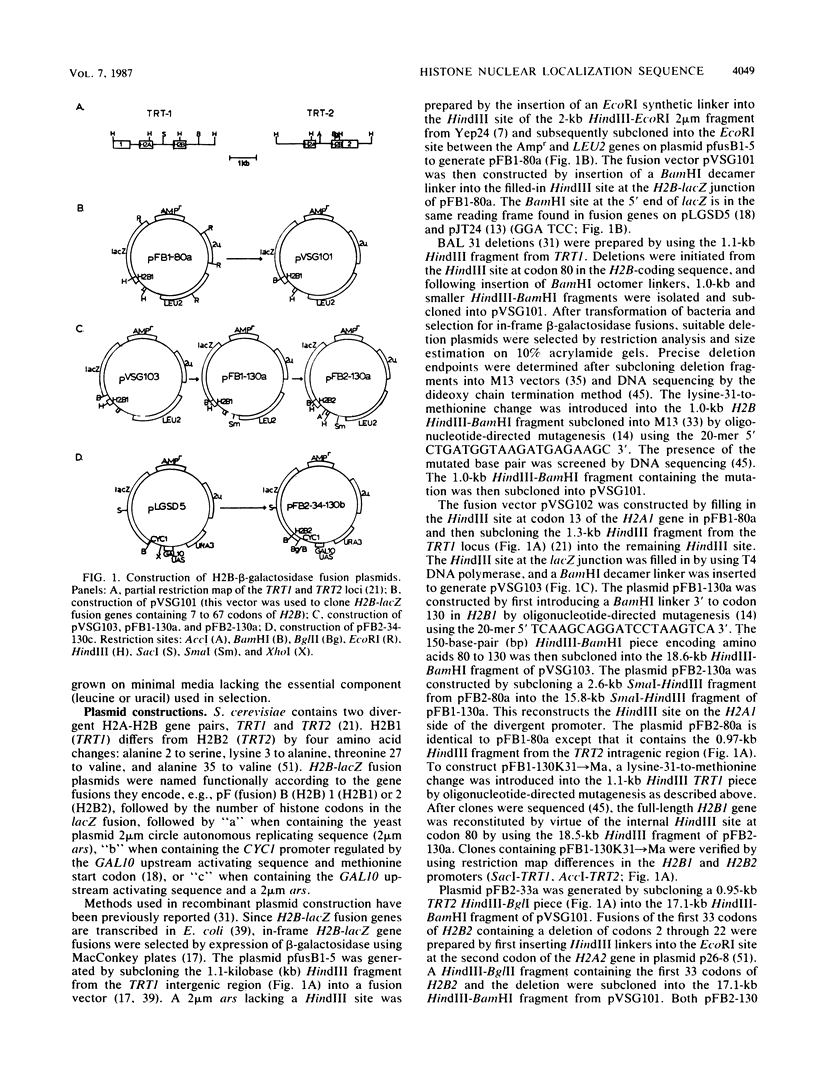
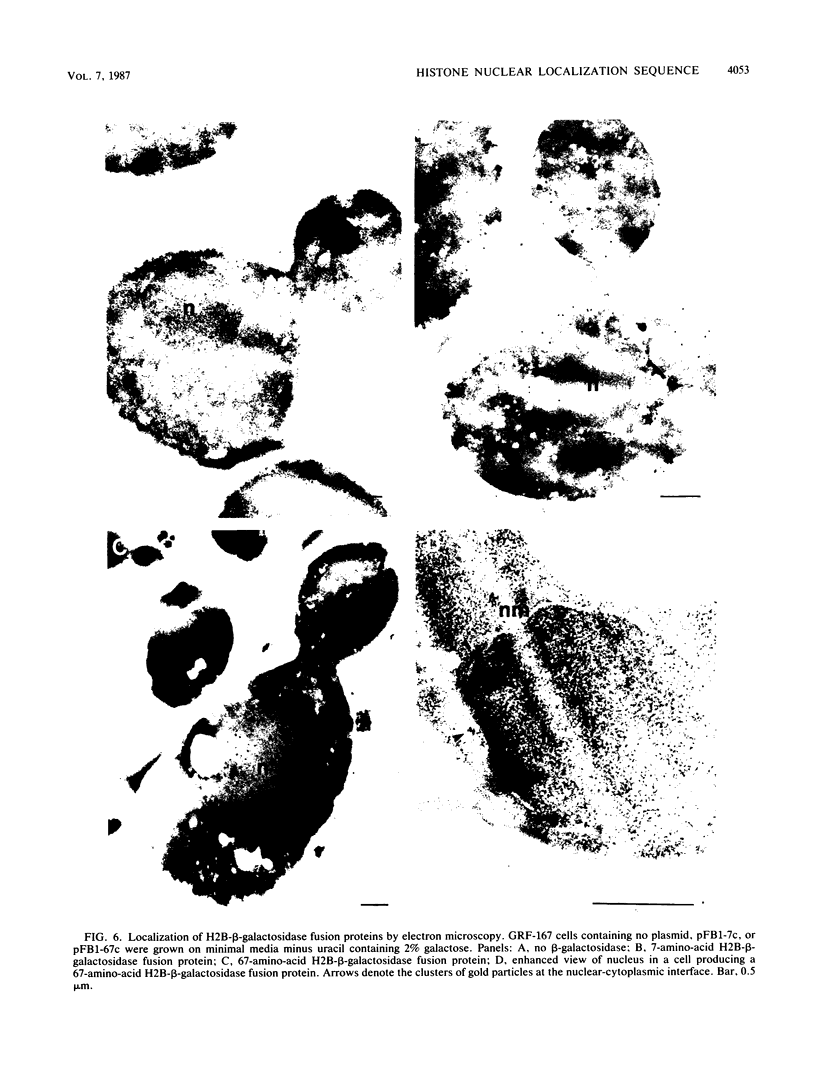
Abstract
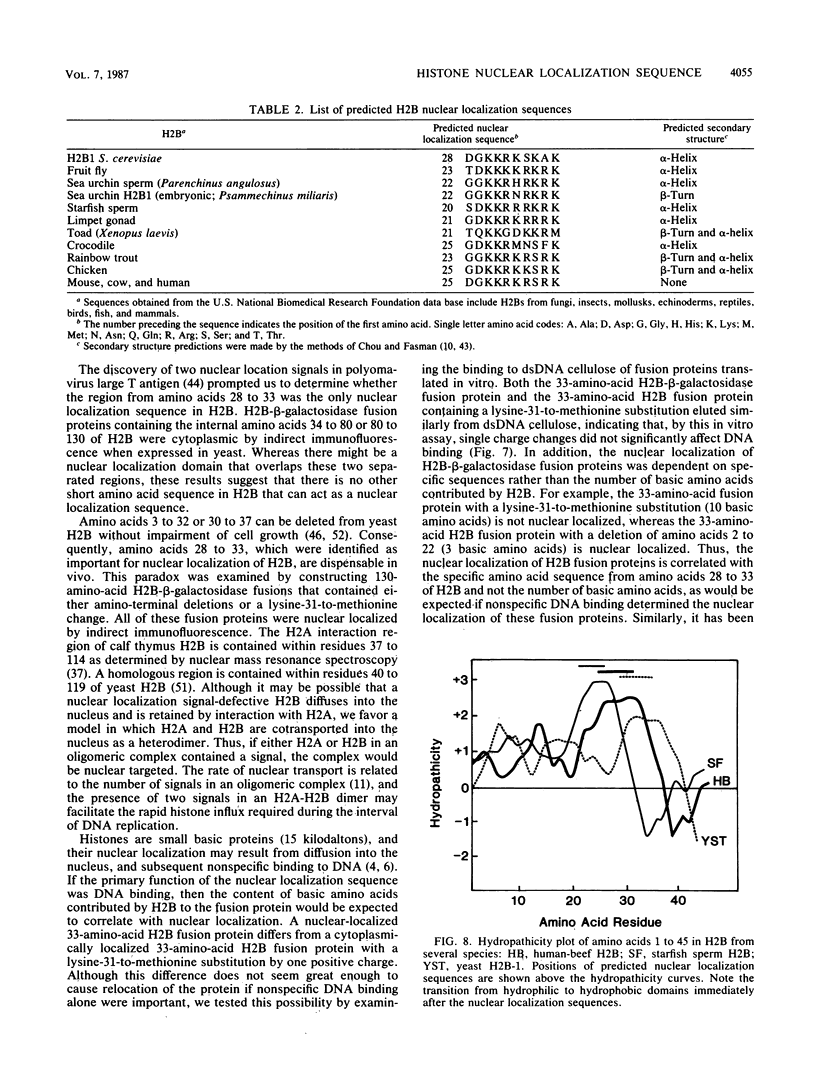
Histone-beta-galactosidase protein fusions were used to identify the domain of yeast histone 2B, which targets this protein to the nucleus. Amino acids 28 to 33 in H2B were required for nuclear localization of such fusion proteins and thus constitute a nuclear localization sequence. The amino acid sequence in this region (Gly-29 Lys Lys Arg Ser Lys Ala) is similar to the nuclear location signal in simian virus 40 large T antigen (Pro-126 Lys Lys Lys Arg Lys Val) (D. Kalderon, B.L. Roberts, W.D. Richardson, and A.E. Smith, Cell 39:499-509, 1984). A point mutation changing lysine 31 to methionine abolished nuclear localization of an H2B-beta-galactosidase fusion protein containing amino acids 1 to 33 of H2B. However, an H2B-beta-galactosidase fusion protein containing both this point mutation and the H2A interaction domain of H2B was nuclear localized. These results suggest that H2A and H2B may be cotransported to the nucleus as a heterodimer.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams A. E., Pringle J. R. Relationship of actin and tubulin distribution to bud growth in wild-type and morphogenetic-mutant Saccharomyces cerevisiae. J Cell Biol. 1984 Mar;98(3):934–945. doi: 10.1083/jcb.98.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendayan M., Zollinger M. Ultrastructural localization of antigenic sites on osmium-fixed tissues applying the protein A-gold technique. J Histochem Cytochem. 1983 Jan;31(1):101–109. doi: 10.1177/31.1.6187796. [DOI] [PubMed] [Google Scholar]
- Blake M. S., Johnston K. H., Russell-Jones G. J., Gotschlich E. C. A rapid, sensitive method for detection of alkaline phosphatase-conjugated anti-antibody on Western blots. Anal Biochem. 1984 Jan;136(1):175–179. doi: 10.1016/0003-2697(84)90320-8. [DOI] [PubMed] [Google Scholar]
- Bonner W. M. Protein migration into nuclei. I. Frog oocyte nuclei in vivo accumulate microinjected histones, allow entry to small proteins, and exclude large proteins. J Cell Biol. 1975 Feb;64(2):421–430. doi: 10.1083/jcb.64.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner W. M. Protein migration into nuclei. II. Frog oocyte nuclei accumulate a class of microinjected oocyte nuclear proteins and exclude a class of microinjected oocyte cytoplasmic proteins. J Cell Biol. 1975 Feb;64(2):431–437. doi: 10.1083/jcb.64.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein D., Falco S. C., Stewart S. E., Brennan M., Scherer S., Stinchcomb D. T., Struhl K., Davis R. W. Sterile host yeasts (SHY): a eukaryotic system of biological containment for recombinant DNA experiments. Gene. 1979 Dec;8(1):17–24. doi: 10.1016/0378-1119(79)90004-0. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Empirical predictions of protein conformation. Annu Rev Biochem. 1978;47:251–276. doi: 10.1146/annurev.bi.47.070178.001343. [DOI] [PubMed] [Google Scholar]
- Davey J., Dimmock N. J., Colman A. Identification of the sequence responsible for the nuclear accumulation of the influenza virus nucleoprotein in Xenopus oocytes. Cell. 1985 Mar;40(3):667–675. doi: 10.1016/0092-8674(85)90215-6. [DOI] [PubMed] [Google Scholar]
- De Robertis E. M., Longthorne R. F., Gurdon J. B. Intracellular migration of nuclear proteins in Xenopus oocytes. Nature. 1978 Mar 16;272(5650):254–256. doi: 10.1038/272254a0. [DOI] [PubMed] [Google Scholar]
- Dingwall C., Sharnick S. V., Laskey R. A. A polypeptide domain that specifies migration of nucleoplasmin into the nucleus. Cell. 1982 Sep;30(2):449–458. doi: 10.1016/0092-8674(82)90242-2. [DOI] [PubMed] [Google Scholar]
- Feldherr C. M., Kallenbach E., Schultz N. Movement of a karyophilic protein through the nuclear pores of oocytes. J Cell Biol. 1984 Dec;99(6):2216–2222. doi: 10.1083/jcb.99.6.2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried H. M., Nam H. G., Loechel S., Teem J. Characterization of yeast strains with conditionally expressed variants of ribosomal protein genes tcm1 and cyh2. Mol Cell Biol. 1985 Jan;5(1):99–108. doi: 10.1128/mcb.5.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillam S., Astell C. R., Smith M. Site-specific mutagenesis using oligodeoxyribonucleotides: isolation of a phenotypically silent phi X174 mutant, with a specific nucleotide deletion, at very high efficiency. Gene. 1980 Dec;12(1-2):129–137. doi: 10.1016/0378-1119(80)90023-2. [DOI] [PubMed] [Google Scholar]
- Gosselin E. J., Sorenson G. D., Dennett J. C., Cate C. C. Unlabeled antibody methods in electron microscopy: a comparison of single and multistep procedures using colloidal gold. J Histochem Cytochem. 1984 Aug;32(8):799–804. doi: 10.1177/32.8.6379035. [DOI] [PubMed] [Google Scholar]
- Guarente L., Lauer G., Roberts T. M., Ptashne M. Improved methods for maximizing expression of a cloned gene: a bacterium that synthesizes rabbit beta-globin. Cell. 1980 Jun;20(2):543–553. doi: 10.1016/0092-8674(80)90640-6. [DOI] [PubMed] [Google Scholar]
- Guarente L., Yocum R. R., Gifford P. A GAL10-CYC1 hybrid yeast promoter identifies the GAL4 regulatory region as an upstream site. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7410–7414. doi: 10.1073/pnas.79.23.7410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall M. N., Hereford L., Herskowitz I. Targeting of E. coli beta-galactosidase to the nucleus in yeast. Cell. 1984 Apr;36(4):1057–1065. doi: 10.1016/0092-8674(84)90055-2. [DOI] [PubMed] [Google Scholar]
- Hereford L. M., Osley M. A., Ludwig T. R., 2nd, McLaughlin C. S. Cell-cycle regulation of yeast histone mRNA. Cell. 1981 May;24(2):367–375. doi: 10.1016/0092-8674(81)90326-3. [DOI] [PubMed] [Google Scholar]
- Hereford L., Bromley S., Osley M. A. Periodic transcription of yeast histone genes. Cell. 1982 Aug;30(1):305–310. doi: 10.1016/0092-8674(82)90036-8. [DOI] [PubMed] [Google Scholar]
- Hereford L., Fahrner K., Woolford J., Jr, Rosbash M., Kaback D. B. Isolation of yeast histone genes H2A and H2B. Cell. 1979 Dec;18(4):1261–1271. doi: 10.1016/0092-8674(79)90237-x. [DOI] [PubMed] [Google Scholar]
- Isenberg I. Histones. Annu Rev Biochem. 1979;48:159–191. doi: 10.1146/annurev.bi.48.070179.001111. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalderon D., Richardson W. D., Markham A. F., Smith A. E. Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature. 1984 Sep 6;311(5981):33–38. doi: 10.1038/311033a0. [DOI] [PubMed] [Google Scholar]
- Kalderon D., Roberts B. L., Richardson W. D., Smith A. E. A short amino acid sequence able to specify nuclear location. Cell. 1984 Dec;39(3 Pt 2):499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lanford R. E., Butel J. S. Construction and characterization of an SV40 mutant defective in nuclear transport of T antigen. Cell. 1984 Jul;37(3):801–813. doi: 10.1016/0092-8674(84)90415-x. [DOI] [PubMed] [Google Scholar]
- Lanford R. E., Kanda P., Kennedy R. C. Induction of nuclear transport with a synthetic peptide homologous to the SV40 T antigen transport signal. Cell. 1986 Aug 15;46(4):575–582. doi: 10.1016/0092-8674(86)90883-4. [DOI] [PubMed] [Google Scholar]
- Leary J. J., Brigati D. J., Ward D. C. Rapid and sensitive colorimetric method for visualizing biotin-labeled DNA probes hybridized to DNA or RNA immobilized on nitrocellulose: Bio-blots. Proc Natl Acad Sci U S A. 1983 Jul;80(13):4045–4049. doi: 10.1073/pnas.80.13.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardian J. K., Isenberg I. Yeast inner histones and the evolutionary conservation of histone-histone interactions. Biochemistry. 1978 Sep 5;17(18):3825–3833. doi: 10.1021/bi00611a023. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., Felsenfeld G. Nucleosome structure. Annu Rev Biochem. 1980;49:1115–1156. doi: 10.1146/annurev.bi.49.070180.005343. [DOI] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreland R. B., Nam H. G., Hereford L. M., Fried H. M. Identification of a nuclear localization signal of a yeast ribosomal protein. Proc Natl Acad Sci U S A. 1985 Oct;82(19):6561–6565. doi: 10.1073/pnas.82.19.6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss T., Cary P. D., Abercrombie B. D., Crane-Robinson C., Bradbury E. M. A pH-dependent interaction between histones H2A and H2B involving secondary and tertiary folding. Eur J Biochem. 1976 Dec 11;71(2):337–350. doi: 10.1111/j.1432-1033.1976.tb11120.x. [DOI] [PubMed] [Google Scholar]
- Osley M. A., Gould J., Kim S., Kane M. Y., Hereford L. Identification of sequences in a yeast histone promoter involved in periodic transcription. Cell. 1986 May 23;45(4):537–544. doi: 10.1016/0092-8674(86)90285-0. [DOI] [PubMed] [Google Scholar]
- Osley M. A., Hereford L. Identification of a sequence responsible for periodic synthesis of yeast histone 2A mRNA. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7689–7693. doi: 10.1073/pnas.79.24.7689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paine P. L., Moore L. C., Horowitz S. B. Nuclear envelope permeability. Nature. 1975 Mar 13;254(5496):109–114. doi: 10.1038/254109a0. [DOI] [PubMed] [Google Scholar]
- Paucha E., Kalderon D., Richardson W. D., Harvey R. W., Smith A. E. The abnormal location of cytoplasmic SV40 large T is not caused by failure to bind to DNA or to p53. EMBO J. 1985 Dec 1;4(12):3235–3240. doi: 10.1002/j.1460-2075.1985.tb04071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters R. Nucleo-cytoplasmic flux and intracellular mobility in single hepatocytes measured by fluorescence microphotolysis. EMBO J. 1984 Aug;3(8):1831–1836. doi: 10.1002/j.1460-2075.1984.tb02055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pustell J., Kafatos F. C. A convenient and adaptable package of computer programs for DNA and protein sequence management, analysis and homology determination. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):643–655. doi: 10.1093/nar/12.1part2.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson W. D., Roberts B. L., Smith A. E. Nuclear location signals in polyoma virus large-T. Cell. 1986 Jan 17;44(1):77–85. doi: 10.1016/0092-8674(86)90486-1. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster T., Han M., Grunstein M. Yeast histone H2A and H2B amino termini have interchangeable functions. Cell. 1986 May 9;45(3):445–451. doi: 10.1016/0092-8674(86)90330-2. [DOI] [PubMed] [Google Scholar]
- Silver P. A., Brent R., Ptashne M. DNA binding is not sufficient for nuclear localization of regulatory proteins in Saccharomyces cerevisiae. Mol Cell Biol. 1986 Dec;6(12):4763–4766. doi: 10.1128/mcb.6.12.4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver P. A., Keegan L. P., Ptashne M. Amino terminus of the yeast GAL4 gene product is sufficient for nuclear localization. Proc Natl Acad Sci U S A. 1984 Oct;81(19):5951–5955. doi: 10.1073/pnas.81.19.5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. E., Kalderon D., Roberts B. L., Colledge W. H., Edge M., Gillett P., Markham A., Paucha E., Richardson W. D. The nuclear location signal. Proc R Soc Lond B Biol Sci. 1985 Oct 22;226(1242):43–58. doi: 10.1098/rspb.1985.0078. [DOI] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis J. W., Hereford L., Grunstein M. Histone H2B genes of yeast encode two different proteins. Cell. 1980 Dec;22(3):799–805. doi: 10.1016/0092-8674(80)90556-5. [DOI] [PubMed] [Google Scholar]
- Wallis J. W., Rykowski M., Grunstein M. Yeast histone H2B containing large amino terminus deletions can function in vivo. Cell. 1983 Dec;35(3 Pt 2):711–719. doi: 10.1016/0092-8674(83)90104-6. [DOI] [PubMed] [Google Scholar]