis showed poor cancer-specific survival and PSA-free survival in patients with lower CTBP1 expression compared with those with higher CTBP1 expression (Figure 2E and Supplementary Figure S2D). In addition, multivariate analysis demonstrated that CTBP1 downregulation was an independent prognostic factor (Supplementary Tables III and IV). Importantly, CTBP1-AS was upregulated in cancer and metastatic cancer samples compared with normal prostate tissues in the available microarray data (Rhodes et al, 2004; Varambally et al, 2005) (Figure 2F). We further analysed CTBP1-AS expression in our prostate cancer samples by performing an in situ hybridization (ISH) study of clinical samples. We did not detect CTBP1-AS expression in benign prostate tissues. However, CTBP1-AS expression was upregulated in the cancer samples (Figure 2G and H), demonstrating inverse correlation between CTBP1-AS and CTBP1 expression (Figure 2I). This correlation was also validated by qRT–PCR analysis (Figure 2J). Interestingly, CTBP1-AS expression increased with disease progression to metastasis. In the analysis of clinicopathological parameters, we found CTBP1-AS expression is significantly correlated with high Gleason scores and AR high expression status (Supplementary Table V). These data indicate the involvement of CTBP1-AS in tumour development and the direct regulation by AR. CTBP1 protein expressions were shown to be concordant with RNA expression detected by CTBP1 ISH analysis (Supplementary Figure S2E), suggesting that CTBP1 is regulated at the mRNA level. Taken together, these results from clinical studies suggest that the sense–antisense transcriptional regulation by androgen is important for prostate cancer.
Figure 2.
CTBP1-AS is upregulated in prostate cancer and negatively correlated with CTBP1. (A, B) qRT–PCR analysis of CTBP1-AS (A) and CTBP1 (B) expression levels in prostate cancer and benign epithelial tissues (n=16). We performed LCM to purify total RNA from each tissue. Paired t-test was performed to obtain P-values. (C) Representative view of CTBP1 expression in prostate tissue or prostate cancer specimens. Bar=100 μm. (D) Decreased CTBP1 expression in prostate cancer. CTBP1 labelling index (LI) is quantified as the immunoreactivity score. Benign, benign prostate epithelium (n=95); Meta, prostate carcinoma in the metastatic site (n=7); PCa, prostate carcinoma in the primary lesion (n=105). (E) Kaplan–Meier analysis using the log-rank test. (F) Expression level of CTBP1-AS in prostate cancer. The values of the probe (1563571_at) in microarray data (GS1439) corresponding to the CTBP1-AS region (AX747592) are shown. *P=0.023, *P=0.015 by the Mann–Whitney test from normal samples. (G) Representative view of CTBP1-AS in situ hybridization. CTBP1-AS was predominantly detected in the nucleus of carcinoma cells (arrows). Bar=100 μm. (H) Summary of CTBP1-AS expression in prostate cancer tissues. The rates of positive expression of CTBP1-AS of patients at each stage of disease progression are shown. (I) Negative correlation between CTBP1-AS expression and the CTBP1 LI. (J) Negative correlation between CTBP1-AS expression and the CTBP1 repression in cancer. We evaluated the expression changes between cancer (Ca) and benign (B) regions by qRT–PCR analysis (data in panels A and B). The association between CTBP1-AS expression and CTBP1-repression was analysed by two-sided t-test.
CTBP1 functions as a novel AR corepressor for inhibiting cell growth
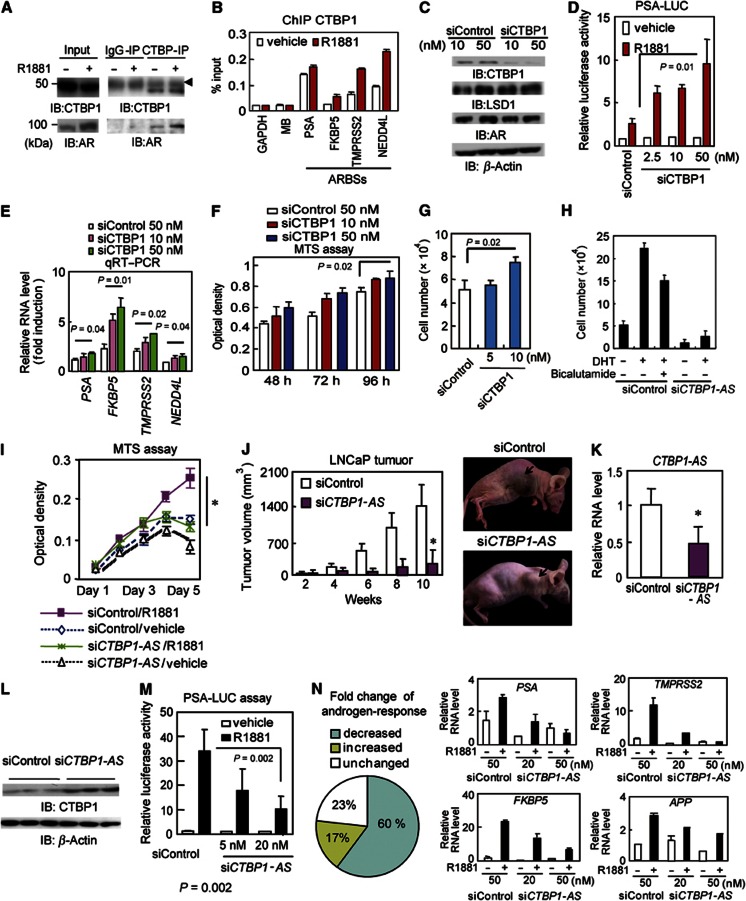
To investigate the significance of the CTBP1-AS-mediated repression of CTBP1, we examined the function of CTBP1 as a corepressor of AR in prostate cancer cells. Coimmunoprecipitation assays demonstrated ligand-dependent interaction of both exogenous and endogenous CTBP1 with AR (Figure 3A and Supplementary Figure S3A and B). ChIP analysis showed that treatment with 10 nM R1881 for 1 h induced ligand-dependent recruitment of CTBP1 to ARBS for at least four ARBSs assayed (Figure 3B). Re-ChIP analysis (Supplementary Figure S3C) showed colocalization of AR and CTBP1 at ARBSs; this colocalization decreased over time due to decreased expression of CTBP1. Next, we investigated the relationship between CTBP1 association and AR transcriptional activity. The expression levels of androgen-responsive genes adjacent to ARBSs are upregulated gradually in a time-dependent manner following R1881 or dihydrotestosterone (DHT) treatment, increasing to maximum at 24 or 48 h (Wang et al, 2005; Takayama et al, 2011). We investigated whether recruitment of CTBP1 at early time points represses induction of androgen-regulated genes by using a siRNA directed against CTBP1 (Figure 3C). Induction of androgen-dependent target genes and promoter activity were upregulated by siCTBP1 transfection (Figure 3D and E). In contrast, addition of exogenous CTBP1 repressed AR transcriptional activity (Supplementary Figure 3D and E). Progressive demethylation of histone H3K9 at the ARBS was observed by siCTBP1 treatment (Supplementary Figure S3F). Demethylation of H3K9 is the representative histone modification in ARBSs induced by LSD1 (Metzger et al, 2005). We also observed that CTBP1 interacts with the histone methyltransferase, G9a, even after androgen treatment, suggesting that H3K9 methylation by CTBP1 is probably mediated by G9a (Supplementary Figure S3G) and presumably opposes LSD1 function (Wang et al, 2007). CTBP1 interaction with histone deacetylase (HDAC) is weak and becomes no longer apparent after androgen treatment. Taken together, our data show that CTBP1 functions as a AR corepressor by inhibiting androgen-mediated demethylation. We found that CTBP1 overexpression reduced cell proliferation with accompanying repression of androgen-regulated genes (Supplementary Figure S4A–E) and that knocking down CTBP1 increased LNCaP cell proliferation (Figure 3F and G). These results demonstrated the importance of CTBP1 in controlling cancer proliferation. We also observed these effects of CTBP1 in VCaP cells (Supplementary Figure S5). However, these tumour-suppressive effects are limited to AR-positive cell lines, suggesting that CTBP1 modulates prostate cancer cell proliferation dependent on AR.
Figure 3.
CTBP1-AS promotes tumour growth by activating AR signalling. (A) Immunoprecipitation by anti-CTBP1 antibody. LNCaP cells were treated with 10 nM R1881 or vehicle for 6 h. Lysates were immunoprecipitated by anti-CTBP1 antibody. The arrowhead indicates IgG heavy chain. (B) ChIP analysis of CTBP1 in ARBSs. Cells were treated with 10 nM R1881 for 1 h. Normalized to IgG-IP control. GAPDH, MB (myoglobin) locus: negative controls. (C) Western blot analysis of LNCaP cells transfected with siControl or siCTBP1. (D) PSA-LUC assay in LNCaP cells transfected with siControl or siCTBP1. (E) LNCaP cells transfected with siControl or siCTBP1 (10 or 50 nM) were treated with 10 nM R1881 for 4 h. Fold induction is shown. mRNA levels were measured by qRT–PCR. (F) MTS assay of LNCaP cells transfected with siCTBP1 (n=4). (G) CTBP1 knockdown accelerates cell proliferation. The numbers of LNCaP cells transfected with siControl or siCTBP1 were counted 3 days after transfection. (H) Cell proliferation assay of LNCaP cells ransfected with siControl or siCTBP1-AS (n=4). The number of viable cells is counted and shown. (I) MTS assay of LNCaP cells transfected with siControl or siCTBP1-AS (n=4). *P<0.05. (J, left) Tumour-growth curves in nude mice inoculated with LNCaP cells transfected with siRNA *P<0.05 (n=7). (J, right) Representative view of LNCaP-derived tumours in nude mice administered siRNA. (K) CTBP1-AS knockdown in tumours transfected with siCTBP1-AS (n=4). *P<0.05. (L) CTBP1 protein expression in tumours transfected with siCTBP1-AS. (M) PSA-LUC assay in LNCaP cells transfected with siControl or siCTBP1-AS (n=3). (N, left) Effect of siCTBP1-AS on androgen-upregulated gene expression (>2.0-fold change by R1881 versus vehicle, 198 genes) in LNCaP cells. (N, right) qRT–PCR analysis of androgen-regulated genes. At 2 days before androgen stimulation, siControl or siCTBP1-AS was transfected to LNCaP cells. LNCaP cells were treated with 10 nM R1881 or vehicle for 24 h. Bar: s.d.
Source data for this figure is available on the online supplementary information page.
CTBP1-AS is associated with prostate cancer castration-resistant tumour growth and activates AR signals
Next, we examined whether CTBP1-AS is associated with tumour growth in prostate cancer. Androgen-dependent LNCaP cell proliferation was inhibited by siCTBP1-AS treatment (Figure 3H and I). In addition, overexpression of the CTBP1-AS transcript stimulated cell proliferation and imparted resistance to growth inhibition by bicalutamide (Supplementary Figures S6A–D and S7). Moreover, CTBP1-AS overexpression accelerates cell cycle (Supplementary Figure S6E). We also examined the potential tumour-promoting effects of CTBP1-AS in vivo. Tumour growth in a xenograph model was reduced after CTBP1-AS knockdown (Figure 3J and K), leading to increased CTBP1 expression (Figure 3L).
Based on these findings, we further examined the effects of CTBP1-AS on AR transcriptional activity. First, androgen-dependent induction of a PSA-luciferase reporter was inhibited in a dose-dependent manner by addition of siCTBP1-AS (Figure 3M and Supplementary Figure S8A and B). In addition, microarray analysis demonstrated that transcriptional activation of androgen-induced genes was diminished by siCTBP1-AS (Figure 3N and Supplementary Figure S8C) in both AR-positive LNCaP and VCaP cells. Furthermore, qRT–PCR analyses confirmed that transcriptional upregulation of representative androgen-regulated genes was repressed by siCTBP1-AS (Figure 3N and Supplementary Figure S9A). This inhibition of AR signalling is at least partially due to continuation of CTBP1 binding and inhibition of demethylation of H3K9 because siCTBP1-AS reversed androgen-mediated CTBP1 repression (Supplementary Figure S9B–D). Thus, our results indicate that CTBP1-AS positively regulates AR signalling.
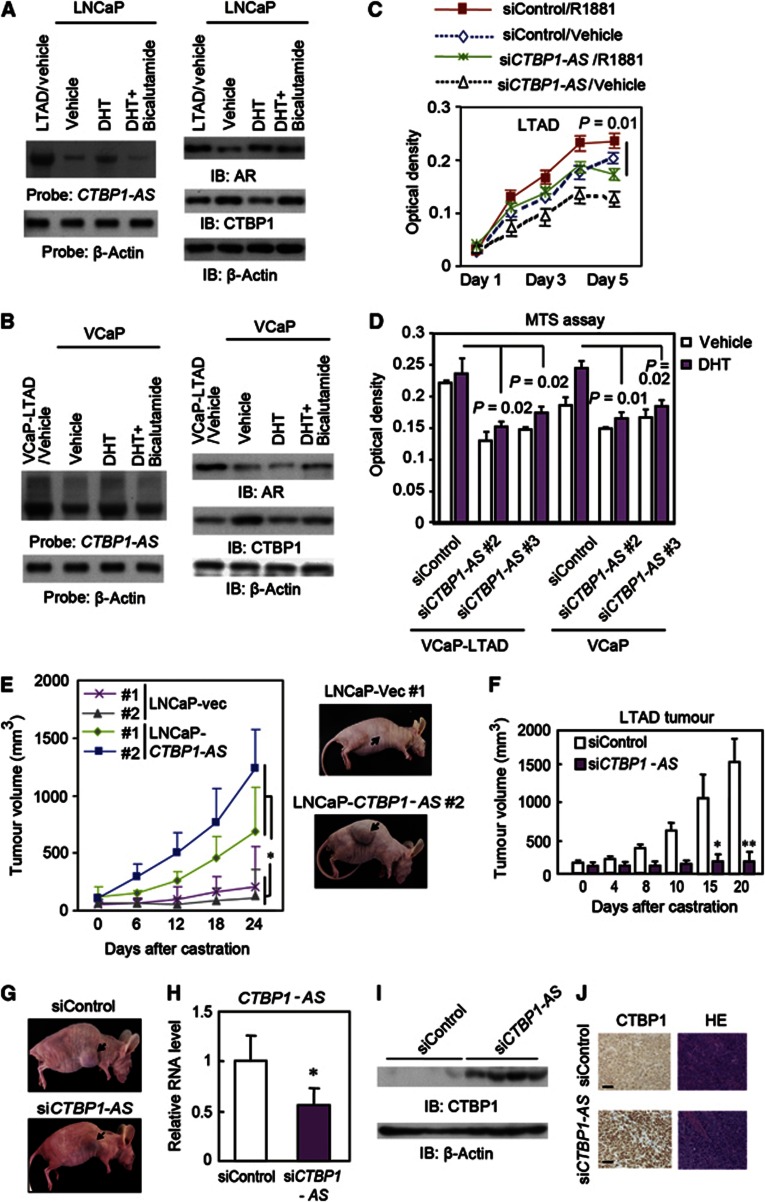
Moreover, northern blot analysis showed that CTBP1-AS is overexpressed in long-term androgen deprivation (LTAD) cells derived from LNCaP, a widely used model of CRPC derived from LNCaP cells (Culig et al, 1999; Takayama et al, 2012) (Figure 4A). In contrast, CTBP1 protein levels were decreased in the absence of androgen. Similar results were also observed in VCaP and VCaP-LTAD (LTAD derived from VCaP) cells (Figure 4B). In addition, CTBP1-AS knockdown inhibited LTAD cell proliferation in hormone-depleted condition in both cell lines (Figure 4C and D). In a xenograph model, CTBP1-AS overexpression induced tumour growth after castration (Figure 4E). We further transplanted nude mice subcutaneously with LTAD cells, castrated the mice after the tumours formed, and then injected siCTBP1-AS or siControl into the tumours (Figure 4F–J). Interestingly, siCTBP1-AS treatment inhibited LTAD tumour growth after castration (Figure 4F–H) and induced CTBP1 (Figure 4I and J). Thus, our results indicate that CTBP1-AS promotes tumour growth in castration resistance.
Figure 4.
CTBP1-AS is associated with castration-resistant tumour growth. (A) Expression levels of CTBP1-AS RNA by northern blot analysis (left), AR and CTBP1 protein by western blot analysis (right) in LNCaP and LTAD cells. (B) Expression levels of CTBP1-AS RNA by northern blot analysis (left), AR and CTBP1 protein by western blot analysis (right) in VCaP and VCaP-LTAD cells. (C) Proliferation of LTAD cells transfected with siRNA (n=4). (D) CTBP1-AS is related to castration-resistant cell proliferation. MTS assay of LTAD-VCaP cells (incubated in androgen-depleted conditions for more than 2 months) and parental VCaP cells. Cells were transfected with siControl, siCTBP1-AS#2, or siCTBP1-AS #3 (10 nM) 72 h before androgen treatment (10 nM DHT or vehicle). MTS assay was performed on day 5 after androgen treatment (n=4). (E) Nude mice inoculated with LNCaP cells stably expressing CTBP1-AS or vector after castration. Tumour growth curves (n=6, 3 for each clone). *P=0.04 (right). Representative clones are shown. (F) Growth curves of LTAD tumours in nude mice treated with siRNAs after castration (n=7). *P=0.02, **P=0.01. (G) Representative view of LTAD tumours in nude mice treated with siRNAs after castration. (H) Knockdown of CTBP1-AS in tumours transfected with siCTBP1-AS (n=4). *P<0.05. (I) CTBP1 protein expression in tumours transfected with siCTBP1-AS. (J) Representative view of immunohistochemical analysis by anti-CTBP1 specific antibody and HE staining in LTAD tumours. Bar=50 μm. Error bar: s.d.
Source data for this figure is available on the online supplementary information page.
CTBP1-AS orchestrated repression by chromatin modification at CTBP1 promoter
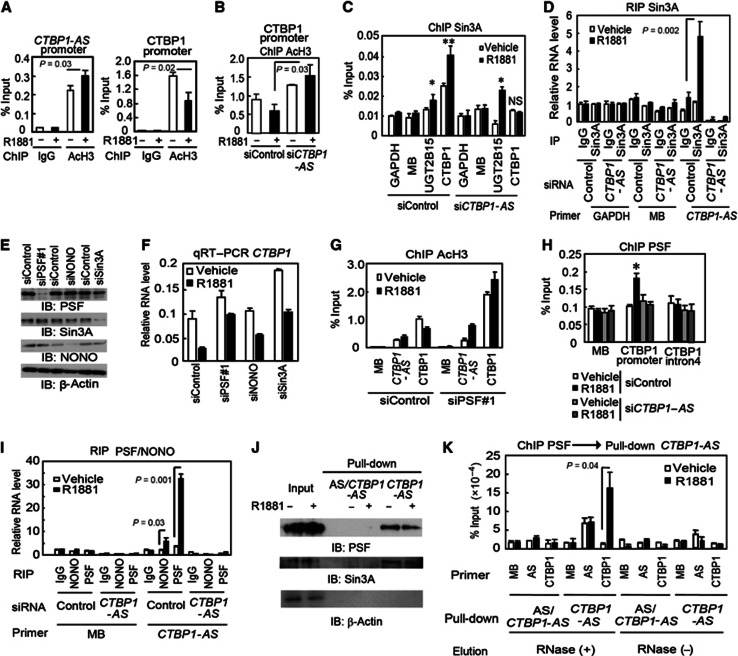
We next questioned how CTBP1 sense–antisense transcript pair contributes to the transcriptional regulation of CTBP1. The canonical AS-mediated post-transcriptional degradation of sense mRNA was not shown in this CTBP1 pair (Supplementary Figure S10A and B). Consistent with the androgen-induced CTBP1 repression in AR-regulated prostate cancer cells, the androgen-dependent repression of RNA polymerase II (pol II) recruitment and luciferase activity was shown for the CTBP1 promoter region (Supplementary Figure S10C and D). Thus, we assumed whether the androgen-dependent CTBP1 repression is epigenetically mediated by chromatin modification. ChIP assay revealed that androgen treatment substantially reduced the chromatin markers for activation, namely histone H3 acetylation and H3K4 methylation levels at the CTBP1 promoter, without altering the levels of repressive markers of H3K9 and H3K27 methylation (Figure 5A and Supplementary Figure S10E–H).
Figure 5.
PSF-induced CTBP1-AS-associated histone modification regulates CTBP1 expression. (A) ChIP analysis (n=3) using anti-AcH3 was performed on CTBP1 and CTBP1-AS promoters in LNCaP cells treated with vehicle or R1881 for 24 h. Bar: s.d. (B) ChIP analysis (n=3) using anti-AcH3 was performed on the CTBP1 promoter after 24 h of vehicle or R1881 treatment in LNCaP cells transfected with siControl or siCTBP1-AS. Bar: s.d. (C) ChIP analysis (n=3) of Sin3A recruitment to the CTBP1 promoter region. LNCaP cells were treated with R1881 or vehicle for 24 h. Bar: s.d. *P<0.05, **P<0.01, NS, P>0.05. (D) RNA immunoprecipitation of Sin3A and detection of associated RNA by qRT–PCR analysis (n=3). Bar: s.d. (E) Expression levels of PSF, NONO, and Sin3A in LNCaP cells transfected with siControl or respective siRNA (50 nM for siPSF#1 and 10 nM for siNONO and siSin3A). (F) CTBP1 mRNA level after R1881 or vehicle treatment in LNCaP cells transfected with siControl, siPSF#1, siNONO, or siSin3A (n=3). Bar: s.d. (G) ChIP analysis using anti-AcH3 was performed on sense CTBP1 and AS CTBP1-AS promoters after 24 h of vehicle or R1881 treatment in LNCaP cells transfected with siControl or siPSF#1 (n=3). Bar: s.d. (H) ChIP analysis using anti-PSF was performed on the CTBP1 promoter after vehicle or R1881 stimulation in LNCaP cells transfected with siControl or siCTBP1-AS (n=3). Bar: s.d. *P<0.05. (I) RNA immunoprecipitation of PSF and NONO, and detection of associated RNA by qRT–PCR analysis (n=3). RNA level was normalized by GAPDH. MB, myoglobin. Error bars: s.d. (J) RNA pull-down assay. Probes for CTBP1-AS fragment and AS fragment to CTBP1-AS (AS/CTBP1-AS) were prepared. (K) RNA pull-down after PSF ChIP analysis (n=3). RNase was used for elution to detect RNA-dependent pull-down. All ChIP assays were normalized to their respective IgG-IP controls. Error bar: s.e.m.
Source data for this figure is available on the online supplementary information page.
In terms of the involvement of HDACs in the androgen-dependent CTBP1 repression mechanism, we next showed that the prototypic HDAC inhibitor trichostatin-A (TSA) abolished this androgen-mediated CTBP1 mRNA reduction (Supplementary Figure S10I). We further demonstrated that CTBP1-AS overexpression decreased histone acetylation in the absence of androgen (Supplementary Figure S10J), suggesting that CTBP1-AS upregulation is responsible for the androgen-mediated repressive histone modification. The critical role of CTBP1-AS in androgen-dependent regulation of CTBP1 was also confirmed by the knockdown study, as the addition of siCTBP1-AS inhibited the androgen-mediated histone H3 deacetylation of the CTBP1 promoter (Figure 5B).
To assess the mechanism of H3 deacetylation, we first verified the ligand-dependent recruitment of HDAC and a HDAC-associated corepressor Sin3A to the promoter region of CTBP1 by ChIP analysis (Figure 5C). We used the UGT2B15 promoter as a positive control for HDAC recruitment because the involvement of direct AR binding has been reported for the downregulation of UGT2B15 (Bao et al, 2008). We found that the recruitment of HDACs to the CTBP1 promoter was abolished by CTBP1-AS knockdown, while the recruitment of HDACs to the UGT2B15 promoter was not affected. With regard to the isoforms of HDACs, we found that HDAC1, HDAC2, HDAC3, and HDAC8 are all required for the full repression of CTBP1 (Supplementary Figure S11). To elucidate the direct interaction of CTBP1-AS with HDAC complex, we performed RNA immunoprecipitation (RIP) assay using a specific antibody against Sin3A, as a HDAC-associated corepressor. RIP assay demonstrates that the ligand-dependent association of CTBP1-AS with Sin3A was significantly increased compared with the control sample immunoprecipitated with normal IgG (Figure 5D). Taken together, these results indicate that CTBP1-AS binds to HDAC–Sin3A complex and orchestrates the HDAC-mediated repression by chromation modification at the CTBP1 promoter in the AR-dependent cell system.
PSF recruitment to CTBP1 promoter is required for CTBP1 repression
We suspected that the transcript–Sin3A interaction might be indirect and mediated by an RNA-binding protein. Interestingly, Sin3A forms complexes with repressors, PTB-associated splicing factor (PSF), or non-POU domain-containing octamer-binding protein (NONO), which has both RNA- and DNA-binding domains (Mathur et al, 2001; Shav-Tal and Zipori, 2002; Song et al, 2004), binds to and represses gene promoter regions. siRNA knockdown (Figure 5E) revealed that PSF is the major component associated with CTBP1 repression since PSF siRNA rescued the androgen-mediated repression of CTBP1 transcript level by ∼75%, while the knockdown of NONO or Sin3A rescued it by ∼50% (Figure 5F). We also confirmed that androgen-mediated histone deacetylation at the CTBP1 promoter was inhibited by siPSF (Figure 5G). In addition, these results were also confirmed in VCaP cells (Supplementary Figure S12B and D).
Next, we investigated how PSF is involved in CTBP1 histone modification and repression. ChIP assay showed that PSF was recruited to the CTBP1 promoter in a CTBP1-AS-dependent manner (Figure 5H). We confirmed the interaction between PSF and CTBP1-AS by RIP (Figure 5I and Supplementary Figure S13A–C) and RNA pull-down (Figure 5J and Supplementary Figure S13D) assays. We also found that short interspersed repetitive element (SINE) repeats within CTBP1-AS were important for the interaction with the RNA-binding domain of PSF (Supplementary Figure S14A–D), extending and providing a mechanism for previous observations (Chen et al, 2008). However, other ncRNAs expressed in the nucleus, which possess SINE sequences, did not interact with PSF (Supplementary Figure S15), suggesting other sequences may also be involved in this interaction and CTBP1 repression (Supplementary Figure S14E and F). To determine whether PSF and CTBP1-AS could be colocalized at the CTBP1 promoter, we performed CTBP1-AS RNA pull-down for the DNA–protein complexes obtained by PSF ChIP. Interestingly, enrichment of the CTBP1 promoter region was observed in the immunoprecipitated DNA after RNase-mediated elution, suggesting pull-down by the CTBP1-AS RNA probe. These results indicate that the association of PSF with CTBP1-AS is important for PSF recruitment to the CTBP1 promoter (Figure 5K and Supplementary Figure S13E and F). We infer from these results that CTBP1-AS induces CTBP1 transcriptional repression by interacting with PSF, which binds at the CTBP1 promoter to induce histone deacetylation by recruiting HDACs.
PSF global function regulates prostate cancer cell cycle progression
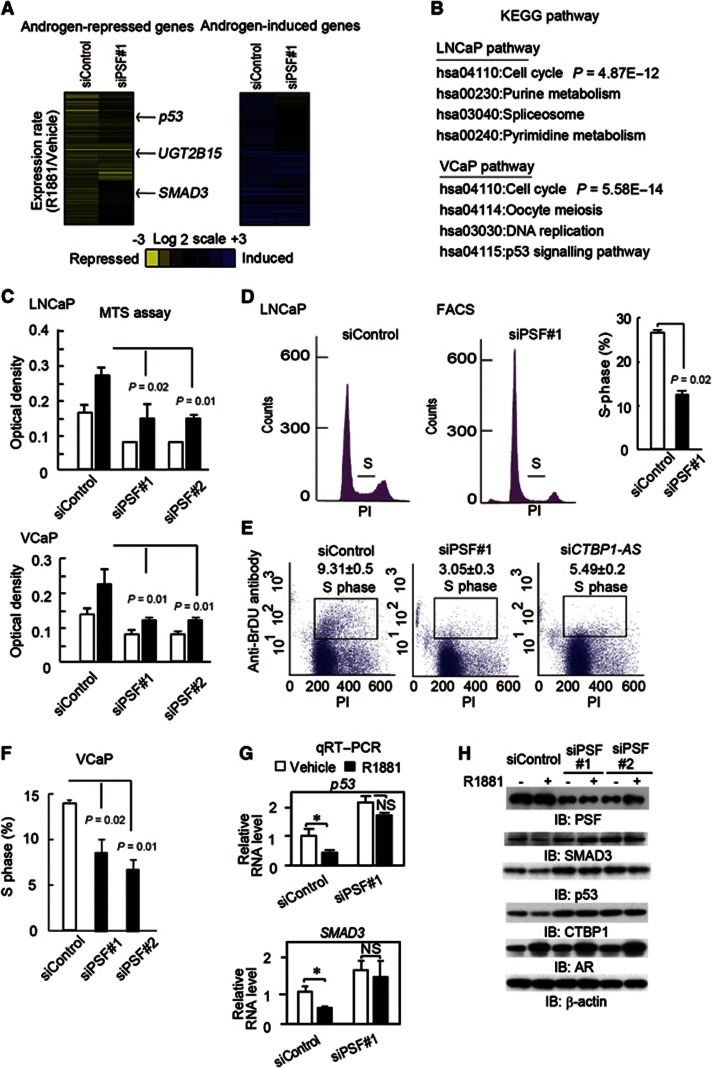
Next, to analyse the impact of PSF on androgen-dependent transcriptional regulation, we performed siPSF knockdown followed by microarray analysis. Surprisingly, siPSF relieved the repression of more than 69.9% (931 of 1331) of the androgen-repressed genes (Figure 6A) in LNCaP cells. This result was also validated in VCaP cells (Supplementary Figure S16A). In addition, most of the genes normally induced by androgen such as APP and TMPRSS2 were also repressed by siPSF (Figure 6A and Supplementary Figure S16A and B), presumably because siPSF abolishes CTBP1 repression.
Figure 6.
Global function of PSF in androgen-mediated repression of cell cycle regulators. (A, (left) Rescue of androgen-dependent gene repression in LNCaP cells by siPSF. Microarray analysis was performed in ligand-treated (24 h) LNCaP cells transfected with siRNAs (10 nM). The heat maps represent fold changes in the expression of androgen-regulated genes. (A, right) Repression of androgen-dependent gene activation in LNCaP cells by siPSF. (B) Cell cycle-related genes are enriched among PSF target genes. Genes that are repressed by androgen and reversed by siPSF treatment in two cell lines were selected. Pathway analysis was performed by DAVID using KEGG pathway. (C) MTS assay of LNCaP and VcaP cells transfected with siControl or siPSF#1 and #2 (n=4). The assay was performed on day 3 after ligand treatment. (D) Percentage of LNCaP treated with siPSF or siControl in the S phase (n=2). (E) Cell cycle analysis of cells treated with siControl, siPSF#1, and siCTBP1-AS by bivariate FACS. Percentages of BrdU-labelled cells are shown as S phase (n=2). (F) Percentage of VCaP cells treated with siPSF or siControl in the S phase (n=2). (G) Expression of p53 and SMAD3 in ligand-treated (24 h) LNCaP cells transfected with siControl or siPSF#1 (10 nM) (n=3). Bar: s.d. *P<0.05, NS, P>0.05. (H) Immunoblots of PSF, AR, CTBP1, p53, and SMAD3 in ligand-treated (24 h) LNCaP cells transfected with siControl, siPSF#1, or #2 (10 nM).
Source data for this figure is available on the online supplementary information page.
Pathway analysis (DAVID; Huang et al, 2009) has shown the enrichment of cell cycle-related genes (Figure 6B) among the PSF targets repressed by androgen. Gain- and loss-of-function experiments have shown that PSF promotes cell growth (Figure 6C and Supplementary Figure S17), raising the question as to whether PSF may regulate the cell cycle in prostate cancer. Flow cytometry analysis revealed that siPSF treatment inhibited cell cycle progression in LNCaP cells (Figure 6D). BrdU-labelled cells were decreased by siPSF treatment, suggesting that S-phase entry is promoted by PSF (Figure 6E). This inhibition was also observed in VCaP cells (Figure 6F and Supplementary Figure S16C).
Among the PSF target genes, we focussed on p53 and SMAD3 because these are well-known cell cycle regulators that are repressed by androgen treatment (Rokhlin et al, 2005; Song et al, 2010) and are expressed at a low level in prostate cancer cells (Schlomn et al, 2008; Song et al, 2010; Taylor et al, 2010). Interestingly, these two genes are also known to be negative regulators of AR (Hayes et al, 2001; Shenk et al, 2001). Androgen-dependent repression of both genes was abolished by siPSF treatment (Figure 6G and H and Supplementary Figure S16B, D, and E). However, this relief of repression of these genes by siPSF was not affected by addition of siCTBP1, suggesting that the repression is dependent on direct function of PSF (Supplementary Figure S18A). In contrast, combining CTBP1 knockdown with PSF knockdown partially relieved the repression of androgen-mediated gene induction (Supplementary Figure S19). Moreover, cell cycle retardation was partially reversed by the addition of siCTBP1 (Supplementary Figure S18B). We hypothesized that the function of PSF for the cell cycle progression is dependent on both androgen-mediated repressed targets by PSF and AR-mediated upregulated genes by CTBP1 repression. Taken together, our results reveal that PSF regulates cell cycle-related genes by promoting androgen-mediated gene regulations.
CTBP1-AS and PSF cooperatively promote cell cycle progression by repressing cell cycle regulators
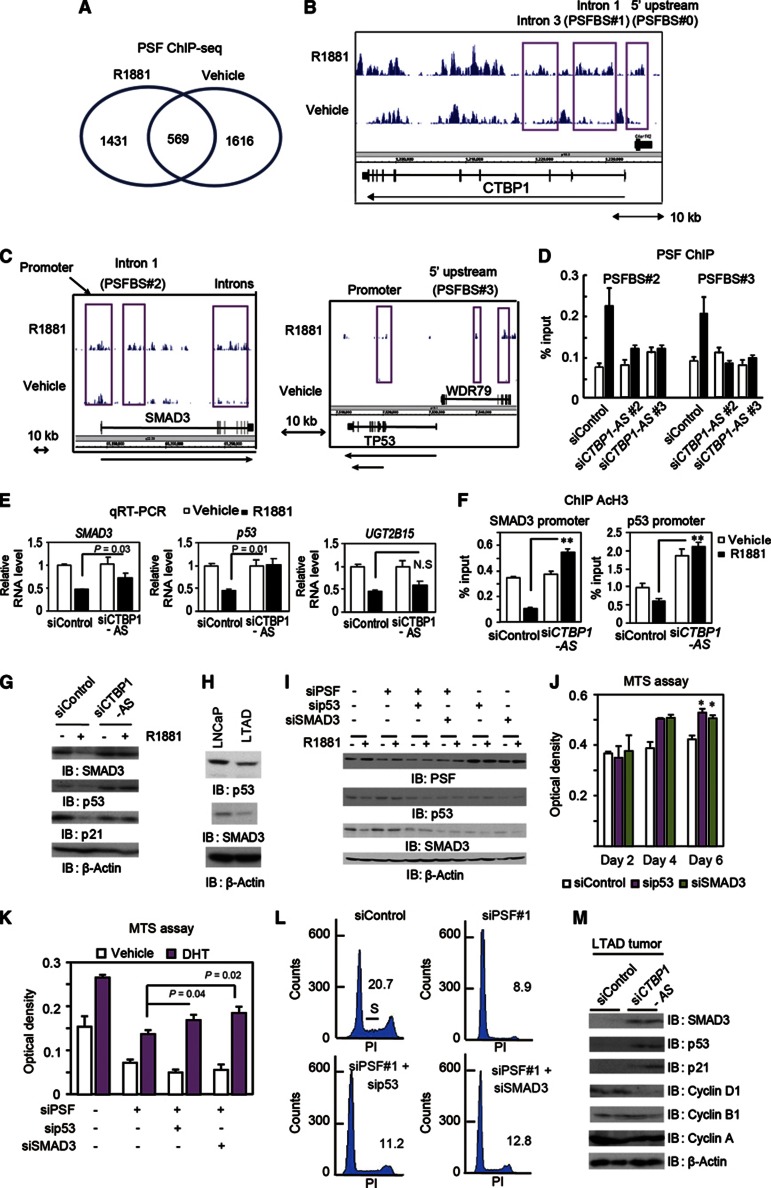
We next explored AR- and PSF-binding sites by ChIP-sequence (ChIP-seq) analysis in LNCaP cells to investigate the PSF genomic function in prostate cancer (Figure 7A–C). The distribution of binding regions was changed by androgen treatment, and in the CTBP1 genomic region, we identified two androgen-dependent PSF-binding sites around the CTBP1 promoter (intron 1 (PSFBS#1) and 5′ upstream (PSFBS#0); Figure 7B). Notably, androgen-dependent PSF occupancy was observed in the vicinity of PSF-regulated genes such as SMAD3 and p53 (Figure 7C). In contrast, no PSF binding was observed in the vicinity of UGT2B15. We validated that these PSF recruitment to the promoters was inhibited by transfection of siCTBP1-AS (Figure 7D and Supplementary Figure S12A). Interestingly, androgen-dependent PSF bindings in vitro was abrogated by CTBP1-AS knockdown (Supplementary Figure S20), suggesting that CTBP1-AS can influence the DNA-binding ability of PSF and subsequent transcriptional change. We confirmed using siCTBP1-AS that p53 and SMAD3 are also regulated by CTBP1-AS (Figure 7E and G and Supplementary Figures S8D and S21A). Although histone acetylation levels were decreased by androgen in the promoters of these two genes, CTBP1-AS knockdown relieved the deacetylation, suggesting epigenetic regulation by PSF recruitment is important for transcriptional repression (Figure 7F). In addition, p21, which is a target of p53 and SMAD3 and is downregulated in AR-dependent CRPC (Wang et al, 2001), was also repressed by 48-h androgen treatment in a CTBP1-AS-dependent manner (Figure 7G and Supplementary Figure S21B).
Figure 7.
Cooperative trans-regulatory function of CTBP1-AS and PSF in inhibiting cell cycle regulators. (A) Identification of PSF binding by ChIP-sequence. LNCaP cells were treated with 10 nM R1881 or vehicle for 24 h. Analysis of PSF-binding sites (PSFBSs, >5-fold) was performed by ChIP-sequence. (B) Androgen-dependent PSFBSs at 5′ upstream region (PSFBS#0), intron 1 (PSFBS#1), and intron 3 are indicated in CTBP1 locus. (C) Analysis of PSF binding in the vicinity of the SMAD3 and p53 loci. PSFBSs in the promoter and intron 1 (PSFBS#2) of SMAD3 (left) and PSFBSs in the promoter and 5′ upstream region (PSFBS#3) of p53 (right) are indicated. Two transcriptional variants of p53 are indicated by arrows. (D) PSF binding was dependent on CTBP1-AS induction. LNCaP cells were transfected with siControl, siCTBP1-AS#2, or siCTBP1-AS#3 (10 nM). PSF ChIP analysis was performed after 24 h treatment with 1 nM R1881 or vehicle. (E) qRT–PCR analysis (n=3) of SMAD3, p53, and UGT2B15 after siCTBP1-AS treatment. LNCaP cells were treated with 10 nM R1881 or vehicle for 24 h and then qRT–PCR was performed. NS, P>0.05. (F) Histone deacetylation of SMAD3 and p53 promoters was reversed by siCTBP1-AS. ChIP-analysis (n=3) of AcH3 in the promoters of p53 and SMAD3. **P<0.01. (G) Immunoblots for p53, SMAD3, and p21 in LNCaP cells transfected with siRNAs after 48-h ligand treatment. (H) Reduced expression of p53 and SMAD3 in LTAD cells. (I) Knockdown of p53 and SMAD3 in LNcaP cells. (J) SMAD3 and p53 knockdowns promote prostate cancer cell proliferation. MTS assay of LNCaP cells transfected with siControl, sip53, and siSMAD3 (n=4). *P<0.01. (K) MTS assay of LNCaP cells transfected with siControl, siPSF#1, siPSF#1+sip53, and siPSF#1+siSMAD3 (n=4). The assay was performed on day 3 after ligand treatment. (L) siPSF-mediated cell cycle repression is partially reversed by p53 and SMAD3 knockdown. Cell cycle analysis of LNCaP cells transfected with siControl, siPSF#1, siPSF#1+sip53, and siPSF#1+siSMAD3. Percentages of cells in the S phase are indicated. (M) Immunoblots for p53, SMAD3, p21, and cyclins in the LTAD tumour lysates shown in Figure 4I. Bar: s.d.
Source data for this figure is available on the online supplementary information page.
We further investigated the function of these cell cycle regulators in prostate cancer cells. Both SMAD3 and p53 are repressed in LTAD cells compared with parental LNCaP cells (Figure 7H). Next, we analysed the role of these two PSF targets in PSF-dependent cell cycle progression by siRNA. Interestingly, knockdown of each gene also reduced the protein level of the other, suggesting that these two genes enhance each other’s expression level by some mechanism (Figure 7I). We found that these cell cycle regulators negatively regulate prostate cancer cell proliferation (Figure 7J). Moreover, siSMAD3 and sip53 reversed the siPSF-mediated cell cycle and growth repression, suggesting that these two target genes facilitate the PSF-dependent cell cycle progression in prostate cancer (Figure 7K and L and Supplementary Figure S21C and D). Notably, both genes interact with AR and negatively control AR activity, suggesting that these genes also modulate AR-dependent transcriptional program (Supplementary Figure S22). Importantly, these cell cycle regulators were strongly induced by siCTBP1-AS in castration-resistant tumours formed by LTAD cells (Figure 7M), indicating that PSF and CTBP1-AS control these genes in CRPC. Thus, the present results indicate that CTBP1-AS and PSF cooperatively promote cell cycle progression by repressing cell cycle regulators under the control of AR.
CTBP1-AS and PSF function cooperatively to modulate global androgen signalling
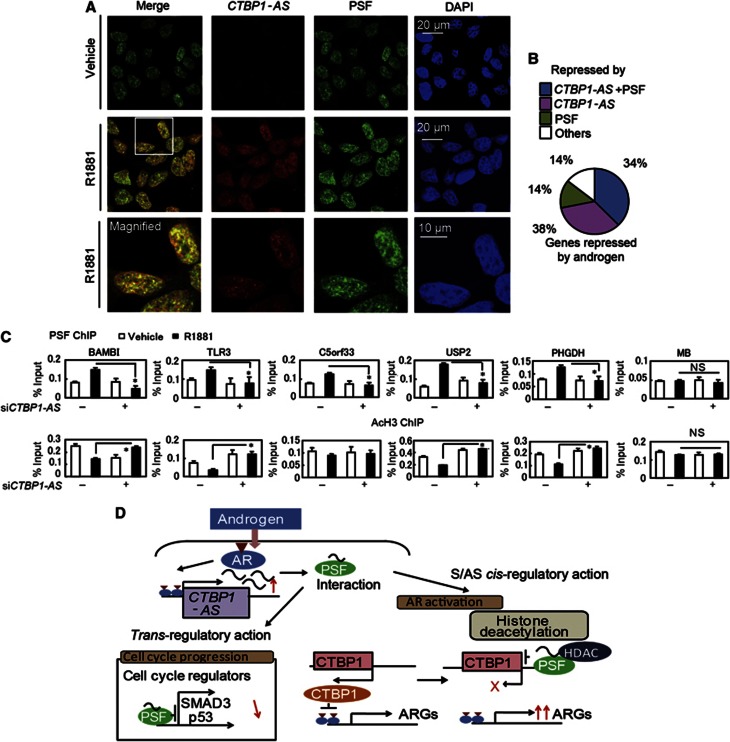
Since CTBP1-AS is localized in diffuse and discrete regions of the nucleus, we examined global CTBP1-AS functions over the entire genome. By combining immunofluorescence and RNA FISH, we have shown that CTBP1-AS and PSF colocalized in the nucleus (Figure 8A and Supplementary Figure S21E). Next, we compared CTBP1-AS and PSF targets. We selected 369 androgen-mediated repressed genes from two microarray analysis data sets of siControl-treated samples. Among these, repression of 139 genes (38%) was reversed by both CTBP1-AS and PSF depletion, suggesting that PSF and CTBP1-AS cooperatively mediate gene repression by androgen at these loci (Figure 8B). Further evidence of cooperation between CTBP1-AS and PSF was supported by microarray analysis in VCaP cells (Supplementary Figure S16F). To analyse the global role of CTBP1-AS in PSF recruitments, we further analysed PSF and histone H3 acetylation by ChIP analysis (Figure 8C). Among androgen-repressed genes by both PSF and CTBP1-AS, we selected five genes and found PSF binding regions by referring to ChIP-seq data. PSF recruitments to these regions and histone deacetylations are abrogated by CTBP1-AS depletion. Taken together, these results indicate that CTBP1-AS/PSF complex-mediated histone deacetylation is a critical machinery that exerts the global androgen-mediated gene repression programme (Figure 8D).
Figure 8.
Global function of PSF and CTBP1-AS governs androgen-dependent transcriptional program. (A) Detection of CTBP1-AS by RNA FISH and PSF immunofluorescence analysis. (B) Summary of androgen-repressed genes. The number of androgen-repressed genes with reduced repression after treatment with siPSF and/or siCTBP1-AS is indicated. (C) ChIP analysis (n=2) of AcH3 and PSF bindings in the vicinity of genes repressed by PSF and CTBP1-AS. Cells were transfected with siControl or siCTBP1-AS. Bar: s.d. *P<0.05, NS, P>0.05. (D) Working model of the role of CTBP1-AS and PSF in prostate cancer.
Discussion
Here we demonstrate novel functions of prostate cancer-associated ncRNA CTBP1-AS, which has been originally identified as an AR-regulated AS transcript of CTBP1 locus by the combinational study of AR-binding sites and CAGE transcriptome analysis. At first, this natural AS transcript of CTBP1-AS associates with the transcriptional repressor PSF and recruits the HDAC–Sin3A complexes to CTBP1 promoter in cis, with the loss of activating histone marks. Secondly, the release of the repressor CTBP1 from the regulatory regions of AR-regulated genes leads to the transcriptional activation with the loss of repressing histone marks. In trans-regulatory pathway, CTBP1-AS also guides PSF complexes to the regulatory regions of their endogenous target genes, leading to the transcriptional repression of genes that have suppressive functions for tumour growth.
As far as we know, CTBP1-AS is the first hormone-regulated natural AS transcript that has been identified to be directly associated with hormone-dependent cancer biology in vivo. In terms of the chromatin-modifying functions of natural AS transcripts, a small number of AS ncRNAs (for example, Tsix (Lee et al, 1999) and p15AS (Yu et al, 2008)) have been identified as transcriptional silencers for their target genes mainly in cis by inactivating chromatin structure. In contrast, long ncRNAs that have been identified to function in trans are predominantly intergenic ncRNAs, as exemplified by HOTAIR in breast cancer (Rinn et al, 2007; Gupta et al, 2010), known as their PRC2 complex-repressing functions hormone independently. Our present findings will also have an impact on the research field of steroid hormone receptors, as we still have limited knowledge with regard to hormone-regulated long ncRNAs including natural AS transcripts, although our group (Takayama et al, 2011) and others (Hah et al, 2011) have identified some of those by high-throughput technology. Moreover, we revealed local and global regulatory functions of this long ncRNA for the androgen-mediated transcriptional regulation. The androgen-activated functions of CTBP1-AS in prostate cancer will have implications in the pathogenesis and the management of the disease including CRPC. The present results suggest that CTBP1-AS and its protein partner PSF could be promising targets for therapeutic options of prostate cancer.
In this study, we also showed that CTBP1 exerts tumour-suppressive effects in AR-positive prostate cancer cells. On the contrary, there is a recent report describing that CTBP1 is rather upregulated in metastatic prostate cancer and CTBP1 exerts growth stimulatory effects in the carcinoma cells (Wang et al, 2012). We consider that one of the reasons for the inconsistency in CTBP1 functions may be due to the differences in tumour samples or cell lines, as functional assays by Wang et al (2012) were performed predominantly in AR-negative prostate cancer cell lines. There is also a difference in the timing of obtaining metastatic samples between their study and ours, the former at autopsy whereas the latter at radical prostatectomy. In some reports, AR transcriptional activity is decreased after in distal metastases relative to primary prostate cancers (Stanbrough et al, 2006). We infer that CTBP1 exerts proto-oncogenic and tumour-suppressive effects through its interactions with various transcriptional factors. Further study in other cohorts will reveal the contribution of androgen-repressed CTBP1 function to prostate cancer progression.
Materials and methods
Cell culture and reagents
VCaP cells were grown in DMEM medium supplemented with 10% FBS. LNCaP, DU145, and RWPE cells were grown in RPMI medium supplemented with 10% FBS, 50 units/ml penicillin, and 50 μg/ml streptomycin. Before androgen treatment, cells were cultured in phenol red-free medium containing 5% dextran charcoal-stripped FBS for 48–72 h, and then treated with 10 nM R1881, 10 μM Bicalutamide, 10 nM DHT, or vehicle. Human PrEC cells (derived from two individuals) were obtained from TAKARA Bio and cultured as instructed by the company. Antibodies used included those against Sin3A (AK-11), HDCA1 (H-51), p53 (Pab-240), p21 (F-5), Cyclin D1 (C-20), Cyclin A (H-432), Cyclin B1 (H-433; Santa Cruz Biotechnology), CTBP1 (#612042), NONO (#611278; BD Bioscience), AcH3K9 (#07-352; Upstate), K9me2 (ab1220), K9me3 (ab8898), K27me3 (ab6002), G9a (#3306; Cell Signaling), PSF (6D7; Sigma; for ChIP), PSF (#61045; Novus Biologicals; for immunofluorescence and western blot), LSD1 (#00518; Bio Matrix Research), K4me3 (#17614), and SMAD3 (#04-1035; Millipore). Antibodies against AR, AcH3, RNA Pol II, FLAG, and Myc were described previously (Takayama et al, 2007, 2009, 2011, 2012). TSA and bicalutamide were purchased from Sigma, actinomycin D from Nakarai, R1881 from Perkin Elmer, and DHT from Wako (Saitama, Japan).
Northern blot analysis
Antisense CTBP1-AS probe preparation and subsequent northern blot analysis were performed using the northern blot starter kit (Roche), according to the manufacturer’s protocol. Total RNA (1 μg) was denatured and loaded onto formaldehyde-mixed agarose gels prepared in 1 × MOPS buffer. RNA was transferred onto Hybond-XL membrane (Roche) and probed with a DIG-labelled RNA probe spanning the middle region of CTBP1-AS. Positive control of nuclear fraction was confirmed by U1 RT–PCR analysis. The primer sequences are as described previously (Rinn et al, 2007).
Western blot analysis and immunoprecipitation
For immunoprecipitation by anti-CTBP1, AR antibodies, 1 mg of cell lysate protein was incubated with anti-CTBP1, AR antibody, or normal rabbit IgG (Sigma) at 4 °C overnight. The mixture of cell extract and antibody was then incubated with protein G-Sepharose beads (Amersham Biosciences) at 4 °C for 2 h, and washed 4 times with NP-40 lysis buffer. The immunoprecipitated proteins were boiled for 5 min in Laemmli sample buffer and separated by SDS–PAGE. Immunoblotting was performed as described previously (Takayama et al, 2007).
Cell cycle analysis
Cells were centrifuged, washed in PBS, and then fixed by slow addition of 3 ml of ice-cold 70% ethanol with mild shaking. Fixed cells were stored at 4 °C until use. On the day of cycle analysis, the cells were centrifuged, washed in PBS, resuspended in 1 ml of PBS containing 100 μg/ml RNase A (TAKARA) per 106 cells, and incubated at 37 °C for 30 min. To determine the DNA content, 30 000 cells were analysed by flow cytometry using a FACS Calibur instrument and CellQuest software (BD Biosciences). To perform bivariate FACS, we used FlowCellect Bivariate Cell Cycle kit for DNA Replication Analysis (Millipore), according to the manufacture’s protocol.
siRNA
For siRNA experiments, we purchased Stealth RNAi siRNAs targeting CTBP1-AS, CTBP1 (HSS102437), HDAC1 (HSS179192), HDAC2 (HSS104729), HDAC3 (HSS189614), HDAC8 (HSS183330), Sin3A (HSS177954), NONO (HSS143135), PSF#1 (HSS109643), PSF#2 (HSS109642), SMAD3 (VHS41114), p53 (VHS40366), and a negative control siRNA from Invitrogen. Cells were transfected with RNA using Hiperfect transfection reagent (Quiagen) 48–72 h before each experiment. The Stealth RNAi siRNA sequence targeting CTBP1-AS (50 nM) was: 5′-UUAUGUCUCCAGCAAGCUUGGUCUU-3′. To exclude the possibility of off-target effect, we used two other siRNAs targeting CTBP1-AS that were purchased from Sigma Genosys Japan (Tokyo, Japan). These two siRNA sequences were siCTBP1-AS#2: 5′-CCAAUUAAUUAGACCACAAAA-3′ and siCTBP1-AS#3: 5′-CAACUGUCAAGAAACAAUUAG-3′.
Patients and tissue samples
We obtained 105 prostate cancer samples from surgeries performed at the University of Tokyo Hospital (Tokyo, Japan). The Tokyo University ethics committee approved this study, and informed consent was obtained from each patient before surgery. The age of the patients ranged from 52 to 78 years (mean, 66.8±6.0 years), and pretreatment serum PSA levels ranged from 2.2 to 136 ng/ml (mean, 16.9±19.5 ng/ml). The prostate tissue sections submitted for this study contained 95 benign and 105 cancerous foci. Besides 105 primary cancer samples excised at the time of operation, lymph node metastatic regions from seven cancer patients were also prepared. Other clinicopathological parameters are shown in Supplementary Table IV.
Immunohistochemistry
Formalin-fixed tissues were embedded in paraffin and sectioned. A Histofine Kit (Nichirei), which employs the streptavidin-biotin amplification method, was used for immunohistochemical analysis of CTBP1, and the antigen–antibody complex was visualized with 3,3′-diaminobenzidine solution (1 mM 3,3′-diaminobenzidine, 50 mM Tris–HCl buffer (pH 7.6), and 0.006% H2O2). In an immunohistochemical analysis, the immunoreactivity was evaluated in more than 1000 carcinoma cells for each case. Subsequently, the percentage of immunoreactivity was determined by two pathologists (TS and K-IT). An association between immunoreactivity and clinicopathological factors was evaluated using Student’s t-test, a cross-table using the χ2 test, or a correlation coefficient (r) and regression equation. Cancer-specific survival curve was generated according to the Kaplan–Meier method, and statistical significance was calculated using the log-rank test. Univariate and multivariate analyses were evaluated using Cox’s proportional hazard model with PROC PHREG in SAS software (V9.2; Cary, NC).
In situ hybridization
DNA fragments including a 500-bp of the CTBP1-3′ UTR (sequences not overlapping with CTBP1-AS, Chr4: 1195228–1195754) or CTBP1-AS (nonrepetitive sequences not overlapping with CTBP1 exons, Chr4: 1200796–1201396) were amplified by PCR and cloned into the Bluescript SK(−) vector. RNA probes for ISH were generated using a digoxigenin (DIG) RNA labelling kit (Roche) according to the manufacturer’s instruction. Their sense RNA probes were also used as negative controls in this study. mRNA ISH was performed in prostate carcinoma tissues fixed in 10% formalin and embedded in paraffin wax using a Discovery XT automated slide-processing system (Roche) (Suzuki et al, 2005). The slides were reacted by hybridization with CTBP1 or CTBP1-AS probe (100 ng per slide) for 3 h at 65 °C and specific signals were chromogemically detected using a BlueMap kit (Roche). Counterstain was performed by Nuclear Fast Red (Roche). The signals of ISH were evaluated by two trained pathologists (TS and K-IT).
Statistical analyses
For the cell proliferation assay, we analysed four wells. For the growth assay in vitro and in vivo of stable cell lines, we performed two-way analysis of variance (ANOVA) at each time point. For other cell line experiments, statistical differences (P-values) among groups were obtained using a two-sided Student’s t-test. All experiments were performed at least twice and similar results were obtained. The P-values of <0.05 were considered to be statistically significant. Statistical procedures were performed using Graphpad Prism 5 software (GraphPad Software, San Diego, CA) or Excel.
Cell proliferation assay
Cells were plated at 3 × 103 cells per well in 96-well plates. For RNAi experiments, cells were transfected with siRNA 24 h after cell plating. The MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt) assay was performed using CellTiter 96 Aqueous MTS reagent (Promega), according to the manufacturer’s protocol. The experiment was performed in quintuplicate. For cell counting, cells were trypsinized and counted using the Trypan blue exclusion method to quantify cell viability.
In vivo tumour formation assay
LNCaP (3 × 106) or LTAD (1 × 107) cells were injected subcutaneously (s.c.) into each side of twenty 5-week-old male nude mice. When the tumour volume reached 100 mm3, castration was performed in mice with LTAD cell tumours. Each tumour was transfected with 5 μg of CTBP1-AS RNAi or control RNA 3 times a week by using Lipofectamine RNAi MAX Transfection Reagent. Tumour volume was determined using the formula 0.5 × r1 × r2 × r3 (r1<r2<r3).
Supplementary Material
Acknowledgments
We are very grateful to Dr T Kanda (Aichi Cancer Center Research Institute) for technical advice about FISH experiment and Dr B Blumberg (University of California, Irvine) for critical reading. We thank N Sasaki for technical assistance. This work was supported by Grants of the Cell Innovation Program (to SI) and P-Direct (to SI) from the MEXT, Japan; by grants (to SI and KT) from the JSPS, Japan; by Grants-in-Aid (to SI) from the MHLW, Japan; by the Program for Promotion of Fundamental Studies in Health Sciences (to SI), NIBIO, Japan.
Author contributions: KT designed and performed biological experiments; SK analysed the CAGE data; TS and KIT performed the clinicopathological analysis; SHT analysed the ChIP-chip and ChIP-seq data; TF, YH, and SAT analysed the tumour samples; YO, YH, and HA supported the study; SI designed and supervised the study; and KT, KH-I, TU, KI, and SI wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bao BY, Chuang BF, Wang Q, Sartor O, Balk SP, Brown M, Kantoff PW, Lee GS (2008) Androgen receptor mediates the expression of UDP-glucuronosyltransferase 2 B15 and B17 genes. Prostate 68: 839–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman LM, Birts CN, Darley M, Gabrielli B, Blaydes JP (2009) CtBPs promote cell survival through the maintenance of mitotic fidelity. Mol Cell Biol 29: 4539–4551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CD, Welsbie DS, Tran C, Baek SH, Chen R, Vessella R, Rosenfeld MG, Sawyers CL (2004) Molecular determinants of resistance to antiandrogen therapy. Nat Med 10: 33–39 [DOI] [PubMed] [Google Scholar]
- Chen LL, DeCerbo JN, Carmichael GG (2008) Alu element-mediated gene silencing. EMBO J 27: 1694–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnadurai G (2007) Transcriptional regulation by C-terminal binding proteins. Int J Biochem Cell Biol 39: 1593–1607 [DOI] [PubMed] [Google Scholar]
- Chinnadurai G (2009) The transcriptional corepressor CtBP: a foe of multiple tumor suppressors. Cancer Res 69: 731–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culig Z, Hoffmann J, Erdel M, Eder IE, Hobisch A, Hittmair A, Bartsch G, Utermann G, Schneider MR, Parczyk K, Klocker H (1999) Switch from antagonist to agonist of the androgen receptor bicalutamide is associated with prostate tumor progression in a new model system. Br J Cancer 82: 242–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debes JD, Tindall DJ (2004) Mechanism of androgen-refractory prostate cancer. N Eng J Med 351: 1488–1490 [DOI] [PubMed] [Google Scholar]
- FANTOM Consortium; RIKEN Genome Exploration Research Group and Genome Science Group (Genome Network Project Core Group). (2005) The transcriptional landscape of the mammalian genome. Science 309: 1559–1563 [DOI] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY (2010) Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464: 1071–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, Lander ES (2009) Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458: 223–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hah N, Danko CG, Core L, Waterfall JJ, Siepel A, Lis JT, Kraus WL (2011) A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell 145: 622–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes SA, Zarnegar M, Sharma M, Yang F, Peehl DM, ten Dijke P, Sun Z (2001) SMAD3 represses androgen receptor-mediated transcription. Cancer Res 61: 2112–2118 [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nature Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, Suzuki H, Carninci P, Hayashizaki Y, Wells C, Frith M, Ravasi T, Pang KC, Hallinan J, Mattick J, Hume DA et al. RIKEN Genome Exploration Research Group; Genome Science Group (Genome Network Project Core Group); FANTOM Consortium. (2005) Antisense transcription in the mammalian transcriptome. Science 309: 1564–1566 [DOI] [PubMed] [Google Scholar]
- Kurokawa R, Rosenfeld MG, Glass CK (2009) Transcriptional regulation through noncoding RNAs and epigenetic modifications. RNA Biol 6: 233–236 [DOI] [PubMed] [Google Scholar]
- Lee JT, Davidow LS, Warshawsky D (1999) Tsix, a gene antisense to Xist at the X-inactivation centre. Nat Genet 21: 400–404 [DOI] [PubMed] [Google Scholar]
- Mathur M, Tucker PW, Samuels HH (2001) PSF is a novel corepressor that mediates its effect through Sin3A and the DNA binding domain of nuclear hormone receptors. Mol Cell Biol 21: 2298–2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger E, Wissmann M, Yin N, Müller JM, Schneider R, Peters AH, Günther T, Buettner R, Schüle R (2005) LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 437: 436–439 [DOI] [PubMed] [Google Scholar]
- Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, Brenner JC, Laxman B, Asangani IA, Grasso CS, Kominsky HD, Cao X, Jing X, Wang X, Siddiqui J, Wei JT, Robinson D, Iyer HK, Palanisamy N, Maher CA, Chinnaiyan AM (2011) Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol 29: 742–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM (2004) ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 6: 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY (2007) Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129: 1311–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokhlin OW, Taghiyev AF, Guseva NV, Glover RA, Chumakov PM, Kravchenko JE, Cohen MB (2005) Androgen regulates apoptosis induced by TNFR family ligands via multiple signal pathway in LNCaP. Oncogene 24: 6733–6784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlomn T, Iwers L, Kirstein P, Jessen B, Kollermann J, Minner S, Passow-Drolet A, Mirlacher M, Milde-Langosch K, Graefen M, Haese A, Steuber T, Simon R, Huland H, Sauter G, Erbersdobler A (2008) Clinical significance of p53 alterations in surgically treated prostate cancers. Mod Pathol 21: 1371–1378 [DOI] [PubMed] [Google Scholar]
- Shav-Tal Y, Zipori D (2002) PSF and p54(nrb)/NonO--multi-functional nuclear proteins. FEBS Lett 531: 109–114 [DOI] [PubMed] [Google Scholar]
- Shenk JL, Fisher CJ, Chen SY, Zhou XF, Tillman K, Shemshedini L (2001) p53 represses androgen-induced transactivation of prostate-specific antigen by disrupting hAR amino- to carboxyl-terminal interaction. J Biol Chem 276: 38472–38479 [DOI] [PubMed] [Google Scholar]
- Shi Y, Sawada J, Sui G, Affar el B, Whetstine JR, Lan F, Ogawa H, Luke MP, Nakatani Y, Shi Y (2003) Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature 422: 735–738 [DOI] [PubMed] [Google Scholar]
- Song K, Wang H, Krebs TL, Wang B, Kelley TJ, Danielpour D (2010) DHT selectively reverses Smad3-mediated/TGF-beta-induced responses through transcriptional down-regulation of Smad3 in prostate epithelial cells. Mol Endocrinol 24: 2019–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Sui A, Garen A (2004) Binding of mouse VL30 retrotransposon RNA to PSF protein induces genes repressed by PSF: effects on steroidogenesis and oncogenesis. Proc Natl Acad Sci USA 101: 621–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanbrough M, Bubley GJ, Ross K, Golub TR, Rubin MA, Penning TM, Febbo PG, Balk SP (2006) Increased expression of genes converting adrenal androgens to testosterone in androgen-independent prostate cancer. Cancer Res 66: 2815–2825 [DOI] [PubMed] [Google Scholar]
- Suzuki T, Miki Y, Fukuda T, Nakata T, Moriya T, Sasano H (2005) Analysis for localization of steroid sulfatase in human tissues. Methods Enzymol 400: 303–316 [DOI] [PubMed] [Google Scholar]
- Takayama K, Kaneshiro K, Tsutsumi S, Horie-Inoue K, Ikeda K, Urano T, Ijichi N, Ouchi Y, Shirahige K, Aburatani H, Inoue S (2007) Identification of novel androgen response genes in prostate cancer cells by coupling chromatin immunoprecipitation and genomic microarray analysis. Oncogene 26: 4453–4463 [DOI] [PubMed] [Google Scholar]
- Takayama K, Tsutsumi S, Suzuki T, Horie-Inoue K, Ikeda K, Kaneshiro K, Fujimura T, Kumagai J, Urano T, Sakaki Y, Shirahige K, Sasano H, Takahashi S, Kitamura T, Ouchi Y, Aburatani H, Inoue S (2009) Amyloid precursor protein is a primary androgen target gene that promotes prostate cancer growth. Cancer Res 69: 137–142 [DOI] [PubMed] [Google Scholar]
- Takayama K, Tsutsumi S, Katayama S, Okayama T, Horie-Inoue K, Ikeda K, Urano T, Kawazu C, Hasegawa A, Ikeo K, Gojyobori T, Ouchi Y, Hayashizaki Y, Aburatani H, Inoue S (2011) Integration of cap analysis of gene expression and chromatin immunoprecipitation analysis on array reveals genome-wide androgen receptor signaling in prostate cancer cells. Oncogene 30: 619–630 [DOI] [PubMed] [Google Scholar]
- Takayama K, Horie-Inoue K, Suzuki T, Urano T, Ikeda K, Fujimura T, Takahashi S, Homma Y, Ouchi Y, Inoue S (2012) TACC2 is an androgen-responsive cell cycle regulator promoting androgen-mediated and castration-resistant growth of prostate cancer. Mol Endocrinol 26: 748–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B, Antipin Y, Mitsiades N, Landers T, Dolgalev I, Major JE, Wilson M, Socci ND, Lash AE, Heguy A, Eastham JA et al. (2010) Integrative genomic profiling of human prostate cancer. Cancer Cell 18: 11–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varambally S, Yu J, Laxman B, Rhodes DR, Mehra R, Tomlins SA, Shah RB, Chandran U, Monzon FA, Becich MJ, Wei JT, Ghosh D, Rubin MA, Chinnaiyan AM (2005) Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell 8: 393–406 [DOI] [PubMed] [Google Scholar]
- Wang J, Scully K, Zhu X, Cai L, Zhang J, Prefontaine GG, Krones A, Ohgi KA, Zhu P, Garcia-Bassets I, Liu F, Taylor H, Lozach J, Jayes FL, Korach KS, Glass CK, Fu XD, Rosenfeld MG (2007) Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature 446: 882–887 [DOI] [PubMed] [Google Scholar]
- Wang LG, Ossowski L, Ferrari AC (2001) Overexpressed androgen receptor linked to p21WAF1 silencing may be responsible for androgen independence and resistance to apoptosis of a prostate cancer cell line. Cancer Res 61: 7544–7551 [PubMed] [Google Scholar]
- Wang Q, Caroll JS, Brown M (2005) Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol Cell 19: 631–642 [DOI] [PubMed] [Google Scholar]
- Wang Q, Li W, Zhang Y, Yuan X, Xu K, Yu J, Chen Z, Beroukhim R, Wang H, Lupien M, Wu T, Regan MM, Meyer CA, Carroll JS, Manrai AK, Jänne OA, Balk SP, Mehra R, Han B, Chinnaiyan AM et al. (2009) Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell 138: 245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Asangani IA, Chakravarthi BV, Ateeq B, Lonigro RJ, Cao Q, Mani RS, Camacho DF, McGregor N, Schumann TE, Jing X, Menawat R, Tomlins SA, Zheng H, Otte AP, Mehra R, Siddiqui J, Dhanasekaran SM, Nyati MK, Pienta KJ et al. (2012) Role of transcriptional corepressor CtBP1 in prostate cancer progression. Neoplasia 14: 905–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelin R, Dahary D, Sorek R, Levanon EY, Goldstein O, Shoshan A, Diber A, Biton S, Tamir Y, Khosravi R, Nemzer S, Pinner E, Walach S, Bernstein J, Savitsky K, Rotman G (2003) Widespread occurrence of antisense transcription in the human genome. Nat Biotechnol 21: 379–386 [DOI] [PubMed] [Google Scholar]
- Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, Cui H. (2008) Epigenetic silencing of tumor suppressor gene p15 by its antisense RNA. Nature 451: 202–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.