Abstract
Human surfactant protein A, an innate immunity molecule, is encoded by two genes: SFTPA1 (SP-A1) and SFTPA2 (SP-A2). The 5′ untranslated (5′UTR) splice variant of SP-A2 (ABD), but not of SP-A1 (AD), contains exon B (eB), which is an enhancer for transcription and translation. We investigated whether eB contains cis-regulatory elements that bind trans-acting factors in a sequence-specific manner as well as the role of the eB mRNA secondary structure. Binding of cytoplasmic NCI-H441 proteins to wild-type eB, eB mutant, AD, and ABD 5′UTR mRNAs were studied by RNA electromobility shift assays (REMSAs). The bound proteins were identified by mass spectroscopy and specific antibodies (Abs). We found that 1) proteins bind eB mRNA in a sequence-specific manner, with two cis-elements identified within eB to be important; 2) eB secondary structure is necessary for binding; 3) mass spectroscopy and specific Abs in REMSAs identified 14-3-3 proteins to bind (directly or indirectly) eB and the natural SP-A2 (ABD) splice variant but not the SP-A1 (AD) splice variant; 4) other ribosomal and cytoskeletal proteins, and translation factors, are also present in the eB mRNA-protein complex; 5) knockdown of 14-3-3 β/α isoform resulted in a downregulation of SP-A2 expression. In conclusion, proteins including the 14-3-3 family bind two cis-elements within eB of hSP-A2 mRNA in a sequence- and secondary structure-specific manner. Differential regulation of SP-A1 and SP-A2 is mediated by the 14-3-3 protein family as well as by a number of other proteins that bind UTRs with or without eB mRNA.
Keywords: 5′ untranslated regions, translation, trans-acting factors, 14-3-3 family of proteins, electromobility shift assays
interindividual variances in predisposition and severity of pulmonary infections and inflammatory lung disease are poorly understood. Alveolar macrophages (AM), the sentinel cells of lung innate immunity (52), play an indispensable role against infection. Inappropriate control or imbalance of AM-mediated immune responses in the lung can cause acute or chronic inflammation. Surfactant protein A (SP-A), aside from its surfactant-related functions (i.e., stability, structure, surfactant secretion), has an essential role in innate host defense. SP-A modulates the phenotype of AM (64) and enhances phagocytosis of pathogens, as well as affects cytokine production and other host defense functions (14, 23, 63, 105).
Human SP-A is encoded by two homologous genes (SFTPA1: SP-A1, and SFTPA2: SP-A2) with several genetic and 5′ untranslated region (5′UTR) splice variants identified for each gene (Fig. 1A). Functional, structural, biophysical, and biochemical differences have been observed between the two SP-A proteins (26, 57, 58, 91, 92, 96–98). Genetic heterogeneity in SP-A genes has been shown to associate with susceptibility to lung infections and predisposition to lung inflammatory diseases in neonates, children, and adults (22, 78). Alterations in total SP-A levels have been observed for several pulmonary diseases: reduced levels in airway infections (7) and inflammatory lung diseases, including cystic fibrosis (55, 65) and lung transplantation (9), and higher levels in sarcoidosis (34) and hypersensitivity pneumonitis (33, 34). The ratio of SP-A1 to total SP-A protein has been shown to differ in bronchoalveolar lavage among individuals, as a function of age and lung health status (85, 100), supporting the notion that imbalance of SP-A1 and SP-A2 content contributes to susceptibility to lung disease.
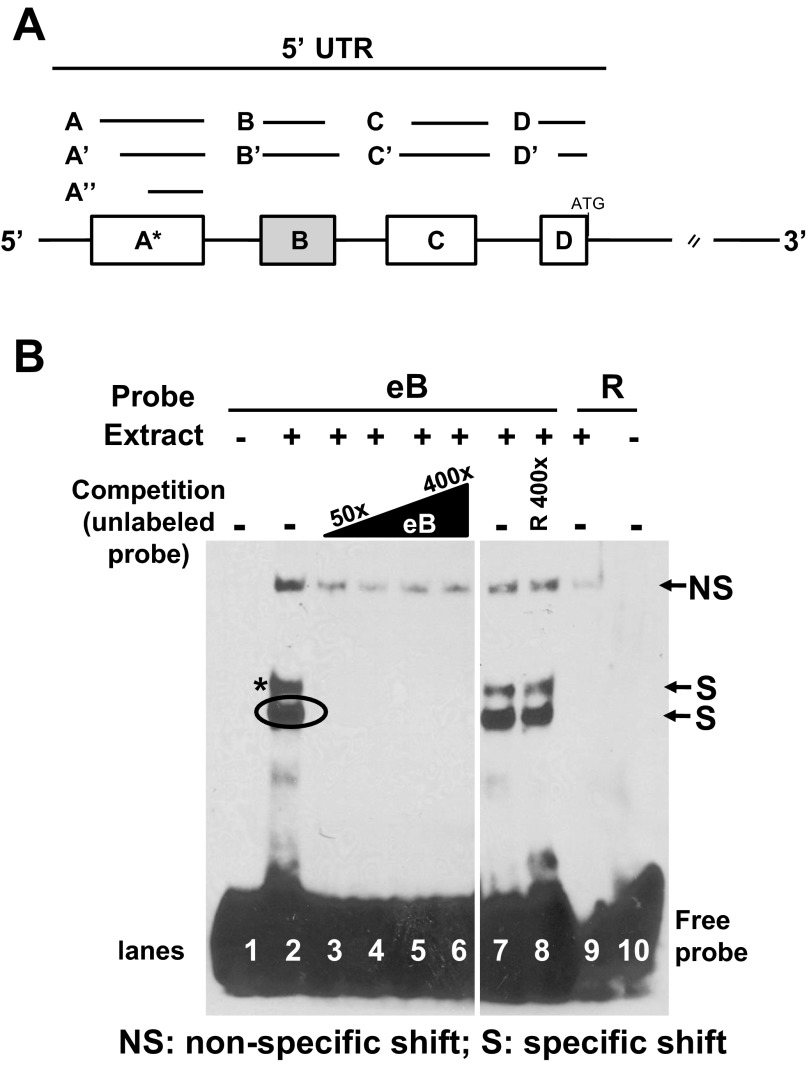
Fig. 1.
5′ Untranslated region (5′UTR) structure of SP-A1 and SP-A2 and exon B (eB) sequence-specific shifts. A: exons A, B, C, D differ in length and are represented by lines and boxes. The sizes of each untranslated exon are as follows: A (44 nt), A′ (39 nt), A″ (34 nt), B (30 nt), B′ (70 nt), C (60 nt), C′ (63 nt), D (26 nt), and D′ (23 nt). Alternative splicing of 5′UTR exons gives rise to several SP-A1 and SP-A2 variants; the most frequent variant for SP-A1 is AD′ and for SP-A2 are ABD and ABD′. B: RNA electromobility shift assays (REMSA) of eB biotinylated probe with cytoplasmic extract of NCI-H441 cells. Lane 1, free probe (negative control); lanes 2 and 7, 200 nM eB riboprobe incubated with 8.5 μg extract (positive controls); lanes 3–6, competition of eB-mediated shifts with 50-, 100-, 200-, and 400-fold molar excess of unlabeled eB RNA, respectively; lane 8, competition of eB shifts with 400-fold molar excess of unlabeled random (R) RNA; lane 9, R biotinylated probe incubated with extract; lane 10, R probe; (+) presence of NCI-H441 cytoplasmic extract; (−) absence of cytoplasmic extract or competitor RNA. The top shift band is a nonspecific shift (NS) band because it appears in the random probe (R) (lane 9) and is not competed. Lanes 1–10 are from the same film but some lanes between lanes 6 and 7 were removed.
In human SP-A mRNAs, a number of untranslated exons (A, B, B′, C, D, D′) splice in different configurations (ABD, AD′, etc.) to give rise to several 5′UTR splice variants (Fig. 1A) (44), with the AD′ variant being the most common for SP-A1 and the ABD and ABD′ for SP-A2. SP-A mRNAs with different 5′UTR splice variants are translated both in vitro and in vivo (44, 45). For the latter, differences were observed among individuals (45) in the relative amount of a given 5′UTR variant in the polysomal (translated) and nonpolysomal (nontranslated) fractions, indicating that SP-A 5′UTR splice variants are subject to differential translational control. Transient transfection of constructs containing SP-A 5′UTR variants (93) showed that constructs with the SP-A2 ABD 5′UTR are translated more efficiently than those with SP-A1 5′UTRs (AD′, AB′D′, or ACD′), indicating a role of 5′UTRs in translational control of SP-A variants. In addition, the SP-A 5′UTR ABD mRNA variant was shown to have a lower rate of mRNA decay, compared with SP-A 5′UTR splice variants that lack the untranslated exon B (eB). eB is included in the 5′UTRs of the common SP-A2 splice variants, but not in those of SP-A1 variants. Furthermore, the eB was shown to contain specific regions that mediate cap-independent translation (95). Recently, eB was shown to be a transcriptional and translational enhancer, and to contain regulatory elements for both transcription and translation (79). eB increases mRNA content regardless of position and orientation, enhances translation when placed in either orientation within its natural 5′UTR sequence or in heterologous 5′UTRs.
SP-A1 and SP-A2 gene regulation is a dynamic, multifaceted, and complex biological process (80, 93, 94) that is achieved through a multitude of mechanisms, most of which are still not well understood. In many genes translational regulation is mediated by many mRNA regulatory elements including the UTRs (2, 29, 47, 49). Important roles have been identified for 5′UTRs in posttranscriptional RNA processing, mRNA stability, and localization, along with ribosomal recycling and translation initiation (62). Differences in SP-A expression among individuals, and/or among variants in model systems, have been described (80, 85, 93, 100). These differences, singly or synergistically, may account for individual susceptibilities to environmental cues, oxidative stress, disease, or other insults.
The translation initiation complex requires the concerted action of numerous initiation factors/proteins to bind correctly and position mRNAs on the 40S ribosome. Specific cis-regulatory elements, frequently located primarily within the untranslated regions, and trans-acting factors (often proteins) bind mRNA or ribosomal subunits to generate the RNA ribosomal complex and enable the translation initiation process usually via binding to specific cis-elements. Regulatory trans-acting proteins may enhance or attenuate translation of certain mRNAs. For example, the 14-3-3 σ isoform has been shown to be a regulator of mitotic translation by binding directly eukaryotic translation/initiation factors such as eIF4A1, eIF4B, eIF4G, eIF3, eIF2α, and eukaryotic elongation factor 1α (104). The 14-3-3 proteins are conserved, abundant, 28- to 33-kDa acidic polypeptides widely expressed in all eukaryotic organisms and are involved in several cellular processes, including apoptosis, intracellular signaling/trafficking, response to environmental cues and actin dynamics, and other interactions (1, 3, 24, 60, 81, 86, 89). In humans, >200 proteins have been found to bind seven isoforms of 14-3-3 protein directly or as part of multiprotein complexes (68). Homo- or heterodimerization of 14-3-3 proteins is important for their function and they bind a large number of partners usually by recognition of a phosphoserine- or phosphothreonine-rich containing motif, resulting in modulation of the activity of the binding partner by conformational changes, masking of phosphorylated regions, binding with other proteins, and phosphorylation (5, 6, 16, 51, 61).
In the present work, we studied the hypothesis that eB contains cis-elements that trans-acting factors bind in a sequence-specific manner. Toward this, 1) we generated several in vitro mRNAs [eB WT, mRNAs containing eB (SP-A2 5′UTR: ABD) or lacking eB (SP-A1 5′UTR: AD)] and used these in shift assays to study whether specific protein factors bind these mRNAs; 2) we generated eB deletion mutant mRNAs as well as eB mutant transcripts maintaining the secondary structure characteristics of the WT eB to study cis-regulatory elements; 3) we used mass spectroscopy and specific antibody (Ab) competition shift assays to identify binding of 14-3-3; other ribosomal, cytoskeletal, and translational proteins were also identified; and 4) we used shRNAs to knock down the YWHAB gene (14-3-3 β/α) in NCI-H441 cell line and correlate 14-3-3 inhibition with SP-A2 levels.
MATERIALS AND METHODS
Recombinant Constructs
Four different SP-A 5′UTRs were cloned into EcoRI and PstI sites of pGEM-3Z vector (Promega, Madison, WI) downstream of T7 RNA polymerase promoter. The 5′UTR inserts included 1) the natural SP-A2 5′UTR ABD (100 nt) (44); 2) the SP-A1 5′UTR AD (70 nt) (44); 3) eB (GUC GCU GAU UUC UUG GAG CCU GAA AAG AAG); and 4) a random (R) sequence (UCG GGC CUC AUC CCG ACG CAG UUC UUC GUC) of the same length as eB (30 nt) obtained from “shuffle DNA” software (84) having as template a coding sequence of the human SP-B gene. After cloning, the EcoRI restriction site was removed and the PstI restriction endonuclease site was substituted with HindIII (to provide a 5′ protruding end necessary for in vitro transcription) by QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA) following the manufacturer's protocol.
Deletion Mutant Constructs
The eB 30-nt sequence was modified by site-directed mutagenesis with specific primers. We generated deletion (Del) mutants by a sequential and overlapping deletion of 10 nucleotides (n = 5) as shown in results. The primers, listed in Table 1, were synthesized in the macromolecular core facility of Pennsylvania State University College of Medicine.
Table 1.
Primers used in this study
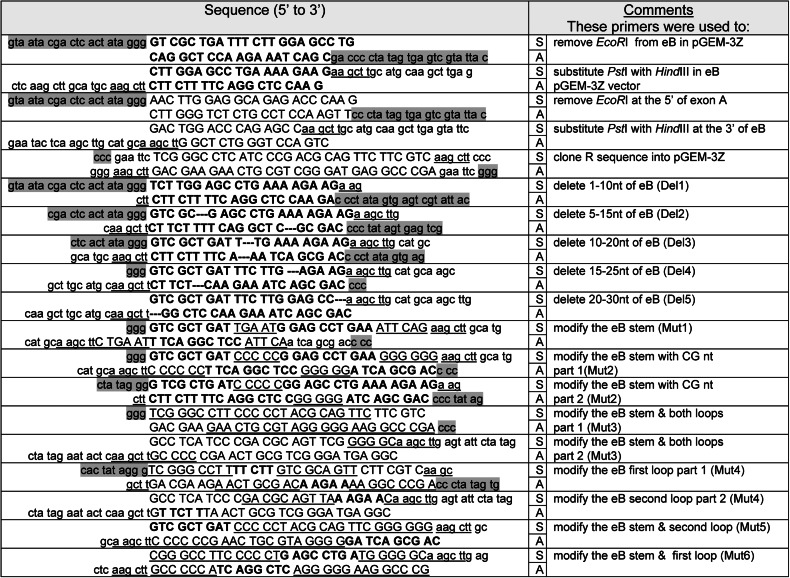
Highlighted lower case on the 5′ part of each sense (S) primer or on the 3′ of antisense (A) primer indicates part of the T7 promoter sequence; lower case (not highlighted) indicates the restriction sites of the vector; the HindIII restriction site or part of it is indicated with underlined lower case; upper case indicates the insert cloned in the pGEM-3Z vector; exon B (eB) sequence or part of it is denoted with bold upper case, and underlined upper case indicates the modified sequences of stem or loops; --- denotes a 10-nt deletion of eB sequence (see materials and methods).
eB Mutant Constructs With Characteristics of Secondary Structure Maintained
The QuikChange Lightning Site-Directed Mutagenesis Kit was used to generate a series of mutant (Mut) constructs (n = 6), by changing sequences corresponding to different domains of eB secondary structure with random (R) sequences (Table 2). RNA secondary structures were obtained by use of the RNAfold web server (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi, Institute for Theoretical Chemistry, University of Vienna, Austria) (31). In the present study, we used the secondary structure with the best possible combination of paired bases and the minimum free energy (dG); dG was normalized for size, by dividing dG by the total number of bases of each experimental construct. The sequences of all experimental constructs were verified by DNA sequencing (Eurofins MWG Operon, Huntsville, AL, and Molecular Genetics Core Facility at Pennsylvania State University, College of Medicine).
Table 2.
Sequences of eB mutants with conserved secondary structure
| Pseudo-Loop | Stem | Loop | Stem | Shift 1 (*) | Shift 2 (circled) | |
|---|---|---|---|---|---|---|
| Exon B (WT) | GUC GCU GAU | UUC UU | G GAG CCU GAA | AAG AAG | + | + |
| Mut 1 | GUC GCU GAU | uga au | G GAG CCU GAA | auu cag | + | + |
| Mut 2 | GUC GCU GAU | ccc cc | G GAG CCU GAA | ggg ggg | + | + |
| Mut 3 | ucg ggc cuu | ccc cc | u acg cag uuc | ggg ggc | − | − |
| Mut 4 | ucg ggc cuu | UUC UU | g ucg cag uua | AAG AAc | − | − |
| Mut 5 | GUC GCU GAU | ccc cc | u acg cag uuc | ggg ggg | + | − |
| Mut 6 | ucg ggc cuu | ccc cc | u GAG CCU GAu | ggg ggc | − | + |
WT, wild-type eB; upper case (bold or regular) indicates eB sequence; upper case and bold indicates the 2 cis-elements within eB; lower case denotes the mutant sequence; Mut, mutant. Pseudo-loop and loop are shown, respectively, with asterisk (*) and circle in shift assays of Figs. 1B, 3A, and 4; presence (+) or absence (−) of specific shift.
Cell Culture
The human lung adenocarcinoma cell line NCI-H441 was purchased from the American Type Culture Collection (ATCC, Manassas, VA). The NCI-H441 cell line expresses endogenous SP-A proteins and thus these cells contain factors required for SP-A protein translation. Cells were grown in RPMI 1640 medium (Invitrogen Life Technologies, Carlsbad, CA) with 10% heat-inactivated fetal bovine serum (FBS; Gemini, Atlanta, GA), 1× antimycotic-antibiotic solution (Sigma, St. Louis, MO), and 1× l-glutamine (Sigma). The cells were cultured at 37°C in 5% (vol/vol) CO2 atmosphere and were subcultured weekly in 10-cm dishes to 50–80% confluence.
Knockdown of 14-3-3 β/α Isoform in NCI-H441 Cell Line
SureSilencing shRNA plasmids for human 14-3-3 β/α isoform (Qiagen, Valencia, CA) were employed to knock down human YWHAB gene by RNA interference. Eight different plasmids with a different shRNA sequence were used. A plasmid encoding a scrambled shRNA that does not target any human, mouse, or rat gene was used as control. NCI-H441 cells were transfected with 0.40 μg of each 14-3-3 β/α-specific shRNA plasmid and 3 μl of Attractene transfection reagent (Qiagen) in Opti-MEM reduced-serum medium (Life Technologies, Grand Island, NY) and then were incubated for 48 h at 37°C in 5% (vol/vol) CO2 atmosphere, followed by 100 μg/ml hygromycin B (Life Technologies) selection for 7 days, per vendor's protocol. Stably transfected NCI-H441 cell lines were isolated, expanded, and studied for 14-3-3 β/α, 14-3-3 τ/θ, SP-A2, SP-A1 and actin expression by Western immunoblot. We used rabbit polyclonal Abs for detection of 14-3-3 β/α and 14-3-3 τ/θ (Cell Signaling, Danvers, MA) in dilution 1:1,000, rabbit polyclonal anti-SP-A2 (Aviva, San Diego, CA) in dilution 1:5,000, chicken SP-A1 gene-specific antibody (IgY) (85) in dilution 1:500, mouse monoclonal anti-actin (Sigma) in dilution 1:2,000, and appropriate secondary horseradish peroxidase-conjugated Abs (Bio-Rad, Hercules, CA). Western Lightning Plus-ECL Chemiluminescence substrate (PerkinElmer, Waltham, MA) was used to detect proteins.
Cytoplasmic Fractionation
Cytoplasmic extracts were obtained by using the NE-PER Nuclear and Cytoplasmic Extraction kit (Thermo Scientific, Rockford, IL), after harvest with PBS and trypsinization. Protein concentration was determined by the BCA protein assay (Thermo Scientific), following the manufacturer's instructions. Potential cross-contamination between cytosolic and nuclear fractions was checked by immunoblot using antibodies MEK1/2 (cytoplasmic) and c-Jun and/or histone H3 (nuclear) (Cell Signaling).
In Vitro Transcription
Plasmids (n = 15) containing WT ABD, experimental AD, eB, R, Del 1–5, and Mut 1–6 were linearized by digestion with HindIII (Invitrogen, Grand Island, NY) and purified by QIAquick PCR purification kit (Qiagen). RNA probes for gel mobility shift experiments were synthesized in vitro with the AmpliScribe T7-Flash Biotin-RNA Transcription Kit (Epicentre, Madison, WI), by using the linearized plasmid DNA as template following the manufacturer's protocol. In brief, a 20-μl reaction, containing 110 ng of linear plasmid DNA, 2 μl of 10× transcription buffer, 9 mM of NTP/Biotin-UTP PreMix, 10 mM DTT, 40 U RiboGuard RNase Inhibitor, and 10 U of AmpliScribe T7-Flash RNA Polymerase (Epicentre, Madison, WI) was incubated at 37°C for 1 h. One unit of RNase-Free DNase I (Epicentre), was added and the reaction was incubated at 37°C for 15 min to remove the DNA template. Unlabeled probes were prepared likewise with 9 mM of nonbiotinylated NTPs (Epicentre). RNA transcripts were purified using RNA-bee (TEL-TEST, Friendswood, TX) in conjunction with Direct-zol RNA MiniPrep columns (Zymo Research, Irvine, CA).
RNA Electromobility Shift Assay
Biotinylated RNA transcripts were heated at 80°C for 20 min and rapidly cooled on ice for 1 min prior to incubation with cytoplasmic protein extracts. Binding reactions (20 μl) contained 10 mM HEPES pH 7.3, 20 mM KCl, 1 mM MgCl2, 1 mM DTT, 1% (vol/vol) glycerol, 40 U of RNaseOUT recombinant ribonuclease inhibitor (Invitrogen, Carlsbad, CA), 2 μg of tRNA nonspecific competitor, 200 nM of in vitro transcribed biotinylated RNA, and 8.5 μg of cytoplasmic NCI-H441 extract. Reaction mixtures were incubated at room temperature (RT) for 20 min prior to loading on the gel. For competition shift assays a 50- to 400-fold molar excess of unlabeled in vitro transcribed RNA was used to compete specific interactions between RNA and cytoplasmic extracts. Unlabeled probes were incubated with protein extracts at RT 10 min prior to addition of biotinylated probe.
Samples were fractionated on native 6% (wt/vol) polyacrylamide gels in 1× Tris-borate-EDTA (TBE) and transferred to positively charged nylon membranes (Roche, Indianapolis, IN) by use of a semidry Amersham Biosciences Nova blot apparatus in 1× TBE at 400 mA for 30 min. Membranes were exposed to UV (254 nm) 120 mJ/cm2 for 45 s (Spectrolinker XL-1000 UV cross-linker; Spectronics, New Cassel, NY) for cross-linking. Band shifts were detected by use of the Chemiluminescent Nucleic Acid Detection Module (Thermo Scientific) and autoradiography (HyBlot CL film, Denville Scientific, Saint-Laurent, QC, Canada).
For identification of 14-3-3 on RNA electromobility shift assay (REMSA), 1–2 μg of 14-3-3 rabbit polyclonal antibody that recognizes all 14-3-3 isoforms (Abcam, Cambridge, MA) was added to the binding reactions and incubated for an additional 20 min at RT, for abrogation of the specific shift and/or supershift formation. As negative control the anti-DDK monoclonal antibody (anti-tag) (Origene, Rockville, MD) was used.
Mass Spectroscopy
RNA-protein complexes were fractionated on native 6% (wt/vol) polyacrylamide gel in 1× TBE buffer and stained with mass spectroscopy compatible SilverQuest Silver Staining Kit (Invitrogen) following the manufacturer's protocol. The stained gel was superimposed on the X-ray film from the REMSA and the bands corresponding to RNA-mediated specific shifts were excised. Silver ions were removed following the destaining protocol of the manufacturer. The destained gel bands were dehydrated in 100% (vol/vol) methanol for 5 min at RT, rehydrated in 30% (vol/vol) methanol for 5 min at RT, washed twice with ultrapure water for 10 min, followed by three washings with 100 mM ammonium bicarbonate in 30% (vol/vol) acetonitrile for 10 min at RT. Gel was then cut into small pieces and washed thoroughly in ultrapure water, dried in SpeedVac for 30 min, and resuspended in 50 mM ammonium bicarbonate. Ten nanograms of trypsin were added to each sample, which was then incubated at 37°C overnight and centrifuged at 12,000 g for 1 min, and the supernatant was transferred to a sterile microcentrifuge tube. The remaining peptides in the gel pellet were extracted with 20 μl 50% (vol/vol) acetonitrile containing 0.1% (vol/vol) trifluoroacetic acid at RT and combined with the previous supernatant. Samples were concentrated to 5 μl by use of SpeedVac and then analyzed with LC-MS/MS (Keck MS and Proteomics Yale University School of Medicine, New Haven, CT and Proteomics Core Facility at Pennsylvania State University, College of Medicine, Hershey, PA). A number of independent LC-MS/MS analyses were done from different REMSAs. For eB, n = 4; AD, n = 3; ABD, n = 3; Del 4, n = 2; Mut 5, n = 2; Mut 6, n = 2; eB competed with ABD, n = 2; eB competed with AD, n = 2.
RESULTS
Our published studies showed that SP-A 5′UTR splice variants differentially affect translation (45, 93) and that eB is an enhancer of transcription and translation (79). In this study, we found that trans-acting factors bind eB specific sequences, supporting the hypothesis that interactions between SP-A 5′UTR splice variants and regulatory proteins differentially affect expression of SP-A1 and SP-A2 (43, 46, 74).
Sequence-Specific Protein Shifts of the Untranslated eB
When eB in vitro transcribed biotinylated RNA as a probe and NCI-H441 cytoplasmic extracts were used, sequence-specific shifts (S) were observed (Fig. 1B, lanes 2 and 7). The eB shifts were competed (lanes 3-6) with eB-unlabeled probe (50- to 400-fold molar excess). However, the shifts remained (lane 8) when a random sequence probe (R) was used for competition. The R probe migrates to the bottom of the gel, and it did not form a sequence-specific shift with NCI-H441 cell extracts (lane 9). A non-sequence-specific (NS) shift was observed with all probes used. Similar observations were made with whole NCI-H441 extract (not shown). We next 1) studied whether specific sequences and/or the secondary structure of eB are required for the formation of the specific shift, 2) studied proteins that bind directly or indirectly and, 3) carried out a proof-of-principle experiment to determine whether inhibition of one of the identified proteins, shown to bind eB alone or within its surrounding sequences, correlated with reduced SP-A2 levels.
Specific eB Sequence Elements and Secondary Structure Contribute to the Sequence-Specific Protein Shifts
Two groups of mutants were generated, as described in materials and methods. One group included five deletion mutants and the other group included six mutants with eB secondary structure characteristics maintained.
Deletion mapping of eB cis-acting elements.
Figure 2 shows that, of the five mutants studied, only deletion mutants 3 and 4 retained the eB-specific shift (A), indicating that the sequence involved in the shift is located in the first half of eB toward the junction with exon A (B).
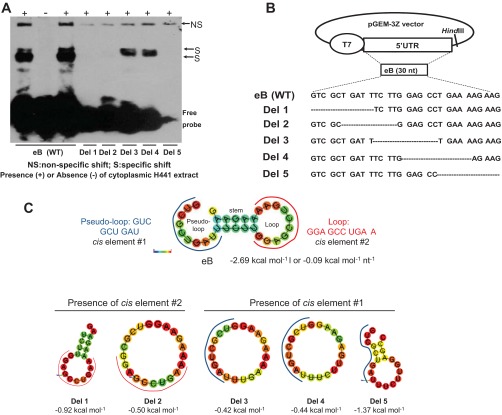
Fig. 2.
Deletion mapping of eB cis-acting elements and predicted secondary structure of WT eB and deletion mutant mRNAs. A: REMSA results of biotinylated wild-type (WT) and deletion (Del) mutant riboprobes with cytoplasmic extract of NCI-H441 cells. Del 3 and Del 4 formed sequence-specific shifts whereas the rest deletion mutants did not. B: the WT eB sequence and the sequences of the deletion mutants. The mutants were generated by site-directed mutagenesis in pGEM-3Z vector by sequentially removing 10 nt. C: the secondary structure of WT eB and deletion mutants, as well as the sequence and location of the two eB cis-elements. The eB secondary structure is described by a pseudo-loop, a stem, and a loop. The secondary structure of Del 5 differs from that of Del 3 and Del 4. In particular, the secondary structures of both Del 3 and Del 4 are single loops whereas Del 5 results in a small loop, a short stem, and a short pseudo-loop. The RNAfold online software was used to obtain the secondary structure of WT eB and deletion mutants. The minimum free energy secondary structure was used, with the best possible combination of paired bases, and the minimum free energy (ΔG) normalized for size, by dividing ΔG by the total number of bases of each experimental construct. The colors indicate the propensity of individual nucleotides to participate in base pairs. The scale ranges from red (highest probability) to blue-violet (lower probability).
eB mutants with eB secondary structure characteristics maintained identify two cis-elements.
The predicted secondary structure of the wild-type (WT) eB is operationally described here as consisting of a pseudo-loop, a stem, and a loop (Fig. 2C). Because the deletion mutants studied above (Fig. 2, A and B) disrupt normal formation of eB RNA secondary structure (Fig. 2C), we generated six mutants (Table 2) where the eB nucleotides were replaced with nucleotides unrelated to eB. These nucleotides were chosen so that the secondary structure of eB WT shown in Fig. 2C was maintained.
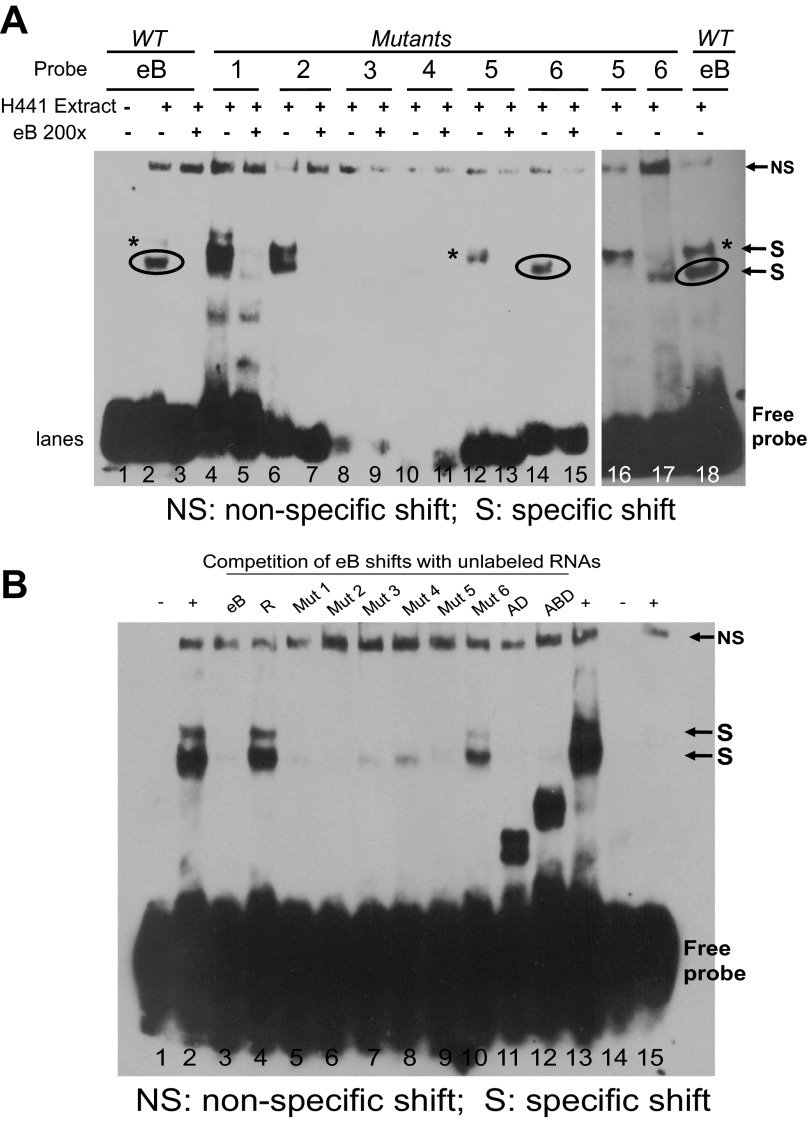
A summary of these mutants is shown in Table 2 and the sequences of the cis-elements in pseudo-loop and loop are marked. REMSAs (Fig. 3A) of Mut 1 and 2 with altered stem sequences produced prominent upper and lower shifts. In contrast, Mut 3 and 4 with altered loop sequences did not form shifts. The sequence of the pseudo-loop connects to exon A and the sequence of the loop is close to exon D, indicating that binding of protein factors occurs near A-B and B-D junctions. Interestingly, mutants with random sequences in either loop (Mut 5) or pseudo-loop (Mut 6) formed upper and lower shifts, respectively, indicating formation of different eB-cytoplasmic protein complexes at A and D junctions. The shift generated by Mut 5 exhibits a higher size (Fig. 3A, lane 12, marked with *) compared with that generated by Mut 6 (lane 14, marked by circle). Both of these are generated by eB (Fig. 1B, lanes 2 and 7, *, circle). The shifts of the eB mutant RNAs were competed with 200× molar excess of eB probe (Fig. 3A, lanes 5, 7, 13, and 15). However, when eB-mediated shifts were competed with 200× molar excess of eB mutant probes (Fig. 3B), these mutants (with secondary structure maintained as WT eB) eliminated or significantly attenuated the intensity of the shift regardless of whether they had partial or no sequence similarity. These observations are consistent with those of the deletion mutants (Fig. 2) and together support the notion that both sequence and secondary structure are important for formation and/or competition of the eB-mediated shift.
Fig. 3.
Shift assays of eB mutants (Mut) with conserved secondary structure and competition of eB-mediated shifts with each mutant. A: shift assays of eB mutants with conserved secondary structure. Lane 1, eB probe (negative control); lanes 2 and 18, eB riboprobe incubated with cytoplasmic NCI-H441 extract (positive controls); lane 3, competition of eB-mediated shifts with 200-fold molar excess of eB unlabeled RNA; lanes 4–17, mutant riboprobes incubated with extracts in the presence (+) or absence (−) of unlabeled WT eB 200-fold molar excess. Mut 1 and 2 formed shifts similar to the WT eB that were competed with excess WT eB unlabeled probe. Mut 1 also formed nonspecific shifts that were not competed with WT eB (nonspecific interactions). Mut 3 and 4 failed to form shifts. Mut 5 gave rise to the upper shift band (*) of the WT eB, whereas Mut 6 formed the lower shift band (circled) of the WT eB. B: competition assays of eB-mediated shifts, with 200-fold molar excess of mutant probes with conserved secondary structure, and excess AD and ABD unlabeled RNAs. Lane 1, eB probe (negative control); lanes 2 and 13, eB riboprobe incubated with cytoplasmic NCI-H441 extract (positive controls); lane 3, competition of eB-mediated shifts with unlabeled eB RNA; lane 4, competition of eB-mediated shifts with unlabeled random (R) RNA; lanes 5–10, competition of eB-mediated shifts with mutant unlabeled RNAs; lane 11, competition with unlabeled AD RNA; lane 12, competition with unlabeled ABD RNA; lane 14, R probe; lane 15, R biotinylated probe incubated with cytoplasmic NCI-H441 extract.
The sequence of the cis-element 1 (marked with a blue line in Fig. 2C) identified in the present manuscript is present in both Del 3 and Del 4. These deletion mutants form a large loop structure, and presumably the nucleotide sequence of the cis-element is free to interact and bind proteins whereas in Del 5 the sequence of this cis-element spans from the pseudo-loop to the stem and the loop (Fig. 2C, Del 5). It is possible that the Del 5 structure does not enable protein interactions as shown by its inability to form a shift. On the other hand, in Del 1 and Del 2 the cis-element 1 is deleted and these did not form any shifts despite the fact that these deletion constructs contain the nucleotide sequence of the cis-element 2. We speculate that the cis-element 2 alone (i.e., without the presence of the cis-element 1) cannot form a shift. However, Mut 6 (Fig. 3A, lane 14) that lacks the cis-element 1 (but contains cis-element 2) forms the lower size shift. Mut 6 in contrast to Del 1 and Del 2 maintains a secondary structure for both cis-elements similar to that of the WT eB. We conclude that the sequence and/or the secondary structure of cis-element 1 are necessary for a shift to form by the cis-element 2.
Mass Spectroscopy Analysis of Bound Proteins
LC-MS/MS analysis of the eB-, eB mutant-, AD-, and ABD-protein complexes identified a number of potential protein factors. The most significant expectation scores identified human 14-3-3 isoforms (β/α, γ, ζ/δ, ε, η, τ/θ, σ) to interact with eB, eB mutants, or ABD (Table 3) but not with the AD riboprobe, indicating that 14-3-3 proteins interact with eB or 5′UTRs containing eB. Of note, the following proteins were also found in REMSA protein shifts of ABD or eB: ribosomal 40S and 60S subunit proteins (SA and S3 for 40S subunit, and L5, L7, L11, L15, L22, and L26 for the 60S subunit), elongation factor 1-α1 (EF 1-α1), eukaryotic initiation factors (eIF4A, eIF4H, eIF6), cytokeratin 17 (KRT17), actin, tubulin, and calreticulin. However, for the AD-specific shift significantly fewer ribosomal or other proteins were present (Table 3 and Supplemental Table S1). Together, these data indicate that a number of proteins directly or indirectly bind eB, or ABD and AD, and that the 14-3-3 family of proteins is one of the groups of proteins that differentially bind eB or ABD but not AD.
Table 3.
Mass-spectroscopy protein identification of WT eB, AD, ABD, and mutant-mediated specific shifts
| eB band 1 (*) | eB band 2 (circled) | Del 4 | Mut 5 | Mut 6 | AD | ABD band 1 | ABD band 2 | eB comp.ABD | eB comp. AD | |
|---|---|---|---|---|---|---|---|---|---|---|
| 14-3-3 ζ/δ | + | + | + | + | + | + | + | |||
| 14-3-3 β/α | + | + | + | + | + | + | ||||
| 14-3-3 τ/θ | + | + | + | + | + | + | ||||
| 14-3-3 ε | + | + | + | + | + | + | + | |||
| 14-3-3 γ | + | + | + | + | ||||||
| 14-3-3 σ | + | + | + | |||||||
| 14-3-3 η | + | + | + | + | + | |||||
| 40S ribosomal protein SA | + | + | ||||||||
| 40S ribosomal protein S3 | + | |||||||||
| ubiquitin-40S ribosomal protein S27a | + | + | ||||||||
| 60S ribosomal protein L5 | + | + | + | + | + | + | ||||
| 60S ribosomal protein L7 | + | |||||||||
| 60S ribosomal protein L11 | + | + | ||||||||
| 60S ribosomal protein L15 | + | |||||||||
| 60S ribosomal protein L22 | + | + | + | + | + | + | + | |||
| 60S ribosomal protein L26 | + | |||||||||
| Elongation factor 1-α1 | + | + | + | + | + | |||||
| eIF4H | + | + | + | |||||||
| eIF4A | + | |||||||||
| eIF6 | + | + | ||||||||
| Actin, cytoplasmic | + | + | + | + | + | + | + | + | ||
| tubulin α | + | + | + | + | + | + | + | |||
| KRT17 | + | + | ||||||||
| calreticulin | + | + | + | + | + | + | + |
+, Presence of the particular protein factor; eB comp. ABD, eB shift was competed with excess ABD; eB comp. AD, eB shift was competed with excess AD. Del, deletion.
The 14-3-3 Protein Family Is Present in the eB- and the SP-A2 ABD-Mediated RNA-Protein Complex
eB-mediated shifts.
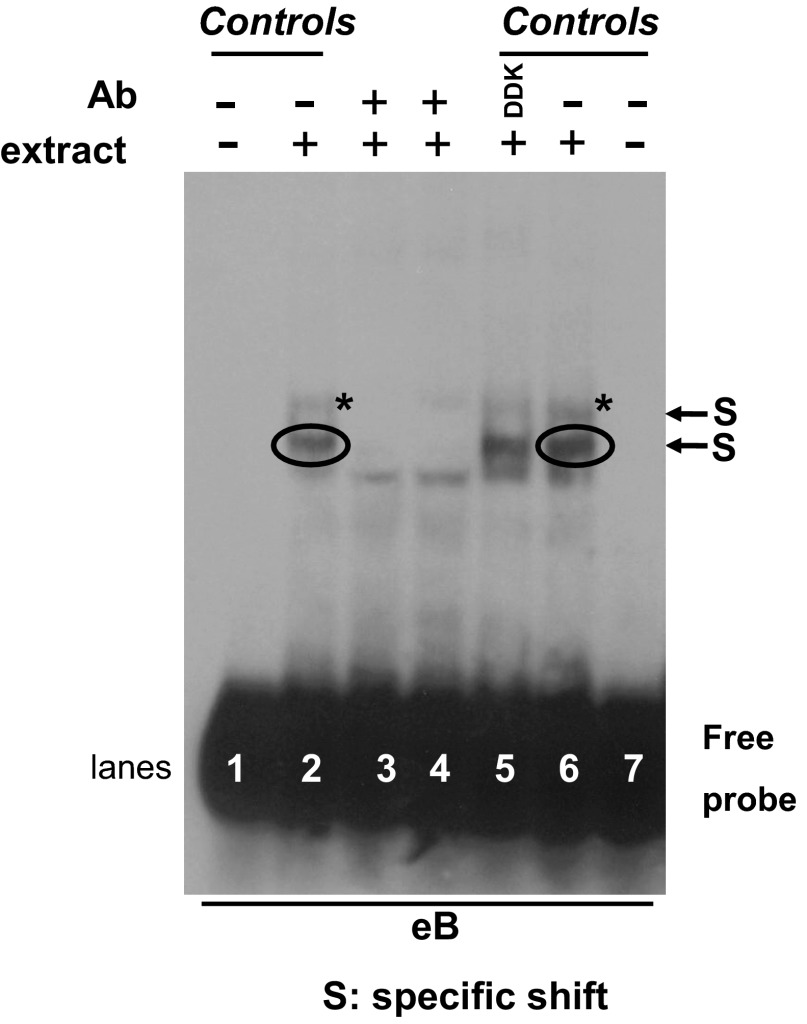
To further determine the presence and importance of the eB-mediated shifts we performed REMSA in the presence of 14-3-3 Ab. Figure 4 shows that the Ab that recognizes all isoforms of 14-3-3 competed the two specific shifts (S) (lanes 3 and 4) whereas an irrelevant control Ab (DDK) did not affect the eB shifts (lane 5). This is consistent with the mass spectroscopy data and provides further support of the presence of 14-3-3 in eB-mediated shift.
Fig. 4.
eB shift competition with specific 14-3-3 antibody (Ab). REMSA of eB biotinylated probe with NCI-H441 cytoplasmic extract in the presence of Ab against all isoforms of the 14-3-3 protein family. Lanes 1 and 7, eB probe (negative controls); lanes 2 and 6, eB riboprobe incubated with extract (positive controls); lanes 3 and 4, 14-3-3 Ab disrupts the eB-specific shifts; lane 5, eB riboprobe incubated with extract and an irrelevant control anti-DDK Ab. S, specific shift.
To further investigate the mass spectroscopy data that identified most human 14-3-3 isoforms and the REMSAs that in the presence of the specific 14-3-3 Ab resulted in abrogation of the eB specific shifts, we performed competition assays of eB shift with natural 5′UTR sequences that either included or did not include eB.
Competition of eB shift with ABD.
When the eB sequence-specific shift was competed with unlabeled ABD molar excess, the shift was not entirely disrupted as it may have been expected (Fig. 3B, lane 12). An altered mobility shift of a lower size was observed and mass spectroscopy analysis of this shift (Fig. 3B, lane 12 and Table 3) identified a subset of the 14-3-3 isoforms, shown previously again by mass spectroscopy to bind eB or eB mutant sequence. In specific, isoforms 14-3-3 ζ/δ, ε, γ, and σ, were present in the altered shift (Table 3).
The absence of isoforms β/α and τ/θ in the altered shift indicates that eB may bind directly or indirectly isoforms β/α and τ/θ regardless whether eB was used as probe, alone or within the context of the surrounding sequences of exons A and D. These two isoforms were found in all eB shifts (WT, Del 4, and Mut 5 and 6, Table 3) as well as in the ABD shift (band 2, Table 3). When the ABD RNA was used as probe in shift assays, two bands were formed as observed with the eB RNA (data not shown). Isoform ζ/δ and ε were present in all eB shifts and in ABD shift (band 2, Table 3) but were not competed when the ABD sequence was used as a competitor for the eB shift. Whether these play a role in SP-A translation remains to be determined. Isoform η was identified in all eB shifts (WT, Del 4, and Mut 5 and 6, Table 3) but not in ABD shifts (bands 1 and 2, Table 3). This indicates that its interaction with eB may be mediated via eB secondary structure of eB sequence characteristics, present when eB is alone in the absence of the surrounding A and D exons. Isoform σ on the other hand was present in the ABD shift (band 1, Table 3) as well as in Mut 6, indicating a potential role of this in SP-A 5′UTR containing eB. Together REMSA and mass spectroscopy data indicate that specific 14-3-3 isoforms directly or indirectly bind eB either alone or within the context of surrounding sequences, with isoforms β/α and τ/θ binding eB in the presence or absence of the surrounding A and D sequences. Other isoforms appear to bind eB (isoform η) when it is by itself in the absence of the A and D sequences, whereas yet others require further experimentation to determine whether these are or are not involved in the regulation of SP-A with 5′UTR containing eB.
Competition of eB with AD.
When the AD 5′UTR was used as competitor for the eB shift an altered shift was also observed (Fig. 3B, lane 11). The mass spectroscopy analysis showed no 14-3-3 isoforms present in the altered shifts (Table 3). Since the RNA-protein complex formed by AD (data not shown) does not contain 14-3-3 proteins (Table 3), this finding was rather unexpected.
To gain insight into this apparent paradox we performed two in silico analyses. In the first we determined the secondary structures of eB (Fig. 2C), ABD (Fig. 5A), and AD (Fig. 5B). The stability of these structures differs (ABD > AD > eB) as assessed by RNAfold software, with ABD being the most stable (ΔG ABD = −37.79 kcal/mol or −0.38 kcal·mol−1·nt−1; ΔG AD = −20.71 kcal/mol or 0.30 kcal·mol−1·nt−1; and ΔG eB = −2.69 kcal/mol or −0.09 kcal·mol−1·nt−1). As shown above, for eB to form a sequence-specific shift, specific sequences (Fig. 2) and a loop structure (Table 2) are necessary. Upon inspection of the ABD secondary structure, we observed that the last three nucleotides of the eB cis-sequence element 1 (GUC GCU GAU) contribute to the formation of a small loop, whereas the second eB cis-element GGA GCC UGA A forms parts of two loops (Fig. 5A). These looplike structures may enable partial protein binding to the two cis-elements, so that when the ABD is used as a competitor for the eB shift, only partial displacement of eB-bound proteins can occur.
Fig. 5.
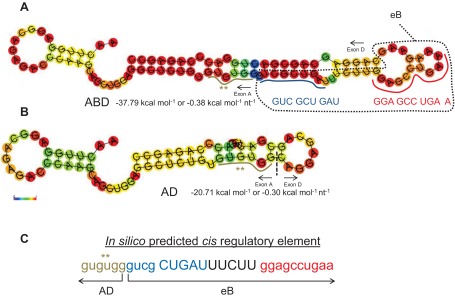
Secondary structure of ABD and AD and cis-elements. A and B depict the secondary structures for ABD and AD, respectively. The sequence and the location of the two eB cis-elements is noted (A), with blue and red colors. The RNAfold online software was used to obtain the secondary structures as described in Fig. 2C. C: the cis-element identified within ABD by the matrix-based motif discovery algorithm. The ** denotes the last 6 nt of exon A that were included in the identified cis-element. The sequence in blue and red denotes the 2 eB cis-elements, the eB pseudo-loop and loop, respectively. The free energy ΔG in kcal/mol and the color scale 0 to 1 (red, most stable) are shown.
In the second in silico analysis using a Matrix-based motif discovery tool algorithm (17), we sought to determine regulatory elements within eB, by analyzing the entire ABD sequence. The eB sequence was identified as an important regulatory element (except for the last 6 nt AAG AAG that constitute one part of the eB stem, Table 2, and comparison of Fig. 2C and Fig. 5C), plus 6 nt of exon A immediately adjacent to eB (Fig. 5C). The in silico identification of a cis-element consisting of adjacent exon A and eB sequences holds the potential for factors to bind both exons A (3′ end) and B (5′ end) within the natural ABD 5′UTR. It is possible that when AD is used in eB shift assays as a competitor (at 200-fold molar excess) it partially disrupts the eB-protein complex by displacing proteins that could bind both the 3′ end of exon A and the 5′ end of eB. On the basis of the collective data above, the AD competition is likely to affect the upper shift (*) of eB (Fig. 1B). The requirement of binding sequences in both exon A and eB may contribute to the instability of this shift when exon A sequences are absent. It is also possible that the AD competition indirectly affects the lower shift band (Fig. 1B, circle) resulting in a shift that lacks 14-3-3 proteins (Fig. 3B, lane 11). Proteins, other than the 14-3-3, ribosomal, and cytoskeleton, were still present in the altered shift, when AD was used as competitor. Moreover, the AD higher secondary structure stability (compared with eB, Figs. 2C and 5B) and perhaps other characteristics (yet to be defined) may contribute to this protein displacement and partial dissociation of the eB complex.
Correlation of 14-3-3 β/α Isoform Inhibition and SP-A2 Expression in NCI-H441 Cell Line
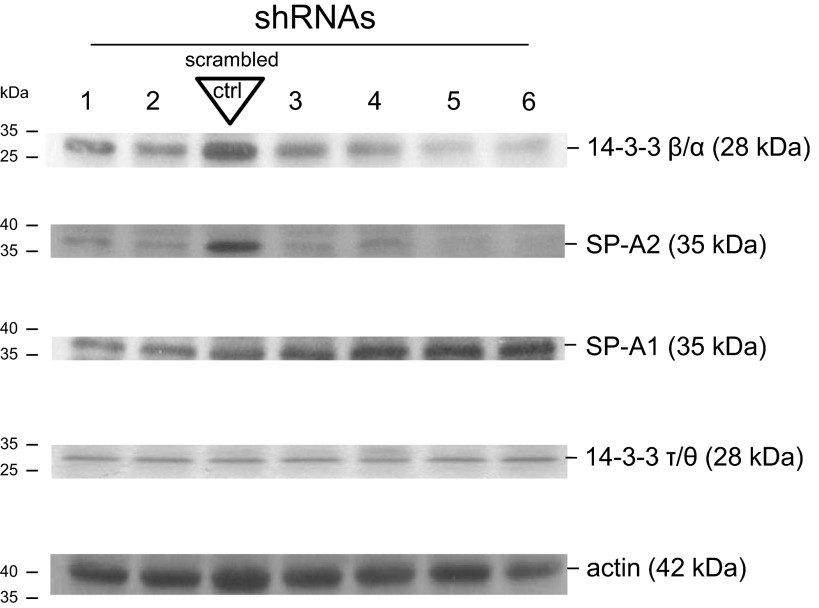
To study the functional role of 14-3-3 β/α protein and correlate it with SP-A2 expression and translation in NCI-H441 cells, as proof of principle, we employed shRNAs that target the human YWHAB gene. The shRNAs knocked down the 14-3-3 β/α isoform to a varying degree, as shown by Western immunoblot (Fig. 6), and this correlated with the expression of SP-A2. The scrambled shRNA (negative control) did not inhibit 14-3-3 β/α protein and the levels of SP-A2 were high compared with experimental shRNA that inhibited 14-3-3 β/α, while the expression of SP-A1 remained unaffected. The levels of 14-3-3 τ/θ and actin (serving as controls) did not change. These data indicate that 14-3-3 β/α protein plays a role in the regulation of SP-A2 expression.
Fig. 6.
Knockdown of 14-3-3 β/α isoform in NCI-H441 cell line and correlation with SP-A2 expression. Six different shRNAs (1–6), plus another 2 (not shown), were used to knock down the expression of 14-3-3 β/α isoform by RNA interference. A scrambled shRNA (negative control) that does not target any human, mouse, or rat gene was used as control. Western immunoblot was employed to monitor the expression of 14-3-3 β/α isoform (28 kDa), SP-A2 (35 kDa), SP-A1 (35 kDa), 14-3-3 τ/θ isoform (28 kDa), and actin (42 kDa). The experimental shRNAs inhibited the 14-3-3 β/α isoform to a varying degree and this correlated with levels of SP-A2. No differences were observed for SP-A1 and 14-3-3 τ/θ; actin was used as internal control.
DISCUSSION
Human SP-A, an important molecule of lung host defense, is encoded by two functional genes, and a number of genetic variants have been reported for each gene (19). Several SP-A1 and SP-A2 5′UTR splice variants have been identified (44, 54), with differences in mRNA stability and translational efficiency (93). We have shown that eB of the SP-A2 5′UTR mRNA is an enhancer for translation and transcription (79). In the present study, we studied the hypothesis that trans-acting factors bind eB alone or within the context of the surrounding sequences to modulate the previously observed translation efficiency. We found that cytoplasmic NCI-H441 cell extract proteins bind eB RNA in a sequence-specific manner and that both specific cis-elements within eB and eB secondary structure are necessary for binding. Members of the 3-monooxygenase/tryptophan 5-monooxygenase activation protein family (14-3-3) bind (directly or indirectly) eB and the natural SP-A2 (ABD) splice variant but not the SP-A1 (AD) splice variant that lacks eB. The knockdown of 14-3-3 β/α isoform in NCI-H441 cells results in a downregulated expression of SP-A2. Besides the 14-3-3, a number of other proteins (ribosomal, cytoskeletal, and translation factors) are also present in the eB mRNA-14-3-3 complex.
The deletion and mutation mapping analyses identified two cis-elements, element 1 GUC GCU GAU immediately next to exon A and element 2 GGA GCC UGA A near exon D, within eB that are important for protein interactions, as assessed by the presence of sequence-specific shifts. These shifts failed to form when both cis-elements were missing. However, if the characteristics of eB's secondary structure were maintained, the presence of one or the other cis-element was sufficient to generate the upper or lower band shift, indicating that both sequence elements and the secondary structure are necessary for the formation of the eB sequence-specific shifts. Thus eB regulatory sequence motifs and eB-mediated secondary structure characteristics may contribute to the higher level of translation observed with transcripts of SP-A 5′UTRs that contain the eB sequence (79, 93). Hence, differences in SP-A1 and SP-A2 translation may result in different immune profiles and responses to bacterial and inflammatory stressors. Understanding these differences is important in determining individual disease response patterns.
The fact that the motif gugugggucg CUGAUUUCUU ggagccugaa, identified by in silico analysis, contained most of the eB sequence supports the notion that the eB sequence is needed for functional and structural integrity of the SP-A2 ABD 5′UTR. Furthermore, this in silico-identified regulatory element contained both eB cis-elements identified by mutation mapping analysis and shown to be both functionally and structurally important. Consistent with this is our published work in which deletion of the pseudo-loop or the loop of eB (described in the present study) resulted in reduced internal ribosome entry site (IRES) activity (95). The importance of both the sequence specificity and the secondary structure in RNA function has been shown to be crucial in other systems of innate host defense (69, 101, 102). For example, the 20S RNA Narnavirus, via its secondary structure can evade the 5′-exonuclease SKI1/XRN1 host defense in Saccharomyces cerevisiae (21), and the human U1 RNA, owing to its double-stranded secondary structure, can induce innate immunity signaling by activating TLR-3 (37).
Mass spectroscopy findings and/or shift assays data with 14-3-3 specific Ab demonstrated that 5′UTR RNA-protein interactions include the 14-3-3 protein family. Isoforms β/α and τ/θ, in particular, bind eB regardless whether eB is found in the presence or absence of the surrounding sequences of exons A and D, as shown by competition of eB shift assays with ABD. Furthermore, knockdown of the 14-3-3 β/α isoform in NCI-H441 cell line results in a downregulation of SP-A2 expression, reinforcing the notion that the interaction between eB mRNA and 14-3-3 β/α is very important for the efficient translation of SP-A.
On the other hand, isoforms ζ/δ and ε, present in all eB shifts and in the ABD shift 2, failed to be competed when the ABD was used as competitor. It is possible that differences in the secondary structure characteristics between eB and ABD, along with potential differences in the other binding proteins, “protect” the binding of these isoforms in eB from the ABD competitor. As a result, these 14-3-3 isoforms are still present in the altered eB shift after competition with ABD.
However, a number of 14-3-3 isoforms (γ, σ, η) were present in shifts when eB WT (γ, η) or various mutant sequences (γ, σ, η) were used as probes. These (γ, σ) were not competed with ABD and were (γ, η) absent in the ABD shift. It is possible that the secondary structure characteristics of eB in the absence of the surrounding sequences of exons A and D are such that promote binding of other 14-3-3 isoforms such as the γ and η isoforms and that these isoforms do not actually bind eB in its natural sequence context (i.e., ABD sequence). However, further experimentation and use of additional methodologies are required to assess the precise interactions and the role of various 14-3-3 isoforms in SP-A variant expression.
The involvement of 14-3-3 in SP-A regulation is rather intriguing. The 14-3-3 protein family is involved in the innate host defense of various species. Isoforms β/α and ζ/δ identified in the present study to bind eB are evolutionary conserved proteins and are required for phagocytosis and microbial resistance in Drosophila, zebrafish larvae, and mouse RAW 264.7 cells (88). SP-A also plays a role in phagocytosis and innate immunity processes, with differences being observed in the ability of SP-A1 and SP-A2 to modulate phagocytosis (56, 57) and regulate inflammatory processes (38, 97, 99) in the alveolar macrophage. Of interest, differences in the ratio of SP-A1 to total SP-A in bronchoalveolar lavage samples have been observed between healthy population and nonhealthy individuals (infection, cystic fibrosis, asthma) (85, 100), raising the possibility that regulatory differences exist between SP-A1 and SP-A2 under various conditions. Moreover, levels of 14-3-3 have been shown to change in disease, with increased levels observed in cerebrospinal fluid after infection with prion in Creutzfeldt-Jakob disease and spongiform encephalopathies (30). It is possible that the 14-3-3 proteins are involved in mechanisms that differentially affect SP-A1 and SP-A2 levels. The fact that inhibition of one of the 14-3-3 isoforms (β/α) shown to differentially bind SP-A1 and SP-A2 5′UTRs results in the downregulation of SP-A2 indicates an important role of this isoform in SP-A translation.
Moreover, isoform β/α has been shown to be a phagosome-associated protein (8) that dimerizes in response to mitogen-activated protein kinase-dependent inflammatory signaling (67). The tyrosine kinase of the epidermal growth factor receptor has been shown to regulate SP-A expression and modulate inflammation (39). Whether 14-3-3 regulates SP-A expression via tyrosine kinase pathways remains to be determined. However, the identification of the 14-3-3 proteins as novel phagosomal proteins (27), and the correlation between inhibition of 14-3-3 (β/α) and the reduced levels of SP-A2, provides, in general, further support for their involvement in innate immunity, and, in specific, the regulation of the innate host defense molecule, SP-A. The τ/θ isoform identified in the present study as an eB-specific binding protein has been shown to play a regulatory role in Toll-like receptor 2 (TLR2) signaling pathway within the innate immune system (75). SP-A has also been demonstrated to interact directly with TLR2, modulating inflammatory responses against bacterial components (59), indicating a potential role of the 14-3-3 in this process.
Ribosomal and cytoskeletal proteins and initiation and elongation translation factors were found by mass spectroscopy to be included in the WT eB, eB mutants, or ABD shifts. These were virtually all competed when ABD was used as a competitor for the eB shift. This indicates that eB, either alone or within the context of the surrounding A and D sequences, is an important element for bringing together protein factors necessary for the enhanced translation observed with SP-A UTRs that contain eB (79, 93).
The AD shift, on the other hand, did not contain any 14-3-3 proteins and not as many of the other group of proteins indicating significant differences in the RNA-protein interactions between AD, and eB or ABD. This is consistent with published data where significant differences in translation and mRNA stability were observed between SP-As with AD and ABD 5′UTRs (93, 95). The 40S ribosomal proteins, SA and S3, appear to bind exon A and/or D but not eB. These are necessary for Met-tRNA to bind 40S and translation initiation (13). The S3 is found in ribonucleoprotein granules (42) and in hepatitis C IRES-mediated translation complex (103). However, the absence of the 60S ribosomal protein L5 in the SP-A1 5′UTR AD splice variant may alter translation efficiency or pose a major disadvantage in terms of translation compared with SP-A2 5′UTR ABD splice variant. L5 is required for rRNA maturation and formation of the 60S ribosomal subunits (28). Furthermore, the AD splice variant does not bind the 60S ribosomal protein L7, shown to play a regulatory role in translation by binding to G-rich structures in mRNAs (35). Another 60S ribosomal protein, L22, a structural constituent of ribosome, was shown to bind all probes except AD and Mut 6, indicating that the nucleotide content of the eB pseudo-loop (GUC GCU GAU) may be the cis-element that elicits the binding of this trans-acting factor. These observations support the notion that differences exist between AD and eB-containing UTRs in their interactions with ribosomal proteins, and that these may differentially impact the translation of SP-As with different UTRs.
Cytoskeletal proteins actin and tubulin-α were present virtually in all probes indicating that a cytoskeleton-mediated stabilization of SP-A RNA is required for all SP-A RNAs with either AD or ABD 5′UTR. This observation is consistent with previous findings in which cytoskeletal proteins were shown to stabilize the surfactant protein mRNAs in rat alveolar type II cells (77). Together these indicate that mechanisms of cytoskeleton-mediated stabilization of AD and ABD mRNAs are similar.
Interestingly, eukaryotic initiation factor 4A (eIF4A) interacted only with the natural ABD variant, and not the eB alone, or the AD; eIF4H was found in eB and two mutants (Del 4 and Mut 5). It seems that this RNA helicase (eIF4A) requires eB to be within its natural context as in the SP-A2 5′UTR ABD variant. The group of eIF4A and eIF4H are DEAD box helicases (18, 53, 83) that help the scanning process of the 40S small ribosomal unit by unwinding the secondary structure (82). Thus these may enable the SP-A2 5′UTR ABD splice variant to be more efficiently translated than the SP-A1 5′UTR AD that does not interact with these helicases.
The direct or indirect binding of a multitude of proteins to eB was also seen in the competition assays. Consistently, in all cases competition of an mRNA-protein complex with molar excess of another mRNA resulted in partial dissociation of this complex. The observations from the competition and mass spectroscopy experiments, when ABD and AD RNAs were used as competitors for the eB-mediated shifts, were unexpected. These are summarized in Fig. 7. One may have expected that the ABD competitor would have displaced the 14-3-3 isoforms, since these were found by mass spectroscopy to be present only in shifts of eB or ABD probes and not of the AD probe. However, on the basis of the in silico analysis in which 6 nt at the 3′ end of exon A were identified as part of the eB regulatory element, we speculate that the ABD (but not the AD) competitor somehow provides a certain level of stability to A-B junction, maintaining thus a partial displaced RNA-protein complex with most of 14-3-3 still present. Another possibility is that an altered 3D structure formed by ABD and eB together creates an artificial platform upon which the disrupted 14-3-3 complexes may bind. Future experiments are warranted to gain insight into these possibilities.
Fig. 7.
Schematic representation of competition of eB-mediated shifts with 200-fold molar excess of unlabeled AD and ABD RNAs. The figure depicts the collective results of eB shift assays, competition shift assays, and mass spectroscopy protein identification of the sequence-specific shifts. The 14-3-3 protein family (β/α, ζ/δ, ε, η, and τ/θ), 40S and 60S ribosomal proteins, along with cytoskeletal proteins (actin and tubulin) bind directly or indirectly eB. Competition with molar excess of ABD gives rise to an altered eB mRNA-protein complex, which includes 14-3-3 ζ/δ, ε, γ, σ isoforms, L22 60S ribosomal protein, S27a 60S ribosomal protein, and actin; the rest of the proteins (including 14-3-3 isoforms β/α and τ/θ) are competed by the ABD probe. Competition with molar excess of AD results in a smaller eB mRNA-protein complex that includes L22 and L26 60S ribosomal proteins, S27a 60S ribosomal protein, eIF6, and actin; the rest of the proteins (including all of the 14-3-3 family of proteins) are competed with the AD probe.
In summary, eB, which was shown previously to be an enhancer for both translation and transcription of SP-A2 (79), contains two cis-elements, and trans-acting factors bind eB in a sequence- and a secondary structure-specific manner. The binding factors include 14-3-3 β/α and τ/θ isoforms, along with ribosomal, cytoskeletal, and translation factors and perhaps additional 14-3-3 isoforms. In particular, 14-3-3 β/α isoform, shown to be part of the eB-protein complex either when eB was alone or in the context of the surrounding sequences of exon A and exon D, is likely to play an important role in the translation of SP-A. The trans-acting proteins that bind eB in the presence or absence of exon A and D sequences probably play important roles in the translation and the differential regulation of SP-As with 5′UTRs that contain or lack eB. Future studies are needed to characterize the specific RNA-protein interactions and delineate the underlying molecular mechanisms involved in the SP-A variant differential regulation.
GRANTS
This work was supported by National Heart, Lung, and Blood Institute Grant HL34788.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
G.T.N., P.S., and J.F. conceived and designed the research; G.T.N. and F.B. performed experiments; G.T.N. and J.F. analyzed data; G.T.N. and J.F. interpreted results of experiments; G.T.N. prepared figures; G.T.N. drafted manuscript; G.T.N. and P.S. edited and revised manuscript; G.T.N., P.S., F.B., and J.F. approved final version of manuscript.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Dr. D. S. Phelps and T. M. Umstead for input and advice on mass spectrometry analysis, and L. Murray for typing.
Present address for F. Bhatti: Neonatal-Perinatal Medicine, University of Oklahoma Health Sciences Center, Oklahoma City, OK.
REFERENCES
- 1. Aitken A. Functional specificity in 14-3-3 isoform interactions through dimer formation and phosphorylation. Chromosome location of mammalian isoforms and variants. Plant Mol Biol 50: 993–1010, 2002 [DOI] [PubMed] [Google Scholar]
- 2. Andreassi C, Riccio A. To localize or not to localize: mRNA fate is in 3′UTR ends. Trends Cell Biol 19: 465–474, 2009 [DOI] [PubMed] [Google Scholar]
- 3. Baldin V. 14-3-3 proteins and growth control. Prog Cell Cycle Res 4: 49–60, 2000 [DOI] [PubMed] [Google Scholar]
- 4. Brands JH, Maassen JA, van Hemert FJ, Amons R, Möller W. The primary structure of the alpha subunit of human elongation factor 1. Structural aspects of guanine-nucleotide-binding sites. Eur J Biochem 155: 167–171, 1986 [DOI] [PubMed] [Google Scholar]
- 5. Braselmann S, McCormick F. Bcr and Raf form a complex in vivo via 14-3-3 proteins. EMBO J 14: 4839–4848, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bridges D, Moorhead GBG. 14-3-3 proteins: a number of functions for a numbered protein. Sci STKE 2004: re10-, 2004 [DOI] [PubMed] [Google Scholar]
- 7. Bruce SR, Atkins CL, Colasurdo GN, Alcorn JL. Respiratory syncytial virus infection alters surfactant protein A expression in human pulmonary epithelial cells by reducing translation efficiency. Am J Physiol Lung Cell Mol Physiol 297: L559–L567, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Burlak C, Whitney AR, Mead DJ, Hackstadt T, Deleo FR. Maturation of human neutrophil phagosomes includes incorporation of molecular chaperones and endoplasmic reticulum quality control machinery. Mol Cell Proteomics 5: 620–634, 2006 [DOI] [PubMed] [Google Scholar]
- 9. Casals C, Varela A, Ruano ML, Valiño F, Pérez-Gil J, Torre N, Jorge E, Tendillo F, Castillo-Olivares JL. Increase of C-reactive protein and decrease of surfactant protein A in surfactant after lung transplantation. Am J Respir Crit Care Med 157: 43–49, 1998 [DOI] [PubMed] [Google Scholar]
- 10. Ceci M, Gaviraghi C, Gorrini C, Sala LA, Offenhäuser N, Marchisio PC, Biffo S. Release of eIF6 (p27BBP) from the 60S subunit allows 80S ribosome assembly. Nature 426: 579–584, 2003 [DOI] [PubMed] [Google Scholar]
- 11. Chang JH, Cho YH, Sohn SY, Choi JM, Kim A, Kim YC, Jang SK, Cho Y. Crystal structure of the eIF4A-PDCD4 complex. Proc Natl Acad Sci USA 106: 3148–3153, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cheng ST, Nguyen TQ, Yang YS, Capra JD, Sontheimer RD. Calreticulin binds hYRNA and the 52-kDa polypeptide component of the Ro/SS-A ribonucleoprotein autoantigen. J Immunol 156: 4484–4491, 1996 [PubMed] [Google Scholar]
- 13. Clemens MJ, Henshaw EC, Rahamimoff H, London IM. Met-tRNAfMet binding to 40S ribosomal subunits: a site for the regulation of initiation of protein synthesis by hemin. Proc Natl Acad Sci USA 71: 2946–2950, 1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Crouch EC. Collectins and pulmonary host defense. Am J Respir Cell Mol Biol 19: 177–201, 1998 [DOI] [PubMed] [Google Scholar]
- 15. Dabiri GA, Young CL, Rosenbloom J, Southwick FS. Molecular cloning of human macrophage capping protein cDNA. A unique member of the gelsolin/villin family expressed primarily in macrophages. J Biol Chem 267: 16545–16552, 1992 [PubMed] [Google Scholar]
- 16. Davezac N, Baldin V, Gabrielli B, Forrest A, Theis-Febvre N, Yashida M, Ducommun B. Regulation of CDC25B phosphatases subcellular localization. Oncogene 19: 2179–2185, 2000 [DOI] [PubMed] [Google Scholar]
- 17. Defrance M, van Helden J. info-gibbs: a motif discovery algorithm that directly optimizes information content during sampling. Bioinformatics 25: 2715–2722, 2009 [DOI] [PubMed] [Google Scholar]
- 18. Dever TE. Gene-specific regulation by general translation factors. Cell 108: 545–556, 2002 [DOI] [PubMed] [Google Scholar]
- 19. DiAngelo S, Lin Z, Wang G, Phillips S, Ramet M, Luo J, Floros J. Novel, non-radioactive, simple and multiplex PCR-cRFLP methods for genotyping human SP-A and SP-D marker alleles. Dis Markers 15: 269–281, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dubois T, Rommel C, Howell S, Steinhussen U, Soneji Y, Morrice N, Moelling K, Aitken A. 14-3-3 is phosphorylated by casein kinase I on residue 233. Phosphorylation at this site in vivo regulates Raf/14-3-3 interaction. J Biol Chem 272: 28882–28888, 1997 [DOI] [PubMed] [Google Scholar]
- 21. Esteban R, Vega L, Fujimura T. 20S RNA narnavirus defies the antiviral activity of SKI1/XRN1 in Saccharomyces cerevisiae. J Biol Chem 283: 25812–25820, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Floros J, Thomas N. Genetic variations of surfactant proteins and lung injury. In: Surfactant in Pathogenesis and Treatment of Lung Disease, edited by Nakos G, Papathanasiou A. Trivandrum, India: Research Signpost, 2009, p. 25–48 [Google Scholar]
- 23. Floros J, Wang G, Mikerov AN. Genetic complexity of the human innate host defense molecules, surfactant protein A1 (SP-A1) and SP-A2—impact on function. Crit Rev Eukaryot Gene Expr 19: 125–137, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fu H, Subramanian RR, Masters SC. 14-3-3 Proteins: structure, function, regulation. Annu Rev Pharmacol Toxicol 40: 617–647, 2000 [DOI] [PubMed] [Google Scholar]
- 25. Fujita Y, Okamoto T, Noshiro M, McKeehan WL, Crabb JW, Whitney RG, Kato Y, Sato JD, Takada K. A novel heparin-binding protein, HBp15, is identified as mammalian ribosomal protein L22. Biochem Biophys Res Commun 199: 706–713, 1994 [DOI] [PubMed] [Google Scholar]
- 26. García-Verdugo I, Wang G, Floros J, Casals C. Structural analysis and lipid-binding properties of recombinant human surfactant protein a derived from one or both genes. Biochemistry 41: 14041–14053, 2002 [DOI] [PubMed] [Google Scholar]
- 27. Garin J, Diez R, Kieffer S, Dermine JF, Duclos S, Gagnon E, Sadoul R, Rondeau C, Desjardins M. The phagosome proteome: insight into phagosome functions. J Cell Biol 152: 165–180, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gazda HT, Sheen MR, Vlachos A, Choesmel V, O'Donohue MF, Schneider H, Darras N, Hasman C, Sieff CA, Newburger PE, Ball SE, Niewiadomska E, Matysiak M, Zaucha JM, Glader B, Niemeyer C, Meerpohl JJ, Atsidaftos E, Lipton JM, Gleizes PE, Beggs AH. Ribosomal protein L5 and L11 mutations are associated with cleft palate and abnormal thumbs in Diamond-Blackfan anemia patients. Am J Hum Genet 83: 769–780, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol 5: 827–835, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gmitterová K, Heinemann U, Bodemer M, Krasnianski A, Meissner B, Kretzschmar HA, Zerr I. 14-3-3 CSF levels in sporadic Creutzfeldt-Jakob disease differ across molecular subtypes. Neurobiol Aging 30: 1842–1850, 2009 [DOI] [PubMed] [Google Scholar]
- 31. Gruber AR, Lorenz R, Bernhart SH, Neuböck R, Hofacker IL. The Vienna RNA websuite. Nucleic Acids Res 36: W70–W74, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gu YM, Jin YH, Choi JK, Baek KH, Yeo CY, Lee KY. Protein kinase A phosphorylates and regulates dimerization of 14-3-3 epsilon. FEBS Lett 580: 305–310, 2006 [DOI] [PubMed] [Google Scholar]
- 33. Guzman J, Wang YM, Kalaycioglu O, Schoenfeld B, Hamm H, Bartsch W, Costabel U. Increased surfactant protein A content in human alveolar macrophages in hypersensitivity pneumonitis. Acta Cytol 36: 668–673, 1992 [PubMed] [Google Scholar]
- 34. Hamm H, Lührs J, Guzman y Rotaeche J, Costabel U, Fabel H, Bartsch W. Elevated surfactant protein A in bronchoalveolar lavage fluids from sarcoidosis and hypersensitivity pneumonitis patients. Chest 106: 1766–1770, 1994 [DOI] [PubMed] [Google Scholar]
- 35. Hemmerich P, von Mikecz A, Neumann F, Sözeri O, Wolff-Vorbeck G, Zoebelein R, Krawinkel U. Structural and functional properties of ribosomal protein L7 from humans and rodents. Nucleic Acids Res 21: 223–231, 1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hermeking H, Lengauer C, Polyak K, He TC, Zhang L, Thiagalingam S, Kinzler KW, Vogelstein B. 14-3-3σ is a p53-regulated inhibitor of G2/M progression. Mol Cell 1: 3–11, 1997 [DOI] [PubMed] [Google Scholar]
- 37. Hoffman RW, Gazitt T, Foecking MF, Ortmann RA, Misfeldt M, Jorgenson R, Young SL, Greidinger EL. U1 RNA induces innate immunity signaling. Arthritis Rheum 50: 2891–2896, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Huang W, Wang G, Phelps DS, Al-Mondhiry H, Floros J. Human SP-A genetic variants and bleomycin-induced cytokine production by THP-1 cells: effect of ozone-induced SP-A oxidation. Am J Physiol Lung Cell Mol Physiol 286: L546–L553, 2004 [DOI] [PubMed] [Google Scholar]
- 39. Inoue A, Xin H, Suzuki T, Kanehira M, Kuroki Y, Fukuhara T, Kikuchi T, Maemondo M, Nukiwa T, Saijo Y. Suppression of surfactant protein A by an epidermal growth factor receptor tyrosine kinase inhibitor exacerbates lung inflammation. Cancer Sci 99: 1679–1684, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jang SW, Liu X, Fu H, Rees H, Yepes M, Levey A, Ye K. Interaction of Akt-phosphorylated SRPK2 with 14-3-3 mediates cell cycle and cell death in neurons. J Biol Chem 284: 24512–24525, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jin Y, Dai MS, Lu SZ, Xu Y, Luo Z, Zhao Y, Lu H. 14-3-3γ binds to MDMX that is phosphorylated by UV-activated Chk1, resulting in p53 activation. EMBO J 25: 1207–1218, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jønson L, Vikesaa J, Krogh A, Nielsen LK, Hansen T, Borup R, Johnsen AH, Christiansen J, Nielsen FC. Molecular composition of IMP1 ribonucleoprotein granules. Mol Cell Proteomics 6: 798–811, 2007 [DOI] [PubMed] [Google Scholar]
- 43. Karinch AM, Deiter G, Ballard PL, Floros J. Regulation of expression of human SP-A1 and SP-A2 genes in fetal lung explant culture. Biochim Biophys Acta 1398: 192–202, 1998 [DOI] [PubMed] [Google Scholar]
- 44. Karinch AM, Floros J. 5′ Splicing and allelic variants of the human pulmonary surfactant protein A genes. Am J Respir Cell Mol Biol 12: 77–88, 1995 [DOI] [PubMed] [Google Scholar]
- 45. Karinch AM, Floros J. Translation in vivo of 5′ untranslated-region splice variants of human surfactant protein-A. Biochem J 307: 327–330, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kumar AR, Snyder JM. Differential regulation of SP-A1 and SP-A2 genes by cAMP, glucocorticoids, and insulin. Am J Physiol Lung Cell Mol Physiol 274: L177–L185, 1998 [DOI] [PubMed] [Google Scholar]
- 47. Le Quesne JP, Spriggs KA, Bushell M, Willis AE. Dysregulation of protein synthesis and disease. J Pathol 220: 140–151, 2010 [DOI] [PubMed] [Google Scholar]
- 48. Liu Y, Ross JF, Bodine PV, Billiard J. Homodimerization of Ror2 tyrosine kinase receptor induces 14-3-3β phosphorylation and promotes osteoblast differentiation and bone formation. Mol Endocrinol 21: 3050–3061, 2007 [DOI] [PubMed] [Google Scholar]
- 49. Liu Y, Wimmer E, Paul AV. Cis-acting RNA elements in human and animal plus-strand RNA viruses. Biochim Biophys Acta 1789: 495–517, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Loh PG, Yang HS, Walsh MA, Wang Q, Wang X, Cheng Z, Liu D, Song H. Structural basis for translational inhibition by the tumour suppressor Pdcd4. EMBO J 28: 274–285, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Margolis SS, Walsh S, Weiser DC, Yoshida M, Shenolikar S, Kornbluth S. PP1 control of M phase entry exerted through 14-3-3-regulated Cdc25 dephosphorylation. EMBO J 22: 5734–5745, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Martin TR, Frevert CW. Innate immunity in the lungs. Proc Am Thorac Soc 2: 403–411, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mathews MB. Lost in translation. Trends Biochem Sci 27: 267–269, 2002 [DOI] [PubMed] [Google Scholar]
- 54. McCormick SM, Boggaram V, Mendelson CR. Characterization of mRNA transcripts and organization of human SP-A1 and SP-A2 genes. Am J Physiol Lung Cell Mol Physiol 266: L354–L366, 1994 [DOI] [PubMed] [Google Scholar]
- 55. Meyer KC, Sharma A, Brown R, Weatherly M, Moya FR, Lewandoski J, Zimmerman JJ. Function and composition of pulmonary surfactant and surfactant-derived fatty acid profiles are altered in young adults with cystic fibrosis. Chest 118: 164–174, 2000 [DOI] [PubMed] [Google Scholar]
- 56. Mikerov A, Wang G, Umstead T, Zacharatos M, Thomas N, Phelps D, Floros J. Surfactant protein A2 (SP-A2) variants expressed in CHO cells stimulate phagocytosis of Pseudomonas aeruginosa more than do SP-A1 variants. Infect Immun 75: 1403–1412, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mikerov AN, Umstead TM, Gan X, Huang W, Guo X, Wang G, Phelps DS, Floros J. Impact of ozone exposure on the phagocytic activity of human surfactant protein A (SP-A) and SP-A variants. Am J Physiol Lung Cell Mol Physiol 294: L121–L130, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mikerov AN, Umstead TM, Huang W, Liu W, Phelps D, Floros J. SP-A1 and SP-A2 variants differentially enhance association of Pseudomonas aeruginosa with rat alveolar macrophages. Am J Physiol Lung Cell Mol Physiol 288: L150–L158, 2005 [DOI] [PubMed] [Google Scholar]
- 59. Murakami S, Iwaki D, Mitsuzawa H, Sano H, Takahashi H, Voelker DR, Akino T, Kuroki Y. Surfactant protein A inhibits peptidoglycan-induced tumor necrosis factor-α secretion in U937 cells and alveolar macrophages by direct interaction with Toll-like receptor 2. J Biol Chem 277: 6830–6837, 2002 [DOI] [PubMed] [Google Scholar]
- 60. Muslin AJ, Xing H. 14-3-3 proteins: regulation of subcellular localization by molecular interference. Cell Signal 12: 703–709, 2000 [DOI] [PubMed] [Google Scholar]
- 61. Obsil T, Ghirlando R, Klein DC, Ganguly S, Dyda F. Crystal structure of the 14-3-3zeta:serotonin N-acetyltransferase complex. A role for scaffolding in enzyme regulation. Cell 105: 257–267, 2001 [DOI] [PubMed] [Google Scholar]
- 62. Pesole G, Mignone F, Gissi C, Grillo G, Licciulli F, Liuni S. Structural and functional features of eukaryotic mRNA untranslated regions. Gene 276: 73–81, 2001 [DOI] [PubMed] [Google Scholar]
- 63. Phelps DS. Surfactant regulation of host defense function in the lung: a question of balance. Pediatr Pathol Mol Med 20: 269–292, 2001 [PubMed] [Google Scholar]
- 64. Phelps DS, Umstead TM, Quintero OA, Yengo CM, Floros J. In vivo rescue of alveolar macrophages from SP-A knockout mice with exogenous SP-A nearly restores a wild type intracellular proteome; actin involvement. Proteome Sci 9: 67, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Postle AD, Mander A, Reid KB, Wang JY, Wright SM, Moustaki M, Warner JO. Deficient hydrophilic lung surfactant proteins A and D with normal surfactant phospholipid molecular species in cystic fibrosis. Am J Respir Cell Mol Biol 20: 90–98, 1999 [DOI] [PubMed] [Google Scholar]
- 66. Powell DW, Rane MJ, Chen Q, Singh S, McLeish KR. Identification of 14-3-3ζ as a protein kinase B/Akt substrate. J Biol Chem 277: 21639–21642, 2002 [DOI] [PubMed] [Google Scholar]
- 67. Powell DW, Rane MJ, Joughin BA, Kalmukova R, Hong JH, Tidor B, Dean WL, Pierce WM, Klein JB, Yaffe MB, McLeish KR. Proteomic identification of 14-3-3zeta as a mitogen-activated protein kinase-activated protein kinase 2 substrate: role in dimer formation and ligand binding. Mol Cell Biol 23: 5376–5387, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Pozuelo Rubio M, Geraghty KM, Wong BH, Wood NT, Campbell DG, Morrice N, Mackintosh C. 14-3-3-affinity purification of over 200 human phosphoproteins reveals new links to regulation of cellular metabolism, proliferation and trafficking. Biochem J 379: 395–408, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Qi Y, Ding B. Inhibition of cell growth and shoot development by a specific nucleotide sequence in a noncoding viroid RNA. Plant Cell 15: 1360–1374, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Richter NJ, Rogers GW, Hensold JO, Merrick WC. Further biochemical and kinetic characterization of human eukaryotic initiation factor 4H. J Biol Chem 274: 35415–35424, 1999 [DOI] [PubMed] [Google Scholar]
- 71. Robledo S, Idol RA, Crimmins DL, Ladenson JH, Mason PJ, Bessler M. The role of human ribosomal proteins in the maturation of rRNA and ribosome production. RNA 14: 1918–1929, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Rogers GW, Richter NJ, Lima WF, Merrick WC. Modulation of the helicase activity of eIF4A by eIF4B, eIF4H, and eIF4F. J Biol Chem 276: 30914–30922, 2001 [DOI] [PubMed] [Google Scholar]
- 73. Sato S, Fujita N, Tsuruo T. Regulation of kinase activity of 3-phosphoinositide-dependent protein kinase-1 by binding to 14-3-3. J Biol Chem 277: 39360–39367, 2002 [DOI] [PubMed] [Google Scholar]
- 74. Scavo LM, Ertsey R, Gao BQ. Human surfactant proteins A1 and A2 are differentially regulated during development and by soluble factors. Am J Physiol Lung Cell Mol Physiol 275: L653–L669, 1998 [DOI] [PubMed] [Google Scholar]
- 75. Schuster TB, Costina V, Findeisen P, Neumaier M, Ahmad-Nejad P. Identification and functional characterization of 14-3-3 in TLR2 signaling. J Proteome Res 10: 4661–4670, 2011 [DOI] [PubMed] [Google Scholar]
- 76. Schütz P, Karlberg T, van den Berg S, Collins R, Lehtiö L, Högbom M, Holmberg-Schiavone L, Tempel W, Park HW, Hammarström M, Moche M, Thorsell AG, Schüler H. Comparative structural analysis of human DEAD-box RNA helicases. PLoS One 5, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Shannon JM, Pan T, Edeen KE, Nielsen LD. Influence of the cytoskeleton on surfactant protein gene expression in cultured rat alveolar type II cells. Am J Physiol Lung Cell Mol Physiol 274: L87–L96, 1998 [DOI] [PubMed] [Google Scholar]
- 78. Silveyra P, Floros J. Genetic variant associations of human SP-A and SP-D with acute and chronic lung injury. Front Biosci 17: 407–429, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Silveyra P, Raval M, Simmons B, Diangelo S, Wang G, Floros J. The untranslated exon B of human surfactant protein A2 mRNAs is an enhancer for transcription and translation. Am J Physiol Lung Cell Mol Physiol 301: L795–L803, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Silveyra P, Wang G, Floros J. Human SP-A1 (SFTPA1) variant-specific 3′ UTRs and poly(A) tail differentially affect the in vitro translation of a reporter gene. Am J Physiol Lung Cell Mol Physiol 299: L523–L534, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Skoulakis EM, Davis RL. 14-3-3 proteins in neuronal development and function. Mol Neurobiol 16: 269–284, 1998 [DOI] [PubMed] [Google Scholar]
- 82. Sonenberg N. Cap-binding proteins of eukaryotic messenger RNA: functions in initiation and control of translation. Prog Nucleic Acid Res Mol Biol 35: 173–207, 1988 [DOI] [PubMed] [Google Scholar]
- 83. Sonenberg N, Dever TE. Eukaryotic translation initiation factors and regulators. Curr Opin Struct Biol 13: 56–63, 2003 [DOI] [PubMed] [Google Scholar]
- 84. Stothard P. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques 28: 1102, 1104, 2000 [DOI] [PubMed] [Google Scholar]
- 85. Tagaram HR, Wang G, Umstead TM, Mikerov AN, Thomas NJ, Graff GR, Hess JC, Thomassen MJ, Kavuru MS, Phelps DS, Floros J. Characterization of a human surfactant protein A1 (SP-A1) gene-specific antibody; SP-A1 content variation among individuals of varying age and pulmonary health. Am J Physiol Lung Cell Mol Physiol 292: L1052–L1063, 2007 [DOI] [PubMed] [Google Scholar]
- 86. Todd A, Cossons N, Aitken A, Price GB, Zannis-Hadjopoulos M. Human cruciform binding protein belongs to the 14-3-3 family. Biochemistry 37: 14317–14325, 1998 [DOI] [PubMed] [Google Scholar]
- 87. Troyanovsky SM, Leube RE, Franke WW. Characterization of the human gene encoding cytokeratin 17 and its expression pattern. Eur J Cell Biol 59: 127–137, 1992 [PubMed] [Google Scholar]
- 88. Ulvila J, Vanha-aho LM, Kleino A, Vähä-Mäkilä M, Vuoksio M, Eskelinen S, Hultmark D, Kocks C, Hallman M, Parikka M, Rämet M. Cofilin regulator 14-3-3ζ is an evolutionarily conserved protein required for phagocytosis and microbial resistance. J Leukoc Biol 89: 649–659, 2011 [DOI] [PubMed] [Google Scholar]
- 89. van Hemert MJ, Steensma HY, van Heusden GP. 14-3-3 proteins: key regulators of cell division, signalling and apoptosis. Bioessays 23: 936–946, 2001 [DOI] [PubMed] [Google Scholar]
- 90. Vladimirov SN, Ivanov AV, Karpova GG, Musolyamov AK, Egorov TA, Thiede B, Wittmann-Liebold B, Otto A. Characterization of the human small-ribosomal-subunit proteins by N-terminal and internal sequencing, and mass spectrometry. Eur J Biochem 239: 144–149, 1996 [DOI] [PubMed] [Google Scholar]
- 91. Wang G, Bates-Kenney SR, Tao JQ, Phelps DS, Floros J. Differences in biochemical properties and in biological function between human SP-A1 and SP-A2 variants, and the impact of ozone-induced oxidation. Biochemistry 43: 4227–4239, 2004 [DOI] [PubMed] [Google Scholar]
- 92. Wang G, Guo X, Diangelo S, Thomas NJ, Floros J. Humanized SFTPA1 and SFTPA2 transgenic mice reveal functional divergence of SP-A1 and SP-A2: formation of tubular myelin in vivo requires both gene products. J Biol Chem 285: 11998–12010, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Wang G, Guo X, Floros J. Differences in the translation efficiency and mRNA stability mediated by 5′-UTR splice variants of human SP-A1 and SP-A2 genes. Am J Physiol Lung Cell Mol Physiol 289: L497–L508, 2005 [DOI] [PubMed] [Google Scholar]
- 94. Wang G, Guo X, Floros J. Human SP-A 3′-UTR variants mediate differential gene expression in basal levels and in response to dexamethasone. Am J Physiol Lung Cell Mol Physiol 284: L738–L748, 2003 [DOI] [PubMed] [Google Scholar]
- 95. Wang G, Guo X, Silveyra P, Kimball SR, Floros J. Cap-independent translation of human SP-A 5′-UTR variants: a double-loop structure and cis-element contribution. Am J Physiol Lung Cell Mol Physiol 296: L635–L647, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Wang G, Myers C, Mikerov A, Floros J. Role of Cys85 of human SP-A variants on their biochemical properties and biological function. FASEB J 21: A552, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Wang G, Phelps DS, Umstead TM, Floros J. Human SP-A protein variants derived from one or both genes stimulate TNF-α production in the THP-1 cell line. Am J Physiol Lung Cell Mol Physiol 278: L946–L954, 2000 [DOI] [PubMed] [Google Scholar]
- 98. Wang G, Taneva S, Keough KM, Floros J. Differential effects of human SP-A1 and SP-A2 variants on phospholipid monolayers containing surfactant protein B. Biochim Biophys Acta 1768: 2060–2069, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Wang JY, Reid KB. The immunoregulatory roles of lung surfactant collectins SP-A, and SP-D, in allergen-induced airway inflammation. Immunobiology 212: 417–425, 2007 [DOI] [PubMed] [Google Scholar]
- 100. Wang Y, Voelker DR, Lugogo NL, Wang G, Floros J, Ingram JL, Chu HW, Church TD, Kandasamy P, Fertel D, Wright JR, Kraft M. Surfactant protein A is defective in abrogating inflammation in asthma. Am J Physiol Lung Cell Mol Physiol 301: L598–L606, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Wassenegger M, Spieker RL, Thalmeir S, Gast FU, Riedel L, Sänger HL. A single nucleotide substitution converts potato spindle tuber viroid (PSTVd) from a noninfectious to an infectious RNA for Nicotiana tabacum. Virology 226: 191–197, 1996 [DOI] [PubMed] [Google Scholar]
- 102. Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW, Swanstrom R, Burch CL, Weeks KM. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature 460: 711–716, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Weinlich S, Hüttelmaier S, Schierhorn A, Behrens SE, Ostareck-Lederer A, Ostareck DH. IGF2BP1 enhances HCV IRES-mediated translation initiation via the 3′UTR. RNA 15: 1528–1542, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Wilker EW, van Vugt MA, Artim SA, Huang PH, Petersen CP, Reinhardt HC, Feng Y, Sharp PA, Sonenberg N, White FM, Yaffe MB. 14-3-3σ controls mitotic translation to facilitate cytokinesis. Nature 446: 329–332, 2007 [DOI] [PubMed] [Google Scholar]
- 105. Wright JR. Immunomodulatory functions of surfactant. Physiol Rev 77: 931–962, 1997 [DOI] [PubMed] [Google Scholar]
- 106. Yu Y, Ji H, Doudna JA, Leary JA. Mass spectrometric analysis of the human 40S ribosomal subunit: native and HCV IRES-bound complexes. Protein Sci 14: 1438–1446, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.