Abstract
It is well recognized that there is sex-dimorphic expression of mRNA and protein in the heart; however, the underlying mechanism is poorly understood. Endothelial nitric oxide synthase (eNOS) is an important regulator of cardiac function, and the expression levels of eNOS differ between male and female hearts. The aim of this study was to examine whether expression of specific microRNA (miRNA, miR) in males and females contributes to changes in the expression of eNOS. miRNA was extracted from the myocardium of male and female C57BL/6 mice and subjected to an Affymetrix miRNA array. Decreased expression of miR-222 was discovered in females and confirmed by qRT-PCR from whole heart or isolated cardiomyocytes. The transcription factor V-ets erythroblastosis virus E26 oncogene homolog-1 (ets-1) was identified as a potential target of miR-222 using TargetScan, and fivefold increased ets-1 protein expression in females was confirmed by Western blot. Targeting of ets-1 by miR-222 was determined in HEK293 cells overexpressing luciferase under regulation of either the ets-1 3′-UTR, a null 3′-UTR control, or a scrambled ets-1 3′-UTR and treated with a small molecule miR-222 mimic or inhibitor. Additionally qRT-PCR confirmed that mRNA levels of the ets-1 transcriptional target, eNOS, were 25% higher in females. Compared with untreated myocyte controls, 50% inhibition of eNOS expression was achieved by treatment with a miR-222 mimic, compared with a 25% increase due to miR-222 inhibitor. Our findings indicate that sex-dependent miR-222 regulation alters the expression of the cardiac regulatory protein eNOS.
Keywords: sexual dimorphism, cardiac, nitric oxide synthase, ets-1, miR-222
it is well known that healthy males and females have differences in cardiac phenotypes at baseline, including heart size (11), calcium handling (10), and electrophysiology (2, 24). Underlying these differences are sex-dependent protein expression profiles that contribute to altered cardiac regulatory mechanisms and response to stimuli. While it is evident that females and males display disparate cardiovascular protein regulation (9, 29, 36), the mechanisms through which these intrinsic genetic changes are effected remain unclear.
Endothelial nitric oxide synthase (eNOS) is a major source of nitric oxide in the heart and contributes to signaling both through classical activation of cyclic GMP and the posttranslational modification, S-nitrosylation (32, 34). In the healthy heart, eNOS contributes to the regulation of contractility and beta-adrenergic signaling (20, 25, 30, 38). eNOS protein expression is known to differ between male and female hearts, with both increased mRNA and protein levels in females (9). This increase has been linked to signaling through the female sex hormone estrogen (28); however, as the eNOS promoter does not contain an estrogen response element (23) it is unlikely that estrogen acts as a transcription factor for eNOS. In support of this, estrogen treatment was found to increase eNOS promoter activity without estrogen receptor binding (23), indicating that the increase in eNOS mRNA mediated by estrogen occurs through an unidentified intermediate regulatory pathway. Identification of this pathway will improve the understanding of sex-dependent cardiac function.
One possible mechanism of gene and translational regulation is through expression of microRNAs (miRNAs). miRNAs are small (20–24 nucleotide) single-stranded pieces of noncoding RNA that bind to the 3′-untranslated region (UTR) of mRNA and inhibit translation. Our study tested the hypothesis that differential miRNA expression in the heart contributes to differences in eNOS levels between males and females. Although estrogen treatment has been shown to regulate the expression of several miRNAs both in vivo (40) and in vitro (6), intrinsic differences in miRNA expression in healthy male and female hearts and how these differences contribute to protein expression have not been studied.
To test our hypothesis, we isolated total RNA (including miRNA) from sexually mature male and female mouse hearts and examined differences in miRNA expression. In females compared with males, miR-222 was significantly decreased and this correlated with increased eNOS expression. We demonstrated that mRNA and protein levels of the eNOS transcription factor and putative miR-222 target, V-ets erythroblastosis virus E26 oncogene homolog 1 (ets-1), were significantly increased in females compared with males, and we confirmed miR-222 targeting of ets-1 in vitro. Additionally, treatment with miR-222 mimic inhibited eNOS expression in cardiomyocytes while inhibition of miR-222 enhanced eNOS expression. These data suggest that females have less miR-222 levels, leading to increased expression of eNOS via increased expression of the ets-1 transcription factor.
MATERIALS AND METHODS
RNA isolation.
Male and female C57BL/6J mice (20–30 g) aged 15 wk were obtained from Jackson Laboratories (Bar Harbor, ME). All animals were treated and cared for in accordance with the Guide for the Care and Use of Laboratory Animals [National Institutes of Health (NIH), 1996]. This study was approved by the Institutional Laboratory Animal Care and Use Committee. Total RNA (including miRNA) was isolated from hearts using the miRNeasy Mini kit from Qiagen (Valencia, CA). In brief, 50 mg of tissue was homogenized in 700 μl of lysis reagent and allowed to sit at room temperature for 5 min. We added 140 μl of chloroform to the tube and shook it for 15 s. After 3 min, the samples were centrifuged for 15 min at 12,000 g at 4°C. The upper aqueous portion was transferred to a new tube, and 525 μl of 100% ethanol was added and mixed thoroughly. The samples were then transferred to a spin column and centrifuged at 8,000 g for 15 s. After discarding the flow-through, we added 700 μl of buffer RNA washing buffer (Buffer RWT) to wash the column followed by 500 μl of mild ethanol-containing RNA washing buffer (buffer RPE). The spin column was then transferred to a new collection tube, and RNA was eluted with RNase-free water. The total amount of RNA was measured using the NanoDrop 2000 (ThermoFisher, Wilmington, DE). RNA samples were stored at −80°C until their use in subsequent assays.
Genisphere FlashTag biotin labeling.
mRNA samples were labeled for the Affymetrix GeneChip miRNA 1.0 array (Santa Clara, CA) using the Genisphere FlashTag Biotin HSR RNA labeling kit (Hatfield, PA). Briefly, the total RNA samples were diluted to 8 μl with nuclease-free water, and 2 μl RNA spike control oligos were added. Next, 1.5 μl 10× reaction buffer, 1.5 μl 25 mM MnCL2, 1 μl ATP, and 1 μl polyadenylate polymerase were added. The samples were mixed gently, microcentrifuged, and incubated at 37°C for 15 min. We then added 4 μl of 5× FlashTag ligation mix biotin and 2 μl of T4 DNA ligase and mixed gently. The samples were incubated at room temperature for 30 min, and the reaction was stopped by the addition of 2.5 μl stop solution. We removed 2 μl of the labeled sample and analyzed it for quality using the Genisphere ELOSA QC assay (enzyme-linked oligo-sorbent assay quality control assay).
Affymetrix GeneChip miRNA array.
To prepare the hybridization cocktail, 50 μl 2× hybridization mix, 15 μl 27.5% formamide, 10 μl DMSO, 5 μl 20× eukaryotic hybridization controls, and 1.7 μl control oligonucleotide B2, 3 nM was added to the remaining 21.5 μl of labeled miRNA (from above), bringing the total volume to 103.2 μl. This mixture was then incubated at 99°C for 5 min and then at 45°C for 5 min. We removed 100 μl of this mixture and injected it into an array. The arrays were then placed into hybridization ovens and allowed to incubate at 48°C and 60 rpm for 16 h. After 16 h, the hybridization cocktail was removed and each array was completely filled with array holding buffer and allowed to come to room temperature. The arrays were then washed and stained using the fluidics station script FS450_0003 and scanned. The data discussed in this publication have been deposited in National Center for Biotechnology Information's Gene Expression Omnibus (GEO) (15a) and are accessible through GEO Series accession number GSE42829. (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=bzidrkieokwqmtg&acc=GSE42829).
Cardiomyocyte isolation.
Adult (12 wk old) C57BL/6 mice were obtained from Jackson Laboratory (Bar Harbor, ME). Adult Sprague-Dawley rats were obtained from Taconic Farms (Germantown, NY). Animals were euthanized by pentobarbital injection. Heparin was injected into the vena cava, and the heart was Langendorff-perfused with calcium-free normal Tyrode's solution bubbled with 100% oxygen (in mM: 136 NaCl, 5.4 KCl, 1 MgCl2, 10 HEPES, 10 d-glucose, pH 7.4). Hearts were equilibrated for 7 min. Mouse hearts were digested with Liberase TM (Roche Applied Science, Mannheim, Germany) until the flow rate increased to 1.5-fold of perfused flow rate and then removed and minced. Mouse hearts were serially digested for 4 min at 37°C, and cells were checked for rod shape prior to RNA and miRNA extraction. Rat hearts were perfused with Blendzyme IV for 7 min then removed and minced. Hearts were serially digested for 3 min at 30°C, and cells were checked for rod shape. Calcium was slowly reintroduced over 30 min, and following gravity settling, cells were resuspended in ACCT media (DMEM; Gibco, Gaithersburg, MD) supplemented with 2 g/l albumin, 2 mM l-carnitine, 5 mM taurine, 5 mM creatine, and 1% penicillin-streptomycin. Cell viability was assessed by trypan blue exclusion, and cells were plated on laminin-coated culture dishes at a density of 3,000 cells/cm2. Media were changed after 1 h. After overnight culture, cells were treated with 20 pmol of miR-222 mimic, anti-miR, mimic negative control, or anti-miR negative control using FuGene transfection reagent (Roche Applied Sciences) in OptiMEM (Gibco). ACCT medium was added after 1 h. Cells were collected after 7 h, lysed in RIPA lysis buffer with protease and phosphatase inhibitor cocktail (Roche Applied Sciences), and stored at −80°C until processing.
miRNA quantitative PCR.
miRNA was quantified using the Applied Biosystems TaqMan MicroRNA Assay (Foster City, CA). Briefly, 5 μl of RNA (1 ng total) was mixed with 7 μl of reverse transcription (RT) master mix. We placed 12 μl of the RNA/master mix in a 0.2 ml reaction tube and added 3 μl of RT primer. The mixture was incubated on ice for 5 min and then placed in a GeneAmp 2700 Thermocycler (Applied Biosystems). The parameters were 30 min at 16°C, 30 min at 42°C, and 5 min at 85°C. After the reverse transcriptase step, 17.76 μl of PCR Master Mix, No AmpErase uracil-N-glycosylase/nuclease-free water was added to 1.33 μl of the RT reaction product and 1 μl of the experimental or housekeeping TaqMan MicroRNA Assay. This solution was added to a PCR reaction plate and run using the Applied Biosystems Real-Time PCR System. Thermal cycling parameters were 10 min at 95°C followed by 40 cycles of 15 s at 95°C and 60 s at 60°C. Taqman assay number 002276 was used for miR-222, and U6 RNA (assay number 001973) was used as a control. The delta-delta cycle threshold (ΔΔCT) method was used to quantify miRNA.
RNA quantitative PCR.
mRNA was quantified using the Applied Biosystems TaqMan Gene Expressions Assay. Briefly, cDNA was synthesized from total RNA using the high-capacity cDNA reverse transcription kit (Applied Biosystems). The thermal cycler conditions were 10 min at 25°C, 120 min at 37°C, and 5 s at 85°C. Then, 4.0 μl (40 ng total) of cDNA, 1 μl of 20× Gene Expression Assay, 10 μl of 2× gene expression master mix, and 5 μl of RNase-free water were mixed. The mixture was transferred to a 96-well reaction plate and was sealed. The plate was added to the Applied Biosystems Real-Time PCR System and run with the following parameters: hold at 50°C for 2 m, hold at 95°C for 10 min, and 40 cycles of 95°C for 15 s and 60°C for 1 min. The ΔΔCT method was used to quantify RNA. Taqman Assay primer ID for eNOS was Mm00435217_m1*, assay for ets-1 was Mm00468970_m1, and for the housekeeping mRNA mouse beta actin was Mm00607939_s1 (Applied Biosystems).
Ets-1 luciferase assay.
The putative target sequence in the ets-1 3′-UTR was identified using TargetScan software (MIT, Cambridge, MA). TargetScan was searched using the mouse database and “mmu-miR-222.” Potential targeting sites were chosen based on 8-mer probability of preferential conservation. The pMIR-REPORT luciferase plasmid (Promega, Fitchburg, WI) and the following oligos containing the target sequence or scramble sequence, respectively (IDT, Coralville, IA), were restriction enzyme cut with PME1 and HindIII (New England Biolabs, Ipswich, MA) (5′-aagcttagaactgaggatccttagagatgtagcgatgctacattaaatgttttgtttaaac-3′,3′-ttcgaatcttgactcctaggaatcttacatcgctacgatgtaatttacaaaacaaatttg-5′, 5′-aagcttagaactgaggatccttagagattgagcgatgctacattaaatgttttgtttaaac-3′ and 3′-ttcgaatcttgactcctaggaatctaactcgctacgatgtaatttacaaaacaaatttg-5′ target sequence in boldface and added BamHI site underlined). The oligo and plasmid were ligated using T4 DNA Ligase (New England Biolabs) overnight at 16°C. Either empty or ets-1-containing pMIR-REPORT luciferase (200 ng) and pRL-TK renilla (Promega) were transiently overexpressed in HEK293 cells (ATCC, Manassas, VA) using FuGene transfection reagent in OptiMEM (Gibco). At the same time, cells were treated with 20 pmol of either miR-222 mimic or anti-miR-222 (Dharmacon, Lafayette, CO). After 24 or 48 h, cells treated with mimic or anti-miR, respectively, were subjected to passive lysis and luciferase measurements according to the Dual Luciferase Assay System (Promega). Luminescence was recorded using an Omega FluorStar plate reader (BMG LabTech, Ortenberg, Germany). Firefly luciferase values were normalized to renilla luciferase and presented as change from control.
Western blot.
Cell extracts were prepared in RIPA buffer (Invitrogen) containing phosphatase and protease inhibitor cocktail (Roche). Myocardial extracts were prepared in HEPES/EDTA/SDS buffer containing protease and phosphatase inhibitor cocktail and homogenized using a Dounce glass homogenizer. Total protein were separated electrophoretically on a 4–12% Bis-Tris gel (Invitrogen, Grand Island, NY) and transferred to nitrocellulose membranes. Membranes were blocked for 1 h at room temperature with 5% nonfat milk in Tris-buffered saline with Tween 20 (TBS-T) followed by 1 h incubation with the primary antibody for either Ets-1 (C-4, 1:1,000; Santa Cruz Biotechnology, Santa Cruz, CA) or eNOS (B-5, 1:1,000; Santa Cruz Biotechnology) in 5% nonfat milk in TBS-T. Membranes were subsequently incubated with horseradish peroxidase conjugated secondary antibody (1:5,000; Cell Signaling Technologies, Beverly, MA) for 1 h at room temperature. Proteins were detected using Western Lightning Enhanced Chemiluminescence solution (Perkin Elmer, Waltham, MA) and visualized on film. Optical densities for each band were obtained using ImageJ (NIH, Bethesda, MD).
Data analysis.
The Affy miRNA QC tool (version 1.0.33) was used to read and normalize the data on mouse GeneChip miRNA 1.0 array containing 609 mouse miRNAs. Sample variability was explored with principle component analysis. GraphPad Prism software (La Jolla, CA) was used to determine significance of all other data. A Student's t-test was used to determine the significance between two samples. An ANOVA with a Bonferroni post hoc test was used to determine significance for multiple comparisons. All data are presented as means ± SE. A P value ≤ 0.05 was considered statistically significant.
RESULTS
Male and female hearts have divergent miRNA expression patterns.
We isolated miRNA from four male and four female mice and subjected it to an Affymetrix miRNA microarray. Of 609 assayed miRNA, eight miRNAs show changes in relative expression between the sexes, with four decreased and four increased in females (Fig. 1A). The heat map (Fig. 1B) depicts changes in miRNA species where there was at least a 20% change between males and females and a P value of < 0.05. Among the four miRNAs with higher relative expression in males, the absolute expression levels of miR-144, miR-34b-3p, and miR-205 in both the male and female groups were low. miR-222 expression levels were significantly higher in males than in females, and absolute levels were well above background noise. miR-1, miR-106b, miR-720, and miR-29b showed higher expression in females than in males and were all well above background noise. We chose to focus on the change in miR-222 because it has been shown to indirectly regulate NOS, and NOS activity is known to be important in imparting cardioprotection in females(8, 9, 27).
Fig. 1.
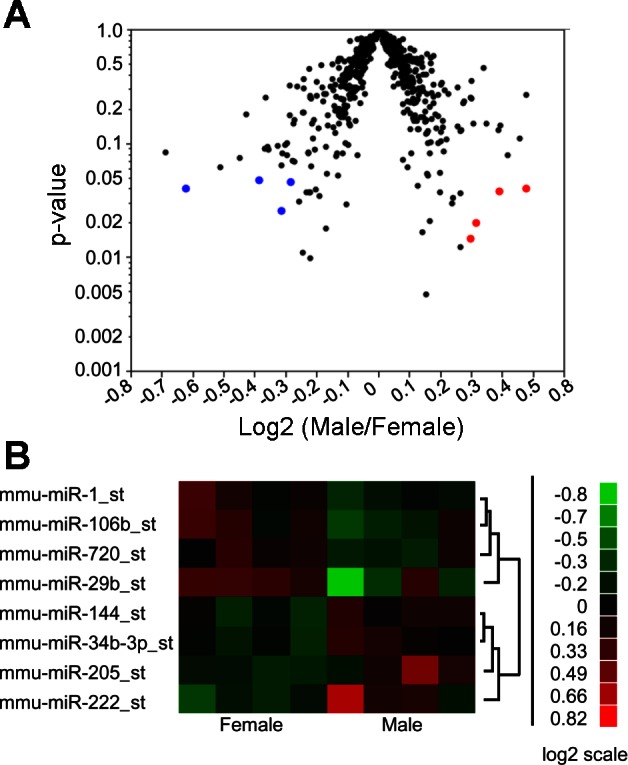
Male and female hearts display differential microRNA (miRNA) expression. A: miRNA expression shown as the ratio of male to female expression levels. miRNA shown in blue represent those that had significantly higher expression in females. miRNA shown in red had significantly higher expression in males. B: heat map of miRNA expression in male and female ventricles where there was a 20% change between male and females.
Female hearts have increased eNOS and ets-1.
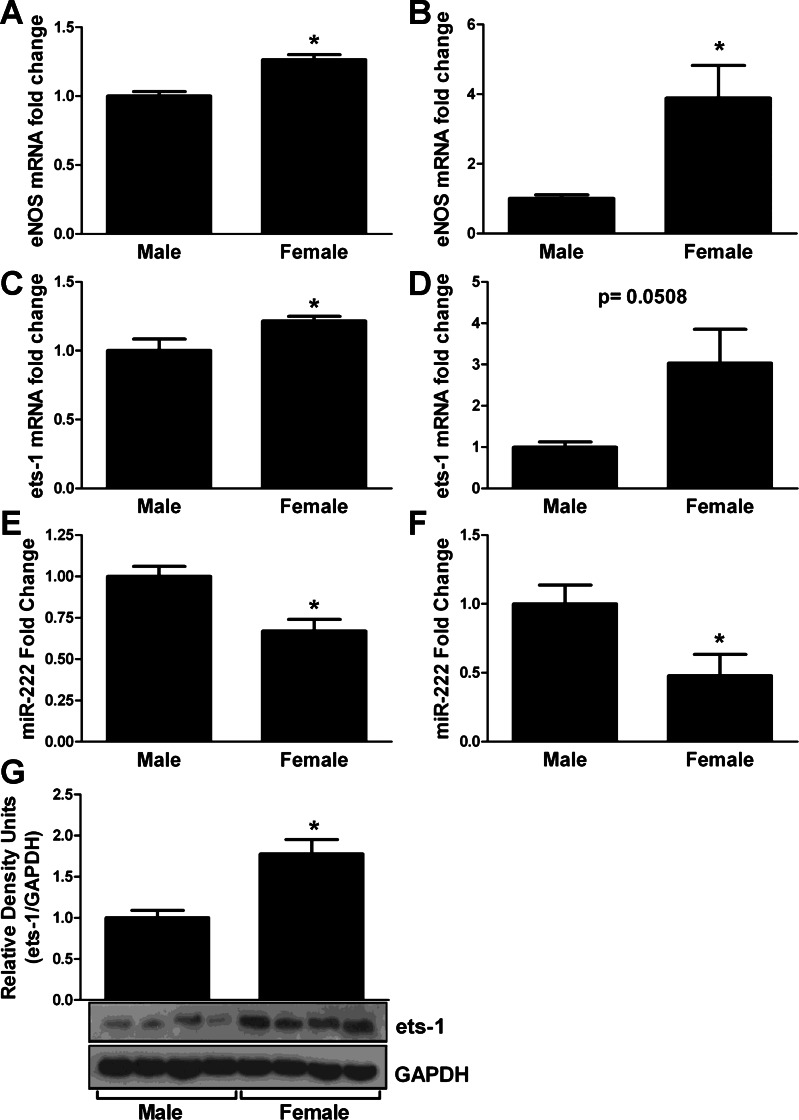
We quantified eNOS mRNA expression between male and female mouse heart samples and demonstrated a significant increase in eNOS mRNA in female whole heart samples or isolated cardiomyocytes (Fig. 2, A and B), which is consistent with previously demonstrated increases in protein levels in females (9). Using http://targetscan.org, we found that eNOS is not a direct target of any of the eight miRNA identified in the microarray screen; however, miR-222 has been previously demonstrated to indirectly alter NOS levels (26, 31). We first validated the decrease in miR-222 in females by quantifying miR-222 in the male and female whole heart or cardiomyocytes samples by real-time PCR (Fig. 2, E and F). Because miR-222 is an indirect regulator of eNOS, we sought to determine the intermediary component. Using http://targetscan.org, we found that an in silico target of miR-222 is the ets-1 transcription factor. ets-1 is known to be regulated by miR-222 in metastatic melanoma (26). In addition, ets-1 has been demonstrated to bind to the promoter region of eNOS and affect transcription in endothelial cells (7, 22). Consistent with this, we found that ets-1 mRNA levels (Fig. 2C) and protein levels (Fig. 2, C, D, and G) were significantly increased in female mice compared with male mice.
Fig. 2.
Endothelial nitric oxide synthase (eNOS) and V-ets erythroblastosis virus E26 oncogene homolog-1 (ets-1) are increased in female hearts. Total mRNA was isolated from adult mouse hearts (A) or isolated mouse cardiomyocytes (B, n = 4) and assessed for eNOS mRNA by qRT-PCR (P < 0.05). Fold change in miR-222 transcript levels as assessed by qRT-PCR of whole heart (E) or isolated cardiomyocytes (F) (n = 4). Total mRNA was isolated from adult mouse hearts (C, n = 4) or cardiomyocytes (D) and assessed for ets-1 mRNA by qRT-PCR (P < 0.05). G: cardiac homogenate was assessed for ets-1 expression in both male and female mice. Data are reported as arbitrary density units; a representative Western blot of cardiac ets-1 protein levels is shown below (n = 4, P < 0.05). *P < 0.05.
miR-222 regulates ets-1 and eNOS.
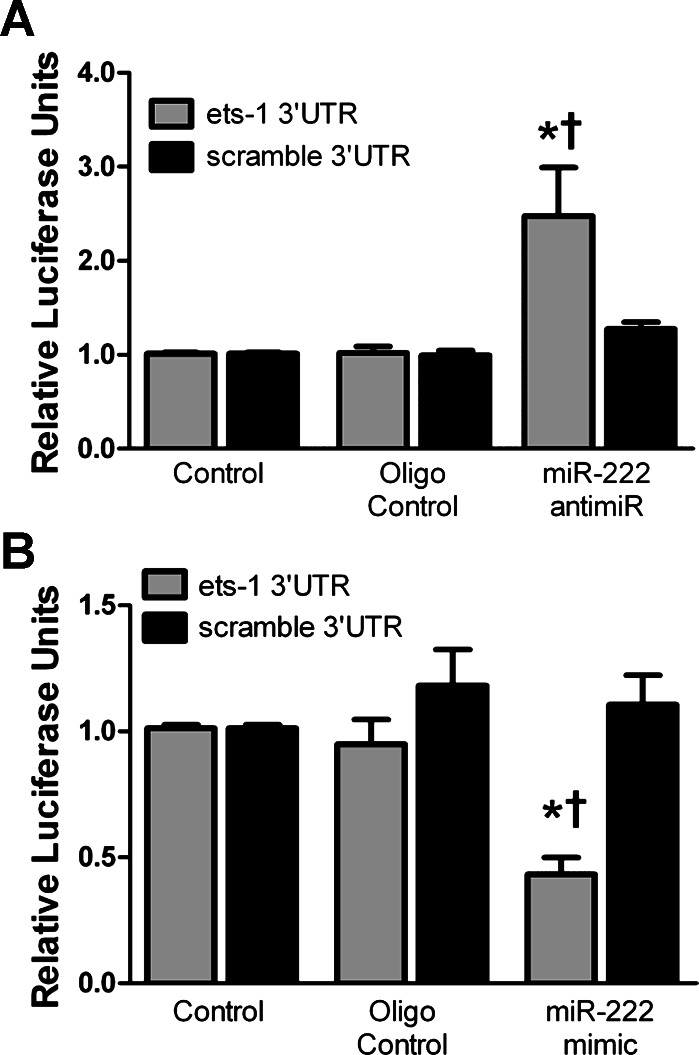
To confirm the interaction of miR-222 with ets-1, we inserted the miR-222 target site in the ets-1 3′-UTR into the 3′-UTR of the Promega pMIR-luciferase Report construct. Luciferase activity was compared in HEK293 cells containing the luciferase construct with the ets-1miR-222 target site or a scrambled ets-1 target site (ets-1 3′-UTR and scramble 3′-UTR, respectively). In the presence of an miR-222 anti-miR, luciferase activity was significantly enhanced in cells containing the ets-1 3′-UTR construct compared with those expressing the scramble 3′-UTR (Fig. 3A). Similarly, in cells expressing ets-1 3′-UTR, but not those with scramble 3′-UTR, treatment with a miR-222 mimic significantly inhibited luciferase activity (Fig. 3B). Cells in which a scrambled mimic or anti-miR (oligo control) was used showed no change in luciferase activity from untreated controls.
Fig. 3.
miR-222 is a direct inhibitor of ets-1. HEK293 cells were transfected with either a luciferase reporter containing the ets-1 3′-untranslated region (UTR) miR-222 target sequence or a scrambled ets-1 3′-UTR sequence. A luciferase activity following treatment of transfected cells with a miR-222 anti-miR inhibitor for 48 h (n = 4). B: luciferase activity following treatment of transfected cells for 24 h with a miR-222 mimic (n = 4; *P < 0.05 vs. control, †P < 0.05 vs. oligo control).
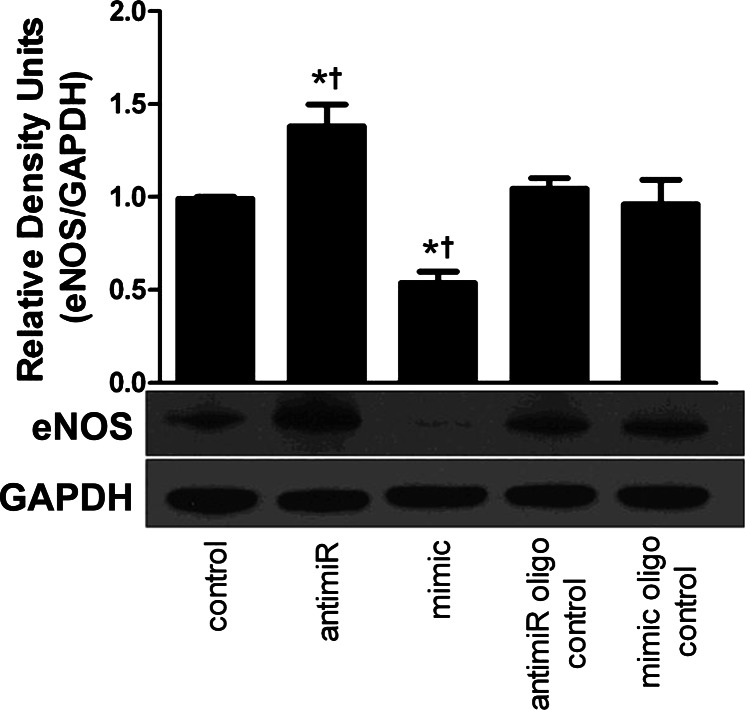
To test a direct link between miR-222 levels and eNOS expression in the heart, we isolated cardiomyocytes from adult rat ventricle and cultured them with a miR-222 mimic or miR-222 anti-miR. Rat myocytes were chosen for these experiments as they showed greater viability in a cell culture system than isolated mouse myocytes. After 7 h, eNOS expression was significantly decreased in cardiomyocytes receiving miR-222 mimic but not a scrambled mimic control oligo (Fig. 4). Conversely, cells treated with the miR-222 anti-miR showed increased eNOS expression compared with untreated cells or cells treated with a scrambled anti-miR control (Fig. 4).
Fig. 4.
miR-222 inhibits eNOS expression in cardiomyocytes. Adult rat ventricular myocytes (ARVM) were cultured overnight then treated with either a miR-222 anti-miR inhibitor or the miR-222 mimic or their respective oligo controls. Top: eNOS protein expression from treated ARVM normalized to GAPDH levels (n = 4, P < 0.05). Bottom: representative Western blot of eNOS protein levels. *P < 0.05 vs. control, †P < 0.05 vs. oligo control.
DISCUSSION
In this study, we tested the hypothesis that differences in miRNA expression are involved in male-female differences in cardiac eNOS expression. miRNAs are powerful endogenous inhibitors of mRNA translation. We found that eight miRNAs were significantly different between males and females, with four increasing and four decreasing in females. Of these eight miRNA, none are reported to directly alter eNOS expression; however, six have documented expression changes during cardiac pathology, including heart failure (miR-1, miR-106b, miR-29b, miR-34b, miR-222)(3, 4, 14, 21), arrhythmias (miR-1)(4), hypertrophy (miR-34b, miR-222)(1, 16), and fibrosis (miR-29b)(37). The potential impact of these miRNAs on sex-dependent protein expression has not been explored.
miR-222 is the only miRNA we identified that is encoded by the X-chromosome and is known to both regulate and be regulated by the estrogen receptor alpha (ER-α) (13, 39). We found that miR-222 was significantly decreased in female hearts compared with male hearts. miR-222 regulation is known to play an important role in vascular inflammation, endothelial cell migration, and angiogenesis, in part through alteration of eNOS levels (12). However, the role of miR-222 in sex-specific cardiac physiology has not been explored. Much of the literature on miR-222 and ER-α is in the context of breast cancer, in which it is known that ER-α signaling inhibits transcription of the miR-221/222 family through recruitment of co-repressors (13). Conversely, miR-222 downregulates ER-α through direct translational inhibition (39). In the context of the heart, this interplay between miR-222 and ER-α represents an uncharacterized pathway through which sex-dependent signaling can mediate cardiac translation.
The role of eNOS signaling in cardiac function has long been recognized. Genetic ablation of eNOS results in increased mortality of both male and female mice (20). Decreased levels of cardiac eNOS have also been linked to increased hypertrophy (5) and inhibition of ischemic preconditioning (35). In contrast, increased eNOS levels, such as in cardiac-specific transgenic mice, have decreased infarct size following ischemia/reperfusion injury (15, 17). In part, eNOS contributes to these effects by increasing the nitric oxide-dependent posttranslational modification S-nitrosylation and altering protein function, including that of the L-type calcium channel (34), ryanodine receptor (19), and SERCA2a (33). Modulation of these proteins contributes to altered calcium signaling, and differences in calcium signaling between males and females have been shown both at baseline (18) and following beta-adrenergic stimulation (9), indicating that eNOS-dependent protein regulation may play an important role in sex-dimorphic cardiac function.
In this study, we found that decreased miR-222 levels corresponded to increased eNOS expression in female hearts. Additionally, we found that the transcription factor ets-1, which activates transcription of eNOS, was also increased in female hearts. Sex-dependent regulation of ets-1 in the heart has not been previously reported. In addition to providing a direct mechanism through which decreased expression of miR-222 leads to increased eNOS expression in females, modulation of ets-1 represents a powerful pathway through which sex-dependent signaling can alter a large number of proteins via diverse transcriptional activation.
Conclusion
By comparing miRNA from male and female hearts, we have demonstrated sex-specific expression patterns of miRNA and examined the impact of decreased miR-222 on expression of eNOS in female hearts. Through inhibition of miR-222 expression, female hearts exhibit increased eNOS expression, which is known to regulate cardiac function. Additionally, we found that miR-222 exerts its effects on eNOS in the cardiomyocyte through direct inhibition of the transcription factor ets-1. These results indicate that altered miRNA expression patterns can alter expression of proteins such as eNOS that have important regulatory effects in heart.
GRANTS
This work was supported, in whole or in part, by NHLBI Intramural Grants Z01HL-006059 and Z01HL-002066.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: A.M.E., A.M.D., D.L., N.R., and E.M. conception and design of research; A.M.E., A.M.D., and D.L. performed experiments; A.M.E., A.M.D., D.L., and E.M. analyzed data; A.M.E., A.M.D., D.L., N.R., and E.M. interpreted results of experiments; A.M.E., A.M.D., and D.L. prepared figures; A.M.E., A.M.D., and E.M. drafted manuscript; A.M.E., A.M.D., D.L., N.R., and E.M. edited and revised manuscript; A.M.E., A.M.D., D.L., N.R., and E.M. approved final version of manuscript.
ACKNOWLEDGMENTS
We acknowledge the help of National Heart, Lung, and Blood Institute genomics core facility for the gene chip experiments.
REFERENCES
- 1.Bernardo BC, Gao XM, Winbanks CE, Boey EJ, Tham YK, Kiriazis H, Gregorevic P, Obad S, Kauppinen S, Du XJ, Lin RC, McMullen JR. Therapeutic inhibition of the miR-34 family attenuates pathological cardiac remodeling and improves heart function. Proc Natl Acad Sci USA 109: 17615–17620, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bidoggia H, Maciel JP, Capalozza N, Mosca S, Blaksley EJ, Valverde E, Bertran G, Arini P, Biagetti MO, Quinteiro RA. Sex-dependent electrocardiographic pattern of cardiac repolarization. Am Heart J 140: 430–436, 2000 [DOI] [PubMed] [Google Scholar]
- 3.Bostjancic E, Zidar N, Stajer D, Glavac D. MicroRNAs miR-1, miR-133a, miR-133b and miR-208 are dysregulated in human myocardial infarction. Cardiology 115: 163–169, 2010 [DOI] [PubMed] [Google Scholar]
- 4.Bostjancic E, Zidar N, Stajner D, Glavac D. MicroRNA miR-1 is up-regulated in remote myocardium in patients with myocardial infarction. Folia Biol (Praha) 56: 27–31, 2010 [PubMed] [Google Scholar]
- 5.Brede M, Roell W, Ritter O, Wiesmann F, Jahns R, Haase A, Fleischmann BK, Hein L. Cardiac hypertrophy is associated with decreased eNOS expression in angiotensin AT2 receptor-deficient mice. Hypertension 42: 1177–1182, 2003 [DOI] [PubMed] [Google Scholar]
- 6.Castellano L, Giamas G, Jacob J, Coombes RC, Lucchesi W, Thiruchelvam P, Barton G, Jiao LR, Wait R, Waxman J, Hannon GJ, Stebbing J. The estrogen receptor-α-induced microRNA signature regulates itself and its transcriptional response. Proc Natl Acad Sci USA 106: 15732–15737, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan Y, Fish JE, D'Abreo C, Lin S, Robb GB, Teichert AM, Karantzoulis-Fegaras F, Keightley A, Steer BM, Marsden PA. The cell-specific expression of endothelial nitric-oxide synthase. J Biol Chem 279: 35087–35100, 2004 [DOI] [PubMed] [Google Scholar]
- 8.Cross HR, Murphy E, Koch WJ, Steenbergen C. Male and female mice overexpressing the β2-adrenergic receptor exhibit differences in ischemia/reperfusion injury: role of nitric oxide. Cardiovasc Res 53: 662–671, 2002 [DOI] [PubMed] [Google Scholar]
- 9.Cross HR, Murphy E, Steenbergen C. Ca2+ loading and adrenergic stimulation reveal male/female differences in susceptibility to ischemia-reperfusion injury. Am J Physiol Heart Circ Physiol 283: H481–H489, 2002 [DOI] [PubMed] [Google Scholar]
- 10.Curl CL, Delbridge LMD, Wendt IR. Sex differences in cardiac muscle responsiveness to Ca2+ and L-type Ca2+ channel modulation. Eur J Pharmacol 586: 288–292, 2008 [DOI] [PubMed] [Google Scholar]
- 11.de Simone G, Devereux RB, Daniels SR, Meyer RA. Gender differences in left ventricular growth. Hypertension 26: 979–983, 1995 [DOI] [PubMed] [Google Scholar]
- 12.Dentelli P, Rosso A, Orso F, Olgasi C, Taverna D, Brizzi MF. microRNA-222 controls neovascularization by regulating signal transducer and activator of transcription 5A expression. Arterioscler Thromb Vasc Biol 30: 1562–1568, 2010 [DOI] [PubMed] [Google Scholar]
- 13.Di Leva G, Gasparini P, Piovan C, Ngankeu A, Garofalo M, Taccioli C, Iorio MV, Li M, Volinia S, Alder H, Nakamura T, Nuovo G, Liu Y, Nephew KP, Croce CM. MicroRNA cluster 221–222 and estrogen receptor α interactions in breast cancer. J Natl Cancer Inst 102: 706–721, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dickinson BA, Semus HM, Montgomery RL, Stack C, Latimer PA, Lewton SM, Lynch JM, Hullinger TG, Seto AG, van Rooij E. Plasma microRNAs serve as biomarkers of therapeutic efficacy and disease progression in hypertension-induced heart failure. Eur J Heart Fail [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 15.du Toit EF, Genade S, Carlini S, Moolman JA, Brunner F, Lochner A. Efficacy of ischaemic preconditioning in the eNOS overexpressed working mouse heart model. Eur J Pharmacol 556: 115–120, 2007 [DOI] [PubMed] [Google Scholar]
- 15a.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucl Acids Res 30: 207–210, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.El-Armouche A, Schwoerer AP, Neuber C, Emmons J, Biermann D, Christalla T, Grundhoff A, Eschenhagen T, Zimmermann WH, Ehmke H. Common microRNA signatures in cardiac hypertrophic and atrophic remodeling induced by changes in hemodynamic load. PLoS One 5: e14263, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elrod JW, Greer JJ, Bryan NS, Langston W, Szot JF, Gebregzlabher H, Janssens S, Feelisch M, Lefer DJ. Cardiomyocyte-specific overexpression of NO synthase-3 protects against myocardial ischemia-reperfusion injury. Arterioscler Thromb Vasc Biol 26: 1517–1523, 2006 [DOI] [PubMed] [Google Scholar]
- 18.Farrell SR, Ross JL, Howlett SE. Sex differences in mechanisms of cardiac excitation-contraction coupling in rat ventricular myocytes. Am J Physiol Heart Circ Physiol 299: H36–H45, 2010 [DOI] [PubMed] [Google Scholar]
- 19.Gonzalez DR, Beigi F, Treuer AV, Hare JM. Deficient ryanodine receptor S-nitrosylation increases sarcoplasmic reticulum calcium leak and arrhythmogenesis in cardiomyocytes. Proc Natl Acad Sci USA 104: 20612–20617, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gyurko R, Kuhlencordt P, Fishman MC, Huang PL. Modulation of mouse cardiac function in vivo by eNOS and ANP. Am J Physiol Heart Circ Physiol 278: H971–H981, 2000 [DOI] [PubMed] [Google Scholar]
- 21.Ikeda S, Kong SW, Lu J, Bisping E, Zhang H, Allen PD, Golub TR, Pieske B, Pu WT. Altered microRNA expression in human heart disease. Physiol Genomics 31: 367–373, 2007 [DOI] [PubMed] [Google Scholar]
- 22.Karantzoulis-Fegaras F, Antoniou H, Lai SLM, Kulkarni G, D'Abreo C, Wong GKT, Miller TL, Chan Y, Atkins J, Wang Y, Marsden PA. Characterization of the human endothelial nitric-oxide synthase promoter. J Biol Chem 274: 3076–3093, 1999 [DOI] [PubMed] [Google Scholar]
- 23.Kleinert H, Wallerath T, Euchenhofer C, Ihrig-Biedert I, Li H, Förstermann U. Estrogens increase transcription of the human endothelial NO synthase gene. Hypertension 31: 582–588, 1998 [DOI] [PubMed] [Google Scholar]
- 24.Kuch B, Hense HW, Sinnreich R, Kark JD, von Eckardstein A, Sapoznikov D, Bolte HD. Determinants of short-period heart rate variability in the general population. Cardiology 95: 131–138, 2001 [DOI] [PubMed] [Google Scholar]
- 25.Massion PB, Dessy C, Desjardins F, Pelat M, Havaux X, Belge C, Moulin P, Guiot Y, Feron O, Janssens S, Balligand JL. Cardiomyocyte-restricted overexpression of endothelial nitric oxide synthase (NOS3) attenuates β-adrenergic stimulation and reinforces vagal inhibition of cardiac contraction. Circulation 110: 2666–2672, 2004 [DOI] [PubMed] [Google Scholar]
- 26.Mattia G, Errico MC, Felicetti F, Petrini M, Bottero L, Tomasello L, Romania P, Boe A, Segnalini P, Di Virgilio A, Colombo MP, Carè A. Constitutive activation of the ETS-1-miR-222 circuitry in metastatic melanoma. Pigm Cell Melanoma Res 24: 953–965, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murphy E, Steenbergen C. Cardioprotection in females: a role for nitric oxide and altered gene expression. Heart Fail Rev 12: 293–300, 2007 [DOI] [PubMed] [Google Scholar]
- 28.Nuedling S, Kahlert S, Loebbert K, Doevendans PA, Meyer R, Vetter H, Grohé C. 17β-Estradiol stimulates expression of endothelial and inducible NO synthase in rat myocardium in-vitro and in-vivo. Cardiovasc Res 43: 666–674, 1999 [DOI] [PubMed] [Google Scholar]
- 29.Rosenkranz-Weiss P, Tomek RJ, Mathew J, Eghbali M. Gender-specific differences in expression mRNAs for functional and structural proteins in rat ventricular myocardium. J Mol Cell Cardiol 26: 261–270, 1994 [DOI] [PubMed] [Google Scholar]
- 30.Sterin-Borda L, Bernabeo G, Ganzinelli S, Joensen L, Borda E. Role of nitric oxide/cyclic GMP and cyclic AMP in β3 adrenoceptor-chronotropic response. J Mol Cell Cardiol 40: 580–588, 2006 [DOI] [PubMed] [Google Scholar]
- 31.Suarez Y, Fernandez-Hernando C, Pober JS, Sessa WC. Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ Res 100: 1164–1173, 2007 [DOI] [PubMed] [Google Scholar]
- 32.Sun J, Kohr MJ, Nguyen T, Aponte AM, Connelly PS, Esfahani SG, Gucek M, Daniels MP, Steenbergen C, Murphy E. Disruption of caveolae blocks ischemic preconditioning-mediated S-nitrosylation of mitochondrial proteins. Antioxid Redox Signal 16: 45–56, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun J, Morgan M, Shen RF, Steenbergen C, Murphy E. Preconditioning results in S-nitrosylation of proteins involved in regulation of mitochondrial energetics and calcium transport. Circ Res 101: 1155–1163, 2007 [DOI] [PubMed] [Google Scholar]
- 34.Sun J, Picht E, Ginsburg KS, Bers DM, Steenbergen C, Murphy E. Hypercontractile female hearts exhibit increased s-nitrosylation of the L-type Ca2+ channel α1 subunit and reduced ischemia/reperfusion injury. Circ Res 98: 403–411, 2006 [DOI] [PubMed] [Google Scholar]
- 35.Talukder MAH, Yang F, Shimokawa H, Zweier JL. eNOS is required for acute in vivo ischemic preconditioning of the heart: effects of ischemic duration and sex. Am J Physiol Heart Circ Physiol 299: H437–H445, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomas NM, Jasmin JF, Lisanti MP, Iacobas DA. Sex differences in expression and subcellular localization of heart rhythm determinant proteins. Biochem Biophys Res Commun 406: 117–122, 2011 [DOI] [PubMed] [Google Scholar]
- 37.van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, Marshall WS, Hill JA, Olson EN. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci USA 105: 13027–13032, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Varghese P, Harrison RW, Lofthouse RA, Georgakopoulos D, Berkowitz DE, Hare JM. β3-Adrenoceptor deficiency blocks nitric oxide-dependent inhibition of myocardial contractility. J Clin Invest 106: 697–703, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao JJ, Lin J, Yang H, Kong W, He L, Ma X, Coppola D, Cheng JQ. MicroRNA-221/222 negatively regulates estrogen receptorα and is associated with tamoxifen resistance in breast cancer. J Biol Chem 283: 31079–31086, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Zhao J, Imbrie GA, Baur WE, Iyer LK, Aronovitz MJ, Kershaw TB, Haselmann GM, Lu Q, Karas RH. Estrogen receptor-mediated regulation of microRNA inhibits proliferation of vascular smooth muscle cells. Arterioscler Thromb Vasc Biol 33: 257–265, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]